Abstract
Amyloids are associated with many neurodegenerative diseases, motivating investigations into their structure and function. Although not linked to a specific disease, albumins have been reported to form many structural aggregates. We were interested in investigating host immune responses to amyloid fibrils assembled from the model protein ovalbumin. Surprisingly, upon subjecting ovalbumin to standard denaturing conditions, we encountered giant protein nanosheets harboring amyloid-like features and hypothesized that these nanosheets might have potential in clinical or therapeutic applications. We found that the nanosheets, without the administration of any additional adjuvant, evoked a strong antibody response in mice that was higher than that observed for native ovalbumin. This suggests that amyloid nanosheets have a self-adjuvanting property. The nanosheet-induced immune response was helper T cell 2 (Th2) biased and negligibly inflammatory. While testing whether the nanosheets might form depots for the sustained release of precursor proteins, we did observe release of ovalbumin that mimicked the conformation of native protein. Moreover, the nanosheets could load the anticancer drug doxorubicin and release it in a slow and sustained manner. Taken together, our results suggest that amyloid nanosheets should be further investigated as either an antigen delivery vehicle or a multifunctional antigen and drug co-delivery system, with potential applications in simultaneous immunotherapy and chemotherapy.
Keywords: amyloid, albumin, nanotechnology, drug delivery, drug delivery system, antigen, antigen delivery, nanosheet, ovalbumin
Introduction
Self-assembly of proteins into amyloid fibrils, the highly ordered nanostructures harboring cross β-cores, has been associated with various neurodegenerative disorders (1–3). On the contrary, amyloids have been reported to perform normal biological functions, including hormone storage and memory persistence (4, 5). Owing to high thermal stability, stiffness, biocompatibility, and controllable self-assembly (6, 7), amyloids are emerging as novel class of biological nanomaterials. They have found applications in the areas of food science, materials science, medicine, and electronics (8–12). Amyloid fibrils have also been hybridized with graphene and DNA origami to develop hybrid biomaterials (7, 13–15). Moreover, amyloid-based supramolecular assemblies are gaining attention as immune adjuvant and antigen delivery systems because of their potential to release the precursor protein/peptide and resistance to proteolytic attack that extends their residence time in circulation (16). This feature allows them to boost the immune system time and again in the absence of external boosters.
Under specific denaturation conditions, ovalbumin has been reported to form aggregates in vitro (8). A decrease in α-helical content and binding of thioflavin T or Congo red to the albumin aggregates suggest formation of β-sheet–type structures (17). Albumin aggregates of diverse morphologies have been reported, and interestingly, the morphologies obtained seem to have direct correlation with the kind of denaturing environment provided to the protein. For example, human serum albumin (the human counterpart of ovalbumin) has been observed to form both fibrillar and nonfibrillar aggregates under various environmental conditions (17). Like fibrillar aggregates, some of its nonfibrillar aggregates (displaying a certain filamentous structure but not the conventional fibril form) have been found to possess a typical negative peak in CD spectra at wavelengths close to 220 nm and bind Congo red. The different precursor states that result from different solution conditions can generate fibrillar or nonfibrillar aggregates with similar β-sheet content but with distinct morphological features. For human serum albumin, the flat ribbons observed are very much different from cylindrical forms of amyloid fibrils and relatively thick rodlike specimens, although each one of them can bind to the β-sheet–binding dye Congo red and exhibits a minimum at 220 nm for CD spectra (17).
One of the denaturing conditions provided to trigger amyloid nucleation is agitation of the protein solutions (18, 19). Once the amyloidal seeds are formed, the aggregates mature rapidly without any detectable lag phase during the incubation period. Agitation of proteins has been reported to form amyloids of very distinct morphologies (19). In the present study, we agitated ovalbumin to induce formation of amyloid fibrils. However, to our surprise, the aggregate obtained upon shaking for 24 h exhibited a nanosheet-like morphology instead of the fibrillar form. This is in analogy with a recent report where shaken but not stirred peptoid monolayer was found to form free-floating, stable nanosheets (20). Because nanosheets were formed in an amyloid-inducing ambience, we investigated their amyloidal character. The various biophysical studies ascertained the amyloid character of nanosheets. Further, the nanosheets were tested for release of the precursor proteins and for whether the released precursors carried the native antigen conformation. The amyloid nanosheets were evaluated for their potential to evoke immune response. The antibodies evoked upon nanosheet immunization were tested for cross-reactivity with native antigen. We anticipated that the availability of both obverse and reverse surfaces in nanosheets may facilitate high drug loading. Therefore, we examined whether the anticancer drug doxorubicin can be loaded onto nanosheets and later be released. Next, we tested whether the amyloid nanosheet formation is a characteristic of only very amyloidogenic proteins like albumins or amyloid nanosheets can be formed by other proteins/peptides, especially cell-surface antigens, as they are the potential targets of immunotherapy. Because they are whole protein bodies, the nanosheets can themselves act as antigen delivery vehicles. Moreover, their free obverse and reverse surfaces can provide ample area for additional drug loading. Hence, these nanosheets may emerge as a novel class of biomaterials with applications in simultaneous immunotherapy and chemotherapy. To the best of our knowledge, this report is the first study to elucidate the discovery of amyloid-like nanosheets for ovalbumin and also illuminates the immune responses evoked upon immunizing amyloid nanosheets.
Results
Continuous shaking of ovalbumin at neutral pH forms amyloid nanosheets
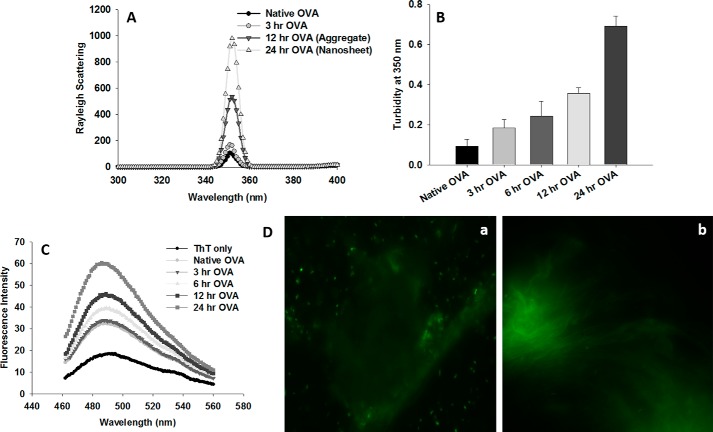
Fluorescence intensity was monitored to detect aggregation in agitated ovalbumin. A marked increase in fluorescence intensity was observed in the ovalbumin samples kept on agitation (Fig. 1A) as compared with native ovalbumin. The 12- and 24-h aggregates exhibited ∼5- and 10-fold increases in fluorescence intensity, respectively, as compared with the native form. The 3-h ovalbumin exhibited only a slight increase in fluorescence intensity compared with the native form, so the chances of aggregation at this state could be ruled out. The turbidity measurements for protein samples agitated for more than 10 h also revealed markedly enhanced turbidity compared with native ovalbumin, thus providing further evidence in support of aggregate formation (Fig. 1B).
Figure 1.
Continuous shaking induces amyloid nanosheet formation. A, Rayleigh scattering measurements at 350 nm for various aggregates. B, turbidity measurements of various aggregates at 350 nm. Error bars, S.E. of three independent experiments. C, thioflavin T–binding assay of various aggregates as analyzed by fluorescence spectroscopy. Thioflavin T emission spectra were obtained by excitation at 450 nm and emission in the range of 460–560 nm. D, thioflavin T binding to ovalbumin nanosheets (i.e. 24-h aggregate (100 μg)) as visualized by fluorescence microscopy. a, aggregate displaying a nanosheet-like structure with seemingly numerous amyloidal seeds in the vicinity. b, a mature nanosheet with numerous folds and crumples. Thioflavin T (30 μm) was incubated with ovalbumin nanosheets for 30 min. Further, 5 μl of the suspension was placed onto the glass slide and observed under a fluorescence microscope. OVA, ovalbumin; ThT, thioflavin T. Data are representative of at least three independent experiments with similar observations.
Thioflavin T is a benzothiazole dye that exhibits enhanced fluorescence emissions upon binding to amyloid fibrils and does not bind to amorphous aggregates as well as nonamyloid structures (21, 22). Thus, a thioflavin T binding assay was performed to examine the amyloid character of aggregates formed by shaking ovalbumin. The agitated protein samples exhibited strong thioflavin T binding, as revealed by markedly enhanced thioflavin T fluorescence compared with unshaken native ovalbumin (Fig. 1C). The results indicate the formation of β-sheet–rich ordered aggregates. Fluorescence microscopy was also exploited to observe thioflavin T binding. The ovalbumin aggregate obtained at 24 h of shaking was incubated with thioflavin T and monitored for thioflavin T fluorescence by fluorescence microscopy. The aggregate exhibits a nanosheet-like structure with seemingly numerous amyloidal seeds in the vicinity (Fig. 1D (a)). Fig. 1D (b) shows a mature nanosheet with numerous folds and crumples.
To monitor the formation of amyloid aggregates, Congo red dye is diagnostically used. Amyloid binding to Congo red causes a red shift in the absorbance spectrum of the dye (23). Aliquots of agitated ovalbumin collected at various time points were mixed with Congo red, and spectra were obtained. Absorbance intensity was found to increase with increase in agitation time; however, a red shift started to appear only from the aggregate formed at 12 h of agitation. Interestingly, a marked increase in absorption and significant red shift was observed for aggregate formed at 24 h, indicating β-sheet–rich amyloid formation (Fig. 2A). Aggregates obtained upon agitation for more than 24 h exhibited negligible change in thioflavin T binding as compared with that observed for 24-h aggregates (data not shown). This indicates that 24 h is the saturation point for nanosheet formation, and shaking beyond this time does not produce any more sheets.
Figure 2.
Ovalbumin forms giant β-sheet–rich amyloid nanosheets. A, absorption spectra of Congo red with various aggregates. The aggregates were scanned in the 400–700-nm range. B, secondary structure determination of nanosheets by UV CD. Native ovalbumin and nanosheet (200 μg/ml) were scanned in the range of 200–250 nm. Data are representative of at least three independent experiments with similar observations. C, nanosheet formation as seen by transmission EM. a, 6-h ovalbumin; b, 12-h ovalbumin; c and d, 24-h ovalbumin (nanosheet). Various aggregates (100 μg) were coated onto the grid. OVA, ovalbumin; CR, Congo red. Images shown are those selected after similar observation of at least two independent experiments performed for each sample.
Once thioflavin T and Congo red binding to the aggregates was ascertained, we also determined secondary structure content of the aggregates by far-UV CD. Native ovalbumin was observed to show double minima at 208 and 222 nm, which is a hallmark of α-helical forms. On the contrary, 24-h aggregates exhibited a single negative peak at around 220 nm, suggesting their transition to the β-sheet conformation (Fig. 2B). The morphology of aggregates formed was further investigated using transmission EM. The 6- and 12-h aggregates did not reveal any typical amyloid morphology (Fig. 2C, a and b); however, the aggregates obtained at 24 h exhibited a unique sheetlike morphology (Fig. 2C, c and d). The sheets are seen to be crumpled and have numerous folds.
We also calculated nanosheet yield in the 24-h aggregates. Nanosheet yield was found to be 67.6%. This indicates that 67.6% of the monomeric ovalbumin converted to nanosheets, and the rest made the unpolymerized ovalbumin pool.
Nanosheets release precursor proteins having native antigen conformation
In our previous study (16), we suggested exploitation of amyloids for vaccine development, considering their property to act as depots for release of precursor proteins and resistance to enzymatic degradation that extends their time in circulation. We wondered whether the nanosheets also release precursor proteins like those observed for fibrils; therefore, we evaluated release of precursor ovalbumin from amyloid nanosheet over a period of 15 days. Interestingly, the ovalbumin nanosheets were found to release the precursor proteins in a time-dependent manner and were found to be superior depots for the protein release as compared with the fibrillar form of ovalbumin (Fig. 3A). This might have happened because the sheets may be formed of laterally assembled fibrils, and several fibrils may be releasing the precursors from their termini in the nanosheets. The fibril form of ovalbumin released ovalbumin precursors in a sustained manner over a period of 15 days; however, the amount released was lower than that observed for the nanosheets.
Figure 3.
Nanosheets release precursor proteins over an extended time period, and the released precursors bear native ovalbumin properties. A, in vitro release kinetics of ovalbumin from amyloid nanosheets over a period of 15 days as determined by taking absorbance at 280 nm. B, size-exclusion chromatography profile of released ovalbumin after 7 days, exhibiting release of monomers, dimers, and oligomers, with monomers being the predominant species. C, light scattering by ovalbumin precursors released from nanosheets at 350 nm. D, thioflavin T–binding assay of ovalbumin released from nanosheets. E, secondary structure determination of released precursors by far-UV CD. F, tertiary structure determination of released precursors by near-UV CD. OVA-R (Fibril) and OVA-R (Nanosheet), ovalbumin released from fibril and nanosheet, respectively. Error bars, S.E. of three independent experiments. At least three independent experiments were carried out for each sample, and data obtained with similar results are presented.
To analyze the state of released precursors from the nanosheet depot, we performed size-exclusion chromatography of the released proteins. Both fibrillar and nanosheet forms of ovalbumin could be seen to release monomers, dimers, and higher-order oligomers (Fig. 3B). However, the dominant species that is released is the monomeric form. It may be possible that oligomeric forms were first released that subsequently released monomeric forms, but we did not get into these details, as our concern was whether the precursor proteins are released or not. The released precursor proteins, whether in monomeric/dimeric/multimeric forms, may serve the purpose of boosting the immune system repeatedly.
We believe that similar to existence of fibril-monomer equilibrium in conditions of fibril depolymerization, a dynamic equilibrium between the released monomers and nanosheets is also established. Concomitantly, we characterized the nanosheet dissociation equilibrium by calculating t½, Kd, and Koff of the nanosheets. The t½, Kd, and Koff of nanosheets are found to be 50 days, 3.3 μm, and 1.64 × 10−7 s−1, respectively (Table 1). The details of these are provided under “Materials and methods.”
Table 1.
Properties of ovalbumin aggregates
| Ovalbumin aggregate | Kd | t½ | Koff |
|---|---|---|---|
| μm | days | s−1 | |
| Fibril | 1.6 | 106 | 7.5 × 10−8 |
| Nanosheet | 3.3 | 50 | 1.6 × 10−7 |
Next, we studied the nature of the precursor proteins released from nanosheets. Intriguingly, as is observed for fibrillar amyloids, the released precursors from amyloid nanosheets mimicked native ovalbumin for light scattering and thioflavin T binding (Fig. 3, C and D). Moreover, the CD spectrum of released precursors was found to be similar to that obtained for native ovalbumin (Fig. 3, E and F). These results indicate that the released ovalbumin from the nanosheet depot may be refolding to native conformation. It is interesting to observe that the near-UV CD spectrum of released monomers, which indicates the tertiary structure of proteins (24), is quite similar to that obtained for native ovalbumin. This demonstrates that not only are the secondary structures conserved (as observed from the far-UV CD spectrum), but so is the tertiary conformation of the released monomers. It has been observed previously that there is a marked difference in the near-UV CD spectrum of disulfide-reduced and disulfide-intact (native) ovalbumin (25). Reckoning with this, the native-like near-UV CD spectrum of released ovalbumin monomers, in the present study, can be said to suggest conservation of native conformation along with the native disulfide bond.
Immunization with nanosheets induces high antibody titers against native antigen
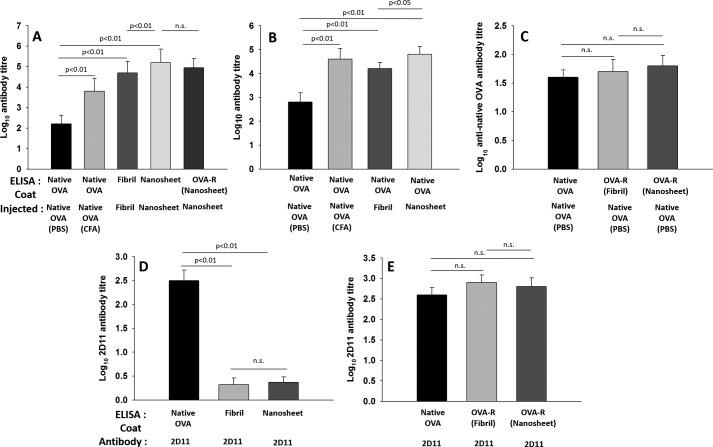
BALB/c mice were immunized with ovalbumin nanosheets, and the immune response evoked was evaluated. Intriguingly, nanosheets were found to induce significantly higher antibody titers compared with that induced by native ovalbumin (Fig. 4A). The antibody titers induced by nanosheets were even higher than those induced by native ovalbumin given in complete Freund's adjuvant. Of note, the strong antibody responses evoked against nanosheets are without co-administration of any additional adjuvant. As the native form of ovalbumin did not produce a strong antibody response, the self-assembly of ovalbumin to amyloid nanosheet structure can be said to be responsible for the robust antibody responses. This indicates that amyloid nanosheets have a self-adjuvanting property as well.
Figure 4.
A, nanosheet immunization induced strong antibody titers against nanosheets and ovalbumin released from nanosheets. ELISA plates were coated with native ovalbumin, fibril, and nanosheet and allowed to react with antisera obtained from animals immunized with the same. Wells coated with ovalbumin released from sheets were probed with anti-nanosheet antisera. B, nanosheet antiserum is cross-reactive to native ovalbumin. ELISA plates were coated with native ovalbumin and allowed to react with antisera obtained from animals immunized with native ovalbumin (both in PBS and complete Freund's adjuvant), fibril, and nanosheet. C, ovalbumin released from nanosheets is reactive to native ovalbumin-specific polyclonal antibodies. ELISA plates were coated with native ovalbumin and ovalbumin released from fibrils and nanosheets and allowed to react with antisera obtained upon native ovalbumin immunization. D, 2D11 anti-ovalbumin mAb recognizes native ovalbumin but not aggregated ovalbumin. ELISA plates were coated with native ovalbumin, fibrils, and nanosheets and allowed to react with 2D11 mAb. E, ovalbumin precursors released from nanosheets are reactive to 2D11 antibody. ELISA plates were coated with native ovalbumin and ovalbumin released from fibrils and nanosheets and allowed to react with 2D11 antibody. Results are presented as the mean of three independent experiments, with error bars representing S.E. of three independent experiments. n.s., not significant. OVA-R (Fibril) and OVA-R (Nanosheet), ovalbumin released from ovalbumin fibril and nanosheet, respectively.
Nanosheet antisera were also found to react with ovalbumin monomers released from nanosheets. There was a practically insignificant difference between the reactivity of anti-nanosheet antibodies with intact nanosheets and released monomers. This indicates that both the multivalency of the sheets and released precursors might be contributing to the overall immunogenicity of the nanosheets. Our purpose of exploiting the nanosheet as a vaccine candidate is fulfilled whether the host system favors multivalency of the sheets or the released precursors for generating immune response. Whatever the source of immunogenicity, as long as nanosheets are able to evoke immune response against native antigen, the purpose of exploiting them as vaccine candidates is accomplished.
The cross-reactivity of the nanosheet antisera with native antigen was also tested. The nanosheet antisera were found to be strongly cross-reactive with native ovalbumin (Fig. 4B), indicating that nanosheet immunization is capable of eliciting high antibody titers against native ovalbumin, by virtue of retaining native antigen epitopes and/or releasing precursors carrying native epitopes.
Precursors released from nanosheets are reactive to both polyclonal and monoclonal anti-native ovalbumin antibodies
Precursors released from nanosheets were analyzed for reactivity with anti-native ovalbumin antibodies. Interestingly, the released precursors from nanosheets were found to react with anti-native ovalbumin polyclonal antibodies (Fig. 4C). This indicates that the released precursors might be harboring native antigen epitopes. To test the conformational integrity of released precursors, we tested the reactivity of released ovalbumin with 2D11 mAb. This antibody reacts with native ovalbumin but not aggregated forms (Fig. 4D), which indicates that it recognizes a conformational epitope. The precursor ovalbumin released from nanosheets was found to be reactive to 2D11 antibody (Fig. 4E), indicating retention of native ovalbumin conformation owing to the possible presence of conformational epitope(s). The biophysical and immunological results described above indicate that released precursors from the nanosheet depot may harbor native protein conformation.
Immunization of BALB/c mice with nanosheets induces a Th2 biased immune response
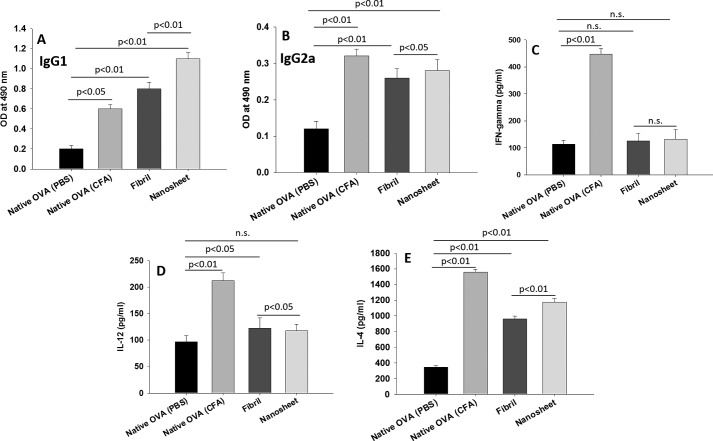
The nature of the immune response was determined by evaluating the isotypes of the responding antibodies in the nanosheet-immunized group. IgG1 was found to be the dominant isotype with lower levels of IgG2a, which supports elicitation of a predominantly Th2 biased immune response (Fig. 5, A and B). We also evaluated levels of pro-inflammatory cytokines IFN-γ4 and IL-12 and anti-inflammatory cytokine IL-4 to examine whether any inflammatory response is also evoked upon nanosheet immunization. Nanosheets were found to produce negligible levels of IFN-γ and IL-12 in nanosheet-immunized mice compared with animals immunized with native ovalbumin (Fig. 5, C and D). On the contrary, IL-4 level was found to be significantly higher in nanosheet-immunized animals than in mice immunized with native ovalbumin (Fig. 5E). This establishes a Th2 biased response and indicates that amyloid nanosheets are noninflammatory despite being immunogenic.
Figure 5.
Nanosheet immunization induces Th2 biased immune response. A and B, IgG1 and IgG2a levels in sera of mice immunized with nanosheets. C–E, IFN-γ, IL-12, and IL-4 production in ovalbumin-challenged splenocyte cultures. Results are presented as the mean of three independent experiments with error bars representing S.E. of three independent experiments. n.s., not significant.
Immune response to ovalbumin amyloid nanosheets in C57BL/6 mice
To ascertain the results of the immune response to ovalbumin amyloid nanosheets in BALB/c mice, a prototypical Th2 mouse strain, we evaluated the immune response to nanosheets in C57BL/6 mice (a prototypical Th1 mouse strain) as well. Similar to that observed in BALB/c mice, a robust antibody response is evoked upon immunizing C57BL/6 mice with amyloid nanosheets (Fig. S1A). In this case as well, nanosheets were found to evoke higher antibody titers than native ovalbumin without co-administration of any adjuvant, confirming the self-adjuvanting property of amyloid nanosheets. Practically similar levels of IgG2a to IgG1 ratio and pro-inflammatory cytokines (IFN-γ and IL-12) are observed in groups immunized with native OVA and amyloid nanosheet (Fig. S1, B–D). However, the level of anti-inflammatory cytokine, IL-4, is found to be higher in the nanosheet-immunized group (Fig. S1E). These results indicate that a negligibly inflammatory and Th2 biased immune response is evoked in C57BL/6 mice as well, validating the results obtained in BALB/c mice. Although the pattern for antibody isotypes and cytokines appears to be similar to that observed in BALB/c mice, when we look at the cytokine levels separately in BALB/c and C57BL/6 mice, we observe that levels of pro-inflammatory cytokines are slightly higher and anti-inflammatory cytokines are lower for each group in C57BL/6 mice than observed in BALB/c mice. This is in agreement with the fact that C57BL/6 mice, being a prototypical Th1 mouse strain, may favor generation of a higher pro-inflammatory response than the Th2 prototypical strain of BALB/c mice.
Nanosheets interact with the anticancer drug doxorubicin and also act as depots for its slow and sustained release over time
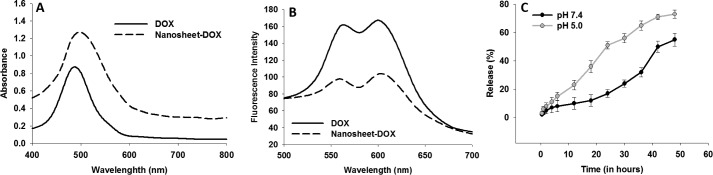
Nanosheets were incubated with doxorubicin and were monitored for possible interaction. An increase in absorption intensity of doxorubicin and a red shift indicate interaction of nanosheets with doxorubicin. Moreover, quenching of fluorescence intensity of doxorubicin upon incubation with nanosheets also indicates possible binding of doxorubicin to nanosheets (Fig. 6, A and B).
Figure 6.
Nanosheets interact with doxorubicin and release it subsequently. A, UV-visible absorption spectra of doxorubicin and nanosheets loaded with doxorubicin. B, fluorescence spectra of doxorubicin and nanosheets loaded with doxorubicin. C, release (%) of doxorubicin from nanosheets at pH 7.4 and 5.0. For C, data are expressed as mean ± S.E. of three independent experiments. DOX, doxorubicin; Nanosheet-dox, doxorubicin-loaded nanosheets.
The release of doxorubicin loaded onto nanosheets was also monitored. Doxorubicin was found to be released from the sheets in a slow and sustained manner over time at pH 7.4 as well as pH 5.0 (Fig. 6C). However, the release was found to be significantly higher at pH 5.0.
Overexpressed tumor antigens form amyloid nanosheets
Overexpressed tumor antigens represent attractive targets for immunotherapy. We selected antigens overexpressed in several cancers and examined their tendency to form amyloid nanosheets. We chose a tandem repeat fragment of mucin 1 (MUC1) (VTSAPDTRPAPGSTAPPAHG) (26), glypican-3 (FVGEFFTDV) (27), and CD45 (KFLDALISL) (27) and analyzed amyloid nanosheet formation by these. MUC1 failed to form nanosheets even after 15 days of agitation in all three of the pH conditions (pH 2, 7, and 10). Also, it did not show any sign of aggregation despite the addition of 1 m NaCl (Fig. S2A). Interestingly, glypican-3 formed nanosheets after 3 days of agitation at neutral pH in high salt concentration (1 m NaCl) (Fig. S2B). Glypican-3 samples that were not agitated could not form amyloid nanosheets (data not shown). CD45 also could not form nanosheets despite vigorous shaking and the addition of concentrated salts, but it did form oligomeric aggregates (Fig. S2C).
MUC1 peptide might have failed to form amyloids because of the presence of numerous proline residues in its sequence, which have been reported to inhibit β-sheet formation (28, 29). If we look at the MUC1 sequence, we find that 25% of the residues are proline, which may have developed kinks in the peptide; therefore, instead of forming β-sheets, they would have formed β-turns. Of note, the lack of aromatic amino acids in the MUC1 peptide might have further contributed to its inability to aggregate. It is suggested that the presence of aromatic residues promotes aggregation as a result of π-π stacking interactions between benzene rings (30).
The rich presence of amino acids, such as valine, threonine, phenylalanine, and tyrosine, which have been reported to favor aggregation (30, 31), might be the reason for easy aggregation of glypican-3 into nanosheets upon shaking. On the other hand, a seemingly lesser number of such amino acids in the CD45 peptide may have prevented the formation of nanosheets. Intriguingly, coupling of a tripeptide KFF to the CD45 peptide sequence induced formation of nanosheets under continuous agitation for 2 days at neutral pH (without the addition of salts) (Fig. S2D). KFF is composed of amino acids having a predilection for aggregation. The peptide or protein sequences reported to form extended 2D nanostructures are phenylalanine-and tyrosine-rich (32–34). Although it would be highly premature to say that these amino acids have a propensity to form nanosheets, such reports and results of the present study certainly indicate that these amino acids may be of importance in inducing nanosheet formation. None of the peptides could form nanosheets when agitated at pH 2 and 10 (data not shown).
Like glypican-3, KFF containing CD45 peptide (KFF-CD45) could not form nanosheets in the absence of shaking (data not shown). Moreover, nanosheet formation could be observed only at neutral pH, irrespective of the protein/peptide taken. In light of these results, it appears that shaking at neutral pH plays a critical role in inducing amyloid nanosheet formation. Despite being composed of amino acids having a propensity to induce aggregation, the inability of these peptides to form amyloid nanosheets in the absence of agitation at neutral pH indicates the importance of shaking and neutral pH conditions in inducing amyloid nanosheet formation.
Moreover, nanosheets formed by both glypican-3 and KFF-CD45 showed a marked increase in fluorescence upon thioflavin T binding, indicating an abundance of β-sheet–rich structures (Fig. S2E).
Discussion
Amyloidal structures are conventionally fibrillar in appearance, but for several proteins/peptides, whether naturally amyloidogenic or designed amphiphilic, many nonfibrillar amyloids have also been reported (15, 35–43). These nonfibrillar amyloids have been exploited for catalysis, biosensing, retroviral gene transfer, and bio-based electronics (15, 44–47). In the present study, ovalbumin dissolved in PBS (pH 7.0) was kept on continuous agitation so as to induce formation of amyloid fibrils. However, after 24 h of incubation, the aggregates obtained exhibited a unique nanosheet-like morphology instead of fibrils (Fig. 2C). Fine nanosheets with folds and crumples could be observed, but their maturation was preceded by unordered oligomeric aggregates, as observed in Fig. 2C. The thickness of nanosheet was found to be 5.2 nm by atomic force microscopy (data not shown). Sanii et al. (20) have reported that shaking plays an important role in the generation of protein nanosheets. It is believed that the careful motion of shaking allows compression from the air–water interface to precisely fold the protein into desired shape. During the shaking process, the proteins compress, pushing chains of the proteins together and squeezing them into a nanosheet. This process is not possible without the air–water interface, which is why the same effect is not observed when the protein solution is stirred (20).
As the nanosheets were observed in conditions usually exploited for the generation of protein fibrils and during the nanosheet maturation, some of the oligomers seen were similar to those conventionally observed in the fibril maturation pathway; we wondered whether the nanosheets too might bear amyloidal features. Various dye-binding assays (thioflavin T and Congo red) and spectroscopic and CD studies (Figs. 1 and 2 (A and B)) indicate the presence of amyloid character in the in-house–synthesized nanosheets. The tendency of the nanosheets to carry high β-sheet content similar to amyloid-like aggregates was interesting but not surprising because it has been recently reported that an amyloid (residues 1–40) solution rich in coil, turn, and α-helix but poor in β-sheet developed monolayers with a high β-sheet content when spread at the air–water interface (48). Initially, shaking can cause structural instability in the protein, inducing it to aggregate with a structural transition from α-helix to β-sheet, as is conventionally seen with most of the protein aggregation reactions. Then the aggregating proteins may coalesce, and due to continuous agitation, the side chains of coalescing aggregates can compress at the air–water interface, pushing them into β-sheet–rich nanosheets.
Interestingly, nanosheets provided a depot effect and released native-like precursors in a manner similar to that observed for fibrils. The precursors released from nanosheets mimicked native protein conformation and appear to be predominantly monomeric in nature (Fig. 3). Therefore, like amyloid fibrils, amyloid nanosheets may also follow thermodynamic equilibrium conditions between nanosheets and released monomers. Considering that the monomer release from nanosheets follows a first order reaction (as detailed under “Materials and methods”), we calculated Koff and t½ of the reaction. Lower t½ and higher Koff of nanosheets as compared with fibrils corroborated the markedly higher release of precursors from nanosheets compared with fibrils (Table 1). The Koff of nanosheets (1.6 × 10−7 s−1) suggests that nanosheets release monomers approximately 10 times faster than fibril (7.5 × 10−8 s−1). The nanosheet–monomer equilibrium is of particular importance for the proposed immunotherapeutic application, because nanosheets must be stable enough to guarantee a long duration of precursor release to boost immune system, and the release of precursors must be sufficient to evoke desirable immune response. In human subjects, this equilibrium may shift in a forward direction, as the monomers released mimic native proteins and therefore may get degraded by the proteolytic machinery of the body. Hence, the half-life of nanosheets in vivo may be lower than in vitro half-life. Therefore, it can be concluded that like any other amyloidal aggregate, nanosheets also have a particular half-life, and after a certain period of time, the whole nanosheet may fully disassemble to monomers. Considering this fate, fibrillar and oligomeric amyloids of gonadotrophin-releasing hormone and insulin, respectively, have been proposed as drug depots (49, 50). The storage of pituitary hormones as amyloids (4) is the best example of nature's choice to use amyloids as natural reservoirs for peptide release. It also elucidates physiological tolerance to amyloids. Moreover, there is plentiful other evidence suggesting that amyloid-based biomaterials are well tolerated in vivo (51–56).
Because ovalbumin nanosheets bear an amyloid character and are capable of releasing precursor proteins, we evaluated immune response evoked upon immunizing the same. In a manner similar to conventional amyloids, they may also withstand proteolytic degradation, which may increase their residence time in circulation. We evaluated immune response to nanosheets considering that if they are found to evoke robust immune response, they could be exploited as antigen delivery system for vaccine development. Moreover, keeping in mind that the extended 2D structure of nanosheets may facilitate loading of small-molecule drugs, we analyzed loading of the anticancer drug doxorubicin onto nanosheets. We set forth with the idea that if nanosheets are immunogenic and can also load small-molecule drugs, they may present a modular platform for the development of a novel simultaneous immunotherapy and chemotherapy system with potential clinical applications.
Nanosheets were found to evoke a robust immune response and interacted with doxorubicin efficiently. Interestingly, robust antibody responses against self-assembled ovalbumin nanosheets were evoked without co-administration of any additional adjuvant (Fig. 4A), suggesting that nanosheet amyloids have a self-adjuvanting property. The strong cross-reactivity of nanosheet antisera with native ovalbumin (Fig. 4B) demonstrates that nanosheet immunization is capable of eliciting high antibody titers against native ovalbumin, by virtue of retaining native antigen epitopes and/or releasing precursors carrying native epitopes. To obtain a clearer picture, ELISA plates coated with precursors released from nanosheets were probed with native ovalbumin antisera (polyclonal antibodies). Intriguingly, released precursors recognized anti-native ovalbumin polyclonal antibodies (Fig. 4C), indicating possible conservation of native ovalbumin epitopes in released precursors. This was further confirmed by probing plates coated with released ovalbumin with 2D11 mAb that recognizes native ovalbumin but not denatured ovalbumin. The ovalbumin precursors released from nanosheets reacted with 2D11 mAb, but the nanosheet itself did not recognize 2D11 antibody (Fig. 4, D and E). This indicates a possible conformational change in released precursors, from the amyloid state to the native form. This phenomenon reiterates our previous finding that the native form of proteins may be released from amyloid depots (16). It further elucidates that this phenomenon is independent of the morphological structure of amyloids; whether they are in oligomeric, fibril, or nanosheet form, they may release precursors harboring native antigen epitopes. In other words, release of native protein may be a generic feature of amyloids.
Considering the levels of IgG1 and IgG2a isotypes in responding antibodies and pro- and anti-inflammatory cytokines evoked (Fig. 5 and Fig. S1), it appears that nanosheets elicit predominantly Th2 biased immune response with robust humoral and insignificant inflammatory responses. This is in contrast to our previous finding that the oligomeric intermediates formed in the process of fibril maturation evoked Th1 biased inflammatory immune responses (16). It indicates that the mature amyloids like fibrils and nanosheets may evoke Th2 biased responses, whereas the intermediates formed in the process of aggregate maturation may evoke Th1 biased inflammatory responses. Interestingly, Aβ oligomers but not fibrils also have been reported to initiate inflammatory responses (57, 58). This provides a platform to tune the immune response, as required. If Th2 biased immune response is desired, mature amyloids (fibrils and nanosheet) can be exploited; on the other hand, for evoking Th1 biased immune response, oligomeric intermediates are seemingly better suited. Nevertheless, further immunological studies with amyloid nanosheets formed by other peptides/proteins are needed to come to any generalized statement.
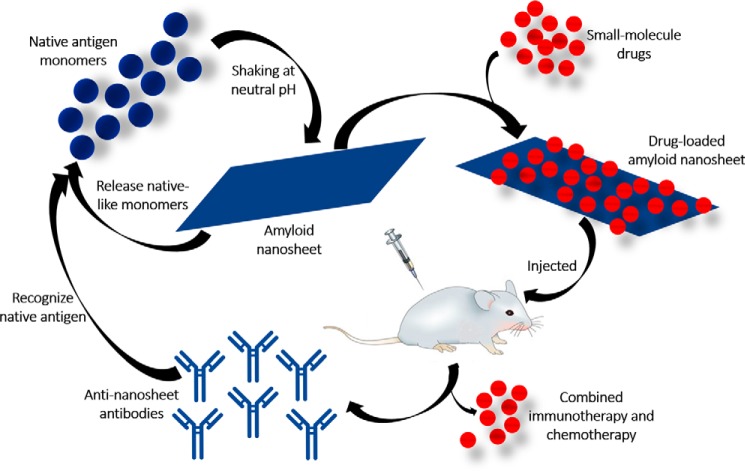
Novel medical applications of 2D nanomaterials in drug delivery, vaccine protein delivery, wound healing, etc. have attracted the attention of the scientific community across the globe (59–62). 2D nanomaterials are thought to be superior drug delivery agents because of the availability of both reverse and obverse surfaces. Synergistic combination therapy using 2D nanomaterials is also one of the popular areas of current research (63, 64). However, developing multifunctional 2D nanomaterials with high loading capacity remains a challenge. We propose exploiting the in-house–produced amyloid nanosheets as a multifunctional antigen and drug delivery vehicle. Amyloid nanosheets, being whole protein bodies capable of releasing precursor proteins and evoking strong antibody response, may themselves act as antigen and delivery vehicle simultaneously. The antigen does not need to be loaded onto the sheets; therefore, the in-house–synthesized nanosheets may load small-molecule drugs with high loading capacity. In the present study, we evaluated loading of the anticancer drug doxorubicin onto ovalbumin amyloid nanosheets. Doxorubicin was found to be successfully loaded onto the sheets and subsequently be released (Fig. 6). This suggests that nanosheets can be used for combined immunotherapy and chemotherapy approaches, where the vehicle itself (nanosheets) may serve as an immunotherapeutic agent and the loaded drug may act as a chemotherapeutic agent. Moreover, as the antigen itself is the vehicle, the other drug(s) can have a high loading capacity, which is one of the biggest challenges with the combined synergistic drug delivery platforms currently in vogue. Fig. 7 shows a schematic illustration of the proposed amyloid nanosheet–based dual-delivery vehicle with applications in simultaneous immunotherapy and chemotherapy.
Figure 7.
Schematic illustration of amyloid nanosheet–based simultaneous immunotherapeutic and chemotherapeutic platform. The model shows that native antigen monomers under continuous agitation at neutral pH (in the presence or absence of salts) form nanosheets harboring amyloid-like character. The sheets are capable of releasing monomers predominantly, carrying a native antigen conformation. These sheets can be loaded with small-molecule drugs. If drug-loaded sheets are injected, they may act as reservoirs for simultaneous release of native-like antigens and loaded drugs; hence, they may have applications in simultaneous immunotherapy and chemotherapy. Immunization with nanosheets leads to generation of native antigen–specific antibodies.
As we propose exploitation of amyloid nanosheets for vaccine development, it becomes indispensable to examine whether amyloid nanosheet formation is limited to amyloidogenic proteins like albumins or if it can be extended to real-world antigens, especially cell-surface antigens, which are the preferred targets for immunotherapy. Considering these facts, we evaluated the formation of amyloid nanosheets by MUC1, a glycosylated transmembrane protein overexpressed in numerous cancers (26); glypican-3, a cell-surface protein overexpressed in hepatocellular carcinoma (27); and CD45, a cell-surface leukocyte common antigen overexpressed in leukemia and lymphoma (27). The reason behind studying these antigens for nanosheet formation is their overexpression in various cancers, which makes them potential targets for immunotherapy. Interestingly, we could successfully make nanosheets for glypican-3 under continuous agitation at neutral pH in the presence of concentrated salt. MUC1 failed to aggregate despite vigorous shaking and the addition of concentrated salts at all of the three pH conditions. CD45 also could not form nanosheets despite vigorous shaking and the addition of concentrated salts, but it did form oligomeric aggregates (Fig. S2). However, when KFF, a tripeptide containing amino acids that favor aggregation, was coupled to CD45 peptide, it successfully formed nanosheets when agitated at neutral pH. Interestingly, unshaken glypican-3 and KFF-CD45 could not form amyloid nanosheets (data not shown). Moreover, in each case, nanosheet formation could be induced only at neutral pH. These results indicate that a confluence of factors, such as aggregation inducing amino acids, continuous agitation, neutral pH, and ionic strength, may be required to induce the formation of nanosheets, although the conditions may vary for different peptides/proteins. The results also show that nanosheet formation is not limited to amyloidogenic proteins like albumins, but it can also be extended to other proteins, including cell-surface proteins, which is important for developing real-world immunoprophylactic/immunotherapeutic platforms.
Short fibrillizing peptides capable of self-assembling to β-sheet–rich nanofibers have been reported (51). It is feasible to add desired amino acid sequences to the N terminus of these fibrillizing peptides, which self-assemble to form fibrils of the desired peptides. In a manner similar to short fibrillizing peptides, we believe that short nanosheet-forming peptides can be designed that could self-assemble to form amyloid nanosheets at specific pH and salt concentrations. The desired peptides can be added to the nanosheet-forming peptides, which will induce formation of nanosheet of the whole amino acid stretch even if the desired peptide is nonamyloidogenic. Although we believe that amyloid nanosheets can be formed for any peptide/protein under specific agitation and pH conditions irrespective of their amyloidogenic nature, designing nanosheet-forming peptides will save time, energy, and resources. An epitope or even a precise combination of epitopes can be added to the N terminus of nanosheet-forming peptides, leading to development of a novel class of immunotherapeutic platform.
Conclusion
A nanosheet-like structure formed by continuous shaking of ovalbumin was observed in conditions usually exploited for the synthesis of fibrillar amyloids. The nanosheets were found to have amyloidal features. Nanosheets released precursor ovalbumin harboring native antigen epitopes and preserving native antigen conformation. Immunization with nanosheets evokes strong antibody titers cross-reactive with native ovalbumin, and the immune response evoked is Th2 biased and negligibly inflammatory. As the obverse and reverse surfaces of nanosheets were free, we evaluated loading of the anticancer drug doxorubicin onto them. The amyloid nanosheets were found to interact with doxorubicin and also acted as a depot for its release over time. Reckoning with these findings, amyloid nanosheets can be exploited as a novel class of antigen delivery system owing to their potential to withstand proteolytic degradation and remain in circulation for longer periods. As they can be successfully loaded with small-molecule drugs, they can also be useful for combined immunotherapy and chemotherapy. The results of the present study also demonstrate that formation of amyloid nanosheets is not limited to amyloidogenic proteins like albumins, but cell-surface antigens (such as tumor antigens) can also form amyloid nanosheets under specific conditions.
Materials and methods
Chemicals and reagents
All of the reagents used were of the highest purity available. Ovalbumin (A2512), thioflavin T, Congo red, and doxorubicin were purchased from Sigma-Aldrich. IgG2a (R35-95) isotype control was procured from eBiosciences (San Diego, CA). IgG1 and IgG2a isotypes (550487) and cytokines (namely IFN-γ, IL-12, and IL-4 cytokine estimation kits) were procured from BD Biosciences, OptEIATM. Anti-ovalbumin mAb 2D11 was purchased from Santa Cruz Biotechnology, Inc. All other reagents used were of analytical grade and procured from local suppliers.
Nanosheet formation
Ovalbumin was dissolved in 1 ml of PBS (pH 7.0) carrying 0.01% azide at a concentration of 1 mg/ml and incubated at room temperature under continuous agitation at 180 rpm. Aliquots were collected at various time points and pelleted down at 15,000 × g for 15 min using a Sigma 3K30 microcentrifuge. Monomeric ovalbumin did not sediment at 15,000 × g and remained in the supernatant. The pellet obtained was characterized by light scattering, thioflavin T fluorescence, Congo red binding, CD studies, atomic force microscopy, and transmission EM.
Fibril formation
Ovalbumin was dissolved in PBS (pH 7.0) carrying 0.01% azide at a concentration of 1 mg/ml and incubated at room temperature under continuous agitation at 90 rpm for 15 days (16). The sample was centrifuged at 15,000 × g for 15 min, and the pellet was collected and analyzed for fibril formation by thioflavin T fluorescence, Congo red binding, and transmission EM (data not shown).
Rayleigh scattering measurements
Rayleigh scattering measurements were performed on a Hitachi F-4500 fluorescence spectrophotometer at room temperature in a 1-cm path length cell. Samples were excited at 350 nm, and spectra were recorded in the range of 300–400 nm. Both excitation and emission slits were fixed at 5 nm.
Turbidity measurements
The turbidity of the incubated samples collected at various time points was monitored by UV absorbance at 350 nm using a PerkinElmer Life Sciences UV-visible spectrometer model λ25 in a cuvette of 1-cm path length at room temperature.
Thioflavin T binding
Various ovalbumin aggregates (100 μg) were incubated with 30 μm thioflavin T solution (30 μl of 1 mm thioflavin T stock solution), and fluorescence was measured after 30 min on a Hitachi F-4500 fluorescence spectrophotometer. Thioflavin T fluorescence was excited at 450 nm, and spectra were recorded from 460 to 560 nm. The excitation and emission slit widths were fixed at 5 and 10 nm, respectively. Thioflavin T solution (30 μm) in PB (5 mm, pH 7.4) was scanned to obtain the thioflavin T–only spectrum. Native ovalbumin (100 μg) incubated with 30 μm thioflavin T was taken as control.
Fluorescence microscopy was also used to establish the amyloid formation by ovalbumin aggregates. Aggregates formed at 24 h were incubated with 20 μm thioflavin T for 30 min at room temperature and further transferred onto a glass slide to be analyzed under a fluorescence microscope (Axio, HBU 50/AC; Zeiss, Gottingen, Germany).
Congo red binding
Congo red solution (20 μm) prepared in PB (5 mm, pH 7.4) from a 1 mm stock solution of Congo red (prepared in ethanol) was incubated with 100 μg of various aggregates for 30 min at room temperature. UV absorbance was measured from 300 to 700 nm on a PerkinElmer Life Sciences UV-visible spectrometer, model λ25. Congo red solution (20 μm) in PB (5 mm, pH 7.4) served as control (for the measurement of the Congo red-only spectrum). Absorbance of 100 μg of native ovalbumin mixed with 20 μm of Congo red solution was also measured.
CD measurements
CD measurements were performed on a JASCO spectropolarimeter (J-815). The instrument was calibrated with d-10-camphorsulfonic acid. All measurements were made at room temperature with a thermostatically controlled cell holder attached to a Peltier device with Multitech water circulator. Spectra were collected with a scan speed of 100 nm/min and a response time of 2 s. Far-UV CD spectra were taken in the range of 200–250 nm in a cell of 0.1-cm path length with protein samples having a final concentration of 200 μg/ml. Near-UV CD spectra were recorded in the range of 250–320 nm.
Transmission EM
Transmission electron micrographs were collected on a JEOL transmission electron microscope operating at an accelerating voltage of 200 kV. Nanosheet formation was assessed by applying 6 μl of sample (100 μg) on a carbon-coated copper grid. Excess fluid was removed after 2 min, and the grids were then negatively stained with 2% (w/v) uranyl acetate.
Atomic force microscopy
An aliquot of aggregates was placed on a silicon disc and dried in a nitrogen atmosphere, and the samples were analyzed using a contact mode atomic force microscope (PerkinElmer Life Sciences).
Nanosheet yield
The nanosheet yield was quantified by measuring the soluble protein concentration in the supernatant (at 280 nm) of ovalbumin solution agitated for 24 h. Ovalbumin solution constantly agitated for 24 h was centrifuged at 15,000 × g for 15 min. The concentration of proteins in supernatant was determined by UV absorption, and the nanosheet yield was calculated by subtracting this value from the initial concentration of monomeric proteins incubated for nanosheet development. We believe the pellet contained only nanosheets, as the transmission electron microscopic images did not show presence of any oligomeric or fibrillar aggregates in samples agitated for 24 h.
In vitro release kinetics
The ovalbumin samples kept for agitation were pelleted at 15,000 × g for 15 min, and the pellets obtained were washed three times with PB (pH 7.4) and resuspended in PB (pH 7.4). The kinetics of ovalbumin release in PB (pH 7.4) was monitored spectrophotometrically at 280 nm (16, 49). The supernatant obtained after centrifugation was analyzed at various time points for 15 days on a PerkinElmer UV-visible spectrometer, model λ25.
Size-exclusion chromatography
Five milliliters of the sample containing ovalbumin released from nanosheet and fibril after 7 days were applied to a 83-cm-long column with an internal diameter of 2.3 cm, filled with about 340 ml of preswollen Bio-Gel P10 (Bio-Rad), and equilibrated with 20 mm phosphate-HCl buffer (pH 3.0) containing 18 mm NaCl. The flow rate was adjusted to 35 ml/h, and 1.0-ml fractions were collected. Absorbance of the collected fractions was measured at 280 nm using a PerkinElmer Life Sciences UV-visible spectrometer, model λ25, and plotted against the elution volume.
Theoretical treatment of release kinetics of monomers from nanosheets
In a manner similar to amyloid fibrils, amyloid nanosheets may follow thermodynamic equilibrium conditions between nanosheet and monomers released. The following equation may be applied to elaborate the monomer release from nanosheets over time.
| (Eq. 1) |
Kd is the equilibrium dissociation constant. In a manner similar to that reported for fibril dissociation (49, 65), at equilibrium, the molar concentration of nanosheets may not change practically, as a comparatively small amount of monomers may be released, so the expression for Kd reduces to the concentration of monomers at dissociation equilibrium (Eq. 2).
| (Eq. 2) |
Monomer release from nanosheets or nanosheet dissociation is governed by the first-order rate constant, Koff, as the dissociation reaction depends exclusively on the nanosheet concentration. To calculate Koff and t½ (half-life), we obtained the concentration of undissociated or remaining nanosheets at various time points of release kinetics. The concentration of undissociated nanosheets was obtained by subtracting the concentration of monomers released at various time points from the initial concentration of nanosheets taken for release kinetics analysis. Natural log concentration of the undissociated nanosheets was plotted as a function of incubation time, and linear regression analysis was performed. The slope of the plot gave the value of Koff, and t½ was calculated using the equation, t½ = ln 2/Koff.
Animals
In-bred female BALB/c mice and C57BL/6 mice (6–8 weeks old, 20 ± 2 g) were obtained from the Animal House Facility of the Interdisciplinary Biotechnology Unit (Aligarh Muslim University) and the Department of Biochemistry (Jawaharlal Nehru Medical College, Aligarh Muslim University), respectively. The mice were housed in commercially available polypropylene cages and maintained under controlled temperature conditions on a 12-h light/dark cycle and had free access to food and water ad libitum. All of the animal experiments were performed according to the National Regulatory Guidelines issued by the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA). The approval IDs for the Interdisciplinary Biotechnology Unit and the Department of Biochemistry were 332/CPCSEA and 401/CPCSEA, respectively. All of the procedures used for the animal experiments were reviewed and approved by the Institutional Animal Ethics Committee of the Interdisciplinary Biotechnology Unit and Department of Biochemistry, Jawaharlal Nehru Medical College, Aligarh Muslim University.
Mode and schedule of immunization
Animals were immunized subcutaneously in the lower abdominal region aseptically with 50 μg of ovalbumin amyloid nanosheets. A booster was given 3 weeks after the first immunization with 25 μg of the nanosheets.
Collection of sera
Blood was collected from mice 5 days after the last booster. Sera were separated from the clotted blood by centrifugation at 1500 × g for 10 min at 4 °C. Finally, the supernatant was collected for ELISA analysis and purification of polyclonal antibodies.
Determination of antigen-specific total IgGs by ELISA
Antigen-specific total IgGs against the ovalbumin nanosheet were determined in the sera of mice immunized following the protocol as described elsewhere (16). Briefly, 100 μl (2 μg) of native ovalbumin as well as ovalbumin amyloids (fibril and nanosheet) were dissolved in carbonate/bicarbonate buffer (0.05 m, pH 9.6) and poured into 96-well microtiter plates that were further incubated overnight at 4 °C. The plates were then incubated with serially diluted sera at 37 °C for 2 h, after the usual washing and blocking steps. Furthermore, the plates were washed again, 100 μl of (1:5000 dilution of stock) horseradish peroxidase–conjugated goat anti-mouse antibodies were added to each well, and the plates were incubated at 37 °C for 1 h. After the usual plate washing, 100 μl of substrate solution (6 mg of o-phenylenediamine) in 12 ml of substrate buffer with 5 μl of 30% H2O2 were added to the wells, and the plates were finally incubated at 37 °C for 40 min. The reaction was terminated by the addition of 50 μl of 7% H2SO4. The absorbance was read at 490 nm with a microtiter plate reader (Bio-Rad).
ELISA of ovalbumin released from nanosheets
Briefly, 96-well microtiter plates were incubated overnight with 100 μl (2 μg) of ovalbumin released from nanosheets in carbonate/bicarbonate buffer (0.05 m, pH 9.6) at 4 °C. After the usual washing and blocking steps, the plates were finally incubated with serially diluted anti-native ovalbumin-specific polyclonal and 2D11 monoclonal antibodies at 37 °C for 2 h. After washing the plates, 100 μl of (1:5000 dilution of stock) horseradish peroxidase-conjugated goat anti-mouse antibodies were added to each well, and the plates were incubated at 37 °C for 1 h. Substrate solution (100 μl) was added to the wells after the usual plate washing. Finally, the plates were incubated at 37 °C for 40 min. For terminating the reaction, 50 μl of 7% H2SO4 were added to the wells. The absorbance was read at 490 nm with a microtiter plate reader (Bio-Rad).
Determination of antibody isotype in sera of immunized mice
Sera from mice immunized with nanosheets were analyzed for antibody isotypes using the protocol described elsewhere (66). Briefly, 2 μg (100 μl) of antigen in carbonate/bicarbonate buffer (0.05 m, pH 9.6) were added to 96-well microtiter plates that were incubated overnight at 4 °C. After washing and blocking steps, the plates were incubated with serially diluted sera at 37 °C for 2 h. After excessive washing of the plates, 100 μl of (1:5000 dilution of stock) goat anti-mouse anti-IgG1 and IgG2a antibodies were added in each well and incubated for 1 h at 37 °C. Plates were again washed, 100 μl of (1:5000 dilution of stock) horseradish peroxidase-conjugated rabbit anti-goat antibodies were added to each well, and each plate was incubated at 37 °C for 1 h. After further washing of the plates, 100 μl of substrate solution were added to the wells, and the plates were finally incubated at 37 °C for 40 min. The reaction was stopped by the addition of 50 μl of 1 m H2SO4. The absorbance was read at 490 nm with a microtiter ELISA plate reader (Bio-Rad).
T lymphocyte isolation from spleens of immunized mice
Mice immunized with native as well as ovalbumin amyloids (fibril and nanosheet) were sacrificed 5 days after the last booster. T lymphocytes were isolated from the spleens of sacrificed mice as described elsewhere (66). Briefly, spleens isolated from animals belonging to various immunized groups were macerated, and the suspension was treated with ACK lysis buffer (0.15 mol/liter ammonium chloride, 10 mmol/liter potassium/bicarbonate, and 88 mmol/liter edetic acid) for lysis of the red blood cells. The cell suspension was centrifuged at 1500 × g for 5 min, and the cell pellet was washed with Hanks' balanced salt solution three times and resuspended in RPMI 1640 medium containing 10% fetal calf serum and 0.1% antimycotic mixture.
Cytokine assay: Determination of IFN-γ, IL-4, and IL-12 by sandwich ELISA
Levels of IFN-γ, IL-12, and IL-4 cytokines induced by lymphocytes upon their culture in the co-presence of ovalbumin nanosheet were quantitated using appropriate and specific biotinylated antibody pairs according to the manufacturer's protocols. Briefly, 96-well microtiter plates were coated with 50 μl of the purified capture antibodies in carbonate/bicarbonate buffer (pH 9.5) at 4 °C. After the usual washing and blocking steps, 50 μl of the supernatant (isolated from cultured splenocytes after 48 h) were poured in each well to determine the level of cytokine induced. Plates were washed and incubated with biotinylated polyclonal goat anti-mouse cytokine detection antibody. Furthermore, after washing the plates, 100 μl of streptavidin-horseradish peroxidase conjugate were added to each well, and plates were incubated for 30 min at room temperature. The plates were then washed, and the colored complex was developed with tetramethylbenzidine. The absorbance was read at 450 nm with a microtiter plate reader (Bio-Rad). A known specific recombinant cytokine was used as a standard for calculating the level of the given cytokine in the samples tested, and the concentration was expressed as pg/ml.
Interaction of ovalbumin amyloid nanosheets with the anticancer drug doxorubicin
Two milliliters of doxorubicin (2.5 mg/ml) aqueous solution were added to 3 ml of ovalbumin nanosheet (50 μg/ml) solution in water. After stirring vigorously overnight, the obtained doxorubicin-loaded nanosheets were gathered by centrifugation and washed with PBS. Afterward, the doxorubicin-loaded nanosheets were redispersed in PBS for further use.
Interaction of doxorubicin with nanosheets was ascertained by recording UV-visible absorbance spectra and fluorescence intensity. To study the fluorescence quenching of doxorubicin by nanosheets, the fluorescence spectra of doxorubicin-loaded nanosheets were measured on a fluorescence spectrophotometer (FL-4600, Hitachi, Tokyo, Japan), using free doxorubicin (47.5 μg/ml) as control. To exclude the inner filter effect, nanosheet (50 μg/ml) and doxorubicin (47.5 μg/ml) were mixed for only 10 s, and then the fluorescence spectrum was measured instantly.
Release of doxorubicin from nanosheets
Doxorubicin-loaded nanosheets were suspended in 10 ml of PBS (pH 7.4) and were poured in a dialysis bag. Then the dialysis bag was placed in 50 ml of PBS. The whole assembly was maintained at 37 ± 0.5 °C, covered by parafilm to avoid evaporation, and shaken at 90 cycles/min. At fixed time intervals, 2 ml of medium was withdrawn and replaced with the same volume of fresh buffer to maintain the required sink condition. This was taken into account while calculating cumulative drug release. The sample was filtered, and drug quantity in filtrate was determined by HPLC analysis.
Peptide synthesis
Tumor peptides were synthesized following the published protocol (67) with slight modifications in our laboratory. Briefly, peptides were synthesized manually via the solid-phase method on Wang resin utilizing standard Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry. The peptides were purified by reverse-phase HPLC on an analytical Waters Symmetry C18 column using a linear gradient of 20–80% acetonitrile for 40 min with a flow rate of 0.6 ml/min. Both acetonitrile and water contained 0.1% TFA. The purity of the peptides was further determined to be ≥95% by reverse-phase analytical chromatography.
Nanosheet formation by overexpressed tumor antigens
Overexpressed antigens represent attractive targets for immunotherapy. We exploited a tandem repeat fragment of MUC1, glypican-3, and CD45, having sequences VTSAPDTRPAPGSTAPPAHG (24), FVGEFFTDV (25), and KFLDALISL (25), respectively, for examining nanosheet synthesis. The in-house–synthesized peptides were dissolved in buffers of pH 2, 7, and 10 (with or without 1 m NaCl) and kept under continuous agitation for various time periods. Aggregates obtained were characterized by thioflavin T binding and transmission EM using the protocols described in this paper.
Statistical analysis
Data were analyzed, and two groups were compared using Student's t test, and one-way analysis of variance (Holm–Sidak method) was used to compare all groups with each other. p values <0.05 were considered significant. SigmaPlot (versions 10 and 11; SigmaPlot Software, San Jose, CA) was used for data presentation.
Author contributions
S. T. and M. A. S. conceptualization; S. T., M. A. S., S. S., and S. A. data curation; S. T. software; S. T., M. O., and N. I. formal analysis; S. T. funding acquisition; S. T., M. A. S., S. S., and S. A. validation; S. T., M. A. S., S. S., S. A., M. O., and N. I. investigation; S. T. and M. A. S. visualization; S. T., M. A. S., S. S., S. A., and M. O. methodology; S. T., M. A. S., and N. I. writing-original draft; S. T. and N. I. writing-review and editing; M. O. and N. I. resources; N. I. supervision.
Supplementary Material
The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and S2.
- IFN
- interferon
- 2D
- two-dimensional
- IL
- interleukin
- PB
- phosphate buffer.
References
- 1. Dogra P., Bhattacharya M., and Mukhopadhyay S. (2017) pH-responsive mechanistic switch regulates the formation of dendritic and fibrillar nanostructures of a functional amyloid. J. Phys. Chem. B 121, 412–419 10.1021/acs.jpcb.6b11281 [DOI] [PubMed] [Google Scholar]
- 2. Jacob R. S., Das S., Ghosh S., Anoop A., Jha N. N., Khan T., Singru P., Kumar A., and Maji S. K. (2016) Amyloid formation of growth hormone in presence of zinc: relevance to its storage in secretory granules. Sci. Rep. 6, 23370 10.1038/srep23370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Greenwald J., and Riek R. (2010) Biology of amyloid: structure, function, and regulation. Structure 18, 1244–1260 10.1016/j.str.2010.08.009 [DOI] [PubMed] [Google Scholar]
- 4. Maji S. K., Perrin M. H., Sawaya M. R., Jessberger S., Vadodaria K., Rissman R. A., Singru P. S., Nilsson K. P., Simon R., Schubert D., Eisenberg D., Rivier J., Sawchenko P., Vale W., and Riek R. (2009) Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science 325, 328–332 10.1126/science.1173155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shorter J., and Lindquist S. (2005) Prions as adaptive conduits of memory and inheritance. Nat. Rev. Genet. 6, 435–450 10.1038/nrg1616 [DOI] [PubMed] [Google Scholar]
- 6. Mesquida P., Riener C. K., MacPhee C. E., and McKendry R. A. (2007) Morphology and mechanical stability of amyloid-like peptide fibrils. J. Mater. Sci. Mater. Med. 18, 1325–1331 10.1007/s10856-006-0075-0 [DOI] [PubMed] [Google Scholar]
- 7. Udomprasert A., Bongiovanni M. N., Sha R., Sherman W. B., Wang T., Arora P. S., Canary J. W., Gras S. L., and Seeman N. C. (2014) Amyloid fibrils nucleated and organized by DNA origami constructions. Nat. Nanotechnol. 9, 537–541 10.1038/nnano.2014.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lara C., Gourdin-Bertin S., Adamcik J., Bolisetty S., and Mezzenga R. (2012) Self-assembly of ovalbumin into amyloid and non-amyloid fibrils. Biomacromolecules 13, 4213–4221 10.1021/bm301481v [DOI] [PubMed] [Google Scholar]
- 9. Humblet-Hua N. P., van der Linden E., and Sagis L. M. (2012) Microcapsules with protein fibril reinforced shells: effect of fibril properties on mechanical strength of the shell. J. Agric. Food Chem. 60, 9502–9511 10.1021/jf3024529 [DOI] [PubMed] [Google Scholar]
- 10. Mine Y. (1995) Recent advances in the understanding of egg white protein functionality. Trends Food Sci. Technol. 6, 225–232 10.1016/S0924-2244(00)89083-4 [DOI] [Google Scholar]
- 11. Knowles T. P. J., and Buehler M. J. (2011) Nanomechanics of functional and pathological amyloid materials. Nat. Nanotechnol. 6, 469–479 10.1038/nnano.2011.102 [DOI] [PubMed] [Google Scholar]
- 12. Bolisetty S., Adamcik J., Heier J., and Mezzenga R. (2012) Amyloid directed synthesis of titanium dioxide nanowires and their applications in hybrid photovoltaic devices. Adv. Funct. Mater. 22, 3424–3428 10.1002/adfm.201103054 [DOI] [Google Scholar]
- 13. Li C., Adamcik J., and Mezzenga R. (2012) Biodegradable nanocomposites of amyloid fibrils and graphene with shape-memory and enzyme-sensing properties. Nat. Nanotechnol. 7, 421–427 10.1038/nnano.2012.62 [DOI] [PubMed] [Google Scholar]
- 14. Li C., and Mezzenga R. (2013) The interplay between carbon nanomaterials and amyloid fibrils in bio-nanotechnology. Nanoscale 5, 6207–6218 10.1039/c3nr01644g [DOI] [PubMed] [Google Scholar]
- 15. Dai B., Li D., Xi W., Luo F., Zhang X., Zou M., Cao M., Hu J., Wang W., Wei G., Zhang Y., and Liu C. (2015) Tunable assembly of amyloid-forming peptides into nanosheets as a retrovirus carrier. Proc. Natl. Acad. Sci. U.S.A. 112, 2996–3001 10.1073/pnas.1416690112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tufail S., Owais M., Kazmi S., Balyan R., Khalsa J. K., Faisal S. M., Sherwani M. A., Gatoo M. A., Umar M. S., and Zubair S. (2015) Amyloid form of ovalbumin evokes native antigen-specific immune response in the host: prospective immuno-prophylactic potential. J. Biol. Chem. 290, 4131–4148 10.1074/jbc.M113.540989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Taboada P., Barbosa S., Castro E., and Mosquera V. (2006) Amyloid fibril formation and other aggregate species formed by human serum albumin association. J. Phys. Chem. B 110, 20733–20736 10.1021/jp064861r [DOI] [PubMed] [Google Scholar]
- 18. Sasahara K., Yagi H., Sakai M., Naiki H., and Goto Y. (2008) Amyloid nucleation triggered by agitation of β2-microglobulin under acidic and neutral pH conditions. Biochemistry 47, 2650–2660 10.1021/bi701968g [DOI] [PubMed] [Google Scholar]
- 19. Buttstedt A., Wostradowski T., Ihling C., Hause G., Sinz A., and Schwarz E. (2013) Different morphology of amyloid fibrils originating from agitated and non-agitated conditions. Amyloid 20, 86–92 10.3109/13506129.2013.784962 [DOI] [PubMed] [Google Scholar]
- 20. Sanii B., Kudirka R., Cho A., Venkateswaran N., Olivier G. K., Olson A. M., Tran H., Harada R. M., Tan L., and Zuckermann R. N. (2011) Shaken, not stirred: collapsing a peptoid monolayer to produce free-floating, stable nanosheets. J. Am. Chem. Soc. 133, 20808–20815 10.1021/ja206199d [DOI] [PubMed] [Google Scholar]
- 21. LeVine H., 3rd (1999) Quantification of β-sheet amyloid fibril structures with thioflavin T. Methods Enzymol. 309, 274–284 10.1016/S0076-6879(99)09020-5 [DOI] [PubMed] [Google Scholar]
- 22. Ban T., Hamada D., Hasegawa K., Naiki H., and Goto Y. (2003) Direct observation of amyloid fibril growth monitored by thioflavin T fluorescence. J. Biol. Chem. 278, 16462–16465 10.1074/jbc.C300049200 [DOI] [PubMed] [Google Scholar]
- 23. Klunk W. E., Jacob R. F., and Mason R. P. (1999) Quantifying amyloid by Congo Red spectral shift assay. Methods Enzymol. 309, 285–305 10.1016/S0076-6879(99)09021-7 [DOI] [PubMed] [Google Scholar]
- 24. Kelly S. M., and Price N. C. (2000) The use of circular dichroism in the investigation of protein structure and function. Curr. Protein Pept. Sci. 1, 349–384 10.2174/1389203003381315 [DOI] [PubMed] [Google Scholar]
- 25. Takahashi N., Koseki T., Doi E., and Hirose M. (1991) Role of an intrachain disulfide bond in the conformation and stability of ovalbumin. J. Biochem. 109, 846–851 10.1093/oxfordjournals.jbchem.a123469 [DOI] [PubMed] [Google Scholar]
- 26. Bánóczi Z., Mezõ G., Windberg E., Uray K., Majer Z., and Hudecz F. (2008) Synthesis and antibody recognition of synthetic antigens from MUC1. J. Pept. Sci. 14, 610–616 10.1002/psc.950 [DOI] [PubMed] [Google Scholar]
- 27. Vigneron N., Stroobant V., Van den Eynde B. J., and van der Bruggen P. (2013) Database of T cell-defined human tumor antigens: the 2013 update. Cancer Immun. 13, 15 [PMC free article] [PubMed] [Google Scholar]
- 28. Steward A., Adhya S., and Clarke J. (2002) Sequence conservation in Ig-like domains: the role of highly conserved proline residues in the fibronectin type III superfamily. J. Mol. Biol. 318, 935–940 10.1016/S0022-2836(02)00184-5 [DOI] [PubMed] [Google Scholar]
- 29. Monsellier E., and Chiti F. (2007) Prevention of amyloid-like aggregation as a driving force of protein evolution. EMBO Rep. 8, 737–742 10.1038/sj.embor.7401034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bemporad F., Taddei N., Stefani M., and Chiti F. (2006) Assessing the role of aromatic residues in the amyloid aggregation of human muscle acylphosphatase. Protein Sci. 15, 862–870 10.1110/ps.051915806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Richardson J. S., and Richardson D. C. (2002) Natural β-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc. Natl. Acad. Sci. U.S.A. 99, 2754–2759 10.1073/pnas.052706099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Min K. I., Yun G., Jang Y., Kim K. R., Ko Y. H., Jang H. S., Lee Y. S., Kim K., and Kim D. P. (2016) Covalent self-assembly and one-step photocrosslinking of tyrosine-rich oligopeptides to form diverse nanostructures. Angew. Chem. Int. Ed. Engl. 55, 6925–6928 10.1002/anie.201601675 [DOI] [PubMed] [Google Scholar]
- 33. Hu Y., Lin R., Zhang P., Fern J., Cheetham A. G., Patel K., Schulman R., Kan C., and Cui H. (2016) Electrostatic-driven lamination and untwisting of β-sheet assemblies. ACS Nano. 10, 880–888 10.1021/acsnano.5b06011 [DOI] [PubMed] [Google Scholar]
- 34. Lin Y., Thomas M. R., Gelmi A., Leonardo V., Pashuck E. T., Maynard S. A., Wang Y., and Stevens M. M. (2017) Self-assembled 2D free-standing Janus nanosheets with single-layer thickness. J. Am. Chem. Soc. 139, 13592–13595 10.1021/jacs.7b06591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lu K., Jacob J., Thiyagarajan P., Conticello V. P., and Lynn D. G. (2003) Exploiting amyloid fibril lamination for nanotube self-assembly. J. Am. Chem. Soc. 125, 6391–6393 10.1021/ja0341642 [DOI] [PubMed] [Google Scholar]
- 36. Reches M., and Gazit E. (2006) Controlled patterning of aligned self-assembled peptide nanotubes. Nat. Nanotechnol. 1, 195–200 10.1038/nnano.2006.139 [DOI] [PubMed] [Google Scholar]
- 37. Koutsopoulos S., Unsworth L. D., Nagai Y., and Zhang S. (2009) Controlled release of functional proteins through designer self-assembling peptide nanofiber hydrogel scaffold. Proc. Natl. Acad. Sci. U.S.A. 106, 4623–4628 10.1073/pnas.0807506106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Childers W. S., Mehta A. K., Ni R., Taylor J. V., and Lynn D. G. (2010) Peptides organized as bilayer membranes. Angew. Chem. Int. Ed. Engl. 49, 4104–4107 10.1002/anie.201000212 [DOI] [PubMed] [Google Scholar]
- 39. Lakshmanan A., Zhang S., and Hauser C. A. (2012) Short self-assembling peptides as building blocks for modern nanodevices. Trends Biotechnol. 30, 155–165 10.1016/j.tibtech.2011.11.001 [DOI] [PubMed] [Google Scholar]
- 40. Zhao X., Pan F., Xu H., Yaseen M., Shan H., Hauser C. A., Zhang S., and Lu J. R. (2010) Molecular self-assembly and applications of designer peptide amphiphiles. Chem. Soc. Rev. 39, 3480–3498 10.1039/b915923c [DOI] [PubMed] [Google Scholar]
- 41. Guo C., Luo Y., Zhou R., and Wei G. (2012) Probing the self-assembly mechanism of diphenylalanine-based peptide nanovesicles and nanotubes. ACS Nano 6, 3907–3918 10.1021/nn300015g [DOI] [PubMed] [Google Scholar]
- 42. Wang D., Ha Y., Gu J., Li Q., Zhang L., and Yang P. (2016) 2D protein supramolecular nanofilm with exceptionally large area and emergent functions. Adv. Mater. 28, 7414–7423 10.1002/adma.201506476 [DOI] [PubMed] [Google Scholar]
- 43. Knowles T. P., Oppenheim T. W., Buell A. K., Chirgadze D. Y., and Welland M. E. (2010) Nanostructured films from hierarchical self-assembly of amyloidogenic proteins. Nat. Nanotechnol. 5, 204–207 10.1038/nnano.2010.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Reches M., and Gazit E. (2003) Casting metal nanowires within discrete self-assembled peptide nanotubes. Science 300, 625–627 10.1126/science.1082387 [DOI] [PubMed] [Google Scholar]
- 45. Rufo C. M., Moroz Y. S., Moroz O. V., Stöhr J., Smith T. A., Hu X., DeGrado W. F., and Korendovych I. V. (2014) Short peptides self-assemble to produce catalytic amyloids. Nat. Chem. 6, 303–309 10.1038/nchem.1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. You J., Li M., Ding B., Wu X., and Li C. (2017) Crab chitin-based 2D soft nanomaterials for fully biobased electric devices. Adv. Mater. 29 10.1002/adma.201606895 [DOI] [PubMed] [Google Scholar]
- 47. Gulseren G., Khalily M. A., Tekinaya A. B., and Guler M. O. (2016) Catalytic supramolecular self-assembled peptide nanostructures for ester hydrolysis. J. Mater. Chem. B 4, 4605–4611 10.1039/C6TB00795C [DOI] [PubMed] [Google Scholar]
- 48. Schladitz C., Vieira E. P., Hermel H., and Möhwald H. (1999) Amyloid-β-sheet formation at the air-water interface. Biophys. J. 77, 3305–3310 10.1016/S0006-3495(99)77161-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Maji S. K., Schubert D., Rivier C., Lee S., Rivier J. E., and Riek R. (2008) Amyloid as a depot for the formulation of long-acting drugs. PLoS Biol. 6, e17.10.1371/journal.pbio.0060017 10.1371/journal.pbio.0060017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gupta S., Chattopadhyay T., Pal Singh M., and Surolia A. (2010) Supramolecular insulin assembly II for a sustained treatment of type 1 diabetes mellitus. Proc. Natl. Acad. Sci. U.S.A. 107, 13246–13251 10.1073/pnas.1005704107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rudra J. S., Tian Y. F., Jung J. P., and Collier J. H. (2010) A self-assembling peptide acting as an immune adjuvant. Proc. Natl. Acad. Sci. U.S.A. 107, 622–627 10.1073/pnas.0912124107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jung J. P., Nagaraj A. K., Fox E. K., Rudra J. S., Devgun J. M., and Collier J. H. (2009) Co-assembling peptides as defined matrices for endothelial cells. Biomaterials 30, 2400–2410 10.1016/j.biomaterials.2009.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Davis M. E., Motion J. P., Narmoneva D. A., Takahashi T., Hakuno D., Kamm R. D., Zhang S., and Lee R. T. (2005) Injectable self-assembling peptide nanofibers create intramyocardial microenvironments for endothelial cells. Circulation 111, 442–450 10.1161/01.CIR.0000153847.47301.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tysseling-Mattiace V. M., Sahni V., Niece K. L., Birch D., Czeisler C., Fehlings M. G., Stupp S. I., and Kessler J. A. (2008) Self-assembling nanofibers inhibit glial scar formation and promote axon elongation after spinal cord injury. J. Neurosci. 28, 3814–3823 10.1523/JNEUROSCI.0143-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Holmes T. C., de Lacalle S., Su X., Liu G., Rich A., and Zhang S. (2000) Extensive neurite outgrowth and active synapse formation on self-assembling peptide scaffolds. Proc. Natl. Acad. Sci. U.S.A. 97, 6728–6733 10.1073/pnas.97.12.6728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hsieh P. C. H., Davis M. E., Gannon J., MacGillivray C., and Lee R. T. (2006) Controlled delivery of PDGF-BB for myocardial protection using injectable self-assembling peptide nanofibers. J. Clin. Invest. 116, 237–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sengupta U., Nilson A. N., and Kayed R. (2016) The role of amyloid-β oligomers in toxicity, propagation, and immunotherapy. EBioMedicine 6, 42–49 10.1016/j.ebiom.2016.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. White J. A., Manelli A. M., Holmberg K. H., Van Eldik L. J., and Ladu M. J. (2005) Differential effects of oligomeric and fibrillar amyloid-β 1–42 on astrocyte-mediated inflammation. Neurobiol. Dis. 18, 459–465 10.1016/j.nbd.2004.12.013 [DOI] [PubMed] [Google Scholar]
- 59. An J., Gou Y., Yang C., Hu F., and Wang C. (2013) Synthesis of a biocompatible gelatin functionalized graphene nanosheets and its application for drug delivery. Mater. Sci. Eng. C Mater. Biol. Appl. 33, 2827–2837 10.1016/j.msec.2013.03.008 [DOI] [PubMed] [Google Scholar]
- 60. Mitra S., Sasmal H. S., Kundu T., Kandambeth S., Illath K., Díaz Díaz D., and Banerjee R. (2017) Targeted drug delivery in covalent organic nanosheets (CONs) via sequential postsynthetic modification. J. Am. Chem. Soc. 139, 4513–4520 10.1021/jacs.7b00925 [DOI] [PubMed] [Google Scholar]
- 61. Li H., Fierens K., Zhang Z., Vanparijs N., Schuijs M. J., Van Steendam K., Feiner Gracia N., De Rycke R., De Beer T., De Beuckelaer A., De Koker S., Deforce D., Albertazzi L., Grooten J., Lambrecht B. N., and De Geest B. G. (2016) Spontaneous protein adsorption on graphene oxide nanosheets allowing efficient intracellular vaccine protein delivery. ACS Appl. Mater. Interfaces 8, 1147–1155 10.1021/acsami.5b08963 [DOI] [PubMed] [Google Scholar]
- 62. Shojaee M., Navaee F., Jalili-Firoozinezhad S., Faturechi R., Majidi M., and Bonakdar S. (2015) Fabrication and characterization of ovalbumin films for wound dressing applications. Mater. Sci. Eng. C Mater. Biol. Appl. 48, 158–164 10.1016/j.msec.2014.11.063 [DOI] [PubMed] [Google Scholar]
- 63. Liu T., Wang C., Gu X., Gong H., Cheng L., Shi X., Feng L., Sun B., and Liu Z. (2014) Drug delivery with PEGylated MoS2 nano-sheets for combined photothermal and chemotherapy of cancer. Adv. Mater. 26, 3433–3440 10.1002/adma.201305256 [DOI] [PubMed] [Google Scholar]
- 64. Chen W., Ouyang J., Liu H., Chen M., Zeng K., Sheng J., Liu Z., Han Y., Wang L., Li J., Deng L., Liu Y. N., and Guo S. (2017) Black phosphorus nanosheet-based drug delivery system for synergistic photodynamic/photothermal/chemotherapy of cancer. Adv. Mater. 29 10.1002/adma.201603864 [DOI] [PubMed] [Google Scholar]
- 65. O'Nuallain B., Shivaprasad S., Kheterpal I., and Wetzel R. (2005) Thermodynamics of Aβ(1–40) amyloid fibril elongation. Biochemistry 44, 12709–12718 10.1021/bi050927h [DOI] [PubMed] [Google Scholar]
- 66. Ansari M. A., Zubair S., Tufail S., Ahmad E., Khan M. R., Quadri Z., and Owais M. (2012) Ether lipid vesicle-based antigens impart protection against experimental listeriosis. Int. J. Nanomedicine 7, 2433–2447 10.2147/IJN.S25875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Azmi S., Srivastava S., Mishra N. N., Tripathi J. K., Shukla P. K., and Ghosh J. K. (2013) Characterization of antimicrobial, cytotoxic, and antiendotoxin properties of short peptides with different hydrophobic amino acids at “a” and “d” positions of a heptad repeat sequence. J. Med. Chem. 56, 924–939 10.1021/jm301407k [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.