Abstract
Crosstalk of signaling pathways play crucial roles in cell proliferation, cell differentiation, and cell fate determination for development. In the case of ventx1.1 in Xenopus embryos, both BMP-4/Smad-1 and FGF/Xbra signaling induce the expression of neural repressor ventx1.1. However, the details of how these two pathways interact and lead to neural inhibition by ventx1.1 remain largely unknown. In the present study, Xbra directly bound to the ventx1.1 promoter region and inhibited neurogenesis in a Ventx1.1-dependent manner. Furthermore, Smad-1 and Xbra physically interacted and regulated ventx1.1 transcription in a synergistic fashion. Xbra and Smad-1 interaction cooperatively enhanced the binding of an interacting partner within the ventx1.1 promoter and maximum cooperation was achieved in presence of intact DNA binding sites for both Smad-1 and Xbra. Collectively, BMP-4/Smad-1 and FGF/Xbra signal crosstalk cooperate to activate the transcription of neural repressor ventx1.1 in Xenopus embryos. This suggests that the crosstalk between BMP-4 and FGF signaling negatively regulates early neurogenesis by synergistic activation of ventx1.1 in Xenopus embryos.
Introduction
Bone morphogenetic protein-4 (BMP-4) signaling is actively involved in germ-layer specification, axis formation and cell differentiation in Xenopus1,2, and blocking of BMP-4 signaling is achieved via organizer-secreted BMP antagonists, including Chordin, Noggin, Follistatin, and Cerberus results in neural induction3. Previous study demonstrated that BMP-4/Smad-1 directly induces the ventx1.1 (PV.1, Xvent-1b) expression in Xenopus embryos and leads to ventral mesoderm and ectoderm formation4. Ventx1.1 is one of the homeobox transcriptional factors of Xvent family5 and locates to Ventx synteny in Xenopus6. Ventx synteny is evolutionary conserved in human, dog, chicken and Xenopus genomes, but it is absent in genomes of rat and mouse6. Studies have reported that ventx1.1 is an endogenous neural inhibitor in Xenopus, inhibiting the expression of organizer and early neural genes, including Chordin, Noggin, FoxD5a/b, gsc and Zic3 via its C-terminal5,7,8. Additionally, ectopic expression of ventx1.1 not only inhibits neural genes expression but also induces the expression of ventral genes, including wnt8 and xhox3 and results in the headless phenotype along with neural inhibition in Xenopus5,7,9. Previous studies have demonstrated that Sox2/3 and geminin regulates the proliferation and differentiation of neural progenitor cells10,11. The expressions for Sox2/3 and geminin are positively regulated by Wnt/b-catenin signaling while being negatively regulated by BMP-4 signaling in Xenopus embryos. Ectopic expression of ventx1/2 inhibits endogenous expression of the early neural genes, namely Sox2/3 and geminin in Xenopus embryos, causing the neural inhibition. Furthermore, knockdown of ventx1/2 triggers the expression of Sox2/3 and geminin in Xenopus embryos, resulting in neural induction.
FGF/Xbra signaling actively participates in cell fate determination and anterior-posterior (A-P) patterning of neural tissue12. FGF leads mesoderm formation through the activation of an autocatalytic loop (FGF/Ras/Xbra/AP-1) in Xenopus12,13. Xbra is a member of the T-box gene family and a downstream transcriptional activator of FGF signaling. Ectopic expression of Xbra triggers mesoderm formation and inhibits neurogenesis in Xenopus embryos. Our previous study demonstrated that the ectopic expression of dominant negative Xbra (DN-Xbra) inhibits ventx1.1 expression and triggers expression of chordin and neural marker genes, including Zic3, Otx2, and N-CAM in animal cap explants14. This study suggested that FGF/Xbra may negatively regulate neurogenesis in a Ventx1.1-dependent manner in Xenopus embryos. However, the detailed molecular mechanism of the Xbra-mediated transcriptional activation of ventx1.1 and its involvement in neural inhibition is not completely understood.
Recent studies documented that FGF signaling leads to both activation and inhibition in neural induction in Xenopus embryos14,15. FGF/MAPK enhanced neural induction by inhibiting BMP-4/Smad-1 signaling in vertebrates15. However, the FGF-mediated BMP-4/Smad-1 inhibition alone was not sufficient to trigger neurogenesis in Xenopus embryos3. Our previous study has demonstrated that the bFGF-treatment, where Xbra expresses (with it being a kind of a mesoderm tissue), induces the ventx1.1 expression in animal cap explants, resulting in neural inhibition14. The knockdown of ventx1.1 by ventx1.1 morpholino causes the neurogenesis by inducing the expression of early and late neural genes, including Zic3, NCAM, Otx2, Rx1 and HoxB9 in bFGF-treated ectodermal explants of Xenopus embryos, suggesting that the BMP-4/Smad-1 and FGF/Xbra may participate in a signaling crosstalk to inhibit neurogenesis in a Ventx1.1-dependent manner4,14. Recent studies have indicated that T-box transcriptional factors bind to the consensus sequence (A/G)(A/T)(A/T)NTN(A/G)CAC(C/T)T and positively regulate expression of genes, including eFGF, Bix4 and Bix116–18. Another study reported that phosphorylated C-terminal of Smad-1 physically interacts with the N-terminal containing HLL(S/N)AV(E/Q) motif of Xbra in Xenopus19. Additionally, the study suggested that Smad-1-Xbra interaction may induce the expression of ventral genes. However, the detailed molecular mechanism that allows BMP-4/Smad-1 and FGF/Xbra-mediated signaling crosstalk to inhibit neurogenesis in a Ventx1.1-dependent manner is largely unknown.
In the present study, we demonstrated that Xbra directly binds to Xbra response elements, ATCACACTT (XbRE, within −70 bp~−62 bp) upstream of the putative transcription initiation site (TSS) of ventx1.1 and positively induces ventx1.1 transcription. The results showed that Xbra and Smad-1 synergistically cooperate to regulate ventx1.1 transcription during Xenopus development. ChIP-PCR assays indicated that Smad-1 and Xbra directly bind to their respective consensus sequences within the proximal region of the endogenous ventx1.1 promoter. Additionally, Xbra and Smad-1 enhanced the binding of their respective interacting partners to the promoter region. We further observed that the Xbra and Smad-1 interaction displays target specificity in the Xvent family as Xbra and Smad-1 did not synergistically induce Xvent2 expression. Taken together, it is concluded that BMP-4/Smad-1 and FGF/Xbra signaling demonstrate a crosstalk for inhibition of neurogenesis in a Ventx1.1-dependent manner in Xenopus embryos. These results suggest that FGF/Xbra and BMP-4/Smad-1 are positively involved in the transcriptional activation of ventx1.1 and synergistically cooperate to inhibit neurogenesis in the ventral and lateral mesoderm in Xenopus embryos.
Results
Ectopic expression of Xbra inhibits neurogenesis in a Ventx1.1-dependent manner in animal cap explants of Xenopus
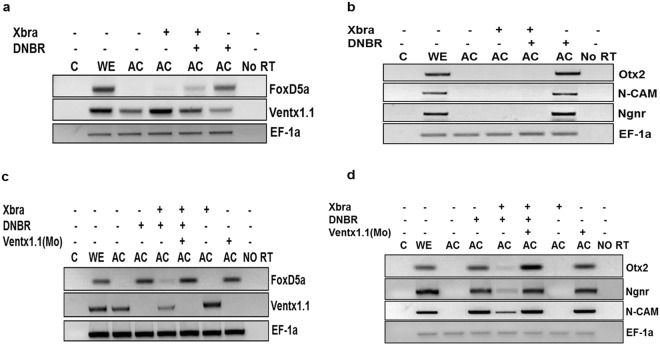
Previous studies demonstrated that both BMP-4/Smad-1 and FGF/Xbra signaling induce ventx1.1 expression which in turn negatively regulates neurogenesis in Xenopus embryos4,14. Studies have documented that Xbra is also induced by BMP-4 in Xenopus and human embryonic stem cells, leading to mesoderm differentiation20. Thus, we examined whether Xbra induces ventx1.1 expression under a BMP-4-inhibited condition and causes neural inhibition. Dominant-negative BMP-4 receptor (DNBR) mRNA was injected for BMP-4 inhibition. RT-PCR results showed Xbra induced ventx1.1 expression under the BMP-4-inhibited condition and suppressed early and late neural genes including FoxD5a, Ngnr, N-CAM and Otx2 in animal cap explants (Fig. 1a,b). The results suggested that Xbra positively regulates ventx1.1 expression in a BMP-inhibited condition and inhibits neurogenesis in animal cap explants of Xenopus embryos. However, it was still questionable whether Xbra-mediated neural inhibition is truly dependent on Ventx1.1. Thus, we performed a knockdown of ventx1.1 by ventx1.1 morpholino (MOs) and found that the knockdown of ventx1.1 strongly augments and recovers the expression of both early and late neural genes including FoxD5a, N-CAM, Ngnr, and Otx2, which were reduced by overexpression of Xbra (Fig. 1c,d). Moreover, ventx1.1 MOs specifically reduced ventx1.1 expression in both the absence and presence of Xbra. These results showed that Xbra negatively regulates neurogenesis in a Ventx1.1-dependent manner. Taken together, these results indicated that Xbra induces ventx1.1 expression in ventral and lateral mesodermal zones, leading to inhibition of expression of early neural genes and causing neural inhibition in part of the mesodermal region. In addition, the 5′-flanking region of ventx1.1 likely contains an Xbra response element (XbRE) which directly regulates ventx1.1 transcription. Moreover, ventx1.1 establishes signaling crosstalk between BMP-4/Smad-1 and FGF/Xbra in order to inhibit neurogenesis in Xenopus embryos.
Figure 1.
Ectopic expression of Xbra inhibits neurogenesis in a Ventx1.1-dependent manner in animal cap explants of Xenopus. Ventx1.1 MOs (1 ng) were injected either with or without DNBR and Xbra mRNA at the 1-cell stage and dissected the animal cap at the stage 8. The dissected animal caps were harvested until stages 11 and 24 in L-15 culture medium. The relative gene expressions were analyzed by RT-PCR. (a,b) Xbra increases ventx1.1 expression and reduces the expression of early and late neural genes, including that of FoxD5a, N-CAM, Ngnr, and Otx2 while DNBR reduces ventx1.1 expression and induces the expression of neural genes, FoxD5a, N-CAM, Ngnr, and Otx2. (c,d) ventx1.1 MOs reduces ventx1.1 expression and increases the expression of neural genes, including for FoxD5a, N-CAM, Ngnr, and Otx2. C: PCR reaction without adding cDNA while No RT means PCR reaction without added reverse transcriptase.
Xbra response element (XbRE) is identified within the proximal region of the ventx1.1 promoter
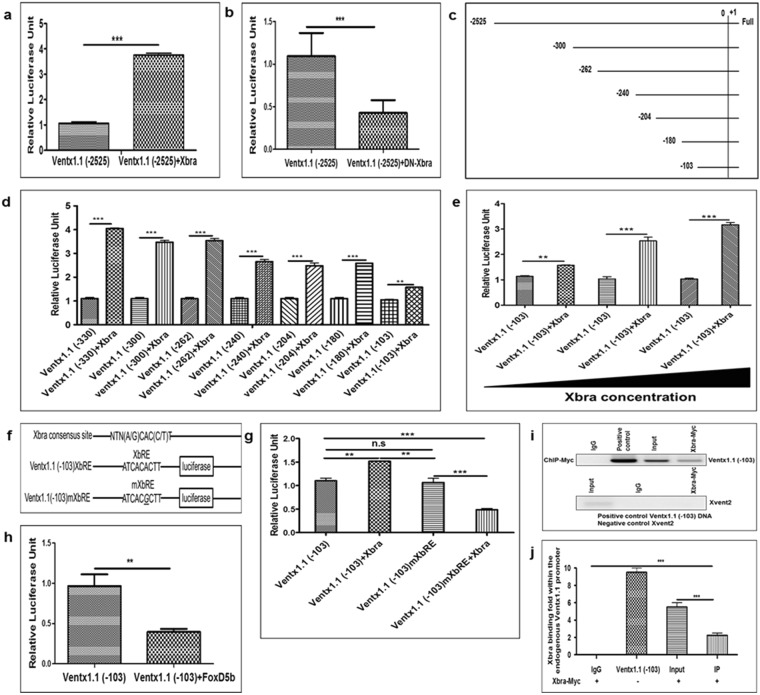
The results (Fig. 1) suggested that the ventx1.1 promoter region may contain a putative cis-acting XbRE which is required for triggering the ventx1.1 transcription. To evaluate the presence of XbRE within the ventx1.1 promoter region, we injected a ventx1.1 (−2525) promoter construct either with or without DN-Xbra, and Xbra, separately. These results showed that DN-Xbra decreased the relative promoter activity of ventx1.1 (−2525) up to 2.5-fold, while Xbra increased the relative promoter activity of ventx1.1 (−2525) up to 3.5-fold compared to that of ventx1.1 (−2525) construct alone (Fig. 2a,b). The results showed that Xbra positively regulates ventx1.1 transcription and the ventx1.1 promoter may contain a putative cis-acting XbRE. Therefore, we generated different serially-deleted ventx1.1 promoter constructs (Fig. 2c), which were co-injected either with or without Xbra, separately. The results showed that Xbra increases the relative promoter activity of serially-deleted ventx1.1 promoter constructs up to 1.5-4.0-fold compared to those of ventx1.1 promoter without Xbra (Fig. 2d). Moreover, Xbra increased the relative promoter activity of ventx1.1 (−103) in a dose-dependent manner (Fig. 2e). These results strongly provided evidence that the ventx1.1 (−103) promoter construct contained a cis-acting XbRE which induces ventx1.1 transcription in Xenopus embryos.
Figure 2.
Identification of Xbra response elements (XbRE) within the ventx1.1 promoter region. Different serially-deleted ventx1.1 promoter constructs were co-injected either with or without DN-Xbra or Xbra at the 1-cell stage and grown until stage 11 in 30% MMR to measure the relative promoter activity at the stage 11. (a) ventx1.1 (−2525) promoter injected either with or without DN-Xbra. (b) ventx1.1 (−2525) promoter injected either with or without Xbra. (c,d) Serially-deleted promoter constructs of ventx1.1 injected either with or without Xbra. (e) ventx1.1 (−103) promoter construct injected either with or without Xbra in a dose-dependent manner. (f) Putative Xbra binding consensus was mutated by site-directed mutagenesis within ventx1.1 (−103) promoter constructs. (g) XbRE-mutated ventx1.1 (−103) mXbRE and ventx1.1 (−103) promoter constructs injected either with or without Xbra. (h) ventx1.1 (−103) promoter construct injected either with or without FoxD5b. (i,j) A ChIP-PCR assay was performed with anti-Myc antibody during gastrula. All ChIP bindings were measured by PCR using specific primers. ventx1.1 (−103) promoter DNA was used as a positive control while Xvent2 coding region primers were used as a negative control for all the ChIP-PCR experiments. All the relative promoter activity data are shown as mean ± SE. The ChIP-PCR band intensities were quantified using a densitometer.
Previous studies have addressed the putative binding consensus sequence (A/G)(A/T)(A/T)NTN(A/G)CAC(C/T)T of T-box transcription factors21 within the promoter region of targeted genes, including eFGF, Bix4, and Bix116–18. Thus, we mapped the ventx1.1 (−103) promoter nucleotide sequence and found a putative XbRE, (ATCACACTT, within −70 bp~−62 bp), upstream of putative TSS of ventx1.1. We mutated one nucleotide (changed ATCACACTT to ATCACGCTT) within the putative XbRE in ventx1.1 (−103) promoter construct and generated XbRE-mutated ventx1.1 (−103) mXbRE (Fig. 2f). We then tested the reporter gene activities of ventx1.1 (−103) and ventx1.1 (−103) mXbRE promoter constructs either with or without Xbra. The results showed that Xbra-mediated transcriptional activation of ventx1.1 (−103) was abolished in ventx1.1 (−103) mXbRE (Fig. 2g). Xbra significantly reduced and did not increase the relative promoter activity of XbRE-mutated ventx1.1 (−103) promoter construct. We also tested whether the ventx1.1 (−103) promoter construct contains the negative response element of early neural protein FoxD5b. Result showed that FoxD5b significantly reduces the relative promoter activity of ventx1.1 (−103) (Fig. 2h).
The results firmly indicated that ventx1.1 (−103) promoter construct contains a cis-acting XbRE in the proximal region (−70 bp~−62 bp) of the ventx1.1 promoter. Previous studies have documented that Xbra directly binds within the promoter regions of Xvent family of genes for Xenopus and human22–24. A genome-wide ChIP-Seq study of T-box transcription factors in X. tropicalis has shown that Ventx family is one of the direct targets of T-box family transcription factors. We then performed ChIP-PCR assay and found that Xbra directly bound within the proximal region of the endogenous ventx1.1 promoter (Fig. 2i,j). These findings collectively concluded that ventx1.1 is one of the direct targets of Xbra and the direct binding of Xbra within the proximal promoter region increases transcription activity of ventx1.1.
Xbra and Smad-1 synergistically regulate ventx1.1 transcription
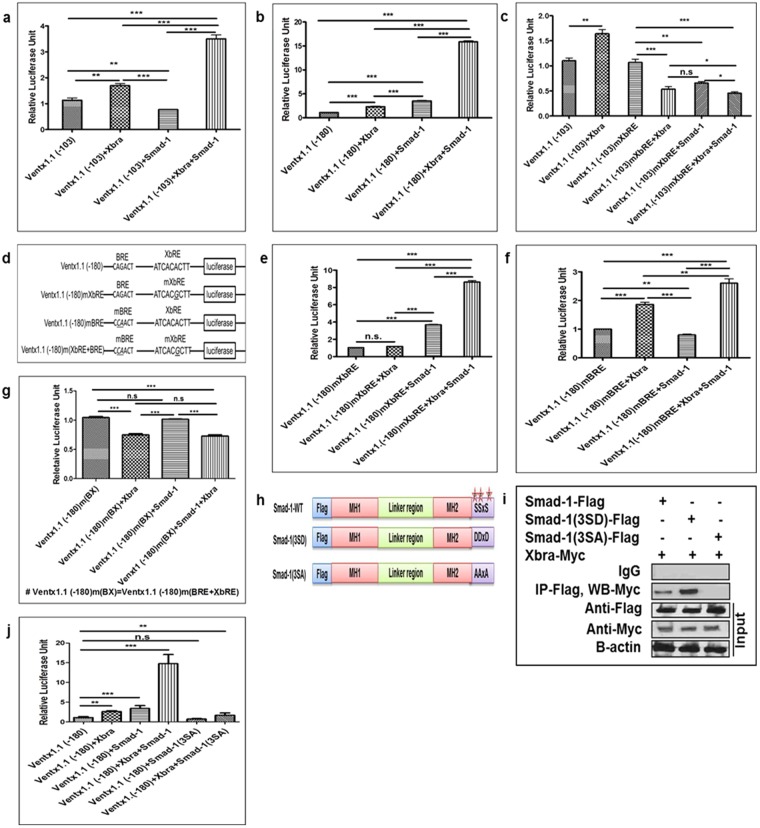
Lee H. S. et al.4 demonstrated that Smad-1 directly binds to cis-acting BMP-4 response elements (BRE; CAGACT, −180 bp to −162 bp) within the ventx1.1 promoter4. Our findings showed that both Xbra and Smad-1 positively regulate ventx1.1 expression4,14 and two cis-acting elements (BRE; −180 bp to −162 bp and XbRE; −70 bp to −62 bp) exist within the proximal promoter region of the ventx1.1. Thus, we further examined whether Xbra and Smad-1 synergistically cooperate to activate ventx1.1 transcription in Xenopus embryos. We co-injected the ventx1.1 (−103 and −180) promoter constructs either with or without Xbra and Smad-1, in a combination or separately. The ventx1.1 (−103) construct contains only the XbRE, while the ventx1.1 (−180) construct contains both the cis-acting elements BRE and XbRE. The results showed that co-injection of Smad-1 and Xbra mRNA increased the relative promoter activities of ventx1.1 (−103 and −180) up to 3.5-fold and 16-fold compared to those of ventx1.1 (−103 and −180) alone, respectively (Fig. 3a,b). Xbra or Smad-1 alone did not lead to the maximal increase (16-fold) in the relative promoter activities neither with ventx1.1 (−103) nor with ventx1.1 (−180) promoter constructs. Collectively, the results strongly indicated that Smad-1 and Xbra synergistically cooperate to activate ventx1.1 transcription. To examine the DNA sequence required for Xbra and Smad-1-mediated synergistic regulation of ventx1.1 transcription, we injected the ventx1.1 (−103) and XbRE-mutated ventx1.1 (−103) promoter (ventx1.1 (−103) mXbRE) constructs with both Xbra and Smad-1 in combination or separately. With the ventx1.1 (−103) mXbRE construct, the Xbra and Smad-1-mediated stimulation of the ventx1.1 (−103) promoter construct was completely abolished (Fig. 3c, bar 4–6 vs. bar 2), indicating that the cis-acting element of XbRE is essential for Xbra-mediated activation as well as Xbra and Smad-1-mediated synergistic activation of ventx1.1 transcription.
Figure 3.
Xbra and Smad-1 synergistically regulate ventx1.1 transcription. Different ventx1.1 promoter constructs were co-injected either with or without Smad-1 and with Xbra, either in combination or separately, at the 1-cell stage and grown until stage 11 to measure the relative promoter activity. (a,b) ventx1.1 (−103 and −180) injected with Xbra and Smad-1, in combination or separately. (c) XbRE-mutated ventx1.1 (−103) mXbRE and ventx1.1 (−103) injected with Xbra and Smad-1, in combination or separately, in separate groups. (d) XbRE and BRE were mutated by site-directed mutagenesis in different ventx1.1 (−180) promoter constructs. (e) XbRE-mutated ventx1.1 (−180) mXbRE injected either with or without Xbra and Smad-1, in combination or separately. (f) BRE-mutated ventx1.1 (−180) mBRE injected either with or without Xbra and Smad-1, in combination or separately. (g) The doubly mutated ventx1.1 (−180) m(BRE + XbRE) injected either with or without Xbra and Smad-1, in combination or separately. (h,i) Flag-Smad-1, Flag-Smad-1 (3SD), and Flag-Smad-1 (3SA) constructs were co-injected with Myc-Xbra and immunoprecipitation was performed with an anti-Flag antibody. (j) ventx1.1 (−180) injected either with or without Myc-Xbra, Flag-Smad-1, and Flag-Smad-1 (3SA) constructs in different groups, in combination or separately. All relative promoter activity data are shown as mean ± SE.
The concomitant overexpression of Xbra and Smad-1 increased the relative promoter activity of ventx1.1 (−103 and −180) for up to 3.5-fold and 16-fold as compared with those of ventx1.1 (−103 and −180) alone, respectively (Fig. 3a,b, bar 4). We then examined whether both of the cis-acting elements (BRE and XbRE) were equally important in the synergistic activation of ventx1.1 transcription. We mutated BRE and XbRE within the ventx1.1 (−180) promoter construct and generated three different mutated promoter constructs of ventx1.1 (−180); these were the XbRE-mutated ventx1.1 (−180) mXbRE, the BRE-mutated ventx1.1 (−180) mBRE and the doubly mutated ventx1.1 (−180) m(BRE + XbRE) (Fig. 3d). The ventx1.1 (−180) mXbRE promoter construct was co-injected either with or without Smad-1and Xbra in combination or separately. As we expected, the results showed that Xbra-mediated stimulation was completely abolished (Fig. 3e, 2nd bar) while Smad-1-mediated stimulation was sustained with a 3.5-fold induction in presence of Smad-1 (Fig. 3e, 3rd bar). Interestingly, the XbRE-mutated ventx1.1 (−180) mXbRE promoter construct still contained synergistic activation with an 8.5-fold induction when Xbra was co-injected with Smad-1 (Fig. 3e, 4th bar). It was noted that the Xbra and Smad-1-mediated relative promoter activity of ventx1.1 (−180) mXbRE was decreased 8.5-fold compared to a 16-fold increased activity with the ventx1.1 (−180) promoter construct (Fig. 3b, 4th bar). The results indicated that the XbRE contributes to achieve a maximal fold increase (16-fold) in the synergistic activation of ventx1.1 transcription with Smad-1 and Xbra. We next examined the ventx1.1 (−180) mBRE either with or without Smad-1 and Xbra in combination or separately. The results showed that the Smad-1-mediated stimulation was completely abolished (Fig. 3f, 3rd bar), but the Xbra-mediated stimulation was sustained with a 1.5-fold increase in presence of Xbra (Fig. 3f, 2nd bar). The ventx1.1 (−180) mBRE promoter construct still displayed a certain synergistic activation with a 2.5-fold induction when Smad-1 was co-injected with Xbra (Fig. 3f, 4th bar). Additionally, the relative promoter activity of ventx1.1 (−180) mBRE was dramatically decreased to 2.5-fold compared to the 16-fold increase which was achieved by the Smad-1 and Xbra with the ventx1.1 (−180) promoter construct (Fig. 3b, 4th bar). These results collectively provide evidence that BMP-4/Smad-1 and BRE play a more crucial role in the synergistic regulation of ventx1.1 transcription activation than with FGF/Xbra and XbRE. We further examined the doubly mutated ventx1.1 (−180) m(BRE + XbRE) promoter construct with Smad-1 and Xbra in combination or separately. The results showed that the Xbra and Smad-1-mediated stimulation of the promoter activity of the ventx1.1 (−180) promoter construct was completely abolished in the doubly mutated ventx1.1 (−180)m(BRE + XbRE) construct. Both the synergy as well as stimulation were completely abolished in the doubly mutated ventx1.1 (−180) m(BRE + XbRE) construct (Fig. 3g). These results collectively indicated that both the consensus cis-acting BRE and XbRE are required for maximal activation of ventx1.1 transcription.
Studies have reported that the N-terminal of Xbra physically interacts with phosphorylated C-terminal MH2 domain of Smad-1 for both human and Xenopus embryos19,22. Thus, we examined the physical interaction of Xbra and Smad-1. We co-injected Xbra with different Smad-1 constructs: Smad-1 (wild-type; wt), the C-terminal phospho-mimic Smad-1 (3SD) (constitutively active form), and the phospho-dead Smad-1 (3SA) (inactive form) (Fig. 3h). The results showed that Xbra physically interacts with Smad-1 and Smad-1 (3SD), while the interaction of Xbra with Smad-1 (3SA) was not detected (Fig. 3i). Moreover, Xbra more strongly bound with Smad-1 (3SD) than Smad-1, suggesting that the C-terminal phosphorylation of Smad-1 plays a critical role in the Xbra-Smad-1 interaction. We then inquired on the effect of Smad-1 and Xbra interaction in synergistic regulation of ventx1.1 transcription. When Smad-1 (wt) was co-injected with Xbra, the relative promoter activity of the ventx1.1 (−180) construct increased up to 15-fold compared to that of ventx1.1 (−180) alone. On the other hand, Smad-1 (3SA) and Xbra did not show synergistic activation of ventx1.1 (−180) (Fig. 3j). These results supported the findings of the previous studies, which showed that the C-terminal phosphorylation of Smad-1 is required for the Smad-1-Xbra interaction and it stimulates the expression of the ventral genes19,25. Moreover, the results indicated that the Xbra-Smad-1 interaction is required for the establishment of BMP-4/Smad-1 and FGF/Xbra-mediated signaling crosstalk which synergistically regulates the transcription of a specific target gene, namely ventx1.1 in early Xenopus embryos.
Xbra enhances Smad-1 binding affinity on its cis-acting element (BRE) within the ventx1.1 promoter
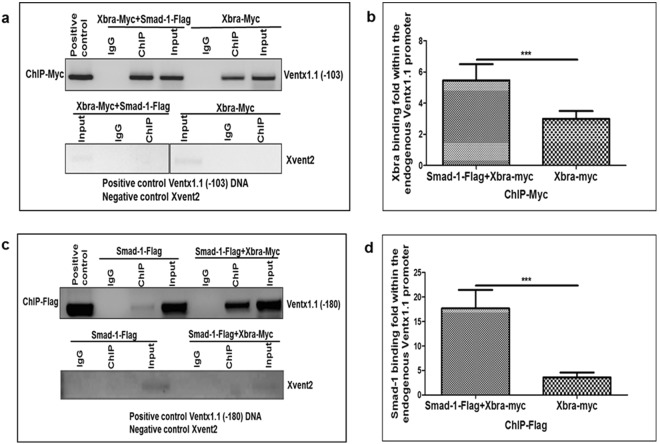
Smad-1 and Xbra synergistically up-regulated the relative promoter activities of ventx1.1 promoter construct. Therefore, we examined whether Smad-1 and Xbra stimulated the DNA binding of their interacting partner within the proximal region of the endogenous ventx1.1 promoter. We co-injected Myc-Xbra either with or without Smad-1 to perform ChIP-PCR assays with an anti-Myc antibody. The results showed that ectopic expression of Smad-1 increased about 2 fold the Xbra binding intensity within the proximal region of the endogenous ventx1.1 promoter (Fig. 4a,b). We further asked whether Xbra stimulated Smad-1 binding within the proximal region of the endogenous ventx1.1 promoter. We co-injected Flag-Smad-1 either with or without Xbra and performed a ChIP-PCR assay with an anti-Flag antibody. The results showed that Xbra strongly enhances the Smad-1 binding affinity within the proximal region of the endogenous ventx1.1 promoter (Fig. 4c,d). These results collectively indicated that both Xbra and Smad-1 directly bind within the proximal region of the endogenous ventx1.1 promoter. Xbra positively regulates ventx1.1 transcription activation as well as strongly stimulates the Smad-1 binding within the proximal region of the endogenous ventx1.1 promoter.
Figure 4.
Xbra enhances Smad-1 binding affinity on its cis-acting element (BRE) within the ventx1.1 promoter. To perform ChIP-PCR assays, mRNA was co-injected either with or without Smad-1 at the 1-cell stage and harvested the embryos until stage 11 in 30% MMR. (a,b) The ChIP-PCR assay was performed with the anti-Myc antibody. (c,d) The ChIP-PCR assay performed with the anti-Flag antibody at the stage 11. All bindings were measured by PCR method using specific primers. ventx1.1 (−103 and −180) promoter DNA served as positive control. The ChIP-PCR band intensity was quantified using a densitometer.
Discussion
In this study, we tried to answer two basic questions. The first concerned the presence and the details of the crosstalk between two main signaling pathways, BMP/Smad1 and FGF/Xbra, in the regulation of ventx1.1 expression. The second one was the role and significance of ventx1.1 expression in neural inhibition. In this discussion, we try to put forward the significance of ventx1.1 expression regulated by BMP-4/Smad-1 and FGF/Xbra signal crosstalk in the germ layer specification and its inhibitory role for early neurogenesis in mesoderm and ectoderm regions. The implications of such a crosstalk in Xenopus between these two pathways in the synergistic activation of ventx1.1 and the negative regulation of early neurogenesis relative to germ layer specification are discussed below.
Our previous studies demonstrated that BMP-4/Smad-1 and FGF/Xbra separately induce ventx1.1 expression in the ectoderm and mesoderm4,14. In the present study, we examined the mechanism of the synergistic activation of ventx1.1 expression due to BMP-4/Smad-1 and FGF/Xbra pathways. Previously, a study had reported that Smad-1 physically interacts with Xbra in Xenopus19. That study suggested that Smad-1-Xbra interaction may induce the expression of ventral genes. However, a direct target gene for Xbra-Smad-1 interaction was not reported. Here, we first tried to find an Xbra response cis-acting element in the ventx1.1 promoter. Through serial deletion constructs of the ventx1.1 promoter (Fig. 2c), we found that the ventx1.1 (−103) promoter construct contained a putative cis-acting XbRE (ATCACACTT, within −70 bp~−62 bp). The site-directed mutagenesis confirmed that the putative XbRE positively regulates ventx1.1 transcription activation in Xenopus embryos (Fig. 2f,g), and the ChIP-PCR assay confirmed that Xbra directly binds and stimulates the transcription of ventx1.1 (Fig. 2i,j). Although, we observe that XbRE-mutated ventx1.1 (−103) mXbRE promoter construct contains at least one negative cis-acting response element(s) where a putative transcription factor increased by Xbra may bind. However, we do not know what it can be at this moment. In the beginning, we suspected that ventx1.1 itself may be the auto-negative regulatory factor (based on ChIP-Seq data and reporter gene assays with ventx1.1 (−2525). However, ventx1.1 negative response was abolished with ventx1.1 (−399 and smaller) constructs. As such, we do not know the identity of the factor. Overexpression of Xbra and Smad-1 synergistically cooperated to activate ventx1.1 transcription in the ventx1.1 (−103) construct which did not contain the BRE (Fig. 3a,b,f). The site-directed mutagenesis showed that an intact BRE without XbRE also plays a significant role in Smad-1 and Xbra-mediated synergistic activation of ventx1.1 transcription (Fig. 3e). We also documented the contribution of each cis-acting acting element (BRE and XbRE) to the ventx1.1 promoter activity by studying signal changes in presence and also absence of the other interacting trans-acting element (Smad-1 and Xbra) (Fig. 5). Xbra and Smad-1 interaction enhanced the binding of an interacting partner on its cis-acting element (Fig. 4) and maximum cooperation was achieved in presence of intact DNA binding sequences for both Smad-1 and Xbra (Fig. 3j). It was also noted that Smad-1 and Xbra did not synergistically cooperate to activate transcription of another direct target of BMP/Smad-1, namely Xvent2, in Xenopus embryos (Fig. S1a and S1b). The present study collectively concludes that both the protein interaction between Xbra and the C-terminal phosphorylated Smad-1 and the DNA binding enhancement of Xbra and Smad1 within the ventx1.1 promoter establish a signaling crosstalk between the BMP-4/Smad-1 and FGF/Xbra pathways.
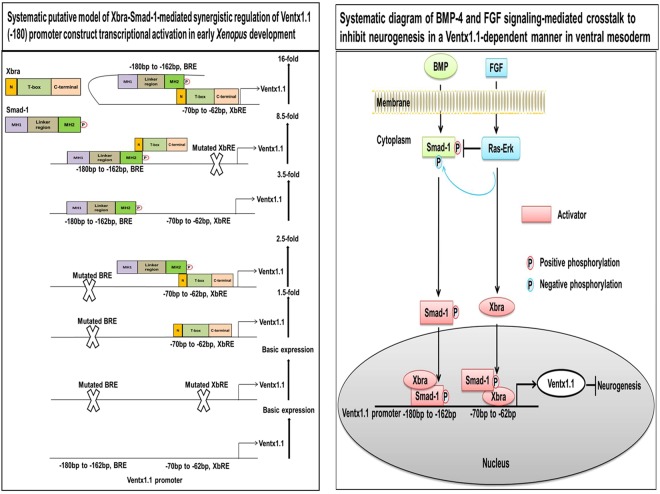
Figure 5.
The putative model of Xbra-Smad-1-mediated synergistic regulation of ventx1.1 transcriptional activation and neurogenesis inhibition in VMZ during early Xenopus development. The putative synergistic transcriptional activation of ventx1.1 (−180) promoter is regulated by the physical interaction of Smad-1 and Xbra in Xenopus embryos. BMP-4/Smad-1 and FGF/Xbra mediate a crosstalk in VMZ, inhibiting neurogenesis in a Ventx1.1-dependent manner during embryonic development of Xenopus.
Our previous studies reported that Ventx1.1 was an endogenous neural inhibitor in Xenopus through the suppression of organizer genes and early neural genes including chordin, noggin, foxD5a/b, gsc and zic3 via its C-terminal5,7,8. In this paper, we demonstrated that Xbra induced ventx1.1 expression in a BMP-4-inhibited condition and suppressed the expression of early and late neural genes including foxD5a, Ngnr, N-CAM and Otx2 in animal cap explants (Fig. 1). Studies have documented that inhibition of Xbra by DN-Xbra reduces ventx1.1 expression and triggers the neurogenesis in animal cap explants of Xenopus embryos14,26. As Xbra is not expressed in the ectodermal animal pole region and does not play a role in the neural gene expression in ectoderm region, the DN-Xbra construct eliciting neural tissue formation in ectoderm was an enigma. Our present results demonstrate that Xbra directly upregulates ventx1.1 expression which is involved in inhibition of neurogenesis in animal cap explants; this suggests that ectopic expression of DN-Xbra may be involved in inhibition of ventx1.1 expression, leading to neural tissue formation in ectoderm.
Presence and absence of Xbra determines whether FGF/MAPK is inhibitory or activating in the target tissue. In mesoderm, FGF/MAPK induces Xbra which activates ventx1.1 expression, resulting in inhibition of neurogenesis14,15. FGF/MAPK enhances neural induction by inhibiting BMP-4/Smad-1 signaling in the ectoderm15. However, FGF-mediated BMP-4/Smad-1 inhibition alone is not sufficient to trigger neurogenesis in Xenopus embryos in animal cap explants3. In our previous and present study, FGF/Xbra induced ventx1.1 expression in animal cap explants, resulting in neural inhibition and the knockdown of ventx1.1 induced neurogenesis in FGF-treated ectodermal explants14. Xbra triggers and maintains the mesoderm and it seems that Xbra plays a role in regulating ventx1.1 expression in the mesoderm and that if ventx1.1 is upregulated, neural tissue formation is inhibited in the mesoderm region. Our study suggests that FGF/Xbra negatively regulate neurogenesis in a Ventx1.1-dependent manner in the mesoderm region. Xbra induces ventx1.1 expression and inhibits the expression of early neural genes, leading to neural inhibition in part of mesodermal region (ventral and lateral mesodermal zone) of Xenopus embryos. Our study suggests that in Xenopus embryos, venxt1.1 plays neural inhibitory roles both in the ectoderm as a target of BMP and in part of the mesoderm as a target gene of FGF with gradient of BMP signaling.
The first main event in embryogenesis is germ layer specification where we hypothesize that the immediate early transcription factors expressed by the gradients of main signaling pathways including TGF-β (Vg1, nodal and activin), BMP and FGF maintain the resultant germ layer and suppress the commitment genes of other germ layers. In the mesoderm, Xbra is a main transcription factor elicited by BMP, FGF and nodals/activin via both direct and indirect signaling cascades. In the present paper, we suggest that BMP-4/Smad-1-mediated Ventx1.1 inhibits neurogenesis in the ectoderm and BMP-4/Smad-1 and FGF/Xbra mediated Ventx1.1 inhibits neural tissue formation in ventral and lateral mesoderm of Xenopus embryos. We also found that Xbra induces ventx1.1 expression in a BMP-4-inhibited condition in animal cap explants of Xenopus embryos (Fig. 1a). Xbra-induced ventx1.1 expression may play an essential role in inhibition of neurogenesis via suppressing the expression of early neural genes, including FoxD5a and Zic3 in animal cap explants (Fig. 1b). As we expected, knockdown of ventx1.1 restored the Ventx1.1-inhibited expression of neural genes, including FoxD5a, Otx2, Ngnr and N-CAM (Fig. 1c,d). These studies strongly suggest that Xbra stimulates ventx1.1 expression and inhibits neurogenesis in a Ventx1.1-dependent manner in the ventral and lateral mesoderm regions.
In this paper, we also hypothesize that the immediate early transcription factors expressed in a specific germ layer suppress the commitment genes of other germ layers. We suggest that Ventx1.1 in the ectoderm is involved in repression of neuroectoderm genes (FoxD5a/b and Zic3) and organizer genes (noggin, chordin, and gsc). Due to the involvement of Ventx1.1 in neural inhibition, we performed a genome-wide ChIP-Seq of Ventx1.1 in Xenopus and found that Ventx1.1 directly binds to the promoter regions of neural and organizer specific genes, including Zic3, noggin, chordin, FoxD5b and gsc, repressing their mRNA expression (unpublished data). Xbra may thus participate in ventx1.1 expression in the ventral and lateral mesoderm and maintain the mesoderm character. On the other hand, in the neuroectoderm, BMP inhibition leads to expression of foxd5 and zic3 as early neural genes. Ventx1.1 (−103) reporter construct contains a negative response element for FoxD5b (Fig. 2f). In genome-wide ChIP-Seq of FoxD5b, FoxD5b directly binds within the promoter regions of ectoderm and mesoderm specific genes including ventx1.1 (unpublished data). It will be interesting to study that the reciprocal repression networks existing between germ layer specific repressor factors. We consider ventx1.1 to be an appealing candidate to investigate as a reciprocal repressor in the ectoderm versus FoxD5b in the neuroectoderm or for ventx1.1 in the ventral and lateral mesoderm versus Gsc in the organizer region. During germ layer specification, a newly transcribed repressor in a specific germ layer preoccupies the region and inhibits the other germ layer specific genes that include essential genes for commitment and maintenance of the germ layers.
Germ layer specification is dependent on various cross talking pathways. In this regard, the FGF pathway is one of these main signaling routes concerning the regulation of germ layer specific genes. Our previous study demonstrated that Activin/Smad-2 and FGF signaling regulate the conversion of dorsal-ventral mesoderm formation in Xenopus embryos27. In presence of FGF signaling, Activin/Smad-2 stimulates DMZ formation and inhibits ventx1.1 and α-globin expression. Meanwhile, in the absence of FGF signaling, Activin/Smad-2 converts the DMZ into VMZ and induces the expression of ventx1.1 and α-globin in animal cap explants27. It will be interesting to investigate ventx1.1 expression and its interaction with various signal cross talks in a context dependent fashion. In this paper, we demonstrated the cooperation of BMP/Smad-1 and FGF/Xbra to positively regulate ventx1.1 expression. Smad-1 is activated by its C-terminal phosphorylation when BMP binds to hetero-dimeric BMP receptors. On the other hand, activation of FGF/Ras/MAPK pathway leads to phosphorylation of the Smad-1 linker region, known to inactivate Smad-1. In this paper, we demonstrated that C-terminal phosphorylation of Smad-1 is required for protein interaction with Xbra protein (Fig. 3i) and that the C-terminal phosphorylated mimic of Smad-1 (Smad-1(3SD)) showed a stronger affinity to Xbra than that of wild type Smad-1 (Fig. 3i, lane 1 and 2). Smad-1 affinity changes to Xbra, a co-Smad (Smad-4) or other transcription factors and whether Smad-1 changes affinity for its cis-acting element(s) in absence or presence of FGF signaling needs to be studied. These could answer how BMP-4 signaling with/without FGF differently regulates the expression of germ layer specific genes including ventx1.1 in Xenopus embryos. The mechanism uncovered in this paper is similar to that reported by Trompouki et al. (2011) and Mullen et al. (2011), in which a master tissue regulator or another transcription factor directs Smad-1 to specific targets depending on the tissue. We found that a mesoderm specific transcription factor Xbra directs stronger binding of Smad-1 onto ventx1.1 promoter in Xbra overexpressed animal cap explants (a mesoderm tissue). Since ventx1.1 transcription is tightly regulated in different germ layers, it will be interesting to investigate the specificity of Smad-1 binding to target genes in different germ layers and under different sets of signaling combinations28,29. In addition, it will be interesting to examine the effects of a master neuro-ectoderm transcription factor, namely FoxD5b, and the dorsal mesoderm or organizer specific transcription factor, Gsc, on interaction and binding of Smad-1 to ventx1.1 promoter.
Recently, a study documented that BMP-4 induces the expression of hematopoietic genes, including ventx1.1, SCL (Stem cell leukemia), Xvent1/2, globin and GATA1/2/3, leading to hematopoiesis in human and Xenopus embryos30. Other studies have demonstrated that elevated levels of GATA2 and Xvent2 are strongly associated with myeloid cell differentiation and acute myeloid leukemia (AML) progression in human cells31,32. Knockdown of GATA2 and Ventx inhibits the AML cell growth and proliferation in vivo and in vitro. In the present study, we found that Ventx synteny and putative Smad-1 and Xbra binding response elements were evolutionarily conserved for the Xvent family members in human and Xenopus cells (Fig. S2a and S2b). The above studies suggested that the interactions of Ventx family genes with master transcription factors (R-Smads, TCF7l1/2, and C/EBPα) may regulate the fate of embryonic stem cells (ESCs) and hematopoietic progenitor cells (HPCs) and direct the differentiation of cells into specific lineages in vertebrates. Taken together, we propose a putative model of Xbra-Smad-1-mediated synergistic regulation and transcriptional activation of ventx1.1 in the ventral and lateral mesoderm, inhibiting the neurogenesis in a Ventx1.1-dependent manner in early Xenopus embryogenesis (Fig. 5).
Materials and Methods
Ethics Statement
Institutional Animal Care and Use Committee (IACUC) approval is not required for the experimental use of amphibians or reptiles in Korea. All the members of our research group attended both the institutional educational and training courses for appropriate care and usage of experimental animals. Adult X. laevis were grown in 12-hour light/dark (LD 12:12 h) cycles at 18 °C according to the guidelines of Institutes of Laboratory Animal Resources for laboratory animal maintenance. Methods of DNA and RNA preparation, embryo injection, animal cap assay, luciferase assay, immunoprecipitation, site-directed mutagenesis, RT-PCR, and ChIP-PCR assay are available as supplementary information accompanying the manuscript.
Electronic supplementary material
Acknowledgements
This research was supported by the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science and Technology (NRF-2016R1D1A1B02008770), (NRF-2016M3A9B8914057) and by the Hallym University Research Fund (HRF-201410-017).
Author Contributions
S.K. performed the study and wrote the manuscript; Z.U. and J.Y. assisted with the experiments; J.B.P., S.C.K., U.L. and J.Y.L. analyzed the data; and K.J.B. designed the study, analyzed the data and corrected the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-29740-9.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dale L, Wardle FC. A gradient of BMP activity specifies dorsal-ventral fates in early Xenopus embryos. Seminars in cell & developmental biology. 1999;10:319–326. doi: 10.1006/scdb.1999.0308. [DOI] [PubMed] [Google Scholar]
- 2.Delaune E, Lemaire P, Kodjabachian L. Neural induction in Xenopus requires early FGF signalling in addition to BMP inhibition. Development. 2005;132:299–310. doi: 10.1242/dev.01582. [DOI] [PubMed] [Google Scholar]
- 3.Rogers CD, Moody SA, Casey ES. Neural induction and factors that stabilize a neural fate. Birth defects research. Part C, Embryo today: reviews. 2009;87:249–262. doi: 10.1002/bdrc.20157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee HS, et al. Direct response elements of BMP within the PV.1A promoter are essential for its transcriptional regulation during early Xenopus development. PloS one. 2011;6:e22621. doi: 10.1371/journal.pone.0022621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hwang YS, et al. Active repression of organizer genes by C-terminal domain of PV.1. Biochemical and biophysical research communications. 2003;308:79–86. doi: 10.1016/S0006-291X(03)01321-4. [DOI] [PubMed] [Google Scholar]
- 6.Zhong YF, Holland PW. The dynamics of vertebrate homeobox gene evolution: gain and loss of genes in mouse and human lineages. BMC evolutionary biology. 2011;11:169. doi: 10.1186/1471-2148-11-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ault KT, Dirksen ML, Jamrich M. A novel homeobox gene PV.1 mediates induction of ventral mesoderm in Xenopus embryos. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:6415–6420. doi: 10.1073/pnas.93.13.6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoon J, et al. PV.1 suppresses the expression of FoxD5b during neural induction in Xenopus embryos. Molecules and cells. 2014;37:220–225. doi: 10.14348/molcells.2014.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hwang YS, et al. Antimorphic PV.1 causes secondary axis by inducing ectopic organizer. Biochemical and biophysical research communications. 2002;292:1081–1086. doi: 10.1006/bbrc.2002.6740. [DOI] [PubMed] [Google Scholar]
- 10.Rogers CD, Archer TC, Cunningham DD, Grammer TC, Casey EM. Sox3 expression is maintained by FGF signaling and restricted to the neural plate by Vent proteins in the Xenopus embryo. Developmental biology. 2008;313:307–319. doi: 10.1016/j.ydbio.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor JJ, Wang T, Kroll KL. Tcf- and Vent-binding sites regulate neural-specific geminin expression in the gastrula embryo. Developmental biology. 2006;289:494–506. doi: 10.1016/j.ydbio.2005.10.047. [DOI] [PubMed] [Google Scholar]
- 12.Gamse J, Sive H. Vertebrate anteroposterior patterning: the Xenopus neurectoderm as a paradigm. BioEssays: news and reviews in molecular, cellular and developmental biology. 2000;22:976–986. doi: 10.1002/1521-1878(200011)22:11<976::AID-BIES4>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 13.Kim J, Lin JJ, Xu RH, Kung HF. Mesoderm induction by heterodimeric AP-1 (c-Jun and c-Fos) and its involvement in mesoderm formation through the embryonic fibroblast growth factor/Xbra autocatalytic loop during the early development of Xenopus embryos. The Journal of biological chemistry. 1998;273:1542–1550. doi: 10.1074/jbc.273.3.1542. [DOI] [PubMed] [Google Scholar]
- 14.Yoon, J. et al. PV.1 induced by FGF-Xbra functions as a repressor of neurogenesis in Xenopus embryos. BMB reports (2014). [DOI] [PMC free article] [PubMed]
- 15.Pera EM, Ikeda A, Eivers E, De Robertis EM. Integration of IGF, FGF, and anti-BMP signals via Smad1 phosphorylation in neural induction. Genes & development. 2003;17:3023–3028. doi: 10.1101/gad.1153603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casey ES, O’Reilly MA, Conlon FL, Smith JC. The T-box transcription factor Brachyury regulates expression of eFGF through binding to a non-palindromic response element. Development. 1998;125:3887–3894. doi: 10.1242/dev.125.19.3887. [DOI] [PubMed] [Google Scholar]
- 17.Casey ES, et al. Bix4 is activated directly by VegT and mediates endoderm formation in Xenopus development. Development. 1999;126:4193–4200. doi: 10.1242/dev.126.19.4193. [DOI] [PubMed] [Google Scholar]
- 18.Tada M, Casey ES, Fairclough L, Smith JC. Bix1, a direct target of Xenopus T-box genes, causes formation of ventral mesoderm and endoderm. Development. 1998;125:3997–4006. doi: 10.1242/dev.125.20.3997. [DOI] [PubMed] [Google Scholar]
- 19.Messenger NJ, et al. Functional specificity of the Xenopus T-domain protein Brachyury is conferred by its ability to interact with Smad1. Developmental cell. 2005;8:599–610. doi: 10.1016/j.devcel.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 20.Haramoto Y, et al. Insulin-like factor regulates neural induction through an IGF1 receptor-independent mechanism. Scientific reports. 2015;5:11603. doi: 10.1038/srep11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kusch T, Storck T, Walldorf U, Reuter R. Brachyury proteins regulate target genes through modular binding sites in a cooperative fashion. Genes & development. 2002;16:518–529. doi: 10.1101/gad.213002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faial T, et al. Brachyury and SMAD signalling collaboratively orchestrate distinct mesoderm and endoderm gene regulatory networks in differentiating human embryonic stem cells. Development. 2015;142:2121–2135. doi: 10.1242/dev.117838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsankov AM, et al. Transcription factor binding dynamics during human ES cell differentiation. Nature. 2015;518:344–349. doi: 10.1038/nature14233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gentsch GE, et al. In vivo T-box transcription factor profiling reveals joint regulation of embryonic neuromesodermal bipotency. Cell reports. 2013;4:1185–1196. doi: 10.1016/j.celrep.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marcellini S. When Brachyury meets Smad1: the evolution of bilateral symmetry during gastrulation. BioEssays: news and reviews in molecular, cellular and developmental biology. 2006;28:413–420. doi: 10.1002/bies.20387. [DOI] [PubMed] [Google Scholar]
- 26.Rao Y. Conversion of a mesodermalizing molecule, the Xenopus Brachyury gene, into a neuralizing factor. Genes & development. 1994;8:939–947. doi: 10.1101/gad.8.8.939. [DOI] [PubMed] [Google Scholar]
- 27.Lee SY, et al. Inhibition of FGF signaling converts dorsal mesoderm to ventral mesoderm in early Xenopus embryos. Differentiation; research in biological diversity. 2011;82:99–107. doi: 10.1016/j.diff.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 28.Mullen AC, et al. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trompouki E, et al. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell. 2011;147:577–589. doi: 10.1016/j.cell.2011.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee SY, et al. The role of heterodimeric AP-1 protein comprised of JunD and c-Fos proteins in hematopoiesis. The Journal of biological chemistry. 2012;287:31342–31348. doi: 10.1074/jbc.M112.387266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rawat VP, et al. The vent-like homeobox gene VENTX promotes human myeloid differentiation and is highly expressed in acute myeloid leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:16946–16951. doi: 10.1073/pnas.1001878107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang L, et al. GATA2 Inhibition Sensitizes Acute Myeloid Leukemia Cells to Chemotherapy. PloS one. 2017;12:e0170630. doi: 10.1371/journal.pone.0170630. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.