Abstract
Profilin 2 (PFN2) functions as an actin cytoskeleton regulator and serves an important role in cell motility. However, a role for PFN2 in the progression of colorectal cancer (CRC), particularly in metastasis, has yet to be clarified. The aim of the present study was to investigate whether PFN2 served specific roles in the progression of human CRC. The results demonstrated that PFN2 was differentially expressed in CRC tissues and cell lines by reverse transcription-quantitative polymerase chain reaction and western blotting. PFN2 expression was also negatively associated with the degree of tumor metastasis. Low PFN2 expression in CRC cells was related with enhanced epithelial-mesenchymal transition (EMT) and, in turn, may increase migratory capabilities. Overexpression of PFN2 in CRC cell lines with a low level of endogenous PFN2 inhibited the EMT process, as well as the associated migration; in addition, myosin light chain (MLC) phosphorylation was upregulated. Inhibition of MLC phosphorylation attenuated the inhibition of EMT and cell migratory abilities induced by PFN2 overexpression in CRC cell lines, the results suggested that PFN2 may suppress cancer EMT and the subsequent metastasis by regulating cytoskeletal reorganization. These results demonstrated that PFN2 may serve a suppressive role in the metastasis of CRC and therefore may provide a new potential target for cancer therapeutics.
Keywords: profilin 2, colorectal cancer, epithelial-mesenchymal transition, migration, myosin light chain
Introduction
Colorectal cancer (CRC) is one of the most common malignancies and is the leading cause of cancer-related mortality worldwide (1). Although advanced therapies are increasingly being developed, the 5-year survival rate for CRC remains <50% (2). In addition, it is notable that 90% of cancer mortalities are attributed to metastasis and not the primary tumor in general (3). Therefore, clarification of the molecular mechanisms underlying metastasis and the identification of key molecules involved in the process are of great clinical value.
Metastasis requires cells to acquire migratory and invasive capabilities in order to travel from the primary tumor to a secondary site (4). Epithelial-mesenchymal transition (EMT) serves a critical role during this process, and primarily involves the following steps: i) Dissociation of points of adhesion between epithelial cells; ii) loss of the apical-basolateral polarity; iii) reorganization of the actin cytoskeleton; and iv) an increase in cell motility (5). A recent study suggested that dysregulation of the actin cytoskeleton may be a hallmark of subsequent metastatic dissemination (6), which may indicate cytoskeletal regulators as potential targets for CRC therapies based on metastasis inhibition.
Profilin 2 (PFN2) belongs to a class of small G-actin-binding proteins and is a well-characterized regulator of actin polymerization (7). PFN2 was previously considered to be expressed mainly in the neurons of vertebrates (8). However, altered expression of PFN2 has been reported in different types of cancer; for example, PFN2 expression was downregulated in oral squamous cell carcinoma (9). These data indicated that PFN2 may serve a role in cancer progression and metastasis; however, its expression and function are largely unknown in CRC.
In the present study, the expression levels and the effects of PFN2 were investigated in the development of human CRC, particularly in regard to metastatic capability. PFN2 expression was observed to be downregulated in patients with CRC with metastasis, as well as in metastatic CRC cell lines. In addition, alterations in the expression of PFN2 affected the EMT process and the metastasis of CRC. Furthermore, the results indicated that cytoskeletal reorganization may be involved in PFN2-regulated metastasis. Finally, clinical data suggested that tumors with low PFN2 expression were significantly associated with poor prognosis. Results from the present study indicated that PFN2 may function as a negative regulator of CRC metastasis and, thus, may represent a potential target for CRC therapy.
Materials and methods
Patients and specimens
The present study was approved by the Biomedical Research Ethics Committee of Jiading District Central Hospital Affiliated Shanghai University of Medicine & Health Sciences (Shanghai, China). Informed consent was obtained from all study subjects before sample collection and these samples were used according to ethical standards. CRC and normal paracancerous tissue samples were collected from 70 patients (38 male, 32 female; age, 48–72) that underwent surgery at the Department of Surgery, Jiading District Central Hospital Affiliated Shanghai University of Medicine & Health Sciences between February 2008 and October 2011; none of the patients received any preoperative treatment. Patients with familial adenomatous polyposis, inflammatory bowel diseases and primary tumors in other tissues were excluded. The histological sections were reviewed by two expert pathologists to verify the histological diagnosis. For reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blot analysis, 3 groups of human colorectal tissues (n=10/group; 21 male, 9 female; age 49–81), including normal colorectal tissues, non-metastatic CRC tissues and metastatic CRC tissues) were freshly collected at the Department of Surgery, Jiading District Central Hospital Affiliated Shanghai University of Medicine & Health Sciences between January 2015 and February 2017. Tumors were staged according to the American Joint Committee on Cancer pathological tumor-node-metastasis classification (10).
Cell culture
The human CRC cell lines SW620 and HCT116 were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Preliminary western blotting experiments were conducted to examine the expression of PFN2 in several CRC cell lines, and it was determined that PFN2 protein expression levels were highest in HCT116 cells and lowest in SW620; subsequently, HCT116 and SW620 were selected as representative CRC cell lines for use in the present study. These cell lines were routinely maintained in the laboratory according to the instructions from ATCC. Briefly, cells were cultured in 6- or 12-well plates (BD Biosciences, Franklin Lakes, NJ, USA) and grown as a monolayer. HCT116 cells were cultured at 37°C in a humidified incubator with 5% CO2 in McCoy's 5A (Modified) medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). SW620 cells were cultured at 37°C in an incubator without CO2 in Leibovitz's L 15 Medium (Gibco; Thermo Fisher Scientific, Inc.). Both of the media were supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.) and 100 U/ml penicillin and 100 µg/ml streptomycin (Thermo Fisher Scientific, Inc.).
In certain experiments, the cell-permeable small molecule Rho-associated kinase inhibitor Y27632 (3 µmol/l; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was added into the cell culture medium for 24 h to inhibit myosin light chain (MLC) phosphorylation.
Cell line transfection and transduction
The PFN2 overexpression vector was constructed as described previously (9). Full-length human PFN2 cDNA was amplified by RT-PCR using the forward primer 5′-GCGGCCGCATGGCCGGTTGGCAGAGCTACG-3′ and the reverse primer 5′-GGATCCTTACACATCAGACCTCCTCAG-3′. To construct the overexpression vector, the PFN2 coding sequence was excised from the pCR-Blunt plasmid (Invitrogen; Thermo Fisher Scientific, Inc.) by NotI and BamHI digestion, and subcloned into the pQCXIH expression vector (Clontech Laboratories, Inc., Mountain View, CA, USA). To characterize the PFN2 expression vector, pQCXIH-PFN2 (PFN2-OE) or empty vector (vector) was transfected into SW620 cells using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Briefly, SW620 cells were seeded (2×105 cells/well) in a 6-well plate 24 h prior to transfection and cultured to ~80% confluency, at 37°C in an incubator without CO2. Subsequently, SW620 cells were transfected with 1.5 µg of plasmid and 2.5 µl of Lipofectamine 2000 in 6-well plates, and then cultured at 37°C in an incubator without CO2 for 24 h, then harvested for further experiments.
Liver and lung metastasis of CRC model mice
Animal experiments were approved by the Animal Experiment Administration Committee of the Soochow University (Suzhou, China), and experiments were conducted in accordance with The National Institutes of Health guidelines for animal care during the study. Male BALB/c nude mice (age, 8–10 weeks; average weight, 24.0 g) were obtained from the Shanghai Laboratory Animal Center at the Chinese Academy of Sciences (Shanghai, China). They were maintained in a specific pathogen free room with a constant temperature (20–26°C), relative humidity (40–70%), normal atmosphere (with 21% O2 and 0.03% CO2; 15 times per hour ventilation), 12-h light-dark cycle and free access to food and water.
To establish the liver cancer metastasis model, mice were anesthetized, a transverse incision was made in the left flank to expose the spleen and 1×106 PFN2-OE-transfected or empty vector-transfected SW620 cells in 100 µl phosphate-buffered saline (PBS; Thermo Fisher Scientific, Inc.) were injected intrasplenically (n=5 mice/group). For the lung cancer metastasis model, the mice were anesthetized and 1×106 PFN2-OE-transfected or empty vector-transfected SW620 cells in 100 µl of PBS were injected via the tail vein (5 mice/group). During the 6 weeks following injections, body weight and survival of the mice was monitored; the body weight of each mouse was measured once per week. The volume of the largest metastatic tumor nodules was calculated using the formula: Volume = (width2 × length) / 2. All mice were euthanized 6 weeks post-injection.
RT-qPCR
Total RNA was extracted from tissues or cells using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) and 1 µg RNA was subsequently reverse transcribed using the AMV Reverse Transcription System (Takara Bio Inc., Otsu, Japan). qPCR was performed using the FastStart Universal SYBR Green Master with Rox (Roche Diagnostics, Basel, Switzerland) and an ABI PRISM 7900HT system (Applied Biosystems; Thermo Fisher Scientific, Inc.), with the following thermocycling conditions: an initial 2-min incubation at 50°C, followed by 10 min at 95°C and 40 cycles of 95°C for 15 sec and 60°C for 60 sec. The following primers were used: PFN2, forward 5′-ATGATTGTAGGAAAAGACCGGGA-3′, reverse 5′-GCAGTCACCATCGACGTATAGAC-3′; epithelial (E)-cadherin, forward 5′-CGAGAGCTACACGTTCACGG-3′, reverse 5′-GGGTGTCGAGGGAAAAATAGG-3′; neural (N)-cadherin, forward 5′-AGCCAACCTTAACTGAGGAGT-3′, reverse 5′-GGCAAGTTGATTGGAGGGATG-3′; vimentin, forward 5′-GACGCCATCAACACCGAGTT-3′, reverse 5′-CTTTGTCGTTGGTTAGCTGGT-3′; snail, forward 5′-ACTGCGACAAGGAGTACACC-3′, reverse 5′-GAGTGCGTTTGCAGATGGG-3′; slug, forward 5′-TGACCTGTCTGCAAATGCTC-3′, reverse 5′-TCGGACCCACACATTACCTT-3′; and GAPDH, forward 5′-GGAGCGAGATCCCTCCAAAAT-3′, reverse 5′-GGCTGTTGTCATACTTCTCATGG-3′. mRNA levels in each group were calculated by the 2−ΔΔCt method and normalized to the internal reference gene GAPDH (11). Independent experiments were repeated three times.
Western blotting
Tissues (~5mg) were homogenized with an electric homogenizer and lysed with 100 µl ice cold T-PER Tissue Protein Extraction Reagent (Pierce; Thermo Fisher Scientific, Inc.). Cells (5×105) were washed twice with PBS and lysed with 100 µl ice cold radioimmunoprecipitation assay lysis and extraction buffer (Pierce; Thermo Fisher Scientific, Inc.). The samples were centrifuged at 10,000 × g for 5 min at 4°C to pellet tissue/cell debris and the supernatant containing protein was collected. Protein concentrations were determined using a Bicinchoninic Acid Protein kit (Pierce; Thermo Fisher scientific, Inc.). Total protein (20 µg) was separated by 10% SDS-PAGE and proteins were transferred to polyvinylidene difluoride membranes. The membranes were blocked with 5% fat-free milk in PBS with 0.1% Tween-20 (Sinopharm Chemical Reagent Co., Ltd., Shanghai, China) for 2 h at room temperature and subsequently incubated with the following primary antibodies (1:2,000) in PBS with 0.1% Tween-20 at 4°C overnight: Anti-PFN2 (cat. no. 60094-2-Ig; ProteinTech Group Inc., Chicago, IL, USA), anti-slug (cat. no. ab51772; Abcam, Cambridge, MA, USA), anti-snail (cat. no. ab82846; Abcam), anti-E-cadherin (cat. no. ab1416; Abcam), anti-N-cadherin (cat. no. ab18203; Abcam), anti-vimentin (cat. no. ab20346; Abcam), anti-GAPDH (cat. no. ab8245; Abcam), anti-phosphorylated (p)-MLC (cat. no. 3675; Cell Signaling Technology, Inc., Danvers, MA, USA), anti-MLC (cat. no. 3672, Cell Signaling Technology, Inc.) and anti-β-tubulin (cat. no. ab6046; Abcam). Following washing with PBS, the membranes were incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse immunoglobulin (Ig)G (cat. no. 32430; Thermo Fisher Scientific Inc.) or HRP-conjugated goat anti-rabbit IgG (cat. no. 31460; Thermo Fisher Scientific Inc.) secondary antibodies (1:5,000) in PBS with 0.1% Tween-20 for 1 h at room temperature. Following subsequent washes with PBS, the immunoreactive bands were visualized with Enhanced Chemiluminescence Plus Western Blotting Detection reagents (cat. no. 2650; Merck KGaA). Densitometric analysis was performed to quantify protein expression using ImageJ software (version 1.48; National Institutes of Health, Bethesda, MA, USA); protein expression levels were normalized to the loading controls GAPDH or β-tubulin. Independent experiments were repeated three times.
Immunohistochemistry (IHC)
Tissues were fixed with 4% paraformaldehyde in PBS for 48 h at room temperature and dehydrated through a series of graded ethanol baths at room temperature (70% for 1 h; 95% for 1 h; 100% for 1 h; 100% for 1.5 h; 100% for 1.5 h; 100% for 2 h), followed by clearing in xylene for 1 h at room temperature and infiltrated with paraffin for 1 h at 60°C. The paraffin embedded tissues were cut into 5 µm sections. The tissue sections were subjected to routine deparaffinization and rehydration at room temperature: 100% Xylene (20 min); 100% xylene (20 min); 100% ethanol (10 min); 95% ethanol (10 min); 90% ethanol (10 min); 80% ethanol (10 min). Antigen retrieval was achieved by microwaving the sections in 0.01 mol/l citrate buffer for 10 min and cooling for 30 min to reach the room temperature. Endogenous peroxidase activity was inhibited by incubating the sections with 3% hydrogen peroxide in methanol for 20 min, and non-specific binding was blocked by incubation with 5% bovine serum albumin (Sigma-Aldrich; Merck KGaA) in PBS at room temperature. Sections were washed three times with PBS and incubated overnight at 4°C with anti-PFN2 primary antibody (cat. no. 60094-2-Ig; ProteinTech Group Inc.). Subsequently, sections were incubated with HRP-conjugated rat anti-mouse-IgG2b (cat. no. ab157293, Abcam, Inc.), and the signal was developed with 3,3′-diaminobenzidine tetrahydrochloride in Tris-HCl buffer (pH 7.6) containing 0.02% hydrogen peroxide. The sections were counterstained with hematoxylin and mounted. Negative controls were obtained by incubating sections with PBS as opposed to the specific primary antibodies. IHC staining was independently examined by two clinical pathologists blinded to the study. For each sample, five high-power fields (magnification, ×100) were randomly selected under a Nikon Eclipse Ti-S light microscope (Nikon Corporation, Tokyo, Japan) and analyzed using NIS-Elements version 4.0 (Nikon Corporation) software. The intensity of cytoplasmic staining was determined semi-quantitatively on a scale of 0–3 as follows: 0 (negative), 1 (weakly positive), 2 (moderately positive) and 3 (strongly positive). A consensus score was assigned for each section following discussion and careful review of all slides by the two pathologists. Tissues with a final staining score of 0 or 1 were grouped into the 'low' expression group, and tissues with a score of 2 were grouped into the 'moderate' expression group.
Wound-healing assay
Cells (5×105 cells/well) were seeded into a 12-well plate and grown until they reached 80% confluency, ~24 h. HCT116 cells were cultured at 37°C in a humidified incubator with 5% CO2 in McCoy's 5A (Modified) medium (Thermo Fisher Scientific, Inc.). SW620 cells were cultured at 37°C in an incubator without CO2 in Leibovitz's L 15 Medium (Thermo Fisher Scientific, Inc.). Both of the media were supplemented with 10% FBS (Thermo Fisher Scientific, Inc.) and 100 U/ml penicillin and 100 µg/ml streptomycin (Thermo Fisher Scientific, Inc.). The cell monolayers were manually wounded by scraping with a 200 µl pipette tip. Cells were washed with PBS to remove the debris and were cultured for 24 h to allow for wound healing. Within each well, five sites of a unique regular wound were selected for analysis using a live cell imaging microscope (Zeiss AG, Oberkochen, Germany); images of the wounds were captured immediately following wounding (0 h) and at the end of the experiment (24 h), and the distance between the two edges of the injury were quantified using Adobe Photoshop CS3 (Adobe Systems, Inc., San Jose, CA, USA). The migration distances were recorded as the percentage of cell migration, and the data are presented as the mean ± sandard error of the mean (SEM) of triplicate assays for each cell line; at least three independent experiments were performed.
Transwell migration assay
24-well Transwell insert chambers (Merck KGaA) were used for cell migration assay. Cells (1×105) were added to the top chambers in serum-free medium (200 ml; McCoy's 5A for HCT116 and L 15 for SW620), and the bottom chambers were filled with medium (McCoy's 5A for HCT116 and L 15 for SW620) containing 10% FBS. Cells were cultured for 24 h at 37°C in a 5% CO2-humidified incubator (HCT116) or non-CO2-humidified incubator (SW620). To quantify migration, cells were removed from the upper membrane by gently scrapping this side with a wet cotton swab. The migrated cells attached to the lower membrane were fixed with 4% paraformaldehyde for 20 min, stained with 0.5% crystal violet solution for 30 min, and washed two times with PBS; all at room temperature. The number of migrated cells was subsequently counted in five randomly selected fields under a light microscope (Zeiss AG), and the migrated cell numbers were quantified using Adobe Photoshop CS3 (Adobe Systems, Inc.). Independent experiments were repeated three times.
Statistical analysis
Statistical differences were evaluated using Statistical Package for Social Science software (version 17.0; SPSS, Inc., Chicago, IL, USA). The association between staining intensity and clinicopathological patterns was assessed using the χ2-test and two-sided Fisher's exact test to determine the significance of the difference between the covariates. All measurement data are presented as mean ± SEM. Statistical significance between two groups was evaluated by the Student's t-test and unpaired nonparametric Mann-Whitney U test. Comparison of multiple groups was performed using analysis of variance and Bonferroni post hoc tests. P<0.05 was considered to indicate a statistically significant difference.
Results
Downregulation of PFN2 expression in CRC patients with metastasis
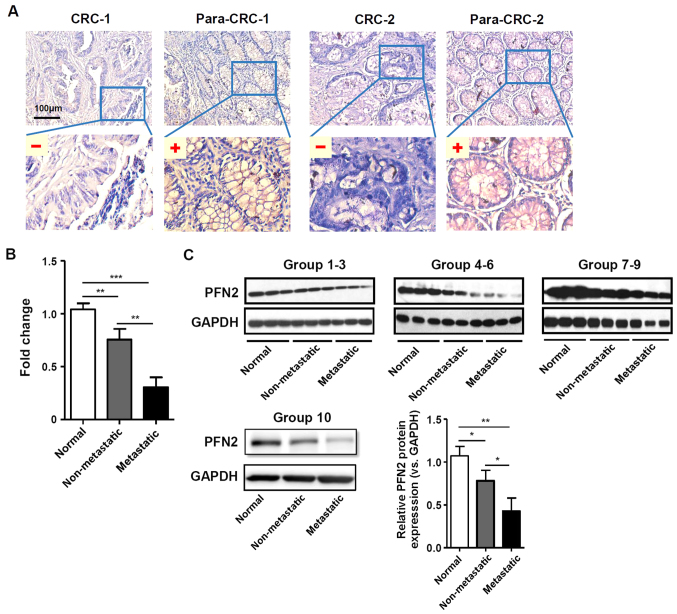
PFN2 expression was examined in CRC and normal paracancerous colon mucosa tissues by IHC staining. The clinical features of the patients and the results of the IHC staining statistical analyses are provided in Table I and Fig. 1A, respectively. The results indicated that PFN2 expression was significantly lower in CRC tissues compared with expression levels in the normal adjacent colorectal tissues. Furthermore, the expression level of PFN2 was significantly lower in CRC patients with lymph node and distant metastasis compared with CRC patients without metastasis (Table I). In addition, RT-qPCR and western blot analyses on freshly collected colorectal tissue samples demonstrated that PFN2 expression was significantly decreased in metastatic tumors compared with either non-metastatic tumor or normal colorectal tissues (Fig. 1B and C, respectively), which suggested that there may be a clinical association between PFN2 expression and CRC progression.
Table I.
Association between PFN2 expression and clinicopathological features in patients with CRC.
| Clinicopathological parameters | n | PFN2 expression
|
P-value | |
|---|---|---|---|---|
| Low | Medium | |||
| Total cases | 70 | 40 | 30 | |
| Age (years) | NS | |||
| ≤60 | 28 | 16 | 12 | |
| >60 | 42 | 24 | 18 | |
| Tissue type | <0.001 | |||
| Normal colorectal tissue | 70 | 0 | 70 | |
| CRC tissue | 70 | 40 | 30 | |
| Sex | NS | |||
| Male | 38 | 22 | 16 | |
| Female | 32 | 18 | 14 | |
| Tumor size (cm) | NS | |||
| ≤5 | 41 | 20 | 21 | |
| >5 | 29 | 20 | 9 | |
| TNM stage | <0.001 | |||
| I | 8 | 0 | 8 | |
| II | 21 | 6 | 15 | |
| III | 26 | 20 | 6 | |
| IV | 15 | 14 | 1 | |
| Lymph node metastasis | <0.001 | |||
| Negative | 29 | 6 | 23 | |
| Positive | 41 | 34 | 7 | |
| Distance metastasis | <0.001 | |||
| Negative | 55 | 26 | 29 | |
| Positive | 15 | 14 | 1 | |
CRC, colorectal cancer; NS, not significant; PFN2, profilin 2; TNM, tumor, node, metastasis.
Figure 1.
PFN2 expression in normal colon mucosa and CRC tissues. (A) Representative examples of PFN2 immunohistochemical staining analysis in human CRC and corresponding para-CRC normal adjacent. (B and C) PFN2 expression levels in freshly collected human normal colorectal, non-metastatic CRC and metastatic CRC tissues was analyzed by (B) reverse transcription-quantitative polymerase chain reaction and (C) western blotting; GAPDH served as the internal reference. Data are presented as the mean ± standard error of the mean of three independent experiments. *P<0.05, **P<0.01 and ***P<0.001. CRC, colorectal cancer; para-CRC, paracancerous; PFN2, profilin 2.
Low PFN2 expression affects CRC cell migration
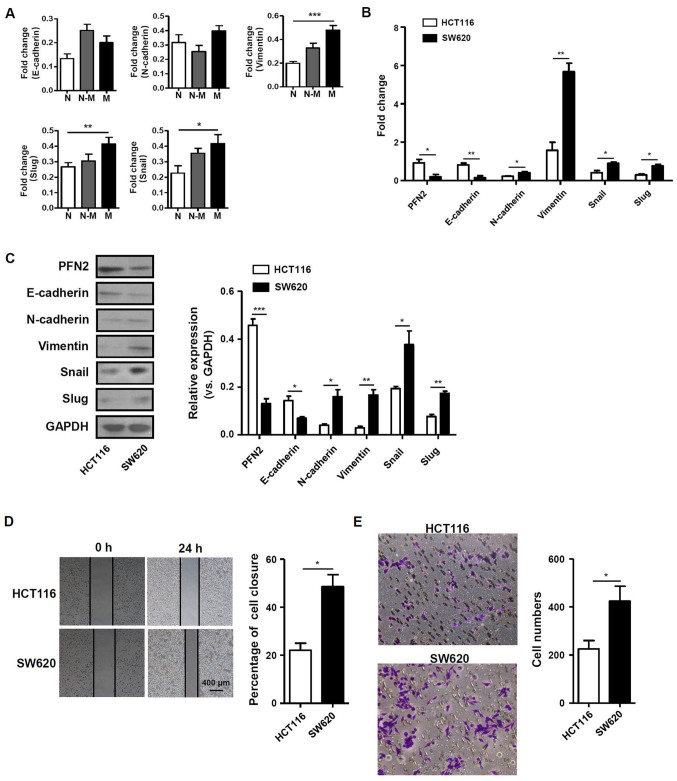
Given the aforementioned differential expression of PFN2, the function of PFN2 in human CRC development was subsequently investigated. CRC metastasis was previously reported to be closely associated with the EMT (12,13). Therefore, the present study examined the mRNA expression levels of EMT-related molecules, including E-cadherin, N-cadherin, vimentin, slug and snail in normal colon, non-metastatic and metastatic CRC tissues. The results demonstrated that the mRNA expression levels of vimentin, slug and snail were significantly higher in metastatic CRC tissues compared with normal colon tissues (Fig. 2A). In addition, PFN2 expression was also determined in the non-metastatic CRC cell line HCT116 and in the SW620 cell line, which was originally derived from a metastatic site of a patient with CRC. PFN2 mRNA was expressed at significantly lower levels in the SW620 cell line compared with expression levels in HCT116 cells. The mRNA expression level of E-cadherin was also lower in SW620 cells, whereas the expression levels of N-cadherin, vimentin, snail and slug were significantly higher in SW620 cells compared with the respective expression levels in HCT116 cells (Fig. 2B). Western blotting results revealed that the epithelial marker E-cadherin and PFN2 protein expression levels were notably decreased, whereas expression levels of the mesenchymal marker proteins vimentin, N-cadherin and related molecules, such as snail and slug, were increased in SW620 cells compare with the respective expression levels in HCT116 cells (Fig. 2C). These results suggested that PFN2 may have negative effects on the EMT process. To confirm the relationship between PFN2 and cancer metastasis, wound-healing and Transwell migration assays were performed in HCT116 and SW620 cells. The migratory ability of SW620 cells was significantly higher compared with that of HCT116 cells (Fig. 2D and E). These data suggested that the reduced level of PFN2 expression in SW620 CRC cells may be associated with higher migratory capability.
Figure 2.
PFN2 expression level is associated with EMT-related molecules and migratory activity in CRC cells. (A) The expression of EMT-related molecules in human normal colorectal, non-metastatic CRC and metastatic CRC tissues. (B and C) mRNA and protein expression levels of PFN2 and EMT related molecules, E-cadherin, N-cadherin, vimentin, slug and snail, were measured in two CRC cell lines, HCT116 and SW620, by (B) reverse transcription-quantitative polymerase chain reaction and (C) western blotting, respectively; GAPDH served as the internal reference. (D and E) The migratory activity of CRC cell lines was analyzed by (D) wound-healing and (E) Transwell migration assays at 24 h. Data are presented as the mean ± sandard error of the mean of three independent experiments. *P<0.05, **P<0.01 and ***P<0.001. CRC, colorectal cancer; EMT, epithelial-mesenchymal transition; N, normal; N-M, non-metastatic; M, metastatic; PFN2, profilin 2.
PFN2 suppresses CRC cell migration with reduced EMT
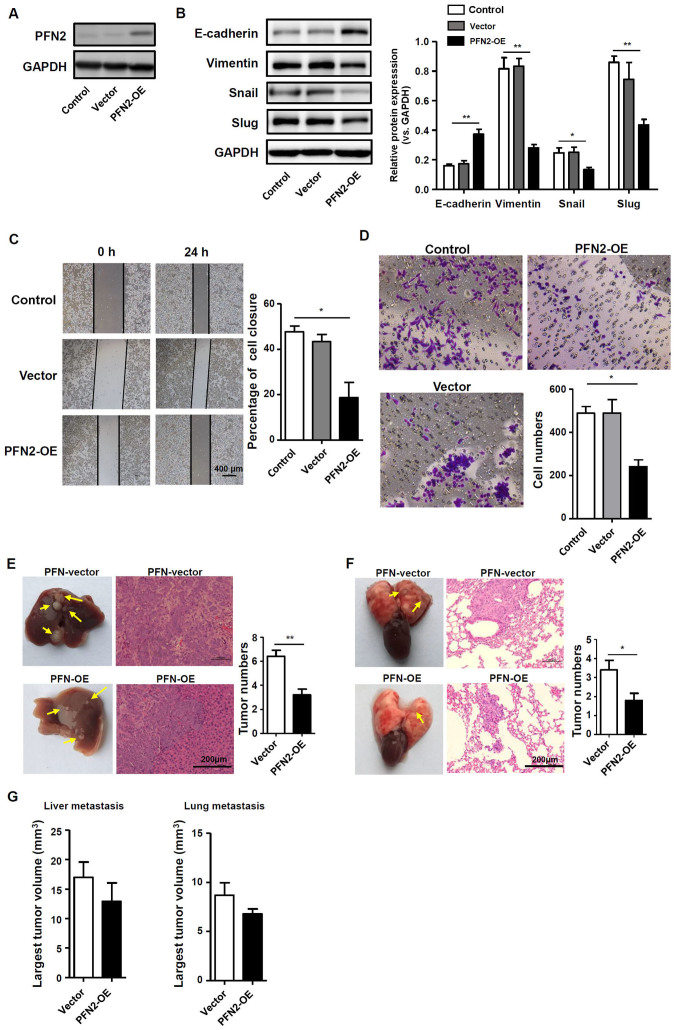
To further determine if PFN2 serves a role in EMT and CRC cell migration, the pQCXIH vector was used to transiently over-express PFN2 in SW620 cells, which express a low level of endogenous PFN2, as confirmed by western blotting (Fig. 3A); untransfected and empty vector-transfected SW620 cells were used as the controls. The potential role of PFN2 as a negative regulator of EMT was further explored by examining the expression of EMT markers and known EMT regulators, slug and snail. Western blotting demonstrated that E-cadherin protein expression was increased, whereas vimentin, slug and snail expression levels were decreased in SW620 cells overexpressing PFN2, which suggested that the inhibition of EMT may be regulated by PFN2 overexpression in SW620 cells (Fig. 3B). Correspondingly, SW620 cell migration was significantly reduced by PFN2 overexpression compared with control cells (Fig. 3C and D). In vivo liver and lung metastasis mouse models were used to evaluate the potential roles of PFN2 in regulating CRC metastasis. The results revealed that PFN2-overexpressing SW620 cells exhibited reduced metastatic potential compared with non-PFN2-overexpressing SW620 cells in liver and lung (Fig. 3E and F, respectively); however, no significant difference in the largest tumor nodule volume was identified between these two groups (Fig. 3G). These results suggested a negative role of PFN2 in CRC cell migration.
Figure 3.
PFN2 overexpression results in decreased EMT in colorectal cancer. PFN2 was transiently overexpressed in SW620 cells using a pQCXIH-PFN2 vector. (A) Western blotting was performed to confirm the overexpression efficiency in PFN2-OE transfected SW620 cells; untransfected and empty vector-transfected cells were used as controls. (B) Protein expression levels of EMT markers and regulators were analyzed by western blotting; GAPDH served as the internal reference. (C and D) Migratory abilities of the transfected SW620 cells were examined by (C) wound-healing and (D) Transwell migration assays, respectively. (E and F) Vector-transfected control SW620 cells or PFN2-OE-transfectd SW620 cells were injected into nude mice (E) via the spleen to induce liver metastasis or (F) via the tail vein to induce lung metastasis. The yellow arrows indicate the metastatic tumor nodules and hematoxylin and eosin staining was performed to confirm the tumor characteristics. (G) The volumes of the largest metastatic tumor nodules were calculated. Data are presented as the mean ± sandard error of the mean of three independent experiments. *P<0.05 and **P<0.01. Ctrl, control; EMT, epithelial-mesenchymal transition; OE, overexpression; PFN2, profilin 2.
PFN2 inhibits CRC EMT by regulating cytoskeletal reorganization
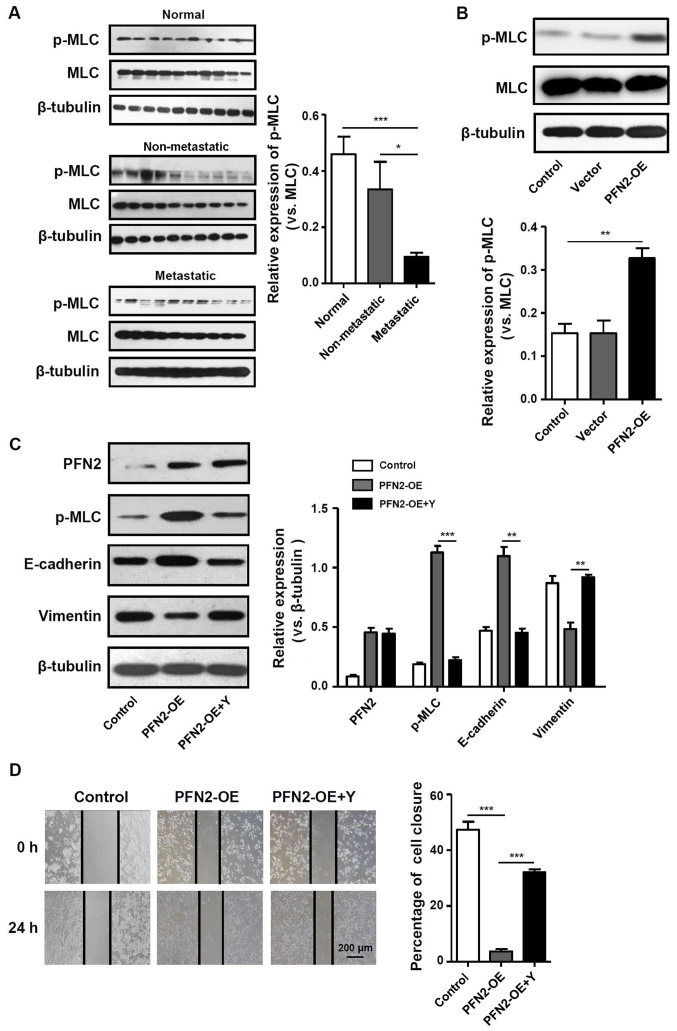
PFN2 triggers various cellular pathways to exert disparate functions. As an actin-binding protein, one of these functions is to regulate cytoskeletal reorganization (14). Notably, contractile actin bundles are thought to be suppressors of cancer protrusive activity, migration and invasion (15). As MLC phosphorylation is a marker of myosin motor contractions (16), the present study examined the level of pMLC in CRC tissues and cell lines. In normal colon tissues, pMLC was expressed at notably higher levels and pMLC expression was significantly lower in metastatic CRC tissues compared with non-metastatic CRC tissues (Fig. 4A). In addition, in PFN2-OE SW620 cells, pMLC expression was significantly increased compared with the untransfected and vector-trans-fected control groups (Fig. 4B), which indicated that PFN2 expression in CRC cells may regulate the contractile actin bundles. To determine the relationship between the generation of contractile actin bundles and PFN2-regulated cancer metastasis, the present study assessed the effects of altering myosin activity on the suppressive effects of PFN2. Therefore, the pharmacological inhibitor of MLC phosphorylation, Y27632, was used in subsequent experiments. Inhibition of MLC phosphorylation attenuated the inhibitory effects of PFN2-OE on EMT, as the expression of E-cadherin decreased whereas the expression of vimentin increased in PFN2-OE SW620 cells treated with Y27632 compared with those cells without Y27632 treatment for 24 h (Fig. 4C). The results of the wound-healing assay demonstrated that with the migratory ability of PFN2-OE SW620 cells treated with Y27632 significantly increased compared with those cells without Y27632 (Fig. 4D). Therefore, it was hypothesized that the suppressive role of PFN2 on CRC metastasis may have resulted from PFN2-regulated cytoskeletal reorganization.
Figure 4.
Cytoskeletal remodeling may be involved in PFN2-regulated EMT. (A and B) p-MLC expression was measured by western blotting in (A) normal colon, non-metastatic CRC and metastatic CRC tissues, and (B) in untransfected, vector- and PFN2-OE-transfected SW620 cells. (C) Protein expression levels of PFN2, p-MLC, E-cadherin and vimentin were examined by western blotting in PFN2-OE-transfected SW620 cells with or without co-treatment with Y27632; β-tubulin served as the internal reference. (D) Wound-healing assays were performed to determine the migratory ability of PFN2-OE-transfected SW620 with or without co-treatment with Y27632. Data are presented as the mean ± sandard error of the mean of three independent experiments. *P<0.05, **P<0.01 and ***P<0.001. CRC, colorectal cancer; EMT, epithelial-mesenchymal transition; MLC, myosin light chain; OE, overexpression; p, phosphorylated; PFN2, profilin 2; Y, Rho-associated kinase inhibitor Y27632.
Discussion
The actin cytoskeleton is a highly dynamic structure, which undergoes constant remodeling in a living cell (5). In general, the regulation of such a structure is a prerequisite for such processes as endocytosis, cell motility and cancer cell invasion. As reported previously, the actin cytoskeleton pathway serves important roles in the development of metastatic CRC (17). Therefore, molecules involved in this pathway may be candidate regulators of the migratory potential of CRC. The aim of the present study was to investigate whether the actin cytoskeleton regulator PFN2 serves specific roles in the progression of human CRC, particularly in the process of metastasis. PFN2 expression was demonstrated to be significantly lower in metastatic CRC tissues, which may function by negatively affecting EMT through the regulation of the actin cytoskeleton.
Although the roles and underlying mechanisms of PFN2 in cytoskeletal regulation has been demonstrated in a number of previous studies (15,18), its relationship with cancer has not been as extensively studied until recently. For example in head and neck squamous cell carcinoma, high PFN2 expression independently predicted poor overall survival; microRNA-30a-5p suppresses EMT by targeting PFN2 in high invasive non-small cell lung cancer cell lines; however, further mechanistic studies are still expected (19–21). However, previous studies have focused on another member of the profilin family in cancer development, PFN1 (22). As a ubiquitously expressed actin polymerization regulator, PFN1 has been studied in different types of cancer, including gastric, bladder, renal and breast cancer (23–26). Considering its inhibitory roles in the progression of carcinomas, it is considered to be a tumor suppressor and may be a novel biomarker for predicting the response to anticancer therapy. However, there may be some limitations in its application owing to its non-specific expression in various tissues: PFN1 is expressed during all embryonic stages, and a previous study reported that it is expressed in nearly all cell and tissue types; by contrast, PFN2 is expressed primarily in the developing nervous system and in adult differentiated neurons (27). Therefore, the neuronal tissue-specific isoform PFN2 may be better candidate for anticancer therapy with fewer side effects on other tissues.
To date, PFN2 has been linked with oral squamous cell carcinoma, breast cancer and inflammatory bowel disease-associated CRC (9,15,17). However, it has not been investigated in depth for its cancer-related functions. To demonstrate the role of PFN2 in CRC, the present study analyzed the expression levels of PFN2 in patients with CRC and in CRC cell lines. The results demonstrated that PFN2 expression was higher in non-metastatic CRC tissues and cell lines, whereas expression in metastatic CRC tissues and cells was comparatively low. These data suggested a suppressive role for PFN2 in cancer metastasis, which was consistent with a previous report (15). Although cancer cell growth has also been suggested to be regulated by PFN2 (28), the present study primarily focused on cell migration, as significant differences in expression were observed between CRC with and without metastasis.
EMT is a key process during cancer metastasis, characterized by a loss of cell-cell adhesion and an increase in cell motility (29). In the present study, the expression levels of several EMT markers were significantly altered by the overexpression of PFN2 in CRC cell lines. In CRC cells expressing low levels of PFN2, higher expression of the mesenchymal markers, including vimentin, snail and slug, was observed, whereas overexpressing PFN2 suppressed the EMT process and, thus, the migration of CRC cells. EMT is regulated by complex signaling pathways, such as transforming growth factor-β, phosphoinositide-3 kinase, mitogen-activated protein kinase, integrin-linked kinase and jagged1 (30). As none of these signaling pathways have been reported in relation to PFN2 functions, the present study detected the expression of the EMT-induced transcription factors, snail and slug (31). Although the signaling of EMT in CRC has been studied previously, whether PFN2-regulated EMT shares similar mechanisms requires further investigation.
As aforementioned, PFN2 is a cytoskeleton regulator. The actin cytoskeleton serves a central role in many carcinogenesis and cancer cell migration processes (32–34). Therefore, it is reasonable to suggest that the actin cytoskeleton may be associated with PFN2-regulated EMT and subsequent cancer migration. MLC phosphorylation is known to activate actomyosin contractility, which is crucial for cytoskeleton remodeling (16). A previous study demonstrated that PFN2 knockdown in breast cancer cells reduced the expression of total p-MLC (15), which was accompanied by reduced cortical actin bundling. The present study overexpressed PFN2 in the CRC cell line SW620 and observed similar effects of PFN2 on MLC phosphorylation. Altered expression levels of the EMT markers indicated that PFN2 may reduce the metastatic ability of CRC by inducing MLC phosphorylation. Furthermore, inhibiting the phosphorylation of MLC using a pharmacological inhibitor reversed the effect of PFN2 on EMT.
In conclusion, results from the present study revealed that PFN2 expression was reduced in metastatic CRC. PFN2 was observed to exert a suppressive effect on EMT, and subsequently on the migratory abilities of CRC cells, and this may be through regulating cytoskeletal reorganization. Therefore, decreased PFN2 expression may have important clinicopathological implications for CRC metastasis and may be a potential target for future CRC therapies.
Acknowledgments
Not applicable.
Abbreviations
- CRC
colorectal cancer
- EMT
epithelial-mesenchymal transition
- IHC
immunohistochemistry
- MLC
myosin light chain
- PFN2
profilin 2
Funding
This study was supported by The Seed Fund Program of Shanghai University of Medicine & Health Sciences (grant no. SFP-18-20-16-004), a grant from The Shanghai Medical Key Specialist Construction Plans (grant no. ZK2015B10), The Project of Nature Science Foundation of China (grant no. 81672348), The Six Major Talent Peak Project of Jiangsu Province, China (grant no. 2015-WSW-014), The Six One Project for Advanced Medical Talent of Jiangsu Province, China (grant no. LGY2016031) and The Jiangsu Provincial Medical Youth Talent, China (grant no. QNRC2016735).
Availability of data and materials
All data generated or analyzed during this study are included in this article.
Authors' contributions
HZ, SH and DL designed the research, analyzed the data and wrote and revised the manuscript. HZ, WY, JY, KZ and BW performed the experiments. PS and YC collected the clinical samples and did the analysis. All authors have read and approved the final version of this manuscript.
Ethics approval and consent to participate
The present study was approved by the Biomedical Research Ethics Committee of Jiading District Central Hospital Affiliated Shanghai University of Medicine & Health Sciences (Shanghai, China). Informed consent was obtained from all study subjects prior to sample collection and the samples were used according to ethical standards.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Iseki Y, Shibutani M, Maeda K, Nagahara H, Ohtani H, Sugano K, Ikeya T, Muguruma K, Tanaka H, Toyokawa T, et al. Impact of the preoperative controlling nutritional status (CONUT) score on the survival after curative surgery for colorectal cancer. PLoS One. 2015;10:e0132488. doi: 10.1371/journal.pone.0132488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu H, Ren G, Wang T, Chen Y, Gong C, Bai Y, Wang B, Qi H, Shen J, Zhu L, et al. Aberrantly expressed Fra-1 by IL-6/STAT3 transactivation promotes colorectal cancer aggressiveness through epithelial-mesenchymal transition. Carcinogenesis. 2015;36:459–468. doi: 10.1093/carcin/bgv017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grünert S, Jechlinger M, Beug H. Diverse cellular and molecular mechanisms contribute to epithelial plasticity and metastasis. Nat Rev Mol Cell Biol. 2003;4:657–665. doi: 10.1038/nrm1175. [DOI] [PubMed] [Google Scholar]
- 5.Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009;28:15–33. doi: 10.1007/s10555-008-9169-0. [DOI] [PubMed] [Google Scholar]
- 6.König K, Meder L, Kröger C, Diehl L, Florin A, Rommerscheidt-Fuss U, Kahl P, Wardelmann E, Magin TM, Buettner R, et al. Loss of the keratin cytoskeleton is not sufficient to induce epithelial mesenchymal transition in a novel KRAS driven sporadic lung cancer mouse model. PLoS One. 2013;8:e57996. doi: 10.1371/journal.pone.0057996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jockusch BM, Murk K, Rothkegel M. The profile of profilins. Rev Physiol Biochem Pharmacol. 2007;159:131–149. doi: 10.1007/112_2007_704. [DOI] [PubMed] [Google Scholar]
- 8.Witke W, Podtelejnikov AV, Di Nardo A, Sutherland JD, Gurniak CB, Dotti C, Mann M. EMBO J; In mouse brain profilin I and profilin II associate with regulators of the endocytic pathway and actin assembly; 1998. pp. 967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma CY, Zhang CP, Zhong LP, Pan HY, Chen WT, Wang LZ, Andrew OW, Ji T, Han W. Decreased expression of profilin 2 in oral squamous cell carcinoma and its clinicopathological implications. Oncol Rep. 2011;26:813–823. doi: 10.3892/or.2011.1365. [DOI] [PubMed] [Google Scholar]
- 10.Greene FL. Current TNM staging of colorectal cancer. Lancet Oncol. 2007;8:572–573. doi: 10.1016/S1470-2045(07)70185-7. [DOI] [PubMed] [Google Scholar]
- 11.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 12.Castosa R, Martinez-Iglesias O, Roca-Lema D, Casas-Pais A, Díaz-Díaz A, Iglesias P, Santamarina I, Graña B, Calvo L, Valladares-Ayerbes M, et al. Hakai overexpression effectively induces tumour progression and metastasis in vivo. Sci Rep. 2018;8:3466–3475. doi: 10.1038/s41598-018-21808-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Findlay VJ, Wang C, Watson DK, Camp ER. Epithelial-to-mesenchymal transition and the cancer stem cell phenotype: Insights from cancer biology with therapeutic implications for colorectal cancer. Cancer Gene Ther. 2014;21:181–187. doi: 10.1038/cgt.2014.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferron F, Rebowski G, Lee SH, Dominguez R. Structural basis for the recruitment of profilin-actin complexes during filament elongation by Ena/VASP. EMBO J. 2007;26:4597–4606. doi: 10.1038/sj.emboj.7601874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mouneimne G, Hansen SD, Selfors LM, Petrak L, Hickey MM, Gallegos LL, Simpson KJ, Lim J, Gertler FB, Hartwig JH, et al. Differential remodeling of actin cytoskeleton architecture by profilin isoforms leads to distinct effects on cell migration and invasion. Cancer Cell. 2012;22:615–630. doi: 10.1016/j.ccr.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takebe T, Enomura M, Yoshizawa E, Kimura M, Koike H, Ueno Y, Matsuzaki T, Yamazaki T, Toyohara T, Osafune K, et al. Vascularized and complex organ buds from diverse tissues via mesenchymal cell-driven condensation. Cell Stem Cell. 2015;16:556–565. doi: 10.1016/j.stem.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 17.Kanaan Z, Qadan M, Eichenberger MR, Galandiuk S. The actin-cytoskeleton pathway and its potential role in inflammatory bowel disease-associated human colorectal cancer. Genet Test Mol Biomarkers. 2010;14:347–353. doi: 10.1089/gtmb.2009.0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schlüter K, Jockusch BM, Rothkegel M. Profilins as regulators of actin dynamics. Biochim Biophys Acta. 1997;1359:97–109. doi: 10.1016/S0167-4889(97)00100-6. [DOI] [PubMed] [Google Scholar]
- 19.Liu J, Wu Y, Wang Q, Liu X, Liao X, Pan J. Bioinformatic analysis of PFN2 dysregulation and its prognostic value in head and neck squamous carcinoma. Future Oncol. 2018;14:449–459. doi: 10.2217/fon-2017-0348. [DOI] [PubMed] [Google Scholar]
- 20.Cui XB, Zhang SM, Xu YX, Dang HW, Liu CX, Wang LH, Yang L, Hu JM, Liang WH, Jiang JF, et al. PFN2, a novel marker of unfavorable prognosis, is a potential therapeutic target involved in esophageal squamous cell carcinoma. J Transl Med. 2016;14:137–152. doi: 10.1186/s12967-016-0884-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan J, Ma C, Gao Y. MicroRNA-30a-5p suppresses epithelial-mesenchymal transition by targeting profilin-2 in high invasive non-small cell lung cancer cell lines. Oncol Rep. 2017;37:3146–3154. doi: 10.3892/or.2017.5566. [DOI] [PubMed] [Google Scholar]
- 22.Ding Z, Roy P. Profilin-1 versus profilin-2: Two faces of the same coin. Breast Cancer Res. 2013;15:311–312. doi: 10.1186/bcr3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng YJ, Zhu ZX, Zhou JS, Hu ZQ, Zhang JP, Cai QP, Wang LH. Silencing profilin-1 inhibits gastric cancer progression via integrin β1/focal adhesion kinase pathway modulation. World J Gastroenterol. 2015;21:2323–2335. doi: 10.3748/wjg.v21.i8.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karamchandani JR, Gabril MY, Ibrahim R, Scorilas A, Filter E, Finelli A, Lee JY, Ordon M, Pasic M, Romaschin AD, et al. Profilin-1 expression is associated with high grade and stage and decreased disease-free survival in renal cell carcinoma. Hum Pathol. 2015;46:673–680. doi: 10.1016/j.humpath.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 25.Coumans JV, Gau D, Poljak A, Wasinger V, Roy P, Moens PD. Profilin-1 overexpression in MDA-MB-231 breast cancer cells is associated with alterations in proteomics biomarkers of cell proliferation, survival, and motility as revealed by global proteomics analyses. OMICS. 2014;18:778–791. doi: 10.1089/omi.2014.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang JW, Shi ZZ, Shen TY, Che X, Wang Z, Shi SS, Xu X, Cai Y, Zhao P, Wang CF, et al. Identification of genomic alterations in pancreatic cancer using array-based comparative genomic hybridization. PLoS One. 2014;9:e114616. doi: 10.1371/journal.pone.0114616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alkam D, Feldman EZ, Singh A, Kiaei M. Profilin1 biology and its mutation, actin(g) in disease. Cell Mol Life Sci. 2017;74:967–981. doi: 10.1007/s00018-016-2372-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang YN, Ding WQ, Guo XJ, Yuan XW, Wang DM, Song JG. Epigenetic regulation of Smad2 and Smad3 by profilin-2 promotes lung cancer growth and metastasis. Nat Commun. 2015;6:8230–8239. doi: 10.1038/ncomms9230. [DOI] [PubMed] [Google Scholar]
- 29.Tsai JH, Yang J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev. 2013;27:2192–2206. doi: 10.1101/gad.225334.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savagner P. Epithelial-mesenchymal transitions: From cell plasticity to concept elasticity. Curr Top Dev Biol. 2015;112:273–300. doi: 10.1016/bs.ctdb.2014.11.021. [DOI] [PubMed] [Google Scholar]
- 31.Medici D, Hay ED, Olsen BR. Snail and Slug promote epithelial-mesenchymal transition through beta-catenin-T-cell factor-4-dependent expression of transforming growth factor-beta3. Mol Biol Cell. 2008;19:4875–4887. doi: 10.1091/mbc.e08-05-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiao Y, Chen J, Lim YB, Finch-Edmondson ML, Seshachalam VP, Qin L, Jiang T, Low BC, Singh H, Lim CT, et al. YAP regulates actin dynamics through ARHGAP29 and promotes metastasis. Cell Reports. 2017;19:1495–1502. doi: 10.1016/j.celrep.2017.04.075. [DOI] [PubMed] [Google Scholar]
- 33.Wieczorek K, Wiktorska M, Sacewicz-Hofman I, Boncela J, Lewiński A, Kowalska MA, Niewiarowska J. Filamin A upregulation correlates with Snail-induced epithelial to mesenchymal transition (EMT) and cell adhesion but its inhibition increases the migration of colon adenocarcinoma HT29 cells. Exp Cell Res. 2017;359:163–170. doi: 10.1016/j.yexcr.2017.07.035. [DOI] [PubMed] [Google Scholar]
- 34.Izdebska M, Gagat M, Grzanka A. Overexpression of lamin B1 induces mitotic catastrophe in colon cancer LoVo cells and is associated with worse clinical outcomes. Int J Oncol. 2018;52:89–102. doi: 10.3892/ijo.2017.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this article.