Abstract
Uterine smooth muscle tumors (USMT) consist of a group of histologically heterogeneous and clinically diverse diseases ranging from malignant leiomyosarcoma (LMS) to benign leiomyoma (ULM). The genetic alterations in LMS are complex with some genetic alterations present in both LMS and other atypical histologic variants of USMT. In this study we reviewed 119 USMT with a diagnosis of LMS, STUMP (smooth muscle tumor of uncertain malignant potential), ALM/LM-BN (atypical leiomyomas/leiomyoma with bizarre nuclei) and CLM (cellular leiomyoma) as well as 46 ULM and 60 myometrial controls. We selected 17 biomarkers highly relevant to LMS in four tumorigenic pathways including steroid hormone receptors (ER and PR), cell cycle/tumor suppressor genes, AKT pathway markers and associated oncogenes. ER and PR expression was significantly lower in LMS than STUMP, ALM/LM-BN, CLM and ULM (p<0.01). 65% of LMS showed complete loss of ER and 75% of LMS showed complete loss of PR. All cell cycle genes were differentially expressed in different types of tumor, but significantly overlap was noted. Over 75% of LMS had ki-67 index greater than 33%, and only 5% in all other types of USMT. Expression of the selected oncogenes varied widely among different types of USMT. PR positivity and p53 had a borderline association with PFS (p=0.055 for PR and p=0.0847 for p53). Furthermore, high PR expression was significantly associated with a longer OS (p=0.0163, HR 0.198). Cell proliferative indices (Ki-67) and sex steroid hormone receptors were the most valuable markers in differentiating LMS from other USMT variants.
Keywords: Uterine smooth muscle tumor, leiomyosarcoma, Immunohistochemistry, clinical correlation
INTRODUCTION
Uterine leiomyosarcomas (LMS) are rare neoplasms representing approximately 1% of all uterine malignancies [1] and 1 in 800 uterine smooth muscle tumors. Although the reported 5-year LMS survival rates are variable, these tumors are clinically aggressive with a high risk of recurrence and an overall poor prognosis [2]. The diagnosis and management of LMS can be challenging since we cannot always predict the tumor behavior based on histologic pattern or differentiation and since the definitive diagnosis of LMS from other uterine smooth muscle tumors with atypical histology (smooth muscle tumors with uncertain malignant potential (STUMP), atypical leiomyoma or leiomyoma with bizarre nuclei (ALM/LM-BN), and cellular leiomyoma (CLM)) can be difficult [3, 4]. The new World Health Organization (WHO 2014) provides the current consensus in classifying these tumor variants based on their histology features and clinical behaviors. Therefore, the diagnosis and prognosis for these major tumor types could be significantly facilitated by histologic and molecular biomarker analysis.
Uterine leiomyoma (ULM) is the most common benign variant of USMT, comprising over 90% of all USMTs [5]. The cause of this disease entity are thought to be mostly related to single gene mutations/alterations, including MED12, HMGA1/2, FH and COL5/6 [6]. While ULM are more well-defined, the molecular features of LMS are less understood and appear to be more complex. Previous studies of the molecular alterations found in LMS have proposed that the common driving pathway may be related to cell cycle, steroid hormones, AKT and RB pathways, and minor metabolites and proteinases [7]. In addition, some oncogenes (c-Kit[8], FASCIN [9], HMGA2 [10] and EGFR [11]) seem to be dysregulated in a portion of LMS. Cross comparison between LMS and other variants are less clear.
In this study, we collected 119 cases of problematic uterine smooth muscle tumors, including LMS, STUMP, ALM/LM-BN, and CLM. We selected ULM and myometrium as controls. We then selected 17 immunohistochemical markers thought to be associated with LMS tumorigenesis and evaluated them for each case in order to determine if any of these markers help in the diagnosis of difficult USMT cases or provide prognostic significance. The objective of this study is to examine the value of each marker in determining a differential diagnosis and to correlate these findings with clinical outcomes.
MATERIALS AND METHODS
Case selection
We reviewed the pathology database from Northwestern Memorial Hospital at Northwestern University from 1993 to 2013 and identified all patients with a diagnosis of LMS, STUMP, ALM/LM-BN or CLM. In total, 119 cases were selected for this study (Table 1). In addition, 46 usual type leiomyoma (ULM) and 60 myometrial samples (20 samples each from LMS, STUMP and ALM/LM-BN hysterectomy cases) were randomly selected as controls. Each case was reviewed by at least two pathologists to confirm the diagnosis based on the Stanford scheme (5) and the 2014 WHO criteria. Each patient chart was then reviewed and patient demographics, clinical findings, treatment modalities, tumor recurrence, and patient survival were recorded up to October 2016. Of the 119 cases, 102 cases had additional follow-up data and the median clinical follow-up was 33 months (range 0–179). The study was approved by the Northwestern University Institutional Review Board.
Table 1.
General information and parameters for 5 different types of USMT
| LMS | STUMP | ALM | CLM | ULM | ||
|---|---|---|---|---|---|---|
| Immunohistochemical analysis | ||||||
| No. Cases | 38 | 18 | 42 | 22 | 46 | |
| Age (yrs) | Mean±sem | 55.3±2.1 | 37.9±1.7 | 46.9±2.1 | 46.9±2.1 | 40.0±2.0 |
| Hysterectomy % cases | 97(37/38) | 56(10/18) | 55(23/42) | 86(19/22) | 32(13/40) | |
| Tumor size | Mean±sem (cm) | 10.5±1.2 | 9.2±1.8 | 7.4±0.6 | 8.1±0.8 | 8.2±0.9 |
| Clinical Follow-up | ||||||
| No. Cases | 37 | 8 | 41 | 15 | 0 | |
| Followup (months) | Mean (Range) | 54(4–179) | 82(12– 218) | 90(13– 234) | 72(3– 232) | NA |
| Survival | Recurrence % | 57(21/37) | 0 | 5 (2/41) | 0 | N/A |
| DWD % | 46(17/37) | 0 | 0 | 0 | N/A |
DWD: Death with disease.
Tissue microarrays
Formalin-fixed paraffin-embedded (FFPE) tissue blocks with the most accurate morphological features were selected for each case and two millimeter tissue cores were taken to create four tissue microarrays (TMAs). The TMAs were sectioned at 4 μm intervals consecutively for 25 unstained slides. The two first and two last slides were hematoxylin and eosin (H&E) stained for quality assurance to confirm the correct tumor types and the presence of viable tumor tissue.
Immunohistochemistry
Seventeen markers were selected for immunohistochemical (IHC) analysis and included steroid hormone receptors (ER and PR), cell cycle and proliferative gene markers (p16, p53, p21, RB, RBL2, CD24, Ki-67), AKT pathway markers (pAKT, pS6, PTEN, BCL2) and selected oncogenes (c-Kit, FASCIN, HMGA2 and EGFR). (Table 2)[12]. All immunohistochemical staining procedures were performed on a Ventana Nexus automated system. All information regarding selected antibodies is summarized in Suppl Table 1.
Table 2.
Analysis of significant IHC markers comparing LMS, ALM STUMP, and CLM by percentage.
| Markers | % | LMS | STUMP | ALM | CLM | ULM | P |
|---|---|---|---|---|---|---|---|
| No. Cases | 38 | 17 | 42 | 22 | 46 | ||
| ER | 0 | 64.9 | 17.6 | 19 | 0 | 14.6 | |
| <33 | 10.8 | 11.8 | 11.9 | 4.5 | 7.3 | ||
| 33–66 | 2.7 | 17.6 | 21.4 | 18.2 | 51.2 | ||
| >66 | 21.6 | 52.9 | 47.6 | 77.3 | 26.8 | <0.01 | |
| PR | 0 | 75.7 | 11.8 | 2.4 | 0 | 7.1 | |
| <33 | 8.1 | 0 | 2.4 | 4.5 | 14.3 | ||
| 33–66 | 5.4 | 0 | 11.9 | 27.3 | 33.3 | ||
| >66 | 10.8 | 88.2 | 83.3 | 68.2 | 45.2 | <0.01 | |
| P16 | 0 | 13.2 | 41.2 | 21.4 | 50 | 55 | |
| <33 | 13.2 | 35.3 | 40.5 | 40.9 | 42.5 | ||
| 33–66 | 2.6 | 0 | 19 | 4.5 | 2.5 | ||
| >66 | 71.1 | 23.5 | 19 | 4.5 | 0 | <0.01 | |
| p53 | 0 | 44.7 | 41.2 | 35.7 | 72.7 | 97.7 | |
| <33 | 10.5 | 17.6 | 45.2 | 4.5 | 2.3 | ||
| 33–66 | 5.3 | 5.9 | 9.5 | 13.6 | 0 | ||
| >66 | 39.5 | 35.3 | 9.5 | 9.1 | 0 | <0.01 | |
| p21 | 0 | 8.3 | 0 | 11.1 | 41.2 | 80 | |
| <33 | 69.4 | 50 | 37 | 52.9 | 20 | ||
| 33–66 | 8.3 | 20 | 11.1 | 5.9 | 0 | ||
| >66 | 13.9 | 30 | 40.7 | 0 | 0 | <0.01 | |
| Ki-67 | 0 | 0 | 0 | 4.8 | 4.5 | 12.8 | |
| <33 | 29.7 | 94.1 | 95.2 | 95.5 | 87.2 | ||
| 33–66 | 24.3 | 5.9 | 0 | 0 | 0 | ||
| >66 | 45.9 | 0 | 0 | 0 | 0 | <0.01 | |
| RBL2 | 0 | 0 | 0 | 0 | 0 | 8.1 | |
| <33 | 2.7 | 0 | 0 | 0 | 8.1 | ||
| 33–66 | 13.5 | 10 | 33.3 | 16.7 | 32.4 | ||
| >66 | 83.8 | 90 | 66.7 | 83.3 | 51.4 | <0.01 | |
| RB | 0 | 65.8 | 40 | 34.8 | 73.3 | 86.1 | |
| ≤33 | 21.1 | 60 | 60.9 | 26.7 | 13.9 | ||
| >33 | 13.2 | 0 | 4.3 | 0 | 0 | <0.01 | |
| CD24 | 0 | 94.6 | 100 | 96 | 100 | 97.5 | |
| ≤33 | 5.4 | 0 | 4 | 0 | 0 | ||
| >33 | 0 | 0 | 0 | 0 | 2.5 | <0.01 | |
| pS6 | 0 | 55.3 | 61.5 | 69.6 | 87.5 | 87.2 | |
| ≤33 | 34.2 | 30.8 | 30.4 | 12.5 | 12.8 | ||
| >33 | 10.5 | 7.7 | <0.05 | ||||
| pAKT | 0 | 69.2 | 75 | 66.0 | 89 | 100 | |
| 1–100 | 30.8 | 25 | 34.0 | 11 | 0 | <0.05 | |
| BCL2 | 0 | 24.3 | 5.9 | 4.8 | 4.5 | 10.3 | |
| <33 | 27 | 0 | 7.1 | 4.5 | 33.3 | ||
| 33–66 | 5.4 | 0 | 14.3 | 4.5 | 25.6 | ||
| >66 | 43.2 | 94.1 | 73.8 | 86.4 | 30.8 | <0.01 | |
| HMGA2 | 0 | 89.5 | 82.4 | 90.5 | 81.8 | 89.3 | |
| >90 | 10.5 | 17.6 | 9.5 | 18.2 | 10.7 | <0.01 | |
| EGFR | 0 | 10.8 | 45.5 | 30.4 | 31.2 | 59.5 | |
| <33 | 5.4 | 9.1 | 17.4 | 25.0 | 24.3 | ||
| 33–66 | 18.9 | 9.1 | 43.5 | 31.2 | 2.7 | ||
| >66 | 64.9 | 36.4 | 8.7 | 12.5 | 13.5 | <0.01 | |
| FASCIN | 0 | 8.3 | 16.7 | 22.5 | 35 | 40.5 | |
| <33 | 5.6 | 5.6 | 5 | 5 | 18.9 | ||
| 33–66 | 8.3 | 16.7 | 45 | 25 | 21.6 | ||
| >66 | 77.8 | 61.1 | 27.5 | 35 | 18.9 | <0.01 | |
| c-Kit | 0 | 89.5 | 100 | 100 | 100 | 100 | |
| ≤33 | 7.9 | 0 | 0 | 0 | 0 | ||
| >33 | 2.6 | 0 | 0 | 0 | 0 | NS | |
| PTEN | Neg | 36.8 | 0 | 9.1 | 0 | 2.8 | |
| Pos | 63.2 | 100 | 90.9 | 100 | 97.2 | <0.01 |
The percent and intensity of each stain were evaluated by two pathologists independently. The intensity was scored as negative (0), weak (1+), moderate (2+), or strong (3+) and the percentage of positive tumor cells was scored from 0% to 100%. The results were then semiquantitatively analyzed, with cut-offs at 0%, 1–9%, 10–32%, 33–66%, and greater than 66%. A Receiving Operator Characteristics (ROC) curve and Youden index were then used to identify the best cut-off values between benign (ULM) and malignant (LMS) tumor types. The intensity The Receiving Operator Characteristics (ROC) curve is the true positive rate (sensitivity) as a function of the false positive rate (1-specificity) for the considered range of cut-off values. The Youden’s index (J), is the difference between the true positive rate and the false positive rate. Maximizing this index allows one to find, from the ROC curve, an optimal cut-off point independently from the prevalence. Score multiplied by percent was used as the final semiquantitative score for each case.
Clinical correlation analysis
A chart review was performed for the 38 LMS, 8 STUMP, 41 ALM/LM-BN and 15 CLM cases. Patient and disease characteristics including age, presenting symptoms, surgical procedure, uterine size and weight, histology, cellular and molecular analyses, and time to recurrence and/or death were recorded and evaluated.
Statistical analysis
The software SPSS version 20.0 was used for statistical analysis. A T-test analyzed differences in age and tumor size. Each marker was analyzed by Kruskal-Wallis test and Mann-Whitney test. P values less than 0.05 were considered statistically significant. R-3.0.1 (http://www.r-project.org/) was utilized to create hierarchical clustering and heat maps for the IHC markers by tumor type. Progression-free survival (PFS) was defined as the time from initial diagnosis to recurrence and overall survival (OS) was defined as the time from initial diagnosis to death.
RESULTS
Case selection
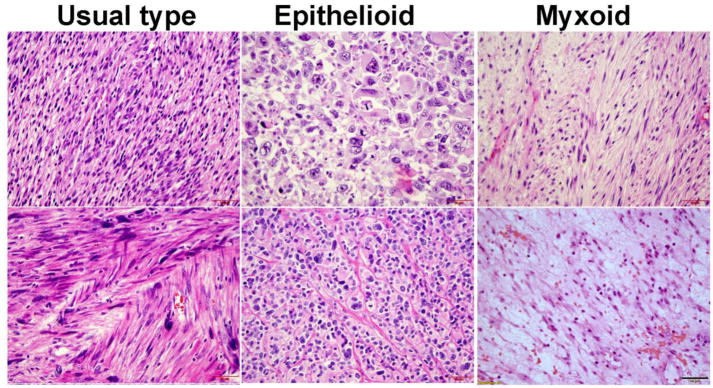
A total of 38 leiomyosarcoma (LMS) cases were included in the study which included 27 classic LMS, 9 epithelioid subtypes, and 2 myxoid subtypes (Figure 1). The average age at LMS diagnosis was 55.3 years which was older than all of the benign variants. Eighteen cases were diagnosed as smooth muscle tumor of uncertain malignant potential (STUMP) which included cases with a wide range of histologic features and clinical presentations. The average age for a STUMP diagnosis was 38 years old and average tumor size was 9.2 cm. There were also 42 atypical leiomyoma/leiomyoma with bizarre nuclei (ALM/LM-BN) that were included. The mean age at diagnosis was 41 years old and the average tumor size was 7.4 cm. A total of 22 cellular leiomyoma (CLM) were randomly selected and included in the study. The mean age for CLM patients was 47 years old at the time of diagnosis and the average tumor size was 8.1 cm. All cases of CLM were confirmed by diffuse immunoreactivity for Desmin. Forty-six usual type leiomyoma (ULM) were also randomly selected for the study and their average tumor size was 8.2 cm. Detailed histologic and clinical information is summarized in Table 1 and Suppl Table 2.
Figure 1.
Photomicrographs (40X) illustrating examples of classic LMS (left: cellular spindled cells with significant nuclear atypia and eosinophilic cytoplasm), epithelioid LMS (middle: large round cells with pleomorphic nuclei) and myxoid LMS (right: less cellular spindled cells with smaller elongated nuclei in the background of myxoid stroma).
Immunohistochemistry patterns
Based on previously published data, we selected 17 biomarkers for this study that appeared to be the most relevant and highly associated with LMS and they constituted four tumorigenic pathways including steroid hormone receptors (ER and PR), cell cycle/tumor suppressor genes (p16, p53, p21, RB, RBL-2, Ki-67), AKT pathway markers (pAKT, pS6, PTEN, BCL2) and associated oncogenes (CD24, c-Kit, FASCIN, HMGA2 and EGFR) (Table 2).
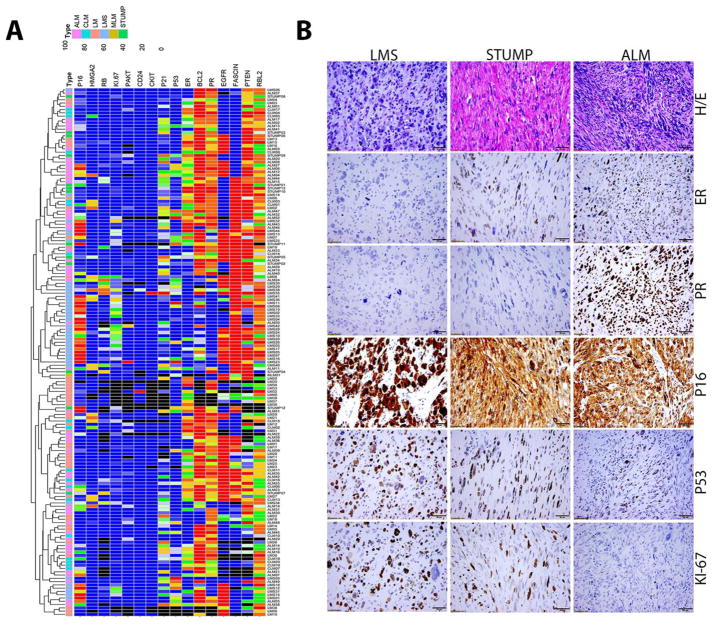
In order to compare the markers across all tumor types, a heat map was prepared based on the semiquantitative scores that combined intensity and percentage for each marker (Figure 2A). Unsupervised cluster analysis of the markers revealed that LMS and ALM/LM-BN tumors as well as some STUMP tumors clustered together while ULM was spread throughout. These findings prompted us to further evaluate these markers using the clinical parameters in relation to the tumor types. Although statistical analysis revealed that some markers were significantly different between tumor types, no single marker completely separated the tumor types by semiquantitative analysis.
Figure 2.
A. Dendragram treeview illustrate the unsupervised cluster analysis for expression levels of 17 selected biomarkers detected in 166 cases from five different subtypes of uterine smooth muscle tumors. Scale bars (blue=0, red=100) indicate the immunopercentage for each markers. Color coding for tumor: ALM=pink, CLM=blue, LMS=light blue, ULM=orange, STUMP=green, and black=no data available. B. Photomicrographs (40X) illustrating H&E and immunohistochemical staining for the five markers ER, PR, p53, p16, and Ki-67 in LMS, STUMP and ALM/LM-BN.
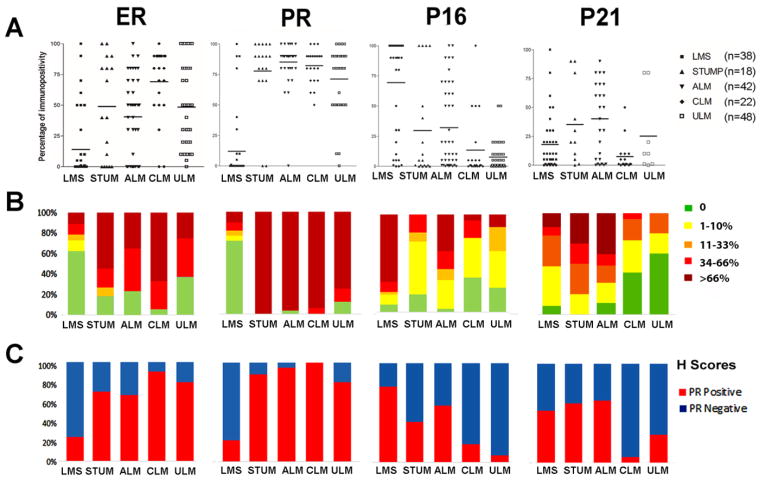
Additionally, we tested several other cutoffs for intensity, percentage, and semiquantitative scores for each marker. We found that the percentage of immunoreactivity in those markers with nuclear staining patterns provided more reproducible results than either those of intensity or semiquantitiave scoring (Suppl Figure 1). We scored the positive immunopercentage of these markers and illustrated by actual values in dot plots (Figure 3A), semiquantitative scores by grouping them into five different levels (Figure 3B) and H-score as positive and negative (Figure 3C). All three methods showed differential expression with statistical significance in the five different tumor types with a p-value <0.001. In particular, the percentage seemed to better separate LMS from other variants and only a small proportion of LMS had any percentage overlap of gene expression with other variants (Figure 3, Suppl Figure 1). The findings suggest that semiquantitative or H-scoring methods can be reliably used to evaluate USMT. For those markers that showed diffuse cytoplasmic staining, we used the intensity rating of 0–3 as the main system. Thus, based on the unique staining characteristics for each IHC marker, we ultimately chose to use one of the different score systems listed above.
Figure 3.
Immunopercentage analysis of four selected markers in five different types of uterine smooth muscle tumors. A. Dot plot analysis of the percentage of ER, PR, p16 and p21 expression in LMS, STUMP, ALM, CLM, and ULM. Each dot represents one case. B. Semiquantitative immunopercentage of the selected markers as shown in A. Colors from light green to dark red indicate the five scale cutoff ranges for immunopercentage as listed on the right. C. Positive (red) and negative (blue) H-scores for each specific cut-off value (as described in Methods section).
Differential expression of 17 biomarkers in different types of USMT
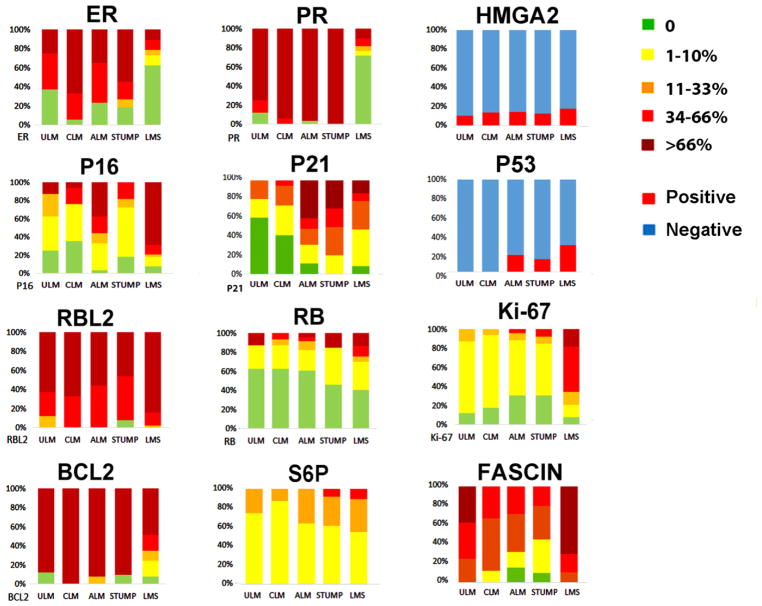
Six cell cycle markers (p16, p21, p53, RB, RBL-2, and Ki-67) were selected for analysis. As shown in Table 2 and Figure 4, the expression of most cell cycle genes and sex hormone receptors were significantly different between LMS and most other variants of USMT (p<0.05–0.01). However, there was overlap between LMS, STUMP and ALM/LM-BN which were similar to the findings reported by Mills et al [13]. When using greater than 66% of immunopositivity as cutoff, p53 overexpression was found in 39% of LMS, 35% of STUMP, 10% of ALM/LM-BN, 9% of CLM and 0% of ULM; diffuse immunoreactivity for p16 was found in 71% of LMS, 23.5% in STUMP, 19% in ALM/LM-BN and 0% in ULM. Overall, these two markers were differentially expressed among different types of USMT and reached statistical significance (p<0.01, Table 2). With >33% of immunopositivity as cutoff, ALM/LM-BN showed diffuse immunoreactivity in nearly 20% for p53 and 40% for p16 compared to LMS which had 45% positive for p53 and 75% positive for p16 and no statistical significance was reached. Therefore, these markers appear to be less valuable in assisting in the differential diagnosis between LMS versus ALM/LM-BN and STUMP (Figure 2B, Figure 4 and Table 2). Of note, more than 40% of ALM/LM-BN showed diffuse immunoreactivity for p21, while only 14% of LMS were positive for p21 (p<0.05). High p21 expression in ALM/LM-BN may suggest a protective role preventing aggressive tumor growth in this tumor type. Strong and diffuse immunoreactivity for RB was found in 13% of LMS, was only rarely seen in ALM/LM-BN (4%) and was not seen in STUMP, CLM or ULM (Table 2). Our findings suggest that although cell cycle genes are often dysregulated in LMS, they should not be used as surrogate markers to differentiate from other smooth muscle tumor variants.
Figure 4.
Semiquantitative values for the selected markers in five different types of uterine smooth muscle tumors. Colors ranging from light green to dark red indicate the five scale cutoff ranges for of immunopercentage as listed on right. P53 and HMGA2 were shown as positive (red) and negative (blue) based on staining patterns.
Ki-67 is a cell proliferative marker that is broadly used in evaluating the cell proliferative activity in any neoplasm. High Ki-67 index can be relatively accurately measured in tumor sections. As shown in Figure 4 and Table 2, Ki-67 index was significantly higher in LMS than in any other variants (p<0.01). Over 45% of LMS had Ki-67 index greater than 66% while only 29% of LMS had index less than 33%. In contrast, almost 95% of all other types of USMT had Ki-67 index less than 33%. Although approximately 10% of ALM/LM-BN, STUMP and MALM had a Ki-67 index over 10%, less than 5% of the cases showed a Ki-67 index over 20% (Suppl Figure 2). Thus, 33% may be used as a cutoff when differentiating LMS from Other variants (Table 2). Only one of 42 ALM/LM-BN had a Ki-67 index of 30%. Therefore, the Ki-67 index remains an important marker in the evaluation of USMT.
As shown in Table 2, the semiquantitative analysis of ER and PR in LMS had much lower rates of expression as compared to STUMP, ALM/LM-BN, CLM and ULM (p<0.01). For ER, we found more than 70% of STUMP, ALM/LM-BN and ULM had immunoreactivity greater than 33% in the tumor cells, while only 24% of LMS had similar rates of ER expression. Diffuse immunoreactivity for PR was found in only 10% of LMS while diffuse immunoreactivity was found in 88% of STUMP and 83% of ALM/LM-BN (p<0.01). Our semiquantitative analysis provides a clear trend in ER and PR expression between LMS and other variants of USMT. ER and PR appear to be valuable markers in evaluation of tumor nature of USMT.
We analyzed several scoring systems including 1) only intensity immunoreactivity using the 0–3+ scale (Suppl Figure 1);, 2) the percentage of immunoreactivity (quantitative);, 3) the percentage of negative (0%), low (<10%), intermediate (11–33%), high (34–65%) and diffuse (>66%) immunoreactivity (semiquantitative); and 4) H-scores (ER positive >40% and PR positive >25%) (Figure 3, Suppl Table 3). The intensity alone showed moderate reproducibility among different observers. We found that LMS had significantly reduced or complete loss of ER and PR expression by both intensity and percentage when compared other tumor variants (p<0.001). When we use the quantitative percentage method, 65% of LMS showed complete loss of ER and 75% of LMS showed complete loss of PR. In contrast, all other variants, including STUMP and ALM/LM-BN, had significantly higher ER expression (>70% of cases) and PR expression (>90% cases) (<0.001). Using an H-score cut-off of 40 for ER and 25 for PR, LMS was positive in 21–24% of cases compared to over 80% in all other variants (Suppl Table 3, Figure 3). Immunopercentage of ER and PR seemed to be more reliable markers in differentiating LMS from the other variants. For practical purposes, H-score may be best and most reproducible ways to evaluate ER and PR expression.
The AKT pathway is also known to be activated in ULM and LMS, but its activity in ALM/LM-BN and STUMP is less understood. In this study, we selected PTEN, pAKT, activated pS6 and the downstream marker BCL-2 for analysis. We noted that pAKT, pS6 and BCL-2 were equally upregulated in approximately one third of tumors in all USMT types (Table 2), indicating a functional role of AKT pathway in most USMT. This shows that its role is less useful for differentiating benign and malignant USMT.
HMGA2 is an oncofetal protein and is known to be overexpressed in approximately 10% of ULM. Its overexpression in ULM is usually caused by chromosomal translocations. In this study, we found HMGA2 overexpression present in 11% LMS, 18% STUMP, 10% in ALM/LM-BN, 18% CLM and 11% ULM (Table 2, Figure 5). These findings suggest that overall 10–15% of all USMT are related to HMGA2 and its expression is not helpful in the differential diagnosis of difficult USMT cases.
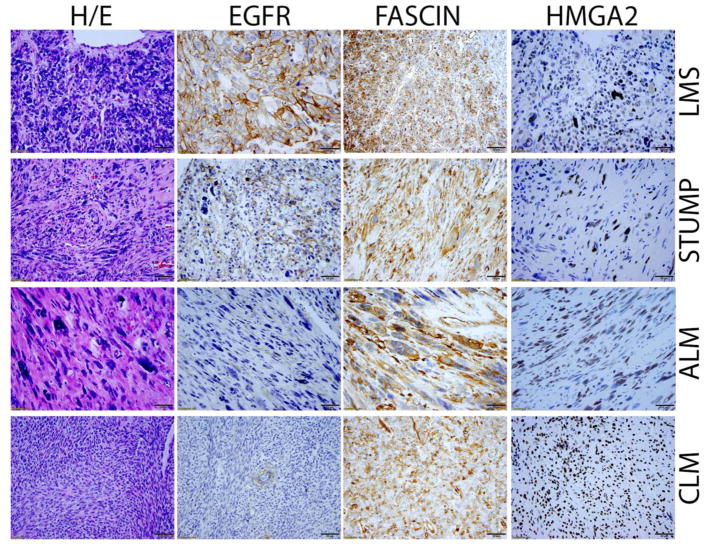
Figure 5.
Photomicrographs (40X) illustrate the examples of immunoreactivity of ERFR, FASCIN, and HMGA2, in LMS, STUMP, ALM, and CLM.
Fascin was recently reported to be highly expressed in LMS. As shown in Figure 4 and Table 2, Fascin was significantly overexpressed in LMS, but is had varied expression among other tumor types making it less significant for clinical application in the differential diagnosis of USMT (Figure 5). EGFR was also significantly overexpressed in LMS (65% of cases), in comparison to STUMP (36%), ALM/LM-BN (9%), CLM (12%) and ULM (13.5%) when using a cutoff of 66% positive tumor cells (Table 2, Figure 5). However, there was great overlapping of EGFR expression in lower cutoffs of immunopercentage across all USMT types (Table 2). C-Kit had limited application as well since only 2.6% of LMS had diffuse immunoreactivity for c-Kit. However, detection of this small percentage of c-Kit positive LMS cases may allow for potential targeted therapies.
Clinical correlation
We wanted to determine whether there was any correlation between LMS-associated biomarkers and the clinical behavior and patient outcome. A total of 102 USMT cases had follow-up data, including 38 LMS, 8 STUMP, 41 ALM/LM-BN and 15 CLM (Table 1). Clinical chart review showed that 57.1% of LMS recurred and 45.9% of LMS patient died of their disease. PFS and OS for all stages were 32 and 65 months respectively. Patients with LMS were also more likely to have a history of another cancer than those with other USMT variants (p<.0004) (Suppl Table 2). There was no association with recurrence for presenting symptoms, tumor size or uterine weight (Suppl Table 2). Hormone use was significantly associated with a shorter OS (p= 00077), but there was no association between survival and tumor size (Suppl Table 2). In the analysis of clinical behavior and biomarker expression in LMS, the level of PR positivity as well as p53 had a borderline association with PFS (p=0.055 for PR and p=0.0847 for p53). Furthermore, high PR expression was significantly associated with a longer OS (p=0.0163, HR 0.198). Low levels of p53 were associated with a shorter OS (p=0.0279, HR 3.805). ER, p16, and Ki-67 were not associated with a different PFS or OS and no marker was associated with risk of LMS recurrence.
DISCUSSION
LMS is characterized by genetic instability and frequent genomic copy number aberrations [14]. Unlike genetically unstable carcinomas, such as high grade serous carcinoma which shows nearly 100% positivity for p53 mutations, the p53 mutation rate in LMS is around 30%. Based on eight separate studies the positivity ranges from 13% to 62% [15, 16]. Strong and diffuse immunoreactivity for p53 can be used as a reliable indicator for mutant p53. In this study, diffusely positive immunohistochemical staining for p53 was found in 39% of LMS cases, but was also present in 35% of STUMP cases and 10% of ALM/LM-BN cases. These findings further suggest that a proportion of STUMP and ALM/LM-BN harbor aberrant p53 expression and has been previously reported to show molecular p53 mutations [15]. Further clinical correlation analysis revealed that LMS with diffuse p53 reactivity had shorter overall survival (p=0.0279, HR 3.805).
A decade ago, gene profiling analysis revealed that upregulation of p16 was present in most cases of LMS [17] and this finding was further supported by many studies analyzing the p16 immunohistochemical patterns [18]. However, many recent studies have shown that p16 overexpression can also be present in a substantial number of other variants of USMT, such as STUMP and ALM/LM-BN [13, 19]. In this study, we found diffuse p16 immunoreactivity in nearly 75% of LMS cases and in no ULM cases (0%), indicating the significant role that the dysfunctional RB/p16 pathway plays in LMS. However, 24% of STUMP and nearly 40% of ALM/LM-BN were also diffusely positive for p16. Therefore, using p16 as surrogate marker for LMS is greatly limited by its overlapping staining patterns with STUMP and ALM/LM-BN. We also found that p16 positivity was not associated with any clinical outcomes.
In LMS, PR has been reported to be positive in 13.3% to 60% of cases [20–24]. PR and its downstream effectors (such as CCND1, WNT and RANKL) are known to promote breast carcinogenesis [25], but the mechanism of PR loss in LMS tumorigenesis was less well understood. Recently, one study proved that PR status was an independent prognosticator in FIGO Stage I LMS and that PR-positive patients had longer overall survival [26]. ER and PR-positive LMS cases showed improved PFS and allow for the potential use adjuvant hormonal therapy [27].[27]. In this study, diffuse immunoreactivity for PR was present in only 10.8% of LMS, while it was present in 88% of STUMP and 83% of ALM/LM-BN (Table 2, Figures 2 and 3). PR also had significant negative correlation with Ki-67 (Pearson Correlation Coefficient=−0.43, p<0.01). PR may be an important biomarker, especially in combination with Ki-67 and ER, for difficult cases to separate LMS from other USMT variant. Also, PR status may allow for additional therapeutic strategy and can provide prognostic clinical information including improved PFS (p=0.055) and longer OS (p=0.0163, HR 0.198) in PR-positive cases. Our results suggest that PR positivity may be beneficial prognostically for recurrence and survival, similar to previously published data[23].
According to the 2014 WHO, uterine smooth muscle tumors are generally classified as benign (leiomyomas and other variants), STUMP (those tumors that do not meet criteria for benign variant or for true malignancy) and LMS. While such classification helps to guide pathologic diagnosis and possibly management of different types of uterine smooth muscle, the molecular and genetic differences among the subtypes remain unclear. The common gene mutations MED12 is found in greater than 60% of ULM but may also be seen in a small proportion of LMS, STUMP and ALM/LM-BN [15]. Fumarate hydratase (FH) mutations can also be found in both leiomyoma and ALM/LM-BN variants, but not in LMS [28, 29]. None of these markers have yet been considered for evaluation in the differential diagnosis of difficult USMT.
Benign variants of USMT are most often successfully treated with surgery alone since recurrence is rare while leiomyosarcoma requires surgical staging including total abdominal hysterectomy, bilateral salpingo-oophorectomy, and excision of all grossly visible tumor by a gynecologic oncologist. Additionally, LMS often requires adjuvant chemotherapy and/or radiation. Unfortunately, there are limited therapeutic options for metastatic or recurrent disease and clinical trials are often the only remaining options.
The results of this study corroborate current clinical practice guidelines. STUMP, ALM/LM-BN, and CLM are considered benign variants with an excellent prognosis. Clinically, CLM and ALM/LM-BN do not require any additional postoperative surveillance or routine imaging. However, the presence of p53 mutations in STUMP and ALM/LM-BN may warrant a heightened concern for the potential of recurrence and previous literature suggests that there is up to a 10% risk of recurrence. Similarly, in our study we identified a 5% rate of recurrence with ALM/LM-BN. With this diagnosis, gynecologists should consider completion hysterectomy after a myomectomy or a second look operation after laparoscopic morcellation. A hysterectomy is the standard of care in those who have completed childbearing which may reduce the risk for tumor progression to true malignancy.
On the other hand, LMS cases require intense surveillance per NCCN guidelines due to their aggressive nature and poor prognosis. Our recurrence rate, PFS, and OS were similar to that of published studies. Overall, recurrence rates range from 53 to 71% and OS for stage I and II disease is 51 and 25% respectively. According to one published review, no patient with extrapelvic disease at the time of diagnosis was alive at 5 years[30]. However, contrary to published literature, we did not identify an association between survival and tumor size.
There are still many questions regarding ALM/LM-BN. While our previous and current studies discovered genetic mutations shared between ALM/LM-BN and LMS [15], there is still no direct evidence proving tumor progression. Also, the best treatment options for ALM/LM-BN remain a challenge. Currently, a hysterectomy is the standard of care in those who have completed childbearing which may reduce the risk for tumor progression to true malignancy. Also, the degree of cytological atypia and presence of gene mutations may influence the risk of future malignancy and should be further pursued in future studies. High-resolution mapping for gene mutations, gene expression, gene methylation, and genomic DNA copy number changes also may be additional areas for investigation.
Conclusions
This study analyzed multiple LMS-associated biomarkers to determine immunohistochemical expression patterns as well as clinical correlation among five different types of USMT variants. We observed some distinct expression patterns between USMT subtypes. In particular, we found that the most significant markers in LMS included ER, PR, and Ki-67 which showed distinct, but also overlapping, expression between LMS, STUMP and ALM/LM-BN. In contrast, CLM and ULM seemed to show drastically different immunoprofile compared to the other three variants. Furthermore, the expression levels by different semiquantitative measurement in some biomarkers may be significantly different among the different types of USMT. Since known oncogenes and tumor suppressors can only detect a small proportion of malignant smooth muscle tumors, their application is limited. In contrast, ER, PR and Ki67 expression are valuable immunohistochemical biomarkers that are clinically useful to differentiate USMT variants in most problematic and difficult cases. Future global gene and molecular study may provide a new inside of genetic basis for the tumorigenesis of leiomyosarcoma and biomarkers for the differential diagnosis.
Supplementary Material
HIGHLIGHTS.
A distinct signature of 17 biomarkers identified in 119 leiomyosarcoma and its mimics;
Immunohistochemistry analysis by H-score can be used in aid of differential diagnosis;
ER, PR & Ki67 are the key markers in differential diagnosis of leiomyosarcoma;
PR and P53 expression is closely associated with leiomyosarcoma outcome.
Acknowledgments
We would like to thank Dr. Haiyang Guo and Mrs. Bella Shmaltsuyeva for their technical support. We would also like to thank the Northwestern University Pathology Core Facility for their help with this study. Part of this work was presented at the 103rd United States and Canadian Academy of Pathology Annual Meeting in San Diego. This study is supported in part by funding from the Edna Foundation of Hope and P01HD057877 (NIH, NICHD).
Footnotes
A conflict of interest disclosure: Authors have nothing to disclose.
Authors’ contributions: JJW and BHK designed study. QZ, MJK, JU, AES and JJW conducted the experiments. QZ, MJK, DL, DMS and JJW analyzed data. BHK and JJW contributed the materials and support. DMS, JRL, SS, BHK and JJW served as mentors. JJW, QZ and MJK prepared the MS. All authors read and approved the final manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Leibsohn S, d’Ablaing G, Mishell DR, Jr, Schlaerth JB. Leiomyosarcoma in a series of hysterectomies performed for presumed uterine leiomyomas. Am J Obstet Gynecol. 1990;162:968–974. doi: 10.1016/0002-9378(90)91298-q. discussion 974–966. [DOI] [PubMed] [Google Scholar]
- 2.Giuntoli RL, 2nd, Metzinger DS, DiMarco CS, Cha SS, Sloan JA, Keeney GL, Gostout BS. Retrospective review of 208 patients with leiomyosarcoma of the uterus: prognostic indicators, surgical management, and adjuvant therapy. Gynecol Oncol. 2003;89:460–469. doi: 10.1016/s0090-8258(03)00137-9. [DOI] [PubMed] [Google Scholar]
- 3.Layfield LJ, Liu K, Dodge R, Barsky SH. Uterine smooth muscle tumors: utility of classification by proliferation, ploidy, and prognostic markers versus traditional histopathology. Arch Pathol Lab Med. 2000;124:221–227. doi: 10.5858/2000-124-0221-USMT. [DOI] [PubMed] [Google Scholar]
- 4.Bell SW, Kempson RL, Hendrickson MR. Problematic uterine smooth muscle neoplasms. A clinicopathologic study of 213 cases. Am J Surg Pathol. 1994;18:535–558. [PubMed] [Google Scholar]
- 5.Rice KE, Secrist JR, Woodrow EL, Hallock LM, Neal JL. Etiology, diagnosis, and management of uterine leiomyomas. J Midwifery Womens Health. 2012;57:241–247. doi: 10.1111/j.1542-2011.2012.00176.x. [DOI] [PubMed] [Google Scholar]
- 6.Mehine M, Kaasinen E, Heinonen HR, Makinen N, Kampjarvi K, Sarvilinna N, Aavikko M, Vaharautio A, Pasanen A, Butzow R, Heikinheimo O, Sjoberg J, Pitkanen E, Vahteristo P, Aaltonen LA. Integrated data analysis reveals uterine leiomyoma subtypes with distinct driver pathways and biomarkers. Proceedings of the National Academy of Sciences of the United States of America. 2016 doi: 10.1073/pnas.1518752113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobayashi H, Uekuri C, Akasaka J, Ito F, Shigemitsu A, Koike N, Shigetomi H. The biology of uterine sarcomas: A review and update. Mol Clin Oncol. 2013;1:599–609. doi: 10.3892/mco.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raspollini MR, Paglierani M, Taddei GL, Villanucci A, Amunni G, Taddei A. The protooncogene c-KIT is expressed in leiomyosarcomas of the uterus. Gynecol Oncol. 2004;93:718. doi: 10.1016/j.ygyno.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 9.Kefeli M, Yildiz L, Kaya FC, Aydin O, Kandemir B. Fascin expression in uterine smooth muscle tumors. Int J Gynecol Pathol. 2009;28:328–333. doi: 10.1097/PGP.0b013e318195da9f. [DOI] [PubMed] [Google Scholar]
- 10.Lu B, Shi H, Zhang X. Myxoid leiomyosarcoma of the uterus: a clinicopathological and immunohistochemical study of 10 cases. Hum Pathol. 2017;59:139–146. doi: 10.1016/j.humpath.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 11.Lee PJ, Yoo NS, Hagemann IS, Pfeifer JD, Cottrell CE, Abel HJ, Duncavage EJ. Spectrum of mutations in leiomyosarcomas identified by clinical targeted next-generation sequencing. Exp Mol Pathol. 2017;102:156–161. doi: 10.1016/j.yexmp.2017.01.012. [DOI] [PubMed] [Google Scholar]
- 12.Bertsch E, Qiang W, Zhang Q, Espona-Fiedler M, Druschitz S, Liu Y, Mittal K, Kong B, Kurita T, Wei JJ. MED12 and HMGA2 mutations: two independent genetic events in uterine leiomyoma and leiomyosarcoma. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2014 doi: 10.1038/modpathol.2013.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mills AM, Ly A, Balzer BL, Hendrickson MR, Kempson RL, McKenney JK, Longacre TA. Cell cycle regulatory markers in uterine atypical leiomyoma and leiomyosarcoma: immunohistochemical study of 68 cases with clinical follow-up. The American journal of surgical pathology. 2013;37:634–642. doi: 10.1097/PAS.0b013e318287779c. [DOI] [PubMed] [Google Scholar]
- 14.Salawu A, Ul-Hassan A, Hammond D, Fernando M, Reed M, Sisley K. High quality genomic copy number data from archival formalin-fixed paraffin-embedded leiomyosarcoma: optimisation of universal linkage system labelling. PLoS One. 2012;7:e50415. doi: 10.1371/journal.pone.0050415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Q, Ubago J, Li L, Guo H, Liu Y, Qiang W, Kim JJ, Kong B, Wei JJ. Molecular analyses of 6 different types of uterine smooth muscle tumors: Emphasis in atypical leiomyoma. Cancer. 2014;120:3165–3177. doi: 10.1002/cncr.28900. [DOI] [PubMed] [Google Scholar]
- 16.Ito M, Barys L, O’Reilly T, Young S, Gorbatcheva B, Monahan J, Zumstein-Mecker S, Choong PF, Dickinson I, Crowe P, Hemmings C, Desai J, Thomas DM, Lisztwan J. Comprehensive mapping of p53 pathway alterations reveals an apparent role for both SNP309 and MDM2 amplification in sarcomagenesis. Clin Cancer Res. 2011;17:416–426. doi: 10.1158/1078-0432.CCR-10-2050. [DOI] [PubMed] [Google Scholar]
- 17.Skubitz KM, Skubitz AP. Differential gene expression in leiomyosarcoma. Cancer. 2003;98:1029–1038. doi: 10.1002/cncr.11586. [DOI] [PubMed] [Google Scholar]
- 18.Gannon BR, Manduch M, Childs TJ. Differential Immunoreactivity of p16 in leiomyosarcomas and leiomyoma variants. Int J Gynecol Pathol. 2008;27:68–73. doi: 10.1097/pgp.0b013e3180ca954f. [DOI] [PubMed] [Google Scholar]
- 19.Chen L, Yang B. Immunohistochemical analysis of p16, p53, and Ki-67 expression in uterine smooth muscle tumors. Int J Gynecol Pathol. 2008;27:326–332. doi: 10.1097/PGP.0b013e31815ea7f5. [DOI] [PubMed] [Google Scholar]
- 20.Raspollini MR, Amunni G, Villanucci A, Boddi V, Simoni A, Taddei A, Taddei GL. Estrogen and progesterone receptors expression in uterine malignant smooth muscle tumors: correlation with clinical outcome. J Chemother. 2003;15:596–602. doi: 10.1179/joc.2003.15.6.596. [DOI] [PubMed] [Google Scholar]
- 21.Akhan SE, Yavuz E, Tecer A, Iyibozkurt CA, Topuz S, Tuzlali S, Bengisu E, Berkman S. The expression of Ki-67, p53, estrogen and progesterone receptors affecting survival in uterine leiomyosarcomas. A clinicopathologic study. Gynecol Oncol. 2005;99:36–42. doi: 10.1016/j.ygyno.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 22.Hewedi IH, Radwan NA, Shash LS. Diagnostic value of progesterone receptor and p53 expression in uterine smooth muscle tumors. Diagn Pathol. 2012;7:1. doi: 10.1186/1746-1596-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leitao MM, Jr, Hensley ML, Barakat RR, Aghajanian C, Gardner GJ, Jewell EL, O’Cearbhaill R, Soslow RA. Immunohistochemical expression of estrogen and progesterone receptors and outcomes in patients with newly diagnosed uterine leiomyosarcoma. Gynecol Oncol. 2012;124:558–562. doi: 10.1016/j.ygyno.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 24.Mittal K, Demopoulos RI. MIB-1 (Ki-67), p53, estrogen receptor, and progesterone receptor expression in uterine smooth muscle tumors. Hum Pathol. 2001;32:984–987. doi: 10.1053/hupa.2001.27113. [DOI] [PubMed] [Google Scholar]
- 25.Brisken C. Progesterone signalling in breast cancer: a neglected hormone coming into the limelight. Nat Rev Cancer. 2013;13:385–396. doi: 10.1038/nrc3518. [DOI] [PubMed] [Google Scholar]
- 26.Davidson B, Kjaereng ML, Forsund M, Danielsen HE, Kristensen GB, Abeler VM. Progesterone Receptor Expression Is an Independent Prognosticator in FIGO Stage I Uterine Leiomyosarcoma. Am J Clin Pathol. 2016;145:449–458. doi: 10.1093/ajcp/aqw030. [DOI] [PubMed] [Google Scholar]
- 27.O’Cearbhaill R, Zhou Q, Iasonos A, Soslow RA, Leitao MM, Aghajanian C, Hensley ML. Treatment of advanced uterine leiomyosarcoma with aromatase inhibitors. Gynecol Oncol. 2010;116:424–429. doi: 10.1016/j.ygyno.2009.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei JJ. Atypical Leiomyoma With Features Suggesting of Fumarate Hydratase Mutation. Int J Gynecol Pathol. 2016;35:531–536. doi: 10.1097/PGP.0000000000000276. [DOI] [PubMed] [Google Scholar]
- 29.Wheeler KC, Warr DJ, Warsetsky SI, Barmat LI. Novel fumarate hydratase mutation in a family with atypical uterine leiomyomas and hereditary leiomyomatosis and renal cell cancer. Fertil Steril. 2016;105:144–148. doi: 10.1016/j.fertnstert.2015.09.034. [DOI] [PubMed] [Google Scholar]
- 30.Gockley AA, Rauh-Hain JA, del Carmen MG. Uterine leiomyosarcoma: a review article. Int J Gynecol Cancer. 2014;24:1538–1542. doi: 10.1097/IGC.0000000000000290. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.