Abstract
Evidence has shown that microRNAs (miRNAs) are vital in cell growth, migration, and invasion by inhibiting their target genes. A previous study demonstrated that miRNA (miR)-155-3p and miR-155-5p exerted opposite effects on cell proliferation, apoptosis, migration and invasion in breast cancer cell lines. An miRNA microarray was used to show that miR-155-3p was downregulated whereas miR-155-5p was upregulated in paclitaxel-resistant (PR) cells compared with parental breast cancer cells. However, the role of miR-155 in breast cancer cell invasion and metastasis remains to be elucidated. A 21-residue peptide derived from the viral macrophage inflammatory protein II (NT21MP), competes with the ligand of CXC chemokine receptor 4 (CXCR4) and its ligand stromal cell-derived factor-1α, inducing cell apoptosis in breast cancer. The present study aimed to identify the underlying mechanism of action of miR-155-3p/5p and NT21MP in PR breast cancer cells. Quantitative polymerase chain reaction, western blotting, wound-healing, cell cycle and apoptosis assays, and Cell Counting kit-8 assay were used to achieve this goal. The combined overexpression of miR-155-3p with NT21MP decreased the migration and invasion ability and increased the number of apoptotic and arrested cells in the G0/G1 phase transition in vitro. The knockdown of miR-155-5p combined with NT21MP had a similar effect on PR breast cancer cells. Furthermore, the ectopic expression of their target gene myeloid differentiation primary response gene 88 (MYD88) or tumor protein 53-induced nuclear protein 1 (TP53INP1) combined with NT21MP enhanced the sensitivity of the breast cancer cells to paclitaxel. Taken together, these findings suggested that miR-155-3p/5p and their target genes MYD88 and TP53INP1 may serve as novel biomarkers for NT21MP therapy through the CXCR4 pathway for improving sensitivity to paclitaxel in breast cancer.
Keywords: breast cancer, drug resistance, 21-residue N-terminal of viral macrophage inflammatory protein II, microRNA-155-3p, microRNA-155-5p, MYD88, TP53INP1
Introduction
Breast cancer is one of the most frequently diagnosed types of cancer and is the leading cause of mortality in Western women (1). However, the specific developmental mechanisms of this cancer remain to be fully elucidated (2). Chemotherapy is a basic strategy for treating breast cancer (3,4), however, this approach has several limitations, including limited drug delivery due to the blood-brain barrier and drug resistance (5,6). Paclitaxel, as one of the most effective chemotherapeutic drugs, is used to treat breast cancer (7,8), however, its failure in treatment and poor prognosis are mainly due to the development of drug resistance (9).
In previous years, small non-coding RNAs, known as microRNAs (miRNAs), have appeared as a pivotal regulators in human tumorigenesis, including breast cancer (10,11). miRNAs bind to the 3′-untranslated regions (3′-UTRs) of the target gene, thus inhibiting expression of the target gene at the transcriptional or posttranscriptional level. Dysregulated miRNAs have significant roles in tumor occurrence (12–14). Additionally, miRNA (miR)-3p and miR-5p, which are processed from the 5′ and 3′ precursors of pre-miRNA, have been identified to be involved in different regulatory loops and exert the same or different effects on tumors (15,16). For example, miR-409-3p/-5p have an oncogenic effect on prostate cancer bone metastasis and can serve as a therapeutic target (17). Deep sequencing revealed that the opposite strands of miR-144-5p/-3p, miR-145-5p/-3p, and miR-139-5p/-3p can function as dual-strand tumor-suppressor miRNAs (18–20). Sakaguchi et al also found that the miR-199 family (miR-199a-3p/5p and miR-199b-3p/5p) may function as tumor suppressors by regulating common the target gene integrin α3 (21). Despite this, miR-3p and miR-5p can have opposite effects on carcinogenesis. For example, a previous study showed that mature miR-96-5p was significantly upregulated in cirrhosis and dysplastic nodules in hepatocellular carcinoma, whereas the expression of passenger strand miR-96-3p was detectable in cirrhosis and dysplastic nodules (22). Based on the previous studies, the miR-155 family was found to be involved in the regulation of corresponding biological activity in breast cancer. miR-155-3p was found to be downregulated whereas miR-155-5p acted as an oncogenic gene in breast cancer cell lines. However, the mechanisms involving 21-residue N-terminal of viral macrophage inflammatory protein II (vMIP-II), termed NT21MP, and the miR-155 family remains to be fully elucidated.
Previous studies have demonstrated that NT21MP, derived from vMIP-II, efficiently inhibits proliferation, invasion, cell cycle, and apoptosis in breast cancer cells by inhibiting CXC chemokine receptor 4 (CXCR4) and its ligand stromal cell-derived factor-1α (SDF-1α; also known as CXCL12) in vitro and in vivo (23–25). Although NT21MP has been shown to reverse breast cancer, the underlying specific molecular mechanism requires further investigation.
The present study aimed to determine whether the miR-155 family can be regulated using NT21MP in breast cancer cells and whether the overexpression of miR-155-3p or downregulation of miR-155-5p combined with NT21MP can reverse paclitaxel-resistant (PR) breast cancer cells more than the single treatment group. In addition, by analyzing the respective target genes of miR-155-3p and miR-155-5p, the present study aimed to verify whether NT21MP combined with the downregulation of myeloid differentiation primary response gene 88 (MYD88), the target gene of miR-155-3p, or upregulation of tumor protein 53-induced nuclear protein 1 (TP53INP1), the target gene of miR-155-5p, can significantly inhibit carcinogenesis in vitro. The findings provided novel insight into the potential efficacy of NT21MP as an adjuvant chemotherapy for breast cancer through regulating the miRNA family.
Materials and methods
Cell culture
The MCF-7 human breast cancer cell line was obtained from the Shanghai Cell Institute of Chinese Academy of Science (Shanghai, China). The corresponding PR cells (MCF-7/PR) were treated with 25 µg/ml paclitaxel. All cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco, Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; HyClone; GE Healthcare Life Sciences, Logan, UT, USA) maintained at 37°C in a saturated humidity atmosphere containing 5% CO2.
Transfection
Cell transfections were performed using Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's protocol. Final concentrations of 50 nM miRNA mimics and 0.75 µg/ml plasmids were added into a 6-well plate with 2 ml of culture medium. MYD88 small interfering (si)RNA (si-MYD88) and pcDNA-TP53INP1 (GenePharma, Shanghai, China) were used for stable transfection. The pcDNA-TP53INP1 was constructed using G418 (200 µg/ml). The sequences for the siRNAs and RNA oligoribonucleotides were as follows: si-MYD88-1, 5′-CCCAUCAGAAGCGACUGAUTTAUCAGUCGCUUCU GAUGG GTT-3′; si-MYD88-2, 5′-GGCAACUGGAACAGAC AAATTUUUGUCUGUUCCAGUUGCCTT-3′; si-MYD88-3, 5′-GCCUGUCUCUGUUCUUGAATTUUCAAGAACAGAG ACAGGCTT-3′.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
The total RNA was isolated from breast cancer cells using TRIzol reagent according to the manufacturer's protocol (Invitrogen; Thermo Fisher Scientific, Inc.). cDNA was synthesized using the First-Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.). The qPCR analysis was performed using the SYBR premix Ex TaqII kit (Takara Biotechnology, Co., Ltd., Dalian, China) through an ABI 7500 fast real-time PCR system (Applied Systems, Thermo Fisher Scientific, Inc.) following the manufacturer's protocol. Additionally, glyceraldehyde 3-phosphate dehydrogenase was used as an internal control. The PCR procedure [SYBR Premix (2X), 10 µl; forward and reverse primer (10 µM), 0.8 µl; ROX reference dye II (50X), 0.4 µl; cDNA, 2 µl; sterile purified water, 6 µl; total volume, 20 µl) was performed under the following conditions: 95.0°C for 30 sec, followed by 40 cycles at 95.0°C for 15 sec, 57°C for 30 sec and 72°C for 34 sec, and a final extension step at 72°C for 5 min. Data were processed using the 2−∆∆Cq method (26). The corresponding primers are listed in Table I.
Table I.
Sequences of primers.
| Name | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| MYD88 | CTGCCTCCTCCTTTCGTTGTAG | GCTCTGCTGGTCCTTCTTAGTC |
| TP53INP1 | GACTTCATAGATACTTGCAC | ATTGGACATGACTCAAACTG |
| Bax | GGGGACGAACTGGACAGTAA | CAGTTGAAGTTGCCGTCAGA |
| Caspase-3 | ACAAATGGACCTGTTGACCTGA | ACACCACTGTCTGTCTCAATGC |
| Bcl-2 | ATGTGTGTGGAGAGCGTCAA | ACAGTTCCACAAAGGCATCC |
| GAPDH | CAGCCTCAAGATCATCAGCA | TGTGGTCATGAGTCCTTCCA |
MYD88, myeloid differentiation primary response gene 88; TP53INP1, tumor protein 53-induced nuclear protein 1; Bcl-2, B-cell lymphoma 2; Bax, Bcl-2-associated X protein; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Western blot analysis
Cell lysates were prepared in lysis buffer containing 150 mM NaCl, 50 mM Tris, 1% Triton X-100, 0.1% sodium dodecyl sulfate (SDS), 0.5% sodium deoxycholate, 0.02% sodium azide, 1 mM sodium vanadate, and protease inhibitors (10 µg/ml leupeptin, 10 µg/ml aprotinin, and 1 mM phenylmethylsulfonyl fluoride). The protein concentration was measured using a Bio-Rad protein assay. Equal quantities of proteins were subjected to 10% SDS-polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The membranes were blocked using phosphate-buffered saline (PBS) with 5% non-fat milk and incubated overnight at 4°C with primary antibodies. Following incubation with peroxidase-conjugated AffiniPure goat anti-mouse immunoglobulin G (IgG) or peroxidase-conjugated affinipure goat anti-rabbit IgG for 2 h at 37°C. The immune complexes were detected using an Enhanced Chemiluminescence Western Blotting kit (EMD Millipore, Billerica, MA, USA) and Bio-Rad Image-Lab software 5.2.1 (Bio-Rad Laboratories, Inc.). The relative protein expression was determined using ImageJ V1.8.0 (National Institutes of Health, Bethesda, MD, USA), with GAPDH used as the internal reference. The following antibodies and dilutions were used: MYD88 (1:2,000; cat. no. ab2068, Abcam, Cambridge, MA, USA), TP53INP1 (1:2,000; cat. no. ab154877, Abcam), B-cell lymphoma 2 (Bcl-2; 1:1,500; cat. no. ab196495, Abcam), caspase-3 (1:5,000; cat. no. ab13586, Abcam), Bcl-2-associated X protein (Bax; 1:1,000; cat. no. 23931-1-AP, ProteinTech Group, Inc., Chicago, IL, USA), β-actin (1:3,000; cat. no. sc-130065, Santa Cruz Biotechnology Co., Ltd., Dallas, TX, USA), goat anti- rabbit IgG-horseradish peroxidase (1:5,000; cat. no. sc-2004, Santa Cruz Biotechnology, Inc.), and goat anti-mouse IgG-horseradish peroxidase (1:5,000; cat. no. sc-2005, Santa Cruz Biotechnology, Inc.).
Wound-healing assay
The transfected breast cancer cells were seeded into 6-well plates and then wounded by scratching with a sterile 10-µl pipette tip. The detached cells were removed by washing twice with PBS, and fresh culture medium without serum was added. The wound closure was monitored at 0 and 24 h using a fluorescence microscope (×40 magnification; IX71; Olympus Corporation, Tokyo, Japan). The wound surface area was quantified by image analysis (Image J V1.8.0; National Institutes of Health).
Cell cycle and apoptosis assays
The cells were seeded into 6-well plates. Following transfection for 24 h, the cells were collected by trypsinization and then analyzed with a flow cytometer (Muse Cell Analyzer, Merck Millipore, Darmstadt, Germany) using an Annexin V and Dead Cell kit and a Cell Cycle Detection kit (Merck Millipore). All experiments were repeated three times.
Cell counting kit-8 (CCK-8) assay
The MCF-7 and MCF-7/PR cells were trypsinized and seeded into 96-well plates at 1×105 cells/ml. After 24 h, various concentrations of paclitaxel (0, 20, 40, 60, 80, and 100 µmol/l) were added, and 50 nmol/l miR-155-3p mimics/miR-155-5p inhibitor/NT21MP was transfected into each well and incubated at 37°C for 72 h. Subsequently, 10 µl of CCK-8 (Beyotime Institute of Biotechnology, Shanghai, China) was added to 100 µl of DMEM medium containing 10% FBS. The values of absorbance were measured at 450 nm.
Statistical analysis
All analyses were performed in triplicate. Statistical analysis was performed using one-way analysis of variance and the post hoc Least Significant Difference test with SPSS 16.0 software (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference. The data are reported as the mean ± standard deviation.
Results
NT21MP reverses the SDF-1α-induced decrease of miR-155-3p and increase of miR-155-5p in MCF-7 and MCF-7/PR cells
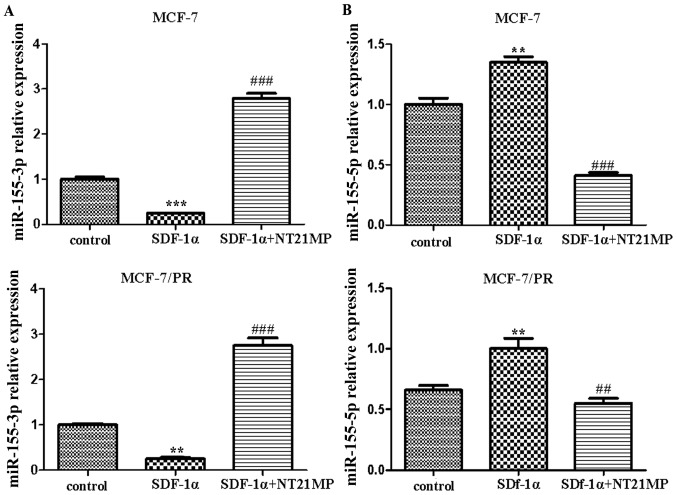
Previous studies have confirmed that NT21MP can inhibit SDF-1α-induced proliferation, migration and invasion, and promote apoptosis by downregulating the expression of CXCR4 in breast cancer cells. The results of the RT-qPCR analysis showed that NT21MP (1 µg/ml) inhibited the SDF-1α-induced decrease of miR-155-3p in the parental and PR cells compared with the control group (Fig. 1A). By contrast, SDF-1α (0.1 µg/ml) promoted the expression level of miR-155-5p, whereas NT21MP inhibited its effect (Fig. 1B). These results suggested that miR-155-3p/5p was involved in the regulation of NT21MP, not only in breast cancer parental cells, but also in drug-resistant cells.
Figure 1.
Effects of NT21MP on the expression level of miR-155-3p/5p in MCF-7 and MCF-7/PR cell lines. (A) Effects of NT21MP on the expression level of miR-155-3p in MCF-7 and MCF-7/PR cells, detected using RT-qPCR analysis, compared with control groups. (B) Effects of NT21MP on the expression level of miR-155-5p in MCF-7 and MCF-7/PR cells, detected using RT-qPCR analysis, compared with control groups. **P<0.01, ***P<0.001, ##P<0.01 and ###P<0.001, compared with SDF-1α treatment. NT21MP; 21-residue peptide derived from viral macrophage inflammatory protein II; miR, microRNA; SDF-1α, stromal cell-derived factor-1α; PR, paclitaxel-resistant; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
NT21MP inhibits the SDF-1α-induced increase of MYD88 and decrease of TP53INP1 in MCF-7 and MCF-7/PR cells
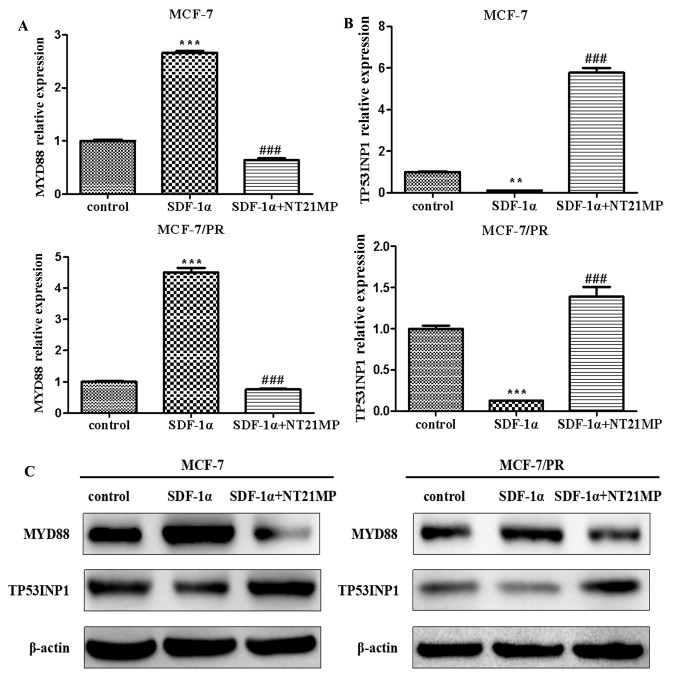
Previous studies have demonstrated that MYD88 functions as the target gene of miR-155-3p and TP53INP1 functions as the target gene of miR-155-5p using TargetScan v7.1, miRanda, and miRTarbase (27). The same experiments for miR-155-3p/5p were performed in the present study to further elucidate whether the targets of miR-155-3p/5p were also involved in the regulatory effect of NT21MP in drug resistance in breast cancer. The results showed that SDF-1α promoted the expression level of MYD88 whereas NT21MP suppressed this effect in the MCF-7 and MCF-7/PR cells (Fig. 2A). Additionally, NT21MP inhibited the SDF-1α-induced decrease of TP53INP1 (Fig. 2B). The corresponding protein levels are shown in Fig. 2C.
Figure 2.
Effects of NT21MP on the expression of MYD88 or TP53INP1 in MCF-7 and MCF-7/PR cells. (A) Effects of NT21MP on the expression of MYD88 using RT-qPCR analysis, compared with the control groups. (B) Effects of NT21MP on the expression of TP53INP1 using RT-PCR analysis, compared with the control groups. (C) Western blot analysis was performed to identify the effects of NT21MP on the expression of MYD88 or TP53INP1 in MCF-7 and MCF-7/PR cells, compared with control groups. The results are representative of three independent experiments. **P<0.01, ***P<0.001 and ###P<0.001, compared with SDF-1α treatment. NT21MP; 21-residue peptide derived from viral macrophage inflammatory protein II; SDF-1α, stromal cell-derived factor-1α; PR, paclitaxel-resistant; MYD88, myeloid differentiation primary response gene 88; TP53INP1, tumor protein 53-induced nuclear protein 1; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
NT21MP, combined with the overexpression of miR-155-3p, inhibits target gene MYD88 and biological activities in MCF-7/PR cells
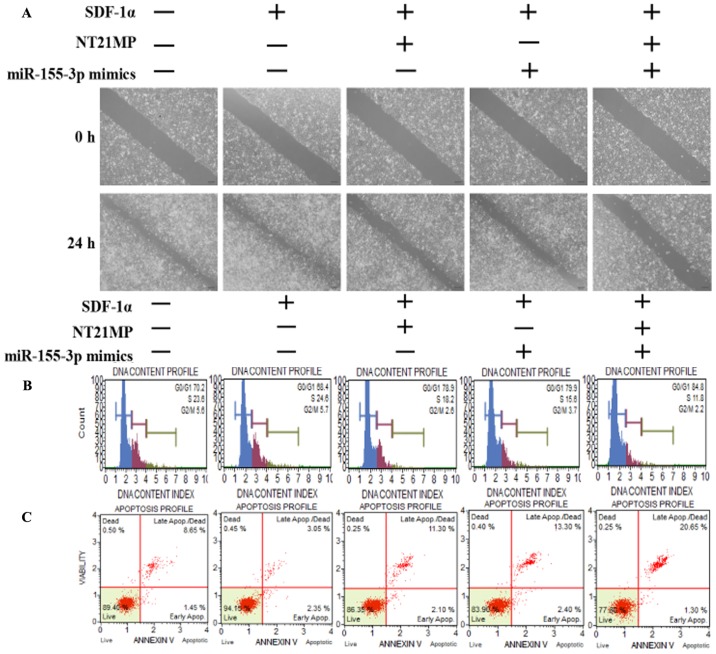
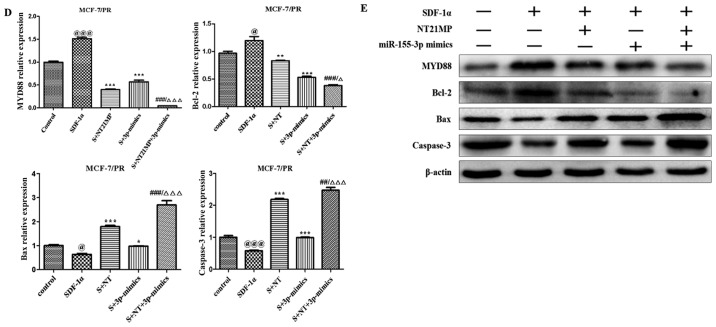
A wound-healing assay was performed to assess the ability of SDF-1α to promote cell migration and the ability of miR-155-3p or NT21MP to weaken this effect, particularly in the combined groups, in order to examine the molecular effect of NT21MP and the overexpression of miR-155-3p in breast cancer-resistant cells (Fig. 3A). Subsequent cell cycle and apoptotic analyses were performed using flow cytometry to detect the combined effect of NT21MP and miR-155-3p. The percentage of cells in the G0/G1 phase decreased from 70.2 to 68.4%, whereas the number of cells in the S phase increased from 23.6 to 24.6% following SDF-1α stimulation, suggesting that SDF-1α promoted cell transformation from the G0/G1 phase to the S phase. When the cells were treated with NT21MP or miR-155-3p mimics, an increased percentage of cells were arrested in the G0/G1 phase, from 68.4 to 78.9% in the NT21MP group, and from 68.4 to 79.9% in the miR-155-3p mimics group, particularly in the combined group (Fig. 3B). Additionally, the changes in apoptotic cells were in line with the cell cycle results (Fig. 3C). RT-qPCR and western blot analyses were used to analyze the mRNA and protein expression levels of cell cycle-related factors. As shown in Fig. 3D and E, SDF-1α upregulated the expression of target gene MYD88 and Bcl-2, and downregu-lated the expression of apoptotic genes Bax and caspase-3. NT21MP inhibited the anti-apoptotic effects of SDF-1α, indicating that NT21MP induced breast cancer-resistant cell apoptosis. Furthermore, MYD88 was significantly attenuated in the NT21MP and overexpression of miR-155-3p treatment group compared with the univariate treatment group. Bcl-2 was also downregulated under the same treatment. By contrast, Bax and caspase-3 exhibited opposite expression trends. These results indicated that NT21MP and miR-155-3p inhibited the stimulatory effect of SDF-1α on the biological activity in PR cells.
Figure 3.
Biological effects of NT21MP and miR-155-3p mimics on PR breast cancer cells. (A) Effects of NT21MP and miR-155-3p mimics on cell migration and invasion were measured using a wound-healing assay. (B) Effects of NT21MP and miR-155-3p mimics on cell cycle were analyzed using PI staining and flow cytometry. (C) Effects of NT21MP and miR-155-3p mimics on cell apoptosis were evaluated using the Annexin V/PI staining and flow cytometry. (D) Reverse transcription-quantitative polymerase chain reaction analysis was used to detect the effects of NT21MP and miR-155-3p mimics on target gene mRNA level, cell cycle, and apoptosis-related factors. (E) Results of western blot analysis of the protein levels of target genes, cell cycle, and apoptosis-related factors in PR cells were consistent with the mRNA results. Data are presented as the mean ± standard deviation of three independent experiments. @P<0.05 and @@@P<0.001, compared with the control group; *P<0.05, **P<0.01 and ***P<0.001, compared with SDF-1α treatment; ##P<0.01 and ###P<0.001, compared with S + NT21MP treatment; ∆P<0.05 and ∆∆∆P<0.001, compared with S + 3p mimics treatment. NT21MP; 21-residue peptide derived from viral macrophage inflammatory protein II; miR, microRNA; S/SDF-1α, stromal cell-derived factor-1α; NT, NT21MP; PR, paclitaxel-resistant; MYD88, myeloid differentiation primary response gene 88; Bcl-2, B-cell lymphoma 2; Bax, Bcl-2-associated X protein; PI, propidium iodide.
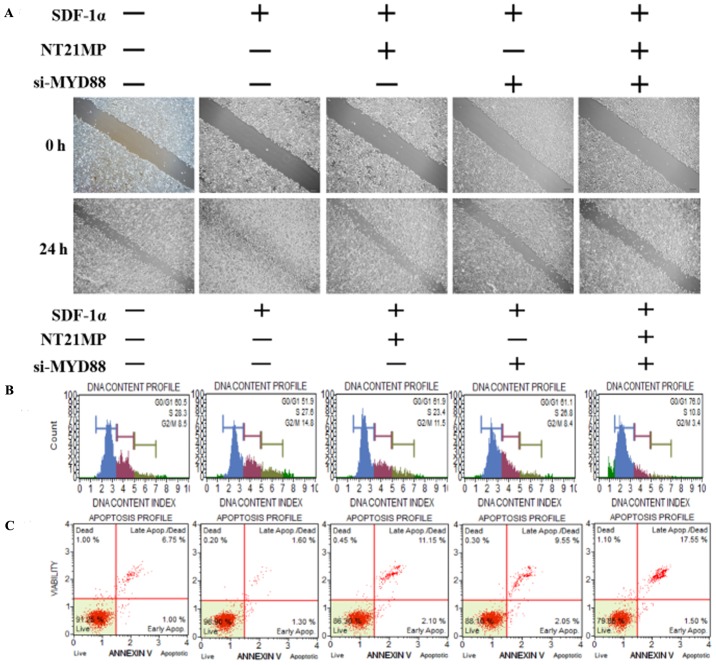
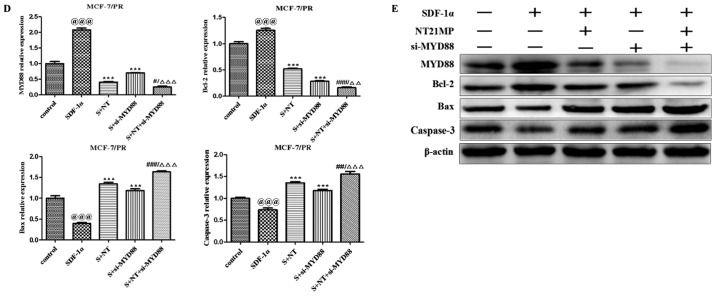
NT21MP, combined with the downregulation of MYD88, inhibited its biological effects in MCF-7/PR cells
The present study further examined the biological effect of MYD88, the target gene of miR-155-3p, when presented in NT21MP. The wound-healing assay also showed that the scratch wounds underwent slower closing following the knockdown of MYD88, particularly when combined with NT21MP (Fig. 4A). The cell cycle and apoptosis assays showed an increased percentage of cells in the G0/G1 phase but a decreased percentage of cells in the S phase. The number of apoptotic cells increased significantly in the si-MYD88 and NT21MP combined group (Fig. 4B and C). In addition, the results of the RT-q PCR and western blot analyses showed that the expression levels of MYD88 and Bcl-2 increased in the SDF-1α group but decreased in the NT21MP and/or downregulation of MYD88 groups. The expression levels of Bax and caspase-3 were decreased in the SDF-1α group but increased in the NT21MP and/or downregulation of MYD88 groups (Fig. 4D and E).
Figure 4.
Biological effects of NT21MP and si-MYD88 on PR cells. (A) Effects of NT21MP and si-MYD88 on cell migration and invasion were measured using a wound-healing assay. (B) Effects of NT21MP and si-MYD88 on cell cycle were analyzed using PI staining and flow cytometry. (C) Effects of NT21MP and si-MYD88 on cell apoptosis were evaluated using Annexin V/PI staining and flow cytometry. (D) Reverse transcription-quantitative polymerase chain reaction analysis was used to detect the effect of NT21MP and si-MYD88 on the mRNA levels of target genes, cell cycle, and apoptosis-related factors. (E) Western blot analysis results of the protein level of target gene, cell cycle, and apoptosis-related factors in PR cells were consistent with the mRNA results. The data are presented as the mean ± standard deviation of three independent experiments. @@@P<0.001, compared with the control group; ***P<0.001, compared with SDF-1α treatment; #P<0.05, ##P<0.01 and ###P<0.001, compared with S + NT21MP treatment; ∆∆P<0.01 and ∆∆∆P<0.001, compared with S + si-MYD88 treatment. NT21MP; 21-residue peptide derived from viral macrophage inflammatory protein II; S/SDF-1α, stromal cell-derived factor-1α; NT, NT21MP; PR, paclitaxel-resistant; MYD88, myeloid differentiation primary response gene 88; Bcl-2, B-cell lymphoma 2; Bax, Bcl-2-associated X protein; PI, propidium iodide; si, small interfering RNA.
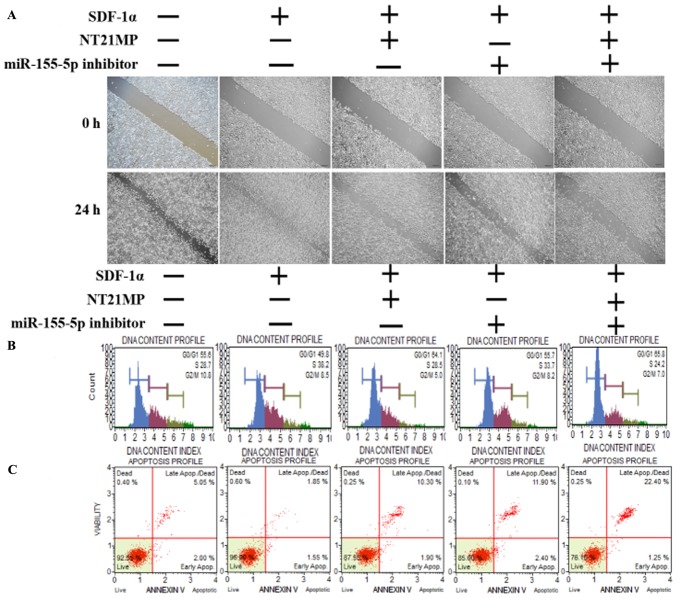
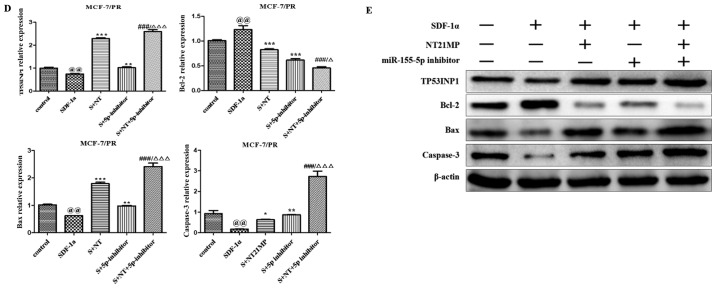
NT21MP, combined with the downregulation of miR-155-5p, induces the expression of its target gene TP53INP1 and biological effects in PR cells
The same functional experiments as in miR-155-3p were performed to examine the association between miR-155-5p and NT21MP in breast cancer. The SDF-1α group underwent faster closing of scratch wounds compared with the negative control group. The NT21MP and miR-155-5p inhibitor groups showed slower closing of scratch wounds compared with the SDF-1α group, particularly in the combination groups (Fig. 5A). The results of cell cycle analysis revealed a decreased number of cells in the G0/G1 phase but an increased number of cells in the S phase when transfected with NT21MP or downregulation of miR-155-5p, compared with the SDF-1α treatment group, and this was more marked in the combination group (Fig. 5B). The ratio of apoptotic cells decreased with SDF-1α treatment but increased with NT21MP or miR-155-5p inhibitor; it also decreased significantly in the combined group (Fig. 5C). The RT-qPCR and western blot analyses verified that the expression level of target gene TP53INP1 decreased with SDF-1α treatment, but increased significantly with NT21MP or downregulation of miR-155-5p (Fig. 5D and E). The expression level of Bcl-2 increased following SDF-1α treatment but decreased following NT21MP treatment or the downregulation of miR-155-5p, whereas Bax and caspase-3 exhibited the opposite results. These results suggested that NT21MP had a more marked inhibitory effect in reversing PR cells when combined with the expression of miR-155-5p.
Figure 5.
Biological effects of NT21MP and miR-155-5p inhibitor on PR cells. (A) Effects of NT21MP and miR-155-5p inhibitor on cell migration and invasion were measured using the wound-healing assay. (B) Effects of NT21MP and miR-155-5p inhibitor on cell cycle were analyzed using PI staining and flow cytom-etry. (C) Effects of NT21MP and miR-155-5p inhibitor on cell apoptosis were evaluated using the Annexin V/PI staining and flow cytometry. (D) Reverse transcription-quantitative polymerase chain reaction analysis was used to detect the effect of NT21MP and miR-155-5p inhibitor on the mRNA levels of target gene, cell cycle, and apoptosis-related factors. (E) Western blot analysis results of the protein levels of target genes, cell cycle, and apoptosis-related factors in PR cells were consistent with the mRNA results. Data are presented as the mean ± standard deviation of three independent experiments. @@P<0.01, compared with the control group; *P<0.05, **P<0.01 and ***P<0.001, compared with SDF-1α treatment; ###P<0.001, compared with S + NT21MP treatment; ∆P<0.05 and ∆∆∆P<0.001, compared with S + 5p inhibitor treatment. NT21MP; 21-residue peptide derived from viral macrophage inflammatory protein II; miR, microRNA; S/SDF-1α, stromal cell-derived factor-1α; NT, NT21MP; PR, paclitaxel-resistant; Bcl-2, B-cell lymphoma 2; Bax, Bcl-2-associated X protein; TP53INP1, tumor protein 53-induced nuclear protein 1; PI, propidium iodide.
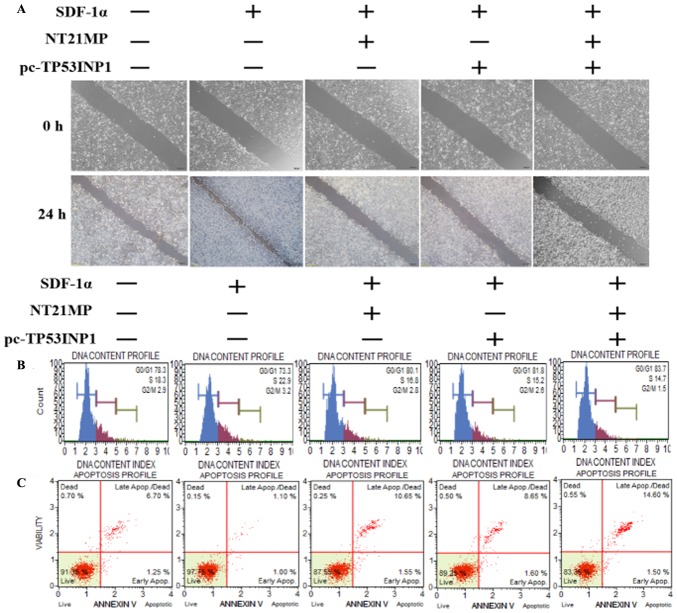
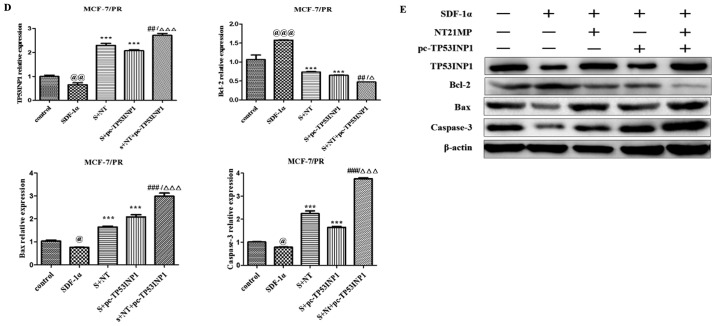
NT21MP, combined with the upregulation of TP53INP1, inhibits its biological effects in MCF-7/PR cells
As shown in Fig. 6A, transfection with NT21MP and/or pC-TP53INP1 led to slower closure of wounds. Changes in cell cycle and cell apoptosis showed that the number of cells in the G0/G1 phase increased but the number of cells in the S phase decreased. In addition, the percentage of apoptotic cells increased with NT21MP and/or pC-TP53INP1 treatment (Fig. 6B and C). The RT-qPCR and western blot analyses showed that the expression levels of TP53INP1, Bax, and caspase-3 increased with NT21MP treatment and/or the overexpression of TP53INP1, whereas the expression level of Bcl-2 showed the opposite effect (Fig. 6D and E). Therefore, these results indicated that miR-155-3p/5p was important in reversing PR, and its target gene had a regulatory effect when combined with NT21MP.
Figure 6.
Biological effects of NT21MP and pc-TP53INP1 on PR cells. (A) Effects of NT21MP and pc-TP53INP1 on cell migration and invasion were measured using the wound-healing assay. (B) Effects of NT21MP and pc-TP53INP1 on cell cycle were analyzed using propidium iodide (PI) staining and flow cytometry. (C) Effects of NT21MP and pc-TP53INP1 on cell apoptosis were evaluated using Annexin V/PI staining and flow cytometry. (D) Reverse transcription-quantitative polymerase chain reaction analysis was used to detect the effect of NT21MP and pc-TP53INP1 on the mRNA levels of target genes, cell cycle, and apoptosis-related factors. (E) Western blot analysis results of the protein levels of target genes, cell cycle, and apoptosis-related factors in PR cells were consistent with the mRNA results. Data are presented as the mean ± standard deviation of three independent experiments. @P<0.05, @@P<0.01 and @@@P<0.001, compared with the control group; ***P<0.001, compared with SDF-1α treatment; ##P<0.01 and ###P<0.001, compared with S + NT21MP treatment; ∆P<0.05 and ∆∆∆P<0.001, compared with S + pc-TP53INP1 treatment. NT21MP; 21-residue peptide derived from viral macrophage inflammatory protein II; S/SDF-1α, stromal cell-derived factor-1α; NT, NT21MP; PR, paclitaxel-resistant; Bcl-2, B-cell lymphoma 2; Bax, Bcl-2-associated X protein; TP53INP1, tumor protein 53-induced nuclear protein 1; PI, propidium iodide.
NT21MP, combined with miR-155-3p/5p, promotes PR cell sensitivity in breast cancer
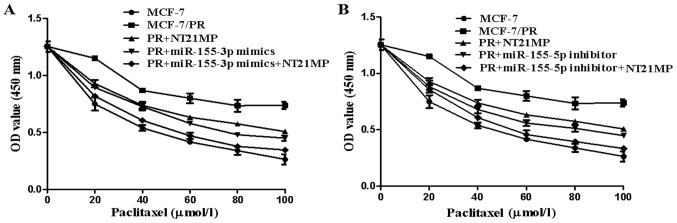
The CCK-8 assay was used to measure the proliferation of cells incubated with paclitaxel (0, 20, 40, 60, 80, and 100 µmol/l) for 72 h to clarify the efficiency of NT21MP in reversing drug resistance. The results showed that NT21MP/miR-155-3p mimics/miR-155-5p inhibitor inhibited the proliferation of PR cells (Fig. 7A and B, P<0.05). The efficiency of NT21MP in reversing drug resistance was found to be more marked when combined with the miR-155-3p mimics or miR-155-5p inhibitor.
Figure 7.
Antiproliferative effects of NT21MP and miR-155-3p/5p on human breast cancer MCF-7 cells. (A) Antiproliferative effects of NT21MP and miR-155-3p mimics on human breast cancer MCF-7 cells. (B) Antiproliferative effects of NT21MP and miR-155-5p inhibitor on human breast cancer MCF-7 cells. The cells were treated with various concentrations of paclitaxel for 72 h, and then analyzed for viability using a Cell Counting kit-8 assay. Absorbance at 450 nm is presented as the mean ± standard deviation. The results are representative of three independent experiments performed in triplicate. NT21MP; 21-residue peptide derived from viral macrophage inflammatory protein II; PR, paclitaxel-resistant; miR, microRNA.
Discussion
Chemotherapy is currently widely used in breast cancer treatment (28). Despite its impressive therapeutic effect, drug resistance has become a hurdle in several clinical cases (29,30). miRNAs have emerged as pivotal regulators of tumorigen-esis, particularly in drug resistance of breast cancer (31–33). However, the molecular mechanism of action of miRNAs in regulating drug resistance remains to be fully elucidated. The present study focused on the mature miR-155 family (miR-155-3p and miR-155-5p), which are expressed abnormally in PR breast cancer cells. A previous study showed that the overexpression of miR-155-3p enhanced cell proliferation and tumorigenesis by inhibiting the expression of F-Box and WD repeat domain containing 7 in hepatocellular carcinoma (34). Yim et al showed that the upregulation of miR-155-3p led to an increased number of sub-G1 apoptotic cells and reduced cellular viability, suggesting its tumor-suppressive effects (35). However, investigations on miR-155-3p in terms of the development of breast cancer or drug resistance have not been performed. miR-155-5p has also been shown to be upregulated in triple-negative breast cancer (36) and involved in cell progression and the epithelial-mesenchymal transition process sponged by myocardial infarction-associated transcript (37). The present study demonstrated that not only miR-155-3p, a tumor-suppressor gene, but also miR-155-5p, an oncogenic gene, were involved in the biological activities of PR breast cancer, including cell invasion, cell cycle and cell apoptosis.
Previous studies have shown that these two miRNAs have opposite effects in breast cancer cells and predicted that their target genes were MYD88 and TP53INP1, using bioinformatics software and dual-luciferase assays, respectively. MYD88 was previously reported to be pivotal in anticancer treatment via activation of the Toll-like receptor-mediated MYD88 signaling pathway in tumor biology, providing a novel potential target for cancer immunotherapy (38–42). Studies have reported that MYD88 is involved in the regulation of drug resistance in breast cancer cells (43,44). For example, using antibody microarrays, MYD88 was found to be differentially expressed as a predictive biomarker of neoadjuvant chemotherapy resistance in breast cancer (45). Xiang et al suggested that the downregulation of MYD88 reduced the proliferation, migration and invasion of breast cancer cells, and increased tumor cell sensitivity to paclitaxel treatment through inhibiting the activation of nuclear factor-κB via the phosphoinositide 3-kinase/Akt pathway (46). TP53INP1, which is located on the chromosome 8q22, acts as a tumor suppressor involved in breast cancer cell proliferation and apoptotic activity, negatively regulated by miR-155 (47,48). Dysregulation on the 3′-UTR of this gene regulates competitive endogenous messenger RNA-mediated migration and invasion and is correlated with drug resistance (49,50). In line with these findings, the present study suggested that MYD88, the target gene of miR-155-3p, and TP53INP1, the target gene of miR-155-5p, were also crucial in promoting and suppressing the biological effects in PR breast tumor cells. Interference of MYD88 or restoration of TP53INP1 enhanced the effects more markedly compared with the single treatment group, which was mediated by NT21MP.
NT21MP, a synthetic 21-mer peptide antagonist of CXCR4, derived from vMIP-II, was previously identified to inhibit cancer growth and metastasis by competitively binding SDF-1α to CXCR4 (23–25,51). The present study verified that SDF-1α inhibited the expression of miR-155-3p and enhanced the level of miR-155-5p, whereas these effects were reversed with NT21MP in MCF-7 parental and PR cells. This indicated that NT21MP acted as a potential antagonist attenuating the effect of the SDF-1α/CXCR4 axis. The combined use of NT21MP with miR-155-3p mimics, miR-155-5p inhibitor, si-MYD88, and pc-TP53INP1 contributed to decreases of cell proliferation, migration, invasion, cell cycle, and apoptosis, compared with that in MCF-7 PR cells. These changes suggested that NT21MP may reverse breast cancer drug resistance and improve the efficacy of paclitaxel through regulating miR-155-3p/5p or MYD88/TP53INP1.
In conclusion, the present study elucidated that, not only miR-155-3p/5p, but also their target genes MYD88/TP53INP1 are involved in the molecular regulation of PR breast cancer cells. Their combination with NT21MP significantly improved the sensitivity to paclitaxel, thus providing a novel clinical therapeutic strategy for the majority of women with breast cancer. However, the specific mechanism underlying how NT21MP regulates the potential miRNAs or the corresponding targets to reverse drug resistance clinically requires further investigation.
Acknowledgments
Not applicable.
Funding
This study was supported by the Major Program of Anhui Educational Committee (grant nos. KJ2015ZD29 and KJ2016SD37), the Key Program of College Discipline (major) Top-Notch Talent Academic Subsidiary of Anhui (grant no. gxbjZD2016069), the Academic And Technical Leaders Backup Candidate Research Funding Projects of Anhui (grant no. 2017H110), the Natural Science Foundation of Anhui province (grant no.1508085MH159), the Key Program of Anhui Educational Committee (grant no. KJ2016A474), the Bengbu Municipal Scientific Research Key Projects (grant no. 20150309), and the Program For Graduate Research of Bengbu Medical College (grant no. Byycx1740).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
LYZ, CJC and QLY designed the research; YYW, LY, LYZ and YL organized, analyzed and interpreted the data; HNX, YYW, WRW, TTC, YL, HFW, SLC and LY performed the experiments. QLY, WRW, CJC and YYW drafted and revised the manuscript; WRW, CJC and QLY were responsible for supervision. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Weigelt B, Peterse JL, van 't Veer LJ. Breast cancer metastasis: Markers and models. Nat Rev Cancer. 2005;5:591–602. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- 2.Li Y, Jin K, van Pelt GW, van Dam H, Yu X, Mesker WE, Ten Dijke P, Zhou F, Zhang L. c-Myb enhances breast cancer invasion and metastasis through the Wnt/β-catenin/Axin2 pathway. Cancer Res. 2016;76:3364–3375. doi: 10.1158/0008-5472.CAN-15-2302. [DOI] [PubMed] [Google Scholar]
- 3.Hackshaw A, Roughton M, Forsyth S, Monson K, Reczko K, Sainsbury R, Baum M. Long-term benefits of 5 years of tamoxifen: 10-year follow-up of a large randomized trial in women at least 50 years of age with early breast cancer. J Clin Oncol. 2011;29:1657–1663. doi: 10.1200/JCO.2010.32.2933. [DOI] [PubMed] [Google Scholar]
- 4.Trail PA, Dubowchik GM, Lowinger TB. Antibody drug conjugates for treatment of breast cancer: Novel targets and diverse approaches in ADC design. Pharmacol Ther. 2018;181:126–142. doi: 10.1016/j.pharmthera.2017.07.013. [DOI] [PubMed] [Google Scholar]
- 5.Phuong NT, Lim SC, Kim YM, Kang KW. Aromatase induction in tamoxifen-resistant breast cancer: Role of phosphoinositide 3-kinase-dependent CREB activation. Cancer Lett. 2014;351:91–99. doi: 10.1016/j.canlet.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Kim YJ, Sung D, Oh E, Cho Y, Cho TM, Farrand L, Seo JH, Kim JY. Flubendazole overcomes trastuzumab resistance by targeting cancer stem-like properties and HER2 signaling in HER2-positive breast cancer. Cancer Lett. 2018;412:118–130. doi: 10.1016/j.canlet.2017.10.020. [DOI] [PubMed] [Google Scholar]
- 7.Vuylsteke P, Huizing M, Petrakova K, Roylance R, Laing R, Chan S, Abell F, Gendreau S, Rooney I, Apt D, et al. Pictilisib PI3Kinase inhibitor (a phosphatidylinositol 3-kinase [PI3K] inhibitor) plus paclitaxel for the treatment of hormone receptor-positive, HER2-negative, locally recurrent, or metastatic breast cancer: Interim analysis of the multicentre, placebo-controlled, phase II randomised PEGGY study. Ann Oncol. 2016;27:2059–2066. doi: 10.1093/annonc/mdw320. [DOI] [PubMed] [Google Scholar]
- 8.Pellegrino B, Boggiani D, Tommasi C, Palli D, Musolino A. Nab-paclitaxel after docetaxel hypersensitivity reaction: Case report and literature review. Acta Biomed. 2017;88:329–333. doi: 10.23750/abm.v%vi%i.6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu T, Sun H, Liu S, Yang Z, Li L, Yao N, Cheng S, Dong X, Liang X, Chen C, et al. The suppression of DUSP5 expression correlates with paclitaxel resistance and poor prognosis in basal-like breast cancer. Int J Med Sci. 2018;15:738–747. doi: 10.7150/ijms.24981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 11.Zhang F, Wang B, Long H, Yu J, Li F, Hou H, Yang Q. Decreased miR-124-3p expression prompted breast cancer cell progression mainly by targeting Beclin-1. Clin Lab. 2016;62:1139–1145. doi: 10.7754/Clin.Lab.2015.151111. [DOI] [PubMed] [Google Scholar]
- 12.Han L, Liu B, Jiang L, Liu J, Han S. MicroRNA-497 downregulation contributes to cell proliferation, migration, and invasion of estrogen receptor alpha negative breast cancer by targeting estrogen-related receptor alpha. Tumour Biol. 2016;37:13205–13214. doi: 10.1007/s13277-016-5200-1. [DOI] [PubMed] [Google Scholar]
- 13.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 14.Samaeekia R, Adorno-Cruz V, Bockhorn J, Chang YF, Huang S, Prat A, Ha N, Kibria G, Huo D, Zheng H, et al. miR-206 inhibits stemness and metastasis of breast cancer by targeting MKL1/IL11 pathway. Clin Cancer Res. 2017;23:1091–1103. doi: 10.1158/1078-0432.CCR-16-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu T, Li J, Yan M, Liu L, Lin H, Zhao F, Sun L, Zhang Y, Cui Y, Zhang F, et al. MicroRNA-193a-3p and -5p suppress the metastasis of human non-small-cell lung cancer by down-regulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway. Oncogene. 2015;34:413–423. doi: 10.1038/onc.2013.574. [DOI] [PubMed] [Google Scholar]
- 16.Zhou H, Huang X, Cui H, Luo X, Tang Y, Chen S, Wu L, Shen N. miR-155 and its star-form partner miR-155* cooperatively regulate type I interferon production by human plasmacytoid dendritic cells. Blood. 2010;116:5885–5894. doi: 10.1182/blood-2010-04-280156. [DOI] [PubMed] [Google Scholar]
- 17.Josson S, Gururajan M, Hu P, Shao C, Chu GY, Zhau HE, Liu C, Lao K, Lu CL, Lu YT, et al. miR-409-3p/-5p promotes tumorigenesis, epithelial-to-mesenchymal transition, and bone metastasis of human prostate cancer. Clin Cancer Res. 2014;20:4636–4646. doi: 10.1158/1078-0432.CCR-14-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsushita R, Seki N, Chiyomaru T, Inoguchi S, Ishihara T, Goto Y, Nishikawa R, Mataki H, Tatarano S, Itesako T, et al. Tumour-suppressive microRNA-144-5p directly targets CCNE1/2 as potential prognostic markers in bladder cancer. Br J Cancer. 2015;113:282–289. doi: 10.1038/bjc.2015.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsushita R, Yoshino H, Enokida H, Goto Y, Miyamoto K, Yonemori M, Inoguchi S, Nakagawa M, Seki N. Regulation of UHRF1 by dual-strand tumor-suppressor microRNA-145 (miR-145-5p and miR-145-3p): Inhibition of bladder cancer cell aggressiveness. Oncotarget. 2016;7:28460–28487. doi: 10.18632/oncotarget.8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yonemori M, Seki N, Yoshino H, Matsushita R, Miyamoto K, Nakagawa M, Enokida H. Dual tumor-suppressors miR-139-5p and miR-139-3p targeting matrix metalloprotease 11 in bladder cancer. Cancer Sci. 2016;107:1233–1242. doi: 10.1111/cas.13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakaguchi T, Yoshino H, Yonemori M, Miyamoto K, Sugita S, Matsushita R, Itesako T, Tatarano S, Nakagawa M, Enokida H. Regulation of ITGA3 by the dual-stranded microRNA-199 family as a potential prognostic marker in bladder cancer. Br J Cancer. 2017;116:1077–1087. doi: 10.1038/bjc.2017.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H, Xing AY, Ma RR, Wang YW, Liu YH, Gao P. Diagnostic value of miRNA-96-5p/3p in dysplastic nodules and well-differentiated small hepatocellular carcinoma. Hepatol Res. 2016;46:784–793. doi: 10.1111/hepr.12628. [DOI] [PubMed] [Google Scholar]
- 23.Yang Q, Zhang F, Ding Y, Huang J, Chen S, Wu Q, Wang Z, Wang Z, Chen C. Antitumour activity of the recombination polypeptide GST-NT21MP is mediated by inhibition of CXCR4 pathway in breast cancer. Br J Cancer. 2014;110:1288–1297. doi: 10.1038/bjc.2014.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Q, Wu H, Wang H, Li Y, Zhang L, Zhu L, Wang W, Zhou J, Fu Y, Chen S, et al. N-terminal polypeptide derived from vMIP-II exerts its antitumor activity by inhibiting the CXCR4 pathway in human glioma. Int J Oncol. 2017;50:1160–1174. doi: 10.3892/ijo.2017.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Wang H, Ding Y, Li Y, Chen S, Zhang L, Wu H, Zhou J, Duan K, Wang W, et al. N-peptide of vMIP-II reverses paclitaxel- resistance by regulating miRNA-335 in breast cancer. Int J Oncol. 2017;51:918–930. doi: 10.3892/ijo.2017.4076. [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Tang B, Xiao B, Liu Z, Li N, Zhu ED, Li BS, Xie QH, Zhuang Y, Zou QM, Mao XH. Identification of MyD88 as a novel target of miR-155, involved in negative regulation of Helicobacter pylori-induced inflammation. FEBS Lett. 2010;584:1481–1486. doi: 10.1016/j.febslet.2010.02.063. [DOI] [PubMed] [Google Scholar]
- 28.Rabanal C, Ruiz R, Neciosup S, Gomez H. Metronomic chemotherapy for non-metastatic triple negative breast cancer: Selection is the key. World J Clin Oncol. 2017;8:437–446. doi: 10.5306/wjco.v8.i6.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones SK, Merkel OM. Tackling breast cancer chemore-sistance with nano-formulated siRNA. Gene Ther. 2016;23:821–828. doi: 10.1038/gt.2016.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monteiro IP, Madureira P, de Vasconscelos A, Pozza DH, de Mello RA. Targeting HER family in HER2-positive metastatic breast cancer: Potential biomarkers and novel targeted therapies. Pharmacogenomics. 2015;16:257–271. doi: 10.2217/pgs.14.133. [DOI] [PubMed] [Google Scholar]
- 31.Mi Y, Zhang D, Jiang W, Weng J, Zhou C, Huang K, Tang H, Yu Y, Liu X, Cui W, et al. miR-181a-5p promotes the progression of gastric cancer via RASSF6-mediated MAPK signalling activation. Cancer Lett. 2017;389:11–22. doi: 10.1016/j.canlet.2016.12.033. [DOI] [PubMed] [Google Scholar]
- 32.Emmrich S, Engeland F, El-Khatib M, Henke K, Obulkasim A, Schöning J, Katsman-Kuipers JE, Michel Zwaan C, Pich A, Stary J, et al. miR-139-5p controls translation in myeloid leukemia through EIF4G2. Oncogene. 2016;35:1822–1831. doi: 10.1038/onc.2015.247. [DOI] [PubMed] [Google Scholar]
- 33.Bahrami A, Aledavood A, Anvari K, Hassanian SM, Maftouh M, Yaghobzade A, Salarzaee O, ShahidSales S, Avan A. The prognostic and therapeutic application of microRNAs in breast cancer: Tissue and circulating microRNAs. J Cell Physiol. 2018;233:774–786. doi: 10.1002/jcp.25813. [DOI] [PubMed] [Google Scholar]
- 34.Tang B, Lei B, Qi G, Liang X, Tang F, Yuan S, Wang Z, Yu S, He S. MicroRNA-155-3p promotes hepatocellular carcinoma formation by suppressing FBXW7 expression. J Exp Clin Cancer Res. 2016;35:93. doi: 10.1186/s13046-016-0371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yim RL, Wong KY, Kwong YL, Loong F, Leung CY, Chu R, Lam WW, Hui PK, Lai R, Chim CS. Methylation of miR-155-3p in mantle cell lymphoma and other non-Hodgkin's lymphomas. Oncotarget. 2014;5:9770–9782. doi: 10.18632/oncotarget.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ouyang M, Li Y, Ye S, Ma J, Lu L, Lv W, Chang G, Li X, Li Q, Wang S, et al. MicroRNA profiling implies new markers of chemoresistance of triple-negative breast cancer. PLoS One. 2014;9:e96228. doi: 10.1371/journal.pone.0096228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luan T, Zhang X, Wang S, Song Y, Zhou S, Lin J, An W, Yuan W, Yang Y, Cai H, et al. Long non-coding RNA MIAT promotes breast cancer progression and functions as ceRNA to regulate DUSP7 expression by sponging miR-155-5p. Oncotarget. 2017;8:76153–76164. doi: 10.18632/oncotarget.19190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hassan F, Islam S, Tumurkhuu G, Naiki Y, Koide N, Mori I, Yoshida T, Yokochi T. Intracellular expression of toll-like receptor 4 in neuroblastoma cells and their unresponsiveness to lipopolysaccharide. BMC Cancer. 2006;6:281. doi: 10.1186/1471-2407-6-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma FJ, Liu ZB, Hu X, Ling H, Li S, Wu J, Shao ZM. Prognostic value of myeloid differentiation primary response 88 and Toll-like receptor 4 in breast cancer patients. PLoS One. 2014;9:e111639. doi: 10.1371/journal.pone.0111639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chow A, Zhou W, Liu L, Fong MY, Champer J, Van Haute D, Chin AR, Ren X, Gugiu BG, Meng Z, et al. Macrophage immunomodulation by breast cancer-derived exosomes requires Toll-like receptor 2-mediated activation of NF-κB. Sci Rep. 2014;4:5750. doi: 10.1038/srep05750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi M, Yao Y, Han F, Li Y, Li Y. MAP1S controls breast cancer cell TLR5 signaling pathway and promotes TLR5 signaling-based tumor suppression. PLoS One. 2014;9:e86839. doi: 10.1371/journal.pone.0086839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou L, Liu Z, Wang Z, Yu S, Long T, Zhou X, Bao Y. Astragalus polysaccharides exerts immunomodulatory effects via TLR4-mediated MyD88-dependent signaling pathway in vitro and in vivo. Sci Rep. 2017;7:44822. doi: 10.1038/srep44822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ge X, Cao Z, Gu Y, Wang F, Li J, Han M, Xia W, Yu Z, Lyu P. PFKFB3 potentially contributes to paclitaxel resistance in breast cancer cells through TLR4 activation by stimulating lactate production. Cell Mol Biol (Noisy-le-grand) 2016;62:119–125. [PubMed] [Google Scholar]
- 44.Edwardson DW, Boudreau J, Mapletoft J, Lanner C, Kovala AT, Parissenti AM. Inflammatory cytokine production in tumor cells upon chemotherapy drug exposure or upon selection for drug resistance. PLoS One. 2017;12:e0183662. doi: 10.1371/journal.pone.0183662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hodgkinson VC, ELFadl D, Agarwal V, Garimella V, Russell C, Long ED, Fox JN, McManus PL, Mahapatra TK, Kneeshaw PJ, et al. Proteomic identification of predictive biomarkers of resistance to neoadjuvant chemotherapy in luminal breast cancer: A possible role for 14-3-3 theta/tau and tBID? J Proteomics. 2012;75:1276–1283. doi: 10.1016/j.jprot.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 46.Xiang F, Ni Z, Zhan Y, Kong Q, Xu J, Jiang J, Wu R, Kang X. Increased expression of MyD88 and association with paclitaxel resistance in breast cancer. Tumour Biol. 2016;37:6017–6025. doi: 10.1007/s13277-015-4436-5. [DOI] [PubMed] [Google Scholar]
- 47.Zhang C, Zhao J, Deng H. 17β-estradiol up-regulates miR-155 expression and reduces TP53INP1 expression in MCF-7 breast cancer cells. Mol Cell Biochem. 2013;379:201–211. doi: 10.1007/s11010-013-1642-6. [DOI] [PubMed] [Google Scholar]
- 48.Zhang CM, Zhao J, Deng HY. miR-155 promotes proliferation of human breast cancer MCF-7 cells through targeting tumor protein 53-induced nuclear protein 1. J Biomed Sci. 2013;20:79. doi: 10.1186/1423-0127-20-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng L, Li X, Chou J, Xiang C, Guo Q, Zhang Z, Guo X, Gao L, Xing Y, Xi T. StarD13 3′-untranslated region functions as a ceRNA for TP53INP1 in prohibiting migration and invasion of breast cancer cells by regulating miR-125b activity. Eur J Cell. 2018;97:23–31. doi: 10.1016/j.ejcb.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 50.Yamamoto Y, Yoshioka Y, Minoura K, Takahashi RU, Takeshita F, Taya T, Horii R, Fukuoka Y, Kato T, Kosaka N, et al. An integrative genomic analysis revealed the relevance of microRNA and gene expression for drug-resistance in human breast cancer cells. Mol Cancer. 2011;10:135. doi: 10.1186/1476-4598-10-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang QL, Zhang LY, Wang HF, Li Y, Wang YY, Chen TT, Dai MF, Wu HH, Chen SL, Wang WR, et al. The N-terminal polypeptide derived from viral macrophage inflammatory protein II reverses breast cancer epithelial-to-mesenchymal transition via a PDGFRα-dependent mechanism. Oncotarget. 2017;8:37448–37463. doi: 10.18632/oncotarget.16394. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.