Significance
Gene expression is controlled by sequence-specific transcription factors (TFs), which bind to regulatory sequences in DNA. The degree to which the arrangement of motif sites within regulatory elements determines their function remains unclear. Here, we show that the positional distribution of TF motif sites within nucleosome-depleted regions of DNA fall into six distinct classes. These patterns are highly consistent across cell types and bring together factors that have similar functional and binding properties. Furthermore, the position of motif sites appears to be related to their known functions. Our results suggest that TFs play distinct roles in forming a functional enhancer, facilitated by their position within a regulatory sequence.
Keywords: transcription factor binding, gene regulation, genomics, chromatin structure
Abstract
Gene expression is controlled by sequence-specific transcription factors (TFs), which bind to regulatory sequences in DNA. TF binding occurs in nucleosome-depleted regions of DNA (NDRs), which generally encompass regions with lengths similar to those protected by nucleosomes. However, less is known about where within these regions specific TFs tend to be found. Here, we characterize the positional bias of inferred binding sites for 103 TFs within ∼500,000 NDRs across 47 cell types. We find that distinct classes of TFs display different binding preferences: Some tend to have binding sites toward the edges, some toward the center, and some at other positions within the NDR. These patterns are highly consistent across cell types, suggesting that they may reflect TF-specific intrinsic structural or functional characteristics. In particular, TF classes with binding sites at NDR edges are enriched for those known to interact with histones and chromatin remodelers, whereas TFs with central enrichment interact with other TFs and cofactors such as p300. Our results suggest distinct regiospecific binding patterns and functions of TF classes within enhancers.
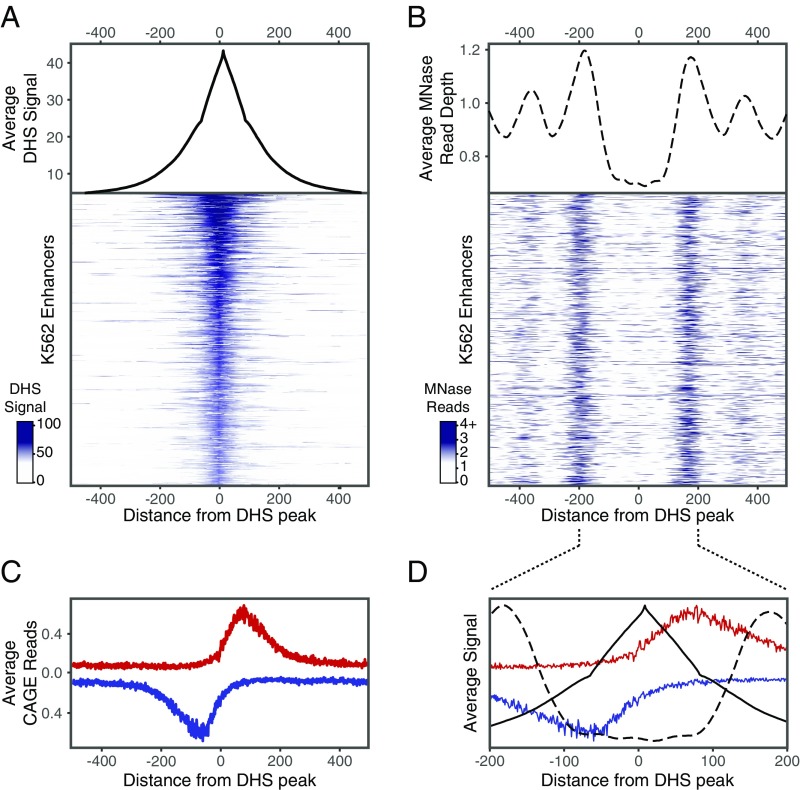
To investigate the characteristic positions of transcription factor (TF)-binding sites in distal regulatory elements (enhancers), we identified active regulatory elements across numerous cell types and characterized predicted functional TF-binding sites within these elements. We defined putative active regulatory elements by first identifying nucleosome-depleted regions of DNA (NDRs) in 47 cell types based on DNaseI-hypersensitive (DHS) sites defined by the Roadmap Epigenomics project (1) and Assay for Transposase-Accessible Chromatin-sequencing (ATAC-seq) experiments performed in each cell type (2–4). We then further selected those NDRs marked by the active chromatin modification H3K27ac using ChIP-sequencing (ChIP-seq) data from the Roadmap Epigenomics project and other studies; two example regions from K562 cells are shown in SI Appendix, Fig. S1A. We and others have previously shown by massively parallel reporter assays (MPRA) that genomic sites satisfying these criteria are highly enriched for enhancer activity compared with other genomic sites and random sequences (5–8). Overall, we identified ∼40,000–160,000 putative active regulatory elements per cell type, together representing a total of ∼500,000 distinct (nonoverlapping) elements (SI Appendix, Fig. S1 B and C). The edges of flanking nucleosomes appear to occur ∼120 ± 50 bp from the peak of the DHS/ATAC-seq signal, as assayed by micrococcal nuclease-digestion assays (MNase-seq) (Fig. 1 A and B). The regions are enriched for transcriptional initiation, consistent with previous reports (9); the peak of transcription initiation is ∼55 bp away from the peak of the DHS/ATAC-seq signal and ∼45 bp before the position of the flanking nucleosome (Fig. 1 C and D). As expected, cell types with similar anatomical and developmental origins tended to have correlated regulatory elements (SI Appendix, Fig. S2). Because developmental enhancers and housekeeping enhancers are typically regulated by distinct sets of TFs (10, 11), in our analysis we distinguished between cell type-restricted enhancers (active in <50% of the cell types) and ubiquitous enhancers (active in >90% of the cell types) (SI Appendix, Fig. S1C).
Fig. 1.
Chromatin structure around putative regulatory NDRs. The nucleosome-depleted region at putative regulatory elements tends to span ∼200 bp centered around the peak of the DHS signal and is generally flanked by well-positioned nucleosomes centered around +200 bp and −200 bp. (A and B) Composite plot (Upper) and heatmap (Lower) of the DHS signal (A) and MNase-seq reads (B) in a 1-kb region aligned around the peak of the DHS signal. Five thousand regions from K562 cells sorted by maximum DHS score are shown in heatmaps. (C) Composite profile of CAGE reads, indicating transcriptional initiation on the plus strand (red) and minus strand (blue) from 14 cell types aligned around the peak of the DHS signal in NDRs. The initiation of gene and enhancer RNA transcription peaks ∼55 bp away from the peak of the DHS signal and is oriented outwards from the accessible region. (D) Overlay of DHS (solid black line), MNase-seq (dashed line), and CAGE (red and blue lines) signals in 400-bp region centered around the peak of the DHS signal.
We next sought to infer functional TF-binding sites within the active regulatory elements. In a recent study (5), we found that TF binding is strongly correlated with the quantitative DNA accessibility of a region. Furthermore, the TF motifs associated with enhancer activity in reporter assays in a cell type corresponded closely to those that are most enriched in the genomic sequences of active regulatory elements in that cell type (5). In these assays, disrupting occurrences of the 20–30 most enriched motifs in such genomic regulatory sequences frequently caused significant changes in enhancer activity, indicating that many represent functional TF-binding sites. Together, these results suggest that occurrences of highly enriched motifs in highly accessible regions very likely represent functional TF-binding sites for a cell type.
We used this approach to define a set of candidate functional TF-binding sites. For each of the 47 cell types, we selected the 7,500 cell-type restricted NDRs (active in <50% of cell types) with the strongest DHS/ATAC-seq signals, with an average of 6% being promoter-proximal regions [<1 kb from an annotated transcription start site (TSS)] and 94% being distal enhancers. Within these regions, we identified all occurrences of 1796 known motifs (corresponding to 777 TFs) and focused on the 20 most enriched motifs in the cell type (after removing highly similar motifs) (Materials and Methods and SI Appendix, Fig. S3A). Overall, these enriched motifs corresponded to 103 different TFs across the 47 cell types. As expected, the motif-enrichment profiles were correlated among related cell types (SI Appendix, Fig. S3B).
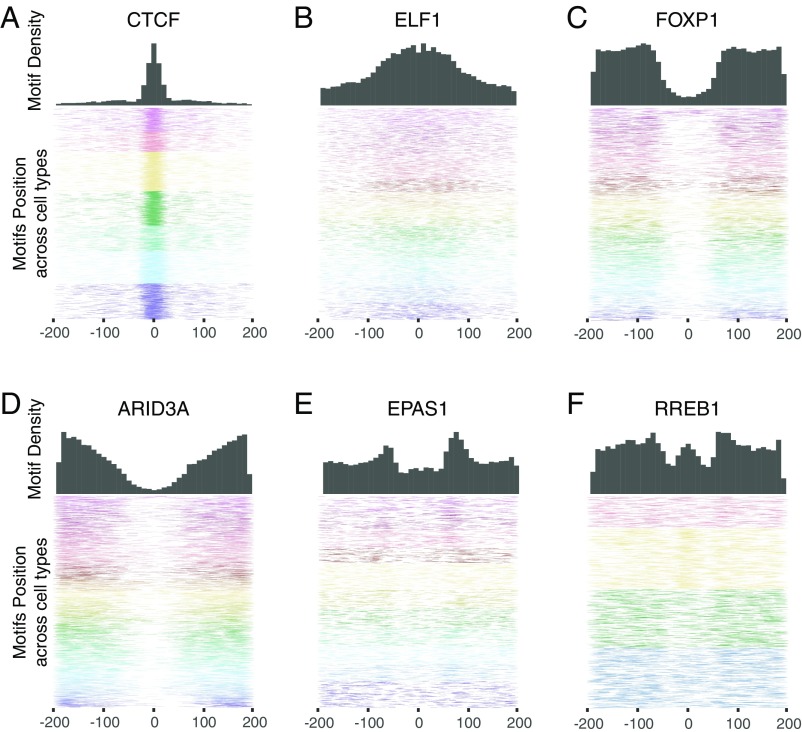
We then studied the positions of inferred binding sites for each of the 103 TFs relative to the peak of the DHS/ATAC-seq signal in the active regulatory elements (SI Appendix, Fig. S4). Different TFs show strikingly different positional binding-site patterns (Fig. 2 and SI Appendix, Figs. S5 and S6). Some are strongly concentrated at the peak of the DNase/ATAC-seq signal (e.g., CTCF); some are enriched over a more widely distributed central region (e.g., ELF1); some are clustered near the edges of the region (e.g., FOXP1 and ARID3A); and some tend to bind at a specific distance from the center of the NDRs (e.g., EPAS1 and RREB1).
Fig. 2.
Positional binding patterns of TF motifs show striking differences. Distribution of the position of motif sites for CTCF (A), ELF1 (B), FOXP1 (C), ARID3A (D), EPAS1 (E), and RREB1 (F) across NDR regions, centered around the peak of the DHS signal. (Upper) Histograms show the density of motif sites in 10-bp bins tiled across the NDR. (Lower) Heatmaps show the position of 10,000 motif sites in NDRs. Colors indicate motif sites in different cell types (see SI Appendix, Fig. S5 for the color key).
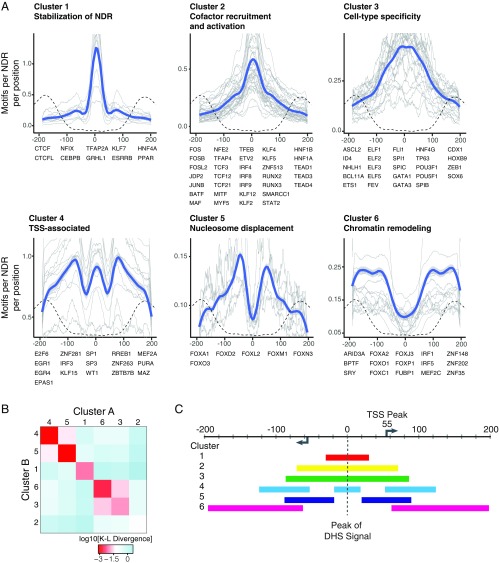
To classify these patterns, we calculated the density profiles in ±200-bp regions around the peak and clustered them using k-medoid clustering (Materials and Methods). The analysis identified six clusters of distinct position patterns (Fig. 3A). The clusters are clearly significant: The mean Kullback–Leibler divergence between density profiles within the same cluster is one to two orders of magnitude smaller than the mean divergence between density profiles in different clusters (Fig. 3B), and the density profiles cannot be explained by local sequence composition (SI Appendix, Fig. S7 A and B). Three of these clusters represent motifs that occur most frequently near the center of NDRs, while the other three clusters tend to occur nearer to the edges (Fig. 3C).
Fig. 3.
TF motif position patterns fall into six distinct clusters. (A) Motif-density profiles in 400-bp regions centered around the peak of the DHS signal (gray lines) were clustered using k-medoids clustering with k = 6. Density profiles were generated by calculating the frequency of motif occurrences in 20-bp bins tiled every 1 bp in the region. Blue lines depict the smoothed overall density profile of the cluster using the LOESS method. MNase-seq read density (indicating the position of the flanking nucleosomes) is shown by dashed dotted lines for context. (B) Average Kullback–Leibler divergence between the motif-density profiles of pairs of motifs in the same cluster (diagonal boxes) and different clusters (off-diagonal boxes). Motif-density profiles within the same cluster are substantially more similar than those in different clusters. (C) Schematic of NDR structure and motif positions. The arrows indicate the peak of transcriptional initiation estimated from CAGE data. The colored bars represent regions for each cluster with motif densities above the mean. Tick marks occur at 20-bp intervals.
Cluster 1 contains 10 TFs with inferred binding sites that are strongly biased toward the peak of highest DNA accessibility at the middle of the NDR, suggesting that their binding directly shapes local chromatin architecture. For six of these TFs (CTCF, NF-I, C/EBPβ, KLF7, GRHL1, and TFAP2) there is clear functional evidence to support this notion: (i) CTCF induces stably positioned arrays of nucleosomes around its genomic binding sites (12); (ii) NF-I, C/EBPβ, KLF7, and GRHL1 can function as pioneer factors that can establish and maintain chromatin accessibility (13–18); (iii) a recent systematic analysis of the TF-dependent changes in chromatin accessibility induced by the binding of 733 TFs identified CTCF, KLF7, and TFAP2 as having some of the strongest effects on local chromatin accessibility during ES cell differentiation (19); (iv) CTCF, NF-I, C/EBPβ, and GRHL1 show unusually stable binding to DNA and long residence times (14, 20–22); and (v) motifs in cluster 1 have especially strong DNaseI footprinting signals (SI Appendix, Fig. S8), a feature associated with a slow DNA-binding off-rate (17, 23). The properties of the six TFs may enable them to serve as central anchor points for displacing the central nucleosome, adapting the surrounding chromatin, and stabilizing the NDR and flanking nucleosomes (14).
The remaining three TFs in cluster 1 are nuclear receptors (ESRRB, HNF4A, and PPAR). Unlike the other TFs in the cluster, nuclear receptors are characterized by transient binding to DNA with short residence times (24, 25) and localize almost exclusively to preaccessible chromatin (16, 25–27). Nuclear receptor binding to genomic motif sites is often aided by assisted loading by a partner factor, which binds to a site overlapping or adjacent to the nuclear receptor motif site and opens the chromatin (28). Notably, two of the pioneer TFs in cluster 1 (C/EBPβ and NF-I) have been shown to catalyze the assisted loading of several nuclear receptors (13, 16, 29–31). The central location of the nuclear receptor motifs may be related to the assisted loading by pioneer TFs in cluster 1.
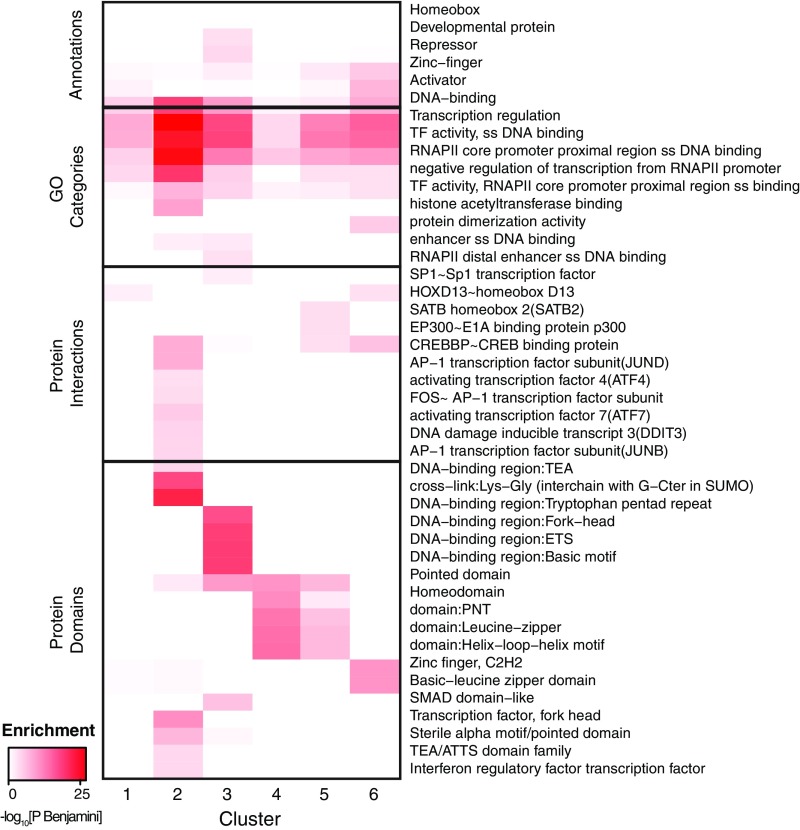
Cluster 2 contains 31 TFs whose binding sites also are peaked at the center of the NDR but with a wider distribution than for cluster 1. The cluster is strongly enriched for transcriptional activators [Gene Ontology (GO) category enrichment, PBenjamini = 3.2 × 10−16], such as the activator protein 1 (AP-1) subunits (JUN, FOS, ATF, and MAF factors) and activating factors from the TCF, TEA, RUNX, IRF, and KLF families. Based on known interactions reported in the bioGRID and IntAct databases (32, 33), these TFs are enriched for interactions with numerous transcriptional coactivators, including p300, CREB-binding protein (CBP), YAP1, KDM1A, KAT2B, and WWTR1 (Fig. 4 and SI Appendix, Table S1). Furthermore, the TFs in this cluster interact frequently with each other [average of 1.8 pairwise interactions among the 32 TFs vs. 0.7–1.4 (mean = 1.0) interactions among the TFs in other clusters], suggesting they could cooperatively activate transcription. For example, studies of the IFNβ enhancer have shown that two TFs from this cluster (ATF2 and Jun) bind overlapping motif sites to form a scaffold that recruits CBP/p300 through multidentate interactions (34), leading to synergistic transcriptional activation in response to viral infection (35, 36). Interestingly, TFs in cluster 2 are twice as likely to participate in signaling pathways as the TFs in other clusters [52% of TFs in cluster 2 vs. 16–30% of TFs in other clusters, based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (37)] (Fig. 4 and SI Appendix, Table S1), and AP-1 factors are required to maintain accessible chromatin to facilitate the binding of stimulus-regulated TFs (38). Therefore the tightly clustered pattern of these motifs in this cluster may promote cooperativity by both facilitating TF–TF interactions and positioning TFs to form complexes that contact multiple sites on cofactors, thereby allowing enhancers to link multiple signaling pathways and respond in a highly synergistic fashion to specific regulatory cues.
Fig. 4.
TF clusters are enriched for distinct functional and structural properties. Selected enrichments for general annotation (Entrez Gene), GO categories, protein–protein interactions, and protein structural domains in the TF clusters. All terms included in the heatmap are significantly enriched (PBenjamini < 0.05) in at least one cluster. See SI Appendix, Table S3 for all significant enrichments.
Cluster 3, which contains 25 TFs, also peaks at the center of the NDR with a broader distribution than cluster 2. These TFs are generally characterized by greater cell-type specificity in expression across the 57 cell types profiled in the Epigenomics Roadmap project and greater motif enrichment than the TFs in other clusters (SI Appendix, Table S1). Consistent with this observation, cluster 3 contains numerous TFs that play critical roles in development, including all the homeobox, POU, SOX, ETS, and GATA factors in our dataset (39–44) (SI Appendix, Table S1). Furthermore, 20 of the 23 TFs have functional annotations in GO related to differentiation and development in a wide range of tissues (Fig. 4 and SI Appendix, Table S2), including erythrocytes (GATA1, GATA3, and ETS1), myeloid and lymphoid cells (SPI1), osteoblasts (TP63 and ID4), keratinocytes (TP63 and POU3F1), blastocysts (SPIC, POU5F1, and ELF3), neurons (ASCL2, FEV, and ZEB1), and more. Although clusters 2 and 3 may represent a continuum of broad-occupancy profiles, the TFs in cluster 3 have fewer annotated interactions with cofactors and other TFs than the TFs in cluster 2 (average 5.2 vs. 10.2 interactions per TF). One possible explanation is that the TFs in cluster 3 participate in fewer physically mediated cooperative interactions and therefore are less tightly clustered in the NDR. These TFs may function more independently or through indirect cooperation with other factors.
Cluster 4, which contains 16 TFs, is unusual in several respects. The motif profiles show both a central peak and flanking peaks at ∼70 bp upstream and downstream. Moreover, many of the motifs in this cluster are asymmetric. When the NDRs are oriented so that the motif occurrences for each TF all appear on the same strand (SI Appendix, Fig. S9A), the motif occurrences in the flanking peaks show a clear preferred orientation relative to the center of the NDRs—that is, one of the two reverse-complementary sequences defining the motifs preferentially points inward (SI Appendix, Figs. S9B and S10A). This bias indicates that one side of the TFs is generally positioned facing the edges of the NDR, while the other side faces the NDR core.
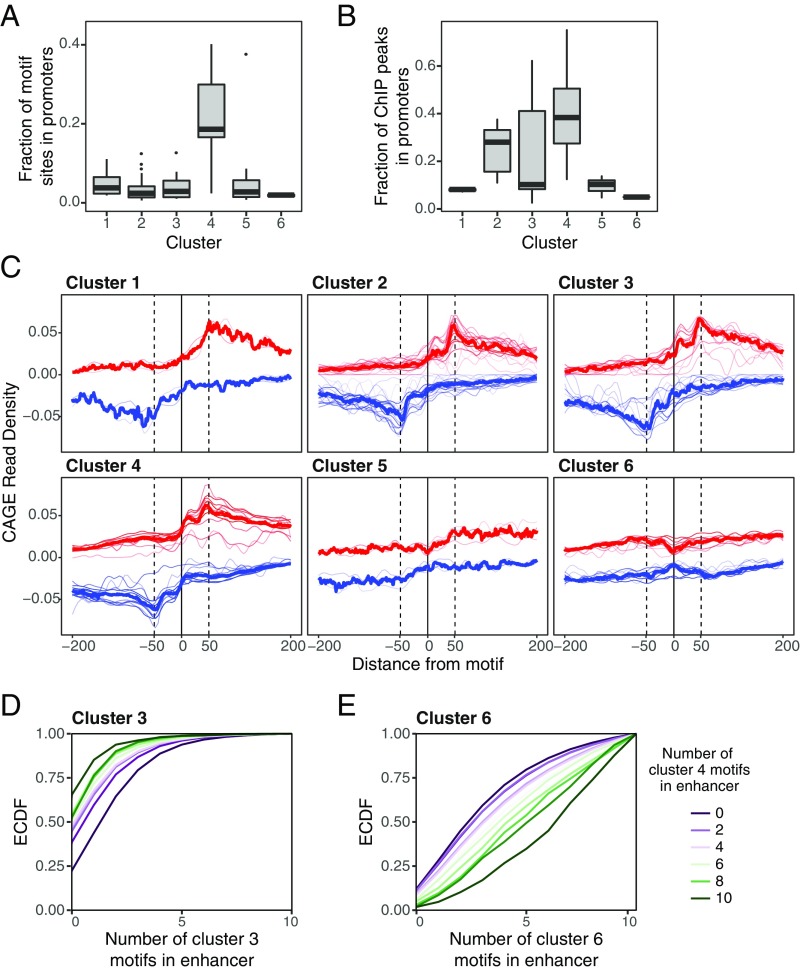
The motifs in cluster 4 are also strongly enriched in promoter-proximal regions (6% of such NDRs contain occurrences for motifs in cluster 4 vs. 1–3% for other clusters) (Fig. 5A). ENCODE ChIP-seq data for 39 TFs in our dataset show greater enrichment in promoter regions for TFs in cluster 4 than for TFs in other clusters (38% of reported peaks within <1 kb of a TSS vs. 8–28% for other clusters) (Fig. 5B). One of the TFs in this cluster, SP1, is a well-characterized promoter-proximal factor that binds GC-rich elements in a wide variety of cellular and viral promoters. Many of the other TFs in the cluster (including SP3, EGR1, EPAS1, ZBTB7B, E2F, KLF15, MEF2C, WT1, and PURA) also bind GC-rich motifs and are known to interact with SP1 at promoters (45–55). We compared the motif-density profiles in NDRs classified as promoter-proximal versus distal enhancers but found them to be indistinguishable (SI Appendix, Fig. S11).
Fig. 5.
TFs in cluster 4 are enriched in promoters and are associated with transcriptional initiation. (A) The fraction of motif sites in NDRs in our analysis that occur in promoters (<1 kb upstream of the annotated TSS) for TFs in each cluster. (B) The fraction of ChIP-seq peaks for TFs in each cluster that overlap promoter (data for 39 TFs profiled in ENCODE are included). Cluster 4 motifs and TF binding occur in promoters far more frequently than do motifs in other clusters. (C) Composite of CAGE reads on the plus strand (red) and minus strand (blue) aligned to the center of each TF motif. Thin red and blue lines correspond to CAGE profiles of individual TF motifs, and thick red and blue lines show the average CAGE profile of all motifs in the cluster. Motifs in clusters 3 and 4 show a peak of transcriptional initiation at the location of the motif site. (D and E) Empirical cumulative distribution function (ECDF) of the number of cluster 3 (D) and cluster 6 (E) motif sites in NDRs, conditional on the number of cluster 4 motifs. NDRs with cluster 4 motif sites are coenriched with cluster 3 motif sites and depleted of cluster 6 motif sites.
TFs in this cluster are also enriched for interactions with p300 [false-discovery rate (FDR) = 2.6 × 10−6] and Dnmt1, a DNA methyltransferase that plays a key role in maintaining CpG island methylation (56) (FDR = 0.03). Notably, functional studies have demonstrated that SP1 stimulates transcription when bound close to the initiation site but not in distal positions (57, 58), unlike distal enhancer-binding factors from clusters 1–3. These results suggest that SP1 and other TFs in cluster 4 may belong to a distinct functional class of TFs with specialized promoter-associated functions.
Because a key function of promoters is transcript initiation, we hypothesized that the flanking peaks and orientation of TFs in cluster 4 might reflect a role in establishing or stabilizing TSSs at both promoters and enhancers. Recent studies have suggested that, in addition to such features as TATA boxes and INR elements, TF-binding sites also contribute to determining the position of the TSS (9, 59). To examine the relationship of TFs in each cluster with the TSS, we examined cap analysis of gene expression (CAGE) data for both enhancer and promoter-proximal NDRs for 14 of the cell lines in our dataset (60). Transcriptional initiation tends to peak at 50–60 bp from the center of the NDRs (as noted above) (Fig. 1C) and ∼50 bp away for TF motif occurrences (Fig. 5C). However, 64% of TFs in cluster 4 and 42% of TFs in cluster 3 (vs. 0–8% in other clusters) show an additional peak of transcriptional initiation ∼10 bp away from the location of motifs sites (EGR1, EGR4, MAZ, PURA, SP1, SP3, ZBTB7B, and ZNF281 from cluster 4 and ELF1, ELF2, ELF5, FLI1, SPI1, and SPIC from cluster 3) (Fig. 5C). This observation suggests these TFs play unique roles in positioning the site of initiation.
Cluster 5 contains six TFs whose binding sites are not enriched at the center of NDRs but have peaks at ∼60 bp upstream or downstream. The TFs in this cluster all belong to the FOX family of TFs and include the two best-characterized pioneer factors, FOXA and FOXO. The DNA-binding domain (DBD) of FOX factors structurally resembles the DBD of linker histones H1 and H5 (61, 62), and FOXA factors can compete for binding to linker histone-binding sites, which are located near the edges of the core nucleosome, ∼65 bp away from its center (61, 63–65). However, whereas linker histone binding leads to the compaction of nucleosomal arrays, FOXA binding destabilizes nucleosomes and opens the region for binding by other TFs (66–68). Since enhancer activation typically entails the elimination of a well-positioned central nucleosome (69), motif sites for FOXA and other FOX factors in cluster 5 may be positioned ±60 bp to displace linker histones and destabilize the central nucleosome, helping other TFs bind their target sites.
Finally, cluster 6 contains 14 TFs with binding sites enriched near the edges of the accessible region (80–200 bp from the center), suggesting these TFs could interact with the surrounding chromatin. As with cluster 4, the TFs in cluster 6 have asymmetric motifs and mostly exhibit a preferred orientation relative to the center of the region (SI Appendix, Figs. S9 and S10B), allowing directional interactions with the surrounding nucleosomes and larger chromatin landscape. Consistent with this notion, 10 of the 14 TFs in cluster 6 are known to play roles in chromatin remodeling. These include BPTF, the DNA-binding subunits of nucleosome remodeling factor (NURF), which recognizes H3K4me3 and facilitates ATP-dependent nucleosome sliding (70–72), ARID3A, which facilitates the opening of the IgH enhancer (73–75), and several FOX factors, which interact with histones and mediate recruitment of chromatin remodeling complexes such as SWI/SNF (68, 76). Many of the motifs in this cluster are A/T-rich (SI Appendix, Fig. S10). It is possible that they also recruit additional members of the ARID (A+T-rich interaction domain) family that binds nonspecifically to A/T sequences and has been implicated in chromatin remodeling, including ARID1A/BAF250, the DNA-binding subunit of the BAF chromatin remodeling complex (77).
The TFs in cluster 6 also play roles in nuclear attachment, DNA bending, and DNA unwinding. These TFs are enriched for interactions with the chromatin organizers SATB1 and SATB2, which induce chromatin looping and tether DNA to the nuclear matrix (78, 79). For example, ARID3A binds to sites on the periphery of the IgH enhancer to mediate the attachment of the nuclear matrix (80). Several of the TFs (ARID3A, SRY, and YY1) induce significant DNA bending (74, 81, 82), facilitating TF binding and TF–TF interactions (83, 84). Finally, some (SRY and FUBP1) unwind the DNA double helix, which can promote transcriptional initiation and attachment to the nuclear matrix (82, 85, 86).
To test directly whether TFs in cluster 6 interact with the surrounding nucleosomes, we used MNase-seq data from two cell types (GM12878 and K562) to infer the position of the flanking nucleosomes for each individual NDR and then aligned the TF motifs. For the TFs present in these cell types, we examined the motif distribution relative to the inferred edge of the flanking nucleosome (rather than to the peak of the DHS signal). The TFs in clusters 1–5 did not show peaks of motif sites adjacent the nucleosome edge, but five of the eight TFs in cluster 6 (FOXC1, FOXJ3, FOXO1, FOXP1, and ARID3A) showed a peak (SI Appendix, Fig. S12). The remaining three TFs (FUBP1, IRF1, and IRF5) are not known to play roles in chromatin remodeling.
Finally, we wondered whether certain classes of TFs tend to co-occur in enhancers. To investigate this, we examined whether the distribution of motif sites from each class in the NDRs varied with the presence or absence of motif sites from each of the other classes (Fig. 5 D and E and SI Appendix, Table S3). We counted the number of nonoverlapping motif sites from each cluster in the NDRs and calculated the odds ratio (OR) for coenrichment between the motif from each pair of clusters. To control for motif similarities, we also calculated the baseline OR for each pair of clusters in shuffled sequences. Significantly coenriched or codepleted cluster pairs were defined as pairs for which the OR falls outside the 95% CI of the OR in shuffled sequences (SI Appendix, Table S3). We found that all six clusters showed significant preferences for coenrichment and codepletion with specific other clusters (SI Appendix, Table S3). For example, regulatory elements with TF motif sites in cluster 4 (associated with TSS-related functions) contain significantly more TF motif sites from cluster 3 (associated with cell type-specific activation) (Fig. 5D) and significantly fewer motif sites from clusters 5 and 6 (associated with nucleosome remodeling and chromatin architecture) (Fig. 5E) than regulatory elements without cluster 4 motifs. Importantly, these cluster associations are consistent across cell types, even though the specific set of TFs active in each cell type differs (SI Appendix, Fig. S13). Thus, the TF clusters may constitute a general regulatory code, with different cell types substituting specific TFs to activate different sets of enhancers.
It has long been suggested that TFs may belong to different functional classes. In some cases, prior biological knowledge of certain TFs has been used to categorize TFs into classes, such as pioneer factors that have the capability to bind motif sites in closed chromatin versus nonpioneer factors that bind motif sites only in open chromatin and cell type-specific versus ubiquitous factors. However, there have been few systematic approaches to recognize distinct classes and properties independent of the known biological properties of the individual TFs. One such functional study was recently performed in Drosophila, in which investigators asked which TFs could substitute for each other across a variety of regulatory contexts (10).
Here, we show that, solely by looking at the positional distribution of motif sites within NDRs, we are able to recognize six distinct classes of TFs. These classes bring together factors that have a number of similar properties, such as binding stability, interactions with other TFs and cofactors, cell-type specificity, and pioneering ability. Furthermore, the position of motif sites appears to be related to their known functions—for example, localizing pioneer factors to the optimal positions to displace nucleosomes and targeting chromatin remodelers in close proximity to flanking nucleosome.
The degree to which the arrangement of motif sites within regulatory elements determines their function remains an open question. At one end of the spectrum, there are examples of enhancesomes, such as the IFNβ enhancer, that are exquisitely sensitive to the spacing and orientation of the motif sites (34, 87, 88). However, the activity of other regulatory elements, referred to as “billboard” enhancers, appears to be relatively insensitive to the arrangement of motif sites (89–91). Instead, our work suggests a different kind of constraint, whereby TFs play distinct roles in forming a functional enhancer, facilitated by their position within a regulatory sequence.
The classes identified here also help shed light on the properties of some less characterized TFs. For example, they suggest that several other FOX factors in cluster 5 may use a mechanism similar to that of FOXA1 to displace nucleosomes and that the uncharacterized zinc finger TFs in cluster 6 (ZNF148, ZNF202, and ZNF35) may have pioneering abilities. In addition, the positional preferences identified may prove useful in building predictors of enhancer activity and recognizing functional enhancers in genomic sequence.
While here we focused on the classes of TFs, these results naturally raise the question of whether different functional classes of enhancers are formed based on these classes of TFs. Identifying such enhancer classes may shed light on the classes of TFs that must come together to accomplish all the functions necessary to build a functional enhancer. Finally, in addition to helping us understand natural enhancers, better knowledge about the constraints and the functional implications of TF positions may aid in creating synthetic enhancers with specific properties that can be used in synthetic biology.
Materials and Methods
ATAC-Seq for Jurkat and U937 Cell Lines.
Cells were washed with ice-cold FACS buffer and were kept on ice until cell sorting. Twenty-five thousand live cells from each condition were sorted into FACS buffer and were pelleted by centrifugation at 500 × g for 5 min at 4 °C in a precooled fixed-angle centrifuge. Cell lines then were tagmented according to the previously described Fast-ATAC protocol (92). Briefly, all supernatant was removed with care taken not to disturb the not-visible cell pellet. Transposase mixture (50 μL: 25 μL of 2× TD, 2.5 μL of TDE1, 0.5 μL of 1% digitonin, 22 μL of nuclease-free water) (catalog no. FC-121-1030, Illumina; catalog no. G9441, Promega) was added to the cells, and the pellet was dissociated by pipetting. Transposition reactions were incubated at 37 °C for 30 min in an Eppendorf ThermoMixer with agitation at 300 rpm. Transposed DNA was purified using a Qiagen MinElute Reaction Cleanup kit (catalog no. 28204), and purified DNA was eluted in 12 μL of elution buffer (10 mM Tris⋅HCl, pH 8). Transposed fragments were amplified and purified as described previously (93) with modified primers (94). Libraries were quantified using qPCR before sequencing. All Fast-ATAC libraries were sequenced using paired-end, dual-index sequencing on a NextSeq sequencer (Illumina) with 76 × 8 × 8 × 76 cycle reads at an average read depth of 30 million reads per sample.
Definition of NDRs.
To define NDRs for our analysis, we used DNaseI-seq and H3K27ac ChIP-seq data for 45 cell types in the Epigenomics Roadmap and ENCODE Projects (1, 60). We supplemented this dataset with ATAC-seq data for Jurkat and U937 cells generated in the N.H. laboratory and H3K27ac ChIP-seq data for Jurkat and U937 cells from studies deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (accession no. SRR1057274) (95) and the European Nucleotide Archive database (accession no. ERR671846), respectively. We aligned the ATAC-seq and H3K27ac data for Jurkat and U937 cells as described in ref. 96 and called peaks using MACS2 (97) with the standard parameters used by the Epigenomics Roadmap Project. To select our initial set of NDRs, we intersected DHS/ATAC-seq narrowPeaks regions and H3K27ac gappedPeaks regions. We then filtered out NDRs that were present in more than 24 (50%) of the cell types in our analysis and selected the top 7,500 cell type-restricted NDRs for motif enrichment and positioning analysis. We defined the coordinates in the NDRs relative to the summit called by MACS2 (i.e., the position with the maximum DHS/ATAC-seq signal). For MNase-seq analysis, we used data from GM12878 and K562 cells generated by the ENCODE project. The center of the nucleosomes flanking the NDRs was estimated by identifying the position with the highest MNase-seq read coverage in the 300 bp upstream and downstream of the peak of the DHS signal.
Motif Enrichment Analysis.
We calculated motif counts for all vertebrate motifs in TRANSFAC (98), JASPAR (99), and CIS-BP (100) in the genomic NDR sequences as well as scrambled genomic NDR sequences (holding dinucleotide frequencies constant). To identify enriched motifs in each cell type, we used AME (101) with the mhg method to calculate the enrichment of the total number of matches of each motif in the genomic sequences compared with the scrambled sequences. When the combined databases contained multiple position weight matrices (PWMs) corresponding to a single TF, we selected the most enriched motif in each cell type corresponding to each TF. To remove highly similar motifs, we calculated the pairwise similarity of the motifs using the R package PWMEnrich and removed motifs that had a similarity of >0.8 with a more highly enriched motif. We then selected the top 20 motifs from the filtered list in each cell type for positioning analysis. We called motif sites in the genomic and scrambled sequences by running FIMO (102) with a P value threshold of 10−4.
Motif-Position Profiles and Clustering.
To analyze the positioning of the motifs with NDRs, we collapsed the motif matches to their central position and calculated the density of each motif in 20-bp windows tiled every 1 bp across the 400 bp centered around the position of maximum DHS/ATAC signal in each NDR. The motif-position profiles were then clustered using the pam function from the R package cluster with k = 6.
To assess how much each motif-position profile is due to the variation in dinucleotide content across the regions, we calculated the background motif-density profiles in shuffled sequences, holding the dinucleotide content at each position constant, and normalized the genomic-density profiles by subtracting out the background motif frequencies (SI Appendix, Fig. S7B).
TF Cluster Feature Enrichment Analysis.
Enrichment analysis was performed using DAVID (103) for each of the six clusters for four types of features: protein domains (PFAM, PIR, and SMART), functional annotations (GO and Entrez Gene), protein–protein interactions (BioGRID interaction and intact databases), and pathways (KEGG and BioCarta). P values were calculated using the Benjamini correction for multiple testing.
TF Coenrichment Analysis.
We tested for coenrichment and codepletion of motifs from the six TF motif clusters in genomic NDR sequences using a Fisher exact test. For each pair of cluster A and cluster B, we calculated the OR that a genomic sequence contains a motif from cluster B, conditional on the presence of a motif from cluster A. To control for motif similarities between motifs in different clusters, we also calculated the same OR in scrambled sequences (holding dinucleotide content constant). To identify significantly coenriched or codepleted pairs, we selected pairs for which the 95% CI of the genomic OR did not overlap the 95% CI of the shuffled OR.
Supplementary Material
Acknowledgments
We thank Karen Adelman, Telmo Henriques, Bradley Bernstein, Aviv Regev, Cigall Kadoch, Seth Cassel, and Kaylyn Williamson for valuable comments and discussion and Ray Louis for help with DNase footprint analysis. This work was supported by National Human Genome Research Institute Grant 2U54HG003067-10 (to E.S.L.) and National Institute of General Medical Sciences Grant T32GM007753) (to S.R.G.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE115438).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804663115/-/DCSupplemental.
References
- 1.Kundaje A, et al. Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Felsenfeld G, Boyes J, Chung J, Clark D, Studitsky V. Chromatin structure and gene expression. Proc Natl Acad Sci USA. 1996;93:9384–9388. doi: 10.1073/pnas.93.18.9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gross DS, Garrard WT. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- 4.Scruggs BS, et al. Bidirectional transcription arises from two distinct hubs of transcription factor binding and active chromatin. Mol Cell. 2015;58:1101–1112. doi: 10.1016/j.molcel.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grossman SR, et al. Systematic dissection of genomic features determining transcription factor binding and enhancer function. Proc Natl Acad Sci USA. 2017;114:E1291–E1300. doi: 10.1073/pnas.1621150114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kheradpour P, et al. Systematic dissection of regulatory motifs in 2000 predicted human enhancers using a massively parallel reporter assay. Genome Res. 2013;23:800–811. doi: 10.1101/gr.144899.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muerdter F, et al. Resolving systematic errors in widely used enhancer activity assays in human cells. Nat Methods. 2018;15:141–149. doi: 10.1038/nmeth.4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.White MA, Myers CA, Corbo JC, Cohen BA. Massively parallel in vivo enhancer assay reveals that highly local features determine the cis-regulatory function of ChIP-seq peaks. Proc Natl Acad Sci USA. 2013;110:11952–11957. doi: 10.1073/pnas.1307449110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Core LJ, et al. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat Genet. 2014;46:1311–1320. doi: 10.1038/ng.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stampfel G, et al. Transcriptional regulators form diverse groups with context-dependent regulatory functions. Nature. 2015;528:147–151. doi: 10.1038/nature15545. [DOI] [PubMed] [Google Scholar]
- 11.Zabidi MA, et al. Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature. 2015;518:556–559. doi: 10.1038/nature13994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu Y, Sinha M, Peterson CL, Weng Z. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 2008;4:e1000138. doi: 10.1371/journal.pgen.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hebbar PB, Archer TK. Chromatin-dependent cooperativity between site-specific transcription factors in vivo. J Biol Chem. 2007;282:8284–8291. doi: 10.1074/jbc.M610554200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denny SK, et al. Nfib promotes metastasis through a widespread increase in chromatin accessibility. Cell. 2016;166:328–342. doi: 10.1016/j.cell.2016.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plachetka A, et al. C/EBPbeta induces chromatin opening at a cell-type-specific enhancer. Mol Cell Biol. 2008;28:2102–2112. doi: 10.1128/MCB.01943-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grøntved L, et al. C/EBP maintains chromatin accessibility in liver and facilitates glucocorticoid receptor recruitment to steroid response elements. EMBO J. 2013;32:1568–1583. doi: 10.1038/emboj.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soufi A, et al. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell. 2015;161:555–568. doi: 10.1016/j.cell.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naval-Sánchez M, Potier D, Hulselmans G, Christiaens V, Aerts S. Identification of lineage-specific cis-regulatory modules associated with variation in transcription factor binding and chromatin activity using Ornstein-Uhlenbeck models. Mol Biol Evol. 2015;32:2441–2455. doi: 10.1093/molbev/msv107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherwood RI, et al. Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nat Biotechnol. 2014;32:171–178. doi: 10.1038/nbt.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao Z, Umek RM, McKnight SL. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 1991;5:1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- 21.Nakahashi H, et al. A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Rep. 2013;3:1678–1689. doi: 10.1016/j.celrep.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nevil M, Bondra ER, Schulz KN, Kaplan T, Harrison MM. Stable binding of the conserved transcription factor grainy head to its target genes throughout Drosophila melanogaster development. Genetics. 2017;205:605–620. doi: 10.1534/genetics.116.195685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sung MH, Guertin MJ, Baek S, Hager GL. DNase footprint signatures are dictated by factor dynamics and DNA sequence. Mol Cell. 2014;56:275–285. doi: 10.1016/j.molcel.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mazza D, Abernathy A, Golob N, Morisaki T, McNally JG. A benchmark for chromatin binding measurements in live cells. Nucleic Acids Res. 2012;40:e119. doi: 10.1093/nar/gks701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharp ZD, et al. Estrogen-receptor-alpha exchange and chromatin dynamics are ligand- and domain-dependent. J Cell Sci. 2006;119:4101–4116, and erratum (2006) 119:4365. doi: 10.1242/jcs.03161. [DOI] [PubMed] [Google Scholar]
- 26.John S, et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siersbæk R, et al. Extensive chromatin remodelling and establishment of transcription factor ‘hotspots’ during early adipogenesis. EMBO J. 2011;30:1459–1472. doi: 10.1038/emboj.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voss TC, et al. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell. 2011;146:544–554. doi: 10.1016/j.cell.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chávez S, Beato M. Nucleosome-mediated synergism between transcription factors on the mouse mammary tumor virus promoter. Proc Natl Acad Sci USA. 1997;94:2885–2890. doi: 10.1073/pnas.94.7.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hebbar PB, Archer TK. Nuclear factor 1 is required for both hormone-dependent chromatin remodeling and transcriptional activation of the mouse mammary tumor virus promoter. Mol Cell Biol. 2003;23:887–898. doi: 10.1128/MCB.23.3.887-898.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lefterova MI, et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008;22:2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chatr-Aryamontri A, et al. The BioGRID interaction database: 2017 update. Nucleic Acids Res. 2017;45:D369–D379. doi: 10.1093/nar/gkw1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orchard S, et al. The MIntAct project–IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014;42:D358–D363. doi: 10.1093/nar/gkt1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thanos D, Maniatis T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 35.Kim TK, Maniatis T. The mechanism of transcriptional synergy of an in vitro assembled interferon-beta enhanceosome. Mol Cell. 1997;1:119–129. doi: 10.1016/s1097-2765(00)80013-1. [DOI] [PubMed] [Google Scholar]
- 36.Carey M. The enhanceosome and transcriptional synergy. Cell. 1998;92:5–8. doi: 10.1016/s0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- 37.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biddie SC, et al. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol Cell. 2011;43:145–155. doi: 10.1016/j.molcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Charron F, Nemer M. GATA transcription factors and cardiac development. Semin Cell Dev Biol. 1999;10:85–91. doi: 10.1006/scdb.1998.0281. [DOI] [PubMed] [Google Scholar]
- 40.Mallo M, Wellik DM, Deschamps J. Hox genes and regional patterning of the vertebrate body plan. Dev Biol. 2010;344:7–15. doi: 10.1016/j.ydbio.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maroulakou IG, Bowe DB. Expression and function of Ets transcription factors in mammalian development: A regulatory network. Oncogene. 2000;19:6432–6442. doi: 10.1038/sj.onc.1204039. [DOI] [PubMed] [Google Scholar]
- 42.Rosenfeld MG. POU-domain transcription factors: Pou-er-ful developmental regulators. Genes Dev. 1991;5:897–907. doi: 10.1101/gad.5.6.897. [DOI] [PubMed] [Google Scholar]
- 43.Sarkar A, Hochedlinger K. The sox family of transcription factors: Versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013;12:15–30. doi: 10.1016/j.stem.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ting CN, Olson MC, Barton KP, Leiden JM. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature. 1996;384:474–478. doi: 10.1038/384474a0. [DOI] [PubMed] [Google Scholar]
- 45.Karlseder J, Rotheneder H, Wintersberger E. Interaction of Sp1 with the growth- and cell cycle-regulated transcription factor E2F. Mol Cell Biol. 1996;16:1659–1667. doi: 10.1128/mcb.16.4.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khachigian LM, Williams AJ, Collins T. Interplay of Sp1 and Egr-1 in the proximal platelet-derived growth factor A-chain promoter in cultured vascular endothelial cells. J Biol Chem. 1995;270:27679–27686. doi: 10.1074/jbc.270.46.27679. [DOI] [PubMed] [Google Scholar]
- 47.Koizume S, et al. HIF2α-Sp1 interaction mediates a deacetylation-dependent FVII-gene activation under hypoxic conditions in ovarian cancer cells. Nucleic Acids Res. 2012;40:5389–5401. doi: 10.1093/nar/gks201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krainc D, et al. Synergistic activation of the N-methyl-D-aspartate receptor subunit 1 promoter by myocyte enhancer factor 2C and Sp1. J Biol Chem. 1998;273:26218–26224. doi: 10.1074/jbc.273.40.26218. [DOI] [PubMed] [Google Scholar]
- 49.Kypriotou M, et al. Human collagen Krox up-regulates type I collagen expression in normal and scleroderma fibroblasts through interaction with Sp1 and Sp3 transcription factors. J Biol Chem. 2007;282:32000–32014. doi: 10.1074/jbc.M705197200. [DOI] [PubMed] [Google Scholar]
- 50.Li J, et al. Sp1 and KLF15 regulate basal transcription of the human LRP5 gene. BMC Genet. 2010;11:12. doi: 10.1186/1471-2156-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Minc E, et al. The human copper-zinc superoxide dismutase gene (SOD1) proximal promoter is regulated by Sp1, Egr-1, and WT1 via non-canonical binding sites. J Biol Chem. 1999;274:503–509. doi: 10.1074/jbc.274.1.503. [DOI] [PubMed] [Google Scholar]
- 52.Nenoi M, Ichimura S, Mita K, Yukawa O, Cartwright IL. Regulation of the catalase gene promoter by Sp1, CCAAT-recognizing factors, and a WT1/Egr-related factor in hydrogen peroxide-resistant HP100 cells. Cancer Res. 2001;61:5885–5894. [PubMed] [Google Scholar]
- 53.Parks CL, Shenk T. Activation of the adenovirus major late promoter by transcription factors MAZ and Sp1. J Virol. 1997;71:9600–9607. doi: 10.1128/jvi.71.12.9600-9607.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tretiakova A, Steplewski A, Johnson EM, Khalili K, Amini S. Regulation of myelin basic protein gene transcription by Sp1 and Puralpha: Evidence for association of Sp1 and Puralpha in brain. J Cell Physiol. 1999;181:160–168. doi: 10.1002/(SICI)1097-4652(199910)181:1<160::AID-JCP17>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 55.Yamamoto J, et al. A Kruppel-like factor KLF15 contributes fasting-induced transcriptional activation of mitochondrial acetyl-CoA synthetase gene AceCS2. J Biol Chem. 2004;279:16954–16962. doi: 10.1074/jbc.M312079200. [DOI] [PubMed] [Google Scholar]
- 56.Robert MF, et al. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat Genet. 2003;33:61–65. doi: 10.1038/ng1068. [DOI] [PubMed] [Google Scholar]
- 57.Courey AJ, Holtzman DA, Jackson SP, Tjian R. Synergistic activation by the glutamine-rich domains of human transcription factor Sp1. Cell. 1989;59:827–836. doi: 10.1016/0092-8674(89)90606-5. [DOI] [PubMed] [Google Scholar]
- 58.Seipel K, Georgiev O, Schaffner W. Different activation domains stimulate transcription from remote (‘enhancer’) and proximal (‘promoter’) positions. EMBO J. 1992;11:4961–4968. doi: 10.1002/j.1460-2075.1992.tb05603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Valen E, Sandelin A. Genomic and chromatin signals underlying transcription start-site selection. Trends Genet. 2011;27:475–485. doi: 10.1016/j.tig.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 60.Consortium EP. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 62.Ramakrishnan V, Finch JT, Graziano V, Lee PL, Sweet RM. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature. 1993;362:219–223. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- 63.Cirillo LA, et al. Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. EMBO J. 1998;17:244–254. doi: 10.1093/emboj/17.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chaya D, Hayamizu T, Bustin M, Zaret KS. Transcription factor FoxA (HNF3) on a nucleosome at an enhancer complex in liver chromatin. J Biol Chem. 2001;276:44385–44389. doi: 10.1074/jbc.M108214200. [DOI] [PubMed] [Google Scholar]
- 65.Cirillo LA, et al. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 66.Iwafuchi-Doi M, et al. The pioneer transcription factor FoxA maintains an accessible nucleosome configuration at enhancers for tissue-specific gene activation. Mol Cell. 2016;62:79–91. doi: 10.1016/j.molcel.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Taube JH, Allton K, Duncan SA, Shen L, Barton MC. Foxa1 functions as a pioneer transcription factor at transposable elements to activate Afp during differentiation of embryonic stem cells. J Biol Chem. 2010;285:16135–16144. doi: 10.1074/jbc.M109.088096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lalmansingh AS, Karmakar S, Jin Y, Nagaich AK. Multiple modes of chromatin remodeling by Forkhead box proteins. Biochim Biophys Acta. 2012;1819:707–715. doi: 10.1016/j.bbagrm.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 69.He HH, et al. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 71.Barak O, et al. Isolation of human NURF: A regulator of Engrailed gene expression. EMBO J. 2003;22:6089–6100. doi: 10.1093/emboj/cdg582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wysocka J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 73.Lin D, et al. Bright/ARID3A contributes to chromatin accessibility of the immunoglobulin heavy chain enhancer. Mol Cancer. 2007;6:23. doi: 10.1186/1476-4598-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kaplan MH, Zong RT, Herrscher RF, Scheuermann RH, Tucker PW. Transcriptional activation by a matrix associating region-binding protein. contextual requirements for the function of bright. J Biol Chem. 2001;276:21325–21330. doi: 10.1074/jbc.M100836200. [DOI] [PubMed] [Google Scholar]
- 75.Webb CF, Das C, Eneff KL, Tucker PW. Identification of a matrix-associated region 5′ of an immunoglobulin heavy chain variable region gene. Mol Cell Biol. 1991;11:5206–5211. doi: 10.1128/mcb.11.10.5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Riedel CG, et al. DAF-16 employs the chromatin remodeller SWI/SNF to promote stress resistance and longevity. Nat Cell Biol. 2013;15:491–501. doi: 10.1038/ncb2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wilsker D, Patsialou A, Dallas PB, Moran E. ARID proteins: A diverse family of DNA binding proteins implicated in the control of cell growth, differentiation, and development. Cell Growth Differ. 2002;13:95–106. [PubMed] [Google Scholar]
- 78.Cai S, Han HJ, Kohwi-Shigematsu T. Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat Genet. 2003;34:42–51. doi: 10.1038/ng1146. [DOI] [PubMed] [Google Scholar]
- 79.Yasui D, Miyano M, Cai S, Varga-Weisz P, Kohwi-Shigematsu T. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature. 2002;419:641–645. doi: 10.1038/nature01084. [DOI] [PubMed] [Google Scholar]
- 80.Herrscher RF, et al. The immunoglobulin heavy-chain matrix-associating regions are bound by Bright: A B cell-specific trans-activator that describes a new DNA-binding protein family. Genes Dev. 1995;9:3067–3082. doi: 10.1101/gad.9.24.3067. [DOI] [PubMed] [Google Scholar]
- 81.Natesan S, Gilman MZ. DNA bending and orientation-dependent function of YY1 in the c-fos promoter. Genes Dev. 1993;7:2497–2509. doi: 10.1101/gad.7.12b.2497. [DOI] [PubMed] [Google Scholar]
- 82.Werner MH, Huth JR, Gronenborn AM, Clore GM. Molecular basis of human 46X,Y sex reversal revealed from the three-dimensional solution structure of the human SRY-DNA complex. Cell. 1995;81:705–714. doi: 10.1016/0092-8674(95)90532-4. [DOI] [PubMed] [Google Scholar]
- 83.Giese K, Cox J, Grosschedl R. The HMG domain of lymphoid enhancer factor 1 bends DNA and facilitates assembly of functional nucleoprotein structures. Cell. 1992;69:185–195. doi: 10.1016/0092-8674(92)90129-z. [DOI] [PubMed] [Google Scholar]
- 84.Giese K, Kingsley C, Kirshner JR, Grosschedl R. Assembly and function of a TCR alpha enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 85.Duncan R, et al. A sequence-specific, single-strand binding protein activates the far upstream element of c-myc and defines a new DNA-binding motif. Genes Dev. 1994;8:465–480. doi: 10.1101/gad.8.4.465. [DOI] [PubMed] [Google Scholar]
- 86.Bode J, et al. Biological significance of unwinding capability of nuclear matrix-associating DNAs. Science. 1992;255:195–197. doi: 10.1126/science.1553545. [DOI] [PubMed] [Google Scholar]
- 87.Arnosti DN, Kulkarni MM. Transcriptional enhancers: Intelligent enhanceosomes or flexible billboards? J Cell Biochem. 2005;94:890–898. doi: 10.1002/jcb.20352. [DOI] [PubMed] [Google Scholar]
- 88.Panne D, Maniatis T, Harrison SC. An atomic model of the interferon-beta enhanceosome. Cell. 2007;129:1111–1123. doi: 10.1016/j.cell.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Arnosti DN, Barolo S, Levine M, Small S. The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development. 1996;122:205–214. doi: 10.1242/dev.122.1.205. [DOI] [PubMed] [Google Scholar]
- 90.Liu F, Posakony JW. Role of architecture in the function and specificity of two Notch-regulated transcriptional enhancer modules. PLoS Genet. 2012;8:e1002796. doi: 10.1371/journal.pgen.1002796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rastegar S, et al. The words of the regulatory code are arranged in a variable manner in highly conserved enhancers. Dev Biol. 2008;318:366–377. doi: 10.1016/j.ydbio.2008.03.034. [DOI] [PubMed] [Google Scholar]
- 92.Corces MR, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet. 2016;48:1193–1203. doi: 10.1038/ng.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Buenrostro JD, et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol. 2015;109:21.29.1–21.29.9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kwiatkowski N, et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature. 2014;511:616–620. doi: 10.1038/nature13393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Engreitz JM, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang Y, et al. Model-based analysis of ChIP-seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Matys V, et al. TRANSFAC and its module TRANSCompel: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mathelier A, et al. JASPAR 2014: An extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42:D142–D147. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Weirauch MT, et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014;158:1431–1443. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.McLeay RC, Bailey TL. Motif enrichment analysis: A unified framework and an evaluation on ChIP data. BMC Bioinformatics. 2010;11:165. doi: 10.1186/1471-2105-11-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Grant CE, Bailey TL, Noble WS. FIMO: Scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Huang dW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.