Abstract
Background
Autosomal dominant polycystic kidney disease (ADPKD) is a ciliopathy caused by mutations in PKD1 and PKD2 that is characterized by renal tubular epithelial cell proliferation and progressive CKD. Although the molecular mechanisms involved in cystogenesis are not established, concurrent inactivating constitutional and somatic mutations in ADPKD genes in cyst epithelium have been proposed as a cellular recessive mechanism.
Methods
We characterized, by whole-exome sequencing (WES) and long-range PCR techniques, the somatic mutations in PKD1 and PKD2 genes in renal epithelial cells from 83 kidney cysts obtained from nine patients with ADPKD, for whom a constitutional mutation in PKD1 or PKD2 was identified.
Results
Complete sequencing data by long-range PCR and WES was available for 63 and 65 cysts, respectively. Private somatic mutations of PKD1 or PKD2 were identified in all patients and in 90% of the cysts analyzed; 90% of these mutations were truncating, splice site, or in-frame variations predicted to be pathogenic mutations. No trans-heterozygous mutations of PKD1 or PKD2 genes were identified. Copy number changes of PKD1 ranging from 151 bp to 28 kb were observed in 12% of the cysts. WES also identified significant mutations in 53 non-PKD1/2 genes, including other ciliopathy genes and cancer-related genes.
Conclusions
These findings support a cellular recessive mechanism for cyst formation in ADPKD caused primarily by inactivating constitutional and somatic mutations of PKD1 or PKD2 in kidney cyst epithelium. The potential interactions of these genes with other ciliopathy- and cancer-related genes to influence ADPKD severity merits further evaluation.
Keywords: ADPKD, molecular genetics, human genetics
Autosomal dominant polycystic kidney disease (ADPKD) is clinically and genetically heterogeneous, caused by mutations in PKD1 or PKD2, and is the most common inherited CKD.1 In 7%–10% of individuals with ADPKD, the pathogenic variant of a PKD gene is not detected, suggesting other genetic causes including hypomorphic alleles, mosaicism, or rare mutations of other genes.2 Inactivating, somatic mutations of PKD1/2 genes that occur in renal epithelial cells, together with a concurrent, constitutional mutation of PKD1 or PKD2, cause clonal proliferation of tubular epithelium and cyst formation.3 In this cellular recessive mechanism of cyst formation in ADPKD, disease severity has been attributed to the affected PKD gene locus and the timing and type of somatic PKD gene mutations. Kidney cyst formation also occurs when there is a deletion of chromosomal regions of the corresponding PKD1 or PKD2 allele, termed loss of heterozygosity (LOH).4–8 These mechanisms have also been identified in malignant neoplasms, suggesting that ADPKD is a benign neoplastic process.9,10
Recent studies, utilizing more technically advanced methods, concluded that modestly reduced levels of PKD1/2 gene expression (i.e., haploinsufficiency), rather than PKD1/2 gene inactivation, caused a spectrum of cystic renal phenotypes, ranging from few, small cysts with relatively well compensated CKD, to markedly enlarged kidneys with early onset ESRD.3,11–14 Although the prevalence of PKD gene haploinsufficiency in the ADPKD population has not been established, it appears to be low.15
Primary cilia are microtubule-based, nonmotile projections of epithelial cells that extend into the lumen of renal tubules and other structures.16 Integral membrane proteins, including polycystin 1 (PC1, TRPP1) and polycystin 2 (PC2, TRPP2), encoded by PKD1 and PKD2, respectively, localize to these cilia.17 Mammalian ciliopathies are inherited disorders characterized by various phenotypes, including renal cyst formation. They are caused by mutations in genes that encode proteins associated with cilia and trafficking pathways.18 The potential interactions of PKD1/2 genes with non-PKD1/2 genes, including other ciliopathy genes, have not been well defined.
Given the complex ADPKD phenotype, and multiple factors affecting cyst development, we characterized the prevalence and diversity of somatic PKD1/2 gene mutations arising in renal cyst epithelium and investigated whether somatic mutations in non-ADPKD genes occur that might contribute to the pathogenesis of kidney cysts. We report a comprehensive genomic analysis of somatic mutations, using whole-exome sequencing (WES) and Sanger sequencing, in cyst epithelial cells that were isolated after native nephrectomy in patients with ADPKD for whom constitutional mutations in PKD1/2 were identified. In this study, inactivating somatic mutations in PKD1/2 genes occurred in 90% of kidney cysts. We also characterized somatic variants in non-PKD1/2 genes (e.g., ciliopathy, cancer) from these cysts, including those associated with other ciliopathies, which might influence cyst formation and disease severity in ADPKD.
Methods
Patients with ADPKD were enrolled in this study if they were scheduled for a living donor kidney transplant, and removal of one or both native kidneys at the New York-Presbyterian Hospital/Weill Cornell Medicine campus. Study eligibility was determined by the transplant surgeon (S.K.) before enrollment of each participant and informed consent was obtained (Supplemental Figure 1). The protocol was approved by the Weill Cornell Medicine Institutional Review Board. Cyst epithelial cells were isolated according to standard procedures.5,19 PKD1/2 genetic analysis was performed by long-range PCR next-generation sequencing and Sanger sequencing.20,21 WES (357,999 exons, >20,000 genes) of renal cyst epithelia and peripheral blood lymphocytes (PBL) DNA was performed using the Agilent HaloPlex Target Enrichment and massive parallel sequencing (Illumina, San Diego, CA). Data analysis of the cyst and matched constitutional DNA was performed for simultaneous detection of single nucleotide variants (SNVs), indels, and copy number variations (CNVs) as previously described.22,23 Schematics of WES data analysis pipeline is shown is Supplemental Figure 2. The pathogenic potential of missense variants was evaluated using the computational analysis tool Combined Annotation–Dependent Depletion (CADD; http://cadd.gs.washington.edu/). A CADD value of ≥15 suggests the variant is likely pathogenic.24 Significantly mutated WES variants were further filtered using a 30% variant allele frequency (VAF) cutoff and a population allele frequency of 5% in The Exome Aggregation Consortium (ExAC) database (http://exac.broadinstitute.org/).
The significance of mutated genes and pathways was analyzed with the Mutational Significance In Cancer (MuSiC) suite of tools (http://gmt.genome.wustl.edu/packages/genome-music/index.html) using default settings, accounting for gene background mutation rates. Potential splice-site effects were evaluated using splicing prediction tools.20 A custom analysis pipeline analyzed constitutional mutations in 211 ciliopathy and 565 cancer genes (http://cancer.sanger.ac.uk/census/; T. Zhang et al., unpublished data). Pathway analysis was performed by Reactome (https://reactome.org/). Statistical analysis to study the association between somatic variant and cyst size was performed using a linear mixed effects model with the R software environment (https://cran.r-project.org/web/packages/nlme/index.html). Additional details are provided in Supplemental Material.
Results
Clinical Cohort
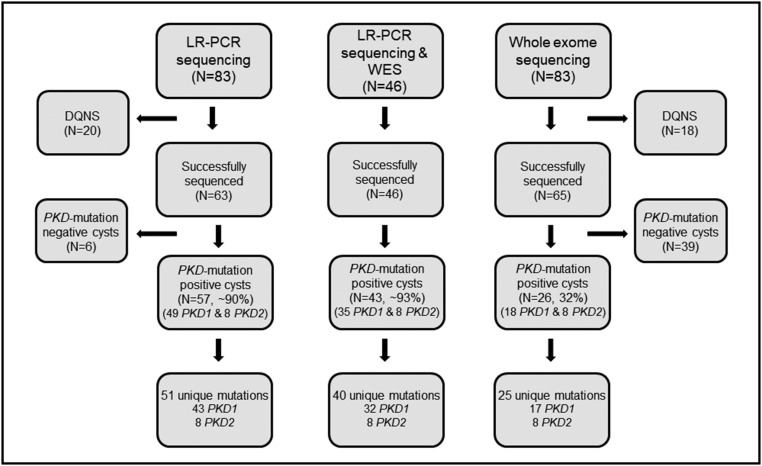
Nine unrelated patients with ADPKD were enrolled. All but one had unilateral nephrectomy during preemptive, living donor transplantation; one underwent bilateral nephrectomy in preparation for future kidney transplantation (Supplemental Figure 1). Each kidney contained innumerable cysts; 237 cysts were collected, with an average of 13 cysts per kidney. The median cyst diameter was 3 cm (range: 1–20 cm; Table 1). A total of 83 cysts were randomly selected for sequencing, where sufficient DNA was available (Figure 1). There was no difference in size of the cysts analyzed and those not analyzed (P=0.94).
Table 1.
ADPKD patient cohort clinical and pathologic characteristics
| Patient | Sex | Age, yr | Age at Diagnosis, yr | Left/Right Kidney | Weight, g | Kidney Size, L×W×H, cm | No. of Cysts Genotypeda | Diameter Range of Cysts, cm |
|---|---|---|---|---|---|---|---|---|
| BS9001424 | F | 73 | 48 | Left | 520 | 16×7×7 | 6 | 2–6 |
| Right | 530 | 15×10×7 | 4 | |||||
| PJ9001456 | F | 41 | 34 | Left | 3460 | 27.5×16×8.5 | 9 | 1.2–6 |
| Right | 2740 | 27.5×15×8.5 | 3 | |||||
| DR9001565 | M | 59 | 26 | Left | 4020 | 35×18×12 | 6 | 1.5–8 |
| Right | 3999 | 31×17×14 | 5 | |||||
| CD001574 | M | 58 | 27 | Left | 1770 | 24×11.3×11 | 4 | 1.5–6 |
| Right | 1680 | 23.5×11.3×11 | 4 | |||||
| BJA001578 | M | 52 | 39 | Left | 4560 | 30.7×15.2×14.5 | 4 | 1.5–10 |
| Right | 4010 | 31×17×13.5 | 6 | |||||
| CMJ0001593 | M | 59 | 53 | Left | 1680 | 24×13×10 | 5 | 2–10 |
| Right | 1500 | 23×12.7×8.3 | 3 | 2–8 | ||||
| RP9001591 | F | 54 | 29 | Left | 2340 | 25.6×14.5×13 | 5 | 1.5–5 |
| Right | 2300 | 23.5×15×10 | 2 | 4–12 | ||||
| JS9001595 | F | 56 | 32 | Left | 1670 | 24×11.5×8.2 | 4 | 2.5–7 |
| Right | 1900 | 27×14×9.3 | 4 | 3–8 | ||||
| BH9002280 | M | 62 | 33 | Left | 6800 | 41.5×25×12 | 5 | 4–12 |
| Right | 8900 | 43.5×31×13 | 4 | 6.5–20 |
L×W×H, length, width, and height; F, female; M, male.
Genotyping performed as described in Figure 1.
Figure 1.
PKD1 and PKD2 mutation analysis was successfully performed using DNA obtained from renal cyst epithelia. Schematic representation of 83 cysts (nine patients) that were subjected to ADPKD gene mutation analysis, broken into subsets by sequencing method is shown. Of the 83 specimens, complete genotyping data by long-range PCR (LR-PCR) Sanger/next generation sequencing and whole exome sequencing (WES) was available for 63 and 65 cysts, respectively. A total of 46 specimens were successfully sequenced by both methods. Mutation detection rates for each sequencing group are denoted in the corresponding boxes. DQNS, DNA quantity not sufficient.
Detection of PKD1 and PKD2 Somatic Mutations in Renal Cyst Epithelia
Using a combination of WES and LR-PCR based methods, PKD1/2 genes were sequenced in epithelial cell DNA isolated from 63 kidney cysts. Constitutional mutations were detected in all PBL samples; PKD1 mutations were detected in 89% (eight out of nine participants); the remaining patient had a mutation in PKD2 (Supplemental Table 1). Of these mutations, six (67%) were truncating and two were missense variants predicted to be likely pathogenic (CADD score ≥15).
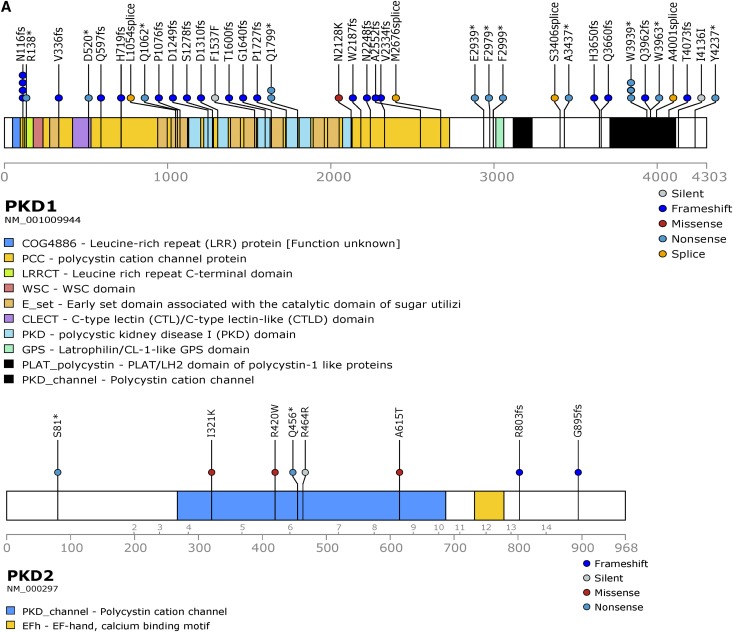
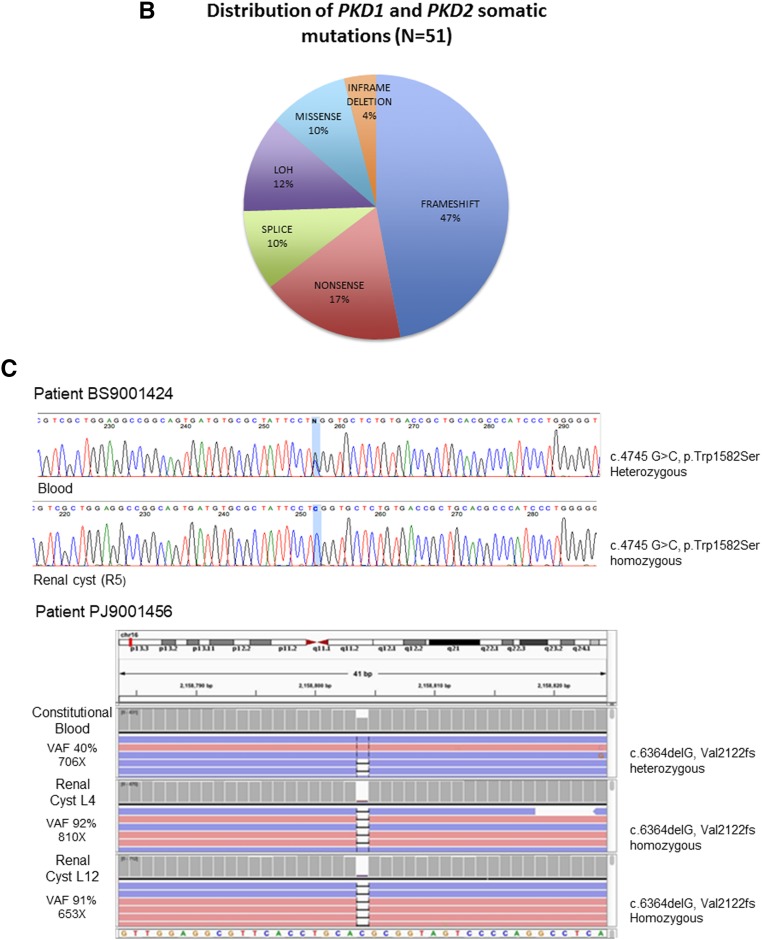
Pathogenic somatic mutations in PKD1/2 genes were identified in all patients and in 90% of cysts (57 out of 63; Supplemental Table 2, Table 2). Long-range PCR Sanger sequencing confirmed all somatic mutations identified by WES. Mutations were distributed across PKD1 and PKD2 (Figure 2A). PKD1 and PKD2 somatic mutations were limited to PKD1 and PKD2 carriers, respectively. There were 51 unique pathogenic/likely pathogenic somatic mutations detected in 49 and eight cysts of patients with constitutional mutations in PKD1 and PKD2, respectively. The vast majority (90%) were truncating, splice site, and in-frame variations predicted to be pathogenic; five (10%) were missense, likely pathogenic variants (Figure 2B). Seven of the 49 PKD1 mutation-positive cysts (14%) were homozygous for constitutional PKD1 mutations, consistent with LOH (Figure 2C, Supplemental Figure 3, Table 2 for representative LOH events identified by long-range PCR Sanger/next-generation sequencing and WES). Mean alternate allele frequencies were 0.48 and 0.96 for the heterozygous (constitutional) and homozygous (somatic) mutations, respectively (Supplemental Table 3). Most (77%; 33 out of 43) PKD1 somatic mutations were located in its duplicated region (exons 1–33); the remaining mutations were located in its single-copy region. Several cysts in different patients shared the same PKD1 truncating mutations (c.5395C>T, p.Gln1799*; c.348_352del, p.Asn116Lysfs*2; c.11817G>A, p.Trp3939*), suggesting a hotspot in PKD1.
Table 2.
Summary of somatic mutations in PKD genes by cyst for nine patients
| Patienta/Gene | Cyst | Constitutional Mutation | Exon | Somatic Mutation | Exon/Intron | Mutation Type | Mutation Designation |
|---|---|---|---|---|---|---|---|
| BS9001424/PKD1 | L1 | c.4745 G>C, p.Trp1582Ser (CADD=26.3) | Exon 15 | ND | |||
| L2 | c.4798dup, p.Thr1600Asnfs*15 | Exon 15 | Frameshift | Pathogenic | |||
| L3 | c.12216_12217del, p.Thr4073Profs*83 | Exon 45 | Frameshift | Pathogenic | |||
| L4 | c.348_352del, p.Asn116Lysfs*2 | Exon 3 | Frameshift | Pathogenic | |||
| L5 | c.3225del, p.Pro1076Argfs*28 | Exon14 | Frameshift | Pathogenic | |||
| L6 | c.348_352del, p.Asn116Lysfs*2 | Exon 3 | Frameshift | Pathogenic | |||
| R1 | c.11888G>A, p.Trp3963* | Exon 43 | Nonsense | Pathogenic | |||
| R2 | c.5395C>T, p.Gln1799* | Exon 15 | Nonsense | Pathogenic | |||
| R4 | c.3162–2A>C | IVS 11 | Splicing | Pathogenic | |||
| R5 | c.4745 G>C,p.Trp1582Ser (LOH) | Exon 15 | LOH | Likely pathogenic | |||
| PJ9001456/PKD1 | L1 | c.6364delG, p.Val2122Lysfs*3 | Exon 15 | ND | |||
| L4 | c.6364delG, p.Val2122Lysfs*3(LOH) | Exon 15 | LOH | Pathogenic | |||
| L5 | c.348_352del, p.Asn116Lysfs*2 | Exon 3 | Frameshift | Pathogenic | |||
| L6 | c.8016+1del | Exon 21 | Splicing | Pathogenic | |||
| L7 | c.12006_12010del, p.Gln4004Alafs*151 | Exon 44 | Frameshift | Pathogenic | |||
| L10 | c.7655_7658 delinsTTG, p.Ala2552Valfs*68 | Exon 19 | Frameshift | Pathogenic | |||
| L11 | c.3745del, p.Asp1249Thrfs*24 | Exon 15 | Frameshift | Pathogenic | |||
| L12 | c.6364delG, p.Val2122Lysfs*3(LOH) | Exon 15 | LOH | Pathogenic | |||
| L15 | c.1284_1292del, p.Trp429_Gln431del | Exon 6 | In-frame deletion | Likely pathogenic | |||
| R4 | c.3831_3847del, p.Ser1278Glyfs*17 | Exon 15 | Frameshift | Pathogenic | |||
| R7 | c.12004–2_12019del (splice_acceptor) | Exon 44 | Splicing | Pathogenic | |||
| R16 | ND | ||||||
| DR9001565/PKD1 | L3 | c.9504C>G, p.Phe3168Leu (CADD=27) | Exon 27 | c.9504C>G,p.Phe3168Leu (LOH) | Exon 27 | LOH | Pathogenic |
| L5 | c.8935_8937delTTC, p.Phe2979del | Exon 24 | In-frame deletion | Likely pathogenic | |||
| L7 | c.5395C>T, p.Gln1799* | Exon 15 | Nonsense | Pathogenic | |||
| R8 | c.2157del, p.His719Glnfs*66 | Exon 11 | Frameshift | Pathogenic | |||
| R9 | c.1789del, p.Gln597Argfs*188 | Exon 9 | Frameshift | Pathogenic | |||
| c.4916dup, p.Gly1640Argfs*18 | Exon 15 | Frameshift | Pathogenic | ||||
| R10 | c.5180del, p.Pro1727Argfs*32 | Exon 15 | Frameshift | Pathogenic | |||
| R13 | c.12711C>A, p.Tyr4237* | Exon 46 | Nonsense | Pathogenic | |||
| CDS001574/PKD1 | L2 | c.10084del, p.Gln3362Serfs*35 | Exon 31 | c.10314_10315insGCTGGCA, p.Arg3439Alafs*34 | Exon 33b | Frameshift | Pathogenic |
| L9 | c.348_352del, p.Asn116Lysfs*2 | Exon 3 | Frameshift | Pathogenic | |||
| R6 | c.11817G>A, p.Trp3939* | Exon 43 | Nonsense | Pathogenic | |||
| R9 | c.11884_11912del, p.Gln3962Alafs*5 | Exon 43 | Frameshift | Pathogenic | |||
| R10 | c.10084del, p.Gln3362Serfs*35 (LOH) | Exon 31 | LOH | Pathogenic | |||
| BJA001578/PKD1 | L3 | c.3929_3930del, p.Asp1310Glyfs*120 | Exon 43 | c.6558_6568del, p.Trp2187Serfs*71 | Exon 15 | Frameshift | Pathogenic |
| L4 | c.10948delC, p.His3650Thrfs*34 | Exon 37 | Frameshift | Pathogenic | |||
| L8 | c.10319del, p.Gly3440Alafs*33/CNV* | Exon 33 | Frameshift | Pathogenic | |||
| R2 | ND | ||||||
| R4 | c.1551_1560dup, p.Leu521* | Exon 7 | Frameshift | Pathogenic | |||
| R10 | c.412C>T, p.Arg138* | Exon 4 | Nonsense | Pathogenic | |||
| R11 | c.3929_3930del, p.Asp1310Glyfs*120 (LOH) | Exon 45 | LOH | Pathogenic | |||
| R13 | ND | ||||||
| R16 | c.12707T>A, p.Val4236Asp | Exon 46 | Missense | Likely pathogenic | |||
| (CADD=21.4) | |||||||
| CMJ001593/PKD1 | L2 | c.10406_10407insG, p.Asp3469Glufs*2 | Exon 34 | c.6994_7000dup, p.Val2334Glyfs*88 | Exon 16 | Frameshift | Pathogenic |
| L3 | c.6384C>A, p.Asn2128Lys | Exon 15 | Missense | Likely pathogenic | |||
| (CADD=26.7) | |||||||
| L10 | c.10977del, p.Glu3660Lysfs*24 | Exon 37 | Frameshift | Pathogenic | |||
| L11 | c.8815G>T, p.Glu2939* | Exon 24 | Nonsense | Pathogenic | |||
| R3 | c.11817G>A, p.Trp3939* | Exon 43 | Nonsense | Pathogenic | |||
| R9 | c.11817G>A, p.Trp3939* | Exon 43 | Nonsense | Pathogenic | |||
| RP9001591/PKD1 | L2 | c.G10549T, p.Glu3517* | Exon 35 | c.8996_8997insG, p.Phe2999Leufs*70 | Exon 25 | Frameshift | Pathogenic |
| L11 | c.G10549T, p.Glu3517* (LOH) | Exon 35 | LOH | Pathogenic | |||
| R17 | c.1005delC, p.Val336Cysfs*129 | Exon 5 | Frameshift | Pathogenic | |||
| JS9001595/PKD1 | R2 | c.6743_6744dup, p.Val2249Metfs*2 | Exon 15 | c.10220+2T>G | IVS 35 | Splicing | Pathogenic |
| R10 | c.3184C>T, p.Gln1062* | Exon 14 | Nonsense | Pathogenic | |||
| BH9002280/PKD2 | L1 | c.923del, p.Phe308Serfs*9 | Exon 4 | c.2409delA, p.Ser804Valfs*40 | Exon 13 | Frameshift | Pathogenic |
| L2 | c.242 C>A, p.Ser81* | Exon 1 | Nonsense | Pathogenic | |||
| L5 | c.962 T>A, p.Ile321Lys (CADD=24.6) | Exon 4 | Missense | Likely pathogenic | |||
| L6 | c.1843G>A, p.Ala615Thr (CADD=33.0) | Exon 8 | Missense | Likely pathogenic | |||
| L9 | c.1258A>T, p.Arg420Trp (CADD=27.7) | Exon 5 | Missense | Likely pathogenic | |||
| R1 | ND | ||||||
| R2 | c.1392A>G, p.=c | Exon 6 | Splicing | Likely pathogenic | |||
| R7 | c.1366 C>T, p.Gln456* | Exon 6 | Nonsense | Pathogenic | |||
| R8 | c.2682del, p.Gly895Valfs*14 | Exon 15 | Frameshift | Pathogenic |
ND, mutation not detected; L, Left kidney; R, Right kidney. LOH. loss of heterozygosity.
Probable pathogenic mutation as evaluated by CADD. A CADD value of ≥15 suggests the variant is likely pathogenic (http://cadd.gs.washington.edu/).
PKD1 exon 33 single-copy region.
Mutation (c.1392A>G, p.=) predicted to create a new donor splice site using splice site prediction tools (ESEfinder3.0, NNSplice and Human splicing finder3.1) with default settings.
Figure 2.
Unique pathogenic somatic mutations in PKD1 and PKD2 were identified in renal cyst epithelia. (A) Prevalence of PKD1 (n=37) and PKD2 (n=8) unique somatic mutations detected in 57 renal cyst DNA samples from patients with ADPKD. Loss of heterozygosity (LOH) mutations not shown. (B) Distribution of SNV and indel mutations in PKD1/2 genes in renal cyst epithelium from all nine patients with ADPKD. Alterations are colored according to mutation type. (C) PKD1 LOH mutations in renal cyst epithelia. Blood and renal cyst DNA heterozygous and homozygous for c.4745 G>C, Trp1582Ser, respectively, detected by long-range PCR (LR-PCR) Sanger sequencing (top panel). Blood and renal cyst DNAs heterozygous and homozygous for c.6364delG, p.Val2122fs, respectively, detected by LR-PCR next-generation sequencing (NGS; bottom panel). PKD1 NGS reads are piled up and are shown in Integrative Genomics Viewer. Allelic total coverage and variant allele frequency (VAF) are displayed for each sample type.
Characterization by WES of Somatic Mutations in Renal Cyst Epithelia
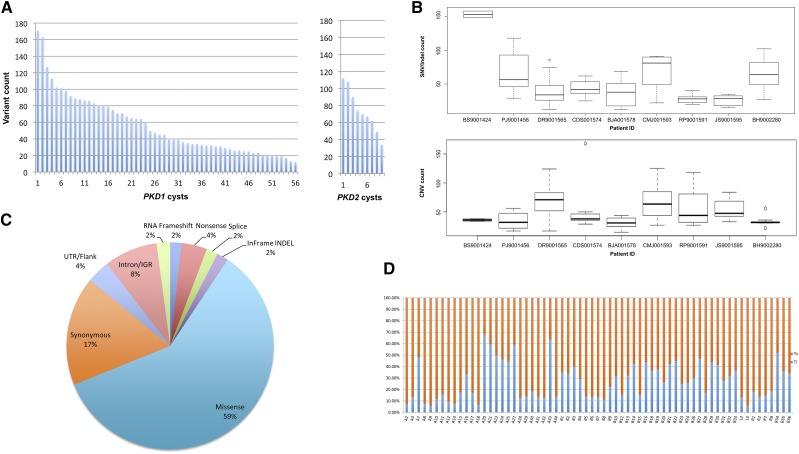
A total of 74 DNA samples (cyst, n=65; PBL, n=9) were analyzed by WES, with an average coverage of 89.5×±8.9×; more than 95% of the exome was covered ≥10× (Supplemental Table 4). Somatic changes were identified in all 56 PKD1 and nine PKD2 cysts analyzed. There were a total of 3263 somatic SNV/indels (2490 genes) and 3333 CNV changes in 2456 genes. All CNVs were focal (i.e., encompassing <25% of a chromosome arm), and at least one CNV change per cyst was found. Of these, 1057 (31.7%) were copy number gains and 2276 (68.3%) were losses. The mutation load varied significantly, with a median of 49 and an interquartile range (IQR) of 81-31=50 mutations per cyst (range: 12–171; Figure 3A). Per patient, a median of 65 and an IQR of 70-39=31 occurred for SNV/indels (range: 25–167), while a median of 38 and an IQR of 47.5-32=15.5 for CNVs (range: 31–71; Figure 3B). Of the SNV/indel mutations, 247 (7.6%) were truncating (frameshift, nonsense, splicing), whereas 1942 (59%) were missense variants (Figure 3C, Supplemental Figure 4, Supplemental Table 5). SNV transition/transversion ratio was with a median of 0.35 and an IQR of 0.67-0.16=0.51 (range: 0.06–2.14; Figure 3D).
Figure 3.
WES of renal cyst epithelia DNA identified a wide spectrum of somatic alterations in all samples. (A) Distribution of mutation (SNV, indel, and CNV) load identified by WES by cyst and by PKD1/2 genes (n=65). (B) The frequency of mutation alterations in cyst epithelia varied significantly among the patients. Alterations are classified by mutation type. SNV/indel and CNV distribution are shown in the top and bottom panels, respectively. (C) Frequency of all somatic SNV substitutions and indel mutations detected in 65 renal cyst epithelia samples from nine patients with ADPKD. Alterations are colored according to mutation type. (D) Transition (Ti) to transversion (Tv) plot of SNVs detected. IGR, intergenic region; UTR, untranslated region.
Patient BS9001424 was incidentally found to have a papillary renal cell carcinoma in the resected kidney, which was not detected preoperatively.22 The mutation load for this patient (153 mutations per cyst) was higher than the median mutation load (65 mutations per cyst) for the cohort (Figure 3B). This patient demonstrated a constitutional heterozygous mutation in ATM (NM_000051.3: c.6228del; p.Leu2077Phefs*5), potentially accounting for the high background mutation rate, and a somatic mutation in MET. No other patient had a constitutional mutation identified in ATM or other cancer genes.
Age was not associated with prevalence of somatic variants (P=0.20) for the cohort. However, in a multivariate analysis of patients aged >50 years, age was significantly associated with a higher variant count (P=0.005; Supplemental Material).
Overall, 1785 variations (1477 genes) with VAF≥5% were classified as pathogenic or likely pathogenic mutations (Supplemental Table 6). Of these, 1473 (83%) were missense mutations (CADD score ≥15) and 312 (17%) were truncating or likely pathogenic in-frame indels. Only 248 variations (175 missense and 73 truncating or in-frame indels) in 191 genes were present at a VAF ≥20%, suggesting they occurred earlier in cyst development than mutations with lower VAFs (Supplemental Table 7 and Supplemental Figure 5). At least one significant pathogenic or likely pathogenic SNV/indel change was found in every cyst. A summary of the principal variants as classified by their potential effect on protein function and VAF (≥30%) is presented in Table 3. No variants with prevalence ≥30% were identified in 16% (nine out of 65) of cysts analyzed.
Table 3.
Pathogenic and probable pathogenic mutations with VAF≥30%, by cyst (n=103)
| Patient | Cyst | Gene | Chr | HGVSc | HGVSp | Read Depth | VAF | CADD Score | Pathway (Reactome) | P Value | rs#/COSMIC (ExAC%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BJA001578 | A23 | PKD1 | chr16 | c.1560_1561insTAACACCGAC | p.Leu521Ter | 10 | 0.8 | VxPx cargo-targeting to cilium | 0.006 | ||
| B13 | B3GNT3 | chr19 | c.156_157insCCCCCC | p.Pro52_Pro53insProPro | 28 | 0.61 | O-linked glycosylation of mucins | 0.000 | |||
| B16 | PKD1 | chr16 | c.6485G>A | p.Arg2162Gln | 753 | 0.35 | 25.8 | VxPx cargo-targeting to cilium | 0.009 | ||
| B16 | SMARCAD1 | chr4 | c.2572T>C | p.Phe858Leu | 66 | 0.48 | 23 | * | * | ||
| B14 | STK19 | chr6 | c.328G>T | p.Asp110Tyr | 42 | 0.31 | 19.46 | * | * | ||
| A23 | C4orf21 | chr4 | c.3919C>A | p.His1307Asn | 15 | 0.4 | 28.3 | * | * | ||
| A24 | PKD1 | chr16 | c.3929_3930del | p.Asp1310Glyfs*120 (LOH) | 12 | 1.00 | VxPx cargo-targeting to cilium | 0.006 | |||
| BS9001424 | A3 | PKD1 | chr16 | c.11888G>A | p.Trp3963Ter | 18 | 0.56 | 41 | VxPx cargo-targeting to cilium | 0.039 | |
| A3 | PSME4 | chr2 | c.4821_4832delCCCAGTGGAAAA | p.Pro1608_Asn1611del | 20 | 0.5 | * | * | |||
| A3 | WARS2 | chr1 | c.404G>T | p.Arg135Leu | 46 | 0.48 | 35 | Mitochondrial tRNA aminoacylation | 0.039 | rs147962776 (0.03) | |
| A3 | PAPD7 | chr5 | c.1371_1373delGTC | p.Met457_Ser458delinsIle | 204 | 0.48 | * | * | |||
| A3 | ITGA7 | chr12 | c.386G>A | p.Arg129Gln | 91 | 0.46 | 19.41 | Laminin interactions | 0.06 | rs376167554 (0.002) | |
| A3 | NCOR1 | chr17 | c.5506G>A | p.Val1836Met | 66 | 0.39 | 24.9 | NR1D1 (REV-ERBA) represses gene expression | 0.006 | ||
| A3 | USP51 | chrX | c.663_666delTGGG | p.Ser221ArgfsTer22 | 32 | 0.59 | * | * | |||
| A3 | INSIG2 | chr2 | c.83T>G | p.Leu28Trp | 61 | 0.57 | 26.5 | * | * | ||
| A3 | NAA15 | chr4 | c.175delA | p.Ile59SerfsTer34 | 48 | 0.56 | * | * | rs375319596 (NA) | ||
| A3 | C1orf74 | chr1 | c.23C>A | p.Ser8Ter | 101 | 0.53 | 36 | * | * | ||
| A3 | BCOR | chrX | c.4690A>G | p.Arg1564Gly | 76 | 0.51 | 31 | * | * | ||
| A3 | EMX2 | chr10 | c.651A>T | p.Glu217Asp | 56 | 0.48 | 19.56 | * | * | ||
| A3 | ELP3 | chr8 | c.1486–1G>T | 77 | 0.48 | 21.1 | * | * | |||
| A3 | SIDT1 | chr3 | c.2269G>A | p.Ala757Thr | 17 | 0.47 | 34 | * | * | COSM201721 | |
| A3 | RAB23 | chr6 | c.398A>T | p.Asn133Ile | 110 | 0.46 | 22.8 | * | * | ||
| A3 | ZBTB40 | chr1 | c.435_436delAG | p.Gln145HisfsTer7 | 47 | 0.45 | * | * | |||
| A3 | DMXL1 | chr5 | c.4099C>A | p.His1367Asn | 52 | 0.44 | 15.66 | * | * | ||
| A3 | CRYZ | chr1 | c.411_414delTCGA | p.Tyr137Ter | 143 | 0.4 | * | * | |||
| A3 | QRICH2 | chr17 | c.4226C>T | p.Ala1409Val | 43 | 0.33 | 21.4 | * | * | ||
| A2 | NOX5 | chr15 | c.844A>C | p.Ser282Arg | 25 | 0.32 | 23.9 | * | * | rs150003957 (1.67) COSM964378 | |
| CDS001574 | A21 | PKD1 | chr16 | c.11817G>A | p.Trp3939Ter | 27 | 0.56 | 39 | VxPx cargo-targeting to cilium | 0.006 | |
| B7 | AKT2 | chr19 | c.1016T>G | p.Val339Gly | 13 | 0.31 | 27.1 | PKB-mediated events | 0.001 | ||
| B12 | PKD1 | chr16 | c.10084del | p.Gln3362Serfs*35 (LOH) | 304 | 0.96 | VxPx cargo-targeting to cilium | 0.006 | |||
| B12 | SNX8 | chr7 | c.910C>T | p.Gln304Ter | 66 | 0.44 | 36 | * | |||
| A20 | SPTY2D1 | chr11 | c.2044C>G | p.Leu682Val | 46 | 0.3 | 20.8 | * | * | ||
| B7 | WDFY3 | chr4 | c.2039A>C | p.Gln680Pro | 13 | 0.38 | 16.39 | * | * | ||
| B7 | SON | chr21 | c.331C>A | p.His111Asn | 32 | 0.38 | 23.3 | * | * | ||
| B8 | PRR12 | chr19 | c.5284G>A | p.Gly1762Arg | 86 | 0.37 | 19.09 | * | * | ||
| B9 | LRCH1 | chr13 | c.705G>T | p.Leu235Phe | 65 | 0.31 | 28.5 | * | * | ||
| B10 | TINAGL1 | chr1 | c.961C>T | p.Arg321Cys | 64 | 0.38 | * | * | |||
| CMJ001593 | A30 | PARP14 | chr3 | c.2768C>A | p.Ser923Tyr | 79 | 0.32 | 26 | Nicotinamide salvaging | 0.006 | |
| A30 | ZG16 | chr16 | c.157C>T | p.Arg53Trp | 64 | 0.31 | 30 | * | * | ||
| A30 | ANKK1 | chr11 | c.754C>T | p.Gln252Ter | 52 | 0.31 | 26.8 | * | * | ||
| A31 | CCT4 | chr2 | c.814A>T | p.Met272Leu | 27 | 0.56 | 25.9 | Folding of actin by CCT/TriC | 0.003 | ||
| A31 | PNISR | chr6 | c.1651T>C | p.Ser551Pro | 12 | 0.67 | 22.9 | * | * | ||
| A31 | EVI5 | chr1 | c.1327A>T | p.Ile443Phe | 41 | 0.44 | 25.1 | * | * | ||
| A33 | MVP | chr16 | c.1807G>A | p.Val603Ile | 51 | 0.45 | 26 | * | * | ||
| A33 | CWF19L2 | chr11 | c.1880G>T | p.Gly627Val | 84 | 0.4 | 23 | * | * | ||
| A34 | SMARCAD1 | chr4 | c.2572T>C | p.Phe858Leu | 22 | 0.41 | 23 | * | * | ||
| DR9001565 | A15 | PKD1a | Chr16 | c.9504C>G | p.Phe3168Leu (LOH) | 66 | 0.88 | VxPx cargo-targeting to cilium | 0.006 | ||
| A15 | C1QL2 | chr2 | c.590G>T | p.Cys197Phe | 10 | 0.3 | 31 | * | * | ||
| A16 | THRAP3 | chr1 | c.2701C>T | p.Arg901Ter | 9 | 0.33 | 41 | Transcriptional regulation of white adipocyte differentiation | 0.008 | ||
| A17 | DOCK4 | chr7 | c.4286_4287delAA | p.Lys1429ArgfsTer3 | 124 | 0.39 | Megakaryocyte development and platelet production | 0.03 | |||
| A17 | CASC5 | chr15 | c.4886C>T | p.Thr1629Ile | 29 | 0.38 | 17.62 | Nucleosome assembly | 0.01 | ||
| A18 | PCDHGB6 | chr5 | c.1415C>A | p.Ala472Glu | 14 | 0.3 | 20.8 | * | * | ||
| B3 | ACSS1 | chr20 | c.500G>A | p.Arg167His | 12 | 0.42 | 31 | Ethanol oxidation | 0.001 | ||
| B4 | PKD1 | chr16 | c.6282G>T | p.Trp2094Cys | 9 | 0.33 | 26.8 | VxPx cargo-targeting to cilium | 0.006 | ||
| B4 | KIF17 | chr1 | c.3002G>C | p.Arg1001Pro | 15 | 0.33 | 34 | Intraflagellar transport | COSM182818 | ||
| B5 | PADI4 | chr1 | c.979G>C | p.Ala327Pro | 69 | 0.3 | 23.8 | Chromatin modifying enzymes | 0.04 | rs145819522 (0.15) | |
| B5 | WRNIP1 | chr6 | c.1120G>A | p.Val374Met | 80 | 0.33 | 32 | * | * | ||
| B6 | UBE3C | chr7 | c.1332G>T | p.Arg444Ser | 16 | 0.31 | 23.2 | Antigen processing: ubiquitination and proteasome degradation | 0.03 | ||
| JS9001595 | B26 | LILRB5 | chr19 | c.1652A>C | p.Gln551Pro | 17 | 0.35 | 16.26 | Immunoregulatory interactions: lymphoid and a nonlymphoid cell | 0.03 | |
| B27 | PNPLA6 | chr19 | c.2966+2T>G | 17 | 0.35 | Glycerophospholipid catabolism | 0.002 | ||||
| B27 | ZNF518B | chr4 | c.641A>C | p.Glu214Ala | 45 | 0.96 | 27.7 | * | * | ||
| B27 | MYOM3 | chr1 | c.3751A>C | p.Lys1251Gln | 20 | 0.3 | 23.2 | * | * | ||
| B29 | PCSK5 | chr9 | c.1312G>T | p.Val438Leu | 13 | 0.31 | 26.8 | NGF processing | 0.001 | ||
| B31 | PKD1 | chr16 | c.10220+2T>G | 39 | 0.46 | VxPx cargo-targeting to cilium | 0.008 | ||||
| B31 | KCNC3 | chr19 | c.*81A>C | 16 | 0.44 | Voltage-gated potassium channels | 0.02 | ||||
| B32 | CRCP | chr7 | c.145–1G>T | 19 | 0.32 | RNA polymerase III chain elongation | 0.003 | ||||
| B33 | PKD1 | chr16 | c.3184C>T | p.Gln1062Ter | 27 | 0.63 | 35 | VxPx cargo-targeting to cilium | 0.006 | ||
| B33 | ANGPTL3 | chr1 | c.124G>C | p.Asp42His | 73 | 0.4 | 28.4 | Assembly of active LPL and LIPC lipase complexes | 0.005 | rs199772471 (0.02) | |
| BH9002280 | B34 | PKD2 | chr4 | c.2409delA | p.Ser804ValfsTer40 | 80 | 0.3 | VxPx cargo-targeting to cilium | 0.004 | ||
| B35 | CEP97 | chr3 | c.1142A>T | p.Asp381Val | 33 | 0.3 | 23.2 | Anchoring of the basal body to the plasma membrane | 0.009 | ||
| L2 | PKD2 | chr4 | c.242C>A | p.Ser81Ter | 14 | 0.57 | 36 | VxPx cargo-targeting to cilium | 0.004 | ||
| L5 | PRKCI | chr3 | c.280G>C | p.Glu94Gln | 35 | 0.34 | 24.3 | p75NTR recruits signaling complexes | 0.003 | ||
| L5 | SCAF1 | chr19 | c.619_620insCCCCCCCCC | p.Ser207_Pro208insProProPro | 12 | 0.33 | * | * | |||
| R2 | PKD2 | chr4 | c.1392A>G | p.= | 32 | 0.34 | VxPx cargo-targeting to cilium | 0.004 | |||
| R7 | PKD2 | chr4 | c.1366C>T | p.Gln456Ter | 98 | 0.38 | 42 | VxPx cargo-targeting to cilium | 0.004 | ||
| B36 | NXNL1 | chr19 | c.577C>G | p.Arg193Gly | 9 | 0.44 | 23.2 | * | * | ||
| R8 | YBX3 | chr12 | c.40_42delACC | p.Thr14del | 16 | 0.31 | * | * | |||
| L5 | SCAF1 | chr19 | c.619_620insCCCCCCCCC | p.Ser207_Pro208insProProPro | 12 | 0.33 | * | * | |||
| PJ9001456 | A10 | PKD1a | chr16 | c.7655_7658 delinsTTG | p.Ala2552Valfs*68 | 779 | 0.32 | VxPx cargo-targeting to cilium | 0.006 | ||
| A11 | MIB2 | chr1 | c.2092G>A | p.Glu698Lys | 10 | 0.3 | 22.8 | Signaling by NOTCH1 HD Domain Mutants in Cancer | 0.003 | ||
| A7 | PKD1a | chr16 | c.6364delG | p.Val2122Lysfs*3 (LOH) | 804 | 0.92 | VxPx cargo-targeting to cilium | 0.008 | |||
| A7 | SVIL | chr10 | c.4121T>G | p.Leu1374Arg | 10 | 0.3 | 25.8 | * | * | ||
| A8 | PKD1a | chr16 | c.348_352del | p.Asn116Lysfs*2 | 793 | 0.3 | VxPx cargo-targeting to cilium | 0.006 | |||
| A9 | PKD1 | chr16 | c.8016+1delG | 12 | 0.5 | VxPx cargo-targeting to cilium | 0.006 | ||||
| B1 | SLC14A2 | chr18 | c.2548A>C | p.Asn850His | 19 | 0.32 | 22.2 | Transport -bile salts, organic acids, metal ions, amine compounds | 0.03 | ||
| B1 | URGCP | chr7 | c.173A>C | p.Asn58Thr | 14 | 0.36 | 26.9 | * | * | ||
| B1 | DNAH5 | chr5 | c.5335G>T | p.Val1779Phe | 28 | 0.32 | 27.9 | * | * | ||
| B2 | EIF3A | chr10 | c.330G>T | p.Gln110His | 33 | 0.36 | 23.5 | Formation of the ternary complex, subsequently 43S complex | 0.01 | ||
| B2 | RGS1 | chr1 | c.553G>T | p.Asp185Tyr | 59 | 0.32 | 33 | Gα(i) signaling events | 0.004 | ||
| RP9001591 | B18 | PKD1 | chr16 | c.8996_8997insG | p.Phe2999LeufsTer70 | 64 | 0.33 | VxPx cargo-targeting to cilium | 0.006 | ||
| B18 | ATP2B3 | chrX | c.2866G>A | p.Gly956Arg | 92 | 0.3 | 27 | Reduction of cytosolic Ca++ levels | 0.003 | ||
| B20 | CACNA2D1 | chr7 | c.668G>T | p.Trp223Leu | 12 | 0.42 | 35 | Phase 2 – plateau phase | 0.00 | COSM747982 | |
| B20 | PRRC2B | chr9 | c.1997A>C | p.Gln666Pro | 11 | 0.36 | 25.6 | * | * | ||
| B21 | PKD1a | chr16 | c.G10549T | p.Glu3517* (LOH) | 140 | 0.97 | VxPx cargo-targeting to cilium | 0.006 | |||
| B21 | KIF26A | chr14 | c.5624C>T | p.Pro1875Leu | 75 | 0.39 | 22 | Kinesins | 0.01 | rs200461988 (0.05) | |
| B21 | KISS1R | chr19 | c.828_832delGGGCC | p.Trp276CysfsTer28 | 72 | 0.35 | Peptide ligand-binding receptors | 0.03 | |||
| B22 | SMARCA2 | chr9 | c.667_675delCAGCAGCAG | p.Gln226_Gln228del | 111 | 0.36 | RUNX1 interacts with cofactors whose precise effect on RUNX1 not known | 0.003 | |||
| B23 | STON2 | chr14 | c.1498C>T | p.Arg500Trp | 22 | 0.36 | 29.3 | Cargo recognition for clathrin-mediated endocytosis | 0.0267 | rs149119701 (0.002) | |
| B23 | ATAD2B | chr2 | c.458_465delATGGGGAC | p.Asp153AlafsTer7 | 29 | 0.52 | * | * | |||
| B23 | SELE | chr1 | c.922C>T | p.Arg308Cys | 76 | 0.41 | 23 | * | * | ||
| B24 | KIF17 | chr1 | c.3002G>C | p.Arg1001Pro | 16 | 0.38 | 34 | Intraflagellar transport | 0.01 | COSM182818 | |
| B24 | ABCA13 | chr7 | c.86A>C | p.Glu29Ala | 11 | 0.36 | 31 | * | * | ||
| B24 | PRRC2B | chr9 | c.1997A>C | p.Gln666Pro | 16 | 0.31 | 25.6 | * | * |
The pathogenic potential of missense variants was evaluated using the computational analysis tool CADD, which enables scoring of deleteriousness of SNVs and indels. A CADD value of ≥15 suggests the variant is likely pathogenic (http://cadd.gs.washington.edu/). Significantly mutated WES variants were further filtered using a 30% VAF cutoff and a population allele frequency of 5% (http://exac.broadinstitute.org/). Variants were further subjected for pathway analysis to identify overrepresented pathways using Reactome (https://reactome.org/) and verified with David (https://david.ncifcrf.gov/). Only significantly overrepresented pathways with low P values (<0.05), i.e., the probability that overlap between the queried variant and pathway occurs by chance, were reported. Cysts/variants for which no pathways were identified are indicated by *. Chr, chromosome; HGVSc, the Human Genome Variation Society coding sequence name; HGVSp, the Human Genome Variation Society protein sequence name; rs#/COSMIC, Catalogue Of Somatic Mutations In Cancer; ExAC, The Exome Aggregation Consortium; tRNA, transfer ribonucleic acid.
PKD1 genotyping performed by long-range PCR next-generation sequencing.
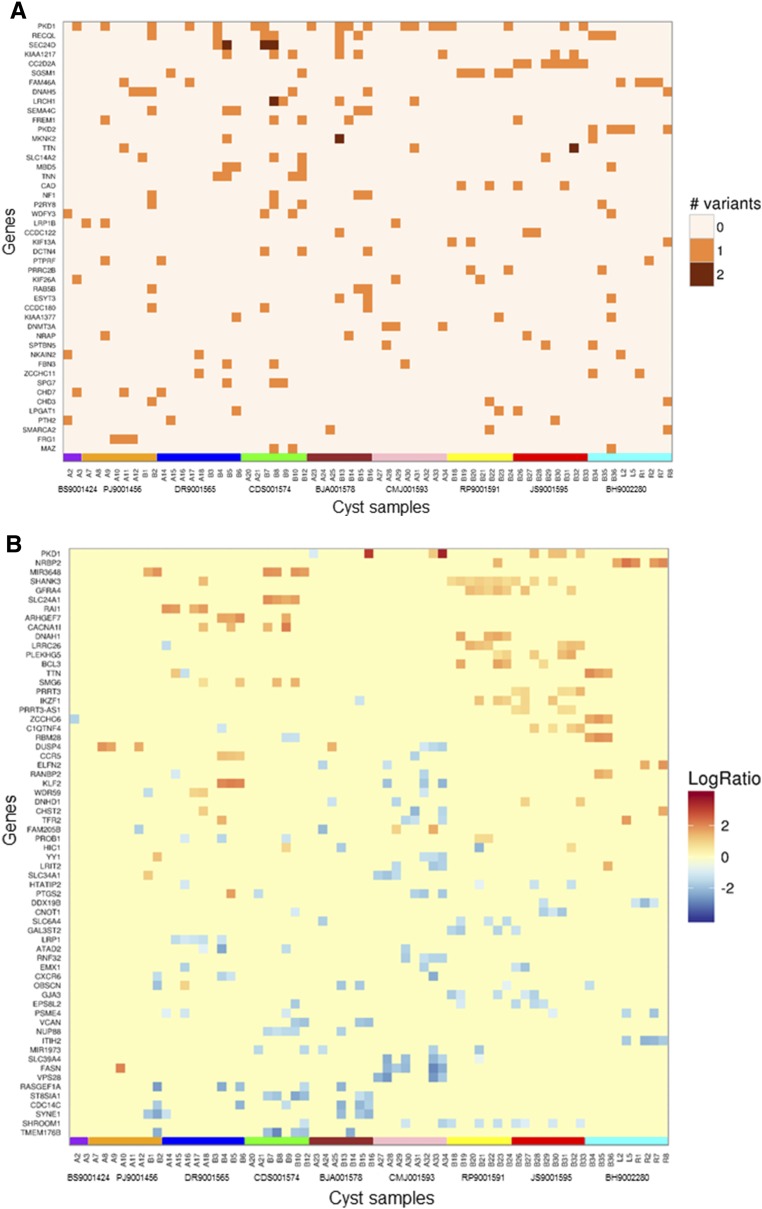
WES analysis of the principal pathogenic or likely pathogenic mutations (n=46) ranked by variant prevalence identified among all cysts and alteration type (i.e., SNV/indel, CNV) in 65 cysts showed that the majority of genes harbored one variation; rare exceptions included PKD1 and SEC24D, in which cysts harboring two variations per gene were found. PKD1 and PKD2 were among the most commonly mutated genes (Figure 4, A and B).
Figure 4.
WES analysis detected significant somatic alterations in commonly mutated genes. WES analysis of the most significantly mutated (pathogenic and likely pathogenic) genes (n=46) ranked by the frequency of all detected (A) SNV/indel and (B) CNV alterations found in the gene among all cysts (n=65). Only genes with three or more SNV/indel and six or more CNV, respectively, are presented. Segments with a log2 ratio (LogR) between renal cyst and PBL (constitutional) read count >0.6 were designated as gains and segments with a log2 value <−0.6 were categorized as losses. Results are grouped by cyst and by patient. Gene data are displayed by patient, indicated by the colored boxes at the bottom of each figure (patients BS9001424, PJ9001456, DR9001565, CDS001574, BJA001578, CMJ001593, RP9001591, JS9001595, and BH9002280).
The average coverage of PKD1 was 32× (61× for PKD1 nonduplicated exons) and 72× for PKD2. Lower mean coverage (<30×) occurred in the duplicated region of PKD1, in PKD1 exons 42–43, and PKD2 exon 1 and exon 8. Mean coverage across all exons in PKD1 and PKD2 was similar for the constitutional and somatic DNA (Supplemental Figure 6). Solely on the basis of WES analysis, PKD1 somatic mutations were identified in only 43% (29 out of 56) of cysts harboring PKD1 constitutional mutations (Supplemental Table 3), significantly below the PKD1 detection rate by LR-PCR based sequencing (91%; Supplemental Table 2). All PKD1 mutations undetected by WES, which were identified by long-range PCR sequencing, were located in its duplicated region, likely reflecting the low coverage by WES in this region. By contrast, WES was 100% sensitive for detecting PKD2 somatic mutations identified by long-range PCR sequencing (Table 2).
We analyzed 211 genes associated with other ciliopathy disorders (Supplemental Table 8) and 565 cancer genes. At least one pathogenic mutation in a ciliopathy or cancer gene was identified in 49% (32 out of 65) and 77% (50 out of 65) of all cysts, respectively. Of the 1477 genes harboring a pathogenic or likely pathogenic SNV or indel mutation, 2.0% (29 out of 1477) were associated with a ciliopathy, whereas 5.6% (83 out of 1477) were cancer-related (Supplemental Tables 9 and 10).
Among the 2456 genes demonstrating CNV changes (Supplemental Table 11), 36 (1.5%) were associated with ciliopathies, whereas 91 (3.7%) were related to cancer. Among the top 64 genes harboring four or more CNV changes (Figure 4B), one (1.6%) was related to ADPKD and cilia dysfunction (PKD1), and four (6.2%) were related to cancer (IKZF1, BCL3, RANBP2, and DUSP4). Copy number changes of PKD1 were observed in 12% of the cysts. PKD1 copy number gains ranging from 151 bp to 28 kb were seen in seven separate cysts obtained from three patients (Supplemental Figures 7, Supplemental Table 12). For the single cyst (patient BJA001578/cystL8) harboring both PKD1/2 copy number changes, the PKD1 amplified region was approximately 20 kb long, spanning exons 5 through 46, whereas the PKD2 amplification was approximately 70 kb long, including the entire gene.
Further computational analysis with MuSiC identified 53 significantly mutated genes (Supplemental Table 13). Of these, 7.5% (four out of 53) were related to ciliopathy disorders (PKD1, PKD2, CC2D2A, and DNAH5) and 9.4% (five out of 53) were related to cancer (RECQL, P2RY8, DNMT3A, SMARCA2, and SMARCAD1). Assessment of constitutional variants in ciliopathy genes identified unique heterozygous pathogenic mutations in CENPF and MYO7A (patient PJ9001456), and CC2D2A and TTC21B (patient CMJ001593) (Supplemental Table 14). No pathogenic constitutional mutation in a non-PKD1/2 ciliopathy gene was identified in BH9002280; this patient had the largest total kidney weight of the cohort (15.7 kg), despite the relatively young age and PKD2 constitutional mutation. Pathway analysis of the most significantly mutated genes with VAF ≥20% by Reactome, on the basis of their biologic function (Table 4), indicated involvement in membrane trafficking, cytoskeleton filament assembly, and chromatin organization.
Table 4.
Significantly altered pathways in cyst epithelia by Reactome (genes with VAF≥0.2)
| Pathway Identifier | Pathway Name | Entities Founda | Entities Totala | Entities P Value | Submitted Entities Found |
|---|---|---|---|---|---|
| R-HSA-390471 | Association of TriC/CCT with target proteins during biosynthesis | 4 | 40 | 0.004 | USP11;KIF13A;TCP1;CCT4 |
| R-HSA-5620920 | Cargo trafficking to the periciliary membrane | 4 | 55 | 0.01 | TCP1;PKD2;PKD1;CCT4 |
| R-HSA-5617833 | Cilium Assembly | 8 | 207 | 0.02 | DYNC2H1;CEP164;TCP1;CEP97;PKD2;KIF17;PKD1;CCT4 |
| R-HSA-1852241 | Organelle biogenesis and maintenance | 11 | 334 | 0.02 | DYNC2H1;NCOR1;CEP164;TCP1;TFAM;CEP97;PKD2;KIF17;PKD1;CCT4 |
| R-HSA-390450 | Folding of actin by CCT/TriC | 2 | 13 | 0.02 | TCP1;CCT4 |
| R-HSA-4839726 | Chromatin organization | 9 | 257 | 0.02 | KDM6B;NCOR1;TRRAP;DNMT3A;EP400;ELP3;PADI4;ARID1A;SMARCA2 |
| R-HSA-3247509 | Chromatin modifying enzymes | 9 | 257 | 0.02 | KDM6B;NCOR1;TRRAP;DNMT3A;EP400;ELP3;PADI4;ARID1A;SMARCA2 |
| R-HSA-5576892 | Phase 0 – rapid depolarization | 3 | 47 | 0.04 | CACNA2D1;SCN4A |
| R-HSA-163282 | Mitochondrial transcription initiation | 1 | 3 | 0.05 | TFAM |
| R-HSA-4085023 | Defective GFPT1 causes CMSTA1 | 1 | 3 | 0.05 | GFPT1 |
| R-HSA-73780 | RNA polymerase III chain elongation | 2 | 22 | 0.05 | CRCP;POLR3E |
| R-HSA-2408550 | Metabolism of ingested H2SeO4 and H2SeO3 into H2Se | 2 | 22 | 0.05 | GSR;PAPSS2 |
| R-HSA-1655829 | Regulation of cholesterol biosynthesis by SREBP (SREBF) | 4 | 86 | 0.05 | INSIG2;SC5D;SEC24D |
| R-HSA-73980 | RNA polymerase III transcription termination | 2 | 23 | 0.05 | CRCP;POLR3E |
| R-HSA-3214858 | RMTs methylate histone arginines | 3 | 53 | 0.05 | DNMT3A;ARID1A;SMARCA2 |
| R-HSA-5620922 | BBSome-mediated cargo-targeting to cilium | 2 | 24 | 0.06 | TCP1;CCT4 |
| R-HSA-5620916 | VxPx cargo-targeting to cilium | 2 | 25 | 0.06 | PKD2;PKD1 |
Pathway analysis was performed by Reactome (https://reactome.org/).
Entities, proteins with an accession sequence in UniPort and Ensemble databases.
Discussion
Inactivating somatic mutations of a PKD gene, which lead to loss of function of the corresponding polycystin, cause clonal expansion of kidney cyst epithelium and cyst formation in ADPKD. However, earlier studies reported a relatively low prevalence (20%–70%) of pathogenic somatic mutations, challenging the primacy of this mechanism in the pathogenesis of ADPKD.3 Methodologic limitations in some of those studies included the required detection of large chromosomal deletions, and in others by the structural complexity of PKD1, particularly in its duplicated region, which limited the detection of smaller, inactivating somatic mutations.21,25–27 In this study, comprehensive genomic analysis of renal cyst epithelium, whereby the exome from individual cysts was screened by WES and long-range PCR and mutations were confirmed by Sanger sequencing, found inactivating somatic mutations of PKD1/2 genes in every patient, encompassing 90% of all cysts. This is consistent with the constitutional mutation detection rate for ADPKD. In our cohort, PKD1 and PKD2 were the most frequently mutated genes in cyst epithelia. The higher somatic mutation detection rate in our study reflects the extensive and sensitive screening methods utilized.4,5,7
A key finding of our study is that approximately 90% of the PKD1/2 gene somatic mutations were truncating in-frame deletions and splicing mutations, implicating them as drivers for cyst development. One example is the rare silent change p.R464R (patient BH9002280), predicted to create a new cryptic donor site and aberrant splicing of PKD2. Splicing defects caused by exonic mutations have been implicated in the pathogenesis in ADPKD.28 No trans-heterozygous mutations of PKD1/2 genes were identified, possibly reflecting the small cohort.
This study was limited to patients in whom constitutional mutations of PKD1/2 genes were identified; as expected, 77% of these were truncating. Altogether, the high prevalence of concurrent truncating constitutional and somatic inactivation of PKD1/2 genes, leading to null/null alleles and kidney cyst development, strongly supports a primary role for the cellular recessive model of cyst formation.29 Although we could not determine variant orientation (i.e., in-cis versus in-trans), previous studies using cloning techniques demonstrated that PKD1/2 constitutional and somatic mutations occurred in-trans.4,5,7,30
PKD1/2 gene haploinsufficiency caused by inadequate gene dosage is an alternative mechanism for ADPKD.3,13,31 In a mouse ortholog of homozygous mutation of Pkd1 alleles in which aberrant splicing of Pkd1 yielded an ADPKD phenotype, 13%–20% of the homozygotes had normally spliced Pkd1 transcripts. The few heterozygotes that survived more than 1 year had Pkd1 expression levels approximately 40%–50% of wild type, suggesting that modestly reduced levels of PC1 sufficiently maintained renal function and prolonged survival.11 Patients with ADPKD who were homozygous or compound heterozygous for hypomorphic alleles reportedly had severe disease phenotypes, whereas milder disease occurred in heterozygotes with hypomorphic alleles.12 Although missense mutations can provide sufficient gene activity to attenuate cyst growth3,32 no PKD1/2 missense/missense allele combination was identified in our study other than LOH in two cysts with likely pathogenic constitutional mutations (patients BS9001424 and DR9001565). Although we did not measure PC1 levels, the vast majority of somatic mutations were truncating, and thus had negligible PKD1/2 gene expression.
PKD gene overexpression in orthologous models can cause kidney cyst development.33 In our study, numerous cysts from three patients had PKD1/2 copy number gains. Although large constitutional rearrangements in PKD1/2 have previously been described,34 to our knowledge, this is the first report of somatic copy number gains in PKD1/2. Interestingly, amplification of both PKD1/2 genes occurred in one patient (BJA001578, cyst L8) with a somatic mutation in PKD1. The significance of these findings is unclear. One consideration is that PKD gene amplification can contribute to cyst development, although our data are insufficient to support this conclusion.
In this study, utilizing WES, we found a median of 65 somatic variations per cyst (0.022 mutations per megabase) and a transition/transversion ratio of 0.35, supporting the preference of transversion mutations. The quantity and ratio of these variations were overall lower than values reported for various cancers.35–37 Although common mechanisms can promote cell proliferation in ADPKD and in cancer, studies associating ADPKD with cancer are conflicting because of methodologic limitations. Their interpretations are confounded by the generally increased risk of renal cell cancer in CKD and the paucity of prospective, adequately controlled clinical trials evaluating cancer in ADPKD.38,39 For example, the report of a lower risk of cancer in patients with or without ADPKD after kidney transplantation may have been confounded by the higher rate of malignancy identified before transplantation in ADPKD as a consequence of more frequent radiographic evaluation of complex kidney cysts, clinical assessment of various symptoms (e.g., gross hematuria, pain), and nephrectomy that may have alerted clinicians to previously undetected malignancies.40 These findings have not been generalized to patients with ADPKD who have not undergone kidney transplantation.35–37
We determined that approximately 5% of the most prevalent mutated genes are designated as cancer genes, including a regulators of gene methylation (i.e., DNMT3A) and chromatin remodeling (i.e., SMARCA2). However, we found no association of mutation rate and disease severity. We detected an association of mutation count with older age, consistent with another report showing that the frequency of Phosphoribosyltransferase (HRPT) gene mutations in 6-thioguanine (TG)-resistant renal cortical epithelial cells from donors increase exponentially with donor age.41 The oldest patient in our cohort, who had the highest mutation load and a constitutional mutation of ATM, which is a cell-cycle checkpoint kinase that regulates various downstream proteins (e.g., p53, BRCA1), also had an incidental finding of papillary renal cell carcinoma caused by a somatic mutation in MET.22 However, a somatic mutation of PKD1 was not identified in the tumor cells, and a somatic MET mutation was not identified in other, nonmalignant renal cyst epithelium in this patient. Although our findings do not suggest a common mechanism for ADPKD and renal cell cancer, this study was not designed to address this issue.
We identified pathogenic somatic mutations in genes associated with other ciliopathies, (i.e., CC2D2A, DNAH5), which were among the most prevalent in this cohort, occurring in approximately 30% of all cysts. Mutations in CC2D2A cause Meckel syndrome and Joubert syndrome, both associated with kidney cysts, whereas mutations in DNAH5 account for primary ciliary dyskinesia, an autosomal recessive disorder involving abnormalities of motile cilia.
Our study was not designed to assess the potential role of variants in non-PKD1/2 genes in the pathogenesis of ADPKD. However, the finding of at least one pathogenic mutation of a non-PKD gene in all the evaluable cysts suggests that these variants might influence cyst development. It is well established that cyst generation involves activation of proliferative signaling pathways.9 Therefore, it is reasonable to hypothesize that the majority of non-PKD1/2 somatic mutations identified herein occurred later in the rapidly dividing population of cyst epithelium, providing a proliferative advantage.
The strengths of this study include its prospective design, well characterized patient cohort, sensitive methods for detecting constitutional and somatic mutations in PKD1/2 genes, and confirmatory Sanger sequencing of PKD1/2 gene mutations detected by WES. To our knowledge, this is the most comprehensive study of WES in the renal epithelium of patients with ADPKD. Limitations of the study include low sensitivity of the WES method to detect mutations in the duplicated region of PKD1, insufficient cyst DNA material to perform long-range PCR studies of the PKD1 WES-negative cysts, and lack of functional studies to evaluate the significance of non-PKD1/2 somatic mutations. Additionally, we cannot exclude cell heterogeneity; specifically, the inclusion of other cell types, leading to an underestimation of the prevalence of these mutations.
In summary, this comprehensive genomic analysis of somatic mutations in cyst epithelial cells of patients with ADPKD strongly supports the primacy of the “second hit” model in the pathogenesis of ADPKD, caused by inactivating constitutional and somatic mutation of the PKD1/2 alleles. This study also characterized cyst-specific somatic variants in non-PKD1/2 genes, which may influence cyst formation and disease severity in patients with ADPKD.
Disclosures
None.
Supplementary Material
Acknowledgments
We are grateful to all patients and their families for their invaluable participation. We thank Moshe Bitton and Andrew Ramnauth for their assistance with data mining and Dr. Peter Harris for critical review of this manuscript.
H.R., J.B., and A.Y.T. designed the study, wrote the manuscript and developed and/or performed computational analyses. A.M., W.Z., G.L., Y.Z., J.X., and Z.Z. planned or performed laboratory experiments. T.Z. and C.M. performed computational analyses. L.R. performed statistical analyses. B.D.R. and S.P.S. performed pathological assessment of the kidneys. S.D. and W.O.B. assisted with patients’ recruitment, S.K. performed the kidney transplantations. Financial support provided by The Starr Foundation.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2017080878/-/DCSupplemental.
References
- 1.Chapman AB, Devuyst O, Eckardt KU, Gansevoort RT, Harris T, Horie S, et al. : Autosomal-dominant polycystic kidney disease (ADPKD): Executive summary from a kidney disease: Improving global outcomes (KDIGO) controversies conference. Kidney Int 88: 17–27, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Porath B, Gainullin VG, Cornec-Le Gall E, Dillinger EK, Heyer CM, Hopp K, et al.; Genkyst Study Group, HALT Progression of Polycystic Kidney Disease Group; Consortium for Radiologic Imaging Studies of Polycystic Kidney Disease : Mutations in GANAB, encoding the glucosidase IIα subunit, cause autosomal-dominant polycystic kidney and liver disease. Am J Hum Genet 98: 1193–1207, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris PC: What is the role of somatic mutation in autosomal dominant polycystic kidney disease? J Am Soc Nephrol 21: 1073–1076, 2010 [DOI] [PubMed] [Google Scholar]
- 4.Watnick TJ, Torres VE, Gandolph MA, Qian F, Onuchic LF, Klinger KW, et al.: Somatic mutation in individual liver cysts supports a two-hit model of cystogenesis in autosomal dominant polycystic kidney disease. Mol Cell 2: 247–251, 1998 [DOI] [PubMed] [Google Scholar]
- 5.Pei Y, Watnick T, He N, Wang K, Liang Y, Parfrey P, et al.: Somatic PKD2 mutations in individual kidney and liver cysts support a “two-hit” model of cystogenesis in type 2 autosomal dominant polycystic kidney disease. J Am Soc Nephrol 10: 1524–1529, 1999 [DOI] [PubMed] [Google Scholar]
- 6.Brasier JL, Henske EP: Loss of the polycystic kidney disease (PKD1) region of chromosome 16p13 in renal cyst cells supports a loss-of-function model for cyst pathogenesis. J Clin Invest 99: 194–199, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watnick T, He N, Wang K, Liang Y, Parfrey P, Hefferton D, et al.: Mutations of PKD1 in ADPKD2 cysts suggest a pathogenic effect of trans-heterozygous mutations. Nat Genet 25: 143–144, 2000 [DOI] [PubMed] [Google Scholar]
- 8.Piontek K, Menezes LF, Garcia-Gonzalez MA, Huso DL, Germino GG: A critical developmental switch defines the kinetics of kidney cyst formation after loss of Pkd1. Nat Med 13: 1490–1495, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seeger-Nukpezah T, Geynisman DM, Nikonova AS, Benzing T, Golemis EA: The hallmarks of cancer: Relevance to the pathogenesis of polycystic kidney disease. Nat Rev Nephrol 11: 515–534, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grantham JJ: Time to treat polycystic kidney diseases like the neoplastic disorders that they are. Kidney Int 57: 339–340, 2000 [DOI] [PubMed] [Google Scholar]
- 11.Lantinga-van Leeuwen IS, Dauwerse JG, Baelde HJ, Leonhard WN, van de Wal A, Ward CJ, et al.: Lowering of Pkd1 expression is sufficient to cause polycystic kidney disease. Hum Mol Genet 13: 3069–3077, 2004 [DOI] [PubMed] [Google Scholar]
- 12.Rossetti S, Kubly VJ, Consugar MB, Hopp K, Roy S, Horsley SW, et al.: Incompletely penetrant PKD1 alleles suggest a role for gene dosage in cyst initiation in polycystic kidney disease. Kidney Int 75: 848–855, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pei Y, Lan Z, Wang K, Garcia-Gonzalez M, He N, Dicks E, et al.: A missense mutation in PKD1 attenuates the severity of renal disease. Kidney Int 81: 412–417, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vujic M, Heyer CM, Ars E, Hopp K, Markoff A, Orndal C, et al.: Incompletely penetrant PKD1 alleles mimic the renal manifestations of ARPKD. J Am Soc Nephrol 21: 1097–1102, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rossetti S, Harris PC: Genotype-phenotype correlations in autosomal dominant and autosomal recessive polycystic kidney disease. J Am Soc Nephrol 18: 1374–1380, 2007 [DOI] [PubMed] [Google Scholar]
- 16.Ma M, Tian X, Igarashi P, Pazour GJ, Somlo S: Loss of cilia suppresses cyst growth in genetic models of autosomal dominant polycystic kidney disease. Nat Genet 45: 1004–1012, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmeister H, Babinger K, Gürster S, Cedzich A, Meese C, Schadendorf K, et al.: Polycystin-2 takes different routes to the somatic and ciliary plasma membrane. J Cell Biol 192: 631–645, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hildebrandt F, Otto E: Cilia and centrosomes: A unifying pathogenic concept for cystic kidney disease? Nat Rev Genet 6: 928–940, 2005 [DOI] [PubMed] [Google Scholar]
- 19.Loghman-Adham M, Nauli SM, Soto CE, Kariuki B, Zhou J: Immortalized epithelial cells from human autosomal dominant polycystic kidney cysts. Am J Physiol Renal Physiol 285: F397–F412, 2003 [DOI] [PubMed] [Google Scholar]
- 20.Tan YC, Blumenfeld JD, Anghel R, Donahue S, Belenkaya R, Balina M, et al.: Novel method for genomic analysis of PKD1 and PKD2 mutations in autosomal dominant polycystic kidney disease. Hum Mutat 30: 264–273, 2009 [DOI] [PubMed] [Google Scholar]
- 21.Tan AY, Michaeel A, Liu G, Elemento O, Blumenfeld J, Donahue S, et al.: Molecular diagnosis of autosomal dominant polycystic kidney disease using next-generation sequencing. J Mol Diagn 16: 216–228, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang W, Tan AY, Blumenfeld J, Liu G, Michaeel A, Zhang T, et al.: Papillary renal cell carcinoma with a somatic mutation in MET in a patient with autosomal dominant polycystic kidney disease. Cancer Genet 209: 11–20, 2016 [DOI] [PubMed] [Google Scholar]
- 23.Rennert H, Eng K, Zhang T, Tan A, Xiang J, Romanel A, et al. : Development and validation of a whole-exome sequencing test for simultaneous detection of point mutations, indels and copy-number alterations for precision cancer care. NPJ Genom Med 1: 16019, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J: A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 46: 310–315, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan YC, Michaeel A, Blumenfeld J, Donahue S, Parker T, Levine D, et al.: A novel long-range PCR sequencing method for genetic analysis of the entire PKD1 gene. J Mol Diagn 14: 305–313, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Audrézet MP, Cornec-Le Gall E, Chen JM, Redon S, Quéré I, Creff J, et al.: Autosomal dominant polycystic kidney disease: Comprehensive mutation analysis of PKD1 and PKD2 in 700 unrelated patients. Hum Mutat 33: 1239–1250, 2012 [DOI] [PubMed] [Google Scholar]
- 27.Rossetti S, Consugar MB, Chapman AB, Torres VE, Guay-Woodford LM, Grantham JJ, et al.; CRISP Consortium : Comprehensive molecular diagnostics in autosomal dominant polycystic kidney disease. J Am Soc Nephrol 18: 2143–2160, 2007 [DOI] [PubMed] [Google Scholar]
- 28.Claverie-Martin F, Gonzalez-Paredes FJ, Ramos-Trujillo E: Splicing defects caused by exonic mutations in PKD1 as a new mechanism of pathogenesis in autosomal dominant polycystic kidney disease. RNA Biol 12: 369–374, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fedeles SV, Gallagher AR, Somlo S: Polycystin-1: A master regulator of intersecting cystic pathways. Trends Mol Med 20: 251–260, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qian F, Watnick TJ, Onuchic LF, Germino GG: The molecular basis of focal cyst formation in human autosomal dominant polycystic kidney disease type I. Cell 87: 979–987, 1996 [DOI] [PubMed] [Google Scholar]
- 31.Pei Y, Paterson AD, Wang KR, He N, Hefferton D, Watnick T, et al.: Bilineal disease and trans-heterozygotes in autosomal dominant polycystic kidney disease. Am J Hum Genet 68: 355–363, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gallagher AR, Germino GG, Somlo S: Molecular advances in autosomal dominant polycystic kidney disease. Adv Chronic Kidney Dis 17: 118–130, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burtey S, Riera M, Ribe E, Pennekamp P, Passage E, Rance R, et al.: Overexpression of PKD2 in the mouse is associated with renal tubulopathy. Nephrol Dial Transplant 23: 1157–1165, 2008 [DOI] [PubMed] [Google Scholar]
- 34.Gogusev J, Murakami I, Doussau M, Telvi L, Stojkoski A, Lesavre P, et al.: Molecular cytogenetic aberrations in autosomal dominant polycystic kidney disease tissue. J Am Soc Nephrol 14: 359–366, 2003 [DOI] [PubMed] [Google Scholar]
- 35.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al.: Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499: 214–218, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al.; Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain : Signatures of mutational processes in human cancer. Nature 500: 415–421, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW: Cancer genome landscapes. Science 339: 1546–1558, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang W, Flemington EK, Zhang K: Mutant TP53 disrupts age-related accumulation patterns of somatic mutations in multiple cancer types. Cancer Genet 209: 376–380, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woldu SL, Weinberg AC, RoyChoudhury A, Chase H, Kalloo SD, McKiernan JM, et al.: Renal insufficiency is associated with an increased risk of papillary renal cell carcinoma histology. Int Urol Nephrol 46: 2127–2132, 2014 [DOI] [PubMed] [Google Scholar]
- 40.Wetmore JB, Calvet JP, Yu AS, Lynch CF, Wang CJ, Kasiske BL, et al.: Polycystic kidney disease and cancer after renal transplantation. J Am Soc Nephrol 25: 2335–2341, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin GM, Ogburn CE, Colgin LM, Gown AM, Edland SD, Monnat RJ Jr: Somatic mutations are frequent and increase with age in human kidney epithelial cells. Hum Mol Genet 5: 215–221, 1996 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.