Synopsis
Clonal hematopoiesis as a hallmark of myelodysplastic syndrome (MDS) is mediated by the selective advantage of clonal hematopoietic stem cells in a context-specific manner. While primary MDS emerges without known predisposing cause and is associated with advanced age, secondary MDS may develop in younger patients with bone marrow failure syndromes or after exposure to chemotherapy, respectively. This article discusses recent advances in our understanding of context-dependent clonal hematopoiesis in MDS with focus on clonal evolution in inherited and acquired bone marrow failure syndromes.
Keywords: Myelodysplastic syndrome, Bone marrow failure syndromes, Genetic predisposition, Clonal hematopoiesis
Introduction
MDS pathogenesis
Myelodysplastic syndrome (MDS) is a heterogeneous group of clonal hematopoietic disorders characterized by dysfunctional hematopoiesis, bone marrow dysplasia and an increased risk of development of acute myeloid leukemia (AML)1. Although MDS is most common in older patients (> 70 years) it can occur in all age groups, including children and young adults. Primary MDS emerges without known predisposing cause and is associated with advanced age, while secondary and therapy-related MDS (t-MDS) are proportionally more common in younger MDS patients and develop in the context of inherited or acquired bone marrow failure or after exposure to chemotherapy, respectively.
Genetic studies have demonstrated that MDS molecular alterations are closely associated with clinical outcomes and disease characteristics2,3. Indeed, the spectrum of genetic alterations in young MDS patients is different than that of older MDS patients, consistent with the distinct age-associated mechanisms of MDS pathogenesis2. Whereas older patients more frequently harbor somatic mutations in genes encoding epigenetic modifiers (TET2 and DNMT3A) or RNA splicing (SRSF2 and SF3B1), younger patients have much higher frequency of genes associated with germline conditions (GATA2 and SBDS) and acquired predispositions (PIGA). Mutations in other genes, such as TP53, RUNX1, or RAS are common across all age groups2.
Clonal hematopoiesis and aging
Advancing age is the most established risk factor for developing clonally-restricted hematopoiesis. During normal aging, individual hematopoietic stem cells (HSCs) steadily accumulate somatic mutations. By age 60, it is estimated that each HSC harbors eight mutations affecting its coding genome4. While most of these mutations do not measurably alter stem cell function, some confer a competitive advantage over normal HSCs and cause preferential contribution to mature hematopoietic cells. This phenomenon, when occurring in otherwise healthy individuals, is termed Clonal Hematopoiesis of Indeterminate Potential (CHIP), which has several key properties:
a strong association with advancing age,
an increased risk of developing frank hematologic malignancy (overall risk = 1% per year), and
an increase in all-cause mortality related to an elevated risk of cardiovascular events5.
The age-dependent accumulation of somatic mutations may underlie the increasing prevalence of MDS among older individuals; the median age at MDS diagnosis is 71–76 years6. The close genetic and epidemiologic concordance between CHIP and primary MDS has engendered a model whereby clinically unapparent clonal HSC expansion is caused by an initiating mutation affecting particular genes, such as DNMT3A, TET2 and ASXL1, while transformation to frank myeloid malignancy is mediated by subsequent stepwise acquisition of additional myeloid driver mutations3,5. The factors that influence the frequency, genetic spectrum, and clinical implications of CHIP remain incompletely understood.
Extrinsic selection and clonal hematopoiesis: CHIP and t-MDS
Changes in cell extrinsic selection pressures due to specific therapeutic exposures or disease characteristics may influence the development and clinical implications of clonal hematopoiesis. For example, CHIP is present in about 30% of patients with non-Hodgkin lymphoma who undergo autologous stem cell transplantation, reflecting a rate more than 5 times higher than healthy adults of similar age spectrum7. Similarly, clonal hematopoiesis is common among patients with non-hematologic cancers8. The genetic spectrum of CHIP that arises in the context of therapeutic exposure is distinct, showing an enrichment of mutations affecting TP53 and PPM1D, genes that are important for the cellular stress response. Mutations in TP53 and PPM1D are also highly associated with t-MDS, compared to primary MDS, suggesting a mechanistic link between CHIP arising in the context of exposure and the subsequent development of t-MDS2.
Clonal hematopoiesis in inherited bone marrow failure and familial MDS/AML predisposition syndromes
The ability of a cell to persist within a specific selective environment defines its “fitness” and reflects aggregate characteristics of cell survival, differentiation and proliferation. Importantly, cellular fitness is functionally defined only relative to surrounding cells. In healthy individuals, clonal hematopoiesis and MDS arise in the competitive backdrop of normal hematopoiesis. In this context, clonal expansion is based on a gain of fitness relative to otherwise fit cells. By contrast, bone marrow failure syndromes can display subtle or profound alterations of HSC function and microenvironment. Therefore, even mutations that cause modest enhancement of fitness may manifest as clonal hematopoiesis or drive myeloid transformation. Moreover, disease-specific cellular defects (Fig. 1) may create context-dependent selective environments that result in distinct opportunities for somatic genetic cooperation (Fig. 2). Below, the authors discuss how inherited bone marrow failure syndromes and MDS/AML predisposition syndromes may exert an influence on the incidence and genetic spectrum of clonal hematopoiesis and myeloid transformation. Further, the authors consider how disease-specific selective pressures, driven by diverse underlying pathogenetic mechanisms, may define distinct selective pressures that are intrinsic and extrinsic to the HSC.
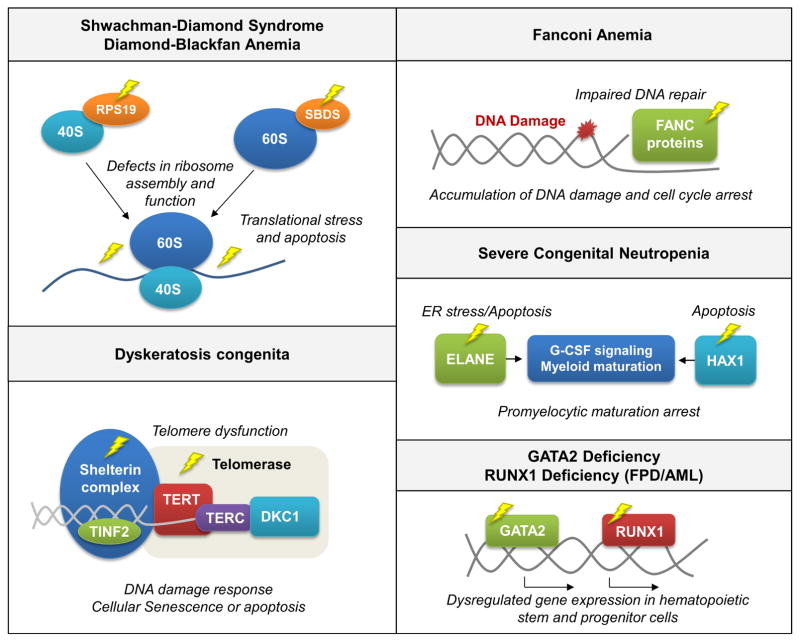
Figure 1.
Pathways affected in inherited bone marrow failure and MDS/AML predisposition syndromes.
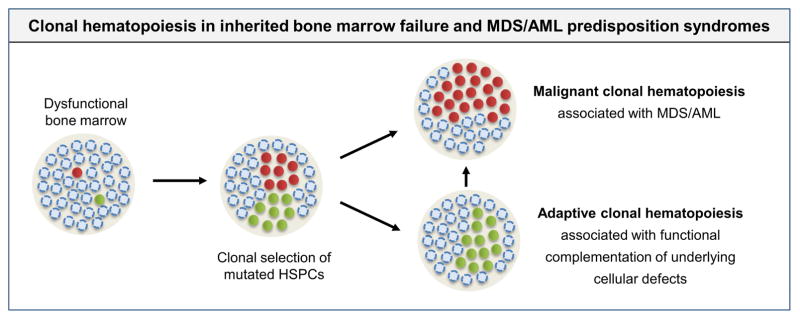
Figure 2. Clonal hematopoiesis in inherited bone marrow failure and MDS/AML predisposition syndromes.
Bone marrow failure syndromes are associated with dysfunctional hematopoiesis that may drive a strong selective pressure that favors mutated hematopoietic stem and progenitor cells with enhanced fitness. While some mutations cause leukemic transformation (red circles), others may enable clonal expansion due to functional complementation of disease specific cellular defects (green circles). The latter may involve biological pathways that are distinct from leukemic transformation and not associated with elevated risk of progression to MDS or AML.
Shwachman-Diamond Syndrome
Shwachman-Diamond Syndrome (SDS) is an autosomal recessive bone marrow failure syndrome that is caused by biallelic inactivating mutations in the SBDS gene on chromosome 7 (q11.21) and associated with short stature, exocrine pancreatic dysfunction and a strong predisposition to MDS/AML transformation9. In a cohort of 55 SDS patients, 36% of patients developed MDS or AML by the age of 30 years10. Cellular deficiency of the SBDS protein results in ribosomal dysfunction and translational inefficiency, which is linked to Fas ligand-induced apoptosis and induction of TP53-mediated cellular senescence pathways11,12.
Recurrent clonal cytogenetic alterations are commonly identified in the bone marrow of SDS patients, including isochromosome 7 (i7(q10)), monosomy 7, and del(20q). While monosomy 7 is associated with rapid clonal progression, del(20q) and i7(q10) correlate with a benign clinical course13,14, suggesting that development of clonal hematopoiesis with somatic genetic alterations may not be deterministic of leukemic transformation.
In SDS patients with compound heterozygosity for the 2 most common SBDS mutations, the presence of i(7)(q10) favored the allele with c.258+2T>C (a splice site mutation causing decreased SBDS levels) over the c.183_184TA>CT allele (a nonsense mutation causing complete SBDS loss). As such, i(7)(q10) may cause a relative increase of SBDS protein, thereby improving the underlying cellular dysfunction and driving a selective clonal advantage14. Similarly, the minimally deleted region of del(20q) involves the EIF6 gene locus. It has been hypothesized that EIF6 haploinsufficiency promotes partial rescue of impaired ribosome biogenesis in SDS cells by favoring ejection of EIF6 from the nascent 60S ribosome13,15. In each of these scenarios, the selective fitness of somatically mutated clones may be driven by functional complementation of underlying cellular defects, rather than by alteration of pathways directly involved in biological transformation13,15. Consistent with this model, neither i(7)(q10) nor del(20q) alterations are associated with an increased risk of leukemic transformation.
In our genetic analysis of samples obtained before transplantation from 1514 patients with MDS, we identified 7 patients with canonical biallelic SBDS mutations2. These SDS patients were significantly younger than other MDS patients (median age 25.1 years) and had poor clinical outcomes (median survival of 1.2 years) compared to other young patients (median survival not reached). Consistent with their poor survival, all patients with biallelic SBDS mutations had at least one somatic TP53 mutation. Moreover, TP53 mutations were significantly more frequent in patients with biallelic SBDS mutations than in patients without SBDS mutations. The high frequency of TP53 mutations in this cohort suggests that there may be a specific cooperative effect between SBDS deficiency and TP53 alterations that mediates clinical progression to MDS.
A subsequent study of 27 patients used a barcoded sequencing approach to confirm that somatic TP53 mutations are highly recurrent in SDS, affecting 48% of patients16. The incidence of TP53 mutations in this SDS cohort correlated with increased age, and several patients harbored multiple TP53 mutations. Importantly, the authors identified TP53 mutations even among patients without clinical or morphologic evidence of transformation, and the presence of TP53 mutations was not associated with worse hematologic function. TP53 mutations were not detected in SCN and cyclic neutropenia patients16, suggesting that TP53 mutations are not broadly associated with neutropenic conditions, but rather may be specifically linked to SDS biology.
TP53 is a central effector of the cellular response to ribosomal dysfunction17. Indeed, TP53 overexpression is observed in bone marrow biopsies from SDS patients12 and Sbds deficiency in mice was shown to induce Tp53 dependent apoptosis in myeloid cells18, suggesting that ribosomal dysfunction and translational inefficiency in SDS patients induce TP53 dependent cellular senescence. Consistent with the role of TP53 in mediating the SDS phenotype, genetic ablation of Trp53 in mice attenuates the atrophy seen in Sbds deficient pancreatic acinar cells11. It is possible that somatic acquisition of TP53 mutations in SBDS-deficient hematopoietic cells similarly rescues inefficient hematopoiesis, resulting in selection and clonal expansion of mutated cells.
Diamond-Blackfan Anemia
Diamond-Blackfan Anemia (DBA) is characterized by red blood cell aplasia and is associated with infantile onset of isolated, severe, macrocytic anemia, as well as short stature and congenital anomalies9. DBA is inherited in an autosomal dominant pattern and is most frequently caused by mutations affecting genes that encode ribosomal proteins which are important for 18S or 28S rRNA maturation and small or large ribosomal subunit synthesis, respectively9. Approximately 25% of DBA patients have a mutation in RPS19, although more than 10 causative genes have been identified9. The Diamond Blackfan Anemia Registry reports that DBA patients have a cumulative incidence for AML of 5% by the age of 46 years19.
In our MDS transplant cohort, only one patient had DBA (defined by the presence of a heterozygous frameshift mutation in the RPS17 gene)2. In this case, we identified 6 distinct somatic mutations involving TP53 and PPM1D, which encodes a serine-threonine protein phosphatase that regulates TP53 and the cellular stress response. This observation is consistent with a mechanistic link between ribosomal dysfunction and TP53 dependent transformation that has been observed in SDS patients that develop MDS.
In bone marrow specimens from DBA patients without transformation, TP53 has been shown to accumulate in erythroid progenitor cells, and TP53 was induced selectively in primary human erythroid progenitor cells after RPS19 knockdown20. Moreover, the erythroid phenotype in mouse models of DBA is rescued by concomitant inactivation of Tp5321. Mutations in the TP53 pathway may thus drive clonal expansion by attenuating DBA-related erythroid apoptosis20,22. However, specific characteristics of clonal evolution in DBA patients and mechanistic links between DBA-related ribosomal dysfunction and TP53-dependent myeloid transformation remain to be elucidated.
Severe Congenital Neutropenia
Severe Congenital Neutropenia (SCN) is most commonly caused by germline mutations in ELANE or HAX1 and leads to promyelocytic maturation arrest, dysfunctional production of neutrophils in the bone marrow, and a heighted risk of life-threatening infections9. Supportive therapy involves administration of G-CSF which results in most cases in significant improvement of neutrophil counts23. The risk of myeloid transformation is high, with a cumulative incidence of 22% after 15 years of G-CSF treatment, and correlates with a poor response to G-CSF therapy23.
In a study of 148 SCN patients, 13 out of 23 patients (78%) who developed MDS or AML carried somatic activating CSF3R receptor mutations24. However, disease latency was highly variable and several patients harbored CSF3R mutations for many years without evidence of transformation. Similarly, serial analysis of one patient showed that 5 different CSF3R mutations were present 15 years prior to transformation, and that eventual development of leukemia was associated with outgrowth of a single clone that had gained additional myeloid driver mutations25. Together, these data suggest that CSF3R mutations may require cooperating genetic events to cause leukemia.
Most CSF3R mutations cause a truncation of the cytoplasmic domain mediating enhanced cell proliferation and survival26. By potentiating G-CSF signaling, CSF3R mutations may thus result in functional compensation for ELANE and HAX1 related defects in neutrophil production, providing a potential explanation for the high prevalence of CSF3R mutations in SCN patients20,27. A definite role for CSF3R mutations in initiating SCN-related leukemogenesis, independent of enabling adaptive hematopoiesis, and the link to G-CSF therapy, remains to be determined.
Somatic RUNX1 mutations were recently identified in 64.5% of SCN patients with MDS or AML, often occurring in clones that had already acquired CSF3R mutations28. In contrast, no RUNX1 mutations were seen in a cohort of 40 SCN patients without leukemic transformation16. These data strongly support a role for RUNX1 mutations as a late step in leukemic transformation in the context of SCN. The variable latency between acquisition of somatic CSF3R mutations and the development of RUNX1 mutations suggests that additional cooperating clinical or genetic variables may not yet be identified25,28.
Fanconi Anemia
Fanconi Anemia (FA) is a disorder of chromosomal instability caused by germline mutations in DNA repair genes of the FA/BRCA pathway9. Clinical features can vary widely, with some individuals manifesting bone marrow failure, short statue, skin and upper limb abnormalities, and others (25–40%) having no abnormal physical findings29. Patients with FA have elevated cumulative incidence of various cancers by the age of 50 years, including MDS (40%), AML (10%) and solid tumors (20–30%)30.
Somatic reversion of germline FANC gene mutations in hematopoietic cells has been reported in 15% of FA patients31. Cells with one functionally corrected allele may have a selective clonal advantage over cells with two pathogenic alleles, thus causing functional rescue with enhanced contribution to the HSC pool and to hematopoiesis, resulting in stabilization of blood counts31. Importantly, reversion events were not detected in FA-related MDS or AML, suggesting that restoration of Fanconi pathway function may drive relative clonal advantage in the context of impaired hematopoiesis, but is biologically distinct from malignant transformation32.
The frequency of somatic chromosomal gains and losses in 57 FA patients was evaluated using high-density genome-wide CGH/SNP arrays32. In this study, alterations were identified in 61% of patients with diverse clinical phenotypes, demonstrating that clonal hematopoiesis is common in FA patients, irrespective of hematologic status. Highly recurrent alterations in this cohort included gains of 1q (45%) and 3q (41%), monosomy 7/del(7q) (17%), 11q- (13.8%) and abnormalities involving RUNX1 (21%), although the distribution of alterations across hematologic phenotypes was distinct. Whereas clonal hematopoiesis involving somatic genetic reversions of mutated FANC genes or 1q+ were associated with an indolent clinical course, the presence of RUNX1 lesions, 3q+, or −7/del(7q) were more common in MDS and AML32–34. Moreover, the presence of molecular genetic alterations associated with non-FA AML, such as oncogenic NRAS mutations, FLT3 internal tandem duplication (FLT3-ITD), or MLL partial tandem duplication (MLL-PTD) were restricted to patients with frank AML32.
Gains of 1q and 3q are specifically enriched in FA patients, suggesting that they may confer a distinct clonal advantage in the context of impaired FA/BRCA pathways32. Moreover, 1q+ can be observed in FA patients with clonal hematopoiesis with and without myeloid transformation and could reflect an uncharacterized mechanism of functional complementation of Fanconi abnormalities in HSCs. Conversely, gain of 3q is highly associated with malignant transformation, possibly due to the amplification of the leukemogenic oncogene EVI132,35.
Dyskeratosis Congenita
Dyskeratosis Congenita (DC) is caused by germline mutations in a set of genes involved in telomere maintenance including TINF2 (12%), TERT (5%), TERC (5%), RTEL1 (2%) and DKC1 (25%)9. Characteristic clinical features include abnormal skin pigmentation, oral leukoplakia and nail dystrophy, hypoplastic bone marrow, pulmonary fibrosis, and liver disease9. DC patients have a high risk of developing hematologic complications such as aplastic anemia, MDS and AML, as well as solid tumors9.
The role of acquired somatic genetic alterations in myeloid transformation in DC patients has not been systematically characterized. In 16 DC patients analyzed using a combination of X-inactivation analyses, comparative whole exome sequencing (WES) and single nucleotide polymorphism arrays (SNP-A), 8 out of 9 female patients showed skewed X-inactivation, suggesting that clonal hematopoiesis is common36. Among 6 patients evaluated by whole exome sequencing, no somatic mutations affecting recurrently mutated genes associated with hematologic cancers were identified. Importantly, one patient showed somatic reversion in DKC1, suggesting that restoration of normal telomere length maintenance affords a selective advantage in hematopoietic cells36. None of the patients in this study had clinical or morphologic evidence of myeloid transformation.
In our analysis of MDS patients receiving allogeneic transplantation, 1% of adults harbored pathogenic germline mutations affecting genes involved in telomere maintenance, including TERT, TERC, or DKC1, while at least another 2% of cases had rare variants of uncertain biological significance2. Among 11 adult patients with germline TERT or TERC mutations, 8 (73%) had somatic mutations in established myeloid driver genes, including 7 with mutations affecting TP53 or PPM1D. TP53 plays a critical role in enforcing senescence and apoptotic responses to telomere dysfunction, while PPM1D is a serine-threonine protein phosphatase that negatively regulates the DNA damage response via dephosphorylation of specific residues on ATM, CHK1, and TP5337. Mutations in PPM1D are localized to exon 6, and cause C-terminal truncations that may cause an increase in phosphatase activity that aberrantly inhibits checkpoint and DNA damage response (DDR) pathways38. Similar to SDS and DBA, where the mechanism of bone marrow failure drives activation of TP53 activity, our data suggest that severe telomere attrition may select for somatic clones with genetic inactivation of TP53.
GATA2 Deficiency
Germline mutations of GATA2 cause a spectrum of clinical phenotypes defined by GATA2 haploinsufficiency with autosomal dominant inheritance. GATA2 regulates HSC function in a dose dependent manner, and mutations lead to inactivation via truncation or impairment of functional DNA-binding39. Patients with GATA2 haploinsufficiency have a 70% risk of progression to early-onset myeloid malignancies along with immune deficiencies and variable systemic features40.
Myeloid transformation in GATA2 deficiency syndromes is associated with somatic mutations in typical myeloid driver genes, including chromosomal abnormalities, such as monosomy 7 and trisomy 841. Among young MDS patients, the association between monosomy 7 and germline GATA2 mutations is particularly striking: 70% of adolescent patients with monosomy 7 have an underlying GATA2 deficiency42. ASXL1 mutations are most common, identified in 14 out of 48 patients (29%) in one study, and associated with monosomy 7 and trisomy 8, young age, female gender and poor survival41. Other smaller studies have seen similar association with ASXL1 mutations, as well as recurrent mutations affecting other hematologic driver genes including SETBP1, RUNX1, NRAS, and STAG243–45.
Familial Platelet Disorder with Predisposition to Acute Myeloid Leukemia
The Familial Platelet Disorder with Predisposition to Acute Myeloid Leukemia (FPD/AML) is an autosomal dominant disease caused by inactivating germline alterations affecting the hematopoietic transcription factor RUNX1. Typical clinical manifestations include thrombocytopenia with defects of platelet function leading to a mild to moderate bleeding tendency and a propensity to develop MDS or AML46. In a study of 10 families with 5 pedigrees harboring RUNX1 germline mutations the median incidence of MDS/AML was 35% 47.
In the context of germline RUNX1 mutations, acquisition of somatic mutations is a common, if not ubiquitous characteristic of MDS/AML transformation. In focused genetic analyses, acquired mutations in FPD/AML patients have been identified in a typical spectrum of myeloid driver genes, but the most frequent progression mutation affects the second RUNX1 allele48,49,50. In a study of 9 asymptomatic individuals with germline RUNX1 mutations, 6 (67%) had evidence of clonal hematopoiesis, reflected by detectable somatic mutations in the blood or bone marrow51. However, only one patient harbored a mutation in a canonical myeloid driver gene, suggesting existence of novel cooperating drivers of clonal hematopoiesis, or other factors that favor development of clonally-restricted hematopoiesis in RUNX1 deficient HSCs. Although these data suggest a high cumulative risk of developing clonal hematopoiesis51, a direct link between clonal hematopoiesis and development of subsequent myeloid malignancies with recurrent drivers has not been established.
Acquired Aplastic Anemia
Acquired aplastic anemia (AA) is caused by human leukocyte antigen (HLA) restricted destruction of HSCs by autoreactive T cells52. Immunosuppression therapy (IST), supportive therapies, and allogeneic HSCT has improved the outcome of the disease. However, even after successful IST, patients remain at high risk of developing clonal disorders such as MDS/AML and PNH, with a 10-year cumulative incidence of 10–15% and 50%, respectively53,54.
Clonal hematopoiesis in AA patients most frequently involves mutations in BCOR, BCORL1, PIGA, DNMT3A, ASXL1, RUNX1 and HLA genes and is often detectable at diagnosis and dynamic over time54–56. ASXL1, RUNX1 and DNMT3A mutations are associated with advanced age, progression to MDS, and an inferior overall survival, while mutations in PIGA and BCOR/BCORL1 correlate with a better response to immunosuppressive therapy and a better overall outcome54,55. Although clonal dynamics detected in serial samples of 35 AA patients were highly variable, clones harboring PIGA or BCOR/BCORL1 mutations tended to remain stable or to decrease over time, suggesting a specific clonal advantage in the setting of autoimmune destruction which is lost during effective IST54.
Frequent uniparental disomy (UPD) involving the HLA locus and recurrent loss-of-function HLA mutations support a model where escape from T-cell mediated destruction can drive selective clonal advantage in some AA patients54,56–58. However, expansion of HLA mutated clones has not been linked to myeloid transformation, suggesting that immune evasion might drive a biologically distinct pathway of clonal dominance (Fig. 3). Clonal hematopoiesis in AA may thus be driven by a range of selection contexts, including immune evasion, aberrant survival in an altered microenvironment, or stem cell attrition.
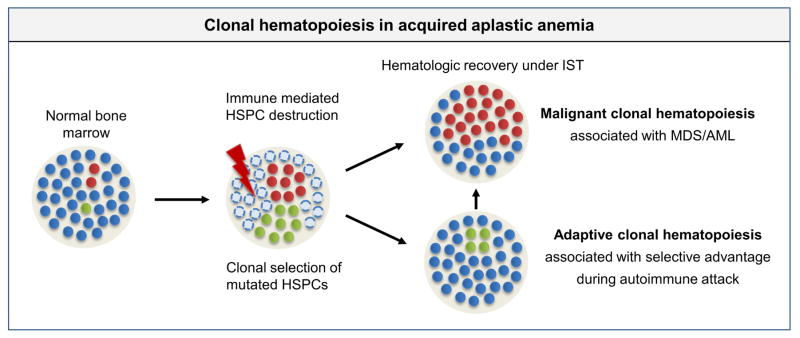
Figure 3. Clonal hematopoiesis in acquired aplastic anemia.
Immune mediated selection pressure drives expansion of HSPCs with context-specific growth advantage. Some clones may expand during the initial phase of disease due to a capacity for immune evasion, but may recede after successful immunosuppressive therapy (IST) and hematologic recovery (green circles). Other clones with typical myeloid driver mutations may display more context-independent expansion (red circles).
Summary
Development of clonal hematopoiesis represents the hallmark initiation of myeloid transformation. However, the timing, genetic spectrum, and clinical implications of clonal hematopoiesis can be influenced by a range of cell-intrinsic and cell-extrinsic factors. Importantly, context-specific variables, such as germline mutations in inherited bone marrow failure or immune mediated cell destruction in AA can exert a strong selection pressure on the development and progression of clonal hematopoiesis. Global hematopoietic dysfunction or bone marrow microenvironmental abnormalities may enable expansion of clones which are better adapted to specific extrinsic or intrinsic selection (Fig. 2, Fig. 3).
Clinical implications
Based on the genetic data outlined above, several outstanding questions remain to be answered. What clinical or genetic factors mediate myeloid transformation in patients with inherited bone marrow diseases? How can adaptive clonal hematopoiesis best be distinguished from incipient malignant degeneration? Can identification of specific somatic genetic characteristics be integrated prospectively into clinical care of individual patients? Unbiased genetic analysis of patient samples using whole exome or whole genome sequencing approaches, paired with systematic longitudinal analysis of samples obtained from patients at multiple times during the course of life may provide answers to these questions. An improved understanding of the role of acquired somatic mutations in clonal progression of bone marrow failure syndromes has the potential to improve outcomes in this high-risk patient group by identifying novel therapeutic vulnerabilities or enabling improved clinical decision-making based on objectively measurable molecular characteristics.
Key Points.
Clonal evolution in myelodysplastic syndromes can be driven by specific extrinsic and intrinsic selective pressures.
Acquired somatic mutations in inherited bone marrow failure syndromes can partially complement underlying cellular defects leading to a selective clonal advantage that might be distinct from myeloid transformation.
Long-term mutational studies with systematic analysis of serial samples in a larger number of patients are required to define prognostic and therapeutic implications of clonal hematopoiesis in patients with bone marrow failure syndromes.
Footnotes
Disclosure statement: the authors have no relationship with a commercial company that has a direct financial interest in subject matter or materials discussed
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–2406. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 2.Lindsley RC, Saber W, Mar BG, et al. Prognostic mutations in myelodysplastic syndrome after stem-cell transplantation. N Engl J Med. 2017;376(6):536–547. doi: 10.1056/NEJMoa1611604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Makishima H, Yoshizato T, Yoshida K, et al. Dynamics of clonal evolution in myelodysplastic syndromes. Nat Genet. 2016;49(2):204–212. doi: 10.1038/ng.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Welch J, Ley T, Link D, Miller C. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150(2):264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sekeres MA. The epidemiology of myelodysplastic syndromes. Hematol Oncol Clin North Am. 2010;24(2):287–294. doi: 10.1016/j.hoc.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 7.Gibson CJ, Lindsley RC, Tchekmedyian V, et al. Clonal hematopoiesis associated with adverse outcomes after autologous stem-cell transplantation for lymphoma. J Clin Oncol. 2017;35(14):1598–1605. doi: 10.1200/JCO.2016.71.6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coombs CC, Zehir A, Devlin SM, et al. Therapy-related clonal hematopoiesis in patients with non-hematologic cancers is common and associated with adverse clinical outcomes. Cell Stem Cell. 2017;21(3):374–382. doi: 10.1016/j.stem.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wegman-Ostrosky T, Savage SA. The genomics of inherited bone marrow failure: from mechanism to the clinic. Br J Haematol. 2017;177(4):526–542. doi: 10.1111/bjh.14535. [DOI] [PubMed] [Google Scholar]
- 10.Donadieu J, Leblanc T, Meunier BB, et al. Analysis of risk factors for myelodysplasias, leukemias and death from infection among patients with congenital neutropenia. Experience of the French Severe Chronic Neutropenia Study Group. Haematologica. 2005;90(1):45–53. [PubMed] [Google Scholar]
- 11.Tourlakis ME, Zhang S, Ball HL, et al. In vivo senescence in the Sbds-deficient murine pancreas: Cell-type specific consequences of translation insufficiency. PLoS Genet. 2015;11(6):e1005288. doi: 10.1371/journal.pgen.1005288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elghetany MT, Alter BP. p53 protein overexpression in bone marrow biopsies of patients with Shwachman-Diamond syndrome has a prevalence similar to that of patients with refractory anemia. Arch Pathol Lab Med. 2002;126(4):452–455. doi: 10.5858/2002-126-0452-PPOIBM. [DOI] [PubMed] [Google Scholar]
- 13.Pressato B, Valli R, Marletta C, et al. Deletion of chromosome 20 in bone marrow of patients with Shwachman-Diamond syndrome, loss of the EIF6 gene and benign prognosis Shwachman-Diamond. Br J Haematol. 2012;157(4):501–503. doi: 10.1111/j.1365-2141.2012.09033.x. [DOI] [PubMed] [Google Scholar]
- 14.Minelli A, Maserati E, Nicolis E, et al. The isochromosome i(7)(q10) carrying c.258+2t>c mutation of the SBDS gene does not promote development of myeloid malignancies in patients with Shwachman syndrome. Leukemia. 2009;23(4):708–711. doi: 10.1038/leu.2008.369. [DOI] [PubMed] [Google Scholar]
- 15.Valli R, Pressato B, Marletta C, et al. Different loss of material in recurrent chromosome 20 interstitial deletions in Shwachman-Diamond syndrome and in myeloid neoplasms. Mol Cytogenet. 2013;6:56. doi: 10.1186/1755-8166-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xia J, Miller CA, Baty J, et al. Somatic mutations and clonal hematopoiesis in congenital neutropenia [published online ahead of print November 1, 2017] Blood. doi: 10.1182/blood-2017-08-801985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGowan KA, Pang WW, Bhardwaj R, et al. Reduced ribosomal protein gene dosage and p53 activation in low-risk myelodysplastic syndrome. Blood. 2011;118(13):3622–3633. doi: 10.1182/blood-2010-11-318584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zambetti NA, Bindels EMJ, Van Strien PMH, et al. Deficiency of the ribosome biogenesis gene Sbds in hematopoietic stem and progenitor cells causes neutropenia in mice by attenuating lineage progression in myelocytes. Haematologica. 2015;100(10):1285–1293. doi: 10.3324/haematol.2015.131573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vlachos A, Rosenberg PS, Atsidaftos E, Alter BP, Lipton JM. Incidence of neoplasia in Diamond Blackfan anemia: A report from the Diamond Blackfan anemia registry. Blood. 2012;119(16):3815–3819. doi: 10.1182/blood-2011-08-375972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dutt S, Narla A, Lin K, et al. Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood. 2011;117(9):2567–2576. doi: 10.1182/blood-2010-07-295238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGowan KA, Li JZ, Park CY, et al. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat Genet. 2008;40(8):963–970. doi: 10.1038/ng.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Danilova N, Sakamoto KM, Lin S. Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood. 2008;112(13):5228–5237. doi: 10.1182/blood-2008-01-132290. [DOI] [PubMed] [Google Scholar]
- 23.Rosenberg PS, Alter BP, Bolyard AA, et al. The incidence of leukemia and mortality from sepsis in patients with severe congenital neutropenia receiving long-term G-CSF therapy. Blood. 2006;107(12):4628–4635. doi: 10.1182/blood-2005-11-4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Germeshausen M, Ballmaier M, Welte K. Incidence of CSF3R mutations in severe congenital neutropenia and relevance for leukemogenesis: results of a long-term survey. Blood. 2007;109:93–99. doi: 10.1182/blood-2006-02-004275. [DOI] [PubMed] [Google Scholar]
- 25.Beekman RE, Valkhof MG, Sanders MA, et al. Sequential gain of mutations in severe congenital neutropenia progressing to acute myeloid leukemia. Blood. 2012;119(22):5071–5077. doi: 10.1182/blood-2012-01-406116. [DOI] [PubMed] [Google Scholar]
- 26.Germeshausen M, Ballmaier M, Welte K. Implications of mutations in hematopoietic growth factor receptor genes in congenital cytopenias. Ann N Y Acad Sci. 2001;938:305–320. doi: 10.1111/j.1749-6632.2001.tb03599.x. discussion 320–301. [DOI] [PubMed] [Google Scholar]
- 27.Qiu Y, Zhang Y, Hu N, Dong F. A truncated granulocyte colony-stimulating factor receptor (G-CSFR) inhibits apoptosis induced by neutrophil elastase G185R mutant. J Biol Chem. 2017;292(8):3496–3505. doi: 10.1074/jbc.M116.755157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skokowa J, Steinemann D, Katsman-Kuipers JE, et al. Cooperativity of RUNX1 and CSF3R mutations in severe congenital neutropenia: a unique pathway in myeloid leukemogenesis. Blood. 2014;123(14):2229–2238. doi: 10.1182/blood-2013-11-538025. [DOI] [PubMed] [Google Scholar]
- 29.Shimamura A, Alter BP. Pathophysiology and management of inherited bone marrow failure syndromes. Blood Rev. 2010;24(3):101–122. doi: 10.1016/j.blre.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alter BP, Giri N, Savage SA, et al. Malignancies and survival patterns in the National Cancer Institute inherited bone marrow failure syndromes cohort study. Br J Haematol. 2010;150(2):179–188. doi: 10.1111/j.1365-2141.2010.08212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soulier J, Leblanc T, Jubert C, et al. Detection of somatic mosaicism and classification of Fanconi anemia patients by analysis of the FA/BRCA pathway. Blood. 2005;105(3):1329–1337. doi: 10.1182/blood-2004-05-1852. [DOI] [PubMed] [Google Scholar]
- 32.Quentin S, Cuccuini W, Ceccaldi R, et al. Myelodysplasia and leukemia of Fanconi anemia are associated with a specific pattern of genomic abnormalities that includes cryptic RUNX1/AML1 lesions. Blood. 2011;117(15):e161–e170. doi: 10.1182/blood-2010-09-308726. [DOI] [PubMed] [Google Scholar]
- 33.Cioc AM, Wagner JE, MacMillan ML, DeFor T, Hirsch B. Diagnosis of myelodysplastic syndrome among a cohort of 119 patients with fanconi anemia: Morphologic and cytogenetic characteristics. Am J Clin Pathol. 2010;133(1):92–100. doi: 10.1309/AJCP7W9VMJENZOVG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tönnies H, Huber S, Kühl JS, Gerlach A, Ebell W, Neitzel H. Clonal chromosomal aberrations in bone marrow cells of Fanconi anemia patients: Gains of the chromosomal segment 3q26q29 as an adverse risk factor. Blood. 2003;101(10):3872–3874. doi: 10.1182/blood-2002-10-3243. [DOI] [PubMed] [Google Scholar]
- 35.Meyer S, Bristow CWM, Pepper S, Whetton AD, Hanenberg H, Neitzel H, Wlodarski MW, Ebell W, Tönnies H. Fanconi anemia (FA)–associated 3q gains in leukemic transformation consistently target EVI1, but do not affect low TERC expression in FA. Blood. 2011;117(22):6047–6050. doi: 10.1182/blood-2011-03-343897. [DOI] [PubMed] [Google Scholar]
- 36.Perdigones N, Perin JC, Schiano I, et al. Clonal hematopoiesis in patients with dyskeratosis congenita. Am J Hematol. 2016;91(12):1227–1233. doi: 10.1002/ajh.24552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu X, Nannenga B, Donehower LA. PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes dev. 2005;19(10):1162–1174. doi: 10.1101/gad.1291305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kleiblova P, Shaltiel IA, Benada J, et al. Gain-of-function mutations of PPM1D/Wip1 impair the p53-dependent G1 checkpoint. J Cell Biol. 2013;201(4):511–521. doi: 10.1083/jcb.201210031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodrigues NP, Janzen V, Forkert R, et al. Haploinsufficiency of GATA-2 perturbs adult hematopoietic stem-cell homeostasis. Blood. 2005;106(2):477–484. doi: 10.1182/blood-2004-08-2989. [DOI] [PubMed] [Google Scholar]
- 40.Micol JB, Abdel-Wahab O. Collaborating constitutive and somatic genetic events in myeloid malignancies: ASXL1 mutations in patients with germline GATA2 mutations. Haematologica. 2014;99(2):201–203. doi: 10.3324/haematol.2013.101303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.West RR, Hsu AP, Holland SM, Cuellar-Rodriguez J, Hickstein DD. Acquired ASXL1 mutations are common in patients with inherited GATA2 mutations and correlate with myeloid transformation. Haematologica. 2014;99(2):276–281. doi: 10.3324/haematol.2013.090217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wlodarski MW, Hirabayashi S, Pastor V, et al. Prevalence, clinical characteristics and prognosis of GATA2-related myelodysplastic syndromes (MDS) in children and adolescents. Blood. 2016;127(11):1387–1398. doi: 10.1182/blood-2015-09-669937. [DOI] [PubMed] [Google Scholar]
- 43.Wang X, Muramatsu H, Okuno Y, et al. GATA2 and secondary mutations in familial myelodysplastic syndromes and pediatric myeloid malignancies. Haematologica. 2015;100(10):e398–e401. doi: 10.3324/haematol.2015.127092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding L, Ikezoe T, Tan K, et al. Mutational profiling of a MonoMAC syndrome family with GATA2 deficiency. Leukemia. 2017;31(1):244–245. doi: 10.1038/leu.2016.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fisher KE, Hsu AP, Williams CL, et al. Somatic mutations in children with GATA2 -associated myelodysplastic syndrome who lack other features of GATA2 deficiency. Blood Adv. 2017;1(7):10–12. doi: 10.1182/bloodadvances.2016002311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Porter CC, Druley TE, Erez A, et al. Recommendations for surveillance for children with leukemia-predisposing conditions. Clin Cancer Res. 2017;23(11):e14–e22. doi: 10.1158/1078-0432.CCR-17-0428. [DOI] [PubMed] [Google Scholar]
- 47.Owen CJ, Toze CL, Koochin A, et al. Five new pedigrees with inherited RUNX1 mutations causing familial platelet disorder with propensity to myeloid malignancy. Blood. 2008;112(12):4639–4645. doi: 10.1182/blood-2008-05-156745. [DOI] [PubMed] [Google Scholar]
- 48.Preudhomme C, Renneville A, Bourdon V, et al. High frequency of RUNX1 biallelic alteration in acute myeloid leukemia secondary to familial platelet disorder. Blood. 2009;113(22):5583–5587. doi: 10.1182/blood-2008-07-168260. [DOI] [PubMed] [Google Scholar]
- 49.Knudson AG. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A. 1971;68(4):820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Antony-Debré I, Duployez N, Bucci M, et al. Somatic mutations associated with leukemic progression of familial platelet disorder with predisposition to acute myeloid leukemia. Leukemia. 2015 Aug;30(2015):999–1002. doi: 10.1038/leu.2015.236. [DOI] [PubMed] [Google Scholar]
- 51.Churpek JE, Pyrtel K, Kanchi K-l, et al. Genomic analysis of germline and somatic variants in familial myelodysplasia/acute myeloid leukemia. Blood. 2015;126(22):2484–2491. doi: 10.1182/blood-2015-04-641100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Risitano AM, Maciejewski JP, Green S, Plasilova M, Zeng W, Young NS. In-vivo dominant immune responses in aplastic anaemia: molecular tracking of putatively pathogenetic T-cell clones by TCR beta-CDR3 sequencing. Lancet. 2004;364(9431):355–364. doi: 10.1016/S0140-6736(04)16724-X. [DOI] [PubMed] [Google Scholar]
- 53.Li Y, Li X, Ge M, et al. Long-term follow-up of clonal evolutions in 802 aplastic anemia patients: a single-center experience. Ann Hematol Oncol. 2011;90(5):529–537. doi: 10.1007/s00277-010-1140-9. [DOI] [PubMed] [Google Scholar]
- 54.Yoshizato T, Dumitriu B, Hosokawa K, et al. Somatic mutations and clonal hematopoiesis in aplastic anemia. N Engl J Med. 2015;373(1):35–47. doi: 10.1056/NEJMoa1414799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Negoro E, Nagata Y, Clemente MJ, et al. Origins of myelodysplastic syndromes after aplastic anemia. Blood. 2011;118(9):2492–2501. doi: 10.1182/blood-2017-02-767731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Babushok DV, Duke JL, Xie HM, et al. Somatic HLA mutations expose the role of class I-mediated autoimmunity in aplastic anemia and its clonal complications. Blood Adv. 2017;1(22):1900–1910. doi: 10.1182/bloodadvances.2017010918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Katagiri T, Sato-Otsubo A, Kashiwase K, et al. Frequent loss of HLA alleles associated with copy number-neutral 6pLOH in acquired aplastic anemia. Blood. 2011;118(25):6601–6609. doi: 10.1182/blood-2011-07-365189. [DOI] [PubMed] [Google Scholar]
- 58.Stanley N, Olson TS, Babushok DV. Recent advances in understanding clonal haematopoiesis in aplastic anaemia. Br J Haematol. 2017;177(4):509–525. doi: 10.1111/bjh.14510. [DOI] [PMC free article] [PubMed] [Google Scholar]