Abstract
Interleukin 4 (IL4) is generally viewed as a Th2 cytokine capable of polarizing macrophages into an anti-inflammatory phenotype, whereas granulocyte macrophage-colony-stimulating factor (GM-CSF) is often viewed as a proinflammatory cytokine with part of this function due to its action on monocytes/macrophages. Paradoxically, these two cytokines act additively to enhance the in vitro differentiation of dendritic cells from precursors such as monocytes. One up-regulated marker of an IL4-polarized M2 macrophage is the chemokine (C–C motif) ligand 17 (CCL17), which we have recently reported to be induced by GM-CSF in monocytes/macrophages in an interferon regulatory factor 4 (IRF4)–dependent manner. In this study, we report that IL4 also induces CCL17 production by acting through IRF4 in human monocytes and murine macrophages. Furthermore, evidence is presented that IL4 up-regulates IRF4 expression at the epigenetic level by enhancing the expression and activity of jumonji domain–containing protein 3 (JMJD3) demethylase. Intriguingly, silencing the signal transducer and activator of transcription 6 (STAT6) gene led to a decrease in not only CCL17 formation, but also in that of its upstream regulators, JMJD3 and IRF4. Moreover, IL4 treatment of human monocytes resulted in an increased association of STAT6 to the promoter regions of the CCL17, IRF4, and JMJD3 genes. Thus, despite their vastly different functions, IL4 and GM-CSF appear to share elements of a common signaling pathway in regulating CCL17 production in human monocytes and murine macrophages.
Keywords: interferon regulatory factor (IRF), STAT transcription factor, signal transduction, epigenetics, monocyte, macrophage, chemokine, cytokine, dendritic cell, interleukin
Introduction
Cells of the monocyte/macrophage lineage are involved in host defense and inflammation, including in the resolution phase leading to homeostasis (1). During inflammation, the numbers of circulating and tissue-recruited monocytes can be elevated (2). Monocytes are highly plastic and they can differentiate into macrophages or monocyte-derived dendritic cells depending on the tissue milieu (3). Plasticity is also evident in the monocyte/macrophage lineage at sites of inflammation as demonstrated by both their proinflammatory and resolving properties, and the concept of macrophage “polarization” into M1 and M2 activation states, respectively, has been developed to help define such properties (4–6).
Following the finding that it could lower inflammatory cytokine formation in human monocytes (7), the cytokine, IL4,2 has been considered to be the M2-polarizing cytokine par excellence (4–6). It has also been proposed as a possible anti-inflammatory therapeutic (8). Its effects on monocyte/macrophage biology have been widely studied although the nature of the response can sometimes exhibit species specificity (9, 10).
The chemokine, CCL17 (also called thymus and activation–regulated chemokine (TARC)), was initially implicated in the recruitment of Th2 lymphocytes; however, it was later shown to be able to recruit Th1 and Th17 lymphocytes (11, 12). It has been viewed as a M2 cytokine because IL4 can induce its formation in macrophages (13). The IFN regulatory factor (IRF) family of transcription factors are broadly implicated in immune cell differentiation and function (14). IRF4 is hemopoietic specific, critical for myeloid and lymphoid lineage development and function (14); in macrophages it has been considered to provide an anti-inflammatory signal (15, 16) and to have a nonredundant role in shaping the phenotype of IL4-treated M2 or “alternatively activated” macrophages (17). Another member of the IRF family, IRF5, has been proposed to be key for IFNγ- and granulocyte macrophage-colony-stimulating factor (GM-CSF)–dependent M1 macrophage polarization (18).
The best characterized function of the histone demethylase, jumonji domain–containing protein 3 (JMJD3) (also known as lysine-specific demethylase 6B (KDM6B)), is to catalyze the demethylation of trimethylated histone, H3K27me3, to its monomethylated state (19), because methylation of lysine at these sites is usually associated with repression of transcription, JMJD3 activity is expected to enhance gene expression. There is disagreement whether IL4 requires JMJD3-mediated H3K27me3 demethylation to promote M2 macrophage polarization (19–21).
GM-CSF is often considered to be a proinflammatory cytokine and clinical trials targeting it or its receptor have met success, for example, in rheumatoid arthritis trials (22, 23). Its action on monocyte/macrophage populations is considered critical in this context (24). A very common method for generating human monocyte–derived dendritic cells and murine DCs in vitro is to culture monocytes or bone marrow cells in GM-CSF and IL4, because these cytokines act together to enhance DC differentiation (25, 26) despite the evidence they can exhibit pro- and anti-inflammatory properties, respectively; also they appear to act via distinct signaling pathways because IL4 transduces signals via signal transducer and activator of transcription (STAT6), whereas GM-CSF activates STAT5 (27, 28).
We have recently shown that GM-CSF can induce CCL17 production in human monocytes and murine macrophages in an IRF4-dependent manner (29); furthermore, we demonstrated that GM-CSF–mediated up-regulation of IRF4 expression was in turn dependent on increased JMJD3 activity. Evidence was also obtained that this pathway could be critical to the proinflammatory and algesic functions of GM-CSF (29). Given the above background literature on IL4 and GM-CSF, either alone or combination, we considered that IL4 might induce CCL17 production in the monocyte/macrophage population via a similar pathway. We provide evidence here for this proposition.
Results
IL4 induces CCL17 expression in human monocytes and murine macrophages
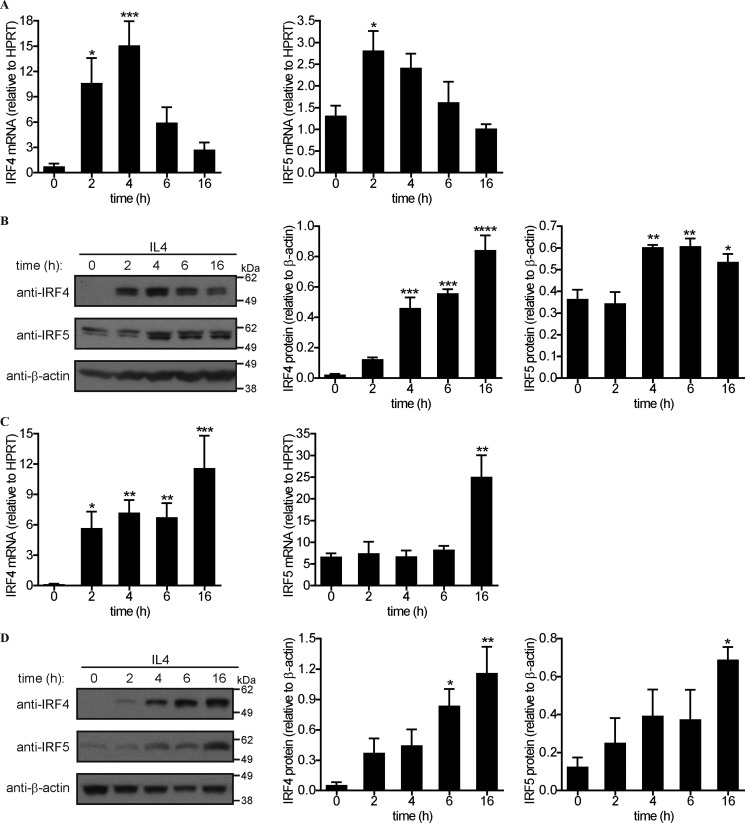
We have reported that GM-CSF can induce CCL17 mRNA and protein secretion in human monocytes and murine bone marrow–derived macrophages (BMM) (29). We first compared the effects of IL4 on the same populations under the same culture conditions as for the prior GM-CSF studies. As we reported before (29), CCL17 mRNA expression and secreted protein could not be detected basally in either of the unstimulated human monocytes and BMM populations (Fig. 1, A–D). IL4 treatment led to a gradual increase in CCL17 mRNA in both cell types, which was maximally expressed at 16 h over the examined period (Fig. 1, A and C); secreted CCL17 protein could not be detected until between 6 and 16 h in the culture supernatants of both cell populations following IL4 treatment (Fig. 1, B and D).
Figure 1.
IL4 induces CCL17 expression in human monocytes and murine macrophages. A–D, human monocytes (A and B) and BMM (C and D) were treated with IL4 (20 ng/ml) for the indicated periods. A and C, CCL17 mRNA expression by qPCR. All the samples were assayed in duplicate and normalized to the housekeeping HPRT mRNA expression. B and D, secreted CCL17 protein in culture supernatants by ELISA. The results were plotted as mean ± S.E. (n = 5 donors/mice). Statistical analyses were performed by one-way ANOVA with Dunnett's multiple comparisons test, where *, p < 0.05; ***, p < 0.001; and ****, p < 0.0001 versus time (h) = 0. ND, not detected.
IL4 up-regulates IRF4 expression in human monocytes and murine macrophages
We have previously shown that GM-CSF–induced CCL17 formation was up-regulated through IRF4 in human monocytes and BMM (29). We therefore checked whether IL4 could up-regulate IRF4 expression in our cultures as has been reported for these cell populations (20, 30). The expression levels of another member of the IRF family, namely IRF5, were also monitored as it can play a key role in monocyte/macrophage activation/polarization (31). IL4 up-regulated IRF4 and IRF5 mRNA expression transiently in human monocytes over the 16-h time period examined with the relative increase being higher for the former mRNA (Fig. 2A). IRF4 protein was expressed basally at very low levels (Fig. 2B) and IL4 led to a gradual increase over the 16-h period. IRF5 protein, on the other hand, was easily detected basally and, as for GM-CSF stimulation (29), its levels were slightly increased by IL4 (Fig. 2B).
Figure 2.
IL4 up-regulates IRF4 expression in human monocytes and murine macrophages. A–D, human monocytes (A and B) and BMM (C and D) were treated with IL4 (20 ng/ml) for the indicated periods. A and C, IRF4 and IRF5 mRNA expression were measured by qPCR. All the samples were assayed in duplicate and normalized to the housekeeping HPRT mRNA expression. B and D, whole cell lysates were subjected to Western blotting with anti-IRF4, anti-IRF5, and anti-β-actin antibodies; IRF4 and IRF5 proteins were quantified relative to β-actin. The graphs were plotted as mean ± S.E. (n = 3 donors/mice). Statistical analyses were performed by one-way ANOVA with Dunnett's multiple comparisons test, where *, p < 0.05; **, p < 0.01; ***, p < 0.001; and ****, p < 0.0001 versus time (h) = 0.
The stimulation of BMM with IL4 also led to an increase in IRF4 mRNA with a more sustained expression than in the human population (Fig. 2C); the basal IRF5 mRNA levels were also increased somewhat by IL4 between 6 and 16 h (Fig. 2C). The basal levels of IRF4 and IRF5 proteins were gradually raised by IL4 with the relative increase in IRF4 formation being higher in this cell type as well (Fig. 2D). Overall, these data confirm that, as for GM-CSF, IL4 can up-regulate IRF4 mRNA expression and protein in both human monocytes and BMM.
IL4-induced CCL17 production is IRF4-dependent in human monocytes and murine macrophages
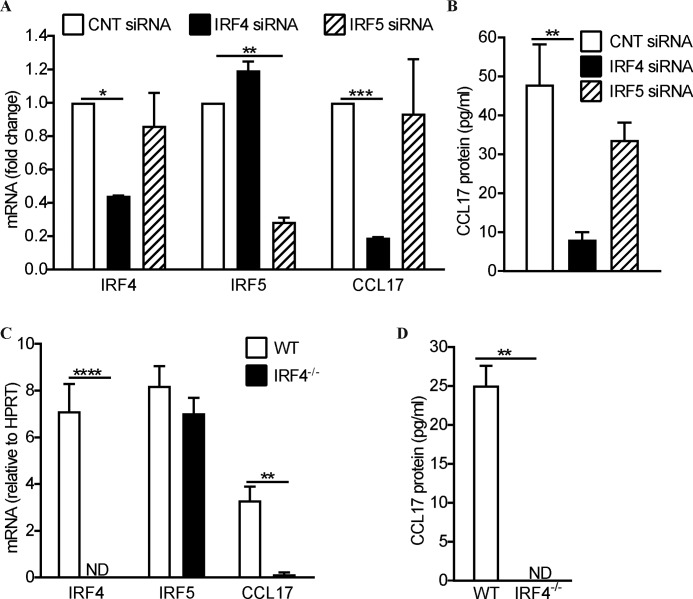
Having identified above that IL4 up-regulated CCL17 levels, as well as those of IRF4 and IRF5, we next explored the requirement of these transcription factors for such up-regulated CCL17 production. As we have found for GM-CSF stimulation (29), silencing the IRF4 gene in human monocytes significantly decreased IL4-induced CCL17 mRNA (Fig. 3A) and secreted protein (Fig. 3B), but not IRF5 mRNA (Fig. 3A). On the other hand, IRF5 gene silencing had no effect on the expression of either CCL17 or IRF4 (Fig. 3, A and B) as we found for GM-CSF stimulation in this cell type (29).
Figure 3.
IL4-induced CCL17 production is IRF4-dependent in human monocytes and murine macrophages. A and B, human monocytes were nucleofected with IRF4 siRNA, IRF5 siRNA, or a nontargeting control siRNA (CNT) before being treated with IL4 (20 ng/ml) for 16 h. C and D, BMM from both wildtype (WT) and IRF4 knockout (IRF4−/−) mice were treated with IL4 (20 ng/ml) for 16 h. A and C, IRF4, IRF5, and CCL17 mRNA were measured by qPCR, in duplicate, and normalized to the housekeeping HPRT mRNA expression. mRNA expression was plotted as fold-change relative to that in CNT siRNA, which was given an arbitrary value of 1.0. B and D, secreted CCL17 protein in culture supernatants was measured by ELISA. The graphs were plotted as mean ± S.E. (n = 3 donors/mice). Statistical analyses were performed by two-way ANOVA with Sidak's multiple comparisons test (A and C), one-way ANOVA with Dunnett's multiple comparisons test (B) or paired t test (D), where *, p < 0.05; **, p < 0.01; ***, p < 0.001; and ****, p < 0.0001. ND, not detected.
Consistent with the human monocyte data, IL4-treated BMM from Irf4−/− mice had lower CCL17 mRNA than IL4-treated wildtype (WT) BMM (Fig. 3C) and no secreted CCL17 (Fig. 3D). In contrast, and again indicating some specificity, the IRF5 mRNA levels were similar in WT and Irf4−/− BMM (Fig. 3C). Thus, the CCL17 induction by IL4 in human monocytes and murine macrophages is IRF4 dependent.
IL4 up-regulates JMJD3 demethylase expression and activity in human monocytes
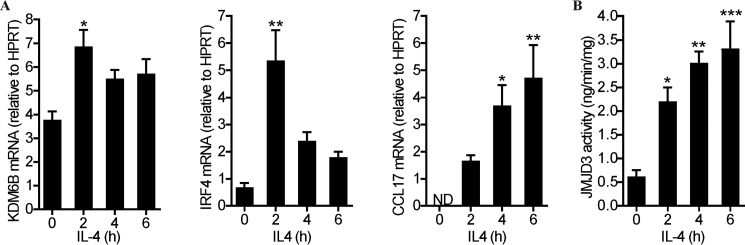
At steady state, the promoter region of IRF4 is enriched in the trimethylated histone, H3Lys27me3, which can be converted by JMJD3 (KDM6B) demethylase to its monomethylated state, thereby promoting transcription (21). We have previously provided evidence that GM-CSF up-regulated JMJD3 expression and activity to control IRF4 transcription in human monocytes (29). Thus, we hypothesized that IL4 might also up-regulate JMJD3 expression and/or activity, thereby controlling IRF4 transcription and eventually CCL17 production. In human monocytes, the basal KDM6B mRNA levels were increased at 2 h by IL4 with similar kinetics to IRF4 mRNA in the same donor monocytes (Fig. 4A). CCL17 mRNA had a similar gradual increase in these donor monocytes (Fig. 4A) as for the donor monocytes in Fig. 1A. In addition to up-regulating JMJD3 (KDM6B) mRNA expression, IL4 treatment of human monocytes also resulted in its increased demethylase activity (Fig. 4B).
Figure 4.
IL4 up-regulates JMJD3 demethylase expression and activity in human monocytes. Human monocytes were treated with IL4 (20 ng/ml) for the indicated periods. A, KDM6B, IRF4, and CCL17 mRNA expression by qPCR. All the samples were assayed in duplicate and normalized to the housekeeping HPRT mRNA expression. B, JMJD3 activity by a colorimetric assay. The graphs were plotted as mean ± S.E. (n = 3 donors). Statistical analyses were performed by one-way ANOVA with Dunnett's multiple comparisons test, where *, p < 0.05; **, p < 0.01; and ***, p < 0.001 versus time (h) = 0. ND, not detected.
JMJD3 activity is required for IL4-induced IRF4 and CCL17 expression in human monocytes and murine macrophages
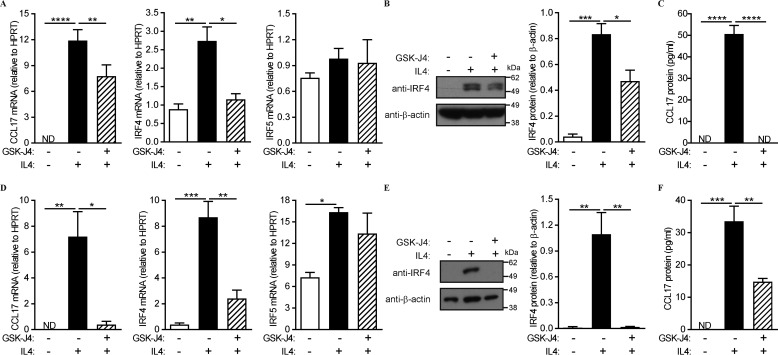
Having shown that IL4 up-regulates JMJD3 demethylase mRNA expression and activity in human monocytes, we next investigated whether a potent inhibitor of JMJD3 activity, GSK-J4 (32), would ameliorate IL4-induced IRF4 and CCL17 formation as we have found previously for their GM-CSF–mediated induction (29). Treatment with GSK-J4 before IL4 stimulation resulted in a decrease in IRF4 and CCL17 mRNA in human monocytes (Fig. 5A) as well as a decrease in the respective proteins (Fig. 5, B and C). In contrast, IRF5 mRNA levels were not affected by GSK-J4 (Fig. 5A).
Figure 5.
JMJD3 activity is required for IL4-induced IRF4 and CCL17 expression in human monocytes and murine macrophages. A–F, human monocytes (A–C) and BMM (D–F) were pre-treated with GSK-J4 (10 μm) for 30 min before treating with IL4 (20 ng/ml) for 16 h. A and D, mRNA expression of CCL17, IRF4, and IRF5 were measured by qPCR. All the samples were assayed in duplicate and normalized to the housekeeping HPRT mRNA expression. B and E, whole cell lysates were subjected to Western blotting with anti-IRF4 and anti-β-actin antibodies; IRF4 protein was quantified relative to β-actin. C and F, secreted CCL17 protein in culture supernatants was measured by ELISA. The graphs were plotted as mean ± S.E. (n = 3 donors/mice). Statistical analyses were performed by one-way ANOVA with Tukey's multiple comparisons test, where *, p < 0.05; **, p < 0.01; ***, p < 0.001; and ****, p < 0.0001. ND, not detected.
Consistent with the human monocyte data, inhibition of JMJD3 activity in BMM also resulted in a decrease in IRF4 and CCL17 mRNA (Fig. 5D) and protein (Fig. 5, E and F). Again, GSK-J4 pre-treatment had no effect on IL4-regulated IRF5 mRNA expression in BMM (Fig. 5D). These data suggest that JMJD3 demethylase activity is required for IL4-induced IRF4 and CCL17 expression in human monocytes and murine macrophages.
IL4-induced CCL17 expression is regulated by JMJD3 demethylase activity at the IRF4 promoter in human monocytes
Having shown that IL4-induced IRF4 and CCL17 expression are dependent on JMJD3 activity, we next explored whether JMJD3 directly regulates the transcription of IRF4 and CCL17. In human monocytes, ChIP analyses demonstrated that GSK-J4 blocked the recruitment of RNA polymerase II to the transcription start sites (TSS) of both IRF4 and CCL17 (Fig. 6, A and B). Significantly, and as reported for GM-CSF–treated monocytes (29), inhibition of JMJD3 activity prevented the IL4-induced loss of H3K27me3 association to the IRF4 TSS, while not altering the total H3 at this site (Fig. 6A); in contrast, IL4 did not alter H3K27me3 association to the CCL17 TSS and GSK-J4 had no effect on this association (Fig. 6B).
Figure 6.
IL4-induced CCL17 expression is regulated by JMJD3 demethylase activity at the IRF4 promoter in human monocytes. A and B, human monocytes were pre-treated with GSK-J4 (10 μm) for 30 min before treating with IL4 (20 ng/ml) for 1 h. A and B, ChIP analysis of the association of RNA Pol II, H3K27me3, and total H3 with: the IRF4 TSS (A), and the CCL17 TSS (B) expressed as percentage of input DNA. The graphs were plotted as mean ± S.E. (n = 3 donors). Statistical analyses were performed by one-way ANOVA with Tukey's multiple comparisons test, where *, p < 0.05 and **, p < 0.01.
IL4-induced CCL17 production is STAT6-dependent in human monocytes
STAT6 is a key transcription factor regulating IL4 signaling (27). Therefore, we investigated whether STAT6 is involved in IL4-induced CCL17 formation in human monocytes. Silencing the STAT6 gene decreased CCL17 mRNA expression (Fig. 7A), as well as secreted CCL17 protein (Fig. 7B). Such silencing also decreased IRF4 and KDM6B mRNA expression (Fig. 7A), consistent with there being a link to the reduced CCL17 mRNA expression given our findings above. As controls, STAT5A and STAT5B genes were not modulated by the STAT6 gene silencing.
Figure 7.
IL4-induced CCL17 production is STAT6-dependent in human monocytes. A and B, human monocytes were nucleofected with either STAT6 siRNA or a nontargeting control siRNA (CNT) before being treated with IL4 (20 ng/ml) for 16 h. A, mRNA expression of STAT6, CCL17, IRF4, KDM6B, STAT5A, and STAT5B were measured by qPCR, in duplicate, and normalized to the housekeeping HPRT mRNA expression. mRNA expression was plotted as fold-change relative to that in CNT siRNA, which was given an arbitrary value of 1.0. B, secreted CCL17 protein in culture supernatants was measured by ELISA. C, human monocytes were treated with IL4 (20 ng/ml) for 1 h. ChIP analyses of the association of STAT6 with the CCL17, IRF4, and KDM6B promoter regions is expressed as percentage of input DNA. The graphs were plotted as mean ± S.E. (n = 4 donors). Statistical analyses were performed by two-way ANOVA with Sidak's multiple comparisons test (A and C) or paired t test (B), where *, p < 0.05; **, p < 0.01; and ****, p < 0.0001. D, a schematic diagram depicting IL4-induced CCL17 production in monocytes/macrophages, incorporating epigenetic and transcriptional regulators, STAT6, JMJD3, and IRF4.
Having identified that STAT6 is required for not only CCL17 expression, but also for that of its upstream regulators, JMJD3 and IRF4, we next explored whether STAT6 can regulate the transcription of each of these genes. Previous studies have identified STAT6-binding sites in the promoter regions of the CCL17, IRF4, and KDM6B genes (20, 33–35). IL4 treatment of human monocytes resulted in increased association of STAT6 with each of these gene promoters (Fig. 7C). Collectively these data suggest that IL4-induced CCL17 production can be transcriptionally regulated by STAT6 in human monocytes.
Discussion
We have reported above that IL4 induces CCL17 formation in monocytes/macrophages and with upstream involvement of JMJD3-regulated IRF4; importantly, this pathway was identified in both human and murine cells indicating a general relevance. Also, silencing the STAT6 gene in human monocytes not only suppressed IL4-induced CCL17 production but also inhibited KDM6B and IRF4 mRNA expression. Furthermore, we have demonstrated that STAT6 can bind to the promoter regions of the CCL17, IRF4, and KDM6B genes. Our finding of IL4-induced CCL17 production in human monocytes is in accord with the literature (36, 37), and our data in BMM are consistent with previous findings in murine peritoneal macrophages (30), wherein CCL17 was considered as one of the M2 polarization markers enhanced in the presence of IL4. The induction of CCL17 by IL4 in both cell types parallels our findings with GM-CSF (29); as we also found for GM-CSF stimulation, in both cell populations IL4 induced IRF4 mRNA expression, which was required for CCL17 induction. We have previously shown that GM-CSF and IL4 do not induce expression of each other in human monocytes (38, 39), suggesting that CCL17 formation is regulated independently by these two cytokines.
In studies with BMM there is disagreement in the literature whether IL4-induced M2 activation is JMJD3 dependent or not (19–21). We have provided evidence above that, again as for GM-CSF stimulation (29), JMJD3 activity is required for IL4-induced IRF4 and ultimately CCL17 expression in BMM and human monocytes. We have also found in human monocytes that IL4-regulated JMJD3 activity was required to remove repressive H3K27me3 at the IRF4 gene locus, but not at the CCL17 gene locus. Collectively, these data suggest that both GM-CSF and IL4 signaling converge at the JMJD3/IRF4 epigenetic/transcriptional levels during regulation of CCL17 formation. It would be of interest to ascertain what other IL4-stimulated downstream mediators whose expression also requires this epigenetic mechanism in each of the cell types in question. Even though the JMJD3 inhibitor, GSK-J4 (32), was not cytotoxic to either monocytes or BMM over the experimental period, the possibility of off-target effects cannot be discounted.
IL4 signaling is mediated via STAT6 phosphorylation by JAK1 and JAK3 kinases (27, 40); when phosphorylated, STAT6 is then translocated to the nucleus where it regulates transcriptional activity of IL4-regulated genes. In line with our siRNA data in human monocytes, it has been reported that IL4 stimulation of BMM from Stat6−/− mice displayed decreased JMJD3 expression compared with their WT littermates (20); furthermore, IL4 treatment of BMM led to enhanced association of STAT6 to the JMJD3 promoter, suggesting that IL4 controls JMJD3 transcription via STAT6 (20). In addition, it has been suggested that the induction of CCL17 by IL4 in macrophages results from cooperative interactions between STAT6-binding sites within the CCL17 promoter (30). Consistent with the literature, we found that STAT6 has a nonredundant role in IL4-induced CCL17 production, as it not only binds to the CCL17 promoter, but also controls the transcription of the upstream regulators, JMJD3 and IRF4 (Fig. 7D). Given that GM-CSF signaling is mediated via the activation of STAT5 (41), it is tempting to speculate that GM-CSF–activated STAT5 may also transcriptionally regulate CCL17, IRF4, and JMJD3 expression in a similar manner as IL4-activated STAT6.
We have mentioned previously (29) that GM-CSF can be added to the list of cytokines, including IL4 and TSLP, which can up-regulate CCL17 expression in monocytes/macrophages, with potential ramifications for pathology. Considering the quite different biologies of GM-CSF and IL4, including their distinct STAT signaling mechanisms, any similarities are not often highlighted even though, as mentioned above, they are widely used in tandem to achieve DC differentiation in vitro. It is quite striking and perhaps surprising that these two different cytokines can share at least one common pathway whose significance for their individual and joint biologies needs to be explored.
Experimental procedures
Isolation and culture of human monocytes
Buffy coats were sourced ethically and their research use was in accord with the terms of the informed consents obtained by the Australian Red Cross Blood Service. Experiments involving human subjects were approved by the Melbourne Health Human Research Ethics Committee and conducted in accordance with the Declaration of Helsinki principles. Human monocytes were isolated as described before (29). Briefly, human monocytes were isolated using the RosetteSep Ab mixture (Stem Cell Technologies, Vancouver, Canada), which negatively selects CD14+ monocytes, followed by Ficoll-Paque density gradient centrifugation. They were cultured in RPMI 1640, supplemented with 10% heat-inactivated FCS, 2 mm GlutaMax-1 (Life Technologies Inc.), 100 units/ml of penicillin, and 100 mg/ml of streptomycin. Monocytes were stimulated with recombinant human IL4 (20 ng/ml, R&D Systems, Minneapolis, MN) for the indicated time periods. Where indicated, human monocytes and mouse macrophages were pre-treated with 10 μm GSK-J4 before treating with IL4. GSK-J4 did not affect the survival of either population compared with vehicle control, as ascertained by an apoptosis assay (42), using an Annexin V/PI detection kit (BD Biosciences) over a 72-h time period.
Isolation and culture of bone marrow–derived mouse macrophages
Experiments involving animals were approved by the University of Melbourne Ethics Committee for Animal Experimentation. Mouse macrophages were prepared as previously described (29, 43). Briefly, bone marrow cells were isolated from femurs of either C57BL/6 WT or Irf4−/− mice and cultured in RPMI 1640 medium supplemented with 10% heat-inactivated FCS, 2 mm GlutaMax-1, 100 units/ml of penicillin, and 100 mg/ml of streptomycin in the presence of M-CSF (5,000 units/ml). At day 4, nonadherent cells were collected and cultured for a further 3 days again in M-CSF (5,000 units/ml) to derive BMM. The BMM were then stimulated with recombinant murine IL4 (20 ng/ml, R&D Systems), in the absence of M-CSF, for the indicated periods of time.
Quantitative PCR
Total RNA was extracted using ISOLATE II RNA Mini Kit (Bioline, London, UK) and reverse transcribed using SuperScript III reverse transcriptase (Invitrogen). Quantitative PCR (qPCR) was performed using the ABI PRISM 7900HT sequence detection system (Applied Biosystems, Carlsbad, CA) and predeveloped TaqMan probe/primer combinations for human CCL17, IRF4, IRF5, KDM6B, STAT6, STAT5A, STAT5B, and HPRT, as well as for murine CCL17, IRF4, and IRF5 (Applied Biosystems). Threshold cycle numbers were transformed to cycle threshold values, and the results were plotted using GraphPad Prism version 7.04.
ELISA
Secreted human and murine CCL17 (R&D Systems) in culture supernatants were measured by ELISA as per the manufacturer's instructions.
Western blotting
Western blotting was performed as described previously (29). Briefly, whole cell extracts were lysed with RIPA buffer. Protein concentrations were determined with a Bio-Rad protein assay kit. Equal amounts of whole cell lysates were run on 10% NuPAGE (Life Technologies). The separated proteins were transferred onto a polyvinylidene difluoride membrane and then Western blotted with appropriate antibodies (Abs). Abs were against human IRF4 (clone M-17) and human IRF5 (clone 10T1) (Santa Cruz Biotechnology, Dallas, TX), and murine β-actin (clone AC-74, Sigma). Western blots were quantified by densitometry using Quantity One version 4.6.9 (Bio-Rad) and the resulting data were plotted as mean ± S.E.
Gene silencing
Human monocytes were nucleofected with 100 nm ON-TARGETplus SMARTpool (GE Dharmacon, Lafayette, CO) human IRF4 (L-019668), IRF5 (L-011706), STAT6 (L-006690) or a nontargeting control (D-001810) siRNA with an Amaxa Kit (Lonza, Basel, Switzerland) according to the manufacturer's instructions. The cells were then cultured for another 16 h in the presence of IL4 (20 ng/ml) before analysis.
JMJD3 activity assay
Human monocytes were lysed following treatment with IL4 (20 ng/ml), and nuclear fractions were enriched with NE-PER Nuclear and cytoplasmic extraction reagents (Thermo Fisher Scientific, Waltham, MA). 10 μg of nuclear extracts was subjected to the JMJD3 activity assay with a 120-min incubation period, using a colorimetric Epigenase JMJD3 Demethylase Activity Assay Kit (Epigentek, Farmingdale, NY), as before (29). The demethylated product was ascertained from the OD at 450 nm using a standard curve, and JMJD3 activity (ng/min/mg) was calculated as the demethylated product (ng) divided by incubation time (min) and input nuclear extract (mg).
ChIP assay
ChIP assays were performed as described previously (29). Briefly, human monocytes were treated with IL4 (20 ng/ml) for 1 h before cross-linking protein–DNA complexes with 1% formaldehyde for 10 min at room temperature. ChIP was performed with a ChIP Assay Kit (17-295, Millipore) as per the manufacturer's instructions. Immunoprecipitation was performed with 1 μg of anti-RNA Pol II (2629, Cell Signaling Technologies), anti-H3K27me3 (07-449, Millipore), anti-histone H3 (05-925, Millipore), anti-STAT6 (5397, Cell Signaling Technologies), or control rabbit IgG (2729, Cell Signaling Technologies) antibodies, followed by reversal of cross-linking. Immunoprecipitated DNA fragments were then amplified by qPCR with a SensiFAST SYBR Hi-ROX Kit (Bioline) and the following specific primers for: human IRF4 TSS (forward, 5′-ccacctcgcactctcagttt-3′ and reverse, 5′-ctggaggtcgaacctctggt-3′) and human CCL17 TSS (forward, 5′-ggaaggatgtgaggaggtga-3′ and reverse, 5′-gtggatgtgctgcagagaag-3′), as well as STAT6-binding sites in the promoter regions of human CCL17 (forward, 5′-cagctgtgcgtggaggcttttca-3′ and reverse, 5′-tccttccctagaccagtgaagttcgaaga-3′), human IRF4 (forward, 5′-ccacatcgctgcagtttagt-3′ and reverse, 5′-ttcggggactgtcactgg-3′) and human KDM6B (forward, 5′-gcgtgaaggaggaagtgaaa-3′ and reverse, 5′-cggaagcggtgtgtgtaaat-3′) genes. Enrichment of histones, RNA Pol II, and STAT6 at the gene loci was expressed as percentage of input DNA.
Statistics
Statistical analyses between groups were performed using one-way ANOVA with Dunnett's/Tukey's multiple comparisons test, two-way ANOVA with Sidak's multiple comparisons test or paired t test, as indicated. The p values < 0.05 indicate significance. Data were plotted as mean ± S.E. from at least three independent experiments using GraphPad Prism version 7.04.
Author contributions
A. T. H., T. J. L., M.-C. L., A. J. F., and A. A. formal analysis; A. T. H., T. J. L., A. J. F., and A. A. investigation; A. T. H., T. J. L., M.-C. L., and A. A. methodology; A. T. H. and A. A. writing-original draft; A. J. F. software; A. D. C., J. A. H., and A. A. supervision; A. D. C., J. A. H., and A. A. funding acquisition; A. D. C., J. A. H., and A. A. writing-review and editing; A. A. conceptualization; A. A. data curation; A. A. project administration.
This work was supported by grants from the National Health and Medical Research Council of Australia (to J. A. H. and A. D. C), and a Research Grant Support Scheme from the University of Melbourne (to A. A.). The authors declare that they have no conflicts of interest with the contents of this article.
- IL4
- interleukin 4
- IRF4
- interferon regulatory factor 4
- GM-CSF
- granulocyte macrophage-colony-stimulating factor
- BMM
- bone marrow–derived macrophages
- TSS
- transcription start site
- STAT6
- signal transducer and activator of transcription 6
- JMJD3
- jumonji domain–containing protein 3
- KDM6B
- lysine-specific demethylase 6B
- qPCR
- quantitative PCR
- HPRT
- hypoxanthine-guanine phosphoribosyltransferase
- Ab
- antibody
- ANOVA
- analysis of variance.
References
- 1. Italiani P., and Boraschi D. (2014) From monocytes to M1/M2 macrophages: phenotypical vs. functional differentiation. Front. Immunol. 5, 514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shi C., and Pamer E. G. (2011) Monocyte recruitment during infection and inflammation. Nat. Rev. Immunol. 11, 762–774 10.1038/nri3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Guilliams M., Ginhoux F., Jakubzick C., Naik S. H., Onai N., Schraml B. U., Segura E., Tussiwand R., and Yona S. (2014) Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat. Rev. Immunol. 14, 571–578 10.1038/nri3712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gordon S., and Martinez F. O. (2010) Alternative activation of macrophages: mechanism and functions. Immunity 32, 593–604 10.1016/j.immuni.2010.05.007 [DOI] [PubMed] [Google Scholar]
- 5. Mantovani A., Sica A., and Locati M. (2005) Macrophage polarization comes of age. Immunity 23, 344–346 10.1016/j.immuni.2005.10.001 [DOI] [PubMed] [Google Scholar]
- 6. Murray P. J., Allen J. E., Biswas S. K., Fisher E. A., Gilroy D. W., Goerdt S., Gordon S., Hamilton J. A., Ivashkiv L. B., Lawrence T., Locati M., Mantovani A., Martinez F. O., Mege J. L., Mosser D. M., et al. (2014) Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 41, 14–20 10.1016/j.immuni.2014.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hart P. H., Vitti G. F., Burgess D. R., Whitty G. A., Piccoli D. S., and Hamilton J. A. (1989) Potential antiinflammatory effects of interleukin 4: suppression of human monocyte tumor necrosis factor α, interleukin 1, and prostaglandin E2. Proc. Natl. Acad. Sci. U.S.A. 86, 3803–3807 10.1073/pnas.86.10.3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hemmerle T., Doll F., and Neri D. (2014) Antibody-based delivery of IL4 to the neovasculature cures mice with arthritis. Proc. Natl. Acad. Sci. U.S.A. 111, 12008–12012 10.1073/pnas.1402783111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheung D. L., and Hamilton J. A. (1992) Regulation of human monocyte DNA synthesis by colony-stimulating factors, cytokines, and cyclic adenosine monophosphate. Blood 79, 1972–1981 [PubMed] [Google Scholar]
- 10. Jenkins S. J., Ruckerl D., Cook P. C., Jones L. H., Finkelman F. D., van Rooijen N., MacDonald A. S., and Allen J. E. (2011) Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science 332, 1284–1288 10.1126/science.1204351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Alferink J., Lieberam I., Reindl W., Behrens A., Weiss S., Hüser N., Gerauer K., Ross R., Reske-Kunz A. B., Ahmad-Nejad P., Wagner H., and Förster I. (2003) Compartmentalized production of CCL17 in vivo: strong inducibility in peripheral dendritic cells contrasts selective absence from the spleen. J. Exp. Med. 197, 585–599 10.1084/jem.20021859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hayashida S., Uchi H., Moroi Y., and Furue M. (2011) Decrease in circulating Th17 cells correlates with increased levels of CCL17, IgE and eosinophils in atopic dermatitis. J. Dermatol. Sci. 61, 180–186 10.1016/j.jdermsci.2010.10.013 [DOI] [PubMed] [Google Scholar]
- 13. Martinez F. O., Gordon S., Locati M., and Mantovani A. (2006) Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J. Immunol. 177, 7303–7311 10.4049/jimmunol.177.10.7303 [DOI] [PubMed] [Google Scholar]
- 14. Tamura T., Yanai H., Savitsky D., and Taniguchi T. (2008) The IRF family transcription factors in immunity and oncogenesis. Annu. Rev. Immunol. 26, 535–584 10.1146/annurev.immunol.26.021607.090400 [DOI] [PubMed] [Google Scholar]
- 15. Negishi H., Ohba Y., Yanai H., Takaoka A., Honma K., Yui K., Matsuyama T., Taniguchi T., and Honda K. (2005) Negative regulation of Toll-like-receptor signaling by IRF-4. Proc. Natl. Acad. Sci. U.S.A. 102, 15989–15994 10.1073/pnas.0508327102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Honma K., Udono H., Kohno T., Yamamoto K., Ogawa A., Takemori T., Kumatori A., Suzuki S., Matsuyama T., and Yui K. (2005) Interferon regulatory factor 4 negatively regulates the production of proinflammatory cytokines by macrophages in response to LPS. Proc. Natl. Acad. Sci. U.S.A. 102, 16001–16006 10.1073/pnas.0504226102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. El Chartouni C., Schwarzfischer L., and Rehli M. (2010) Interleukin-4 induced interferon regulatory factor (Irf) 4 participates in the regulation of alternative macrophage priming. Immunobiology 215, 821–825 10.1016/j.imbio.2010.05.031 [DOI] [PubMed] [Google Scholar]
- 18. Krausgruber T., Blazek K., Smallie T., Alzabin S., Lockstone H., Sahgal N., Hussell T., Feldmann M., and Udalova I. A. (2011) IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat. Immunol. 12, 231–238 10.1038/ni.1990 [DOI] [PubMed] [Google Scholar]
- 19. Bowdridge S., and Gause W. C. (2010) Regulation of alternative macrophage activation by chromatin remodeling. Nat. Immunol. 11, 879–881 10.1038/ni1010-879 [DOI] [PubMed] [Google Scholar]
- 20. Ishii M., Wen H., Corsa C. A., Liu T., Coelho A. L., Allen R. M., Carson W. F. 4th, Cavassani K. A., Li X., Lukacs N. W., Hogaboam C. M., Dou Y., and Kunkel S. L. (2009) Epigenetic regulation of the alternatively activated macrophage phenotype. Blood 114, 3244–3254 10.1182/blood-2009-04-217620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Satoh T., Takeuchi O., Vandenbon A., Yasuda K., Tanaka Y., Kumagai Y., Miyake T., Matsushita K., Okazaki T., Saitoh T., Honma K., Matsuyama T., Yui K., Tsujimura T., Standley D. M., Nakanishi K., Nakai K., and Akira S. (2010) The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat. Immunol. 11, 936–944 10.1038/ni.1920 [DOI] [PubMed] [Google Scholar]
- 22. Hamilton J. A. (2015) GM-CSF as a target in inflammatory/autoimmune disease: current evidence and future therapeutic potential. Expert Rev. Clin. Immunol. 11, 457–465 10.1586/1744666X.2015.1024110 [DOI] [PubMed] [Google Scholar]
- 23. Hamilton J. A., Cook A. D., and Tak P. P. (2016) Anti-colony-stimulating factor therapies for inflammatory and autoimmune diseases. Nat. Rev. Drug Discov. 16, 53–70 [DOI] [PubMed] [Google Scholar]
- 24. Hamilton J. A. (2008) Colony-stimulating factors in inflammation and autoimmunity. Nat. Rev. Immunol. 8, 533–544 10.1038/nri2356 [DOI] [PubMed] [Google Scholar]
- 25. Sallusto F., and Lanzavecchia A. (1994) Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and down-regulated by tumor necrosis factor α. J. Exp. Med. 179, 1109–1118 10.1084/jem.179.4.1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Weigel B. J., Nath N., Taylor P. A., Panoskaltsis-Mortari A., Chen W., Krieg A. M., Brasel K., and Blazar B. R. (2002) Comparative analysis of murine marrow-derived dendritic cells generated by Flt3L or GM-CSF/IL-4 and matured with immune stimulatory agents on the in vivo induction of antileukemia responses. Blood 100, 4169–4176 10.1182/blood-2002-04-1063 [DOI] [PubMed] [Google Scholar]
- 27. Vento-Tormo R., Company C., Rodríguez-Ubreva J., de la Rica L., Urquiza J. M., Javierre B. M., Sabarinathan R., Luque A., Esteller M., Aran J. M., Álvarez-Errico D., and Ballestar E. (2016) IL-4 orchestrates STAT6-mediated DNA demethylation leading to dendritic cell differentiation. Genome Biol. 17, 4 10.1186/s13059-015-0863-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mui A. L., Wakao H., Harada N., O'Farrell A. M., and Miyajima A. (1995) Interleukin-3, granulocyte-macrophage colony-stimulating factor, and interleukin-5 transduce signals through two forms of STAT5. J. Leukoc. Biol. 57, 799–803 10.1002/jlb.57.5.799 [DOI] [PubMed] [Google Scholar]
- 29. Achuthan A., Cook A. D., Lee M. C., Saleh R., Khiew H. W., Chang M. W., Louis C., Fleetwood A. J., Lacey D. C., Christensen A. D., Frye A. T., Lam P. Y., Kusano H., Nomura K., Steiner N., et al. (2016) Granulocyte macrophage colony-stimulating factor induces CCL17 production via IRF4 to mediate inflammation. J. Clin. Invest. 126, 3453–3466 10.1172/JCI87828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liddiard K., Welch J. S., Lozach J., Heinz S., Glass C. K., and Greaves D. R. (2006) Interleukin-4 induction of the CC chemokine TARC (CCL17) in murine macrophages is mediated by multiple STAT6 sites in the TARC gene promoter. BMC Mol. Biol. 7, 45 10.1186/1471-2199-7-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Weiss M., Byrne A. J., Blazek K., Saliba D. G., Pease J. E., Perocheau D., Feldmann M., and Udalova I. A. (2015) IRF5 controls both acute and chronic inflammation. Proc. Natl. Acad. Sci. U.S.A. 112, 11001–11006 10.1073/pnas.1506254112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kruidenier L., Chung C. W., Cheng Z., Liddle J., Che K., Joberty G., Bantscheff M., Bountra C., Bridges A., Diallo H., Eberhard D., Hutchinson S., Jones E., Katso R., Leveridge M., et al. (2012) A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature 488, 404–408 10.1038/nature11262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. De Santa F., Totaro M. G., Prosperini E., Notarbartolo S., Testa G., and Natoli G. (2007) The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 130, 1083–1094 10.1016/j.cell.2007.08.019 [DOI] [PubMed] [Google Scholar]
- 34. Lehtonen A., Veckman V., Nikula T., Lahesmaa R., Kinnunen L., Matikainen S., and Julkunen I. (2005) Differential expression of IFN regulatory factor 4 gene in human monocyte-derived dendritic cells and macrophages. J. Immunol. 175, 6570–6579 10.4049/jimmunol.175.10.6570 [DOI] [PubMed] [Google Scholar]
- 35. Wirnsberger G., Hebenstreit D., Posselt G., Horejs-Hoeck J., and Duschl A. (2006) IL-4 induces expression of TARC/CCL17 via two STAT6 binding sites. Eur. J. Immunol. 36, 1882–1891 10.1002/eji.200635972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Semnani R. T., Mahapatra L., Moore V., Sanprasert V., and Nutman T. B. (2011) Functional and phenotypic characteristics of alternative activation induced in human monocytes by interleukin-4 or the parasitic nematode Brugia malayi. Infect. Immun. 79, 3957–3965 10.1128/IAI.05191-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pechkovsky D. V., Prasse A., Kollert F., Engel K. M., Dentler J., Luttmann W., Friedrich K., Müller-Quernheim J., and Zissel G. (2010) Alternatively activated alveolar macrophages in pulmonary fibrosis-mediator production and intracellular signal transduction. Clin. Immunol. 137, 89–101 10.1016/j.clim.2010.06.017 [DOI] [PubMed] [Google Scholar]
- 38. Hamilton J. A. (1994) Coordinate and noncoordinate colony stimulating factor formation by human monocytes. J. Leukoc. Biol. 55, 355–361 10.1002/jlb.55.3.355 [DOI] [PubMed] [Google Scholar]
- 39. Hamilton J. A., Whitty G. A., Royston A. K., Cebon J., and Layton J. E. (1992) Interleukin-4 suppresses granulocyte colony-stimulating factor and granulocyte-macrophage colony-stimulating factor levels in stimulated human monocytes. Immunology 76, 566–571 [PMC free article] [PubMed] [Google Scholar]
- 40. Borriello F., Longo M., Spinelli R., Pecoraro A., Granata F., Staiano R. I., Loffredo S., Spadaro G., Beguinot F., Schroeder J., and Marone G. (2015) IL-3 synergises with basophil-derived IL-4 and IL-13 to promote the alternative activation of human monocytes. Eur. J. Immunol. 45, 2042–2051 10.1002/eji.201445303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lehtonen A., Matikainen S., Miettinen M., and Julkunen I. (2002) Granulocyte-macrophage colony-stimulating factor (GM-CSF)-induced STAT5 activation and target-gene expression during human monocyte/macrophage differentiation. J. Leukoc. Biol. 71, 511–519 [PubMed] [Google Scholar]
- 42. Achuthan A., Aslam A. S. M., Nguyen Q., Lam P. Y., Fleetwood A. J., Frye A. T., Louis C., Lee M. C., Smith J. E., Cook A. D., Olshansky M., Turner S. J., and Hamilton J. A. (2018) Glucocorticoids promote apoptosis of proinflammatory monocytes by inhibiting ERK activity. Cell Death Dis. 9, 267 10.1038/s41419-018-0332-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lacey D. C., Achuthan A., Fleetwood A. J., Dinh H., Roiniotis J., Scholz G. M., Chang M. W., Beckman S. K., Cook A. D., and Hamilton J. A. (2012) Defining GM-CSF- and macrophage-CSF-dependent macrophage responses by in vitro models. J. Immunol. 188, 5752–5765 10.4049/jimmunol.1103426 [DOI] [PubMed] [Google Scholar]