Abstract
Organ morphogenesis is a complex process coordinated by cell specification, epithelial-mesenchymal interactions, and tissue polarity. A striking example is the pattern of regularly spaced, globally aligned mammalian hair follicles, which emerges through epidermal-dermal signaling and planar polarized morphogenesis. Here, using live-imaging, we discover that developing hair follicles polarize through dramatic cell rearrangements organized in a counter-rotational pattern of cell flows. Upon hair placode induction, Shh signaling specifies a radial pattern of progenitor fates that, together with planar cell polarity (PCP), induce counter-rotational rearrangements through myosin and ROCK-dependent polarized neighbor exchanges. Importantly, these cell rearrangements also establish cell fate asymmetry by repositioning radial progenitors along the anterior-posterior axis. These movements concurrently displace associated mesenchymal cells, which then signal asymmetrically to maintain polarized cell fates. Our results demonstrate how spatial patterning and tissue polarity generate an unexpected collective cell behavior that in turn, establishes both morphological and cell fate asymmetry.
INTRODUCTION
The development of epithelial organs involves a complex interplay between epithelial-mesenchymal interactions, cell fate induction, and tissue polarity. In epithelial organs, intercellular signaling and mesenchymal interactions specify the spatial patterns of tubes, branches, and other specialized cell types1,2. Cell polarity, which defines the apical-basal and planar axes of epithelia, provides directionality to cell signaling, division, and specification events3–5. Polarity thereby positions differentiated cell types into their proper orientation, and thus establishes overall tissue architecture. How cell polarity cooperates with spatial patterning and mesenchymal interactions to establish globally coordinated tissue architecture is unclear.
The polarization and alignment of cellular structures across a tissue plane is a characteristic feature of most epithelial organs known as planar cell polarity (PCP)6–9. PCP directs collective cell behaviors such as unidirectional cilia beating and collective cell motility, without which severe developmental abnormalities arise, including neural tube closure defects, hydrocephalus, infertility, deafness and congenital heart defects9. PCP is evident in a vast array of diverse epithelial structures, from simple protrusions that emanate from individual cells, such as Drosophila wing hairs, to elaborate multicellular structures such as mammalian hair follicles (HFs)5,10. How the principals governing polarization of individual cells apply to complex multicellular structures is poorly understood.
The mammalian skin, which is decorated with spatially patterned, globally aligned HFs is an excellent system to explore how polarized architecture is established in multicellular structures. HFs develop from multicellular placodes that emerge from the embryonic epidermis in waves of evenly spaced epithelial clusters. Epithelial-mesenchymal crosstalk specifies placode fate11–13, while PCP directs the polarized distribution of progenitors and orients the direction of HF growth14,15. Prior to HF induction, PCP is established within basal epidermal progenitors through asymmetric partitioning of core PCP components Frizzled-6 (Fz6), Vangl2, and Celsr1, along the epithelial plane14. However, the cellular mechanisms that direct either morphological or cell fate asymmetry in HFs have not been identified.
Using a combination of long-term live imaging, automated cell tracking, mouse genetics, and laser ablation, we discovered a PCP-dependent program of cell rearrangements that drives planar polarization and cell fate asymmetry of mammalian HFs. The polarization of initially circular hair placodes is driven by dramatic cell rearrangements coordinated in a counter-rotational pattern of cell flows. Counter-rotational movements reposition placode cells within the epithelial plane, displacing centrally-positioned cells forward to lead placode growth, while sweeping outer cells in the opposite direction toward the placode rear. To generate the pattern of cell flow, spatial patterning of radial cell fates cooperates with PCP to direct polarized cell neighbor exchanges, in part, through myosin-dependent junction disassembly. These cell rearrangements generate not only morphological asymmetry, but also reposition HF progenitors from a radial to planar polarized organization. In addition, these movements displace a crucial mesenchymal signaling center - the dermal condensate (DC) – which signals asymmetrically to maintain polarized progenitor fates. This study defines the cellular mechanism that generates planar polarity in complex multicellular structures and demonstrates how polarized cell rearrangements generate not only morphological, but also cell fate asymmetry.
RESULTS
Counter-rotational cell movements accompany hair placode polarization
Following their initial invagination into the underlying dermis, hair placodes of the dorsal epidermis adopt an anterior-directed tilt as they shift their direction of growth from vertical to anterior14. This transition, which we refer to as placode polarization, can be observed using a Shh-Cre reporter driving membrane GFP expression16–18. During embryogenesis, hair placodes are specified in three consecutive waves. Thus, HFs at different developmental stages can be observed simultaneously (Fig. 1a). Initially, Shh expressing cells are positioned at the center of the circular placode, but after invagination, this population is located anteriorly at the leading tip of the budding structure (Fig. 1b). To observe this transition in real time, we established a live imaging protocol to monitor embryonic skin development in E15.5 explants over 16–24 hours. With this method, we observed growth of existing follicles, specification of new placodes, and polarization of early placodes (Supplementary video 1). Early placodes were identified as circular clusters of highly constricted epithelial cells with sparse GFP labeling. Through time, the centrally positioned GFP cells were progressively displaced toward the anterior via a complete reorganization of the placode epithelium (Fig. 1c, Supplementary video 1, 2). During this transition, placode epithelial cells were highly dynamic and motile, while the surrounding interfollicular epidermal cells remained stationary.
Figure 1. Hair follicle placodes polarize through counter rotational cell flows.
(A–B) Scanning confocal images of embryonic skin showing placodes at different stages of development. (A–C) All cells express membrane Tomato (magenta) and Shh-Cre expressing cells are converted to express mGFP to highlight cells within the placode (green). (A) E17.5 skin highlighting an early, unpolarized placode (circle), and polarized germ (square), and peg stage (rectangle). Representative image from four embryos. Scale bar, 100µm. (B) Representative images of an early placode and a polarized placode from three E15.5 embryos. Scale bar, 10µm. (C) Spinning disk confocal images from a time-series showing placode polarization in cultured explants. Representative placode from four embryos. mGFP cells are displaced anteriorly through time. The z plane changes 9µm through time in 3µm steps to follow the base of the placode into the dermis. Scale bar, 10µm. (D) Cell movements during placode polarization. See Supplemental Video 4. Spinning disk confocal images from a time series of placode cells expressing mGFP driven by K14-Cre (top). The z plane changes 3µm in one step to follow the base of the placode into the dermis. Cells were segmented and false colored in a rainbow pattern of vertical lines prior to polarization. Cell tracks show the movement of cells during the designated time window (bottom). Overall cell trajectories are shown in the schematic (right). Cells at the center of the placode move anteriorly, cells in the posterior converge toward the midline (blue and purple) and anterior cells move away from the midline and then posteriorly (red). Representative placode from five embryos. Additional examples of placode cell movements are shown in Supplemental Figure 1 and Supplemental Video 3. Scale bar, 10µm. Anterior is to the left.
To define the morphogenetic process that drives placode polarization, epithelial cell movements were monitored using automated segmentation and cell tracking19. To complement these data, mosaically labeled placode cells were followed through time to monitor the movement of small clones (Supplementary Figure 1, Supplementary video 3). This analysis revealed that placode epithelial cells rearrange in a highly reproducible, planar polarized pattern of counter-rotational cell flows. Color-coding placode epithelial cells in a rainbow pattern of vertical stripes highlights the emergence of this counter-rotational pattern (Fig. 1d, Supplementary video 4). Cells initially positioned near the posterior converge toward the placode midline and are displaced in an anterior direction (blue and purple shaded cells, Supplementary Figure 1a,b) while cells positioned near the anterior extend outward and away from the placode midline (red shaded cells). Cells at the lateral edges are displaced posteriorly (Supplementary Figure 1c), eventually flowing toward the midline. These directionally opposed movements cause clusters of lateral cells to rotate (Supplementary Figure 1d). Collectively, these cell rearrangements reposition placode cells along the AP axis such that the cells that were centrally located during placode invagination are repositioned toward the anterior, while anterior and lateral cells are displaced in the reverse direction, becoming incorporated into the posterior half of the growing placode.
Polarized shrinkage and growth of intercellular junctions directs cell rearrangements within the placode
Epithelial cells rearrange by remodeling their intercellular junctions to drive neighbor exchange20,21. To understand the spatial and directional patterns of junctional remodeling that may contribute to counter-rotational cell flows, we measured the angle of lost and newly formed junctions and mapped their locations over the course of placode polarization (Fig. 2a). In the posterior of the placode, vertically oriented junctions primarily shrank into multicellular vertices that resolved into new, horizontally oriented junctions (Fig. 2b). This collective cell behavior drove cell intercalation of laterally positioned cells toward the placode midline. In contrast, in the anterior of the placode horizontal junctions were lost while new vertical junctions formed causing anterior cells to move outward and away from the midline. Finally, at the lateral edges of the placode we observed more complex junctional remodeling events where both the angles of lost and newly formed junctions aligned roughly parallel to the placode edge. These junctional rearrangements allowed the outermost cells to slide posteriorly past their interior neighbors. Altogether, these spatially polarized remodeling events locally displace cells concurrent with counter-rotational cell flows.
Figure 2. Polarized shrinkage and growth of intercellular junctions directs cell rearrangements within the placode.
(A–B) Lost junctions are shown in red while new junctions are shown in green. (A) Quantification of the angle and position of lost and new junctions during placode polarization. Lines representing the location and angle of individual remodeling events (n=213) from 3 placodes (including the placode shown in Figure 1D and Supplemental Video 4) over 19 hours were combined in a single image (top). Anterior events are shown in a lighter shade while posterior events are shown in a darker shade. The angular frequency of junctional modifications is plotted below where the AP axis is 0 degrees. (B) Examples of neighbor exchange quantified in A from two of three placodes. Anterior junctions are lost horizontally and form vertically (top). Posterior junctions are mainly lost vertically and form horizontally (center). Neighbor exchange causes outer cells (1,3) to slide past inner cells (2,4,5,6) at the lateral edge of the placode (bottom). (C) Celsr polarity within the interfollicular epithelium, early, and polarizing placodes. The orientation of the line shows the direction of Celsr polarity. 0–44 degrees is shown in cyan and 45–90 degrees is shown in yellow. Representative of 15 measured images from four embryos. Additional examples are shown in Supplemental Figure 2. Scale bar, 10µm. Anterior is to the left.
We sought to understand how the complex pattern of cell rearrangements relates to the planar polarity of individual cells. We therefore quantified the orientation of Celsr1 asymmetry in placode epithelial cells to determine how PCP protein enrichment correlates with junctional remodeling events. In the interfollicular epidermis, Celsr1 localizes asymmetrically to vertical junctions, a pattern that is maintained in early placodes (Fig. 2c, Supplementary Figure 2)14,22,23. During placode polarization, Celsr1 asymmetry reorganizes predominantly in the anterior and lateral edges of the placode, becoming rotated up to 90 degrees. In the posterior of the placode, Celsr1 remained asymmetrically localized to vertical junctions. The spatial pattern of Celsr1 localization during placode polarization closely correlated with the pattern of lost junctions, suggesting that PCP proteins may actively drive polarized contractions and neighbor exchange.
Counter-rotational cell flows require planar cell polarity
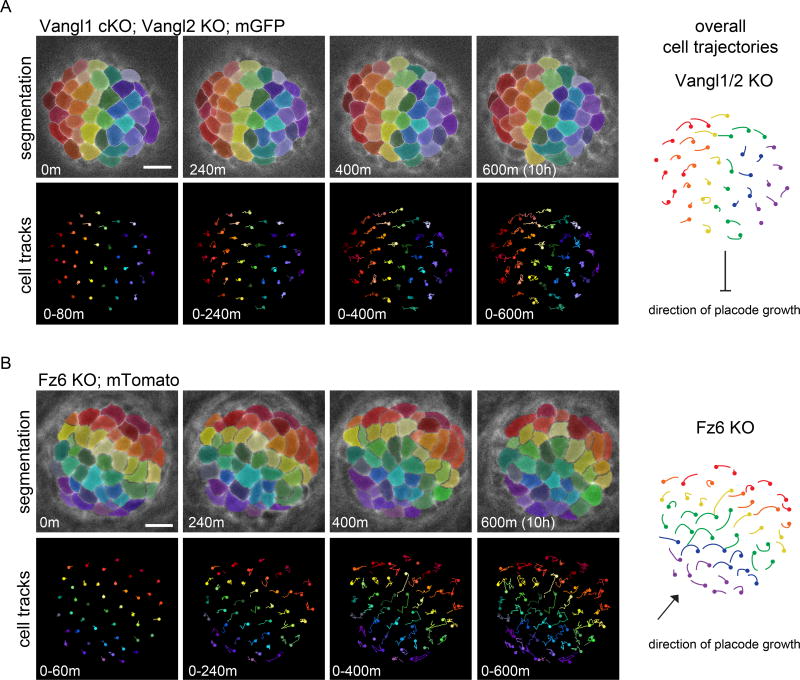
To investigate a potential role for PCP in directing cell flow within the polarizing placode, we performed live imaging on embryonic explants of two different PCP mutants. Vangl1;Vangl2 double mutants produce HFs that predominately grow vertically into the underlying dermis whereas Fz6 mutants display a less severe phenotype where ~45% of HFs emerge at random orientations23,24. Budding placodes were imaged for approximately 10 hours, the time during which counter-rotational cell flows normally occurred in control placodes (Supplementary videos 5, 6). Cells in Vangl mutant placodes failed to undergo extensive neighbor exchange, and thus maintained their relative AP positions through time (Fig. 3a, Supplementary video 5). Fz6 mutant placodes were slightly more dynamic and displayed counter-rotational cell movements, albeit with reduced cell displacement compared to controls. These rotational movements occurred off axis and their collective orientation correlated with direction of placode growth. To highlight these movements, the rainbow pattern of vertical stripes was rotated perpendicular to the axis of placode growth (Fig 3b, Supplementary Figure 3 Supplementary video 6). We conclude from these data that PCP is essential for counter-rotational cell flows and that the direction of cell flow dictates the orientation of placode growth.
Figure 3. Counter-rotational cell movements require planar cell polarity.
(A–B) Vangl1 cKO; Vangl2 KO cells do not rearrange while Fz6 KO cells undergo counter-rotational cell movements in the direction of placode growth rather than the AP axis. See Supplemental Videos 5 and 6. Representative placodes from two and three embryos, respectively. Spinning disk confocal images from a time series of placode cells expressing mGFP driven by K14-Cre (A) or mTomato (B). Cells were segmented and false colored in a rainbow pattern of vertical lines perpendicular to the AP axis (A) or to the direction of growth (B) at the start of the movie. Cell tracks show the movement of cells during the designated time window (bottom). Overall cell trajectories are shown in the schematic (right). An additional example of a polarizing Fz6 KO placode is shown in Supplemental Figure 3. Scale bar, 10µm. Anterior is to the left.
Placode polarization and counter rotational movements require Rho kinase and myosin II activity downstream of PCP
During convergent extension in Drosophila and vertebrate embryos, polarized acto-myosin activity at cell boundaries provides the local force for epithelial cell rearrangements25–29. To address whether placode polarization depends on force production through myosin II activity, we cultured E15.5 epidermal explants with a pharmacological inhibitor of myosin II, blebbistatin. After 24 hours in culture, the number of placodes and HFs at each stage was indistinguishable from controls showing blebbistatin did not inhibit placode formation (Supplementary Figure 4a,b,d). Myosin inhibition did, however, strongly affect placode polarization, which was assessed by placode morphology and the location of Shh-Cre expression. Whereas mature, first and second wave follicles remained planar polarized upon addition of blebbistatin, the polarity of new, third wave placodes was strongly reduced compared to controls (Fig. 4a, b). Similar results were obtained upon inhibition of Rho Kinase (ROCK), a key upstream regulator of myosin activity (Fig 4a, b, Supplementary Figure 4c,d). The failure of myosin and ROCK-inhibited placodes to polarize was not due to a loss of PCP protein localization, as tissue-level Celsr1 asymmetry was maintained in the presence of either inhibitor (Fig. 4c,d). These data suggest that myosin inhibition alters placode planar polarity specifically during the polarization phase. We therefore performed live imaging in the presence of Rho Kinase (ROCK) inhibitor and monitored cell behaviors in both early and polarizing placodes. Similar to what we observed in PCP mutants, cell rearrangements were sharply reduced and cells maintained their AP positions through time (Fig. 4e, Supplementary Figure 4e, Supplementary video 7). These results indicate that myosin activity is required downstream of PCP for the collective cell rearrangements that drive placode polarization.
Figure 4. Placode polarization and counter rotational movements require Rho kinase and myosin II activity downstream of PCP.
(A–D) Skin explants from E15.5 embryos were cultured for 24 hours in the presence of Y-27632 (ROCK inhibitor) or blebbistatin. (A) Representative images of 3rd wave placodes quantified in B. All cells express mTomato (magenta) and Shh-Cre expressing cells express mGFP (green). Additional examples are shown in Supplemental Figure 6. (B) Quantification of 3rd wave placode polarity. Control, n=348 follicles from 7 embryos: blebbistatin, n=337 follicles from 7 embryos (p=4.26×10, two-sided unpaired t-test); Control, n = 174 follicles from 4 embryos; Y27632, n=172 follicles from 4 embryos (p=0.002, two-sided unpaired t-test). (C) Representative immunofluorescence images of Celsr1 polarization within the IFE quantified in D. (D) Quantification of the angular distribution of Celsr1 in the IFE. Mp = magnitude of average Celsr1 polarity (see methods). Control, n=3623 cells from 3 embryos (gray, p=0) and blebbistatin, n=4923 cells from 3 embryos (red, p=0). Control, n=1512 cells from 3 embryos (gray, p=0) and Y27632, n=1828 cells from 3 embryos (red, p=0). (E) Counter-rotational cell flows are inhibited in the presence of Y27632. Representative placode from five treated explants. Spinning disk confocal images from a time series of placode cells expressing mTomato (top). Cells were segmented and false colored in a rainbow pattern of vertical lines at the beginning of the movie. Cell tracks show the movement of cells during the designated time window (bottom). Overall cell trajectories are shown in the schematic (right). Cells remain in their original pattern after 9.7h of imaging. See Supplemental Video 7. An additional example is shown in Supplemental Figure 4E. Scale bar, 10µm. Anterior is to the left.
Specification of radial cell fates patterns counter-rotational movements
Morphogenetic movements are specified by earlier induction events that generate cell fate diversity in spatial patterns. In the placode, centrally located cells display different patterns of movement than peripheral cells suggesting they possess distinct properties that contribute to their behaviors. Early hair follicles are comprised of at least two distinct progenitor populations. P-Cadherin (P-Cad), Shh-expressing cells occupy the leading tip of the follicle, contact the mesenchyme and go on to form the hair matrix, while E-Cadherin (E-Cad), Sox9-expressing cells trail behind and become the future stem cell population30–34. We found that initially, these two progenitor populations were arranged in a radially symmetric pattern where Sox9-expressing cells formed a ring around a central P-Cad, Shh-positive cluster (Fig. 5a, Supplementary Figure 5a). To investigate how radial patterning contributes to morphogenetic behaviors in the placode, we first examined cell fate specification in the absence of Shh signaling, which was previously shown to specify Sox9+ stem cell precursors33,35. Accordingly, we found that the outer ring of Sox9 expression was lost in placodes of Shh KO embryos, while the inner domain, monitored by P-Cad expression and a GFP reporter of Shh promotor activity17, was significantly expanded (Fig5b, Supplementary Figure 5a,b). The resulting mutant placodes displayed several morphological abnormalities suggesting a defect in compartmentalization from the surrounding epithelium. In control placodes, Shh-GFP expressing cells were well-segregated from surrounding cells, forming circular clusters with smooth perimeters. By contrast, the cells of Shh mutant placodes were arranged in irregular, anisotropic shapes with jagged perimeters that were poorly segregated from their neighbors (Supplementary Figure 5a–c). These observations suggested that one of the functions of Shh signaling and radially patterned cell fates is to form well-defined compartments to segregate placode cells from the surrounding interfollicular epidermis.
Figure 5. Planar cell fate asymmetry arises from directional cell rearrangements.
(A) Representative immunofluorescence images of early placodes showing radial symmetry with inner tip cells expressing P-Cadherin and ShhGFP (green) and outer SC progenitors expressing Sox9 (magenta) from one of two embryos (top) and one of four embryos (bottom). (B) Representative image of Sox9 (magenta) expression in control and Shh KO placodes from one of two embryos (het) and one of three embryos (KO). All cells express mTomato (blue). Additional examples of Shh KO placode morphology are shown in Supplemental Figure 5A–C. (C) Shh mutant placode cells undergo atypical cell rearrangements. Representative placode from one of two embryos. Spinning disk confocal images from a time series of placode cells expressing mGFP (top). Overall cell trajectories are shown in the schematic (right). Cells were segmented and false colored in a rainbow pattern of vertical lines perpendicular to the AP axis at t=0. Cell tracks show the movement of cells during the designated time window (bottom). See Supplemental Video 8. Additional example shown in Supplemental Figure 5D. (D) Representative immunofluorescence images of early, mid, and late/germ wild-type placodes (left) stained for Hoechst (blue) and asymmetry markers Sox9 (magenta), P-Cadherin (green) quantified in E. Placodes are initially radially symmetric before becoming planar polarized. Asymmetry markers in Vangl1;Vangl2 dcKO and Fz6 KO placodes fail to polarize or polarize in random orientations (right) quantified in E. (E) Quantification of Sox9 asymmetry in early (n=7), mid (n=11), and late/germ (n=9) wild-type placodes (p=0.048 early vs. mid; p=0.0002 early vs. late; unpaired t-test, from four embryos), and Vangl2 KO (p=7.22×10−5 vs. late, n=7 placodes from three embryos) and Fz6 KO (p=n.s. vs. late, n=9 placodes from three embryos) germ placodes represented as the posterior/anterior ratio of Sox9 immunofluorescence intensity (mean+SD). Randomly oriented, polarized Fz6 KO placodes were chosen for analysis. Fz6 KO hair germ orientation was determined based on morphology. Scale bar, 10µm. Anterior is to the left.
Shh mutants provided a tool to interrogate whether proper specification of radially-arranged cell fates is needed for counter-rotational movements. We therefore performed live imaging of Shh mutant explants, and found that in stark contrast to placodes lacking PCP function or myosin activity, Shh mutant placodes displayed extensive, but uncoordinated cell rearrangements (Fig 5c, Supplementary Figure 5d, Supplementary video 8). Cells rearranged with zig-zag trajectories along the vertical (ML) plane that failed to arrange in a counter rotational pattern and generated minimal displacement along the AP axis. This phenotype was not due to a defect in PCP protein localization as Celsr1 is localized normally to AP boundaries in Shh mutants14. Although Shh signaling also promotes proliferation and dermal condensate formation in developing HFs36–39, our results suggest that its role in specifying radially-arranged cell fates contributes to the pattern of counter-rotational flow, perhaps by creating confined epithelial compartments for neighbor exchange.
Planar cell fate asymmetry arises from directional cell rearrangements
In morphologically polarized HFs, cell fates are asymmetrically positioned along the AP axis, a process that requires PCP14. The PCP pathway is known to establish cell fate asymmetry through asymmetric cell division or polarized signal transduction40–45, but whether cell fate asymmetry in the placode occurs through either of these mechanisms is not clear. We therefore examined how cell fate asymmetry emerges and found that radially arranged, P-Cad+ tip cells and Sox9+ stem cell progenitors progressively shift asymmetrically over time. Midway through polarization, the Sox9 expression domain was shifted toward the posterior and formed a semicircle around anteriorly-displaced tip cells. In later staged, fully polarized hair placodes, the two populations were segregated along the AP axis with tip cells at the anterior, growing front of the placode and stem cell progenitors at the posterior. The redistribution of tip cells and stem cell progenitors from a radial to planar polarized pattern strongly suggested that the two populations are polarized not by asymmetric division or polarized signaling, but are repositioned via PCP-dependent, counter-rotational cell rearrangements.
To test this hypothesis, we examined cell fate asymmetry under conditions where cell rearrangements were disrupted. In Vangl mutants, P-Cad and Sox9-positive populations remained in a radially symmetric configuration, whereas in Fz6 mutant placodes that orient off axis, the two populations polarized in random orientations (Fig. 5d,e). Additionally, newly formed placodes from explants cultured with blebbistatin or the ROCK inhibitor remained radially symmetric (Supplementary Figure 6). We conclude from these data that PCP governs both morphological and cell fate asymmetry in the developing follicle through oriented cell rearrangements.
Asymmetric positioning of the dermal condensate maintains cell fate asymmetry
PCP establishes the planar polarized architecture upon which the future HF will be built by repositioning two HF progenitors that provide distinct functional contributions to follicle morphogenesis. Anterior tip cells contact the DC, divide asymmetrically, and lead anterior-directed growth14,33. Posterior, Sox9 cells trail behind, proliferating symmetrically to expand the future stem cell population32,33. Following polarization, HFs maintain compartmentalization of AP cell fates and continue to grow with an anterior trajectory. Key to continued follicle growth is the DC, a cluster of specialized mesenchymal cells that sends growth promoting signals to the overlying epithelium11,46. Given the close apposition of placode epithelial cells to the DC, we wondered how the DC contributes to the maintenance of asymmetric cell fates.
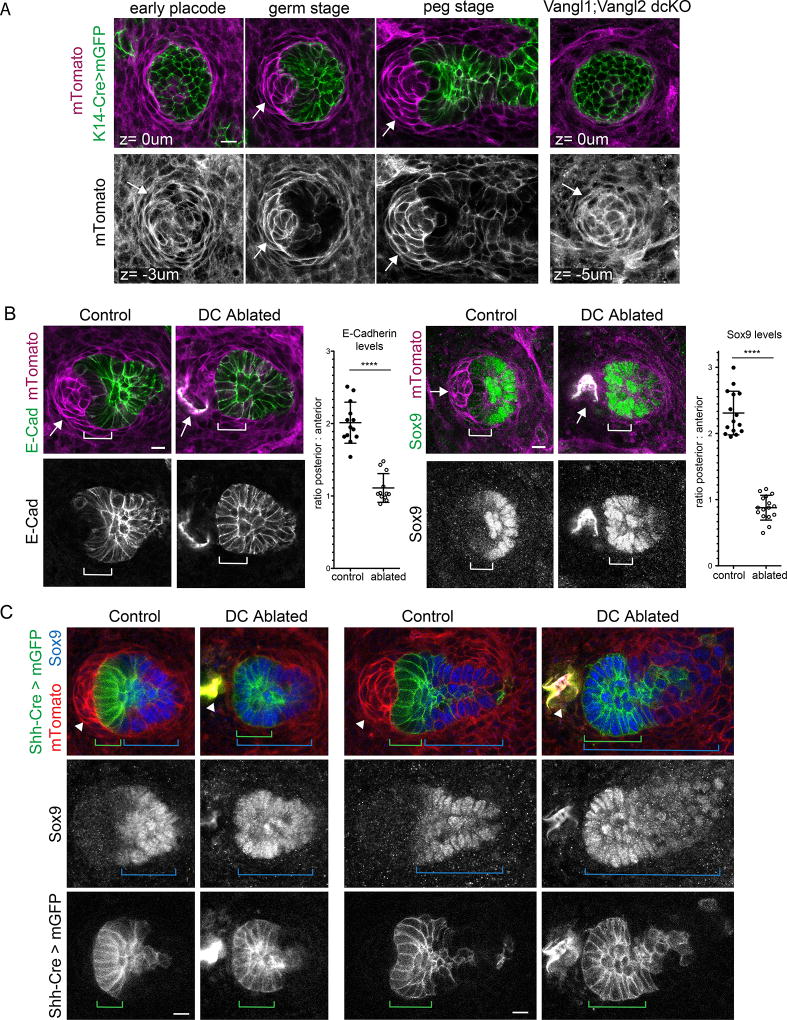
First, we monitored the position of DCs over the course of placode polarization and found they were displaced anteriorly, closely tracking with the position of migrating tip cells (Fig. 6a, Supplementary video 9). Following polarization, the DC remained in close contact with the anterior tip of the hair bud where it eventually becomes enveloped by the epithelium. In Vangl2 mutant embryos, the DC failed to move anteriorly and remained associated with the follicle tip as it grew vertically into the dermis (Fig 6a). We conclude from this data that the DC, by maintaining close contact with tip cells, is physically displaced anteriorly during placode polarization. This positions the DC such that anterior tip cells should receive higher concentrations of DC derived cues compared to cells positioned more posteriorly.
Figure 6. Asymmetric positioning of the dermal condensate maintains cell fate asymmetry.
(A) Dermal condensate position (marked with arrow) before and after placode polarization. K14-Cre was used to drive mGFP expression in the follicular epidermis (green). All dermal cells, including the dermal condensate, express mTomato (magenta). The DC is positioned directly beneath the prepolarized placode but becomes displaced anteriorly as the follicle polarizes. An example of this process is shown in Supplemental Video 9. In the absence of placode polarization (Vangl1;Vangl2 dcKO) the DC remains directly below the base of the follicle (right). Representative images from three control and three mutant embryos. (B,C) Follicle asymmetry markers after laser ablation of the DC. (B) mTomato labels dermal cells (magenta) while posterior fate markers E-cadherin and Sox-9 are shown in green. Representative images from four embryonic explants. Quantification of the ratio of posterior to anterior E-cadherin (n=13 unablated and 12 ablated HFs), and Sox-9 (n=15 unablated and n=16 ablated HFs) levels showing that posterior cell fates expand anteriorly after DC ablation (mean+SD, p=4.12×10−9 for E-cad and p=3.48×10−15 for Sox9, two-tailed unpaired t-test). Bracket indicates anterior cells. (C) Hair follicle asymmetry after laser ablation of the DC at the germ (left) and peg stage (right). All mGFP cells are derived from ShhCre expressing progenitors (green). All cells express mTomato (red). Sox9 expressing cells mark posterior follicle fates (blue). Control hair follicles maintain AP separation of mGFP and Sox9 (green and blue brackets) while mGFP cells are converted to Sox9 expressing cells after DC ablation. Representative images from six control and six DC ablated follicles from two explants. Arrow labels DC. Scale bar, 10µm. Anterior is to the left.
To test if signals from the DC are required to maintain cell fate asymmetry, we laser ablated the DCs of post polarization-stage follicles and 24 hours later, examined the expression of cell fate markers. In unablated control follicles, E-Cadherin and Sox9 expression was confined to the posterior half of the follicle, showing approximately 2-fold higher expression in the posterior compared to the anterior. Strikingly, in DC-ablated follicles, anterior cells upregulated E-Cadherin and Sox9 expression to equivalent levels compared to the posterior (Fig. 6b). Lineage tracing tip cells using the Shh-Cre reporter demonstrated that these cells are not lost or destroyed upon DC ablation, rather they gain Sox9 expression (Fig. 6c). These data indicate that the maintenance of anterior, tip cell fate requires a signal from the neighboring mesenchyme, which actively represses stem cell progenitor fate.
DISCUSSION
Although planar polarization is a near ubiquitous feature of epithelia, only the mechanisms that polarize cells at the individual level were well understood, and how those principals might apply to multicellular structures was not clear. Building upon the live imaging and cell tracking methods previously employed to monitor placode formation in the embryonic skin epidermis33,47,48, we have identified the cellular mechanism that polarizes multicellular HFs. Placode epithelial cells engage in a counter-rotational pattern of cell flows that converts radially-symmetric epithelial clusters to planar polarized hair buds with anterior-directed growth. This elaborate pattern of cell movement shares features with other collective migration events including cell intercalation during convergent extension20,21, and ommatidia rotation in the Drosophila eye55, processes that depend on PCP, myosin II, and Rho Kinase49–51,56–60. Most strikingly, placode polarization resembles the remarkable ‘polonaise’ movements of avian embryos that occur during formation of the primitive streak, where epiblast cells undergo large scale tissue flows organized in two counter-rotating streams52–54. However, placode polarization is distinct in that the movements involve a much smaller number of cells and occur in a repeating pattern of evenly spaced units confined within a continuous, stationary epithelium. Given the ubiquity of PCP expression across epithelial tissues, we suspect that this pattern of cellular movements may be broadly employed in morphogenesis and could explain how epithelial buds emerge with stereotyped angles, not only in feathers and scales, but also in branched epithelial organs.
Taken together, our data suggest the following model for how spatial epidermal patterning and PCP cooperate to generate a counter rotational pattern of cell flows. Induction of the hair placode primordium specifies a radially symmetric developmental program that initiates junctional remodeling and cell intercalation. PCP asymmetry dictates the orientation of cell intercalations, most likely by localizing myosin activity to induce junctional shrinkage27,29. We propose the initial PCP asymmetry combined with radial compartmentalization can impart directionality to these movements. For example, the differential enrichment of Vangl2 and Fz6 to anterior and posterior cell borders, respectively, could bias junction shrinkage to posterior edges, perhaps through polarized recruitment of junctional remodeling factors. Reduced levels of these factors along the anterior margin would decrease the likelihood that these edges undergo contraction. This slight bias along with radial restrictions to cell mixing could initiate directional cell flow. Once initiated, junction loss and assembly can cause rotation of neighboring cell borders22,61, which could explain the rotation of PCP-enriched borders that we observe. These rotations would generate new axes of cell intercalation that reinforce opposing cell flows. Ultimately, counter-rotational cell flows reposition radially symmetric progenitors into a planar polarized distribution while displacing the DC. The DC then maintains polarized cellular diversity by signaling asymmetrically to repress stem cell progenitor fate. Given that Sox9 expression is also expanded in HFs lacking canonical Wnt signaling34, we suspect Wnt ligands are the signals secreted from the DC that inhibit stem cell progenitor fate. Collectively, our findings illustrate how a single mechanism of polarized cell movements can restructure the morphology, epithelial-mesenchymal interactions, and cell specification of an epithelial primordium to establish tissue-wide organ asymmetry.
EXPERIMENTAL PROCEDURES
Mouse lines and breeding
All procedures involving animals were approved by Princeton University's Institutional Animal Care and Use Committee (IACUC). Mice were housed in an AAALAC-accredited facility in accordance with the Guide for the Care and Use of Laboratory Animals. This study was compliant with all relevant ethical regulations regarding animal research. E15.5–E17.5 embryos from C57BL6 backgrounds were used unless otherwise indicated. Both sexes were used as sex was not determined in embryos. Please see Supplementary Table 1 for full genotypes.
Whole-mount immunostaining
For immunostaining, embryos were dissected in PBS and fixed in 4% paraformaldehyde. E15.5 embryos and cultured embryonic explants were fixed at room temperature for 1 h and e17.5 embryos were fixed for 2 h following decapitation. Backskins were dissected from fixed embryos and blocked for 1 h at room temperature or overnight at 4°C in 2% normal goat serum, 2% normal donkey serum, 1% bovine serum albumin and 1% fish gelatin in PBT2 (PBS with 0.2% Triton X-100). Skins were washed in PBT2 and incubated with secondary antibodies for 2 h at room temperature or overnight at 4°C in PBT2. When samples were stained using the P-Cadherin antibody, TBS with 0.2% Triton X-100 was used instead of PBT2 for all steps. Samples were mounted in Prolong Gold. The following primary antibodies were used: guinea pig anti-Celsr 1 (1:1000, D. Devenport), anti-Sox9 (mouse,1:250, Santa Cruz, discontinued; rabbit, 1:1000, Millipore, Cat: AB5535), mouse anti-P-Cadherin (1:250, Clontech, Cat: AB36999), rat anti-E-Cadherin (1:1000, DECMA-1, Thermo Pierce, Cat: MA1-25160), rhodamine phalloidin (1:1000, Cytoskeleton, Inc, Cat: PHDR1) and Acti-stain 488 (1:1000, Cytoskeleton, Inc, Cat: PHDG1). Alexa Fluor-488,-555, and -647 secondary antibodies were used at 1:1000. Hoescht (Invitrogen, Cat: H1399) was used at 1:1000. Images were acquired on an inverted Nikon A1 or A1R-Si confocal microscope controlled by NIS Elements software using a Plan Apo 60/1.4NA or Plan Fluor 40/1.3NA oil immersion objective, or a Plan Apo 20/0.75NA air objective. ImageJ, Improvision Volocity and Photoshop were used for image processing.
Live imaging
E15.5 dorsal skin explants were dissected in PBS and transferred to a 1% agarose gel with F-media containing 10% fetal bovine serum. Explants were sandwiched between the gel on the dermal side and a 35-mm lummox membrane dish (Sarstedt) on the epidermal side. Z-series with a 3 micron step size were acquired at 10 minute intervals for 16–24 h. Images were acquired using a Nikon Ti-E Spinning Disc with Perfect Focus using a Plan Apo 20/0.75 NA air objective and 1.5× optical zoom. Explants were cultured in a humid imaging chamber at 37°C with 5% CO2 during the course of imaging. To avoid potential tissue distortions, explants were dissected from E15.5 embryos, which have been shown to maintain PCP polarization and produce polarized placodes ex vivo14. Cultured explants were fixed and stained for Celsr1 to ensure polarity was maintained. Placodes close to the edge of the explants were not imaged to avoid morphological changes that could be induced by a wound healing response. ImageJ was used for movie processing.
Movie Processing and cell tracking
Time-lapse movies were corrected for drift using the ImageJ plugin, MultiStackReg. To monitor placode cells as they grew further into the dermis, movies were assembled from the z-series using a single z plane that contained the base of the placode. Placode cells expressing membrane GFP or Tomato were segmented and tracked through time using Tissue Analyzer19. Segmentation or tracking mistakes were identified using neighboring z planes from the original z series and hand corrected. Cells were color-coded in a rainbow pattern of vertical stripes prior to placode polarization for visualization of the cell rearrangements. This pattern was rotated perpendicular to the trajectories of centrally positioned cells in Fz6 mutants. Cell trajectories were produced from Tissue Analyzer using the centroid position of cells at 40 minute intervals. Maximum projections of cell tracks were produced in ImageJ to show cell movements in specified periods of time.
Quantification of junctional remodeling
Overlapping groups of 5–10 placode cells were followed through time using manual tracking at all z planes. The position and angle of lost and new junctions within these groups were measured in ImageJ. The angle of lost junctions was measured just before the junction disappeared and the angle of the new junction was measured just after the junction appeared. Lines of equivalent lengths representing the location and angle of individual remodeling events from 3 placodes over 19 hours were combined in a single image. The base of the placode was bisected down the middle, perpendicular to the AP axis, at each time point to divide the placode into anterior and posterior halves. As the placode polarizes, the division shifts slightly anteriorly. Remodeling events were sorted by their AP position and their angular frequency was determined.
Quantification of Celsr1 polarity
Celsr1 polarity was calculated using Packing Analyzer V2 software as previously described61. Cells were segmented using a membrane signal. The software calculates the axis and magnitude (nematic order) of junctional polarity. For visual representation, these values are indicated by a single line within each cell with an angle that corresponds to the axis of polarity and a length that corresponds to the magnitude of polarity. For comparison of inhibitor treated samples across multiple experiments, Celsr polarity within a cell was normalized by the corresponding average cell boundary Celsr intensity as previously described22. Data were plotted in a circular histogram using the polar plot function in Matlab. Histograms were weighted by the average magnitude of polarity in each bin to reflect both the angle and strength of polarity. The magnitude of average Celsr1 polarity (Mp) was defined as in Aigouy et al61. For statistical analysis of Celsr1 polarity, a permutation test was performed to determine whether random resampling of the data could achieve a similar distribution22. Individual components of the nematic tensor for each cell were randomly permuted 1,000 times, and for each permutation, Mp was calculated. The P-value is given by the proportion of permutations with Mp > or = 1/2 standard deviation below that of the original data. A distribution was considered significant if P ≤ 0.05, which was the case for all control and inhibitor treated samples.
Inhibitor Treatments
E15.5 dorsal skin explants were dissected in PBS and mounted on a 13mm Whatman Nucleopore membrane with 8.0µM pore size. Explants were floated on culture of F-media containing 10% fetal bovine serum. 50µM Y-27632 (ROCK inhibitor, Calbiochem, InSolution Y-27632 in DMSO, Cat:688002) or 25µM blebbistatin (Calbiochem Cat:203390) was added to the F-media. Equivalent concentrations of DMSO were used as controls. Explants were cultured for 24 hours and then fixed and stained. For live imaging of Y-27632 treated explants, 100µM was added to the 1% agarose gel. Explants were submerged in 100µM Y-27632/F-media for 2 minutes prior to being transferred onto the agarose gel.
Quantification of hair follicle asymmetry
For inhibitor treated explants, follicles were classified into three groups based on their size. All 3rd wave follicles were scored as either polarized or unpolarized using Sox9 and/or Shh as morphological markers. For wild-type and PCP mutant follicles, maximum intensity Z-projections were created from the basal layer to the base of the follicle. Placodes were staged (pre-polarized, mid-polarized, post-polarized) according to their morphology using only cell membranes and actin. After staging, ImageJ was used to make a square encompassing the entire Sox9 expressing region. The square was bisected along the AP axis and the average Sox9 fluorescence intensity of the anterior and posterior region was measured. Sox9 asymmetry was calculated as a ratio of the posterior/anterior signal of each follicle after subtracting the background signal. For Fz6 KO and Vangl mutants, only placodes that have developed past the point where they should have acquired a tilt were quantified. For Fz6 KO, anterior/posterior was defined relative to the individual follicle rather than the overall anterior-posterior axis.
Quantification of placode morphology
Placode images were processed and analyzed using ImageJ. Images of the Shh-GFP placode signal were converted to 8bit, a mean filter with a 10-pixel radius was applied, placodes were outlined using the freehand tool and the average gray value was measured. The images were thresholded by setting the lower limit to 60% of the average signal and the wand tool was used to detect the outer boundary of the Shh-GFP signal. The area, perimeter, aspect ratio and circularity of the outer boundary were then measured.
Dermal condensate ablation
E15.5 dorsal skin explants expressing ubiquitous membrane-Tomato (see Table for full genotypes) were dissected in PBS and sandwiched between a Lumox membrane and 1% agarose, F-media pad as above. Imaging and ablation was performed on a Prairie Ultima multi-photon microscope with a Coherent Chameleon Ultima II pulse laser, tunable from 680 to 1080 nms, and a Plan ApoCHROMAT 40×/1.0 N.A. water immersion objective. Prior to ablation, a z-stack was collected of the germ- or peg-stage follicle with the laser tuned to 960nm. A rectangular region of interest was drawn around the dermal condensate, and was ablated using an 860nm laser at maximum power for 10–15 seconds. Z-stacks were collected following each ablation. Explants were then cultured at 37 degrees and 5% CO2 for 24 hours following ablation. Cultured explants were fixed and processed for immunofluorescence as described above.
Statistics and Reproducibility
Data between two groups were compared using a two-tailed, unpaired Student’s t-test. An F-test was performed to compare variances, and if significantly different, the t-test was performed with Welsh’s correction. Prism (GraphPad) was used for these analyses and to plot the data. See figure legends for specific p-values and n-values. P-values > 0.05 were considered not to be significant (n.s.). All experiments presented in this study were performed on placodes from at least 2 embryos with the same results. Sample size was limited by the complexity of the mutant genotype and litter size. All mutant, inhibitor treated or laser ablated phenotypes reported contrast greatly with the controls. Therefore, no calculations were used to determine sample size. Representative images have associated quantification and statistical analysis included in the legends. Representative examples of independent repeats of key figures are also provided in the Supplementary data: Supplementary Figure 1 shows 3 independent movies related to Figure 1D and 2A–B. Supplementary Figure 2 shows independent repeats of Figure 2C. Supplementary Figure 3 shows independent repeats of Figure 3B. Supplementary Figure 4E shows independent repeats of Figure 4E. Supplementary Figure 5 shows independent repeats of Figure 5A–C. Supplementary Figure 6 shows independent validation of Figure 4A.
Data availability
Statistics source data for Figures 2, 4, 5, 6 and Supplementary Figures 4, 5 are available in Supplementary Table 2. All other data supporting the findings of the study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
We gratefully acknowledge those who provide mouse lines, technical support, and valuable discussions that contributed to this project. We thank Saori Haigo and Jeremy Reiter for generous donation of Vangl2 and Fz6 alleles in mT/mG backgrounds. Michael Deans and Jeremy Nathans kindly provided the Vangl2 and Vangl1 floxed mouse lines. Beniot Aiguoy developed and distributed Packing Analyzer v2 and Tissue Analyzer software for image analysis. We thank Katie Little for assistance with genetic crosses and genotyping, and members of the Devenport lab for insightful comments and suggestions. Finally, we thank Gary Laevsky for imaging support and expertise. The Confocal Facility at Princeton University is a Nikon Center of Excellence.
FUNDING
Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under award number R01AR066070. L.L. was supported by NIH training grant T32 GM007388.
Footnotes
AUTHOR CONTRIBUTIONS
Conceptualization, M.C., B.J., and D.D.; Investigation, M.C., L.L., B.J., and D.D..; Writing, M.C., L.L and D.D.; Funding Acquisition and Supervision, D.D.
References
- 1.Biggs LC, Mikkola ML. Early inductive events in ectodermal appendage morphogenesis. Seminars in cell & developmental biology. 2014;25–26:11–21. doi: 10.1016/j.semcdb.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 2.Ochoa-Espinosa A, Affolter M. Branching morphogenesis: from cells to organs and back. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a008243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.St Johnston D, Ahringer J. Cell polarity in eggs and epithelia: parallels and diversity. Cell. 2010;141:757–774. doi: 10.1016/j.cell.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 4.Campanale JP, Sun TY, Montell DJ. Development and dynamics of cell polarity at a glance. J Cell Sci. 2017;130:1201–1207. doi: 10.1242/jcs.188599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devenport D. Tissue morphodynamics: Translating planar polarity cues into polarized cell behaviors. Seminars in cell & developmental biology. 2016;55:99–110. doi: 10.1016/j.semcdb.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodrich LV, Strutt D. Principles of planar polarity in animal development. Development. 2011;138:1877–1892. doi: 10.1242/dev.054080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Y, Mlodzik M. Wnt-Frizzled/Planar Cell Polarity Signaling: Cellular Orientation by Facing the Wind (Wnt) Annu Rev Cell Dev Biol. 2015;31:623–646. doi: 10.1146/annurev-cellbio-100814-125315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devenport D. The cell biology of planar cell polarity. J Cell Biol. 2014;207:171–179. doi: 10.1083/jcb.201408039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butler MT, Wallingford JB. Planar cell polarity in development and disease. Nat Rev Mol Cell Biol. 2017;18:375–388. doi: 10.1038/nrm.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adler PN. The frizzled/stan pathway and planar cell polarity in the Drosophila wing. Curr Top Dev Biol. 2012;101:1–31. doi: 10.1016/B978-0-12-394592-1.00001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sennett R, Rendl M. Mesenchymal-epithelial interactions during hair follicle morphogenesis and cycling. Seminars in cell & developmental biology. 2012;23:917–927. doi: 10.1016/j.semcdb.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duverger O, Morasso MI. Epidermal patterning and induction of different hair types during mouse embryonic development. Birth Defects Res C Embryo Today. 2009;87:263–272. doi: 10.1002/bdrc.20158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Millar SE. Molecular mechanisms regulating hair follicle development. J Invest Dermatol. 2002;118:216–225. doi: 10.1046/j.0022-202x.2001.01670.x. [DOI] [PubMed] [Google Scholar]
- 14.Devenport D, Fuchs E. Planar polarization in embryonic epidermis orchestrates global asymmetric morphogenesis of hair follicles. Nat Cell Biol. 2008;10:1257–1268. doi: 10.1038/ncb1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Chang H, Nathans J. When whorls collide: the development of hair patterns in frizzled 6 mutant mice. Development. 2010;137:4091–4099. doi: 10.1242/dev.057455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levy V, Lindon C, Harfe BD, Morgan BA. Distinct stem cell populations regenerate the follicle and interfollicular epidermis. Dev Cell. 2005;9:855–861. doi: 10.1016/j.devcel.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 17.Harfe BD, et al. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118:517–528. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- 18.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 19.Aigouy B, Umetsu D, Eaton S. Segmentation and Quantitative Analysis of Epithelial Tissues. Methods Mol Biol. 2016;1478:227–239. doi: 10.1007/978-1-4939-6371-3_13. [DOI] [PubMed] [Google Scholar]
- 20.Guillot C, Lecuit T. Mechanics of epithelial tissue homeostasis and morphogenesis. Science. 2013;340:1185–1189. doi: 10.1126/science.1235249. [DOI] [PubMed] [Google Scholar]
- 21.Zallen JA, Blankenship JT. Multicellular dynamics during epithelial elongation. Seminars in cell & developmental biology. 2008;19:263–270. doi: 10.1016/j.semcdb.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aw WY, Heck BW, Joyce B, Devenport D. Transient Tissue-Scale Deformation Coordinates Alignment of Planar Cell Polarity Junctions in the Mammalian Skin. Curr Biol. 2016;26:2090–2100. doi: 10.1016/j.cub.2016.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cetera M, Leybova L, Woo FW, Deans M, Devenport D. Planar cell polarity-dependent and independent functions in the emergence of tissue-scale hair follicle patterns. Dev Biol. 2017;428:188–203. doi: 10.1016/j.ydbio.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang H, Smallwood PM, Williams J, Nathans J. The spatio-temporal domains of Frizzled6 action in planar polarity control of hair follicle orientation. Dev Biol. 2016;409:181–193. doi: 10.1016/j.ydbio.2015.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bertet C, Sulak L, Lecuit T. Myosin-dependent junction remodelling controls planar cell intercalation and axis elongation. Nature. 2004;429:667–671. doi: 10.1038/nature02590. [DOI] [PubMed] [Google Scholar]
- 26.Blankenship JT, Backovic ST, Sanny JS, Weitz O, Zallen JA. Multicellular rosette formation links planar cell polarity to tissue morphogenesis. Dev Cell. 2006;11:459–470. doi: 10.1016/j.devcel.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 27.Nishimura T, Honda H, Takeichi M. Planar cell polarity links axes of spatial dynamics in neural-tube closure. Cell. 2012;149:1084–1097. doi: 10.1016/j.cell.2012.04.021. [DOI] [PubMed] [Google Scholar]
- 28.Williams M, Yen W, Lu X, Sutherland A. Distinct apical and basolateral mechanisms drive planar cell polarity-dependent convergent extension of the mouse neural plate. Dev Cell. 2014;29:34–46. doi: 10.1016/j.devcel.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shindo A, Wallingford JB. PCP and septins compartmentalize cortical actomyosin to direct collective cell movement. Science. 2014;343:649–652. doi: 10.1126/science.1243126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jamora C, DasGupta R, Kocieniewski P, Fuchs E. Links between signal transduction, transcription and adhesion in epithelial bud development. Nature. 2003;422:317–322. doi: 10.1038/nature01458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bitgood MJ, McMahon AP. Hedgehog and Bmp genes are coexpressed at many diverse sites of cell-cell interaction in the mouse embryo. Dev Biol. 1995;172:126–138. doi: 10.1006/dbio.1995.0010. [DOI] [PubMed] [Google Scholar]
- 32.Nowak JA, Polak L, Pasolli HA, Fuchs E. Hair follicle stem cells are specified and function in early skin morphogenesis. Cell Stem Cell. 2008;3:33–43. doi: 10.1016/j.stem.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ouspenskaia T, Matos I, Mertz AF, Fiore VF, Fuchs E. WNT-SHH Antagonism Specifies and Expands Stem Cells prior to Niche Formation. Cell. 2016;164:156–169. doi: 10.1016/j.cell.2015.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Z, et al. Embryonic attenuated Wnt/beta-catenin signaling defines niche location and long-term stem cell fate in hair follicle. eLife. 2015;4:e10567. doi: 10.7554/eLife.10567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vidal VP, et al. Sox9 is essential for outer root sheath differentiation and the formation of the hair stem cell compartment. Curr Biol. 2005;15:1340–1351. doi: 10.1016/j.cub.2005.06.064. [DOI] [PubMed] [Google Scholar]
- 36.St-Jacques B, et al. Sonic hedgehog signaling is essential for hair development. Curr Biol. 1998;8:1058–1068. doi: 10.1016/s0960-9822(98)70443-9. [DOI] [PubMed] [Google Scholar]
- 37.Karlsson L, Bondjers C, Betsholtz C. Roles for PDGF-A and sonic hedgehog in development of mesenchymal components of the hair follicle. Development. 1999;126:2611–2621. doi: 10.1242/dev.126.12.2611. [DOI] [PubMed] [Google Scholar]
- 38.Chiang C, et al. Essential role for Sonic hedgehog during hair follicle morphogenesis. Dev Biol. 1999;205:1–9. doi: 10.1006/dbio.1998.9103. [DOI] [PubMed] [Google Scholar]
- 39.Mill P, et al. Sonic hedgehog-dependent activation of Gli2 is essential for embryonic hair follicle development. Genes & development. 2003;17:282–294. doi: 10.1101/gad.1038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schweisguth F. Asymmetric cell division in the Drosophila bristle lineage: from the polarization of sensory organ precursor cells to Notch-mediated binary fate decision. Wiley Interdiscip Rev Dev Biol. 2015;4:299–309. doi: 10.1002/wdev.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng Y, Han C, Axelrod JD. Planar polarized protrusions break the symmetry of EGFR signaling during Drosophila bract cell fate induction. Dev Cell. 2012;23:507–518. doi: 10.1016/j.devcel.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Capilla A, et al. Planar cell polarity controls directional Notch signaling in the Drosophila leg. Development. 2012;139:2584–2593. doi: 10.1242/dev.077446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cooper MT, Bray SJ. Frizzled regulation of Notch signalling polarizes cell fate in the Drosophila eye. Nature. 1999;397:526–530. doi: 10.1038/17395. [DOI] [PubMed] [Google Scholar]
- 44.Fanto M, Mlodzik M. Asymmetric Notch activation specifies photoreceptors R3 and R4 and planar polarity in the Drosophila eye. Nature. 1999;397:523–526. doi: 10.1038/17389. [DOI] [PubMed] [Google Scholar]
- 45.Tomlinson A, Struhl G. Decoding vectorial information from a gradient: sequential roles of the receptors Frizzled and Notch in establishing planar polarity in the Drosophila eye. Development. 1999;126:5725–5738. doi: 10.1242/dev.126.24.5725. [DOI] [PubMed] [Google Scholar]
- 46.Driskell RR, Clavel C, Rendl M, Watt FM. Hair follicle dermal papilla cells at a glance. J Cell Sci. 2011;124:1179–1182. doi: 10.1242/jcs.082446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ahtiainen L, et al. Directional cell migration, but not proliferation, drives hair placode morphogenesis. Dev Cell. 2014;28:588–602. doi: 10.1016/j.devcel.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 48.Glover JD, et al. Hierarchical patterning modes orchestrate hair follicle morphogenesis. PLoS Biol. 2017;15:e2002117. doi: 10.1371/journal.pbio.2002117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keller R. Shaping the vertebrate body plan by polarized embryonic cell movements. Science. 2002;298:1950–1954. doi: 10.1126/science.1079478. [DOI] [PubMed] [Google Scholar]
- 50.Tada M, Heisenberg CP. Convergent extension: using collective cell migration and cell intercalation to shape embryos. Development. 2012;139:3897–3904. doi: 10.1242/dev.073007. [DOI] [PubMed] [Google Scholar]
- 51.Shindo A. Models of convergent extension during morphogenesis. Wiley Interdiscip Rev Dev Biol. 2017 doi: 10.1002/wdev.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chuai M, Weijer CJ. The mechanisms underlying primitive streak formation in the chick embryo. Curr Top Dev Biol. 2008;81:135–156. doi: 10.1016/S0070-2153(07)81004-0. [DOI] [PubMed] [Google Scholar]
- 53.Rozbicki E, et al. Myosin-II-mediated cell shape changes and cell intercalation contribute to primitive streak formation. Nat Cell Biol. 2015;17:397–408. doi: 10.1038/ncb3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voiculescu O, Bertocchini F, Wolpert L, Keller RE, Stern CD. The amniote primitive streak is defined by epithelial cell intercalation before gastrulation. Nature. 2007;449:1049–1052. doi: 10.1038/nature06211. [DOI] [PubMed] [Google Scholar]
- 55.Mlodzik M. Planar polarity in the Drosophila eye: a multifaceted view of signaling specificity and cross-talk. EMBO J. 1999;18:6873–6879. doi: 10.1093/emboj/18.24.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng L, Zhang J, Carthew RW. frizzled regulates mirror-symmetric pattern formation in the Drosophila eye. Development. 1995;121:3045–3055. doi: 10.1242/dev.121.9.3045. [DOI] [PubMed] [Google Scholar]
- 57.Theisen H, et al. dishevelled is required during wingless signaling to establish both cell polarity and cell identity. Development. 1994;120:347–360. doi: 10.1242/dev.120.2.347. [DOI] [PubMed] [Google Scholar]
- 58.Wolff T, Rubin GM. Strabismus, a novel gene that regulates tissue polarity and cell fate decisions in Drosophila. Development. 1998;125:1149–1159. doi: 10.1242/dev.125.6.1149. [DOI] [PubMed] [Google Scholar]
- 59.Winter CG, et al. Drosophila Rho-associated kinase (Drok) links Frizzled-mediated planar cell polarity signaling to the actin cytoskeleton. Cell. 2001;105:81–91. doi: 10.1016/s0092-8674(01)00298-7. [DOI] [PubMed] [Google Scholar]
- 60.Fiehler RW, Wolff T. Drosophila Myosin II, Zipper, is essential for ommatidial rotation. Dev Biol. 2007;310:348–362. doi: 10.1016/j.ydbio.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aigouy B, et al. Cell flow reorients the axis of planar polarity in the wing epithelium of Drosophila. Cell. 2010;142:773–786. doi: 10.1016/j.cell.2010.07.042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Statistics source data for Figures 2, 4, 5, 6 and Supplementary Figures 4, 5 are available in Supplementary Table 2. All other data supporting the findings of the study are available from the corresponding author upon reasonable request.