Significance
Honey bee pollination is required to sustain the biodiversity of wild flora and for agricultural production; however, honey bee populations in Europe and North America are declining due to virus infections. Sacbrood virus (SBV) infection is lethal to honey bee larvae and decreases the fitness of honey bee colonies. Here we present the structure of the SBV particle and show that it contains 60 copies of a minor capsid protein attached to its surface. No similar minor capsid proteins have been previously observed in any of the related viruses. We also present a structural analysis of the genome release of SBV. The possibility of blocking virus genome delivery may provide a tool to prevent the spread of this honey bee pathogen.
Keywords: honeybee, virus, structure, genome, release
Abstract
Infection by sacbrood virus (SBV) from the family Iflaviridae is lethal to honey bee larvae but only rarely causes the collapse of honey bee colonies. Despite the negative effect of SBV on honey bees, the structure of its particles and mechanism of its genome delivery are unknown. Here we present the crystal structure of SBV virion and show that it contains 60 copies of a minor capsid protein (MiCP) attached to the virion surface. No similar MiCPs have been previously reported in any of the related viruses from the order Picornavirales. The location of the MiCP coding sequence within the SBV genome indicates that the MiCP evolved from a C-terminal extension of a major capsid protein by the introduction of a cleavage site for a virus protease. The exposure of SBV to acidic pH, which the virus likely encounters during cell entry, induces the formation of pores at threefold and fivefold axes of the capsid that are 7 Å and 12 Å in diameter, respectively. This is in contrast to vertebrate picornaviruses, in which the pores along twofold icosahedral symmetry axes are currently considered the most likely sites for genome release. SBV virions lack VP4 subunits that facilitate the genome delivery of many related dicistroviruses and picornaviruses. MiCP subunits induce liposome disruption in vitro, indicating that they are functional analogs of VP4 subunits and enable the virus genome to escape across the endosome membrane into the cell cytoplasm.
The pollination services provided by the western honey bee (Apis mellifera) are required for agricultural production and to maintain the diversity of wild flowering plants (1). Over the last few decades, the combination of environmental pollution, habitat loss, and pathogens has resulted in a decrease in honey bee populations in North America and Europe (2, 3). Symptoms of the sacbrood virus (SBV) infection of pupae include the accumulation of ecdysial fluid, cuticle discoloration, and death, resulting in a typical gondola-shaped dry cadaver. Although SBV is lethal for infected larvae, infected colonies rarely collapse, and thus SBV poses only a limited threat to managed honey bees (3). SBV is distributed worldwide, with specific strains also infecting Apis cerana (4).
SBV has a positive-sense single-stranded RNA genome that is 8,832 nt long and contains an additional poly-A sequence at its 3′ end (5). The genome encodes a single 2,858-aa-long polyprotein that is cotranslationally and posttranslationally cleaved by virus proteases into functional protein subunits. The capsid proteins VP1, VP2, and VP3 from one polyprotein precursor form a protomer, an elementary building block of the virus capsid. Based on homology with vertebrate picornaviruses, SBV protomers are expected to assemble into pentamers, and, subsequently, 12 pentamers associate with the genome to form a virion (6). The capsid proteins of iflaviruses have jelly roll β-sandwich folds shared by all picornaviruses and numerous other viruses from other families (7–10). To date, three iflaviruses have been structurally characterized: slow bee paralysis virus (SBPV), deformed wing virus (DWV), and Chinese SBV (CSBV) (8, 11, 12).
Here we present the structure of the SBV virion and show that it contains 60 copies of a minor capsid protein (MiCP) attached at the virion surface. No similar proteins have been observed in any virus from the order Picornavirales. A comparison of the structure of SBV with the structures of other iflaviruses indicates that the protein evolved from an ancestral C-terminal extension of the VP3 subunit through the introduction of a cleavage site for virus-encoded protease. In addition, we show that the MiCP induces the disruption of liposome membranes and thus may facilitate delivery of the SBV genome into the cytoplasm.
Results and Discussion
SBV Virion Structure and Its Comparison with Iflaviruses Containing P-Domains.
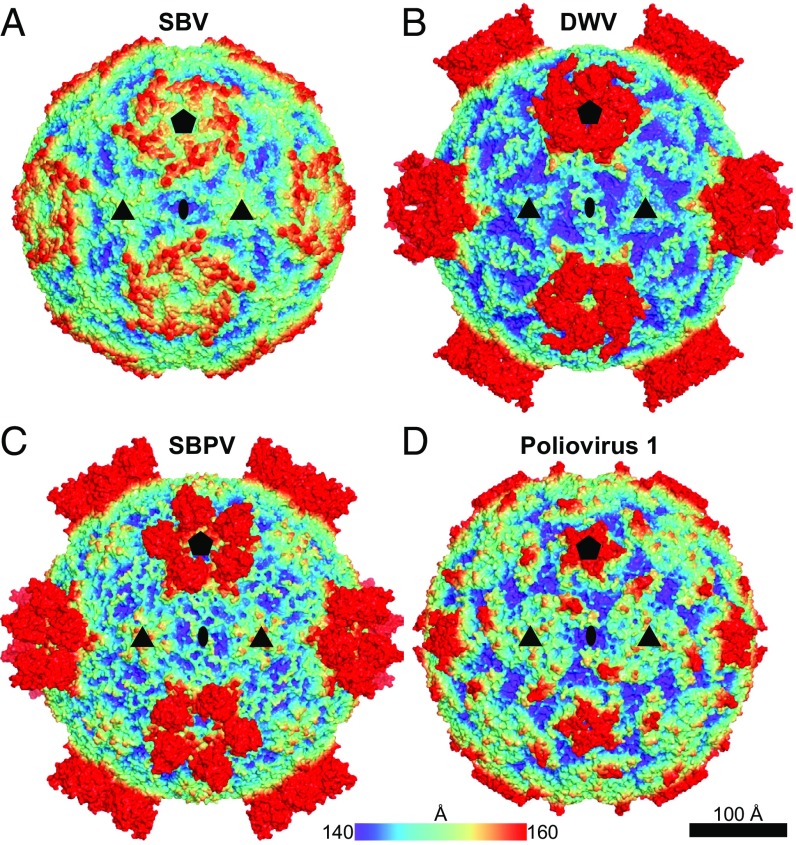
The virion structure of SBV has been determined to a resolution of 2.1 Å using X-ray crystallography (SI Appendix, Table S1). The maximum diameter of the SBV virion is 312 Å, which is 90 Å less than the maximum diameters of related iflaviruses DWV and SBPV (7, 8, 13). The virions of DWV and SBPV are larger because their VP3 subunits contain 160-residue-long C-terminal extensions, which fold into globular P-domains positioned at the virion surface (Figs. 1 and 2). The capsid of SBV is of spherical shape, with plateaus around icosahedral fivefold symmetry axes and shallow depressions at twofold symmetry axes (Fig. 1A). SBV lacks the typical canyon depressions that are dominant surface features of the virions of enteroviruses and some other picornaviruses (Fig. 1D) (9).
Fig. 1.
Virion structures of SBV (PDB ID code 5LSF) (A), DWV (PDB ID code 5L7Q) (B), SBPV (PDB ID code 5J96) (C), and poliovirus 1 (PDB ID code 1ASJ) (D). The molecular surfaces of the respective virions are rainbow-colored according to their distance from the center. The locations of the selected symmetry axes are denoted by pentagons for fivefold, triangles for threefold, and ellipses for twofold.
Fig. 2.
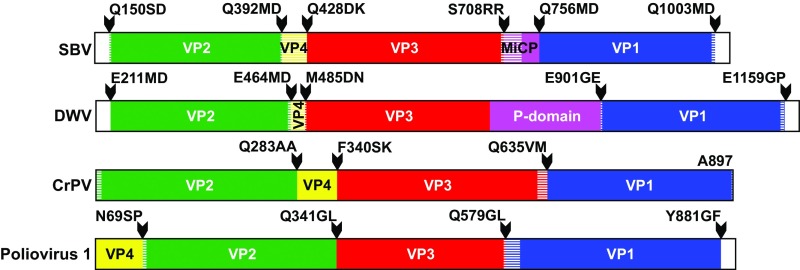
Organization of P1 polyproteins of iflaviruses SBV and DWV, dicistrovirus cricket paralysis virus, and picornavirus poliovirus-1. Capsid proteins within P1 are labeled and colored according to the picornavirus convention: VP1 in blue, VP2 in green, VP3 in red, and VP4 in yellow. The MiCP of SBV and P-domain of DWV are highlighted in magenta. Note that translation of the P1 sequence of CrPV is initiated from an independent internal ribosomal entry site located after the coding sequence for the virus polymerase. Arrowheads indicate positions of protease cleavage sites, and the target cleavage sequences are shown. Parts of the proteins resolved in experimentally determined structures are shown in bright colors; the parts highlighted with hatching are not structured.
There are two alternative conventions for naming the capsid proteins of picorna-like viruses. In some previous annotations of the iflavirus genomes and in the structural characterization of DWV by Organtini et al. (13), the virus proteins were named according to a molecular weight convention from the largest subunit, VP1, to the smallest, VP4. An alternative approach is to label the capsid proteins according to their homology with picornavirus proteins (14). The homology-based convention was used in the previous structural studies of SBPV and DWV (7, 8, 15). Application of the molecular weight convention results in different gene orders of SBV, DWV, and SBPV. Therefore, to simplify the structural comparison of SBV, DWV, and SBPV with one another and to picornaviruses, we use the homology-based convention. The order of the major capsid proteins in the P1 polyprotein is VP2, VP3, and VP1 (Fig. 2). The capsid proteins labeled according to the picornavirus convention occupy homologous positions in the virus capsid (Fig. 3).
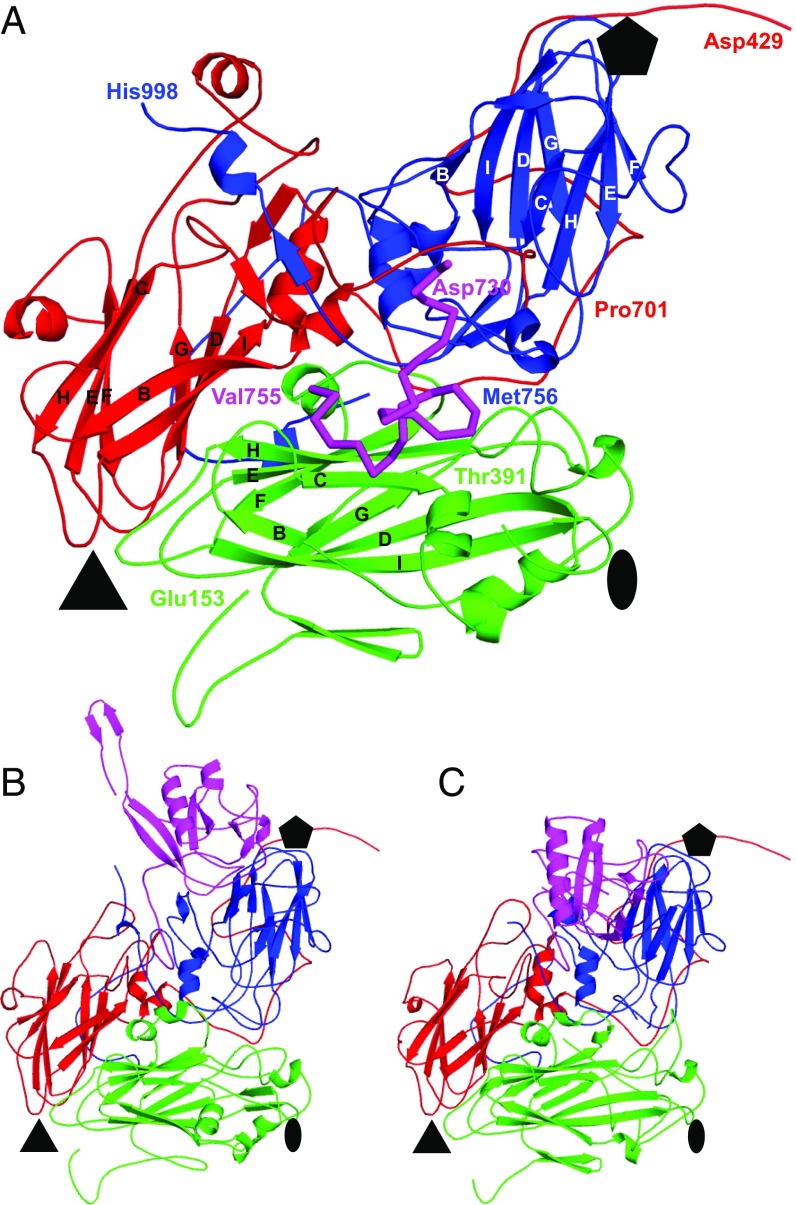
Fig. 3.
Comparison of icosahedral asymmetric units of SBV, DWV, and SBPV. Icosahedral asymmetric unit of SBV (A), DWV (B), and SBPV (C) in cartoon representation with major capsid protein VP1 in blue, VP2 in green, and VP3 in red. The MiCP of SBV is highlighted as a magenta ribbon (A). β-strands of the major capsid proteins are labeled. P-domains of VP3 subunits of DWV and SBPV are highlighted in magenta (B and C). The locations of the fivefold, threefold, and twofold symmetry axes are denoted by pentagons, triangles, and ovals, respectively.
The SBV capsid is built from the major capsid proteins VP1, VP2, and VP3 organized with pseudo-T3 icosahedral symmetry (Fig. 3A). The VP1 subunits form pentamers around fivefold axes, whereas the VP2 and VP3 subunits constitute heterohexamers positioned at the icosahedral threefold axes. The major capsid proteins have jelly roll β-sandwich folds with β-strands named according to the virus capsid protein convention B–I (6). The two antiparallel β-sheets forming the core of each of the capsid proteins contain the strands BIDG and CHEF, respectively (Fig. 3A). The N termini of the capsid proteins are located on the inside of the capsid, whereas their C termini are exposed at the virion surface. The crystallographic electron density map of the SBV virion enabled the building of residues 1–243 out of 247 residues of VP1; residues 3–241 out of 242 residues of VP2; and residues 1–272 out of 280 residues of VP3. SBV encodes a 36-residue-long VP4 subunit (Fig. 2); however, the electron density map of SBV virion does not contain resolved features that could be interpreted as residues of VP4.
In contrast to all other viruses from the order Picornavirales that have been structurally characterized at high resolution so far (32 picornaviruses, 4 dicistroviruses, and 2 iflaviruses), the SBV virion contains a short protein attached to the capsid surface (Fig. 3A). We termed the peptide the MiCP. The structure of the MiCP could be built for residues 22–47 out of 48 residues. The amino acid sequence of the MiCP is located between the C terminus of VP3 and the N terminus of VP1 in the P1 polypeptide of SBV (Fig. 2). LC-MS-MS was used to verify the protein cleavage sites that separate the MiCP from VP3 and VP1 (SI Appendix, Fig. S1). The identified cleavage site, Ser708/Arg709Arg710, is in agreement with the previously characterized target sequences of picornavirus-like proteases (16, 17).
VP1 subunits of enteroviruses have been shown to form hydrophobic pockets that can be targeted by artificial compounds that prevent virus–receptor interaction or genome release (18–20). Similar to SBPV and DWV (7, 8), the VP1 of SBV does not contain such a pocket, and it is unlikely that the virus could be inhibited by capsid-binding inhibitors targeting the hydrophobic core of VP1.
SBV Virions Do Not Contain VP4 Subunits.
The formation of mature infectious virions of most picorna-like viruses requires the cleavage of VP4 subunits from the N terminus of their VP0 precursors that is not performed by the virus proteases (21–23). It has been speculated that the proteolysis of the VP4 subunits of picornaviruses is catalyzed by the RNA genome (24, 25). In dicistroviruses, a conserved motif, Asp-Asp-Phe (DDF), in VP1 was suggested to catalyze the VP4 cleavage (22). Triatoma virus and black queen cell virus, from the family Dicistroviridae, do not contain structured VP4 subunits (26, 27), but the VP4 peptides are present in the virions and likely function in the transport of the genome across the endosome membrane into the cell cytoplasm (27). Some iflaviruses, including SBV and SBPV, also contain the DDF motif in their VP1 subunits (7, 8). The DDF motif of VP1 of SBV is located at the inner face of the SBV virion next to the N terminus of the VP3 subunit of another protomer from the same pentamer (SI Appendix, Fig. S2). Therefore, the DDF motif might function in the cleavage of VP0 subunits of SBV. Nevertheless, VP4 subunits are not resolved in the electron density maps of SBV virions and could not be detected in the particles by MS analysis (SI Appendix, Fig. S1). Similarly, VP4 subunits were not resolved in the structures of SBPV and DWV (7, 8), and they likely were not present in the virions. It is possible that the short 38-residue VP4 subunits of SBV diffuse from the virions after the maturation cleavage of VP0. Iflaviruses lacking the VP4 subunits need to use a different mechanism to penetrate the host membrane than that used by the previously studied picornaviruses and dicistroviruses.
Structure and Putative Receptor-Binding Function of the MiCP.
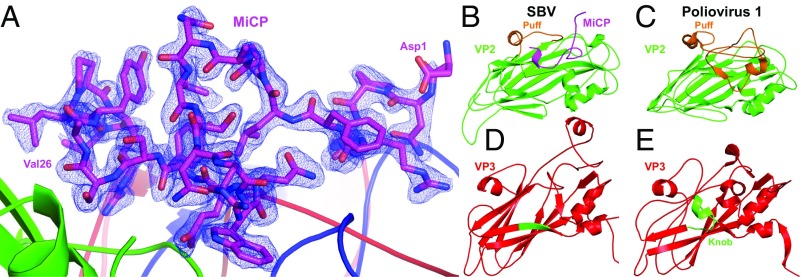
The MiCP is a 48-aa-long protein with a molecular weight of 5.4 kDa. Residues 1–21 of the MiCP, which are not resolved in the SBV virion structure, are more varied among the different SBV isolates than residues 22–47, which form the structured part of the peptide (SI Appendix, Fig. S3). The higher tolerance of the flexible part of the MiCP to mutations indicates that its biological function might not depend on a specific structure. In contrast, the structured part is more conserved, as it is required for binding to the virion surface. The structured 26 residues of the MiCP form a loop that turns 360° on itself (Fig. 3A). The MiCP is positioned above the core of subunit VP2 almost equidistant from the icosahedral twofold, threefold, and fivefold symmetry axes of the capsid (Fig. 3A). The N-terminal arm of the MiCP extends toward the icosahedral fivefold axis and interacts with subunits VP1 and VP3. The MiCP binds to the capsid through an interface area of 1,100 Å2 formed by 24 residues of VP2, 7 residues of VP1, and 6 residues of VP3 (Fig. 4A). An occupancy refinement showed that MiCP peptides are present in all 60 positions at the virion surface (Table 1).
Fig. 4.
Structure of the MiCP and its interaction with other capsid proteins. (A) Structure of the MiCP shown in stick representation with carbon atoms in magenta. The first and last structured residues of MiCP are labeled. The electron density map of the MiCP contoured at 2σ is shown as a blue mesh. Major capsid proteins are shown in cartoon representation with VP1 in blue, VP2 in green, and VP3 in red. (B and C) Comparison of capsid proteins VP2 of SBV (B) and poliovirus 1 (C). The MiCP of SBV, which is highlighted in magenta (B), occupies the volume of the puff loop in VP2 of poliovirus 1, highlighted in orange (C). (D and E) The VP3 subunit of SBV (D) lacks the knob loop present in poliovirus 1 (E), highlighted in green.
Table 1.
Comparison of SBV structures under various pH and expansion states
| Parameter | Crystal structure | Virion, pH 7.4 | Empty particle, pH 7.4 | Virion, pH 5.8 | Empty particle, pH 5.8, expansion state I | Empty particle, pH 5.8, expansion state II |
| Resolution, Å | 2.10 | 3.22 | 3.87 | 3.18 | 4.06 | 7.25 |
| MiCP occupancy | 1.02 | 0.86 | 0.77 | 0.94 | 0.93 | 0.79 |
| Particle radius,* Å | 136.4 | 136.1 | 137.4 | 136.1 | 137.9 | 139.8 |
| Pentamer distance,† Å | 135.8 | 134.4 | 135.7 | 134.4 | 136.2 | 138.4 |
| Pentamer contacts, Å2 | 4,250 | 4,550 | 1,750 | 4,450 | 1,750 | 700 |
| Average B-factor, Å2 | 25 | 39 | 88 | 43 | 118 | 200 |
| Diameter of pore on threefold axis, Å | 4.4 | 3.6 | 4.4 | 3.6 | 5.0 | 7.4 |
| Average B-factor of residues close to threefold axis, Å2 | 23 | 35 | 104 | 40 | 143 | 200 |
| Diameter of pore on fivefold axis, Å | 5.2 | 5.2 | 5.6 | 5.6 | 5.0 | 12.0 |
| Average B-factor of residues close to fivefold axis, Å2 | 19 | 36 | 102 | 41 | 137 | 200 |
| Diameter of pore on twofold axis, Å | 2.6 | 2.6 | 2.8 | 2.4 | 3.0 | 2.8 |
| Average B-factor of residues close to twofold axis, Å2 | 21 | 35 | 55 | 37 | 73 | 200 |
Particle radius is defined as the distance of the center of mass of the icosahedral asymmetric unit from the particle center.
Pentamer distance is defined as the distance of the centers of mass of two neighboring pentamers.
The volume at the surface of the SBV virion occupied by the MiCP is taken up by the EF loop known as the “puff” loop from the VP2 subunit in enteroviruses (Fig. 4 B and C) (9, 28). In poliovirus 1, the 65-residue-long puff loop forms the outer rim of the canyon (Fig. 4C) (28). In contrast, the puff loop of SBV is only 18 residues long (Fig. 4B). In addition, subunit VP3 of SBV lacks a loop known as a “knob” formed by residues before the β-strand B of enteroviruses (Fig. 4 D and E). It has been shown that residues from the puff and knob of many enteroviruses participate in receptor binding (29). Therefore, because of its location at the virion surface, it is possible that the MiCP is involved in SBV receptor recognition.
Evolutionary Relationships Within the Family Iflaviridae.
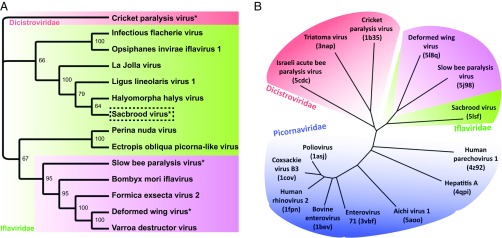
There are no structural or sequence similarities between the 48-residue-long MiCP of SBV and the 160-residue-long P-domains of SBPV and DWV. Nevertheless, the locations of the sequences coding the MiCP and P-domains in Iflavirus genomes show that both the MiCP and P-domains evolved as C-terminal extensions of VP3 subunits (Fig. 2). The evolution of the MiCP required the introduction of a cleavage site for virus protease 3C. The phylogenetic tree based on a comparison of the P1 sequences of iflaviruses shows that the family can be divided into two groups: viruses that lack P-domains and viruses with P-domains (Fig. 5A and SI Appendix, Fig. S4). The P-domain appears to be a filial feature. Based on sequence analysis, it is not clear whether all the iflaviruses that lack the P-domains contain MiCPs. A structure-based phylogenetic tree shows branching inside the Iflaviridae family of viruses with and without P-domains (Fig. 5B), corroborating the sequence-based separation of P-domain–containing iflaviruses into a subgroup within the family Iflaviridae.
Fig. 5.
Phylogenetic analyses of viruses from the families Iflaviridae, Dicistroviridae, and Picornaviridae. (A) Maximum likelihood tree constructed from whole polyprotein sequences of selected iflaviruses with cricket paralysis virus as an outgroup (highlighted in red). Iflaviruses with P-domain are highlighted in the magenta box. The dashed box indicates viruses with observed MiCP; the asterisk indicates viruses with an experimentally determined virion structure. Values at nodes represent thorough bootstrap support. (B) Structure-based neighbor-joining phylogeny tree with picornaviruses highlighted in blue, dicistroviruses in red, and iflaviruses in green. The subgroup of iflaviruses with P-domains is highlighted in magenta.
Genome Release Mechanism of SBV.
The cell entry mechanism of iflaviruses is unknown, but the process likely involves receptor-mediated endocytosis, as is the case for related picornaviruses (29). Endosomal entry involves exposure of the virions to an environment with acidic pH (30, 31). In picornaviruses, the acidic pH triggers the formation of activated (A)-particles that are expanded, contain pores in capsids, and spontaneously release their genomes (29, 31–33). Freshly purified samples of SBV contain 5% empty particles (SI Appendix, Fig. S5A). We used cryo-EM to determine the structures of the full virions and empty particles to resolutions of 3.2 Å and 3.8 Å, respectively (SI Appendix, Figs. S6 and S7 and Table S1). The structure of the full virion is nearly identical to that determined by X-ray crystallography, with an rmsd of the corresponding Cα atoms of the two structures of 0.44 Å (SI Appendix, Table S2). The empty particle is expanded 1.2 Å in radius relative to the genome-containing virion (Table 1). The capsid enlargement is accompanied by movements of pentamers of capsid protein protomers away from one another (Table 1) and by a reduction in interpentamer contacts, as residues 1–14 of VP1 and residues 1–40 of VP2 are not resolved in the empty particles.
The sample of SBV incubated in PBS with pH 5.8, which mimics the environment in endosomes (34), contained 80% empty particles (SI Appendix, Fig. S5B). The structure of the full virions at acidic pH, which was determined to a resolution of 3.2 Å (SI Appendix, Figs. S6 and S7 and Table S1), is similar to that of the virus at neutral pH, as determined by both X-ray crystallography and cryo-EM (rmsd of 0.32 Å and 0.27 Å, respectively) (SI Appendix, Table S2). The structural similarity of the virions, together with the observation that empty SBV particles are expanded, as discussed below, suggests that the SBV RNA genome contributes to the stability of the virion; however, direct protein–RNA interactions are not resolved in the electron density maps.
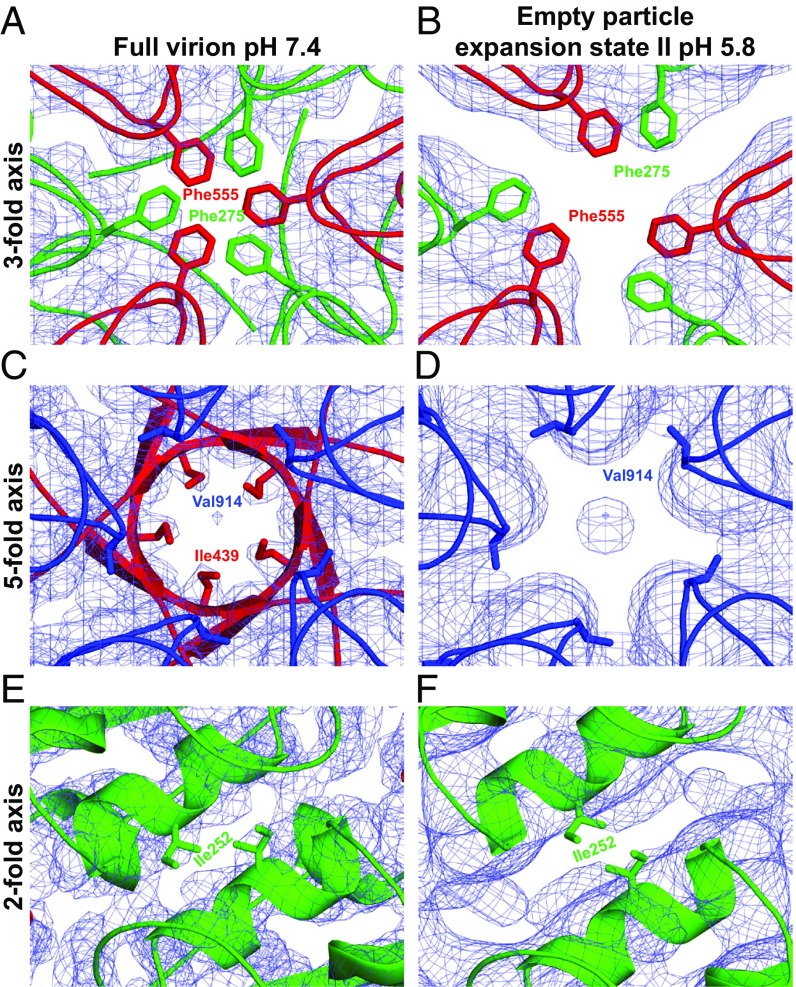
Classification of the empty particles from the acidic pH sample identified two capsid expansion intermediates. The smaller of the two capsids was radially expanded by 1.5 Å relative to the native virions, and determined to a resolution of 4.1 Å, whereas the larger one was radially expanded by 3.4 Å and determined to a resolution of 7.3 Å (SI Appendix, Figs. S6 and S7 and Table S1). The smaller particle contains pores 5.0 Å in diameter located at the icosahedral threefold axes, whereas the pores in the more expanded particle are 7.4 Å in diameter (Fig. 6 A and B, Table 1, and SI Appendix, Fig. S8). These pores are not of sufficient size to allow passage of the single-stranded SBV RNA genome. However, amino acids that form the loops of capsid proteins adjacent to the pores have higher temperature factors than the rest of the structure (Table 1). This indicates that the loops are flexible. Furthermore, the presence of two expansion intermediates suggests that the empty SBV capsids are dynamic. The possible role of pores at threefold axes of SBV capsids as channels for genome release is consistent with our previous study of the genome release of SBPV (15). In contrast, Organtini et al. (13) speculated that the genome of DWV may escape from particles through a channel in a fivefold vertex of its capsid. In native SBV virions, the N termini of VP3 subunits form a 5.2-Å narrow iris-like constriction of the channel along the fivefold axis (Fig. 6C, Table 1, and SI Appendix, Fig. S8). However, in the more expanded acidic pH empty capsid of SBV, residues 1–48 of VP3 are not structured (Fig. 6D and SI Appendix, Fig. S8). Thus, the more expanded empty SBV particles also contain 12-Å-diameter pores along the fivefold axes (Fig. 6D, Table 1, and SI Appendix, Fig. S8). The putative release of the SBV genome through pores along threefold or fivefold symmetry axes differs from the currently accepted genome release mechanism of picornaviruses, in which the pores along twofold icosahedral symmetry axes have been implicated (35–37). Pores along the twofold axes of SBV particles are not expanded after genome release, as is the case in enteroviruses (Fig. 6 E and F, Table 1, and SI Appendix, Fig. S8).
Fig. 6.
Comparison of capsid protein conformations close to the threefold, fivefold, and twofold axes of SBV virions and empty particles. The capsid protein loops that form contacts in the vicinity of rotation axes of the capsid are shown in cartoon representation. VP1, VP2, and VP3 are shown in blue, green, and red, respectively. Side chains of residues closest to the rotation axes are shown as sticks; the density map is depicted as a blue mesh. Details of native virion at pH 7.4 (A, C, and E) and the more expanded empty particle at pH 5.8 (B, D, and F) are shown.
The MiCP May Facilitate Delivery of SBV Genome into Cytoplasm.
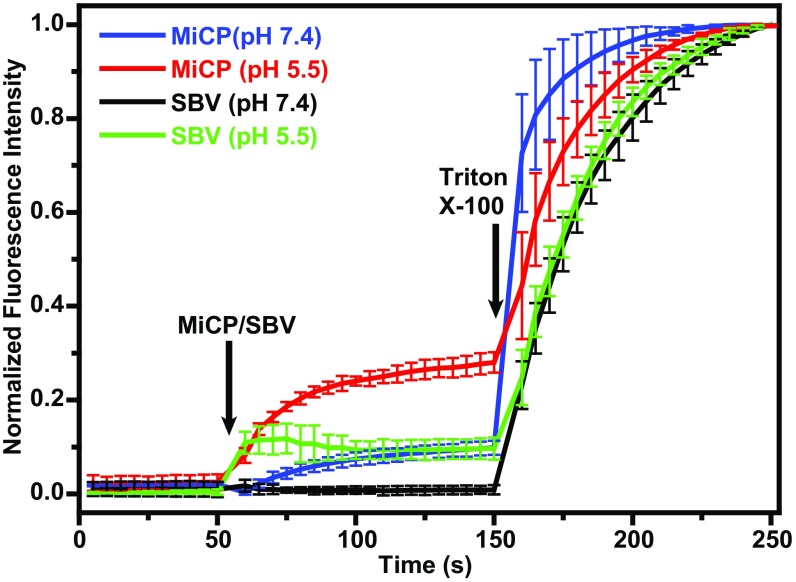
MiCP subunits are tightly associated with the native virions of SBV (Table 1). In contrast, in empty SBV particles, particularly those exposed to acidic pH, the electron density corresponding to the MiCP is visible only at lower contour levels than the rest of the capsid. The occupancy refinement indicates that some of the MiCP subunits detach from empty SBV particles at pH 5.8 (Table 1). The dissociation of MiCPs from the SBV capsid occurs in similar conditions as the release of VP4 subunits from the virions of dicistroviruses and picornaviruses before genome egress (29, 31–33). Furthermore, the hydrophobicity profile of the MiCP is similar to that of the VP4 of HRV16 (SI Appendix, Fig. S3B). The MiCP with an N-terminal His6-SUMO tag is expressed as soluble protein in Escherichia coli and remains soluble after cleavage of the tag (SI Appendix, Fig. S9). However, MiCP expression blocks growth of the bacterial culture at 1 h after induction (SI Appendix, Fig. S9). In buffer with pH 5.5, MiCP subunits at micromolar concentrations induce the disruption of liposomes with phospholipid content mimicking that of endosomes (Fig. 7 and SI Appendix, Fig. S10) (38). The effect of SBV virions on liposomes in the same conditions is limited (Fig. 7 and SI Appendix, Fig. S10); however, in native conditions, receptor binding might contribute to more efficient release of MiCP subunits from virions and thus allow the protein to disrupt the endosome membranes. The biological function of the MiCP may be similar to that of the VP4 of picornaviruses, to facilitate transport of the virus genome across the endosome membrane into the cell cytoplasm (39, 40).
Fig. 7.
The MiCP induces disruption of liposomes at pH 5.5. Liposomes containing carboxyfluorescein were mixed with MiCP at a final concentration of 2 μM in a solution at pH 7.4 (blue line) at the 50-s time point (MiCP/SBV arrow). Liposomes were mixed with SBV virions at a final concentration of 2 nM, corresponding to an MiCP concentration of 120 nM in a solution at pH 7.4 (black line). Liposomes were mixed with MiCP at a final concentration of 2 μM in a solution at pH 5.5 (red line). Liposomes were mixed with SBV virions at a final concentration of 2 nM in a solution at pH 5.5 (green line). The liposomes were disrupted with 1% Triton X-100 at the 150-s time point (arrow). The fluorescence of carboxyfluorescein was increased due to the dequenching effect of fluorophore dilution after the liposomes dissolved. Measured arbitrary fluorescence values were normalized and plotted as an average of three independent experiments with SDs.
In summary, our results show that SBV virions are structurally distinct from virions of DWV and SBPV. We demonstrate that acidic pH triggers expansion of SBV capsids, genome release, and the partial dissociation of MiCP subunits from the virion surface. Subsequently, the MiCP subunits may disrupt the endosome membrane to allow the virus genome to reach the cell cytoplasm and initiate infection.
Methods
The propagation of SBV in honey bee larvae was performed as described in The COLOSS BEEBOOK (41). Purified SBV was applied onto holey carbon grids and vitrified by plunge-freezing in liquid ethane. Micrographs were recorded with a Falcon II camera in a Titan Krios transmission electron microscope (Thermo Fisher Scientific). Acquired data were processed using the RELION package (42). Crystals of SBV were obtained from the identical material with the hanging-drop technique in crystallization conditions containing 0.05 M magnesium chloride, 0.1 M MES, 8% isopropanol, and 4% PEG 4000. The MiCP was expressed in E. coli, purified with Ni-NTA chromatography, and diluted in two types of buffer (pH 8.0 and pH 5.5). A suspension of liposomes filled with carboxyfluorescein dye was distributed to a 96-well plate, and the MiCP was added to wells with respective buffer conditions. Resulting changes in fluorescence levels were recorded before and after the addition of 1% Triton X-100. The methodology is described in detail in SI Appendix, Methods.
Supplementary Material
Acknowledgments
X-ray data were collected at Soleil Synchrotron, beamline Proxima 1. We acknowledge the Central European Institute of Technology (CEITEC) Core Facilities Cryo-Electron Microscopy and Tomography, Proteomics, and Biomolecular Interactions, supported by Czech Infrastructure for Integrative Structural Biology Project LM2015043, funded by the Ministry of Education, Youth, and Sports (MEYS) of the Czech Republic. This research was carried out under the project CEITEC 2020 (LQ1601), with financial support from the MEYS under National Sustainability Program II. This work was supported by the IT4I Project (CZ.1.05/1.1.00/02.0070), funded by the European Regional Development Fund and the national budget of the Czech Republic via the Research and Development for Innovations Operational Program, as well as the MEYS via Grant LM2011033. The research leading to these results received funding from the European Research Council under Grant Agreement 335855 (to P.P.) and EMBO Grant Agreement IG3041 (to P.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Cryo-EM maps of the SBV virions from the different conditions have been deposited in the Electron Microscopy Data Bank [accession nos. 3863 (native virion, pH 7.4); 3881 (empty particle, pH 7.4); 3865 (virion, pH 5.8); 3866 (empty particle, pH 5.8, expansion state I); and 3867 (empty particle, pH 5.8, expansion state II)], and the corresponding coordinates have been deposited in the Protein Data Bank [PDB ID codes 5OYP (native virion, pH 7.4); 6EIW (empty particle, pH 7.4); 6EGV (virion, pH 5.8); 6EGX (empty particle, pH 5.8, expansion state I); and 6EH1 (empty particle, pH 5.8, expansion state II)]. The crystal structure of the SBV virion also has been deposited in the Protein Data Bank (PDB ID code 5LSF). The consensus nucleotide sequence of the SBV capsid proteins has been deposited in GenBank (accession no. KY617033).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1722018115/-/DCSupplemental.
References
- 1.Ellis JD, Munn PA. The worldwide health status of honey bees. Bee World. 2005;86:88–101. [Google Scholar]
- 2.Breeze TD, Bailey AP, Balcombe KG, Potts SG. Pollination services in the UK: How important are honeybees? Agric Ecosyst Environ. 2011;142:137–143. [Google Scholar]
- 3.Gisder S, Genersch E. Viruses of commercialized insect pollinators. J Invertebr Pathol. 2017;147:51–59. doi: 10.1016/j.jip.2016.07.010. [DOI] [PubMed] [Google Scholar]
- 4.Choe S-E, et al. Genetic and phylogenetic analysis of South Korean sacbrood virus isolates from infected honey bees (Apis cerana) Vet Microbiol. 2012;157:32–40. doi: 10.1016/j.vetmic.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Ghosh RC, Ball BV, Willcocks MM, Carter MJ. The nucleotide sequence of sacbrood virus of the honey bee: An insect picorna-like virus. J Gen Virol. 1999;80:1541–1549. doi: 10.1099/0022-1317-80-6-1541. [DOI] [PubMed] [Google Scholar]
- 6.Rossmann MG, Johnson JE. Icosahedral RNA virus structure. Annu Rev Biochem. 1989;58:533–573. doi: 10.1146/annurev.bi.58.070189.002533. [DOI] [PubMed] [Google Scholar]
- 7.Kalynych S, et al. Virion structure of iflavirus slow bee paralysis virus at 2.6-Angstrom resolution. J Virol. 2016;90:7444–7455. doi: 10.1128/JVI.00680-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Škubník K, et al. Structure of deformed wing virus, a major honey bee pathogen. Proc Natl Acad Sci USA. 2017;114:3210–3215. doi: 10.1073/pnas.1615695114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rossmann MG, et al. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature. 1985;317:145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- 10.Harrison SC, Olson AJ, Schutt CE, Winkler FK, Bricogne G. Tomato bushy stunt virus at 2.9-Å resolution. Nature. 1978;276:368–373. doi: 10.1038/276368a0. [DOI] [PubMed] [Google Scholar]
- 11.de Miranda JR, et al. Genetic characterization of slow bee paralysis virus of the honeybee (Apis mellifera L.) J Gen Virol. 2010;91:2524–2530. doi: 10.1099/vir.0.022434-0. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J, et al. Three-dimensional structure of the Chinese sacbrood bee virus. Sci China C Life Sci. 2001;44:443–448. doi: 10.1007/BF02879612. [DOI] [PubMed] [Google Scholar]
- 13.Organtini LJ, et al. Honey bee deformed wing virus structures reveal that conformational changes accompany genome release. J Virol. 2017;91:e01795-16. doi: 10.1128/JVI.01795-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rueckert RR, Wimmer E. Systematic nomenclature of picornavirus proteins. J Virol. 1984;50:957–959. doi: 10.1128/jvi.50.3.957-959.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalynych S, Füzik T, Přidal A, de Miranda J, Plevka P. Cryo-EM study of slow bee paralysis virus at low pH reveals iflavirus genome release mechanism. Proc Natl Acad Sci USA. 2017;114:598–603. doi: 10.1073/pnas.1616562114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicklin MJ, Kräusslich HG, Toyoda H, Dunn JJ, Wimmer E. Poliovirus polypeptide precursors: Expression in vitro and processing by exogenous 3C and 2A proteinases. Proc Natl Acad Sci USA. 1987;84:4002–4006. doi: 10.1073/pnas.84.12.4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pallai PV, et al. Cleavage of synthetic peptides by purified poliovirus 3C proteinase. J Biol Chem. 1989;264:9738–9741. [PubMed] [Google Scholar]
- 18.Plevka P, et al. Structure of human enterovirus 71 in complex with a capsid-binding inhibitor. Proc Natl Acad Sci USA. 2013;110:5463–5467. doi: 10.1073/pnas.1222379110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith TJ, et al. The site of attachment in human rhinovirus 14 for antiviral agents that inhibit uncoating. Science. 1986;233:1286–1293. doi: 10.1126/science.3018924. [DOI] [PubMed] [Google Scholar]
- 20.Badger J, et al. Structural analysis of a series of antiviral agents complexed with human rhinovirus 14. Proc Natl Acad Sci USA. 1988;85:3304–3308. doi: 10.1073/pnas.85.10.3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Oers MM. Genomics and biology of iflaviruses. In: Johnson K, Agari S, editors. Insect Virology. Caister Academic Press; Poole, UK: 2010. pp. 231–250. [Google Scholar]
- 22.Tate J, et al. The crystal structure of cricket paralysis virus: The first view of a new virus family. Nat Struct Biol. 1999;6:765–774. doi: 10.1038/11543. [DOI] [PubMed] [Google Scholar]
- 23.Agirre J, et al. Capsid protein identification and analysis of mature Triatoma virus (TrV) virions and naturally occurring empty particles. Virology. 2011;409:91–101. doi: 10.1016/j.virol.2010.09.034. [DOI] [PubMed] [Google Scholar]
- 24.Arnold E, et al. Implications of the picornavirus capsid structure for polyprotein processing. Proc Natl Acad Sci USA. 1987;84:21–25. doi: 10.1073/pnas.84.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harber JJ, Bradley J, Anderson CW, Wimmer E. Catalysis of poliovirus VP0 maturation cleavage is not mediated by serine 10 of VP2. J Virol. 1991;65:326–334. doi: 10.1128/jvi.65.1.326-334.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spurny R, et al. Virion structure of black queen cell virus, a common honeybee pathogen. J Virol. 2017;91:e02100–e02116. doi: 10.1128/JVI.02100-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Squires G, et al. Structure of the Triatoma virus capsid. Acta Crystallogr D Biol Crystallogr. 2013;69:1026–1037. doi: 10.1107/S0907444913004617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hogle JM, Chow M, Filman DJ. Three-dimensional structure of poliovirus at 2.9-Å resolution. Science. 1985;229:1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- 29.Rossmann MG. Viral cell recognition and entry. Protein Sci. 1994;3:1712–1725. doi: 10.1002/pro.5560031010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fuchs R, Blaas D. Uncoating of human rhinoviruses. Rev Med Virol. 2010;20:281–297. doi: 10.1002/rmv.654. [DOI] [PubMed] [Google Scholar]
- 31.Neubauer C, Frasel L, Kuechler E, Blaas D. Mechanism of entry of human rhinovirus 2 into HeLa cells. Virology. 1987;158:255–258. doi: 10.1016/0042-6822(87)90264-9. [DOI] [PubMed] [Google Scholar]
- 32.Garriga D, et al. Insights into minor group rhinovirus uncoating: The X-ray structure of the HRV2 empty capsid. PLoS Pathog. 2012;8:e1002473. doi: 10.1371/journal.ppat.1002473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, et al. A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71. Nat Struct Mol Biol. 2012;19:424–429. doi: 10.1038/nsmb.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geisow MJ, Evans WH. pH in the endosome: Measurements during pinocytosis and receptor-mediated endocytosis. Exp Cell Res. 1984;150:36–46. doi: 10.1016/0014-4827(84)90699-2. [DOI] [PubMed] [Google Scholar]
- 35.Shingler KL, et al. The enterovirus 71 a-particle forms a gateway to allow genome release: A cryoEM study of picornavirus uncoating. PLoS Pathog. 2013;9:e1003240. doi: 10.1371/journal.ppat.1003240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lyu K, et al. Human enterovirus 71 uncoating captured at atomic resolution. J Virol. 2014;88:3114–3126. doi: 10.1128/JVI.03029-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bostina M, Levy H, Filman DJ, Hogle JM. Poliovirus RNA is released from the capsid near a twofold symmetry axis. J Virol. 2011;85:776–783. doi: 10.1128/JVI.00531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: Where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Panjwani A, et al. Capsid protein VP4 of human rhinovirus induces membrane permeability by the formation of a size-selective multimeric pore. PLoS Pathog. 2014;10:e1004294. doi: 10.1371/journal.ppat.1004294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Danthi P, Tosteson M, Li Q-H, Chow M. Genome delivery and ion channel properties are altered in VP4 mutants of poliovirus. J Virol. 2003;77:5266–5274. doi: 10.1128/JVI.77.9.5266-5274.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Miranda JR, et al. 2013. The COLLOS BEEBOOK: Standard Methods for Apis mellifera Pest and Pathogen Research (IBRA, Treforest, UK)
- 42.Scheres SHW. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.