Abstract
Interleukin-like EMT inducer (ILEI, FAM3C) is a secreted factor that contributes to the epithelial-to-mesenchymal transition (EMT), a cell-biological process that confers metastatic properties to a tumor cell. However, very little is known about how ILEI is regulated. Here we demonstrate that ILEI is an in vivo regulator of melanoma invasiveness and is transcriptionally up-regulated by the upstream stimulatory factor-1 (USF-1), an E-box–binding, basic-helix-loop-helix family transcription factor. shRNA-mediated knockdown of ILEI in melanoma cell lines attenuated lung colonization but not primary tumor formation. We also identified the mechanism underlying ILEI transcriptional regulation, which was through a direct interaction of USF-1 with the ILEI promoter. Of note, stimulation of endogenous USF-1 by UV-mediated activation increased ILEI expression, whereas shRNA-mediated USF-1 knockdown decreased ILEI gene transcription. Finally, we report that knocking down USF-1 decreases tumor cell migration. In summary, our work reveals that ILEI contributes to melanoma cell invasiveness in vivo without affecting primary tumor growth and is transcriptionally up-regulated by USF-1.
Keywords: epithelial-mesenchymal transition (EMT), melanoma, transcription factor, mRNA, cytokine, FAM3C, ILEI, interleukin-like EMT inducer, phenotype switching, USF-1
Introduction
The three most commonly mutated genes in melanoma are BRAF, NRAS, and NF1, all components of the RAS–RAF–MEK2–ERK signaling pathway (subsequently referred to as the MEK signaling pathway) (1). Accordingly, MEK signaling plays a major role in melanoma biology by regulating diverse processes such as pigmentation, apoptosis, and senescence (2–9). At a molecular level, MEK signaling affects many transcription factors including the basic-helix-loop-helix leucine zipper (bHLH LZip) transcription factor micropthalmia-associated transcription factor (MITF) (3, 7). MITF binds to E-box motifs (CATGTG) and activates the transcription of the pigment producing gene PMEL, which encodes the premelanosome protein, and cell cycle genes such as CDK2, which encodes the cyclin-dependent kinase 2 (10, 11). In addition to MITF, MEK signaling affects other bHLH LZip family transcription factors such as upstream stimulatory factor 1 (USF-1) (12, 13). USF-1 binds to E-box motifs (CACGTG) and activates pigmentation genes in response to UV (14, 15).
Interleukin-like EMT inducer (ILEI, FAM3C) is a secreted cytokine-like molecule that contributes to the epithelial-to-mesenchymal transition (EMT) (16). EMT is a cell-biological process in which epithelial cells with apical–basal polarity undergo cytoskeletal rearrangement to become motile mesenchymal cells (17). This process is thought to contribute to chemoresistance and metastasis. Although melanoma cells do not undergo a traditional EMT, they utilize a similar process known as phenotype switching. This is a process in which melanoma cells interconvert between a proliferative MITF-high state and an invasive MITF-low state (18–24). Recently, we have described a contribution of ILEI to the invasive MITF-low phenotype in vitro (25). Additionally, we showed that phenotype switching between the proliferative MITF-high and invasive MITF-low state modulates ILEI mRNA expression. The molecular regulation of ILEI has focused on post-transcriptional mechanisms including translational regulation by hnRNP-E1–TGF-β and proteolytic processing by plasmin (26–30). However, not much is known about transcriptional regulation of ILEI.
Here, we report that ILEI contributes to melanoma cell line invasiveness in vivo and build on our previous finding that vemurafenib (BRAFi) decreases ILEI mRNA expression to characterize the molecular details of ILEI transcriptional regulation by USF-1 (25).
Results and discussion
ILEI regulates lung colonization but not primary tumor growth
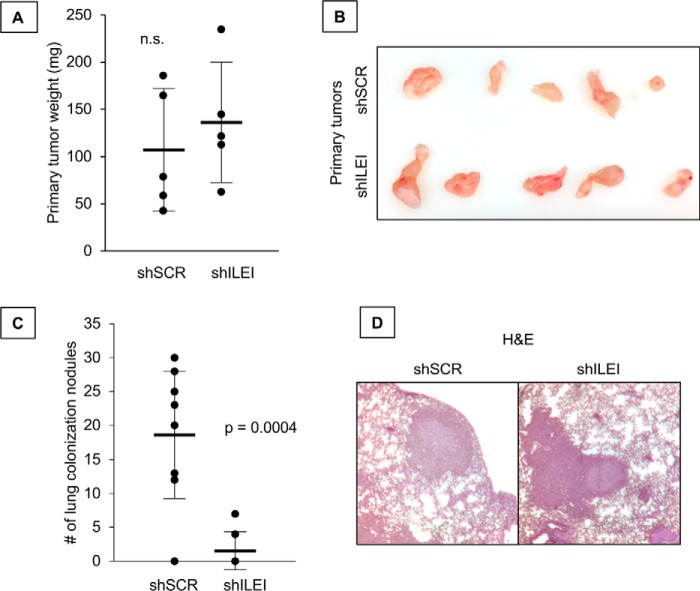
Previously we found that knockdown of ILEI contributes to melanoma invasiveness in vitro, whereas ILEI had no effect on proliferation (25). Here we used the same ILEI-modulated cell lines and conducted in vivo flank and tail vein injections to measure primary tumor formation or lung colonization, respectively. In the flank injection experiment we found no significant difference in primary tumor growth (Fig. 1, A and B), but we observed in the tail vein injection experiment that ILEI knockdown significantly attenuated lung colonization (Fig. 1C). We did not observe a difference in the size of the lung colonies formed by either shSCR or shILEI cell lines, suggesting that the ability to invade the lung was affected rather than the ability to grow metastatic colonies in the lung (Fig. 1D). Based on these results we conclude that ILEI specifically regulates melanoma invasiveness in vivo without affecting primary tumor growth.
Figure 1.
ILEI regulates lung colonization but not primary tumor growth. A, flank injection of 1205Lu cells expressing shSCR or shILEI. n = 5, bar indicates mean ± S.D., n.s. indicates p > 0.05 by Student's t test as compared with shSCR. B, primary tumors from flank injection experiments. C, tail vein injection of 1205Lu cells expressing shSCR or shILEI. n = 9 for shSCR and n = 7 for shILEI, bar indicates mean ± S.D., p value indicated by Student's t test as compared with shSCR. D, representative images of H&E-stained lung nodules from either shSCR or shILEI.
Vemurafenib inhibits ILEI expression
To date there are three known mechanisms of ILEI regulation: the first is translational regulation of ILEI by TGF-β–AKT2–hnRNP-E1, the second is degradation of ILEI by the ubiquitin/proteasome system, and the third is an autophagy-mediated increase in ILEI protein expression (16, 25, 26, 30–33). Nothing has been reported about the mechanistic basis of ILEI transcription. None of the previously established mechanisms appears to be responsible for ILEI regulation by vemurafenib (25). Herein, we sought to determine the mechanism of ILEI transcriptional regulation.
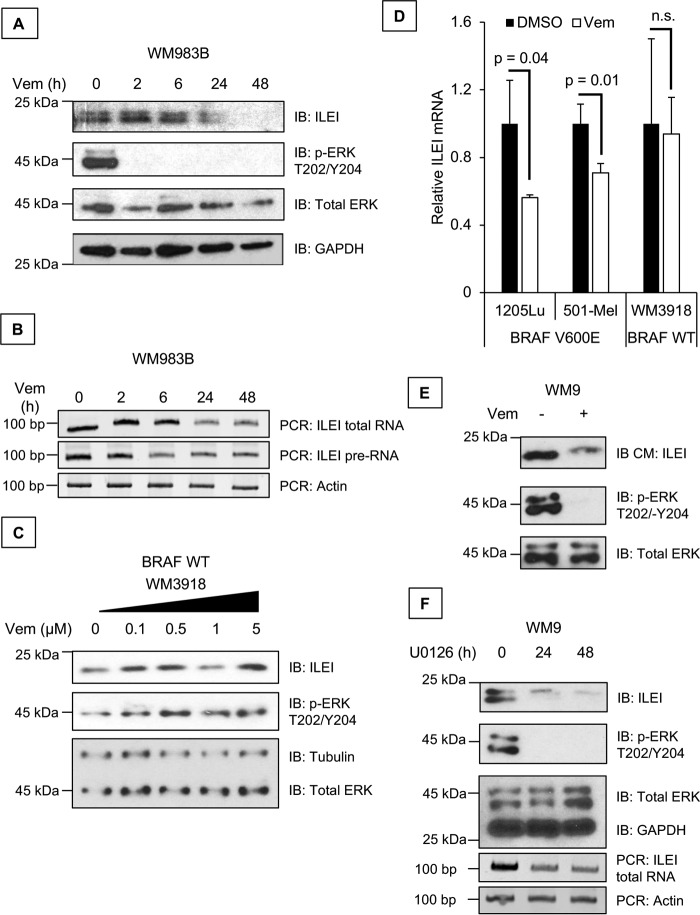
First, we treated WM983B melanoma cell lines with vemurafenib, an inhibitor of V600E BRAF, and observed that ILEI protein and mRNA expression decreased at 24 h (Fig. 2, A and B) (25). We also conducted RT-PCR using primers targeting an intronic sequence of ILEI to amplify ILEI pre-RNA (Fig. 2B). If ILEI mRNA levels are regulated post-transcriptionally by miRNAs, we expect that ILEI pre-RNA should not be affected by vemurafenib. However, if ILEI mRNA levels are regulated transcriptionally, we expect that ILEI pre-RNA should go down upon vemurafenib treatment. We saw that vemurafenib treatment decreased both ILEI total RNA and pre-RNA, suggesting that vemurafenib affected ILEI transcription (Fig. 2B). To confirm that these findings were not due to off-target effects of vemurafenib we conducted a control experiment in which we used the BRAF WT WM3918 melanoma cells. Vemurafenib is a specific inhibitor of mutant BRAF, and it does not affect WT BRAF (34). Therefore, we treated BRAF WT WM3918 melanoma cells with vemurafenib, which should retain all the nonspecific effects of vemurafenib without the BRAF-specific effects. We found that vemurafenib did not affect ILEI protein expression or ERK phosphorylation in BRAF WT WM3918 melanoma cells (Fig. 2C). We confirmed these findings by a quantitative method using real-time qPCR and found that vemurafenib decreased ILEI expression in BRAF mutant 501-Mel and 1205Lu cells but not in BRAF WT WM3918 cells (Fig. 2D). Considering that ILEI is a secreted cytokine, we wanted to confirm the physiological relevance of vemurafenib-mediated ILEI inhibition by conducting immunoblot analysis of the conditioned medium. These results confirmed that vemurafenib decreased secreted ILEI levels (Fig. 2E). Finally, to further determine whether this effect is specific for the MEK pathway we used a second inhibitor of the MEK pathway (U0126, MEKi) and observed decreased ILEI expression by U0126 (Fig. 2F). Based on these results we conclude that vemurafenib inhibits ILEI mRNA expression, and that this effect depends on the presence of oncogenic BRAF V600E mutation.
Figure 2.
Vemurafenib inhibits ILEI expression. A, immunoblot (IB) analysis of ILEI, p-ERK, ERK, and GAPDH levels in WM983B melanoma cell lines treated for 0 to 48 h with vemurafenib (1 μm). B, RT-PCR analysis of ILEI, ILEI intron indicates that PCR primers targeted the pre-RNA but not mRNA of the ILEI gene, and Actin levels in WM983B melanoma cell lines treated for 0 to 48 h with vemurafenib (1 μm). C, immunoblot analysis of ILEI, p-ERK, ERK, and Tubulin levels in WM3918 melanoma cell lines treated for 24 h with vemurafenib (0–5 μm). D, bar diagram showing quantitative RT-PCR analysis of ILEI levels in 501-Mel, 1205Lu, or WM3918 melanoma cell lines treated for 24 h with vemurafenib (1 μm). n = 3, mean ± S.D., p value indicated by Student's t test as compared with vehicle treatment, transcript values are normalized to GAPDH. E, immunoblot analysis of ILEI, p-ERK, and ERK levels in WM9 melanoma cell lines treated for 0 or 24 h with vemurafenib (1 μm). IB CM indicates that the serum-free medium condition for 24 h during vemurafenib treatment was harvested and TCA precipitated for immunoblot analysis. F, immunoblot and RT-PCR analysis of ILEI, p-ERK, ERK, GAPDH, and actin levels in WM9 melanoma cell lines treated for 0 to 48 h with U0126 (MEKi, 10 μm).
Vemurafenib inhibits ILEI at the transcriptional level
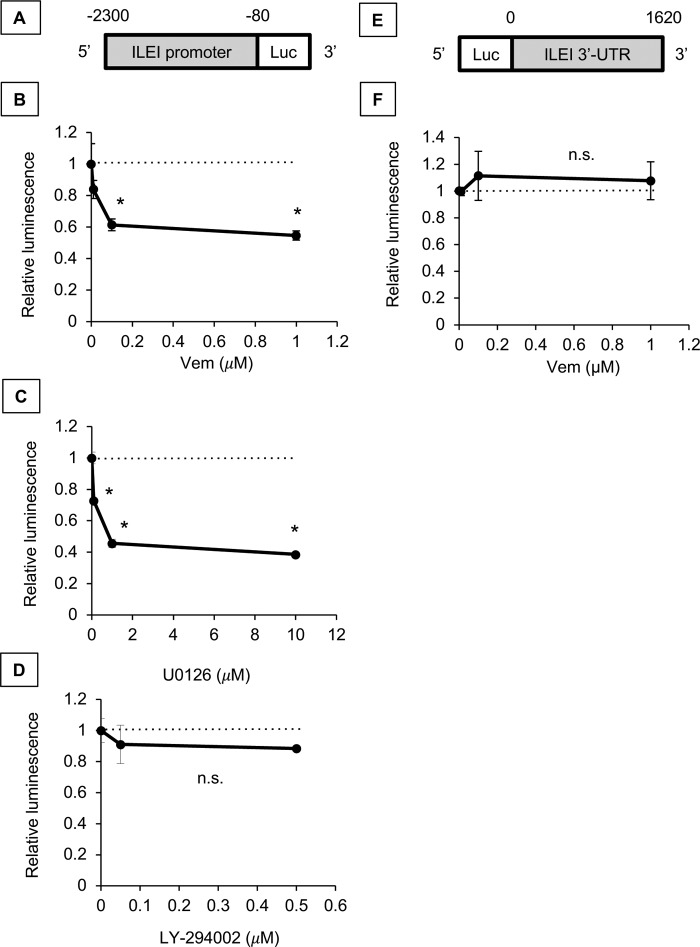
We cloned the ILEI promoter from 2,300 bp upstream of the transcription start site (TSS) to 80 bp upstream of the TSS into pBV-Luc (Fig. 3A) (35). We transfected 501-Mel cells with the ILEI promoter luciferase reporter construct along with a control Renilla construct and treated the cells with vemurafenib (Fig. 3B). We found that vemurafenib decreased the ILEI promoter activity in a dose-dependent manner. As further controls we tested the ILEI promoter construct with the MEK inhibitor U0126 and found that U0126 decreased promoter activity (Fig. 3C). However, when we tested the ILEI promoter construct with the PI3K inhibitor LY-294002, we found that promoter activity was not affected (Fig. 3D). Additionally, we cloned the ILEI 3′-UTR from the stop codon to the end of the mRNA at 1,620 bp into pmirGLO Dual Luciferase (Fig. 3E). We transfected 501-Mel cells with the ILEI 3′-UTR construct and treated the cells with vemurafenib (Fig. 3F). We found that vemurafenib did not affect the ILEI 3′-UTR construct. From these experiments we concluded that vemurafenib regulates ILEI mRNA at the transcriptional level.
Figure 3.
Vemurafenib inhibits ILEI expression at the transcriptional level. A, design of ILEI promoter reporter construct. The promoter reporter construct spans 2300 bp upstream of the TSS to 80 bp upstream of the TSS. B, luciferase assay of ILEI promoter reporter construct in 501-Mel melanoma cell lines treated for 24 h with vemurafenib (BRAFi, indicated concentration). n = 3, mean ± S.D., * indicates p < 0.01 by Student's t test as compared with vehicle treatment. C, luciferase assay of ILEI promoter reporter construct in 501-Mel melanoma cell lines treated for 24 h with U0126 (MEKi, indicated concentration). n = 3, mean ± S.D., * indicates p < 0.01 by Student's t test as compared with vehicle treatment. D, luciferase assay of ILEI promoter reporter construct in 501-Mel melanoma cell lines treated for 24 h with U0126 (MEKi, indicated concentration). n = 3, mean ± S.D., * indicates p < 0.01 by Student's t test as compared with vehicle treatment. E, design of ILEI 3′-UTR reporter construct. The 3′-UTR reporter construct spans from the end of the coding sequence to 1620 bp downstream. F, luciferase assay of the ILEI 3′-UTR reporter construct in 501-Mel melanoma cell lines treated for 24 h with vemurafenib (indicated concentration). n = 3, mean ± S.D., n.s. indicates p > 0.05 by Student's t test as compared with vehicle treatment.
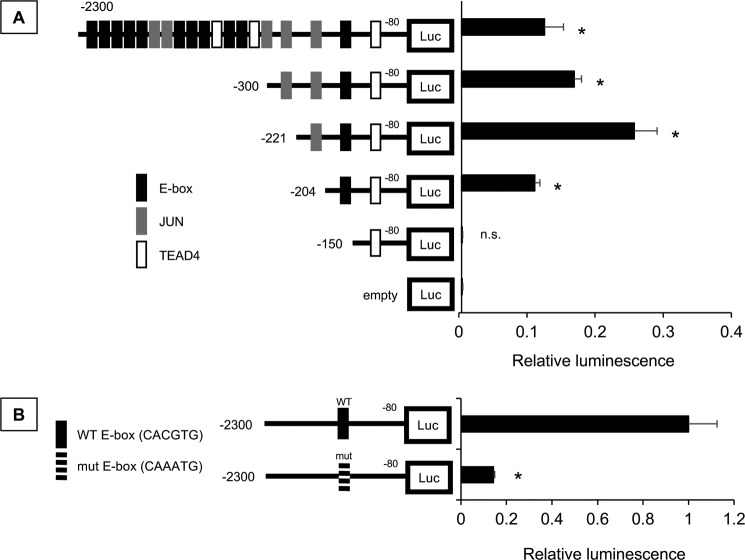
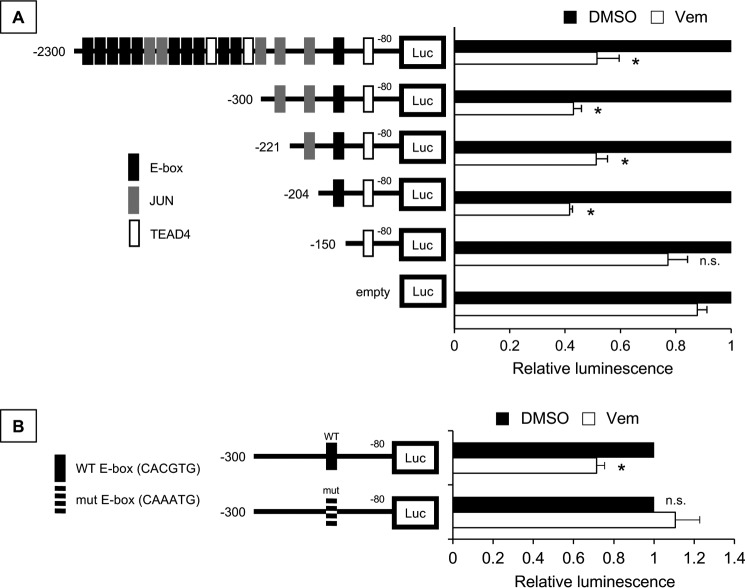
Because vemurafenib regulates ILEI transcription, we hypothesized that a cis element in the ILEI promoter region regulates this phenomenon. We analyzed a 2,300-bp sequence at the ILEI promoter from the TSS for various transcription factor motifs using the JASPAR database (36). Previously we had found that ILEI is highly expressed in MITF-low invasive melanoma cell lines, so we focused our transcription factor search on regulators of the MITF-low invasive state (E-box (ZEB1/2), JUN, and TEAD4) (20, 21, 23, 37–39). We found 11 putative E-box, 5 JUN, and 3 TEAD4 sites in the ILEI promoter, and generated successive 5′-deletions of the ILEI promoter reporter (Fig. 4A). We transfected 501-Mel cells with these constructs and observed that the truncation from −204 to −150 reduced the luciferase activity to empty promoter control levels. There was an E-box motif in this region, so we next wanted to know if this E-box was specifically important for ILEI promoter activity or if any E-box would suffice. Thus, we used our longest ILEI promoter reporter (−2300 to −80) and specifically mutated the E-box consensus site at −163 from CACGTG to CAAATG. Again, we observed a marked inhibition in luciferase activity (Fig. 4B). From these experiments we concluded that an E-box 163 bp upstream of the ILEI TSS is critical to basal ILEI promoter activity.
Figure 4.
Proximal E-box contributes ILEI promoter activity. A, luciferase assay of various length ILEI promoter reporter constructs in 501-Mel melanoma cell lines. n = 3, mean ± S.D., p value indicated by Student's t test as compared with the empty promoterless reporter. B, luciferase assay of WT and E-box mutant (CACGTG to CAAATG) ILEI promoter reporter constructs (−2300/−80) in 501-Mel melanoma cell lines. Black bars indicate WT and white bars E-box mutant. n = 3, mean ± S.D., p value was indicated by Student's t test comparing E-box mutant to WT.
Next, we wanted to know the role of this E-box motif in the vemurafenib-mediated regulation of ILEI expression. We transfected 501-Mel cells with the ILEI promoter truncation constructs and treated the cells with vemurafenib (Fig. 5A). We found that truncation of bp −204 to −150, which includes an E-box motif, eliminated ILEI promoter vemurafenib responsiveness. The −204 to −150 region of the ILEI promoter contains 48 bp in addition to the E-box, so we mutated the E-box in the −300/−80 construct. We sequenced the construct and analyzed the sequence in the JASPAR database to find that our mutations abolished all putative binding sites in this region (data not shown). We conducted luciferase assays and observed that mutation of the E-box similarly eliminated vemurafenib responsiveness (Fig. 5B). From these experiments we concluded that vemurafenib inhibits ILEI at the transcriptional level through an E-box 163 bp upstream of the ILEI TSS.
Figure 5.
Proximal E-box contributes ILEI promoter vemurafenib responsiveness. A, luciferase assay of various length ILEI promoter reporter constructs in 501-Mel melanoma cell lines. Black bars indicate control DMSO treatment and white bars indicate 24 h treatment with vemurafenib (1 μm). Values are normalized to DMSO treatment of the particular reporter construct. n = 3, mean ± S.D., p value was indicated by Student's t test as compared with vemurafenib treatment of the empty promoterless reporter. B, luciferase assay of WT and E-box mutant (CACGTG to CAAATG) ILEI promoter reporter constructs (−300/−80) in 501-Mel melanoma cell lines. Black bars indicate control DMSO treatment and white bars indicate 24 h treatment with vemurafenib (1 μm). n = 3, mean ± S.D., p value was indicated by Student's t test comparing vemurafenib to DMSO treatment.
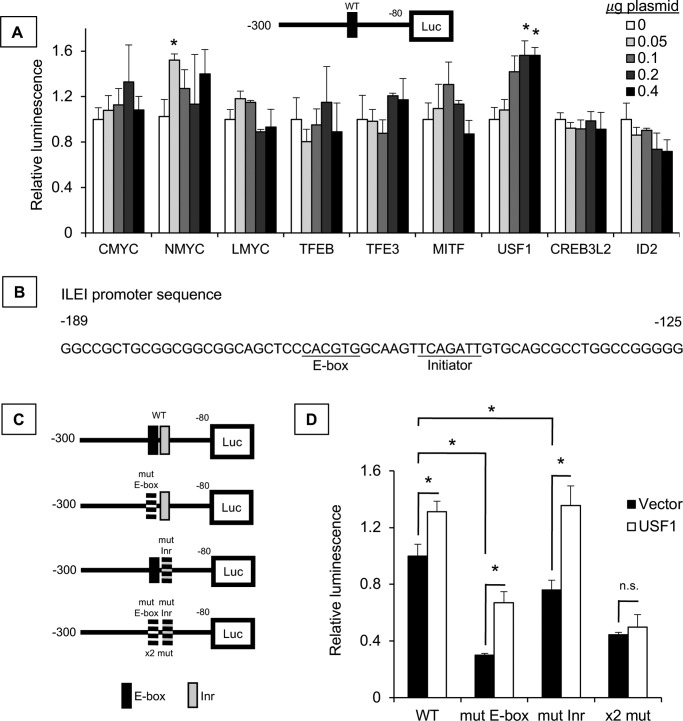
USF-1 directly regulates ILEI transcription
Considering the importance of the E-box motif to ILEI expression, we wanted to identify the trans-acting factor regulating this effect. To this end, we overexpressed different transcription factors known to bind E-box motifs and found that USF-1 induced ILEI promoter activity, but not c-MYC, N-MYC, l-MYC, TFEB, TFE3, MITF, CREB3L2, and ID2 (Fig. 6A). USF-1 activates transcription through the E-box motif, but also through a pyrimidine-rich initiator sequence (Inr) (40). The ILEI promoter sequence has an Inr sequence 6 bp downstream of the E-box (Fig. 6B). Thus, we generated Inr mutant ILEI promoter constructs in addition to our E-box mutants to see if USF-1 regulated ILEI promoter activity directly through the USF-1–binding sites (Fig. 6C). We observed that USF-1 induced ILEI promoter activity in the WT promoter, the E-box mutant promoter, the initiator mutant promoter, but not the E-box and Inr double mutant promoter (Fig. 6D). From these experiments we concluded that USF-1 directly regulates ILEI promoter activity.
Figure 6.
USF-1 directly regulates ILEI transcription. A, luciferase assay of the ILEI promoter reporter construct (−300/−80) in 501-Mel melanoma cell lines. 0, 0.05, 0.1, 0.2, and 0.4 μg of experimental vector and a corresponding 0.4, 0.2, 0.15, 0.1, or 0 μg of empty vector were transfected for 24 h. The increased darkness of the bars indicates increased experimental vector. Luminescence is normalized to vector control for each transcription factor. n = 3, mean ± S.D., * indicates p < 0.01 by Student's t test as compared with vector transfection. B, ILEI promoter sequence from −189 to −125 highlighting E-box and initiator (Inr) motifs. C, various ILEI promoter luciferase reporter constructs either WT or mutant for the E-box or the Inr. D, luciferase assay of ILEI promoter reporter constructs in 501-Mel melanoma cell lines. Black bars indicate vector overexpression and white bars indicate USF-1 overexpression. n = 3, mean ± S.D., p value indicated by Student's t test as compared with vector.
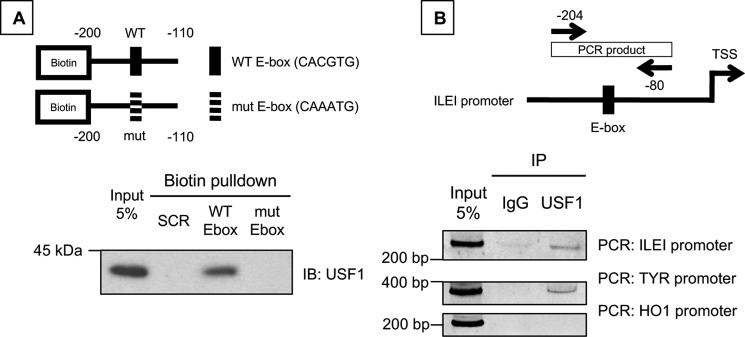
We wanted to further prove a direct role of USF-1 in ILEI regulation so we pursued a potential interaction between the ILEI promoter and USF-1. We conducted streptavidin pulldown experiments using 5′-biotin–tagged ILEI promoter constructs either WT or mutant for the E-box (200 to 110 bp upstream of ILEI TSS, Fig. 7A) and 501-Mel melanoma cell nuclear extract. We found that USF-1 can be isolated from cell nuclear extract using the WT ILEI promoter, but not the E-box mutant ILEI promoter, or a control random 60-bp oligonucleotide (SCR, Fig. 7A).
Figure 7.
USF-1 interacts with the ILEI promoter sequence. A, 5′-biotin–tagged ILEI promoter oligonucleotide constructs with WT or mutant E-box. Biotin pulldown analysis of nuclear extracts from 501-Mel melanoma cells, followed by immunoblot for USF-1. B, PCR primers flanking the ILEI promoter E-box used in ChIP analysis. ChIP analysis of 501-Mel melanoma cell lines immunoprecipitated with control IgG or USF-1 antibody. PCR analysis conducted with primers targeting FAM3C, TYR, or HO1 promoter.
Interestingly, the E-box mutation was sufficient to abolish binding by the streptavidin pulldown experiment, but the E-box mutation was not sufficient to abolish USF-1-mediated induction of the ILEI promoter reporter (Fig. 6D). We speculate either that 1) USF-1 still binds the ILEI promoter at levels that are undetectable by our streptavidin pulldown assay, or 2) endogenous USF-1 binds only to the E-box but overexpressed exogenous USF-1 provides excess USF-1 to bind both the E-box and the Inr. This is supported by the finding that basal ILEI promoter activity is affected more by the mutation of the E-box than the Inr (Fig. 6D). This is also consistent with previously published findings that show the E-box is the high-affinity USF-1–binding site, whereas the Inr is the low-affinity binding site (41).
To confirm that the interaction between USF-1 and the ILEI promoter was physiologically relevant, we conducted ChIP using USF-1 antibody and PCR primers flanking the ILEI promoter E-box (Fig. 7B). We confirmed the efficacy of our assay by showing that USF-1 IP can detect a segment of the TYR promoter (15), and we detected an interaction between USF-1 and the ILEI promoter (Fig. 7B). However, we found that a segment of the HO1 promoter, which is a USF-1 interaction reported in epithelial cells, was not detected in our melanoma cells (42). From these experiments we concluded that USF-1 directly interacts with the ILEI promoter and this interaction is E-box dependent.
USF-1 in vemurafenib-regulated ILEI expression
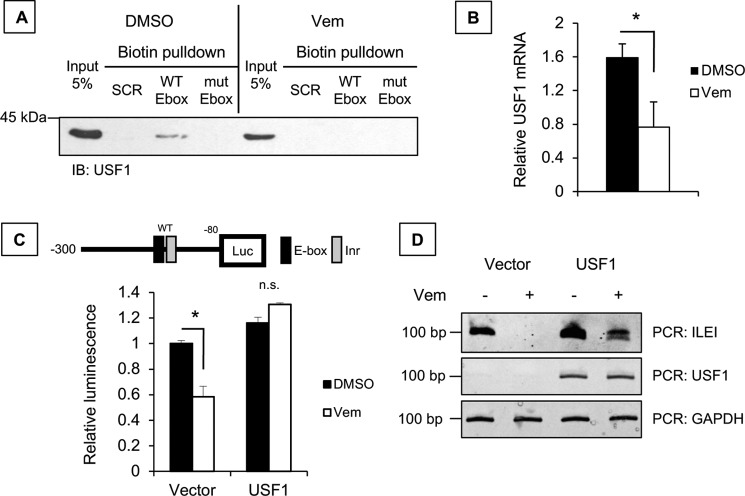
Thus far we have shown that vemurafenib inhibits ILEI transcription through an E-box motif, and that USF-1 directly regulates ILEI transcription through the same E-box motif. Next, we wanted to understand the role of USF-1 in vemurafenib-mediated regulation of ILEI transcription.
For this we employed the streptavidin pulldown assay using 5′-biotin–tagged ILEI promoter constructs WT or E-box mutant in cells treated with vemurafenib. First, we validated the binding of USF-1 to the ILEI promoter (DMSO condition) and showed that vemurafenib treatment abolished USF-1 interaction with the ILEI promoter (Fig. 8A). Importantly, we noticed by comparing the input lanes that USF-1 expression is decreased by vemurafenib treatment. We used qPCR to confirm that vemurafenib represses the mRNA levels of USF-1 (Fig. 8B).
Figure 8.
USF-1 in vemurafenib-regulated ILEI expression. A, biotin pulldown analysis of nuclear extracts from DMSO or vemurafenib-treated 501-Mel melanoma cells, followed by immunoblot for USF-1. B, qPCR analysis of USF-1 in Sk-Mel-28 melanoma cells treated with DMSO or vemurafenib (24 h, 0 μm). C, luciferase assay of the WT ILEI promoter reporter construct (−300/−80) in 501-Mel melanoma cell lines with vector or USF-1 overexpression and DMSO or vemurafenib treatment (1 μm). Black bars indicate DMSO and white bars indicate vemurafenib. n = 3, mean ± S.D., p value indicated by Student's t test as compared with DMSO treatment. D, PCR analysis of ILEI, USF-1, and GAPDH in 501-Mel melanoma cell lines overexpressing vector or USF-1 and treated with vemurafenib (24 h, 5 μm).
If vemurafenib inhibits ILEI expression by repressing USF-1, overexpression of USF-1 should rescue vemurafenib-mediated inhibition of ILEI. Thus, we overexpressed USF-1, treated with vemurafenib, and tested ILEI promoter activity. We found vemurafenib regulation of ILEI expression in vector–overexpressing cells, but not in USF-1–overexpressing cells (Fig. 8C). In fact, we observed that vemurafenib treatment actually induced ILEI promoter activity in the context of USF-1 overexpression. We propose the following explanation: in the absence of exogenous USF-1, vemurafenib decreases USF-1 expression to inhibit ILEI transcription (Fig. 8B), but in the presence of exogenous USF-1, vemurafenib activates p38 kinase to activate USF-1 and counterintuitively activate ILEI transcription (15, 20).
Finally, we confirmed these results with the endogenous ILEI by overexpressing USF-1, treating with vemurafenib, and conducting PCR analysis. Although the rescue was not complete, we observed that USF-1 overexpression rescues vemurafenib inhibition of ILEI (Fig. 8D). From these experiments we concluded that the mechanism of vemurafenib-mediated inhibition of ILEI is through down-regulation of USF-1 mRNA.
Role of endogenous USF-1 in ILEI expression
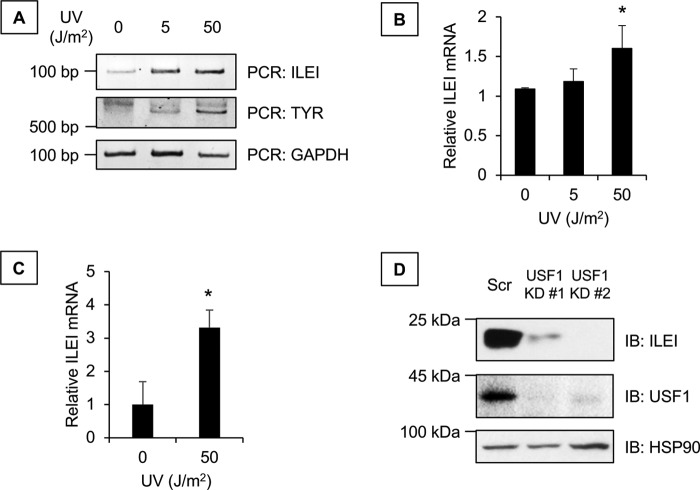
We wanted to assess if endogenous USF-1 had a role in ILEI expression. USF-1 is known to be regulated by stress-mediated p38 MAPK. Mechanistically, p38 phosphorylates USF-1 on threonine 153, which activates its transcriptional activity for target genes, including TYR (15). Thus, we used UV treatment as a model of endogenous USF-1 activation and observed that UV increased ILEI mRNA both by RT- and qPCR (Fig. 9, A and B). Furthermore, we wanted to know if UV–USF-1–ILEI regulation was intact in nonmelanoma cell lines. We used HMLE human mammary epithelial cells and observed that UV increased ILEI mRNA by qPCR (Fig. 9C). In addition to activation of endogenous USF-1, we wanted to test the effect of inhibiting endogenous USF-1. We used two different shRNA molecules specific for USF1 and observed that knockdown of endogenous USF-1 inhibits ILEI expression (Fig. 9D). From these experiments we concluded that endogenous USF-1 regulates ILEI expression.
Figure 9.
Effect of endogenous USF-1 on ILEI expression. A, PCR analysis for ILEI, tyrosinase, or GAPDH of 501-Mel melanoma cells 24 h after treatment with 0, 5, or 50 J/m2 of UV. B, qPCR analysis of ILEI in 501-Mel melanoma cells 24 h after treatment with 0, 5, or 50 J/m2 of UV. n = 3, mean ± S.D., * indicates p < 0.05 compared with 0 UV by Student's t test. C, qPCR analysis of ILEI in HMLE human mammary epithelial cells 24 h after treatment with 0 or 50 J/m2 of UV. n = 3, mean ± S.D., * indicates p < 0.05 compared with 0 UV by Student's t test. D, immunoblot analysis of ILEI, USF-1, or HSP90 in 501-Mel melanoma cells transduced with lentivirus containing scrambled shRNA or two different sequences targeting USF1.
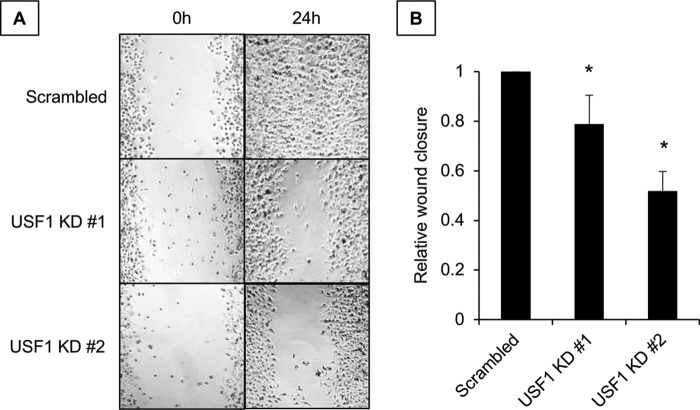
Finally, we wanted to know if USF-1 regulation of ILEI had any biological significance. Given the importance of ILEI to the invasive melanoma phenotype (Fig. 1), we tested the contribution of USF-1 to wound healing. We observed that USF-1 knockdown in 501-Mel melanoma cells attenuates wound healing (Fig. 10, A and B). From these experiments we concluded that USF-1 regulates migration in melanoma cell lines.
Figure 10.
Biological role of the USF-1–ILEI axis in melanoma cells. A, representative images of wound-healing assays of 501-Mel melanoma cells transduced with lentivirus containing scrambled shRNA or two different sequences targeting USF1. B, quantification of A using ImageJ software. n = 3, mean ± S.E., and * indicates p < 0.05 compared with scrambled by unpaired Student's t test.
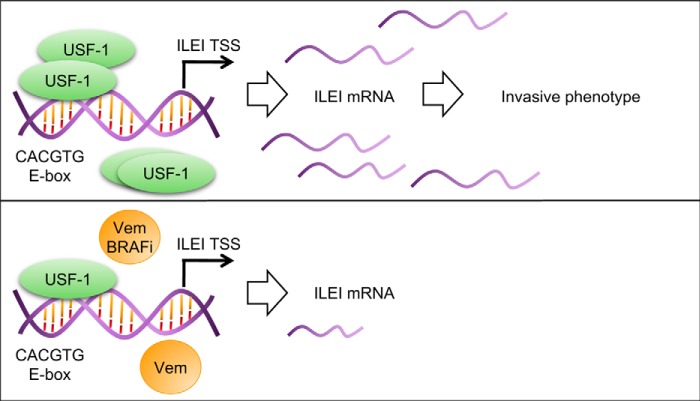
In summary, we have described the following novel findings: ILEI regulates melanoma invasiveness in vivo, vemurafenib inhibits ILEI at the transcriptional level through a specific E-box sequence, USF-1 directly interacts with this E-box in the ILEI promoter and that this interaction is abolished upon vemurafenib treatment, and finally that endogenous USF-1 contributes to ILEI expression and the invasive melanoma phenotype. Through these findings we have established a novel regulatory mechanism for ILEI expression by USF-1, and based on this previously unknown context for ILEI expression we speculate on the following new biological functions for USF-1 and ILEI (Fig. 11).
Figure 11.
Model of USF-1 regulation of ILEI. A model depicting basal versus vemurafenib-treated conditions is shown. BRAFi, B-Raf kinase inhibitor. ILEI: interleukin-like EMT inducer, FAM3C. Vem, vemurafenib.
Although ILEI has been thoroughly described as a tumor autonomous regulator of EMT and invasion, the role of ILEI in paracrine signaling is still unclear (25, 28, 29, 33, 43–45). Our characterization of the novel vemurafenib–USF-1–ILEI regulatory axis has implications for paracrine ILEI signaling. Given the impact of vemurafenib on BRAF mutant melanoma, the mechanisms of vemurafenib-mediated tumor killing have been thoroughly characterized. For instance, mouse models have shown that vemurafenib induces apoptosis of tumor suppressive regulatory T cells while simultaneously promoting CD8 T cell-mediated killing of tumor cells (46). Other models have shown that vemurafenib increases the recruitment of macrophages to BRAF mutant melanomas (47). Combining our finding that vemurafenib inhibits USF-1–ILEI with the established finding that vemurafenib regulates the tumor microenvironment, we suggest that both USF-1 and ILEI could be regulators of the tumor immune microenvironment. Future studies should seek to elucidate the role of USF-1/ILEI in this paracrine signaling context.
Additionally, USF-1 is primarily known as a regulator of glucose and lipid metabolism and is poorly described as an oncogene (48–50). In melanoma USF-1 is known as a stress responsive transcription factor involved in UV-induced, but not MITF-mediated constitutive pigmentation (51). The non-overlapping function of USF-1 with MITF in pigmentation suggests that USF-1 could play other roles in melanoma biology when MITF is poorly expressed, such as in MITF-low invasive melanoma cells. These cells are characterized by the lack of MITF but not by the presence of any major transcription factor, although ZEB1 and JUN have been proposed (20, 21, 23, 37, 38). Considering that ILEI is a regulator of MITF-low cells we speculate that USF-1 could be a novel transcription factor regulating the MITF-low invasive state (Fig. 11) (25). Future work should address the role of USF-1 in melanoma phenotype switching.
Experimental procedures
Animal studies
All procedures were approved by the Institutional Animal Care and Use Committee at the Medical University of South Carolina. Lung colonization experiments were conducted as follows: tail vein injection into 6–8–week-old mice were performed with 1205Lu cells expressing shSCR or shILEI 3 (1 × 105 cells in 100 μl of PBS). Cells were previously characterized (25). After 8 weeks, mice were sacrificed, and lungs were harvested. Organs were formalin-fixed and paraffin-embedded. Histopathological analysis was conducted by Hollings Cancer Center Biorepository & Tissue Analysis Shared Resource. Lung colonies were counted by two individuals.
Constructs
ILEI reporter constructs were cloned as follows: the ILEI promoter sequence from 2,300 bp upstream of the predicted transcription start site to 80 bp upstream was amplified by PCR from 501-Mel genomic DNA with 5′ KpnI and 3′ NheI primers and ligated into pBV-Luc (pBV-Luc vector (a gift from Bert Vogelstein (Addgene plasmid number 16539)) (35). Primers are listed in Table 1. Subsequent ILEI promoter truncation constructs were cloned by PCR using an alternate 5′ KpnI primer and the same 3′ NheI primer, or using a Q5 SDM kit from New England Biolabs (Ipswitch, MA). The ILEI 3′ UTR construct from stop codon to 1,620 bp downstream of the stop codon was amplified by PCR from 501-Mel cDNA with 5′ NheI and 3′ SalI primers and ligated into pmirGLO Dual Luciferase vector (RAB14 3′ UTR WT was a gift from Curt Civin (Addgene plasmid number 61489)) (52). The following constructs were used in this study: pcDNA3-cmyc (a gift from Wafik El-Deiry (Addgene plasmid number 16011) (53), pCDNA3-HA-human MYCN (a gift from Martine Roussel (Addgene plasmid number 74163) (54), pMXs-Hu-l-Myc (a gift from Shinya Yamanaka (Addgene plasmid number 26022) (55), pEGFP-N1-TFEB (a gift from Shawn Ferguson (Addgene plasmid number 38119) (56), pEGFP-N1-TFE3 (a gift from Shawn Ferguson (Addgene plasmid number 38120) (56), pCMV-Tag4A-MITF-M (wt) (a gift from Yardena Samuels (Addgene plasmid number 31151)), and USF1, CREB3L2, and ID2 coding sequence constructs in pLX304 were generated by David Root and supplied by DNASU (57–60). Corresponding empty vector for human l-MYC was constructed by digesting the l-MYC plasmid with NotI to remove the l-MYC coding sequence and religating using T4 ligase.
Table 1.
Primer sequences
| Primer name | Sequence |
|---|---|
| Cloning −150 KpnI ILEI promoter F | GGC GGT ACC AAG TTC AGA TTG TGC AGC G |
| Cloning −204 KpnI ILEI promoter F | GGC GGT ACC ACG TGG CAA GTT CAG ATT |
| Cloning −221 KpnI ILEI promoter F | GGC GGT ACC CAT TTT TCT CCC TCC CGT AGI |
| Cloning −300 KpnI ILEI promoter F | GGC GGT ACC ATG GGAT GGG TCA TTT AAA ATG TTC TGC |
| Cloning −2303 KpnI ILEI promoter F | GGC GGT ACC GGA TTC TCC AAA TAC TCC ATC AGT G |
| Cloning −80 NheI ILEI promoter R | TTA GCT AGC AAG GGC CGG AGA GCG GA |
| SDM E-box ILEI promoter F | CGG CAG CTC CCA AAT GGC AAG TTC AGA TTG TGC AGC GCC TGG C |
| SDM E-box ILEI promoter R | CCG CCG CAG CGG CCC TGC |
| SDM Inr ILEI promoter F | GTA AAA AAA GTG CAG CGC CTG GCC GGG |
| SDM Inr ILEI promoter R | TTG CTT TTT TGG AGC TGC CGC CGC CGC A |
| Cloning NheI ILEI 3′ UTR F | ATT GCT AGC TGG AAA TGT GGA GAG AAT TGA AG |
| Cloning ILEI 3′ UTR SalI R | TTA GTC GAC CTG CAA CAT TTA TTT CAC AAT CCC T |
| RT-PCR ACTB F | ATG CTT CTA GGC GGA CTA TG |
| RT-PCR ACTB R | ACA AAT AAA GCC ATG CCA AT |
| RT-PCR FAM3C F | GCA ACC AAA CTC AAT GAT GA |
| RT-PCR FAM3C R | ACC ACA GAA GAC CCA GTT GT |
| RT-PCR FAM3C Intron F | TTG CCCTAA TGC AGATCA TA |
| RT-PCR FAM3C Intron R | CAA CAA AGA AAC CCA CAA CA |
| RT-PCR GAPDH F | CTC CTC ACA GTT GCC ATG TA |
| RT-PCR GAPDH R | GGT TGA GCA CAG GGT ACT TT |
| RT-PCR TYR F | ACC TCT CAT TTG CAA GGT CAA A |
| RT-PCR TYR R | AG AGG AAC CTC TGC CTG AAA GC |
| RT-PCR USF1 F | GCA CTG GTC AAT TCT TTG TG |
| RT-PCR USF1 R | TTC TGA CTT CGG GGA ATA AG |
Cell culture conditions
The following human melanoma cell lines were used: WM3918, 501-Mel, Sk-Mel-28, WM983B, 1205Lu, and WM9. These cell lines were purchased from ATCC, Coriell, or were a generous gift from Dr. J. Alan Diehl or Dr. Alain Mauviel. All melanoma cell lines were cultured at 37 °C, 5% CO2 in RPMI 1640 medium (HyClone, Logan, UT) supplemented with 10% FBS (Atlanta Biologicals, Flowery Branch, GA), antibiotic–antimycotic (×100; ThermoFisher, Waltham, MA), and prophylactic plasmocin (InvivoGen, San Diego, CA) at 37 °C and in 5% CO2. The HMLE human mammary cell line was obtained from Dr. Sendurai Mani and cultured in Dulbecco's modified Eagle's medium/F-12 supplemented with 5% calf serum, 0.5 μg/ml of hydrocortisone, 10 μg/ml of insulin, 20 ng/ml of EGF, and 1% antibiotic–antimycotic.
Stable cell lines were generated by lentiviral transduction with Polybrene (8 μg/ml; Sigma). 24 h post-transduction the medium was changed, and 48 h post-transduction the cells were selected and cultured with 0.125–0.5 μg/ml of puromycin (InvivoGen). Pools of stably transduced cells were analyzed. shRNA sequences are listed in Table 2. UV treatments were conducted using Fisher Scientific UV Cross-linker FB-UVXL-1000 at 0, 5, or 50 J/m2 in room air, and then incubating in normal cell culture conditions for 24 h.
Table 2.
shRNA sequences
| shRNA name | Target | Sequence |
|---|---|---|
| pLKO.1-puro Non-mammalian shRNA | Scrambled | CCGGCAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTGTTTTT |
| TRC2-pLKO-puro TRCN000020679 | USF1 | CCGGGCTGGATACTGGACACACTAACTCGAGTTAGTGTGTCCAGTATCCAGCTTTTT |
| TRC2-pLKO-puro TRCN000020681 | USF1 | CCGGCACTGGTCAATTCTTTGTGATCTCGAGATCACAAAGAATTGACCAGTGTTTTT |
Lentivirus
Lentiviral shRNAs were obtained from the MUSC Hollings Cancer Center shRNA Shared Resource Technology. All vectors used in this study are listed in Table 2. Lentivirus was generated by seeding 293T (1,000,000 cells; Takara Bio, Mountainview, CA) to a 60-mm cell culture dish, and transfecting with 6 μl of Lipofectamine 2000 (ThermoFisher), 1 μg of pLKO vector, 0.75 μg of psPAX2, and 0.25 μg of pMD2.G. 24 h post-transfection the media was changed, and 48 and 72 h post-transfection the media was harvested. Viral supernatant was cleared by centrifugation, filtered through a 0.22-μm filter, and stored at −80 °C until use.
Luciferase analysis
Cell lines were seeded at 50,000–75,000 cells per well in 24-well plate in 0.5 ml of complete medium. At 24 h the cells were transfected using X-tremeGENE 9 (Roche Applied Science, Switzerland; 100 ng of firefly experimental luciferase, 5 ng of Renilla control luciferase, 0.3 μl of X-tremeGENE reagent, in 10 μl of Opti-MEM, 200 ng of experimental plasmid, where indicated (i.e. USF1 overexpression construct), 0.6 μl of X-tremeGENE reagent, in 20 μl of Opti-MEM). At this time the cells were treated with DMSO vehicle or 1 μm vemurafenib. At 24 h post-transfection the cells were harvested with passive lysis buffer and analyzed with Dual-Glo Luciferase Assay System (Promega, Madison, WI).
Immunoblot analysis
Whole cell lysates were extracted as follows: 100 μl of Tris-Triton lysis buffer (20 mm Tris, pH 7.5, 1% Triton X-100, 10% glycerol, 137 mm NaCl, 2 mm EDTA, and Halt Protease and Phosphatase Inhibitor mixture (ThermoFisher)) was added to 6-well cell culture plates, cells were immediately scraped, incubated on ice for 30 min, and cleared by centrifugation for 20 min at 16,000 × g. Protein concentrations were measured with Bradford Protein Assay (Bio-Rad). For conditioned medium immunoblots, cells were serum starved in RPMI, 0% FBS overnight, medium was harvested, and precipitated using TCA/acetone. Protein samples were denatured by incubating at 95 °C for 5 min with 1× Laemmli Reducing Denaturing Sample Buffer (300 mm Tris-Cl, pH 6.8, 10% SDS, 50% glycerol, 25% β-mercaptoethanol). 1–20 μg of whole cell lysate was resolved on an 8, 10, or 12% polyacrylamide-SDS gel, and transferred onto polyvinylidene difluoride membrane. Membranes were blocked for 1 h at room temperature in 5% skim milk, TBS with 0.01% Tween 20 (TBST) and incubated overnight at 4 °C on primary antibody + 5% skim milk/TBST. The following primary antibodies were used: ILEI (ab72182; Abcam, 1:1,000; specificity for the band between 25 and 20 kDa confirmed in Ref. 25)), α-tubulin (2144; Cell Signaling, Danvers, MA; 1:10,000), p-ERK T202/Y204 (4370; Cell Signaling; 1:2,000), total ERK (9120; Cell Signaling; 1:1,000), USF1 (C-20; sc-229; Santa Cruz, Dallas, TX; 1:1,000), GAPDH (sc-32233; Santa Cruz; 1:10,000), and HSP90 (sc-13119; Santa Cruz; 1:10,000). After primary antibody incubation, membranes were washed 4 × 15 min in TBST and incubated for 1 h at room temperature with secondary antibody in TBST. The following secondary antibodies were used: goat anti-mouse IgG (31430; ThermoFisher; 1:10,000) and goat anti-rabbit IgG3 (31460; ThermoFisher; 1:10,000). After secondary antibody incubation, membranes were washed 4 × 15 min in TBST and detected using Luminata Forte Western HRP substrate (EMD Millipore, Darmstadt, Germany) and HyBlot CL Autoradiography Film (Denville, Holliston, MA) or CCD camera (Bio-Rad ChemiDoc System; Bio-Rad).
PCR analysis
Total RNA was isolated using TRIzol (ThermoFisher Scientific). Reverse transcription was performed using oligo(dT) primers and Moloney- murine leukemia virus Reverse Transcriptase (New England BioLabs). Semi-quantitative PCR was conducted on 10 ng of cDNA using Maxima Hot Start PCR Master Mix (ThermoFisher Scientific). Real-time quantitative PCR was conducted using iQ SYBR Green Supermix (Bio-Rad) using CFX384 Real-Time System (Bio-Rad). Reactions were conducted on 50 to 10 ng of cDNA. Primers are listed in Table 1. Relative gene expression was calculated using RFX Manager software, and genes were normalized to GAPDH internal control.
Chromatin immunoprecipitation (ChIP)
ChIP protocol was modified from Carey et al. (61). Briefly, 1.5 × 107 cells were fixed in 1% formaldehyde for 10 min at room temperature and quenched in 125 mm glycine. Cells were harvested in lysis buffer (5 mm PIPES, pH 8, 85 mm KCl, 0.5% Nonidet P-40), and centrifuged for 10 min (3,000 rpm at 4 °C). Supernatant (cytosolic fraction) was removed, and the pellet was resuspended in nuclei lysis buffer (50 mm Tris, pH 8, 10 mm EDTA, 1% SDS, protease and phosphatase inhibitor tablets). Nuclear extracts were sonicated for 10 min (30 s on and 30 s off), and cleared by centrifugation (2 times, 10 min, 13,000 rpm at 4 °C). The supernatant was considered the chromatin fraction. 100 μg of chromatin samples were precleared for 2 h at 4 °C with 30 μl of slurry ChIP-Grade Protein G-Agarose Beads (9007; Cell Signaling). The resulting supernatants were incubated overnight on a rotator at 4 °C with 5 μg of control Mouse (G3A1) mAb IgG1 Isotype Control (5415; Cell Signaling) or USF1 antibody (C-20). 30 μl of slurry ChIP-Grade Protein G-Agarose Beads was added to each sample for 2 h on a rotator at 4 °C. Beads were washed 4 times in high-salt wash buffer (50 mm HEPES, pH 7.9, 500 mm NaCl, 1 mm EDTA, 0.1% SDS, 1% Triton X-100, 0.1% deoxycholate) each time incubating for 10 min on a rotator at room temperature. Beads were next washed 2 times in TE buffer each time incubating for 10 min on a rotator at room temperature. Beads were resuspended in 300 μl of elution buffer (50 mm Tris, pH 8, 10 mm EDTA, 1% SDS) + 20 μg of RNase A (R1253; ThermoFisher Scientific) and incubated on a 55 °C heat block for 2 h. 1.6 milliunits of proteinase K (P8107S; New England BioLabs) were added prior to another incubation on a 55 °C heat block for 2 h. The samples were transferred to a 65 °C heat block for overnight elution. The samples were purified in 30 μl of H2O using GeneJET PCR Purification Kit (K0702; ThermoFisher Scientific). PCR was conducted with primers used for FAM3C promoter cloning specific for human FAM3C promoter from 300 to 80 bp upstream of TSS, as listed in Table 1, human TYR promoter or human HMOX1 promoter (15, 42).
Biotin DNA pulldown
DNA pulldown assays were conducted with 5′-biotinylated double-stranded annealed oligonucleotides corresponding to FAM3C promoter from −200 to −110 upstream of the TSS. A mutant E-box construct was used in which the E-box at −162 to −156 was mutated from CACGTG to CAAATG. A 5′-biotinylated double-stranded annealed oligonucleotide of a random sequence was used as a negative control (SCR).
Nuclear extracts were isolated by harvesting cells in lysis buffer (10 mm HEPES, 1.5 mm MgCl2, 10 mm KCl, 0.5 mm DTT, 0.05% Nonidet P-40, pH 7.9, protease and phosphatase inhibitor tablets) and pelleting cell nuclei. Pellets were incubated for 30 min on ice in nuclei extraction buffer (5 mm HEPES, 1.5 mm MgCl2, 0.2 mm EDTA, 0.5 mm DTT, 26% glycerol, pH 7.9, protease and phosphatase inhibitor) and NaCl was added to a final concentration of 300 mm. Samples were homogenized by passing 20 times through a 28-guage needle. Samples were incubated on ice for 30 min and centrifuged. The supernatants were considered the nuclear extract. Nuclear extract (20 μg) was reserved as 5% input.
Preclear beads were prepared as follows: 5′-biotinylated random oligonucleotide (SCR) was bound to streptavidin-agarose beads (50 μl of slurry + 10 μg of oligo; ThermoFisher) for 1 h on a rotator at room temperature. IP beads were prepared as follows: 5′-biotinylated FAM3C promoter WT or E-box mutant (CACGTG to CAAATG) was bound to streptavidin-agarose beads (50 μl of slurry + 1 μg of oligo; ThermoFisher) for 1 h on a rotator at room temperature. Preclear was conducted as follows: 400 μg of nuclear extract was incubated with preclear beads for 1 h on a rotator at room temperature. The resulting sample was centrifuged and the supernatant was precleared again for a total of three preclears. The final supernatant was incubated on IP beads overnight on a rotator at 4 °C. The beads were washed in PBS, 0.1% Triton X-100 and resolved by SDS-PAGE, transferred to nitrocellulose, and probed using antibody against USF1 (C-20; sc-229; Santa Cruz; 1:1,000).
Statistical analyses
Data are mean ± S.D. unless indicated otherwise. p < 0.05 by unpaired two sample Student's t test is considered significant. Representative experiments are repeated at least twice.
Wound-healing assays
Cells (3 × 105, 0.5 ml of complete medium) were seeded in a 24-well plate, and a 1-ml pipette tip was used to scratch the cells. Images were recorded from 0 to 24 h, and analyzed using ImageJ (National Institutes of Health, Bethesda, MD).
Author contributions
K. N. and P. H. H. conceptualization; K. N., T. A. D., A. C. D., B. V. H., B. J. M., and B. K. M. data curation; K. N. formal analysis; K. N. and P. H. H. funding acquisition; K. N., T. A. D., A. C. D., B. V. H., B. J. M., and B. K. M. investigation; K. N. methodology; K. N. and P. H. H. writing-original draft; K. N., T. A. D., A. C. D., B. V. H., B. K. M., and P. H. H. writing-review and editing; P. H. H. supervision; P. H. H. project administration.
Acknowledgment
We acknowledge Simon Grelet for critical reading of the manuscript.
This work was supported by National Institutes of Health NCI Grants F30CA203269, T32CA193201, and R01CA154663 and U.S. Department of Defense (DOD) Grant BC170301. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- MEK
- mitogen-activated protein kinase
- ERK
- extracellular signal-regulated kinase kinase
- bHLH
- basic-helix-loop-helix
- LZip
- leucine zipper
- MITF
- micropthalmia-associated transcription factor
- USF-1
- upstream stimulatory factor 1
- ILEI
- interleukin-like EMT inducer
- EMT
- epithelial-to-mesenchymal transition
- TSS
- transcription start site
- Inr
- initiator sequence
- TGF
- transforming growth factor
- qPCR
- quantitative PCR
- UTR
- untranslated region
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- EGFP
- enhanced green fluorescent protein
- IP
- immunoprecipitation
- hnRNP
- heterogeneous nuclear ribonucleoprotein.
References
- 1. Cancer Genome Atlas Network (2015) Genomic classification of cutaneous melanoma. Cell 161, 1681–1696 10.1016/j.cell.2015.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Goodall J., Wellbrock C., Dexter T. J., Roberts K., Marais R., and Goding C. R. (2004) The Brn-2 transcription factor links activated BRAF to melanoma proliferation. Mol. Cell. Biol. 24, 2923–2931 10.1128/MCB.24.7.2923-2931.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wellbrock C., and Marais R. (2005) Elevated expression of MITF counteracts B-RAF-stimulated melanocyte and melanoma cell proliferation. J. Cell Biol. 170, 703–708 10.1083/jcb.200505059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Patton E. E., Widlund H. R., Kutok J. L., Kopani K. R., Amatruda J. F., Murphey R. D., Berghmans S., Mayhall E. A., Traver D., Fletcher C. D., Aster J. C., Granter S. R., Look A. T., Lee C., Fisher D. E., and Zon L. I. (2005) BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr. Biol. 15, 249–254 10.1016/j.cub.2005.01.031 [DOI] [PubMed] [Google Scholar]
- 5. Michaloglou C., Vredeveld L. C., Soengas M. S., Denoyelle C., Kuilman T., van der Horst C. M., Majoor D. M., Shay J. W., Mooi W. J., and Peeper D. S. (2005) BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 436, 720–724 10.1038/nature03890 [DOI] [PubMed] [Google Scholar]
- 6. Sheridan C., Brumatti G., and Martin S. J. (2008) Oncogenic B-RafV600E inhibits apoptosis and promotes ERK-dependent inactivation of Bad and Bim. J. Biol. Chem. 283, 22128–22135 10.1074/jbc.M800271200 [DOI] [PubMed] [Google Scholar]
- 7. Wellbrock C., Rana S., Paterson H., Pickersgill H., Brummelkamp T., and Marais R. (2008) Oncogenic BRAF regulates melanoma proliferation through the lineage specific factor MITF. PLoS ONE 3, e2734 10.1371/journal.pone.0002734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wajapeyee N., Serra R. W., Zhu X., Mahalingam M., and Green M. R. (2008) Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell 132, 363–374 10.1016/j.cell.2007.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pratilas C. A., Taylor B. S., Ye Q., Viale A., Sander C., Solit D. B., and Rosen N. (2009) (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc. Natl. Acad. Sci. U.S.A. 106, 4519–4524 10.1073/pnas.0900780106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Du J., Widlund H. R., Horstmann M. A., Ramaswamy S., Ross K., Huber W. E., Nishimura E. K., Golub T. R., and Fisher D. E. (2004) Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer Cell 6, 565–576 10.1016/j.ccr.2004.10.014 [DOI] [PubMed] [Google Scholar]
- 11. Du J., Miller A. J., Widlund H. R., Horstmann M. A., Ramaswamy S., and Fisher D. E. (2003) MLANA/MART1 and SILV/PMEL17/GP100 are transcriptionally regulated by MITF in melanocytes and melanoma. Am. J. Pathol. 163, 333–343 10.1016/S0002-9440(10)63657-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sawadogo M., and Roeder R. G. (1985) Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell 43, 165–175 10.1016/0092-8674(85)90021-2 [DOI] [PubMed] [Google Scholar]
- 13. Imagawa S., Fujii S., Dong J., Furumoto T., Kaneko T., Zaman T., Satoh Y., Tsutsui H., and Sobel B. E. (2006) Hepatocyte growth factor regulates E box-dependent plasminogen activator inhibitor type 1 gene expression in HepG2 liver cells. Arterioscler. Thromb. Vasc. Biol. 26, 2407–2413 10.1161/01.ATV.0000240318.61359.e3 [DOI] [PubMed] [Google Scholar]
- 14. Corre S., Primot A., Baron Y., Le Seyec J., Goding C., and Galibert M. D. (2009) Target gene specificity of USF-1 is directed via p38-mediated phosphorylation-dependent acetylation. J. Biol. Chem. 284, 18851–18862 10.1074/jbc.M808605200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Galibert M. D., Carreira S., and Goding C. R. (2001) The Usf-1 transcription factor is a novel target for the stress-responsive p38 kinase and mediates UV-induced tyrosinase expression. EMBO J. 20, 5022–5031 10.1093/emboj/20.17.5022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Waerner T., Alacakaptan M., Tamir I., Oberauer R., Gal A., Brabletz T., Schreiber M., Jechlinger M., and Beug H. (2006) ILEI: a cytokine essential for EMT, tumor formation, and late events in metastasis in epithelial cells. Cancer Cell 10, 227–239 10.1016/j.ccr.2006.07.020 [DOI] [PubMed] [Google Scholar]
- 17. Nieto M. A., Huang R. Y., Jackson R. A., and Thiery J. P. (2016) Emt: 2016. Cell 166, 21–45 10.1016/j.cell.2016.06.028 [DOI] [PubMed] [Google Scholar]
- 18. Johannessen C. M., Johnson L. A., Piccioni F., Townes A., Frederick D. T., Donahue M. K., Narayan R., Flaherty K. T., Wargo J. A., Root D. E., and Garraway L. A. (2013) A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature 504, 138–142 10.1038/nature12688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Konieczkowski D. J., Johannessen C. M., Abudayyeh O., Kim J. W., Cooper Z. A., Piris A., Frederick D. T., Barzily-Rokni M., Straussman R., Haq R., Fisher D. E., Mesirov J. P., Hahn W. C., Flaherty K. T., Wargo J. A., Tamayo P., and Garraway L. A. (2014) A melanoma cell state distinction influences sensitivity to MAPK pathway inhibitors. Cancer Discov. 4, 816–827 10.1158/2159-8290.CD-13-0424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ramsdale R., Jorissen R. N., Li F. Z., Al-Obaidi S., Ward T., Sheppard K. E., Bukczynska P. E., Young R. J., Boyle S. E., Shackleton M., Bollag G., Long G. V., Tulchinsky E., Rizos H., Pearson R. B., et al. (2015) The transcription cofactor c-JUN mediates phenotype switching and BRAF inhibitor resistance in melanoma. Sci. Signal. 8, ra82 10.1126/scisignal.aab1111 [DOI] [PubMed] [Google Scholar]
- 21. Riesenberg S., Groetchen A., Siddaway R., Bald T., Reinhardt J., Smorra D., Kohlmeyer J., Renn M., Phung B., Aymans P., Schmidt T., Hornung V., Davidson I., Goding C. R., Jönsson G., et al. (2015) MITF and c-Jun antagonism interconnects melanoma dedifferentiation with pro-inflammatory cytokine responsiveness and myeloid cell recruitment. Nat. Commun. 6, 8755 10.1038/ncomms9755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wellbrock C., and Arozarena I. (2015) Microphthalmia-associated transcription factor in melanoma development and MAP-kinase pathway targeted therapy. Pigment Cell Melanoma Res. 28, 390–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Richard G., Dalle S., Monet M. A., Ligier M., Boespflug A., Pommier R. M., de la Fouchardiere A., Perier-Muzet M., Depaepe L., Barnault R., Tondeur G., Ansieau S., Thomas E., Bertolotto C., Ballotti R., et al. (2016) ZEB1-mediated melanoma cell plasticity enhances resistance to MAPK inhibitors. EMBO Mol. Med. 8, 1143–1161 10.15252/emmm.201505971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Landsberg J., Kohlmeyer J., Renn M., Bald T., Rogava M., Cron M., Fatho M., Lennerz V., Wolfel T., Holzel M., and Tuting T. (2012) Melanomas resist T-cell therapy through inflammation-induced reversible dedifferentiation. Nature 490, 412–416 10.1038/nature11538 [DOI] [PubMed] [Google Scholar]
- 25. Noguchi K., Dalton A. C., Howley B. V., McCall B. J., Yoshida A., Diehl J. A., and Howe P. H. (2017) Interleukin-like EMT inducer regulates partial phenotype switching in MITF-low melanoma cell lines. PLoS ONE 12, e0177830 10.1371/journal.pone.0177830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Song Q., Sheng W., Zhang X., Jiao S., and Li F. (2014) ILEI drives epithelial to mesenchymal transition and metastatic progression in the lung cancer cell line A549. Tumour Biol. 35, 1377–1382 10.1007/s13277-013-1188-y [DOI] [PubMed] [Google Scholar]
- 27. Hussey G. S., Link L. A., Brown A. S., Howley B. V., Chaudhury A., and Howe P. H. (2012) Establishment of a TGFβ-induced post-transcriptional EMT gene signature. PloS One 7, e52624–e52624 10.1371/journal.pone.0052624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hussey G. S., Chaudhury A., Dawson A. E., Lindner D. J., Knudsen C. R., Wilce M. C. J., Merrick W. C., and Howe P. H. (2011) Identification of an mRNP complex regulating tumorigenesis at the translational elongation step. Mol. Cell 41, 419–431 10.1016/j.molcel.2011.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chaudhury A., Hussey G. S., Ray P. S., Jin G., Fox P. L., and Howe P. H. (2010) TGF-β-mediated phosphorylation of hnRNP-E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat. Cell Biol. 12, 286–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Csiszar A., Kutay B., Wirth S., Schmidt U., Macho-Maschler S., Schreiber M., Alacakaptan M., Vogel G. F., Aumayr K., Huber L. A., and Beug H. (2014) Interleukin-like epithelial-to-mesenchymal transition inducer activity is controlled by proteolytic processing and plasminogen-urokinase plasminogen activator receptor system-regulated secretion during breast cancer progression. Breast Cancer Res. 16, 433–433 10.1186/s13058-014-0433-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhu Y. H., Zhang B., Li M., Huang P., Sun J., Fu J., and Guan X. Y. (2015) Prognostic significance of FAM3C in esophageal squamous cell carcinoma. Diagn. Pathol. 10, 192 10.1186/s13000-015-0424-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gao Z., Lu C., Wang Z., Song Y., Zhu J., Gao P., Sun J., Chen X., Wang M., Dong Y., and Xu H. (2014) ILEI: A novel target for epithelial-mesenchymal transition and poor prognosis in colorectal cancer. Histopathology 65, 527–538 10.1111/his.12435 [DOI] [PubMed] [Google Scholar]
- 33. Lahsnig C., Mikula M., Petz M., Zulehner G., Schneller D., van Zijl F., Huber H., Csiszar A., Beug H., and Mikulits W. (2009) ILEI requires oncogenic Ras for the epithelial to mesenchymal transition of hepatocytes and liver carcinoma progression. Oncogene 28, 638–650 10.1038/onc.2008.418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tsai J., Lee J. T., Wang W., Zhang J., Cho H., Mamo S., Bremer R., Gillette S., Kong J., Haass N. K., Sproesser K., Li L., Smalley K. S., Fong D., Zhu Y. L., et al. (2008) Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc. Natl. Acad. Sci. U.S.A. 105, 3041–3046 10.1073/pnas.0711741105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. He T. C., Chan T. A., Vogelstein B., and Kinzler K. W. (1999) PPARdelta is an APC-regulated target of nonsteroidal anti-inflammatory drugs. Cell 99, 335–345 10.1016/S0092-8674(00)81664-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mathelier A., Fornes O., Arenillas D. J., Chen C. Y., Denay G., Lee J., Shi W., Shyr C., Tan G., Worsley-Hunt R., Zhang A. W., Parcy F., Lenhard B., Sandelin A., and Wasserman W. W. (2016) JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 44, D110–115 10.1093/nar/gkv1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Denecker G., Vandamme N., Akay O., Koludrovic D., Taminau J., Lemeire K., Gheldof A., De Craene B., Van Gele M., Brochez L., Udupi G. M., Rafferty M., Balint B., Gallagher W. M., Ghanem G., et al. (2014) Identification of a ZEB2-MITF-ZEB1 transcriptional network that controls melanogenesis and melanoma progression. Cell Death Differ. 21, 1250–1261 10.1038/cdd.2014.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Caramel J., Papadogeorgakis E., Hill L., Browne G. J., Richard G., Wierinckx A., Saldanha G., Osborne J., Hutchinson P., Tse G., Lachuer J., Puisieux A., Pringle J. H., Ansieau S., and Tulchinsky E. (2013) A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell 24, 466–480 10.1016/j.ccr.2013.08.018 [DOI] [PubMed] [Google Scholar]
- 39. Verfaillie A., Imrichova H., Atak Z. K., Dewaele M., Rambow F., Hulselmans G., Christiaens V., Svetlichnyy D., Luciani F., Van den Mooter L., Claerhout S., Fiers M., Journe F., Ghanem G. E., Herrmann C., et al. (2015) Decoding the regulatory landscape of melanoma reveals TEADS as regulators of the invasive cell state. Nat. Commun. 6, 6683 10.1038/ncomms7683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Roy A. L., Meisterernst M., Pognonec P., and Roeder R. G. (1991) Cooperative interaction of an initiator-binding transcription initiation factor and the helix-loop-helix activator USF. Nature 354, 245–248 10.1038/354245a0 [DOI] [PubMed] [Google Scholar]
- 41. Bendall A. J., and Molloy P. L. (1994) Base preferences for DNA binding by the bHLH-Zip protein USF: effects of MgCl2 on specificity and comparison with binding of Myc family members. Nucleic Acids Res. 22, 2801–2810 10.1093/nar/22.14.2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hock T. D., Nick H. S., and Agarwal A. (2004) Upstream stimulatory factors, USF1 and USF2, bind to the human haem oxygenase-1 proximal promoter in vivo and regulate its transcription. Biochem. J. 383, 209–218 10.1042/BJ20040794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhao X., Luo G., Fan Y., Ma X., Zhou J., and Jiang H. (2018) ILEI is an important intermediate participating in the formation of TGF-β1-induced renal tubular EMT. Cell Biochem. Funct. 36, 46–55 10.1002/cbf.3316 [DOI] [PubMed] [Google Scholar]
- 44. Kral M., Klimek C., Kutay B., Timelthaler G., Lendl T., Neuditschko B., Gerner C., Sibilia M., and Csiszar A. (2017) Covalent dimerization of interleukin-like epithelial-to-mesenchymal transition (EMT) inducer (ILEI) facilitates EMT, invasion, and late aspects of metastasis. FEBS J. 284, 3484–3505 10.1111/febs.14207 [DOI] [PubMed] [Google Scholar]
- 45. Jansson A. M., Csiszar A., Maier J., Nyström A. C., Ax E., Johansson P., and Schiavone L. H. (2017) The interleukin-like epithelial-mesenchymal transition inducer ILEI exhibits a non-interleukin-like fold and is active as a domain-swapped dimer. J. Biol. Chem. 292, 15501–15511 10.1074/jbc.M117.782904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Steinberg S. M., Zhang P., Malik B. T., Boni A., Shabaneh T. B., Byrne K. T., Mullins D. W., Brinckerhoff C. E., Ernstoff M. S., Bosenberg M. W., and Turk M. J. (2014) BRAF inhibition alleviates immune suppression in murine autochthonous melanoma. Cancer Immunol. Res. 2, 1044–1050 10.1158/2326-6066.CIR-14-0074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Smith M. P., Sanchez-Laorden B., O'Brien K., Brunton H., Ferguson J., Young H., Dhomen N., Flaherty K. T., Frederick D. T., Cooper Z. A., Wargo J. A., Marais R., and Wellbrock C. (2014) The immune microenvironment confers resistance to MAPK pathway inhibitors through macrophage-derived TNFα. Cancer Discov. 4, 1214–1229 10.1158/2159-8290.CD-13-1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Laurila P. P., Soronen J., Kooijman S., Forsstrom S., Boon M. R., Surakka I., Kaiharju E., Coomans C. P., Van Den Berg S. A., Autio A., Sarin A. P., Kettunen J., Tikkanen E., Manninen T., Metso J., et al. (2016) USF1 deficiency activates brown adipose tissue and improves cardiometabolic health. Sci. Transl. Med. 8, 323ra313 [DOI] [PubMed] [Google Scholar]
- 49. Wu S., Mar-Heyming R., Dugum E. Z., Kolaitis N. A., Qi H., Pajukanta P., Castellani L. W., Lusis A. J., and Drake T. A. (2010) Upstream transcription factor 1 influences plasma lipid and metabolic traits in mice. Hum. Mol. Genet. 19, 597–608 10.1093/hmg/ddp526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lefrançois-Martinez A. M., Martinez A., Antoine B., Raymondjean M., and Kahn A. (1995) Upstream stimulatory factor proteins are major components of the glucose response complex of the l-type pyruvate kinase gene promoter. J. Biol. Chem. 270, 2640–2643 10.1074/jbc.270.6.2640 [DOI] [PubMed] [Google Scholar]
- 51. Corre S., and Galibert M. D. (2005) Upstream stimulating factors: highly versatile stress-responsive transcription factors. Pigment Cell Res. 18, 337–348 10.1111/j.1600-0749.2005.00262.x [DOI] [PubMed] [Google Scholar]
- 52. Kim M., Tan Y. S., Cheng W. C., Kingsbury T. J., Heimfeld S., and Civin C. I. (2015) MIR144 and MIR451 regulate human erythropoiesis via RAB14. Br. J. Haematol. 168, 583–597 10.1111/bjh.13164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ricci M. S., Jin Z., Dews M., Yu D., Thomas-Tikhonenko A., Dicker D. T., and El-Deiry W. S. (2004) Direct repression of FLIP expression by c-myc is a major determinant of TRAIL sensitivity. Mol. Cell. Biol. 24, 8541–8555 10.1128/MCB.24.19.8541-8555.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vo B. T., Wolf E., Kawauchi D., Gebhardt A., Rehg J. E., Finkelstein D., Walz S., Murphy B. L., Youn Y. H., Han Y. G., Eilers M., and Roussel M. F. (2016) The interaction of Myc with Miz1 defines medulloblastoma subgroup identity. Cancer Cell 29, 5–16 10.1016/j.ccell.2015.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nakagawa M., Takizawa N., Narita M., Ichisaka T., and Yamanaka S. (2010) Promotion of direct reprogramming by transformation-deficient Myc. Proc. Natl. Acad. Sci. U.S.A. 107, 14152–14157 10.1073/pnas.1009374107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Roczniak-Ferguson A., Petit C. S., Froehlich F., Qian S., Ky J., Angarola B., Walther T. C., and Ferguson S. M. (2012) The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 5, ra42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Seiler C. Y., Park J. G., Sharma A., Hunter P., Surapaneni P., Sedillo C., Field J., Algar R., Price A., Steel J., Throop A., Fiacco M., and LaBaer J. (2014) DNASU plasmid and PSI:Biology-Materials repositories: resources to accelerate biological research. Nucleic Acids Res. 42, D1253–1260 10.1093/nar/gkt1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cormier C. Y., Mohr S. E., Zuo D., Hu Y., Rolfs A., Kramer J., Taycher E., Kelley F., Fiacco M., Turnbull G., and LaBaer J. (2010) Protein structure initiative material repository: an open shared public resource of structural genomics plasmids for the biological community. Nucleic Acids Res. 38, D743–749 10.1093/nar/gkp999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cormier C. Y., Park J. G., Fiacco M., Steel J., Hunter P., Kramer J., Singla R., and LaBaer J. (2011) PSI:Biology-materials repository: a biologist's resource for protein expression plasmids. J. Struct. Funct. Genomics 12, 55–62 10.1007/s10969-011-9100-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yang X., Boehm J. S., Yang X., Salehi-Ashtiani K., Hao T., Shen Y., Lubonja R., Thomas S. R., Alkan O., Bhimdi T., Green T. M., Johannessen C. M., Silver S. J., Nguyen C., Murray R. R., et al. (2011) A public genome-scale lentiviral expression library of human ORFs. Nat. Methods 8, 659–661 10.1038/nmeth.1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Carey M. F., Peterson C. L., and Smale S. T. (2009) Chromatin immunoprecipitation (ChIP). Cold Spring Harb. Protocols 2009, 10.1101/pdb.prot5279 [DOI] [PubMed] [Google Scholar]