VOLUME 289 (2017) PAGES 2745–2754
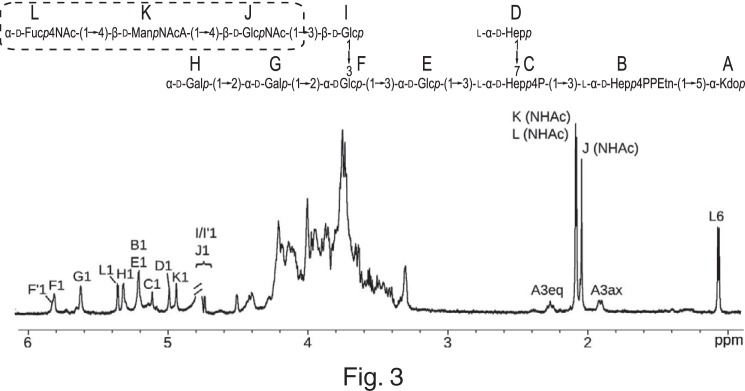
There was an error in the structure of Shigella sonnei phase II ECALPS. The anomeric configuration of residue J should be assigned as a β anomer [→4)-β-d-GlcpNAc-(1→] based on its chemical shifts (Tables 1 and 2) and JH1,C1 (162 Hz). The correction results in an inverted anomeric configuration of the d-GlcNAc residue in the first ECA unit linked to the core oligosaccharide, whereas an α-configuration is characteristic for polymeric chain (Fig. 3). Chemical shifts of the residue J were in agreement with predictions carried out by the CASPER program (http://www.casper.organ.su.se/casper/),1 where professor Göran Widmalm is kindly acknowledged for the error identification (1, 2). This correction does not affect the general results and conclusions of this work.
Table 1.
1H and 13C NMR chemical shifts of fraction IV containing core OS substituted by ECA trisaccharide ([ECA]-dLOS) isolated from S. sonnei phase II LOS
| Residue | Description | Chemical shift (ppm) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| H-1/C-1 | H-2/C-2 | H-3(H3ax,eq)/C-3 | H-4/C-4 | H-5/C-5 | H-6a, H-6b/C-6 | H-7a, H-7b/C-7 (NHAc) | H-8a, H-8b/C-8 [C(O)] | ||
| A | →5)-α-Kdop | NDa | −/96.3 | (1.90, 2.25)/34.1 | 4.11/66.3 | 4.17/73.3 | 3.69/69.7 | 3.80/72.6 | 3.47, 3.93/64.7 |
| B | →3)-l-α-d-Hepp4PPEtn-(1→ | 5.20/100.1 | 4.01/71.6 | 4.08/78.5 | 4.61/72.3 | 4.22/72.0 | 4.10/69.3 | 3.72, 3.72/63.8 | |
| C | →3,7)-l-α-d-Hepp4P-(1→ | 5.10/103.5 | 4.38/70.6 | 4.12/79.8 | 4.40/69.4 | 3.80/73.2 | 4.23/68.5 | 3.58, 3.75/68.4 | |
| D | l-α-d-Hepp-(1→ | 4.98/100.2 | 3.93/70.7 | 3.87/71.4 | 3.84/66.9 | 3.61/71.9 | 4.04/69.5 | 3.65, 3.72/63.7 | |
| E | →3)-α-d-Glcp-(1→ | 5.20/102.0 | 3.66/71.0 | 4.07/76.7 | 3.77/71.2 | 3.91/73.1 | 3.79,3.92/60.5 | ||
| F | →2,3)-α-d-Glcp-(1→ | 5.80/95.3 | 3.87/73.3 | 4.17/78.7 | 3.56/68.7 | 4.10/71.9 | 3.78,3.95/61.0 | ||
| F′b | →2,3)-α-d-Glcp-(1→ | 5.81/95.1 | 3.88/73.3 | 4.19/78.8 | 3.57/68.7 | 4.11/72.0 | 3.79,3.96/61.0 | ||
| G | →2)-α-d-Galp-(1→ | 5.61/92.1 | 3.98/73.2 | 4.19/68.9 | 3.98/70.7 | 4.13/72.0 | 3.74,3.74/61.9 | ||
| H | α-d-Galp-(1→ | 5.31/96.6 | 3.85/69.0 | 3.95/70.1 | 3.99/70.1 | 4.13/72.0 | 3.75,3.75/61.9 | ||
| I | →3)-β-d-Glcp-(1→ | 4.73/103.1 | 3.39/73.6 | 3.68/85.4 | 3.49/68.9 | 3.44/76.3 | 3.72,3.89/61.4 | ||
| I′c | β-d-Glcp-(1→ | 4.75/103.1 | 3.33/73.9 | 3.51/76.6 | 3.40/70.4 | 3.45/76.6 | 3.73,3.91/61.4 | ||
| J | →4)-β-d-GlcpNAc-(1→ | 4.78/102.3 | 3.75/56.3 | 3.74/72.7 | 3.68/79.5 | 3.54/75.2 | 3.86,3.70/60.9 | (2.03/23.0) | [175.5] |
| K | →4)-β-d-ManpNAcA-(1→ | 4.93/99.7 | 4.49/54.2 | 4.07/73.2 | 3.82/74.8 | 3.86/77.2 | −/175.1 | (2.07/22.6) | [176.2] |
| L | α-d-Fucp4NAc-(1→ | 5.35/99.5 | 3.64/69.3 | 4.00/69.1 | 4.20/54.6 | 4.18/66.5 | 1.06/16.2 | (2.07/22.6) | [176.3] |
| PPEtn | 4.20/63.1 | 3.29/40.7 | |||||||
a ND, not determined.
b Residue F′ is a variant of residue F present in the core OS that is devoid of ECA trisaccharide.
c Residue I′ is a terminal residue I present in the core OS that is devoid of ECA trisaccharide.
Table 2.
Selected inter-residue NOE and 3JH,C connectivities from the anomeric atoms of ([ECA]-dLOS) dodecasaccharide isolated from S. sonnei phase II LOS
Data indicating the covalent linkage between ECA and LOS are shown in boldface type.
| Residue | Description | Atom δH/δC (ppm) | Connectivity to |
Inter-residue atom/residue | |
|---|---|---|---|---|---|
| δC | δH | ||||
| B | →3)-l-α-d-Hepp4PPEtn-(1→ | 5.20/100.1 | − | 4.17a | H-5 of A |
| C | →3,7)-l-α-d-Hepp4P-(1→ | 5.10/103.5 | 78.5 | 4.08a | C-3, H-3 of B |
| D | l-α-d-Hepp-(1→ | 4.98/100.2 | 68.5 | 3.59/3.74a | C-7, H-7a, H-7b of C |
| E | →3)-α-d-Glcp-(1→ | 5.20/102.0 | − | 4.12 | H-3 of C |
| F | →2,3)-α-d-Glcp-(1→ | 5.80/95.3 | − | 4.07 | H-3 of E |
| F′ | →2,3)-α-d-Glcp-(1→b | 5.81/95.1 | − | 4.07 | H-3 of E |
| G | →2)-α-d-Galp-(1→ | 5.61/92.1 | − | 3.87a | H-2 of F |
| H | α-d-Galp-(1→ | 5.31/96.6 | − | 3.97a | H-2 of G |
| I | →3)-β-d-Glcp-(1→ | 4.73/103.1 | 78.7 | 4.17 | H-3 of F |
| I′ | β-d-Glcp-(1→c | 4.75/103.1 | 78.8 | 4.19 | H-3 of F′ |
| J | →4)-β-d-GlcpNAc-(1→ | 4.78/102.3 | 85.3 | 3.68 | H-3 of I |
| K | →4)-β-d-ManpNAcA-(1→ | 4.93/99.7 | 79.4 | 3.69a | H-4 of J |
| L | α-d-Fucp4NAc-(1→ | 5.35/99.5 | 74.7 | 3.81 | H-4 of K |
a Value represents NOE connectivities only.
b Residue F′ is a variant of residue F present in the core OS that is devoid of ECA trisaccharide.
c Residue I′ is a terminal residue I present in the core OS that is devoid of ECA trisaccharide.
Footnotes
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party-hosted site.
References
- 1. Lundborg M., and Widmalm G. (2011) Structure analysis of glycans by NMR chemical shift prediction. Anal. Chem. 83, 1514–1517 10.1021/ac1032534 [DOI] [PubMed] [Google Scholar]
- 2. Jansson P. E., Stenutz R., and Widmalm G. (2006) Sequence determination of oligosaccharides and regular polysaccharides using NMR spectroscopy and a novel Web-based version of the computer program CASPER. Carbohydr. Res. 341, 1003–1010 10.1016/j.carres.2006.02.034 [DOI] [PubMed] [Google Scholar]