Abstract
Recent research suggests that intestinal microbiota affect the aging process. Glucagon-like peptide 2 (GLP-2), a growth factor found in the intestinal mucosal epithelium, reduces intestinal permeability and affects intestinal microbiota. The relationship between aging, GLP-2, and intestinal microbiota are still not well understood. The current study examined the influence of aging and GLP-2 on the intestinal microbiota of rats. Twelve 3-month old male SD rats were randomly divided into two groups: a young control group (group C) and a young GLP-2 treatment group (group G). Twelve 26-month old male SD rats were randomly divided into two groups: an aged control group (group L) and an aged GLP-2 treatment group (group T). GLP-2 was intraperitoneally injected into rats from group G and group T for 14 days. Plasma GLP-2 concentration was evaluated by ELISA tests. Fresh intestinal stool samples were collected from each group and total fecal bacterial genomic DNA was extracted from the associated rats. The bacterial composition of fecal samples was analyzed by Miseq high-throughput sequencing and comparison with SRA databases. Overall, the diversity of intestinal microbiota significantly decreases with age in SD rats, while GLP-2 has no significant effect on the diversity of intestinal microbiota. Upon aging, there is a reduction in probiotic bacteria and a concomitant increase in pathogenic bacteria in rats. Treatment with GLP-2 results in a significant reduction in the prevalence of pathogenic bacterial genera and an increase in some potential benefit bacteria in aged rats. In addition, treatment with GLP-2 results in an increase in several probiotics and a reduction in the prevalence of pathogenic bacterial genera in young rats.
Keywords: aging, GLP-2, intestinal microbiota, high-throughput sequencing
Intestinal microbiotas are not only composed of parasitic microorganisms, but also of symbiotic bacterial species that affect the health and coevolution of humans and animals [1]. The composition of the symbiotic bacterial populations that inhabit the gastrointestinal tract is extremely diverse (up to 500-1000 species). Furthermore, there are approximately 100 trillion bacterial cells in the gastrointestinal tract, which is over 10 times the number of host cells [2]. The number of genes encoded by members of the intestinal microbiota is approximately 150 times that encoded by the host [3]. It is well known that the intestinal microbiota plays an important role in host nutrition, glucose and lipid metabolism, anti-inflammation and immune regulation [4]. In recent years, it has also been suggested that the composition of the microbiota also affects the process of aging [5]. Some studies have shown that there is a reduction in Bacteroidetes/Firmicutes (B/F) in the intestinal microbiota of aging rats. Furthermore, the diversity of the intestinal microbiota of aging rats is also reduced [6]. Other studies have revealed that aging causes an increase in the serum concentration of LPS, IL-6, IL-2, IL-1β and TNF-α and a decrease in the serum concentration of IL-10 [7]. It is speculated that the latter occurrence is caused by an intestinal microbiota imbalance that is promoted by intestinal barrier imbalances and the upregulation of chronic low-grade inflammation. However, in most of the studies that analyzed the latter phenomenon, models that simulated aging were generated following the administration of drugs [8]. Hence, it is likely that these models did not accurately reflect changes in the intestinal microbiota experienced by naturally aging rats.
Glucagon-like peptide 2, a specific growth factor found in the intestinal mucosal epithelium, can promote intestinal growth and nutrient absorption. This protein is also involved in reducing intestinal permeability, maintaining the intestinal barrier function and repairing injuries associated with intestinal diseases [9, 10]. Furthermore, Cani PD et al. reported a correlation between intestinal microbiota and GLP-2 following the administration of probiotics to obese mice. The same study confirmed that Bifidobacterium could increase the production of endogenous GLP-2 in mice [11]. A separate study by the same authors revealed similar results in obese rats [12]. In recent years, additional studies have confirmed that probiotics can increase the production of endogenous GLP-2 in healthy populations [13] and glucose-tolerant humans [14]. It has also been suggested that GLP-2 can influence the composition of the intestinal microbiota due to its role in nutrient absorption and barrier protection in the intestinal tract. If GLP-2 has an effect on the intestinal microbiota, treatment with GLP-2 may be useful for improving the change in intestinal microbiota caused by aging.
In order to study the influence of aging and GLP-2 on the intestinal microbiota of rats, aging rats were utilized in this study. Following administration with GLP-2, changes to the diversity and relative abundance of the intestinal microbiota of rats were monitored using Miseq high throughput sequencing. This experiment was performed to preliminarily explore the relationship between aging, GLP-2 and the intestinal microbiota.
MATERIALS AND METHODS
Animals and sample collection
Twelve 3-month-old male SPF SD rats and twelve 26-month-old male SPF SD rats were used in this study. All animal care procedures were approved by the Laboratory Animal Center of Fudan University, Shanghai Medical College and the Animal Studies Committee of Fudan University, China. All of the rats were housed together in groups of six in a specific pathogen-free standard animal laboratory (with a 12 h: 12 h light/dark cycle, 22-25°C ambient temperature and 45-70% relative humidity) and fed ad libitum with standard chow for 1 week prior to treatment initiation. Commercial rat chow (SLAC; Shanghai, China) and water were autoclaved prior to use.
The 3-month old rats were randomly divided into 2 groups with 6 rats per group. The 26-month-old rats were also randomly divided into 2 groups with 6 rats each:
Group C: Rats aged 3 months were injected intraperitoneally with normal saline twice a day for 14 days.
Group G: Rats aged 3 months were injected intraperitoneally with GLP-2 (250 μg·kg-1·d-1) (APC; Rhode Island, USA) twice a day for 14 days.
Group L: Rats aged 26 months were injected intraperitoneally with normal saline twice a day for 14 days.
Group T: Rats aged 26 months were injected intraperitoneally with GLP-2 (250 μg·kg-1·d-1) twice a day for 14 days.
All of the treated rats were housed, fed and watered in an identical manner prior to and after treatment initiation.
The rats were euthanized prior to extraction of fecal samples from the terminal ileum and collection of blood by direct cardiac puncture on the 15th day of treatment. The fecal samples (24 in total) were immediately frozen in liquid nitrogen. After the fecal samples were thoroughly frozen, they were stored at -80°C until DNA was extracted. Blood samples were collected in tubes with EDTA plus Sigma diprotin. The blood samples were centrifuged at 3000 × g for 15 min at 4°C and the separated plasma was stored in a freezer at -80 °C for later analysis of GLP-2.
DNA extraction, PCR amplification and Miseq sequencing
Total genomic DNA from each sample (100 mg) was extracted using the E.Z.N.A. DNA stool purification kit (Omega Bio-tek Norcross; GA, USA) according to the manufacturer’s instructions. The V4-V5 region of the bacterial 16S ribosomal RNA gene was amplified by PCR (95°C for 2 min, followed by 25 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s and a final extension step at 72°C for 5 min) using the following primers (SBC; Shanghai, China): 338F, 5’-barcode-ACTCCTACGG GAGGCAGCA-3’ and 806R, 5’-GGACTACHVGGG TWTCTAAT-3’, where the barcode represents an eight-base sequence unique to each sample. PCR reactions were performed in triplicate in a 20 μl final volume containing 4 μl of 5 × FastPfu Buffer (SGMB; Shanghai, China), 2 μl of 2.5 mM dNTPs and 0.8 μl of 5 μM sequence-specific primers. PCR reactions were performed using 10 ng of template DNA.
Amplicons were extracted from 2% agarose gels (Bio Rad; CA, USA) and purified using a DNA Gel Extraction Kit (AxyGen; CA, USA) according to the manufacturer’s instructions. The resultant gel extracts were quantified using QuantiFluor™-ST (Promega Corporation; Madison, USA). Purified amplicons were pooled in equimolar amounts and paired-end sequenced (2 × 250 bp reads) using an Illumina Miseq high-throughput PE 300 Sequencing Platform (Illumina, USA) in accordance with standard protocols. The raw reads were deposited into the NCBI Sequence Read Archive (SRA) database.
Raw fast files were demultiplexed and quality-filtered using QIIME (version 1.9.1) according to the following criteria: (i) the 300 base pairs reads were truncated at any site receiving an average quality score of less than 20 over a 50 base pair (bp) sliding window and truncated reads that were shorter than 50 bp were discarded, (ii) exact barcode matches, reads containing 2 nucleotide mismatches following primer matching or reads containing ambiguous characters were removed, and (iii) only sequences containing overlaps that were longer than 10 base pairs were assembled according to their overlap sequence. Reads which could not be assembled were discarded.
Bioinformatic analysis
Operational Taxonomic Units (OTUs) were clustered with 97% similarity cutoff using UPARSE (version 7.1, http://drive5.com/uparse/) and chimeric sequences were identified and removed using UCHIME. Taxonomical classification of each 16S rRNA gene sequence was performed using RDP Classifier (http://rdp.cme.msu.edu/) and the SILVA (SSU123) 16S rRNA database using a confidence threshold of 70%.
The OTUs that reached at a 97% nucleotide similarity level were used for alpha diversity (Shannon, Simpson, and Coverage), richness (ACE and Chao), and rarefaction curve analyses. Phylogenetic beta diversity measures such as unweighted UniFrac principal coordinate analysis (PCoA) were performed using OTUs for each sample [15]. The RDP classifier Bayesian algorithm for 97% similarity levels of OTUs was used for representative sequences during phylum and genus taxonomic analysis.
Plasma analysis
Plasma samples of 10 ul were added to a 1.5 ml tube of polypropylene containing 450 ul sample diluent. The plasma was acidified with 1 N hydrogen chloride at 4°C and placed for 60 min. Then the tube was added with 1 N sodium hydroxide and blended. The plasma levels of GLP-2 were measured by ABC ELISA kits (Xinqidi Bio, Wuhan, China) according to the manufacturer’s instructions.
Statistical analysis
The Kruskal-Wallis sum-rank test, Mann-Whitney U test, ANOVA analysis and Pearson correlation coefficients were performed using SPSS version 17.0 for Windows. All of the optimized OTU sequences that were similar to the OTU sequences from selected representatives (greater than 97% similarity) were produced in the OTU forms.
RESULTS
Diversity of the gut microbiota in rats
Miseq sequencing resulted in 226,798,684 raw sequences including 127,924,964 high-quality raw sequences with a median read length of 438.13 base pairs (ranging from 401 to 500 base pairs). Repetitive sequences were extracted using the Usearch platform and non-repetitive sequences were clustered into OTUs at 97% similarity.
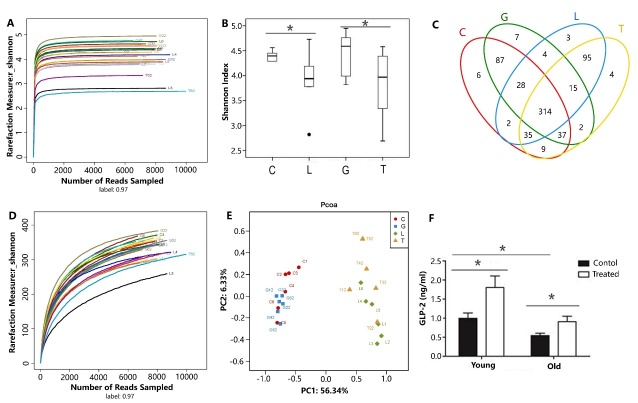
The summary information and detailed characteristics of each sample are shown in Table 1. All of the diversity indices (including OTU numbers, Ace index values, Chao index values, and Simpson indices) revealed that gut microbiota diversity was significantly lower in group L than in group C (P < 0.05). Similarly, these indices revealed that gut microbiota diversity was significantly lower in group T than in group G (P < 0.05). However, there was no significant difference in gut microbiota diversity between young rats and aged rats (Fig. 1B, Table 1). The diversity indices revealed that the abundance of intestinal microorganisms in young rats was higher than that in old rats.
Table 1.
Diversity estimation of the intestinal microbiota in SD rats.
| Group | No. of reads | No. of OTUs | Richness estimator | α-Diversity index | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Ace | 95% CI | Chao | 95% CI | Coverage | Shannon | |||||
| Group C | 69927 | 2124 | 413 | 393.1-431.9 | 415 | 384.9-445.8 | 0.9893 | 4.40 | ||
| Group G | 72390 | 2111 | 400 | 386.5-414.1 | 406 | 392.3-419.7 | 0.9909 | 4.55 | ||
| Group L | 78031 | 1848 | 367 | 345.3-388.4 | 372 | 336.0-408.0 | 0.9916 | 3.90 | ||
| Group T | 71638 | 1912 | 371 | 339.0-402.7 | 385 | 345.7-424.3 | 0.9910 | 3.82 | ||
Figure 1. Comparison of diversity indices, Venn diagram, rarefaction curve, PCoA, and plasma concentration of GLP-2 among the 4 groups.
Comparison of diversity indices including the Shannon Curve (A) and Shannon Index (B). (C) Venn diagram illustrating overlap between the 4 groups. (D) Rarefaction curve. (E) PCoA was analyzed by unweighted Unifrac. (F) Plasma concentration of GLP-2 among the 4 groups. C denotes group C; G denotes group G; L denotes group L; T denotes group T; young denotes group C and group G; old denotes group L and group T; treat denotes treatment with GLP-2; * denotes P<0.05.
Rarefaction curve analysis demonstrated that the curves tended to approach the saturation plateau, while no new OTUs were produced by increasing the number of sequences. This indicated that the sequencing numbers were sufficient (Fig. 1D).
The beta diversity index was calculated to measure the extent of the similarity in the microbial communities and principal coordinate analysis was performed using unweighted UniFrac. Despite significant inter-individual variation, the gut microbiotas of young rats (including rats from group C and group G) and aged rats (including rats from group L and group T) could be divided into two different clusters and separated clearly by principal coordinates analysis (Fig. 1E).
Bacterial community structure at phylum level
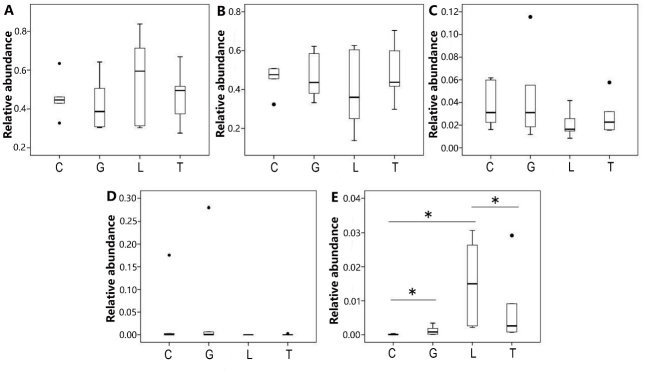
Taxonomically, nine different bacterial phyla including Bacteroidetes, Firmicutes, Proteobacteria, Verruco-microbia, Spirochaetae, Candidate-divison-TM7, Actinobacteria, Tenericutes and Cyanobacteria were observed in the fecal microbiota of the samples analyzed (Fig. 2A). Almost all the analyzed sequences (> 95%) belonged to the following five phyla; Bacteroidetes, Firmicutes, Proteobacteria, Verrucomicrobia and Spirochaetae. The majority of the sequences obtained belonged to Bacteroidetes and Firmicutes, although the relative abundance of each group differed. The relative abundance of Firmicutes was highest in groups C, G and T, while the relative abundance of Bacteroidetes was highest in group L.
Figure 2. Relative abundance at the phylum level and the genus level in fecal microbiota.
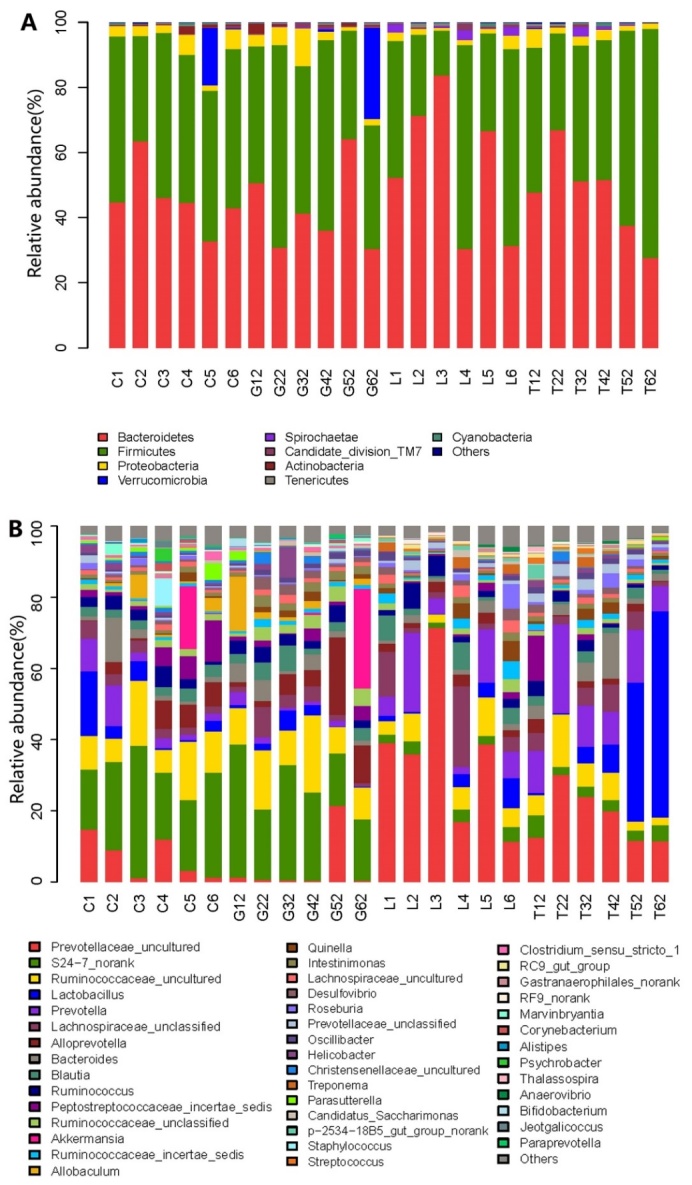
(A) Relative abundance at the phylum level in fecal microbiota. (B) Relative abundance at the genus level in fecal microbiota.
Apart from Spirochaetae, no statistically significant difference was observed among the microbial communities of the 4 groups at the phylum level (Fig. 3). The relative abundance of Spirochaetae was significantly higher (P < 0.05) in group L than in group C, and significantly lower (P < 0.05) in group T than in group L (Fig. 3E).
Figure 3. Comparison relative abundance of microbiota at the phylum level among the 4 groups.
Relative abundance (at the phylum level) of Bacteroidetes (A), Firmicutes (B), Proteobacteria (C), Verrucomicrobia (D), and Spirochaetae (E) among the 4 groups. C denotes group C; G denotes group G; L denotes group L; T denotes group T; * denotes P<0.05.
Table 2.
The relative abundance of microbial phyla in the microbiota of Groups C, G, L and T.
| Group | Bacteroidetes | Firmicutes | Proteobacteria | Verrucomicrobia | Spirochaetae |
|---|---|---|---|---|---|
| (%) | (%) | (%) | (%) | (%) | |
| Group C | 0.457487617 | 0.45781883 | 0.037045467 | 0.029836242 | 0.000116522 |
| Group G | 0.422228642 | 0.465488435 | 0.043870402 | 0.047868783 | 0.001192522 |
| Group L | 0.559329883 | 0.389632182 | 0.020549755 | 0.000000000 | 0.015300716 |
| Group T | 0.470808005 | 0.482163385 | 0.027743444 | 0.0004983 | 0.007512288 |
Bacterial community structure at genus level
At the genus level, Prevotellaceae_uncultured, S24-7_norank, Ruminococcaceae_uncultured, Lactobacillus, Prevotella, Lachnospiraceae_unclassified, Alloprevotella, Bacteroides, Blautia, Ruminococcus, Peptostrepto-coccaceae_incertae_sedis, Ruminococcaceae-unclassified, Akkermansia, Ruminococcaceae_ incertae_ sedis and Allobaculum were the major fecal genera observed in all of the samples analyzed (Fig. 2B).
The 4 groups shared similar intestinal microbiotas at the taxonomic level, although the relative abundance of the constituents varied. To identify the specific bacterial taxa associated with aging, we compared the fecal microbiota of young rats (group C) with aged rats (group L) using the Mann-Whitney U test. The results showed that there were significant differences in relation to the presence of 22 genera between the young and the old groups. The relative abundance of Anaerovibrio, Thalassospira, Prevotellaceae_unclassified, RC9_gut_group, Streptococcus, Treponema and p-2534-18B5_gut_group_norank (Table 3) was significantly higher (P < 0.05) in group L than in group C, while the relative abundance of Alistipes, Allobaculum, Bifidobacterium, Christensenellaceae_uncultured, Clostridium_sensu_stricto_1, Corynebacterium, Helicobacter, Jeotgalicoccus, Marvinbryantia, Para-prevotella, Parasutterella, Peptococcaceae_uncultured, Psychrobacter, Ruminococcaceae_uncultured and S24-7_norank (Table 3) was significantly lower (P < 0.05) in group L than in group C.
Table 3.
The relative abundance of microbial genera in the microbiota of Group C and Group L.
| Sequence | Genus | Abundance in Group C (%) | Abundance in Group L (%) | Trends in Group L |
|---|---|---|---|---|
| 1 | Alistipes * | 0.004228 | 0.000141 | ↓ |
| 2 | Allobaculum * | 0.020827 | 0.000000 | ↓ |
| 3 | Anaerovibrio * | 0.000000 | 0.003652 | ↑ |
| 4 | Bifidobacterium * | 0.000574 | 0.000000 | ↓ |
| 5 | Christensenellaceae_uncultured * | 0.008520 | 0.001827 | ↓ |
| 6 | Clostridium_sensu_stricto_1 * | 0.007729 | 0.000410 | ↓ |
| 7 | Corynebacterium * | 0.005729 | 0.000000 | ↓ |
| 8 | Helicobacter * | 0.010086 | 0.002609 | ↓ |
| 9 | Jeotgalicoccus * | 0.003966 | 0.000000 | ↓ |
| 10 | Marvinbryantia * | 0.006685 | 0.000059 | ↓ |
| 11 | Paraprevotella * | 0.001615 | 0.000000 | ↓ |
| 12 | Parasutterella * | 0.013060 | 0.000063 | ↓ |
| 13 | Peptococcaceae_uncultured * | 0.002393 | 0.000247 | ↓ |
| 14 | Prevotellaceae_unclassified * | 0.068351 | 0.355279 | ↑ |
| 15 | Psychrobacter * | 0.006387 | 0.000000 | ↓ |
| 16 | RC9_gut_group * | 0.000000 | 0.005918 | ↑ |
| 17 | Ruminococcaceae_uncultured * | 0.114876 | 0.060653 | ↓ |
| 18 | S24-7_norank * | 0.244996 | 0.029386 | ↓ |
| 19 | Streptococcus * | 0.001172 | 0.003484 | ↑ |
| 20 | Thalassospira * | 0.000341 | 0.001554 | ↑ |
| 21 | Treponema * | 0.000117 | 0.015301 | ↑ |
| 22 | p-2534-18B5_gut_group_norank * | 0.000000 | 0.007516 | ↑ |
denotes significantly different from Group C to Group L, P<0.05
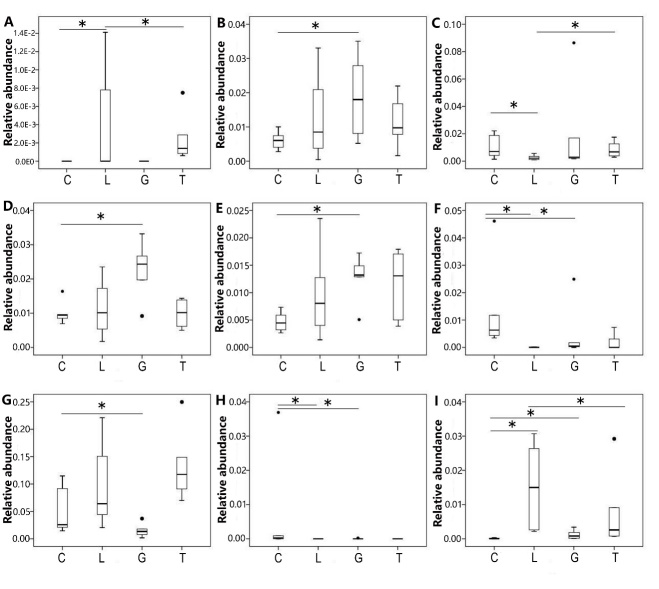
In order to identify if specific bacterial taxa were associated with groups that were administered GLP-2, we compared the fecal microbiota in young rats (groups C and G) and aged rats (groups L and T) using the Mann-Whitney U test. The relative abundance of Anaerovibrio, Desulfovibrio, Helicobacter, Intestinimonas, Oscilli-bacter, Parasutterella, Prevotella, Psychrobacter and Treponema was significantly different (P < 0.05) between the two groups (Fig. 4). Compared to group C, the relative abundance of Desulfovibrio, Intestinimonas, and Oscillibacter was significantly higher (P < 0.05) in group G, while the relative abundance of Parasutterella, Prevotella and Psychrobacter was significantly lower (P < 0.05) in group G. Compared to group L, the relative abundance of Anaerovibrio and Helicobacter was significantly higher (P < 0.05) in group T. The relative abundance of Treponema was significantly lower (P < 0.05) in group T.
Figure 4. Comparison relative abundance of microbiota at the genus level among the 4 groups.
Relative abundance (at the genus level) of Anaerovibrio (A), Desulfovibrio (B), Helicobacter (C), Intestinimonas (D), Oscillibacter (E), Parasutterella (F), Prevotella (G), Psychrobacter (H), and Treponema (I) among the 4 groups. C denotes group C; G denotes group G; L denotes group L; T denotes group T; * denotes P<0.05.
Plasma concentration of GLP-2
To differentiate the circulating levels of GLP-2 in each group, we compared the plasma concentration of GLP-2 among 4 groups using the ANOVA analysis. We found a significant aging × GLP-2 interaction on plasma concentrations of GLP-2 (Fig. 1F). With aging, the plasma concentration of GLP-2 was significantly lower (P < 0.05) in group L than in group C. With treatment with exogenous GLP-2, the plasma concentration of GLP-2 was significantly higher (P < 0.05) in group G than in group C, the plasma concentration of GLP-2 was significantly higher (P < 0.05) in group T than in group L.
Correlation between plasma concentration of GLP-2 and gut microbiota
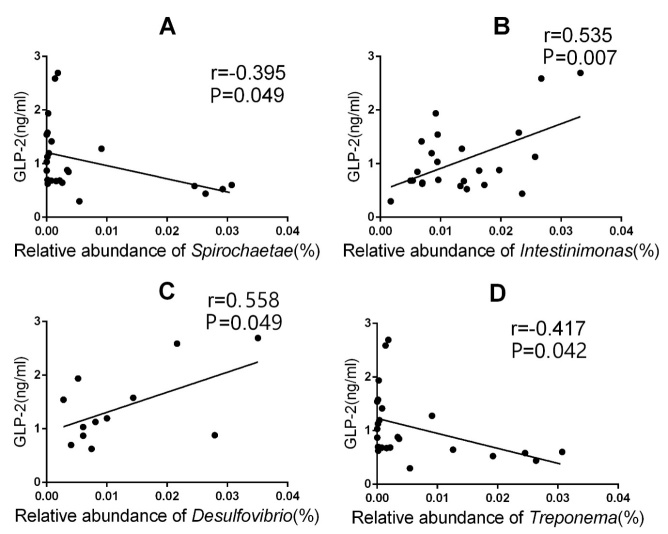
We analyzed the liner correlation between plasma concentration of GLP-2 and the relation abundance of gut microbiota using Pearson correlation coefficient (r). There was a negative correlation relationship between the plasma concentration of GLP-2 and the relation abundance of Spirochaetae (r=–0.395, P<0.05,Fig. 5A), Treponema (r=–0.417, P<0.05,Fig. 5D). When the plasma concentration of GLP-2 increased, the relative abundance of Spirochaetae, Treponema was reduced accordingly. On the other hand, there was a positive correlation relationship between the plasma concentration of GLP-2 and the relation abundance of Intestinimonas (r=0.535, P<0.05; Fig. 5B), Desulfovibrio (r=558, P<0.05; Fig. 5C). When the plasma concentration of GLP-2 increased, the relative abundance of Intestinimonas, Desulfovibrio was increased accordingly.
Figure 5. The linear correlation between plasma concentrations of GLP-2 and relative abundance of microbiota.
The linear correlation between plasma concentrations of GLP-2 and relative abundance of Spirochaetae (A), Intestinimonas (B), Desulfovibrio (C), Treponema (D).
DISCUSSION
Entering into the 21st century, the life expectancy of humans is rapidly increasing. This dramatic increase in life expectancy has resulted in a concomitant increase in aging-associated diseases. Thus, it is imperative that we investigate the many factors that are believed to underpin aging. One of the factors that has generated a lot of interest in recent years in relation to its role in the aging process is the composition of the intestinal microbiota.
In this current study, we observed significant differences in the diversity and structure of the intestinal microbiota of young and aged rats. The diversity of the intestinal microbiota of rats was significantly reduced in the aged group compared with the young group. This result is consistent with previous reports that demonstrated that aging correlates with a reduction in intestinal microbiota diversity in humans and rats [6, 16]. In this study, we confirmed that aging results in a reduction in the diversity of the intestinal microbiota. Apart from Spirochaetae, which was relatively more abundant in rats from group L compared with those from group C, no significant differences were observed at the phylum level in the intestinal microbiota of rats from all groups. A study by Distrutii et al. (2014) suggested that the abundance of Firmicutes and Proteobacteria was significantly reduced in rats aged 22 months compared with 3-month-old rats. Conversely, the relative abundance of Bacteroidetes [17] was greater in 22-month-old Wistar rats than in 3-month-old Wistar rats. It is has been suggested that the proportion of Bacteroidetes/Firmicutes (B/F) increases with age. However, a number of studies have also demonstrated that the proportion of B/F diminishes upon aging. Thus, we cannot currently draw a conclusion from these results.
At the genus level, Allobaculum, Bifidobacterium and other beneficial bacteria (in rats) were significantly lower in group L than in group C. Conversely, Anaerovibrio, Thalassospira, Streptococcus, Treponema and other pathogenic bacteria or opportunistic pathogens (in rats) were more abundant in group L than in group C. Guigoz et al. suggested that autoimmune tolerance is reduced as a consequence of changes to the intestinal microbiota upon aging. This report also suggested that these changes result in the activation of abnormal immune reactions which can cause intestinal mucous membrane inflammation [18]. An article by Schiffrin et al. demonstrated that aging correlates with an increase in Enterobacteria and other gram-negative bacteria in the intestinal tract. The higher prevalence of gram-negative bacteria can lead to an increased release of lipopolysaccharides. Lipopolysaccharides are absorbed into the bloodstream across the gastrointestinal barrier, resulting in endotoxemia and the synthesis of a large number of inflammatory factors [19]. The release of these factors is increasingly important when the barrier function associated with the intestinal mucous membranes is compromised. Aging is also closely associated with chronic low-grade inflammation [20]. As suggested by the results of this current study, aging could result in an obvious increase in enterogenous opportunistic pathogens including Thalassospira, Streptococcus and Treponema. Conversely, the number of beneficial bacteria such as Allobaculum and Bifidobacterium is significantly reduced upon aging. These alterations to the intestinal microbiota are likely to result in chronic low-grade inflammation.
GLP-2 is a beneficial factor involved in nutrient uptake and barrier protection in intestinal mucous membranes. As a nutritional factor of intestinal epithelial cells, secretion of GLP-2 is reduced with aging in previous studies [21]. In this current study, we observed significant reduce of endogenous secretion of GLP-2 in old rats, which was consistent with previous reports. The circulating concentrations of GLP-2 in young and old rats were increased significantly after intraperitoneal injection of GLP-2 to rats.
A study conducted by Cani et al. suggested that Bifidobacterium could increase the production of endogenous GLP-2 in mice. This genus is also involved in improving the intestinal barrier function in diabetic and obese mice [11]. Intestinal pathogens cause chronic low-grade inflammation of the intestinal barrier following age-induced injury. Therefore, the question remains as to whether alterations to the intestinal barrier function affect the composition of the intestinal microbiota. Up until now, very little research has been conducted to investigate this phenomenon. In this current study, we observed that GLP-2 does not significantly affect the diversity of the intestinal microbiota in rats. It is possible that the intestinal microbiota remains relatively constant over time and treatment with GLP-2 for 2 weeks is not enough to facilitate a change in the diversity of the intestinal microbiota. However, we did reveal that GLP-2 does influence the structure of the intestinal microbiota of rats. At the phylum level, the abundance of Spirochaetae was significantly reduced in group T compared with group L; while upon aging, there was an increased abundance of Spirochaetae in group L compared with group C. Interestingly, treatment with GLP-2 results in a reduction in Spirochaetae in aging rats; this result is in contrast to the increase in abundance in Spirochaetae caused by aging in non-treated rats. Some members of the genus Spirochaetae including Leptospira, Borrelia and Treponema are pathogenic, causing diseases such as syphilis [22], leptospirosis, relapsing fever, dilated cardiomyopathy [23] and gingivitis [24]. Thus, GLP-2 is capable of reducing age-induced increases in the prevalence of some pathogens in rats. At the genus level, the abundance of Anaerovibrio and Helicobacter was significantly higher in group T than in group L, the abundance of Treponema was significantly lower in group T than in group L. Treponema is belong to the phylum of Spirochaetae which is harmful for rats. GLP-2 is capable of reducing Treponema like the pathogens Spirochaetae in rats. The harmful bacteria including Spirochaetae and Treponema was negative correlated with the plasma concentration of GLP-2. Upon aging, the abundance of Helicobacter in group L was significantly lower than that in the young rat group. It is possible that GLP-2 administration can cause an increase in Helicobacter in aging rats. Helicobacter is affiliated to Proteobacteria, and several Helicobacter strains are associated with gastritis, enteritis, liver cancer and other diseases associated with both humans and animals [25]. Thus, it is possible that aging can reduce the abundance of Helicobacter, and GLP-2 could alter the extent of this age-related reduction. Anaerovibrio is a kind of bacteria involved in lipid metabolism, including the decomposition of triglycerides to propionic acid, acetic acid and succinic acid [26]. GLP-2 administration could cause an increase in Anaerovibrio of aging rats, which is possibly a potentially beneficial bacterium.
In addition, GLP-2 could increase the abundance of beneficial bacteria such as Desulfovibrio, Intestinimonas and Oscillibacter in young rats, while reducing the abundance of opportunistic bacteria such as Parasutterella, Prevotella, and Psychrobacter. The beneficial bacteria including Desulfovibrio and Intestinimonas was positive correlated with the plasma concentration of GLP-2.
In summary, treatment with GLP-2 appears to be especially beneficial to young rats by a modest increase in the abundance of some beneficial bacteria and a decrease in the abundance of some harmful bacteria. Meanwhile, GLP-2 obviously changes the intestinal microbiota of aging rats by decreasing the abundance of some harmful bacteria and increasing some potentially beneficial bacteria. Therefore, this study confirms that GLP-2 can moderately change the intestinal microbiota of rats. However, further studies are required to more extensively assess the role of GLP-2 during aging.
Acknowledgments
This research was supported by grant from National Science Foundation of China (81570795) to Yu Hu.
References
- [1].Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett C, Knight R, Gordon JI (2007). The human microbiome project: exploring the microbial part of ourselves in a changing world. Nature, 449:804-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R (2009). Bacterial community variation in human body habitats across space and time. Science, 326:1694-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature, 464:59-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jeremy KN, Elaine H, James K, Remy B, Glenn G, Wei J, et al. (2012). Host-gut microbiota metabolic interactions. Science, 336:1262-1267. [DOI] [PubMed] [Google Scholar]
- [5].Biagi E, Nylund L, Candela M, Ostan R, Bucci L, Pini E, et al. (2010). Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One, 5:e10667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lees H, Swann J, Poucher SM, Nicholson JK, Holmes E, Wilson ID, et al. (2014). Age and microenvironment outweigh genetic influence on the Zucker rat microbiome. PLoS One, 9:e100916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ouwehand AC, Bergsma N, Parhiala R, Lahtinen S, Gueimonde M, Finne-Soveri H, et al. (2008). Bifidobacterium microbiota and parameters of immune function in elderly subjects. FEMS Immunol Med Microbiol, 53:18-25. [DOI] [PubMed] [Google Scholar]
- [8].Jeong JJ, Kim KA, Jang SE, Woo JY, Han MJ, Kim DH (2015). Orally administrated Lactobacillus pentosus var. plantarum C29 ameliorates age-dependent colitis by inhibiting the nuclear factor-kappa B signaling pathway via the regulation of lipopolysaccharide production by gut microbiota. PLoS one, 10:e0116533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chen X, Zhao HX, Fu XS, Li CP, Zhong XL (2012). Glucagonlike peptide 2 protects intestinal barrier in severe acute pancreatitis through regulating intestinal epithelial cell proliferation and apoptosis. Pancreas, 41:1080-1085. [DOI] [PubMed] [Google Scholar]
- [10].Litvak DA, Hellmich MR, Evers BM, Banker NA, Townsend CM Jr (1998). Glucagon-like peptide 2 is a potent growth factor for small intestine and colon. J Gastrointest Surg, 2:146-150. [DOI] [PubMed] [Google Scholar]
- [11].Cani PD, Possemiers S, Van de Wiele T, Guiot Y, Everard A, Rottier O, et al. (2009). Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2- driven improvement of gut permeability. Gut, 58:1091-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bomhof MR, Saha DC, Reid DT, Paul HA, Reimer RA (2014). Combined effects of oligofructose and Bifidobacterium animalis on gut microbiota and glycemia in obese rats. Obesity (Silver Spring), 22:763-771. [DOI] [PubMed] [Google Scholar]
- [13].Russo F, Linsalata M, Clemente C, Chiloiro M, Orlando A, Marconi E, et al. (2012). Inulin-enriched pasta improves intestinal permeability and modifies the circulating levels of zonulin and glucagon-like peptide 2 in healthy young volunteers. Nutr Res, 32:940-946. [DOI] [PubMed] [Google Scholar]
- [14].Simon C, Strassburger K, Nowotny B, Kolb H, Nowotny P, Burkart V, et al. (2015). Intake of Lactobacillus reuteri improves incretin and insulin secretion in glucose-tolerant humans: a proof of concept. Diabetes Care, 38:1827-1834. [DOI] [PubMed] [Google Scholar]
- [15].Lozupone C, Knight R (2015). UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol, 71: 8228-8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zwielehner J, Liszt K, Handschur M, Lassl C, Lapin A, Haslberger AG (2009). Combined PCR-DGGE fingerprinting and quantitative-PCR indicates shifts in fecal population sizes and diversity of Bacteroides, bifidobacteria and Clostridium cluster IV in institutionalized elderly. Experimen Geronto, 44:440-446. [DOI] [PubMed] [Google Scholar]
- [17].Distrutii E, Reilly JA, McDonald C, Cipriani S, Renga B, Lynch MA, et al. (2014). Modulation of intestinal microbiota by the probiotic VSL#3 resets brain gene expression and ameliorates the age-related deficit in LTP. PLoS one, 9:e106503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Guigoz Y, Doré J, Schiffrin J (2008). The inflammatory status of old age can be nurtured from the intestinal environment. Curr Opin Clin Nutr Metab Care, 11:13-20. [DOI] [PubMed] [Google Scholar]
- [19].Schiffrin EJ, Morley JE, Donnet-Hughes A, Guigoz Y (2009). The inflammatory status of the elderly: the intestinal contribution. Mutat Res, 690:50-56. [DOI] [PubMed] [Google Scholar]
- [20].Biagi E, Candela M, Turroni S, Garagnani P, Franceschi C, Brigidi P (2013). Ageing and gut microbes: Perspectives for health maintenance and longevity. Pharmacological Research, 69:11-20. [DOI] [PubMed] [Google Scholar]
- [21].Drozdowski L, Thomson AB (2006). Aging and the intestine. World J Gastroenterol, 12:7578-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].McWeeney EJ (1905). Spirochaetae in syphilis. Br Med J, 1:1262-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bartůnĕk P, Gorican K, Veiser T, Taborsky M, Hulínská D (2007). Significance of Borrelia infection in development of dilated cardiomypathy (a pilot study). Prague Med Rep, 108:339-347. [PubMed] [Google Scholar]
- [24].Dapeci A, Zapletal L (1972). Quantitative changes of spirochaetae in ulcerative gingivitis after general therapy with penicillin and after local treatment with hydrogen peroxide. Cesk Stomatol, 72:96-101. [PubMed] [Google Scholar]
- [25].Rimbara E, Mori S, Kim H, Matsui M, Suzuki S, Takahashi S, et al. (2013). Helicobacter cinaedi and Helicobacter fennelliae transmission in a hospital from 2008 to 2012. J Clin Microbiol, 51:2439-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Prins RA, Lankhorst A, van der Meer P, Van Nevel CJ (1975). Some characteristics of Anaerovibrio lipolytica a rumen lipolytic organism. Antonie Van Leeuwenhoek, 41:1-11. [DOI] [PubMed] [Google Scholar]