Significance
Nontypeable Haemophilus influenzae (NTHi) is a major cause of community acquired pneumonia and exacerbation of chronic obstructive pulmonary disease. Current effort in NTHi vaccine development has focused on generating antibody responses to surface antigens and has had limited success, largely because of antigenic diversity among the numerous circulating NTHi strains. In this study, we have uncovered Th17 responses as the immune mechanism of cross-protection and identified Th17 antigens that are conserved and induce protection against NTHi lung infection, thus making a critical first step toward development of a broadly protective “universal” vaccine—the Holy Grail of vaccine development against respiratory pathogens.
Keywords: Haemophilus influenzae, antigenic diversity, pneumonia, Th17 responses, vaccine
Abstract
Nontypeable Haemophilus influenzae (NTHi) is a major cause of community acquired pneumonia and exacerbation of chronic obstructive pulmonary disease. A current effort in NTHi vaccine development has focused on generating humoral responses and has been greatly impeded by antigenic variation among the numerous circulating NTHi strains. In this study, we showed that pulmonary immunization of mice with killed NTHi generated broad protection against lung infection by different strains. While passive transfer of immune antibodies protected only against the homologous strain, transfer of immune T cells conferred protection against both homologous and heterologous strains. Further characterization revealed a strong Th17 response that was cross-reactive with different NTHi strains. Responding Th17 cells recognized both cytosolic and membrane-associated antigens, while immune antibodies preferentially responded to surface antigens and were highly strain specific. We further identified several conserved proteins recognized by lung Th17 cells during NTHi infection. Two proteins yielding the strongest responses were tested as vaccine candidates by immunization of mice with purified proteins plus an adjuvant. Immunization induced antigen-specific Th17 cells that recognized different strains and, upon adoptive transfer, conferred protection. Furthermore, immunized mice were protected against challenge with not only NTHi strains but also a fully virulent, encapsulated strain. Together, these results show that the immune mechanism of cross-protection against pneumonia involves Th17 cells, which respond to a broad spectrum of antigens, including those that are highly conserved among NTHi strains. These mechanistic insights suggest that inclusion of Th17 antigens in subunit vaccines offers the advantage of inducing broad protection and complements the current antibody-based approaches.
The Gram-negative coccobacillus Haemophilus influenzae colonizes assymptomatically in the upper respiratory tract, yet because of its prevalence, is also a significant cause of disease. When host immunity is compromised, H. influenzae can disseminate into privileged anatomical locations and cause a wide spectrum of diseases, including otitis media, conjunctivitis, sinusitis, pneumonia, and meningitis. Some strains of H. influenzae express a polysaccharide capsule, which is the major target of the antibody response. Based on antibody specificity to the capsule, these strains are classified into six different serotypes (a–f). The type b serotype (Hib) is the most virulent and a significant cause of invasive diseases, such as meningitis worldwide. In addition to encapsulated strains, there is a genetically diverse group of H. influenzae strains that expresses no capsule, and they are termed nontypeable H. influenzae (NTHi) (1, 2). With the introduction of highly effective conjugate vaccines against Hib and Streptococcus pneumoniae, NTHi has emerged as a leading cause of otitis media in children, community-acquired pneumonia (CAP), and exacerbation of chronic obstructive pulmonary disease (COPD) (3–5). NTHi is identified in 20–94% of sputum and bronchoalveolar lavage samples taken from patients with CAP and is frequently found in the airways of patients with COPD (6). Recurring NTHi infection by a new strain is strongly associated with exacerbation in patients with COPD, leading to high rates of hospitalization and worsening of symptoms (7). Although antibiotic therapies are effective at reducing severity of both CAP and exacerbations of COPD, treatment failures are becoming more frequent, in large part, due to increasing resistance to the front-line β-lactam antibiotics (8). Moreover, frequent use of antibiotics to treat recurring infections disrupts the normal microbiome, leading to dysbiosis and accompanying disease susceptibility (9). Therefore, preventative strategies such as vaccination against pulmonary NTHi infection are urgently needed.
Current vaccine development effort has been focused on subunit vaccines that are targeted at eliciting antibody responses to bacterial surface proteins and lipooligosaccharide (LOS) antigens (10, 11). Several surface proteins, including outer-membrane proteins (OMPs) OMP26, P6, and protein F, have been identified that elicit bactericidal antibodies and induce limited protective immunity against otitis media and pneumonia in animal models (12–14). Protein D from NTHi is included in the licensed 10-valent PhiD pneumococcal vaccine (Synflorix; GlaxoSmithKline) as the carrier protein while also serving as an immunogen for NTHi. Initial clinical trial data suggest that the Synflorix vaccine could prevent 35% of NTHi acute otitis media episodes; however, a subsequent study shows no significant protection against NTHi in otitis media nor a reduction in NTHi nasopharyngeal carriage (15, 16). In the mouse model, the Synflorix vaccine neither augments the pulmonary clearance of NTHi nor protects against NTHi superinfection, despite the induction of high levels of protein D antibodies (17). The limited success in the development of antibody-based NTHi vaccines has been due, in large part, to enormous sequence and structural variation of NTHi surface antigens (18). In addition to antigenic variation, NTHi is highly adapted to colonize the human respiratory tract and consequently has evolved numerous molecular mechanisms to evade antibody-mediated protection. These mechanisms include LOS-mediated mimicry of host glycans; expression of secreted IgA proteases; recruitment of numerous complement inhibitors (19); formation of biofilms within the host tissues (20); and possibly transient intracellular survival in macrophages, B cells, and epithelial cells (21). Thus, incorporation of additional antigens and engagement of immune mechanisms other than antibodies are likely needed for broad and effective protection against invasive NTHi infections.
A growing body of evidence indicates that T cells are important in defense against respiratory infections caused by bacterial pathogens, including NTHi. Children suffering from recurring otitis media have lower cellular immune responses to the P6 protein in adenoidal lymphocytes (22). In adults with COPD, a lymphocyte proliferative response to P6 is associated with less severe disease exacerbations from NTHi infection (23). IL-17–producing CD4 T cells (Th17), first described as initiators of proinflammatory responses in autoimmune diseases, have now been shown to play an important role in several bacterial and fungal infections at mucosal sites. In murine models, it has been shown that signaling via the IL-1/IL-17R axis is important for eliminating primary lung infection by several bacterial pathogens, including Mycobacterium tuberculosis, Klebsiella pneumoniae, and Legionella pneumophila (24–26). Humans with a mutation in STAT3 leading to impaired Th17 differentiation are highly susceptible to H. influenzae infection (27). While the role of Th17 response in controlling primary bacterial infection in the lung is well established, the role of Th17 memory in protection against reinfection and in vaccine-induced immunity is not well defined.
In this study, we asked whether broad protection against different NTHi strains could be induced by vaccination and investigated the underlying mechanisms of cross-protection. Through these studies, we identified several conserved antigens that induce Th17-mediated protective immunity against genetically diverse H. influenzae strains, including NTHi clinical isolates and encapsulated serotypes, thus making a critical first step toward development of a broadly protective vaccine.
Results
Protection Against Homologous and Heterologous Strains by Immunization with Heat-Killed Bacteria.
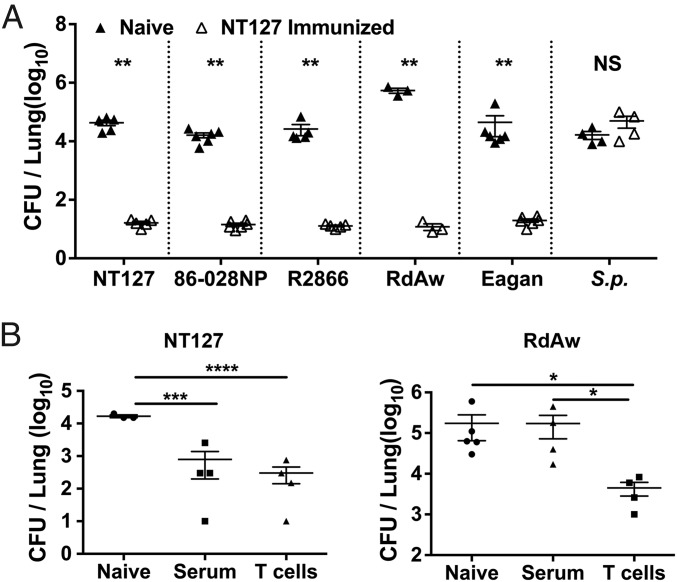
To test whether broad protection against NTHi lung infection can be induced, we immunized C57BL/6 mice intranasally with heat-killed NT127, a clinical NTHi isolate that has been well characterized genetically and in the murine model of lung infection (28, 29). Three weeks later, mice were challenged with the homologous NT127 or heterologous strains by the intranasal route under anesthesia, which results in direct infection of the lower respiratory tract and acute bacterial pneumonia (17, 30). Immunized animals had ∼1,000-fold lower bacterial burdens in the lung on day 2 postchallenge (Fig. 1A), compared with the unimmunized controls. This protection was observed not only in mice challenged with the homologous NT127 strain, but also after heterologous challenge with NTHi strains 86-028NP and R2866, and even strains of encapsulated lineages, Eagan (serotype b) and RdAW (serotype d derived). However, the NT127-immunized mice were not protected from challenge with S. pneumoniae, another common respiratory bacterial pathogen (Fig. 1A). Together, these results show that immunization with heat-killed NT127 induces H. influenzae-specific immunity that is broadly protective against pulmonary infection by different H. influenzae strains.
Fig. 1.
Vaccination induces broad protective immunity against lung infection by different strains. (A) B6 mice were immunized intranasally with heat-killed NT127. Three weeks after immunization, immune mice and naïve controls were challenged intranasally with different H. influenzae strains, or with S. pneumoniae. Two days after challenge, bacterial loads in the lung were determined. (B) Immune sera or T cells from heat-killed NT127 immunized mice were transferred to recipient mice. Twenty-four hours later, recipient mice and naïve controls were challenged intranasally with strain NT127 or RdAW. Two days after challenge, bacterial loads in the lung were determined. Each dot represents an individual mouse. Error bars = means ± SEM. ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05; NS, not significant.
We next investigated the role of immune antibodies and T cells in mediating the homologous and heterologous protection. Sera and purified T cells from NT127-immunized mice were adoptively transferred into naïve mice, which were then challenged with NT127 or RdAW. Both sera and T cells conferred comparable protection against homologous challenge with NT127. In contrast, only T-cell recipient mice were protected from heterologous RdAW challenge (Fig. 1B). These data indicate that T cells play a predominant role in mediating heterologous protection in pulmonary H. influenzae infection.
A Robust Th17 Response in the Lung and Its Role in Heterologous Protection.
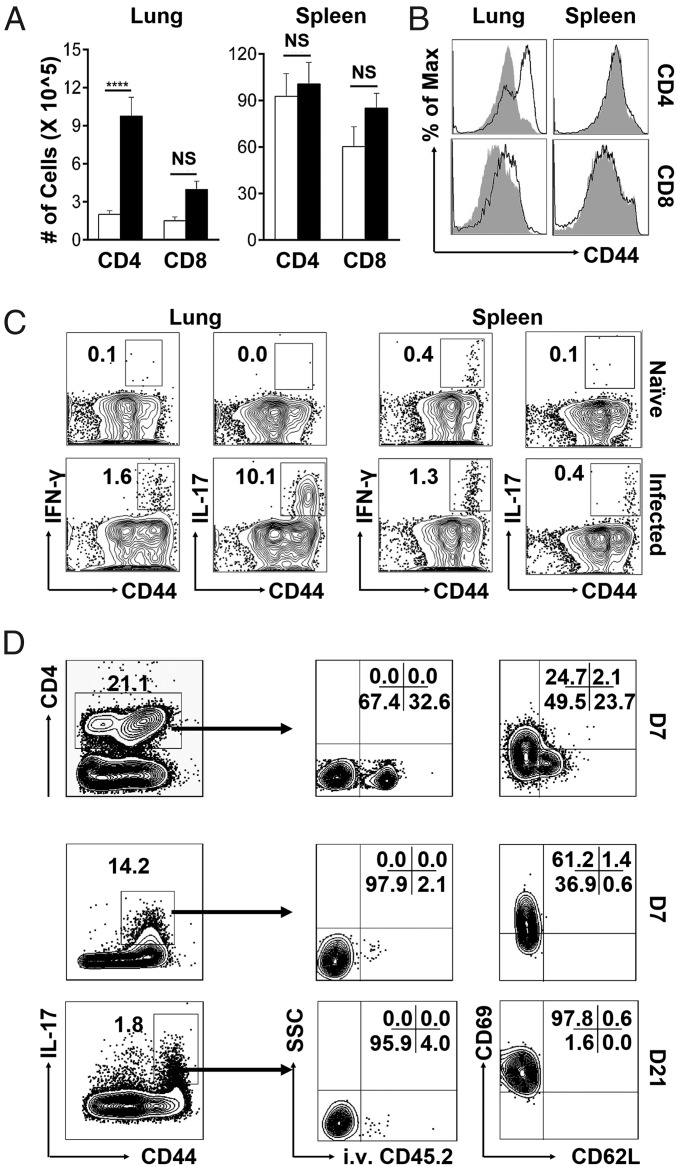
Since T cells were capable of mediating heterologous protection, we characterized the T-cell response to pulmonary NTHi infection. Mice were infected with a sublethal dose of NT127, and the body weight, bacterial loads, NT127-specific antibody, and T-cell responses were analyzed on different days after infection (SI Appendix, Fig. S1). By day 7 when the NT127-specific T-cell response reached the maximum, there were substantial increases in the percentage and number of total CD4 but not CD8 T cells in the lungs of infected mice compared with naïve controls. The majority of CD4 T cells in the lungs of infected mice had an activated CD44hi phenotype. In the spleen, there were no differences in either the quantities or CD44 expression levels of CD4 and CD8 T cells between infected and naïve mice (Fig. 2 A and B). Thus, NT127 infection induces a strong CD4 but not CD8 T-cell response that is localized in the lung.
Fig. 2.
NTHi infection induces a robust Th17 response in the lung. B6 mice were intranasally infected with NT127. On day 7, lymphocytes from the lung and spleen were isolated for analyses. (A) Absolute numbers of CD4 and CD8 T cells from the lung and spleen of naïve (open bars) and NT127-infected (black filled) mice. (B) CD44 expression of CD4 and CD8 T cells from the lung and spleen of naïve (gray shaded) and NT127-infected (solid lines) mice. (C) Production of IFN-γ and IL-17 by CD4 T cells from the lung and spleen of naïve and NT127-infected mice was measured by ICS after in vitro stimulation with heat-killed NT127. (D) B6 mice were intranasally immunized with NT127. On day 7 (D7) and D21, the phenotypes of NT127-specific Th17 cells in the lung were examined and representative data were shown. n = 5, error bars = means ± SEM, ****P < 0.0001; NS, not significant.
To study the functions of responding T cells, we stimulated lymphocytes from lung and spleen (day 7 after NT127 infection) with heat-killed NT127 followed by intracellular cytokine staining (ICS). A small percentage (∼1.6%) of CD4 T cells from the lungs of immunized mice produced IFN-γ, while a high percentage of them (>10%) produced IL-17 (Fig. 2C). A small percentage of CD4 T cells in the spleens of immunized mice produced IFN-γ (1.3%) or IL-17 (0.4%). All cytokine-producing CD4 T cells had an activated CD44hi phenotype and produced either IL-17 or IFN-γ; very few of them, if any, were IL-17/IFN-γ coproducers (SI Appendix, Fig. S2A). CD4 T cells from the lungs or spleens of naïve mice produced little IFN-γ or IL-17 when stimulated in vitro with heat-killed bacteria. Unlike CD4 T cells, CD8 T cells from both lung and spleen produced only IFN-γ (3% and 4.6%, respectively) and little IL-17 (SI Appendix, Fig. S2B). These results show that NT127 infection induces a strong, bacteria-specific CD4 T-cell response in the lung that consists of predominantly IL-17–producing Th17 cells mixed with a weak IFN-γ–producing Th1 response.
To characterize the phenotypes of responding Th17 cells in the lung, we used intravascular staining with FITC-conjugated anti-CD45.2 mAb, which allows staining of intravascular but not parenchymal lymphocytes (31). While total CD4 T cells in the lung were present in both intravascular and parenchymal sites, the bacteria-specific Th17 cells were predominantly in the lung parenchyma on both day 7 and day 21 after infection (Fig. 2D). On day 7, many of the bacteria-specific Th17 cells were effectors (CD69lo/CD62Llo), with some of them being TRM (CD69hi/CD62Llo). By day 21, all of the bacteria-specific Th17 cells had differentiated into the TRM-expressing CD69hi/CD62Llo phenotype.
To test the role of Th17 in heterologous protection, IL-17 KO mice were immunized intranasally with heat-killed NT127 and then challenged with a heterologous strain 86-028NP. There were similar numbers of bacteria detected in the lung of immunized and unimmunized IL-17 KO mice, indicating a critical role for IL-17 in vaccine-induced protective immunity (SI Appendix, Fig. S3B). IL-17 also played a role in host resistance to a primary NTHi lung infection, as there were more bacteria (∼10-fold) in the lungs of IL-17 KO than WT mice. Furthermore, we found that i.p. immunization in WT mice provided minimal protection against challenge with a heterologous strain (SI Appendix, Fig. S3). Further analyses showed that i.p. immunization induced only a weak Th1 response in the spleen but did not induce a detectable Th17 response in the lung. These results indicate the importance of a local Th17 response in heterologous protection, consistent with our previous findings with S. pneumoniae (32).
Broadly Reactive Th17 Cells but Highly Strain-Specific Antibody Responses to NTHi Infection.
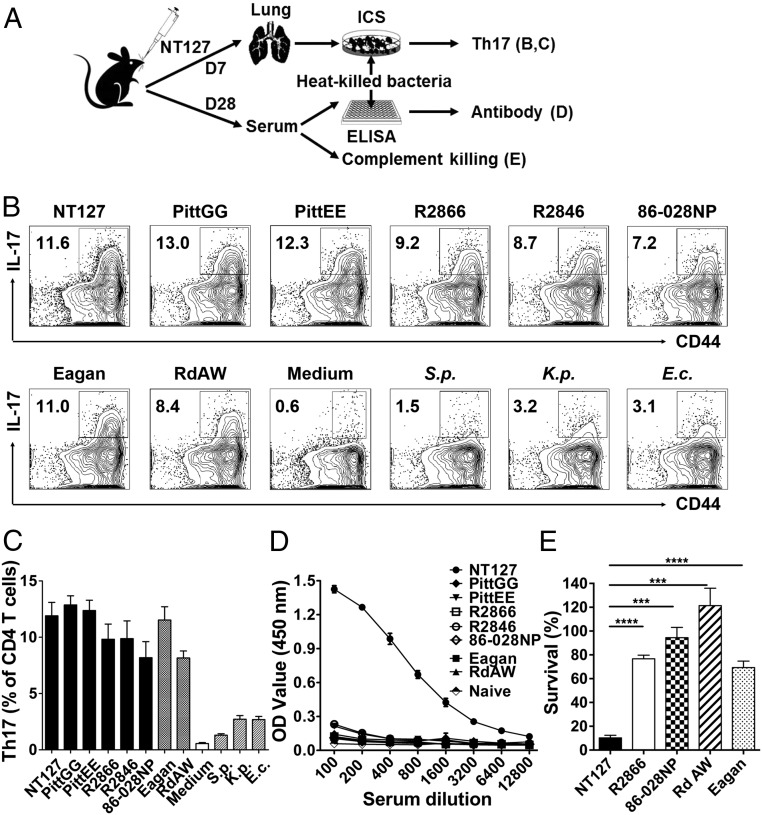
To assess the specificity and cross-reactivity of Th17 cells, we stimulated T cells from the lungs of NT127-infected mice with heat-killed NT127 as well as heterologous NTHi strains. Similar levels of IL-17–producing CD4 T cells were detected when stimulated with NT127 or any of the other five NTHi strains tested, indicating that CD4 T cells from NT127-infected mice were capable of recognizing antigens from heterologous NTHi strains. Interestingly, Th17 cells from NT127-infected mice recognized not only heterologous NTHi strains, but also H. influenzae strains of encapsulated lineages (Eagan and RdAW). Similarly, Th17 cells induced by another NTHi (86-028NP) or H. influenzae strains of encapsulated lineages (RdAW and Eagan) were cross-reactive with all NTHi and typeable H. influenzae strains tested (SI Appendix, Fig. S4). There were low levels of cross-reactivities to other bacterial species, such as S. pneumoniae, K. pneumoniae, and Escherichia coli that were above the background level in the medium control (Fig. 3 B and C). However, the cross-reactivities to other bacterial species were much lower than the responses cross-reactive to NTHi/H. influenzae strains, indicating a strong NTHi/H. influenzae-specific Th17 response induced by NTHi infection.
Fig. 3.
Th17 cells, not antibodies, recognize heterologous strains. (A) Experimental design. (B and C) Lung lymphocytes from NT127-infected mice (day 7) were stimulated with heat-killed bacteria, and production of IL-17 by CD4 T cells was measured by ICS. In addition to different H. influenzae strains, S. pneumoniae (S.p.), E. coli (E.c.), K. pneumoniae (K.p.), and medium alone were included as controls. (D) Immune sera from NT127-infected mice (3 wk) were reacted with various heat-killed bacteria as coating antigens in ELISA. Naïve sera reacting with NT127 were included as a control. (E) Bactericidal activity of immune sera from NT127-infected mice against different H. influenzae strains was determined. n = 3–4, error bars = means ± SEM, ****P < 0.0001; ***P < 0.001.
For comparison, we also examined specificity and cross-reactivity of antibodies induced by NT127 infection. Immune sera from NT127-infected mice were reacted with various heat-killed bacteria used as coating antigens in ELISA. While high levels of IgG specific to the homologous strain NT127 were detected, there was little cross-reactivity to the other five NTHi strains or to the two other H. influenzae strains (Eagan and RdAW), indicating that the antibody responses to NTHi were highly strain specific (Fig. 3D). We further tested bactericidal activity of NT127-immune sera against different strains in complement-dependent killing assays. Anti-NT127 sera sensitized the homologous NT127 strain for complement-mediated killing (90% cfu reduction compared with naïve sera). In contrast, heterologous strains were not killed at all (for 86-028NP and RdAW strains), or only slightly (20% reduction for R2866 and Eagan strains, Fig. 3E). Together, these data indicate that the antibody response to NTHi is highly strain specific, while Th17 cells are broadly reactive, recognizing various heterologous NTHi and even encapsulated H. influenzae.
A Broader Spectrum of NTHi Antigens Recognized by Th17 Cells than by Antibodies.
To understand why antibody responses are highly strain specific while Th17 cells are broadly reactive, we tested the possibility that Th17 cells may recognize a broader spectrum of antigens than antibodies. We separated bacterial proteins into outer-membrane vesicles (OMVs), membrane, and cytosolic fractions, and evaluated antibody and Th17 response to these fractions. Convalescent serum from NT127-immunized mice reacted strongly with outer-membrane vesicles and membrane fractions of NT127, to levels higher than that with whole-cell lysate (WCL), while reacting weakly with cytosolic fractions. Furthermore, we prepared protein fractions from heterologous strains (86-028NP and Eagan), and tested them against serum from NT127-immunized mice. The NT127-immune serum did not react with protein fractions of either 86-028NP or Eagan (Fig. 4B), indicating little cross-recognition of conserved antigens by antibodies. Together, these results indicate that antibody recognition is focused on membrane-associated, surface antigens that are not conserved between NTHi strains, thus explaining why antibodies are highly strain specific.
Fig. 4.
Th17 cells recognize a broader spectrum of bacterial antigens than antibodies. (A) Experimental design. Whole-cell lysate (WCL), cytosol (Cyto), and membrane (Mem) fractions, as well as outer membrane vesicles (OMVs) were isolated from NT127, Eagan, and 86-028NP. Hi, H. influenzae. (B) Reactivity of pooled immune sera from NT127-infected mice to different bacterial fractions was analyzed by ELISA. (C) Lung lymphocytes from NT127-infected mice were stimulated with different bacterial fractions, and IL-17 production by CD4 T cells was measured by ICS.
In contrast to the antibody responses, Th17 cells from NT127-immunized mice responded comparably to all three fractions (outer-membrane vesicles, membrane, and cytosolic fractions) isolated from NT127. Furthermore, Th17 cells from NT127-immunized mice responded to all three fractions isolated from heterologous strains (86-028NP and Eagan, Fig. 4C). Broad reactivity of Th17 cells could be due to recognition of numerous protein antigens that are conserved among NTHi strains, or nonspecific stimulation of Th17 cells by bacterial products, such as TLR ligands (lipid, cell wall components, and nucleic acids). To differentiate these two possibilities, whole-cell lysates of NT127 were treated with protease K, which resulted in complete protein digestion (SI Appendix, Fig. S5A), and then used to stimulate Th17 cells from NT127-immunized mice. The protease treatment resulted in substantial loss of IL-17 induction (SI Appendix, Fig. S5 B and C), indicating that the majority of the Th17 responses were induced by protein antigens. Together, these results show that Th17 cells recognize a broad spectrum of cytosolic and membrane-associated antigens that are conserved between NTHi strains.
Identification of Conserved Bacterial Antigens Recognized by Th17 Cells.
To identify conserved NTHi antigens recognized by Th17 cells, we performed comparative genomic analyses to identify a list of candidate antigens based on: (i) at least 90% amino acid similarity across sequenced NTHi genomes and (ii) no significant homology to human proteins. The list was further refined to prioritize proteins that were either previously reported to be associated with protective immunity or were identified as genes required for growth or survival of H. influenzae in the lung (33). In a continuous effort, we expressed and purified candidate protein antigens and tested them for recognition by Th17 cells from NT127-immunized mice. To date, 20 candidate proteins were successfully expressed to high levels in E. coli and purified to homogeneity by Ni2+ affinity columns (SI Appendix, Fig. S6 A and B). Relevant characteristics of these proteins were described in SI Appendix, Table S2.
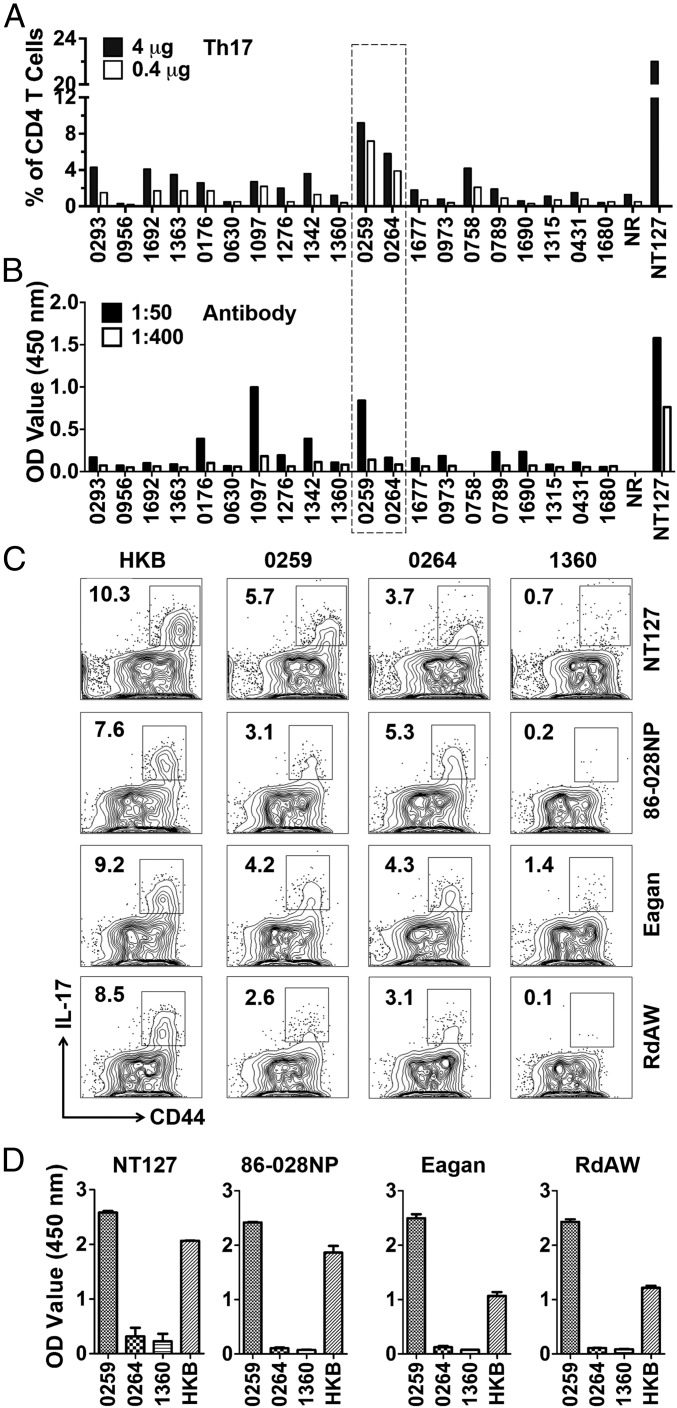
To screen for Th17 antigens, purified proteins were used to stimulate lung lymphocytes from NT127-infected mice in vitro and IL-17 production by CD4 T cells was determined by ICS (SI Appendix, Fig. S6 C and D). Th17 responses to several proteins were detected (Fig. 5A), and the highest level of Th17 responses was induced by two proteins encoded by NT127 genes HIAG_0259 and HIAG_0264 (SI Appendix, Table S2). The protein 0259 but not 0264 was also a target of antibody responses (Fig. 5B). On the other hand, protein 1097 induced a high level of the antibody response but a low level of Th17 response. Overall, among the 20 conserved proteins we selected for testing, there were more proteins recognized by Th17 cells than by antibodies.
Fig. 5.
Identification of conserved bacterial antigens recognized by Th17 cells. (A) Purified bacterial proteins (4 and 0.4 μg) were used to stimulate pooled lung lymphocytes from NT127-infected mice, followed by ICS of IL-17. (B) Pooled NT127-immune sera (1:50 and 1:400 dilutions) were reacted with purified individual proteins in ELISA. Heat-killed NT127 (NT127) and a nonrelated protein (NR) were used as a positive and negative control, respectively. (C) Lung lymphocytes from mice infected with NT127, 86-028NP, Eagan, and RdAW were stimulated with purified proteins (0259, 0264, and 1360) or heat-killed bacteria (HKB) of the corresponding strains. (D) Antibody responses were also determined on day 21 after infection. n = 3, error bars = means ± SEM.
The two strong Th17 antigens identified (proteins 0259 and 0264) are highly conserved among NTHi strains, with >97% homology in all strains that have been sequenced. We further tested whether these two conserved protein antigens could be recognized by Th17 cells and antibodies from mice infected with different strains, including homologous (NT127) and heterologous strains (86-028NP, Eagan, and RdAW). In all cases, both proteins were recognized by Th17, although the responses to protein 0259 were slightly lower in mice infected with the 86-028NP and RdAW strains (Fig. 5C). In contrast, only 0259 was the target of antibody responses in mice infected with the four strains tested (Fig. 5D). As controls, we included protein 1360 that was negative in our screen for Th17 antigens. As expected, protein 1360 was not recognized by lung lymphocytes from infected mice. Together, these results validate our identification of proteins 0259 and 0264 as two highly conserved antigens that are recognized by Th17 cells induced by different NTHi and encapsulated H. influenzae.
Broad Protection Against H. influenzae Infection Following Immunization with Conserved Th17 Antigens.
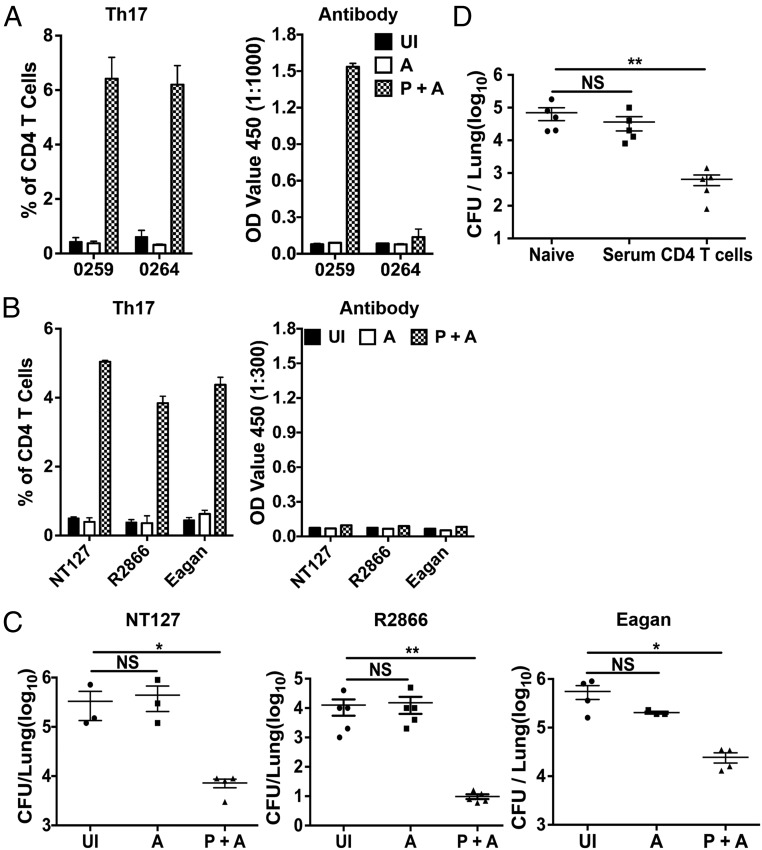
We investigated the potential of identified Th17 antigens as vaccine candidates for inducing broadly protective immunity. As the first step, we tested a combination of the two proteins (0259 and 0264) identified to be highly immunogenic for inducing Th17 responses (Fig. 5C). Mice were immunized three times, at 1-wk intervals, intranasally with a mix of the two purified proteins and curdlan, an experimental Th17-inducing adjuvant (34, 35). Mice immunized with adjuvant only as well as unimmunized mice were included as controls. Three weeks after the final immunization, antigen-specific Th17 and antibody responses were measured by in vitro stimulation of lung lymphocytes and by ELISA using purified individual proteins as reacting antigens. Immunization with the two purified proteins plus adjuvant (P+A) induced high levels of Th17 cells specific to both 0259 and 0264 proteins. Interestingly, mice immunized with P+A had specific antibodies against protein 0259, but not against protein 0264 (Fig. 6A). We next asked whether Th17 and antibodies induced by P+A immunization could recognize different bacterial strains, using heat-killed bacteria as reacting antigens in ICS and ELISA. Lung lymphocytes from P+A-immunized mice responded to all three strains tested (NT127, R2866, and Eagan) and produced IL-17, as measured by ICS. In contrast, sera from P+A-immunized mice did not react with heat-killed intact bacteria of NT127, R2866, and Eagan used as coating antigens in ELISA. As expected, no antigen-specific Th17 and antibody responses were detected in unimmunized mice and mice immunized with adjuvant only (Fig. 6B). Together, these results show that immunization with protein antigens of 0259 and 0264 induces antigen-specific Th17 cells to both proteins, and these Th17 cells are capable of recognizing different bacterial strains. On the other hand, only protein 0259-specific antibodies are induced, yet these antibodies fail to bind to intact bacteria of any strains tested, presumably because antibody epitopes of protein 0259 are not exposed on the surface of intact bacteria.
Fig. 6.
Immunization with identified Th17 antigens induces broadly protective immunity. B6 mice were intranasally immunized, three times at 1-wk intervals, with a mixture of purified protein 0259 and 0264 plus the curdlan adjuvant (P+A). Unimmunized (UI) and immunization with adjuvant only (A) were included as controls. (A) Induction of antigen-specific Th17 responses was measured by ICS following in vitro stimulation of lung lymphocyte (7 d after final immunization) with purified individual proteins. Protein-specific antibody responses in the serum were measured by ELISA, using plates coated with purified individual proteins, n = 3. (B) Recognition of different bacterial strains by Th17 cells and serum antibodies from immunized mice was determined by ICS and ELISA, respectively, using heat-killed bacteria (NT127, R2866, and Eagan) as reacting antigens, n = 3. (C) Three weeks after final immunization, immunized mice were challenged with NT127, R2866, or Eagan and the bacterial loads in the lung were determined 2 d later. (D) Naïve and recipient mice that received serum or purified CD4 T cells from immunized mice (P+A) were challenged with NT127, and bacterial loads in the lung were determined 24 h later. Each dot represents an individual mouse. Error bars = means ± SEM, **P < 0.01; *P < 0.05; NS, not significant.
To test protective immunity, immunized mice were challenged with NT127, R2866, or Eagan by intranasal inoculation under anesthesia, resulting in direct infection of the lower respiratory tract. Two days after bacterial challenge, P+A-immunized mice had significantly lower numbers of bacteria (100- to 1,000-fold) in the lung compared with animals immunized with the adjuvant only or left unimmunized (Fig. 6C). Importantly, protection was evident against not only NTHi strains (NT127 and R2866), but also the encapsulated, highly virulent Eagan strain. To determine the role of antibodies and T cells in vaccine-induced protection, we transferred sera and CD4 T cells harvested from P+A-immunized mice to naïve recipients and then challenged recipient mice with NT127. On day 2 after NT127 challenge, bacterial loads in mice that received immune sera were not significantly different from those in control mice infected with NT127. On the other hand, mice that received CD4 T cells were protected, with 2-log lower bacterial loads in the lung (Fig. 6D). Together, these results show that conserved antigens recognized by Th17 cells induce protection against lung infection by different NTHi and encapsulated H. influenzae strains. Inclusion of Th17 antigens in subunit vaccines may help overcome the limitation of current antibody-based approaches by inducing broadly protective immunity.
Discussion
Antibody responses are readily monitored and have been shown to be critical effectors of protective responses against encapsulated H. influenzae. Therefore, antibody responses have been used to evaluate clinical correlates of immunity, and vaccine design has focused on optimization of such responses. These strategies have been effective against respiratory pathogens that are not commonly carried asymptomatically, as was demonstrated with the successful vaccine against Hib strains, which are essentially of clonal descent and all express the same capsular antigen. However, pathogens that are more capable of generating a large number of asymptomatic carriers have developed sophisticated immune evasion mechanisms and are relatively intractable to this traditional approach. NTHi exemplifies this class of pathogens and exhibits extensive antigenic diversity that is highly concentrated in its abundant cell surface proteins, against which the bulk of the humoral response is directed during infection (Fig. 4). Thus, the most potent antibody responses are generated against the most variable of targets, which limits the ability of vaccines based on antibodies induced by these antigens to protect against the genetically diverse group of circulating NTHi strains. Therefore, intensive efforts have focused on identifying conserved surface proteins or epitopes capable of inducing cross-reactive antibody responses (12–14, 36–44). Despite these efforts, development of a vaccine that can provide broad and highly effective protection against diverse NTHi strains has yet to be attained (11).
Our results show that immunization with heat-killed NTHi induces cross-strain protection in a murine lung infection model. This appears paradoxical since immunizations with protein antigens recognized by antibodies during natural infection have thus far failed to confer similarly broad efficacy. Our further analyses provide a reconciling explanation: the antigens conferring cross-specificity in natural infection are those recognized instead by T cells, specifically of the Th17 subset (Figs. 1 and 2). Th17 cells are known to recruit neutrophils and inflammatory monocytes while antibodies can increase their ability to phagocytose and kill bacteria by opsonization. Thus, Th17 can work in concert with antibodies to mediate optimal protection. Our further analyses show that responding Th17 cells are cross-reactive to different strains and recognize both cytosolic and membrane-associated antigens, while the antibodies that are induced preferentially recognize surface antigens that are variable between strains and thus are highly strain specific (Figs. 3 and 4). Unbiased recognition by Th17 cells of bacterial proteins from different cellular compartments is consistent with findings that several nonsurface proteins are recognized by Th17 cells in the model of pneumococcal colonization (45). The broad spectrum of Th17 targets likely reflects the nature of antigen presentation to T cells whereby all bacterial proteins are accessible to the MHC-II antigen processing machinery following phagocytosis of bacteria by professional antigen presenting cells.
In contrast to the traditional approach of focusing on surface antigens that induce strong antibody responses, we sought to identify relevant Th17 antigens by screening a set of conserved proteins regardless of their subcellular localization, but with a focus on those implicated in allowing NTHi to survive in the lung (29). Several NTHi proteins were identified as targets of the Th17 response. Interestingly, while there are some overlaps, many proteins recognized by Th17 are not targets of antibody responses (Fig. 5). Consistent with our finding of different proteins recognized by Th17 cells versus antibodies, many known targets recognized by antibodies did not score positive in a screen for Th17 antigens in S. pneumoniae, whereas two of the identified Th17 antigens have never been reported as antibody targets (46). Similarly, two proteins (0259 and 0264) that induced the strongest Th17 responses from our screen have not been identified or studied before as potential vaccine candidates by traditional approaches targeting antibody responses to H. influenzae. Together, these results indicate that current vaccine efforts focused on generating antibody responses may fail to include protective antigens recognized by Th17 cells, and that identification and inclusion of Th17 antigens in multisubunit vaccines will induce not only protective antibody responses but also cross-reactive Th17 immunity.
We selected two of the identified Th17 antigens (0259 and 0264) inducing the strongest responses during NTHi infection and tested them as candidate vaccine antigens. Proteins 0259 and 0264 are annotated as ABC transporter protein OppA and a putative membrane protein, respectively. Interestingly, despite protein 0264 being annotated as a putative membrane protein, it lacks any recognizable signal sequence, transmembrane domain, and noncanonical secreted protein characteristics as determined by bioinformatic analysis (47, 48). Thus, it is likely cytosolic, consistent with lack of antibody responses to this protein (Fig. 5). The biological functions have not been identified for 0264 in H. influenzae, or for its orthologs in other bacterial species. The function of OppA (0259) has also not been defined in H. influenzae, but its orthologs in other species are well characterized as binding proteins that deliver substrate to the membrane-bound OppBCDF complex, mediating oligopeptide uptake (49). OppA proteins elicit antibodies during infection despite their periplasmic localization and are under investigation as vaccine candidates in another pathogen of the lung and middle ear, Moraxella catarrhalis (50). Interestingly, we also observed an antibody response to OppA (0259) during NTHi infection. Immunization with the two proteins (0259 and 0264) induced a strong OppA-specific antibody response. However, OppA-specific antibodies did not react with intact bacteria in ELISA (Fig. 6), suggesting that the OppA protein or epitopes may not be abundantly displayed on bacterial cell surface, consistent with its predicted periplasmic localization, and thus are not ideal targets for antibody-mediated protection. On the other hand, our results showed that the two proteins were able to elicit cross-reactive Th17 responses against diverse NTHi strains, and conferred cross-strain protection in the lung infection model. Furthermore, adoptive transfer of memory Th17 cells from immunized mice conferred cross-strain protection while transfer of immune serum failed to protect, supporting a critical role for Th17 in mediating cross-protection following immunization. With these results, we have uncovered Th17 responses as the primary immune mechanism of cross-protection, and the broad spectrum of Th17 targets as the reason for their ability to mediate cross-protection against lung infection by different strains of NTHi. We further identified Th17 antigens that are highly conserved and protective, thus making a critical first step toward development of a broadly protective “universal” vaccine—the Holy Grail in our battle against respiratory pathogens.
Materials and Methods
Mice.
Specific pathogen-free, 6- to 8-wk-old C57BL/6 mice were purchased from the National Cancer Institute. IL-17A–deficient (IL-17 KO) mice were bred in house at the University of Pennsylvania (32). All experiments were performed in accordance with Institutional Animal Care and Use Committee-approved protocols in the University of Pennsylvania animal facility.
Bacteria Strains and Growth Conditions.
NTHi strains were cultured in brain heart infusion (BHI) broth supplemented with 2.0% (vol/vol) Fildes enrichment (BD) and 10 μg/mL NAD (Sigma) (sBHI) or on sBHI agar plates at 37 °C. For preparation of nonviable NTHi, bacterial suspension was heated to 65 °C for 30 min and plated to ensure 100% bacterial killing.
Expression and Purification of Recombinant NTHi Proteins from E. coli.
The His-tagged proteins were expressed and purified as described (46) with slight modifications. Briefly, DNA sequences encoding the selected NTHi proteins were amplified by PCR using primers (SI Appendix, Table S1) designed based on the published genomic sequence of NT127 (accession no. PRJNA39125). Purified PCR products were cloned into the pET28a expression vectors, and the fusion constructs were confirmed by DNA sequencing. E. coli-containing expression vectors were grown in 200 mL LB broth in the presence of 0.1 mM IPTG and cultured for 4 h at 30 °C. Proteins were purified over Ni columns. Inclusion bodies, when present, were solubilized with 8 M urea before being loaded onto Ni columns. The resulting proteins were analyzed by SDS/PAGE with Coomassie staining to confirm purity.
Fractionation of H. influenzae Proteins and Immunoblot Analysis.
Fractionated proteins were prepared according to the protocol of Roier et al. (51). Briefly, bacteria were harvested from overnight cultures and washed with PBS. Cells were then resuspended in PBS with protease inhibitor and disrupted by sonification. After removal of unbroken cells, the whole-cell lysates were separated into cytoplasmic (supernatant) and membrane (pellet) fractions by ultracentrifugation. The membrane pellet was washed three times and resuspended in PBS. OMVs were harvested from supernatants of overnight culture by filtration and ultracentrifugation and then resuspended in PBS. All fractions were stored at −20 °C before use.
Animal Immunization and Infection.
For whole-cell antigen (WCA) immunization, mice were anesthetized by i.p. injection of 100 μL ketamine/xylazine (100 mg/3.8 mg/kg) and inoculated with 1 × 108 heat-killed NT127 in 30 μL PBS (i.n.). For immunization with proteins, mice were immunized i.n. three times, at weekly intervals, with 40 μL PBS containing 5 μg of each NTHi protein (0.125 mg/mL) plus the adjuvant curdlan (5 mg/mL) (Sigma-Aldrich) (52). On day 21 after the final immunization, while anesthetized, mice were challenged i.n. with 30 μL of PBS containing ∼108 cfu of the indicated H. influenzae strain. Forty-eight hours after challenge, the number of H. influenzae in lung homogenates was determined.
Serum Bactericidal Assay.
The sensitivity of heterologous H. influenzae strains to anti-NT127 serum was determined as previously described, using mouse naïve or anti-NT127 immune sera and complement from infant rabbit (53). Mouse naïve and anti-NT127 immune sera were heat inactivated by incubating at 56 °C for 30 min. Results were reported as percent survival (calculated by dividing total colony-forming units of immune serum-treated samples by total colony-forming unit counts of samples treated with naïve serum).
IgG Measurement by ELISA.
Diluted heat-killed bacteria (∼3 × 108 cfu), cellular fraction (1 mg/mL in PBS), or purified proteins (1 mg/mL in PBS) were coated onto 96-well plates by incubation at 37 °C for 1 h. Wells were then blocked with 2% nonfat milk in PBS. Twofold serial dilutions of pooled mouse sera were applied to the wells in duplicate at appropriate dilutions. Plates were incubated with sera for 1 h at 37 °C. Antibodies were detected with goat anti-mouse IgG-HRP (H+L). For detection, the 3,3′,5,5′-tetramethylbenzidine (TMB) peroxidase substrate reagent set (BioLegend) was used according to the manufacturer’s instructions. Optical densities were read at 450 nm with a microplate reader.
Intracellular Cytokine Staining and Flow Cytometry.
Lung and spleen cells were isolated and stimulated with heat-killed H. influenzae strains for 16 h at 37 °C with Golgi block added for the last 5 h. Cells were then stained as described in our previous study (32). All samples were analyzed with FACSCanto. FACS data were analyzed with FlowJo software.
Adoptive Transfer Experiments.
For serum transfer, each naïve mouse was injected i.v. with 200 μL sera from immunized mice. Total T cells and CD4 T cells were purified from the lung and spleen cell preparations by using MACS Pan T Cell Isolation Kit II or CD4 microbeads (Miltenyi Biotec). Approximately 5 × 106 cells were adoptively transferred into naïve recipients via the i.v. route. Mice were then infected with H. influenzae 24 h following sera or T-cell transfer. Two days after infection, bacteria from lung homogenates were enumerated as described above.
In Vivo Staining with Anti-CD45.2.
Anti-mouse CD45.2-FITC (clone 104; BD Biosciences) was diluted to 10 μg/mL in sterile PBS and 250 μL of the solution was injected i.v. via the tail vein 3 min before killing and sample harvest as described by Christensen et al. (31).
Statistical Analysis.
All analyses were performed using Prism software (GraphPad Software). Unpaired, one-tailed, Student’s t tests were used to calculate statistical significance between two groups. The colony-forming unit data that fell below the limit of detection were assigned a value below that limit. The Kruskal–Wallis test was used to evaluate variance among all groups. If a significant variance was found, the Mann–Whitney test was used to identify significant differences between individual groups.
Supplementary Material
Acknowledgments
We thank Dr. Xin Fan for providing H. influenzae strains, Eagan and RdAW, and Drs. Jeffrey N. Weiser, Jun Zhu, and Paul Bates for helpful discussion. This work was supported by National Institutes of Health/National Institute of Allergy and Infectious Diseases Multiple Principal Investigator Grant R01AI095740 (to B.J.A. and H.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1802261115/-/DCSupplemental.
References
- 1.Price EP, et al. Haemophilus influenzae: Using comparative genomics to accurately identify a highly recombinogenic human pathogen. BMC Genomics. 2015;16:641. doi: 10.1186/s12864-015-1857-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Chiara M, et al. Genome sequencing of disease and carriage isolates of nontypeable Haemophilus influenzae identifies discrete population structure. Proc Natl Acad Sci USA. 2014;111:5439–5444. doi: 10.1073/pnas.1403353111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Puig C, et al. Clinical and molecular epidemiology of haemophilus influenzae causing invasive disease in adult patients. PLoS One. 2014;9:e112711. doi: 10.1371/journal.pone.0112711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agrawal A, Murphy TF. Haemophilus influenzae infections in the H. influenzae type b conjugate vaccine era. J Clin Microbiol. 2011;49:3728–3732. doi: 10.1128/JCM.05476-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leibovitz E, Jacobs MR, Dagan R. Haemophilus influenzae: A significant pathogen in acute otitis media. Pediatr Infect Dis J. 2004;23:1142–1152. [PubMed] [Google Scholar]
- 6.Van Eldere J, Slack MP, Ladhani S, Cripps AW. Nontypeable Haemophilus influenzae, an under-recognised pathogen. Lancet Infect Dis. 2014;14:1281–1292. doi: 10.1016/S1473-3099(14)70734-0. [DOI] [PubMed] [Google Scholar]
- 7.Sethi S, Evans N, Grant BJ, Murphy TF. New strains of bacteria and exacerbations of chronic obstructive pulmonary disease. N Engl J Med. 2002;347:465–471. doi: 10.1056/NEJMoa012561. [DOI] [PubMed] [Google Scholar]
- 8.Sethi S, et al. Determinants of bacteriological outcomes in exacerbations of chronic obstructive pulmonary disease. Infection. 2016;44:65–76. doi: 10.1007/s15010-015-0833-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaura E, et al. Same exposure but two radically different responses to antibiotics: Resilience of the salivary microbiome versus long-term microbial shifts in feces. MBio. 2015;6:e01693-15. doi: 10.1128/mBio.01693-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poolman JT, et al. Developing a nontypeable Haemophilus influenzae (NTHi) vaccine. Vaccine. 2000;19(Suppl 1):S108–S115. doi: 10.1016/s0264-410x(00)00288-7. [DOI] [PubMed] [Google Scholar]
- 11.Murphy TF. Vaccines for nontypeable Haemophilus influenzae: The future is now. Clin Vaccine Immunol. 2015;22:459–466. doi: 10.1128/CVI.00089-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kyd JM, Cripps AW. Potential of a novel protein, OMP26, from nontypeable Haemophilus influenzae to enhance pulmonary clearance in a rat model. Infect Immun. 1998;66:2272–2278. doi: 10.1128/iai.66.5.2272-2278.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kyd JM, Dunkley ML, Cripps AW. Enhanced respiratory clearance of nontypeable Haemophilus influenzae following mucosal immunization with P6 in a rat model. Infect Immun. 1995;63:2931–2940. doi: 10.1128/iai.63.8.2931-2940.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jalalvand F, Littorin N, Su YC, Riesbeck K. Impact of immunization with Protein F on pulmonary clearance of nontypeable Haemophilus influenzae. Vaccine. 2014;32:2261–2264. doi: 10.1016/j.vaccine.2014.02.082. [DOI] [PubMed] [Google Scholar]
- 15.Prymula R, et al. Pneumococcal capsular polysaccharides conjugated to protein D for prevention of acute otitis media caused by both Streptococcus pneumoniae and non-typable Haemophilus influenzae: A randomised double-blind efficacy study. Lancet. 2006;367:740–748. doi: 10.1016/S0140-6736(06)68304-9. [DOI] [PubMed] [Google Scholar]
- 16.Vesikari T, et al. Effectiveness of the 10-valent pneumococcal nontypeable Haemophilus influenzae protein D-conjugated vaccine (PHiD-CV) against carriage and acute otitis media-a double-blind randomized clinical trial in Finland. J Pediatric Infect Dis Soc. 2016;5:237–248. doi: 10.1093/jpids/piw010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siggins MK, et al. PHiD-CV induces anti-Protein D antibodies but does not augment pulmonary clearance of nontypeable Haemophilus influenzae in mice. Vaccine. 2015;33:4954–4961. doi: 10.1016/j.vaccine.2015.07.034. [DOI] [PubMed] [Google Scholar]
- 18.Gilsdorf JR. Antigenic diversity and gene polymorphisms in Haemophilus influenzae. Infect Immun. 1998;66:5053–5059. doi: 10.1128/iai.66.11.5053-5059.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clementi CF, Murphy TF. Non-typeable Haemophilus influenzae invasion and persistence in the human respiratory tract. Front Cell Infect Microbiol. 2011;1:1. doi: 10.3389/fcimb.2011.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swords WE. Nontypeable Haemophilus influenzae biofilms: Role in chronic airway infections. Front Cell Infect Microbiol. 2012;2:97. doi: 10.3389/fcimb.2012.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bandi V, et al. Nontypeable Haemophilus influenzae in the lower respiratory tract of patients with chronic bronchitis. Am J Respir Crit Care Med. 2001;164:2114–2119. doi: 10.1164/ajrccm.164.11.2104093. [DOI] [PubMed] [Google Scholar]
- 22.Sharma SK, Casey JR, Pichichero ME. Reduced memory CD4+ T-cell generation in the circulation of young children may contribute to the otitis-prone condition. J Infect Dis. 2011;204:645–653. doi: 10.1093/infdis/jir340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abe Y, et al. Lymphocyte proliferative response to P6 of Haemophilus influenzae is associated with relative protection from exacerbations of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2002;165:967–971. doi: 10.1164/ajrccm.165.7.2109009. [DOI] [PubMed] [Google Scholar]
- 24.Ye P, et al. Requirement of interleukin 17 receptor signaling for lung CXC chemokine and granulocyte colony-stimulating factor expression, neutrophil recruitment, and host defense. J Exp Med. 2001;194:519–527. doi: 10.1084/jem.194.4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freches D, et al. Mice genetically inactivated in interleukin-17A receptor are defective in long-term control of Mycobacterium tuberculosis infection. Immunology. 2013;140:220–231. doi: 10.1111/imm.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimizuka Y, et al. Roles of interleukin-17 in an experimental Legionella pneumophila pneumonia model. Infect Immun. 2012;80:1121–1127. doi: 10.1128/IAI.05544-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milner JD, et al. Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature. 2008;452:773–776. doi: 10.1038/nature06764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gawronski JD, Wong SM, Giannoukos G, Ward DV, Akerley BJ. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc Natl Acad Sci USA. 2009;106:16422–16427. doi: 10.1073/pnas.0906627106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong SM, Bernui M, Shen H, Akerley BJ. Genome-wide fitness profiling reveals adaptations required by Haemophilus in coinfection with influenza A virus in the murine lung. Proc Natl Acad Sci USA. 2013;110:15413–15418. doi: 10.1073/pnas.1311217110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong SM, Shaughnessy J, Ram S, Akerley BJ. Defining the binding region in factor H to develop a therapeutic factor H-Fc fusion protein against non-typeable Haemophilus influenzae. Front Cell Infect Microbiol. 2016;6:40. doi: 10.3389/fcimb.2016.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christensen D, Mortensen R, Rosenkrands I, Dietrich J, Andersen P. Vaccine-induced Th17 cells are established as resident memory cells in the lung and promote local IgA responses. Mucosal Immunol. 2017;10:260–270. doi: 10.1038/mi.2016.28. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, et al. Cross-protective mucosal immunity mediated by memory Th17 cells against Streptococcus pneumoniae lung infection. Mucosal Immunol. 2017;10:250–259. doi: 10.1038/mi.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong SM, Akerley BJ. Genome-scale approaches to identify genes essential for Haemophilus influenzae pathogenesis. Front Cell Infect Microbiol. 2012;2:23. doi: 10.3389/fcimb.2012.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zygmunt BM, Rharbaoui F, Groebe L, Guzman CA. Intranasal immunization promotes th17 immune responses. J Immunol. 2009;183:6933–6938. doi: 10.4049/jimmunol.0901144. [DOI] [PubMed] [Google Scholar]
- 35.LeibundGut-Landmann S, et al. Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat Immunol. 2007;8:630–638. doi: 10.1038/ni1460. [DOI] [PubMed] [Google Scholar]
- 36.Liu DF, et al. The C-terminal fragment of the internal 110-kilodalton passenger domain of the Hap protein of nontypeable Haemophilus influenzae is a potential vaccine candidate. Infect Immun. 2004;72:6961–6968. doi: 10.1128/IAI.72.12.6961-6968.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winter LE, Barenkamp SJ. Antibodies specific for the high-molecular-weight adhesion proteins of nontypeable Haemophilus influenzae are opsonophagocytic for both homologous and heterologous strains. Clin Vaccine Immunol. 2006;13:1333–1342. doi: 10.1128/CVI.00221-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Yp, Thomas WR, Chong P, Loosmore SM, Klein MH. A 20-kilodalton N-terminal fragment of the D15 protein contains a protective epitope(s) against Haemophilus influenzae type a and type b. Infect Immun. 1998;66:3349–3354. doi: 10.1128/iai.66.7.3349-3354.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loosmore SM, et al. The Haemophilus influenzae HtrA protein is a protective antigen. Infect Immun. 1998;66:899–906. doi: 10.1128/iai.66.3.899-906.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neary JM, Yi K, Karalus RJ, Murphy TF. Antibodies to loop 6 of the P2 porin protein of nontypeable Haemophilus influenzae are bactericidal against multiple strains. Infect Immun. 2001;69:773–778. doi: 10.1128/IAI.69.2.773-778.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Novotny LA, Jurcisek JA, Pichichero ME, Bakaletz LO. Epitope mapping of the outer membrane protein P5-homologous fimbrin adhesin of nontypeable Haemophilus influenzae. Infect Immun. 2000;68:2119–2128. doi: 10.1128/iai.68.4.2119-2128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hotomi M, et al. A recombinant P4 protein of Haemophilus influenzae induces specific immune responses biologically active against nasopharyngeal colonization in mice after intranasal immunization. Vaccine. 2005;23:1294–1300. doi: 10.1016/j.vaccine.2004.08.042. [DOI] [PubMed] [Google Scholar]
- 43.Novotny LA, et al. Epitope mapping immunodominant regions of the PilA protein of nontypeable Haemophilus influenzae (NTHI) to facilitate the design of two novel chimeric vaccine candidates. Vaccine. 2009;28:279–289. doi: 10.1016/j.vaccine.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hirano T, Hou Y, Jiao X, Gu XX. Intranasal immunization with a lipooligosaccharide-based conjugate vaccine from nontypeable Haemophilus influenzae enhances bacterial clearance in mouse nasopharynx. FEMS Immunol Med Microbiol. 2003;35:1–10. doi: 10.1111/j.1574-695X.2003.tb00642.x. [DOI] [PubMed] [Google Scholar]
- 45.Moffitt KL, Malley R, Lu YJ. Identification of protective pneumococcal T(H)17 antigens from the soluble fraction of a killed whole cell vaccine. PLoS One. 2012;7:e43445. doi: 10.1371/journal.pone.0043445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moffitt KL, et al. T(H)17-based vaccine design for prevention of Streptococcus pneumoniae colonization. Cell Host Microbe. 2011;9:158–165. doi: 10.1016/j.chom.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nielsen H. Predicting secretory proteins with SignalP. Methods Mol Biol. 2017;1611:59–73. doi: 10.1007/978-1-4939-7015-5_6. [DOI] [PubMed] [Google Scholar]
- 48.Bendtsen JD, Kiemer L, Fausbøll A, Brunak S. Non-classical protein secretion in bacteria. BMC Microbiol. 2005;5:58. doi: 10.1186/1471-2180-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goodell EW, Higgins CF. Uptake of cell wall peptides by Salmonella typhimurium and Escherichia coli. J Bacteriol. 1987;169:3861–3865. doi: 10.1128/jb.169.8.3861-3865.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ren D, et al. Serum antibody response to Moraxella catarrhalis proteins in stringently defined otitis prone children. Vaccine. July 26, 2017 doi: 10.1016/j.vaccine.2017.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roier S, et al. Intranasal immunization with nontypeable Haemophilus influenzae outer membrane vesicles induces cross-protective immunity in mice. PLoS One. 2012;7:e42664. doi: 10.1371/journal.pone.0042664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu W, et al. Th17-stimulating protein vaccines confer protection against Pseudomonas aeruginosa pneumonia. Am J Respir Crit Care Med. 2012;186:420–427. doi: 10.1164/rccm.201202-0182OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Romero-Steiner S, et al. Measurement of serum bactericidal activity specific for Haemophilus influenzae type b by using a chromogenic and fluorescent metabolic indicator. Clin Diagn Lab Immunol. 2004;11:89–93. doi: 10.1128/CDLI.11.1.89-93.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.