Abstract
The critical long non-coding RNAs (lncRNAs) involved in the carcinogenesis and progression of malignant melanoma (MM) have not been fully investigated. In the present study, it was identified that lncRNA activated by transforming growth factor-β (lncRNA-ATB) was upregulated in MM tissues and cells compared with benign nevus cells and human melanocytes, via comparative lncRNA screening from Gene Expression Omnibus datasets and reverse transcription-quantitative polymerase chain reaction analysis. Furthermore, lncRNA-ATB promoted the cell proliferation, cell migration, and cell invasion of MM cells in vitro, and tumor growth in vivo. It was additionally identified that lncRNA-ATB attenuated cell cycle arrest and inhibited cellular apoptosis in MM cells. Finally, it was demonstrated that lncRNA-ATB functions as a competing endogenous RNA (ceRNA) to enhance Yes associated protein 1 expression by competitively sponging microRNA miR-590-5p in MM cells. In conclusion, the present study revealed the expression and roles of lncRNA-ATB in MM, and indicated that lncRNA-ATB functions as a ceRNA to promote MM proliferation and invasion by sponging miR-590-5p.
Keywords: malignant melanoma, lncRNA-ATB, microRNA-590-5p, YAP1, competing endogenous RNA
Introduction
Malignant melanoma (MM) is one of the most aggressive forms of cutaneous neoplasm, the incidence of which has increased markedly in recent decades (1). An estimated 87,110 newly diagnosed MM cases and 9,730 MM-associated mortalities were predicted for 2017 in the United States (1). Despite the notable improvements in diagnosis and treatment made in recent years, the prognosis remains poor for patients diagnosed at the metastatic stage, with a median survival time of 6-9 months and a 5-year survival rate of only 10% (2-4). Thus, it is necessary to establish effective biomarkers for the early diagnosis and efficient evaluation of prognosis following surgery.
Long non-coding RNAs (lncRNAs) are a class of transcripts >200 nucleotides in length with limited protein coding potential (5-7). Recently, studies have demonstrated that lncRNAs have multiple functions in a wide range of biological process, including proliferation (8), apoptosis (9), cell migration (10) and cell invasion (11). A novel regulatory mechanism whereby lncRNAs function as competing endogenous (ce)RNAs to sponge microRNAs (miRNAs) and regulate their downstream signaling pathways has been proposed and confirmed preliminarily (12-14). Although it has been reported that a number of lncRNA transcripts are involved in the carcinogenesis and development of MM (15-17), the miRNA sponge roles of lncRNAs in the modulation of cell proliferation and migration in MM remain unclear.
The present study screened for candidate lncRNAs responsible for the malignant phenotype of MM from the Gene Expression Omnibus (GEO) datasets. It was identified that long non-coding RNA activated by transforming growth factor (TGF)-β (lncRNA-ATB) was the most markedly upregulated lncRNA in MM tissues and A375 cells compared with benign nevus cells and human melanocytes. The roles and underlying mechanism of lncRNA-ATB in the proliferation, apoptosis and invasion-metastasis cascade of MM cells were investigated and identified in vivo and in vitro.
Materials and methods
Cell culture
Highly invasive human MM A2058 cells, moderately invasive human MM A375 cells, normal epidermal melanocyte HEMa-LP cells, 293 and B16/F10 were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China), where they were characterized by mycoplasma detection, DNA-fingerprinting, isozyme detection and cell vitality detection. A2058, A375, 293 and B16/F10 cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) at 37°C in a humidified atmosphere of 95% air and 5% CO2. HEMa-LP cells were cultured in Medium 254 (Invitrogen; Thermo Fisher Scientific, Inc.) suplemented with human melanocyte growth supplement (Hyclone; GE Healthcare Life Sciences) at 37°C in a humidified atmosphere of 95% air and 5% CO2.
Oligonucleotide transfection
pGMLV-lncRNA-ATB negative control (NC), pGMLV-lncRNA-ATB short hairpin (sh)RNA, miRNA (miR)-590-5p NC, miR-590-5p mimics and miR-590-5p inhibitors were purchased from GenePharma Co. Ltd. (Shanghai, China). The sequence for miR-590-5p was GAGCUUAUUCAUAAAAUGCAG; miR-NC was GUCCAGUGAAUUCCCAG; and miR-590-5p inhibitors was GACGUAAAAUACUUAUUCGAG. The lncRNA-ATB overexpressing plasmid was constructed by Shanghai GeneChem Co., Ltd. (Shanghai, China) using pCDNA3.1. An empty vector was used as the control (Shanghai GeneChem Co., Ltd.). When the cell confluence of A375 and A2058 cells reached 50-60%, oligonucleotide transfections were performed using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Oligonucleotides (50 nmol) were mixed with 5 µl Lipofectamine® 2000 in 500 µl medium. The transfection solutions were added to each well containing 500 µl medium. Following transfection, cell samples were collected at 48 h for further analyses.
RNA/miRNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
For lncRNA-ATB quantification, total RNA was extracted from the cells or tissues using TRI reagent (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), according to the manufacturer's protocol. RNA samples were reverse transcribed into cDNA using RT regeants (Takara Bio, Inc., Otsu, Japan) with lncRNA specific primers, and β-actin was used as internal loading controls. RT was performed at 45°C for 60 min and 70°C for 10 min. SYBR Mix (Takara Bio, Inc.) was used to detect and quantify lncRNA-ATB and β-actin expression. For miR-590-5p quantification, total miRNA was extracted from the cells or tissues using the miRNeasy RNA isolation kit (Qiagen, Inc., Valencia, CA, USA), according to the manufacturer's protocol. Total miRNA samples (1 µl; 1 µg/µl) were reverse transcribed into cDNA using the miScript RT kit (Qiagen GmbH, Hilden, Germany) with miR-590-5p specific primers, and universal small nuclear U6 RNA was used as an internal loading control. An Applied Biosystems™ SYBR™ Green dye miRNA RT-qPCR kit (Thermo Fisher Scientific, Inc.) was used to detect and quantify miR-590-5p and U6 expression. Data were analyzed with 7500 software v.2.0.1 (Applied Biosystems; Thermo Fisher Scientific, Inc.), with the automatic Cq setting for adapting baseline and threshold for Cq determination (18). Each sample was examined in triplicate. The primer sequences used in the present study are listed in Table I. RT-qPCR assays were performed under the following thermocycling conditions: 95°C for 5 min, followed by 45 cycles of 95°C for 15 sec, 60°C for 30 sec and 72°C for 30 sec.
Table I.
Primers used for reverse transcription-quantitative polymerase chain reaction analysis.
| Gene name | Forward primer (5′→3′) | Reverse primer (5′→3′) |
|---|---|---|
| lncRNA-ATB | CTTCACCAGCACCCAGAGA | AAGACAGAAAAACAGTTCCGAGTC |
| β-actin | CGTCTTCCCCTCCATCGT | GAAGGTGTGGTGCCAGATTT |
| miR-590-5p | GGAATTCTTCAGTTGTAACCCAG | CGGGATCCTTGAGATGTCACCAA |
| U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
lncRNA-ATB, long non-coding RNA-activated by transforming growth factor-β; miR, microRNA.
Western blotting
Cells were washed in PBS three times prior to the proteins being extracted. Cells were lysed using radioimmunoprecipitation assay buffer (JingCai Technologies, Inc., Xi'an, China) and the protein concentration was determined using the bicinchoninic acid method. Each protein sample (30 µg) was denatured in SDS sample buffer and separated via 10% SDS-PAGE. Separated proteins were transferred to polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA, USA), blocked with 5% bovine serum albumin (BSA; Beyotime Institute of Biotechnology, Haimen, China) for 2 h at room temperature, and incubated overnight with primary antibodies at 4°C. Blotting was performed with primary antibodies against Yes associated protein 1 (YAP1; 1:300; cat. no. 14074; Cell Signaling Technology, Inc., Danvers, MA, USA). Goat anti-rabbit immunoglobulin horseradish peroxidase-linked F(ab)2 fragments (1:5,000; cat. no. TA130071; OriGene Technologies, Inc., Beijing, China) were used as secondary antibodies for 2 h at room temperature. β-actin (1:4,000; cat. no. ab8226; Abcam, Cambridge, UK) was used as a loading control. Blots were then washed three times (10 min/wash) in TBS-Tween-20 and developed using an enhanced chemiluminescence system (JingCai Technologies, Inc.). ImageJ (National Institutes of Health, Bethesda, MD, USA) was used to analyze the gray values of each blot.
Cell counting kit-8 (CCK-8) assay
A2058 cells transfected with lncRNA-ATB NC or lncRNA-ATB shRNA 24 h previously, and A375 cells transfected with empty vector or lncRNA-ATB overexpressing vector 24 h previously, were seeded in a 96-well culture plate at 2,000 cells/well. Each group was established in nine wells. CCK-8 reagents (cat. no. 40203ES60; Yeasen Biological Technology, Inc., Shanghai, China) were added into each well at 24, 48, 72, 96 and 120 h post-seeding, and each group was cultured for a further 50 min at 37°C in a humidified atmosphere of 95% air and 5% CO2. The optical density values was measured at 490 nm using a microplate reader.
Flow cytometry
MM cells in a 6-well culture plate were harvested by trypsinization, and washed three times with PBS. For cellular apoptosis analysis, cells were suspended in 500 µl binding buffer at a density of 2×106 cells/ml, and incubated with Annexin V-fluoroscein isothiocyanate and propidium iodide (PI; BD Biosciences, San Jose, CA, USA) for 15 min in the dark at room temperature. For cell cycle analysis, PI was added into A2058 cells tranfected with lncRNA-ATB NC or lncRNA-ATB, and A375 cells transfected with empty vector or lncRNA-ATB overexpressing vector, and incubated for 20 min in the dark at room temperature. Cellular apoptosis and the cell cycle were analyzed using a flow cytometer and Kaluza analysis software version 2.0 (both Beckman Coulter, Inc., Brea, CA, USA).
Cell migration and invasion
Cell migration and invasion capacity were measured in vitro using Transwell migration assays (EMD Millipore). A2058 cells transfected with lncRNA-ATB NC or lncRNA-ATB, and A375 cells transfected with empty vector or lncRNA-ATB overexpressing vector were suspended in DMEM with 10 g/l BSA at a density of 5×105 cells/ml. Cell suspensions (200 µl) were seeded in the upper chamber with membrane coated with (for the Transwell invasion assay) or without (for the migration assay) Matrigel (BD Biosciences). To attract the cells, 600 µl DMEM with 10% serum was added to the bottom chamber. When the cells had been allowed to migrate for 24 h or to invade for 48 h, the penetrated cells on the filters were fixed in dried methanol for 1 min and stained with 4 g/l crystal violet for 10 min at room temperature. The numbers of migrated or invasive cells were determined from five random fields using a light microscope (Olympus Corporation, Tokyo, Japan) at ×10 magnification.
Ethics statement
All animals were treated in accordance with the Guide for the Care and Use of Laboratory Animals, 1996, by the National Research Council (US) Institute for Laboratory Animal Research (19). Animal experiments were approved by and carried out according to the guidelines of the Ethics Committee of the First Affiliated Hospital of Xi'an Jiaotong University (Xi'an, China).
RNA pull-down assay
The full length lncRNA-ATB sequence or NC sequence was amplified by PCR kit (Takara Bio, Inc.) using a T7-containing primer. The following primer sequences were used: lncRNA-ATB forward, 5′-TCCCTGACTCCTCTATGGCATCTGTGG-3′; lncRNA-ATB reverse, 5′-CCTTTGCTTCCTCTTTTCTCATCTACTC-3′. The thermocycling conditions were as stated above. The DNA fragments were cloned into pcDNA3.1. The restriction enzyme XhoI was used to linearize the plasmids. T7 RNA polymerase (Takara Bio, Inc.) and Biotin RNA Labeling Mix (Roche Diagnostics, Indianapolis, IN, USA) was used to make biotin-labeled RNAs that were reverse transcribed. The products were treated with RNase-free DNase I (Roche Diagnostics) and purified with an RNeasy Mini kit (Qiagen, Inc.), and subsequently mixed with RNA extract from A2058 cells and incubated at room temperature for 1 h. The pull-down complexes were analyzed by RT-qPCR as previously stated following subsequent washes.
Tumor xenograft assays
All mice were housed and maintained under specific pathogen-free conditions at 18-22°C, with 20% humidity, a 12-h light and 12-h dark cycle, with feeding ad libitum. All experiments were approved by the Animal Care and Use Committee of Xi'an Jiaotong University and performed in accordance with institutional guidelines. For the tumorigenesis assays, xenograft tumors were generated via subcutaneous injection of A2058 cells (5×106), including A2058 lncRNA-ATB NC and A2058 lncRNA-ATB shRNA, into the hind limbs of 18-20-g 4-6-week-old Balb/C female athymic nude mice (nu/nu; Animal Center of Xi'an Jiaotong University, Xi'an, China; total number, 10 mice; n=5 mice/group). Tumor size was measured using a vernier caliper. Tumor volume was determined by the following formula: 0.5× A× B2, where A represents the diameter of the base of the tumor and B represents the corresponding perpendicular value. After 35 days, the mice were sacrificed and the tumors were collected and weighed. For the lung colonization assay, B16/F10 cells (5×106), which included lncRNA-ATB NC-luciferase and lncRNA-ATB shRNA-luciferase were intravenously injected into 18-20-g 6-week-old female C57/B6 mice (total number, 10 mice; n=5 mice/group). Lung colonization was measured using Xenogen IVIS Kinetic imaging systems (PerkinElmer, Inc., Waltham, MA, USA) on days 0, 7 and 14 post-injection.
Luciferase assays
The putative binding sites of miR-590-5p for lncRNA-ATB were predicted using RNA hybrid (20) and miRDB (http://www.mirdb.org/). Two hundreds and ninety-three cells were seeded in a 96-well plate at 70% confluence. The lncRNA-ATB was cloned into pMir-Report (Ambion; Thermo Fisher Scientific, Inc.), yielding pMir-Report-lncRNA-ATB. Mutations were introduced in potential miR-590-5p binding sites using the QuikChange site-directed mutagenesis kit (Stratagene; Agilent Technologies, Inc., Santa Clara, CA, USA). miR-590-5p NC or mimics were transfected into 293 cells with 30 ng wild-type (WT) or mutant (Mut) 3′-untranslated region (UTR) of lncRNA-ATB using Lipofectamine® 2000, as previously stated. Cells were collected 48 h post-transfection, and luciferase assays were performed using a Photinus pyralis-Renilla reniformis dual luciferase reporter assay system, according to the manufacturer's protocol (Promega Corporation, Madison, WI, USA). The luciferase activity of each lysate was normalized to Renilla luciferase activity.
Statistical analysis
Differentially expressed lncRNAs were identified from the GEO database GSE3189 with false discovery rate (FDR) <0.01 and |logFC| >1 using the R package (21). The raw P-value was corrected using the Benjamini and Hochberg method (22) to circumvent the multi-test bias. A fold-change value >2 or <0.25 and FDR <0.01 were selected as cutoff criteria for differentially expressed lncRNAs. Statistical analysis was performed using SPSS statistical software (version 21.0; IBM Corp., Armonk, NY, USA). The differences in characteristics between two groups were examined by the Student's t-test. The differences in characteristics between three groups were examined by analysis of variance, and the least significant difference test was applied to detect the differences between each pair of groups. The correlation between miR-590-5p expression and YAP1 expression was analyzed by Pearson correlation analysis. All P-values were determined from two-sided tests, and P<0.05 was considered to indicate a statistically significant difference. All experiments were repeated three times and the data are presented as the mean ± standard deviation from three independent experiments.
Results
lncRNA-ATB is upregulated in MM cell lines
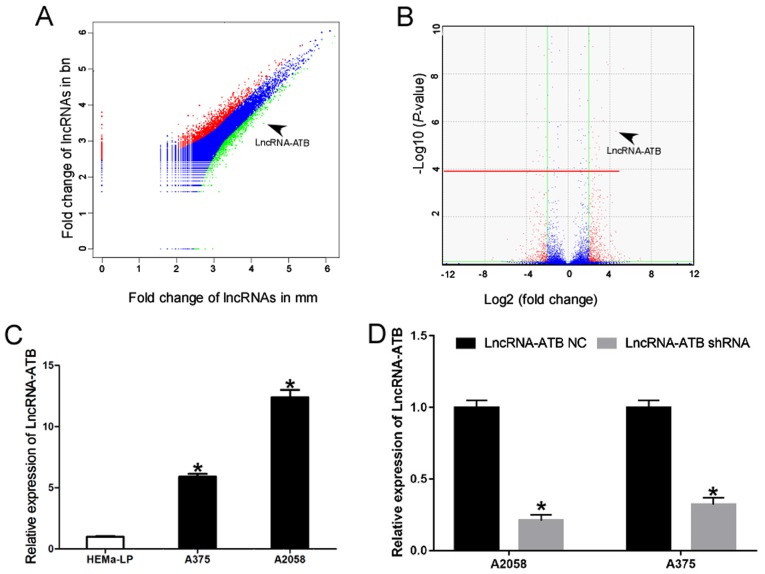
To elucidate the critical lncRNAs involved in the carcinogenesis and progression of MM, comparative lncRNA profiling was performed in 45 MM, 18 benign skin nevus cell (BN), and seven normal skin tissue specimens from the GEO dataset GSE3189 (23). Vectorgram analysis further identified that lncRNA-ATB was upregulated in MM specimens compared with BN (Fig. 1A and Table II). The volcano plot illustrates that the lncRNA-ATB expression level was >4-fold increased between cases and controls (P<0.001; Fig. 1B). The significant upregulation of lncRNA-ATB was further confirmed using RT-qPCR in the human epidermal melanocyte cell line HEMa-LP and in MM cell lines (Fig. 1C).
Figure 1.
lncRNA-ATB is upregulated in MM cell lines of malignant melanoma. (A) Vectorgram analysis of lncRNAs in MM and BN specimens. The x-axis indicates the Log2 fold-change in lncRNA expression in MM tissues, and the y-axis indicates the Log2 fold-change of lncRNA expression in BN tissues. Red, lncRNAs upregulated by ≥2-fold in MM compared with BN. Green, lncRNAs upregulated by ≥2-fold in BN compared with MM. Blue, lncRNAs upregulated by <2-fold in MM compared with BN or lncRNAs upregulated by <2-fold in BN compared with MM. Data from the GEO datasets (GSE3189): Differentially expressed lncRNAs from the GEO dataset GSE3189 with FDR <0.01 and |logFC| >2 were identified using the R package. The raw P-value was corrected using the Benjamini and Hochberg method to circumvent the multi-test bias. A fold-change value >2 or <0.25 and FDR <0.01 were selected as cutoff criteria for differentially expressed lncRNAs. (B) Volcano plot of lncRNAs in MM and BN. The x-axis indicates the Log2 fold-change in lncRNA expression between MM and BN tissues, while the y-axis indicates the Log10 of the adjusted P-value for each lncRNA. Values above the red line were identified to be statistically significant. (P<0.01) following application of the Benjamini and Hochberg method. lncRNA-ATB expression level was >4-fold increased between cases and controls (P<0.001 vs. BN). (C) Relative expression of lncRNA-ATB in human epidermal melanocytes and MM cells. *P<0.05 vs. HEMa-LP cells. (D) Knockdown efficiency of lncRNA-shRNA in MM cells. Data are from three experiments and are presented as the mean ± standard deviation. *P<0.05 vs. respective lncRNA-ATB NC group (Student's t-test). MM, malignant melanoma; BN, benign nevi; lncRNA, long noncoding RNA; ATB, activated by transforming growth factor-β; GEO, Gene Expression Omnibus; FDR, false discovery rate; shRNA, short hairpin RNA; NC, negative control.
Table II.
Top 30 lncRNAs upregulated in malignant melanoma.
| lncRNA name | Log2 fold-change |
|---|---|
| LncRNA-ATB | 4.585437915 |
| Linc00662 | 3.029954694 |
| AC144652.1 | 3.057751736 |
| RP11-654D12.2 | 3.081628575 |
| PVT1 | 3.139985752 |
| PGM5-AS1 | 3.151644278 |
| MIR24-2 | 3.1683935 |
| SNHG19 | 3.233861413 |
| AC112721.2 | 3.301811151 |
| CTD-2066L21.3 | 3.34462291 |
| MIR378D2 | 3.356163777 |
| NUP50-AS1 | 3.420937034 |
| SNHG9 | 3.427925853 |
| AP006621.5 | 3.441188157 |
| LINC00973 | 3.515399942 |
| LUCAT | 3.567557456 |
| LINC01588 | 3.582647048 |
| YTHDF3-AS1 | 3.589774049 |
| BANCR | 3.678548044 |
| TUG1 | 3.78577717 |
| SPRY4-IT1 | 3.826319081 |
| H19 | 3.855857029 |
| HOXD-AS1 | 3.879582756 |
| FALEC | 3.881343407 |
| FTH1P3 | 3.915488817 |
| ILF3-AS1 | 3.980602573 |
| HOXA11-AS | 4.004958111 |
| HEIH | 4.061621096 |
| CCAT1 | 4.155603912 |
| MHENCR | 10.18103013 |
Data from the Gene Expression Omnibus dataset GSE3189. lncRNA, long non-coding RNA.
Effects of lncRNA-ATB on the cell proliferation and apoptosis of MM cells
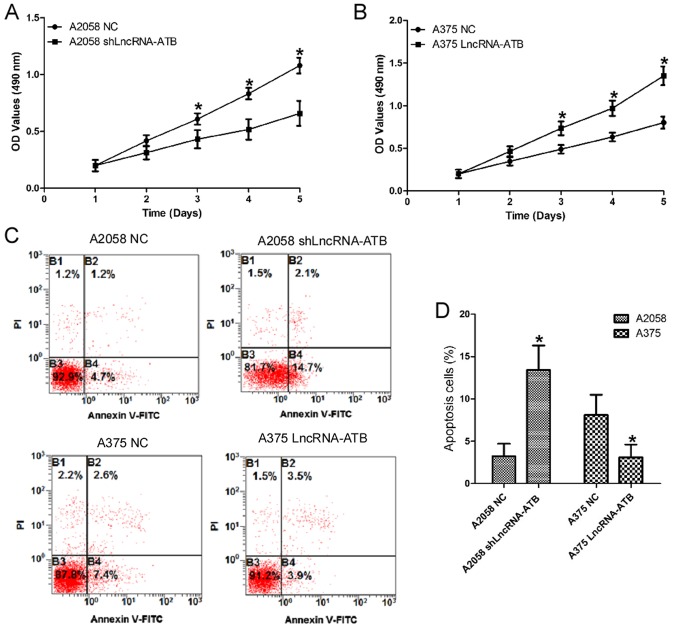
To investigate the functional role of lncRNA-ATB in MM cells, gain- and loss-of function experiments were performed by transfecting an shRNA, shLncRNA-ATB, into the highly invasive MM cell line A2058 to knock down lncRNA-ATB expression (Fig. 1D; P<0.05). An lncRNA-ATB overexpressing plasmid was also transfected into the moderately invasive MM cell line A375 to stably overexpress lncRNA-ATB. CCK-8 assays indicated that the proliferation of A2058 cells transfected with shLncRNA-ATB was inhibited compared with that in the control (Fig. 2A). However, the proliferation of A375 cells transfected with the lncRNA-ATB overexpressing plasmid was enhanced compared with that in the control (Fig. 2B). Cellular apoptosis assays demonstrated that the percentage of early apoptotic cells increased in A2058 cells transfected with shLncRNA-ATB. Whereas, the percentage of early apoptotic cells decreased in A375 cells transfected with the lncRNA-ATB overexpressing plasmid (Fig. 2C and D).
Figure 2.
Effects of lncRNA-ATB on the cell proliferation and apoptosis of MM cells. (A) The effects of lncRNA-ATB shRNA on the proliferation of A2058 cells were detected by CCK-8 assays. (B) Effects of lncRNA-ATB on the proliferation of A375 cells were detected by CCK-8 assays. (C) Effects of lncRNA-ATB on the cellular apoptosis of MM cells were detected by flow cytometry using an Annexin V/PI kit, and (D) the results were quantified. Data are from three experiments and are presented as the mean ± standard deviation. *P<0.05 vs. respective NC group (Student's t-test). MM, malignant melanoma; lncRNA, long noncoding RNA; shRNA, short hairpin RNA; NC, negative control; PI, propidium iodide; FITC, fluorescein isothiocyanate; OD, optical density; CCK-8, cell counting kit-8; ATB, activated by transforming growth factor-β.
Effects of lncRNA-ATB on the cell cycle distribution of MM cells
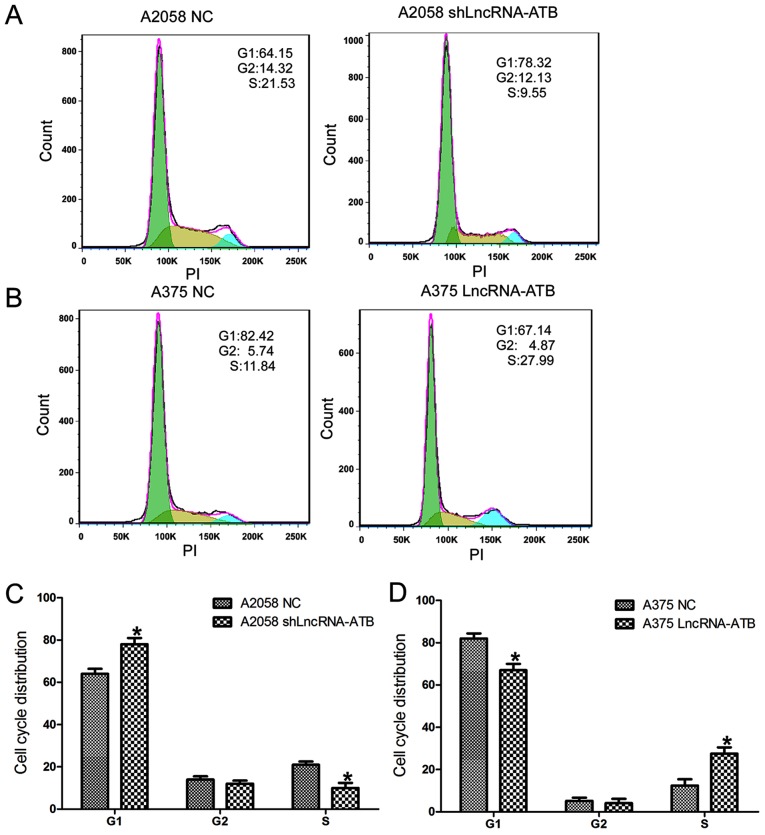
Cell cycle assays (Fig. 3) demonstrated that A2058 cells transfected with shLncRNA-ATB displayed a significant increase in the percentage of cells in the G1 phase and a decrease in the percentage of cells in the S phase (Fig. 3A and C). By contrast, the percentage of cells in the G1 phase decreased and the percentage of cells in the S phase increased in A375 cells transfected with the lncRNA-ATB overexpressing plasmid compared with that in the control (Fig. 3B and D).
Figure 3.
Effects of lncRNA-ATB on the cell cycle distribution of malignant melanoma cells. (A) Effects of lncRNA-ATB shRNA on the cell cycle distribution of A2058 cells were detected by flow cytometry using PI. (B) Effects of lncRNA-ATB on the cell cycle distribution of A375 cells were detected by flow cytometry using PI. (C) Quantification of the results in A2058 cells. (D) Quantification of the results in A375 cells. Data are from three experiments and presented as mean ± SD. *P<0.05 vs. respective NC group (Student's t-test). PI, propidium iodide; lncRNA, long noncoding RNA; shRNA, short hairpin RNA; ATB, activated by transforming growth factor-β; NC, negative control.
Effects of lncRNA-ATB on the cell migration and invasion of MM cells
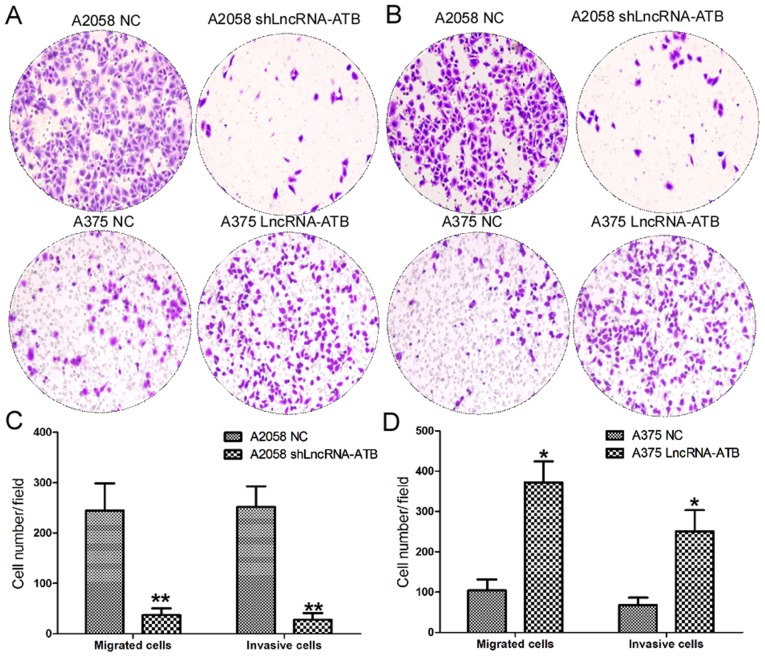
The present study further investigated the effects of lncRNA-ATB on the cell migration and invasion of MM cells using Transwell assays (Fig. 4). The cell migration assays indicated a significant decrease in the number of migratory cells among A2058 cells transfected with shLncRNA-ATB compared with the control (Fig. 4A and C). By contrast, ectopic expression of lncRNA-ATB significantly increased the number of migratory A375 cells compared with the normal control (Fig. 4A and D). As expected, cell invasion assays also confirmed a decrease in the number of invasive cells among A2058 cells transfected with shLncRNA-ATB compared with the control (Fig. 4B and C). Ectopic expression of lncRNA-ATB significantly enhanced the invasive ability of A375 cells compared with the control (Fig. 4B and D).
Figure 4.
Effects of lncRNA-ATB on the cell migration and invasion of MM cells. (A) Effects of lncRNA-ATB shRNA on the cell migration of A2058 and A375 cells were detected by Transwell assays (magnification, ×200). (B) Effects of lncRNA-ATB shRNA on the cell invasion of A2058 and A375 cells were detected by Transwell assays (magnification, x200). (C) Analysis of the migration and invasion data from A2058 cells. (D) Analysis of the migration and invasion data from A375 cells. Data are from three experiments and are presented as the mean ± standard deviation. *P<0.05 and **P<0.01 vs. respective NC group (Student's t-test). NC, negative control; shRNA, short hairpin RNA; lncRNA, long noncoding RNA; ATB, activated by transforming growth factor-β.
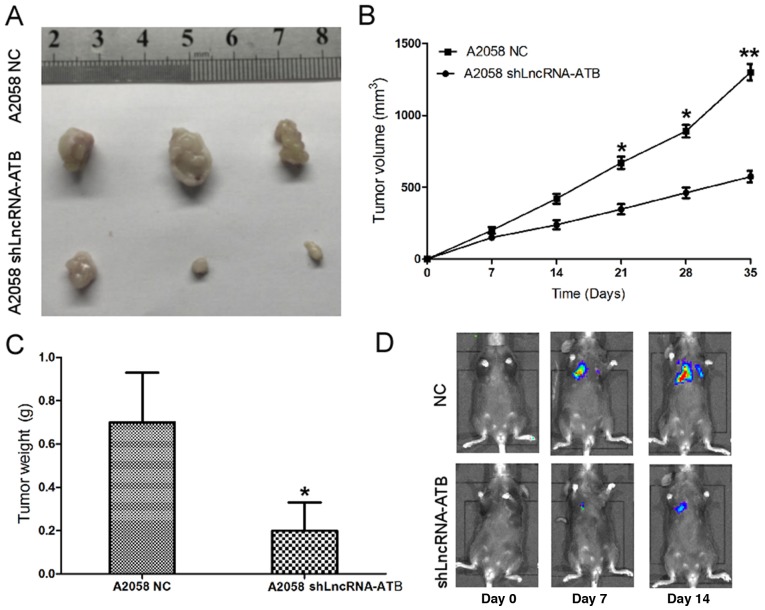
Effects of lncRNA-ATB on the tumorigenic ability of MM cells
The effect of lncRNA-ATB on the tumorigenic ability of MM cells was detected in vivo. shLncRNA-ATB and luciferase were transfected into A2058 cells and tumorigenesis assays were performed in nude mice. Cells (5×106) were injected into the flanks of the nude mice. Tumor sizes were measured using the Xenogen IVIS Kinetic imaging system and Vernier calipers every 7 days. After 35 days, the mice were sacrificed, and the tumors were collected and weighed. It was identified that shLncRNA-ATB had a significant tumor growth-inhibitory effect, causing significant reductions in tumor size (Fig. 5A and B) and weight (Fig. 5C) in the shLncRNA-ATB group compared with the control group. B16/F10 melanoma cells transfected with shLncRNA-ATB or controls were injected intravenously into C57/B6 mice, and lung colonization was assessed using the Xenogen IVIS200 System. It was identified that lung metastasis was inhibited in mice injected with B16/F10 cells transfected with shLn-cRNA-ATB, in comparison with the A2058 cells transfected with a control shRNA (Fig. 5D). Taken together, these results indicated that lncRNA-ATB was upregulated in MM cells, and that knockdown of lncRNA-ATB was able to repress cell proliferation, cell migration, cell invasion and tumor growth in vitro and in vivo.
Figure 5.
Effects of lncRNA-ATB on the tumorigenic ability and lung colonization of MM cells. (A) Tumors collected from the lncRNA-ATB shRNA-luciferase groups and A2058 lncRNA-ATB NC-luciferase groups. (B) Tumor growth curve of A2058 lncRNA-ATB shRNA-luciferase cells and A2058 lncRNA-ATB NC-luciferase cells in nude mice. (C) Tumor weights of lncRNA-ATB shRNA-luciferase and A2058 lncRNA-ATB NC-luciferase groups at day 35. (D) Effects of lncRNA-ATB shRNA on lung metastasis were measured using a Xenogene IVIS Kinetic imaging system every 7 days subsequent to injecting B16/F10 lncRNA-ATB shRNA-luciferase cells and B16/F10 lncRNA-ATB NC-luciferase cells intravenously into C57/B6 mice. Data are from three experiments and are presented as the mean ± standard deviation. *P<0.05 vs. respective NC group (Student's t-test). NC, negative control; shRNA, short hairpin RNA; lncRNA, long noncoding RNA; ATB, activated by transforming growth factor-β.
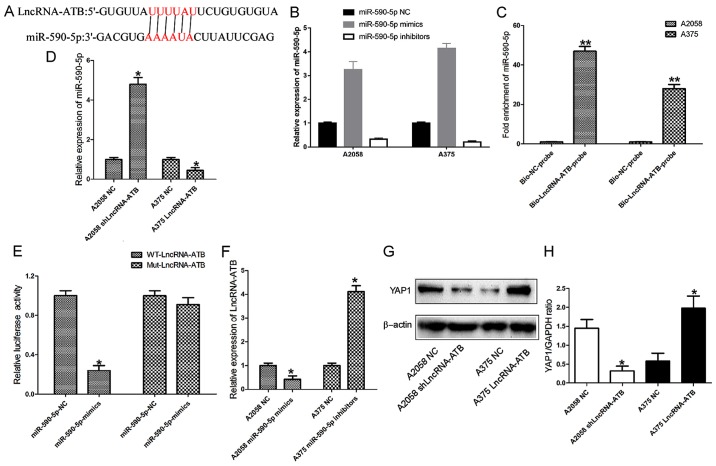
lncRNA-ATB directly sponges miR-590-5p to upregulate YAP1 expression
To examine the potential interaction between YAP1 and its targeting miRNA, miR-590-5p was identified as a potential targeting miRNA that has putative binding sites for lncRNA-ATB (Fig. 6A; data from RNA hybrid and miRDB), by searching in online bioinformatics databases. RT-qPCR assays revealed that miR-590-5p mimics and inhibitors were able to effectively upregulate and downregulate miR-590-5p expression, respectively, in MM cells (Fig. 6B; P<0.01). The biotin-labeled pulldown assay indicated that lncRNA-ATB was able to directly pull down miR-590-5p, which suggested that lncRNA-ATB may directly and specifically sponge miR-590-5p (Fig. 6C). Accordingly, miR-590-5p expression was significantly increased in A2058 cells transfected with shLncRNA-ATB, and significantly downregulated in A375 cells transfected with the lncRNA-ATB overexpressing plasmid (Fig. 6D; P<0.05). In addition, a dual-luciferase reporter assay was performed to verify whether lncRNA-ATB was the functional target of miR-590-5p. It was observed that miR-590-5p was able to reduce the relative luciferase activity of WT-LncRNA-ATB. Conversely, co-transfection of 293 cells with Mut-LncRNA-ATB and miR-590-5p resulted in a non-significant alteration in the relative luciferase activity (Fig. 6E). The RT-qPCR results showed that ectopic expression of miR-590-5p in A2058 cells inhibited lncRNA-ATB expression, while lncRNA-ATB expression was upregulated in A375 cells transfected with miR-590-5p inhibitors (Fig. 6F). These results demonstrated that miR-590-5p was also able to directly bind to lncRNA-ATB at specific recognition sites. A previous study reported miR-590-5p may function as a tumor suppressing miRNA in A375 cells (24). In addition, it may inhibit the cell migration and invasion of A375 by directly targeting YAP1. Thus, the present study investigated the effects of lncRNA-ATB on the YAP1 protein expression level. The western blotting results demonstrated that the expression of YAP1 protein was significantly downregulated in A2058 cells transfected with shLncRNA-ATB. By contrast, ectopic expression of lncRNA-ATB in A375 cells resulted in increased YAP1 protein expression levels (Fig. 6G and H).
Figure 6.
lncRNA-ATB sponges miR-590-5p to promote YAP1 expression in MM cells. (A) lncRNA-ATB and its putative binding sequence in miR-50-5p. (B) Efficacy of miR-590-5p mimics or inhibitors in MM cells. (C) Relative expression of miR-590-5p in the same sample pulled down by biotinylated lncRNA-ATB and NC probe. (D) Relative expression of miR-590-5p in MM cells transfected with lncRNA-ATB shRNA and lncRNA-ATB overexpressing plasmid. (E) Relative luciferase activity in 293 cells transfected with WT or Mut lncRNA-ATB plasmid. (F) Relative expression of lncRNA-ATB in MM cells transfected with miR-590-5p mimics or inhibitors. (G) Immunoblotting analysis and (H) relative quantification of YAP1 expression levels in MM cells transfected with lncRNA-ATB shRNA or overexpressing plasmid. Data are from three experiments and are presented as the mean ± standard deviation. *P<0.05 and **P<0.01 vs. respective NC group (Student's t-test). MM, malignant melanoma; NC, negative control; shRNA, short hairpin RNA; lncRNA, long noncoding RNA; ATB, activated by transforming growth factor-β; WT, wild-type; Mut, mutant; YAP1, Yes associated protein 1; miR, microRNA; Bio, biotinylated.
Discussion
In the present study, it was demonstrated that lncRNA-ATB, which had been reported to be activated by TGF-β (25), was upregulated in A375 cells compared with human melanocytes. In addition, it was observed that lncRNA-ATB was able to promote MM cell proliferation, migration and invasion in vitro, and MM tumor growth in vivo. Furthermore, it was demonstrated that lncRNA-ATB attenuated cell cycle arrest and inhibited cellular apoptosis in MM cells. Finally, the present study provided the first evidence, to the best of our knowledge, that lncRNA-ATB functions as a ceRNA to enhance YAP1 expression by competitively sponging miR-590-5p in MM cells. Taken together, these results demonstrated that lncRNA-ATB may function as a ceRNA to upregulate YAP1 expression, and promote cell proliferation and invasion, in MM cells by sponging miR-590-5p to reduce the binding of miR-590-5p to YAP1.
lncRNA-ATB has been reported to be upregulated in a number of types of cancer, including papillary thyroid cancer (26), renal cell carcinoma (27) and gastric cancer (28). lncRNA-ATB was identified as a TGF-β-activated lncRNA (25); thus, the function of lncRNA-ATB may be consistent with TGF-β pathways in different tumors, although certain lncRNAs, including HOX transcript antisense RNA, serve entirely opposite function in different types of cancer cells (29,30). It is possible that lncRNA-ATB is an example of this type of non-coding RNA. However, according to previous studies (25,31), it was noted that lncRNA-ATB serves an oncogenic role in the majority of types of cancers. In line with these previous studies, the present study identified that lncRNA-ATB was significantly upregulated in MM cell lines compared with normal human epidermal melanocyte cell lines. In non-small cell lung cancer, lncRNA-ATB promotes cellular apoptosis, and inhibits cell viability, cell migration and invasion (31). Furthermore, lncRNA-ATB was identified as a potential oncogene in osteosarcoma (32), glioma (33), prostate cancer (34) and colon cancer (35); the upregulation of lncRNA-ATB was able to promote carcinogenesis and progression in these cancer types. Notably, through searching electronic databases, a meta-analysis reported that lncRNA-ATB was associated with adverse clinical features, overall survival, progression-free survival and prognosis in human cancer (36). Studies have also confirmed that increased lncRNA-ATB expression is an independent prognostic predictor in a number of types of cancer, including non-small cell lung cancer (37), gastric cancer (38) and colorectal cancer (39).
To further determine the biological role of lncRNAs in MM, gain or loss-of- function experiments were performed. The results revealed that lncRNA-ATB promoted cell proliferation, migration and invasion, and attenuated cell cycle arrest in vitro. It was additionally demonstrated that knockdown of lncRNA-ATB was able to inhibit tumor growth in vivo. In addition to lncRNA-ATB, a number of other lncRNAs have been demonstrated to be potential oncogenes or tumor suppressors in MM, including homeobox A11-antisense (40), hepatocellular carcinoma upregulated EZH2-associated lncRNA (41) and colon cancer associated transcript 1 (42). In agreement with these studies, lncRNA-ATB was identified in the present study to be a potential oncogene in MM, and the critical roles of lncRNAs in the carcinogenesis and progression of MM were further confirmed. As it is difficult to perform gain-or-loss of function experiments using normal epidermal melanocytes, the present study did not investigate the role of lncRNA-ATB in HEMa-LP cells. Although lncRNA-ATB serves an important role in carcinogenesis, it was considered likely that the proliferation of HEMa-LP may not exhibit a marked alteration, due to the number of tumor suppressor genes in HEMa-LP cells to suppress the function of lncRNA-ATB. It was also observed that the basal measurements of A2058 cells and A375 cells with respect to the cell cycle, cell migration and cell invasion were different. There may be a number of reasons to account for this discrepancy. Firstly, the source of A2058 and A375 is different. Compared with A375, A2058 is a more invasive cell line. Thus, the number of migrated and invasive A2058 cells transfected with lncRNA-ATB NC was greater compared with the A375 cells transfected with lncRNA-ATB NC. Secondly, the different levels of lncRNA-ATB may be one of the factors accounting for the increased invasive ability of A2058 cells.
Recently, studies identified lncRNAs that were able to act as ceRNAs to interfere with miRNAs and their downstream pathways, which affect post-transcriptional regulation (43,44). ceRNA regulatory networks are implicated in numerous biological processes in cancer, including tumorigenesis (45), epithelialmesenchymal transition (46) and the invasionmetastasis cascade (47). For example, lncRNA metastasis associated lung adenocarcinoma transcript 1 is overexpressed in MM and upregulates the expression levels of matrix metal-lopeptidase 14 and snail family transcriptional repressor 1 by competitively sponging miR-22, thus promoting tumor growth and metastasis (48). Similarly, the lncRNA melanoma highly expressed competing endogenous lncRNA for miR-425 and miR-489 also functions as a ceRNA to upregulate insulin like growth factor 1 and spindling 1 expression by sponging miR-425 and miR-489, thereby promoting tumor growth and metastasis in MM (49). lncRNA-ATB was also demonstrated to be a ceRNA in hepatocellular carcinoma, upregulating zinc finger E-box binding homeobox 1 and 2 expression, and promoting the invasion-metastasis cascade by competitively binding miR-200 (50). The present study further increased understanding of the ceRNA function of lncRNA-ATB. The potential miR-590-5p binding sites were identified in lncRNA-ATB transcripts, and it was demonstrated that lncRNA-ATB expression was negatively regulated by miR-590-5p. Notably, it was confirmed that lncRNA-ATB may function as a ceRNA by sponging miR-590-5p to upregulate YAP1 expression in MM cells.
In conclusion, the present study demonstrated that lncRNA-ATB is a potential oncogene in MM. lncRNA-ATB was able to promote the tumor growth and metastasis of MM cells in vivo and in vitro by sponging miR-590-5p, functionally releasing YAP1 mRNA transcripts that are normally targeted by miR-590-5p. The present study helped to reveal the regulatory mechanism of lncRNA-ATB in MM and may lead to novel therapeutic strategies for MM.
Acknowledgments
Not applicable.
Abbreviations
- MM
malignant melanoma
- lncRNA
long noncoding RNA
- lncRNA-ATB
long noncoding RNA activated by transforming growth factor-β
- YAP1
Yes associated protein 1
Funding
The present study was supported by Hospital Fund of the First Affiliated Hospital of Xi'an JiaoTong University (Xi'an, China; grant no. 2016QN-06).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LW conceived the experiments and drafted the manuscript. KM and BL conducted the experiments and helped to analyze the data. MD performed data analysis. XM, YZ and DH performed data analysis and revised the manuscript. All authors contributed to revise the manuscript and approved the final version for publication.
Ethics approval and consent to participate
Animal experiments were approved by and carried out according to the guidelines of the Ethics Committee of the First Affiliated Hospital of Xi'an Jiaotong University (Xi'an, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics. CA Cancer J Clin. 2017;2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Eggermont AM, Spatz A, Robert C. Cutaneous melanoma. Lancet. 2014;383:816–827. doi: 10.1016/S0140-6736(13)60802-8. [DOI] [PubMed] [Google Scholar]
- 3.Roukos DH. PLX4032 and melanoma: Resistance, expectations and uncertainty. Expert Rev Anticancer Ther. 2011;11:325–328. doi: 10.1586/era.11.3. [DOI] [PubMed] [Google Scholar]
- 4.Balch CM, Gershenwald JE, Soong SJ, Thompson JF, Atkins MB, Byrd DR, Buzaid AC, Cochran AJ, Coit DG, Ding S, et al. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol. 2009;27:6199–6206. doi: 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kondo Y, Shinjo K, Katsushima K. Long non-coding RNAs as an epigenetic regulator in human cancers. Cancer Sci. 2017;108:1927–1933. doi: 10.1111/cas.13342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: A new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolha L, Ravnik-Glavač M, Glavač D. Long noncoding RNAs as biomarkers in cancer. Dis Markers. 2017;2017;7243968 doi: 10.1155/2017/7243968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao L, Sun H, Kong H, Chen Z, Chen B, Zhou M. The Lncrna-TUG1/EZH2 axis promotes pancreatic cancer cell proliferation, migration and EMT phenotype formation through sponging Mir-382. Cell Physiol Biochem. 2017;42:2145–2158. doi: 10.1159/000479990. [DOI] [PubMed] [Google Scholar]
- 9.Zhang BY, Jin Z, Zhao Z. Long intergenic noncoding RNA 00305 sponges miR-136 to regulate the hypoxia induced apoptosis of vascular endothelial cells. Biomed Pharmacother. 2017;94:238–243. doi: 10.1016/j.biopha.2017.07.099. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y, Peng Y, Xu Z, Ge B, Xiang X, Zhang T, Gao L, Shi H, Wang C, Huang J. LncROR promotes bladder cancer cell proliferation, migration, and epithelial-mesenchymal transition. Cell Physiol Biochem. 2017;41:2399–2410. doi: 10.1159/000475910. [DOI] [PubMed] [Google Scholar]
- 11.Wang C, Mao ZP, Wang L, Wu GH, Zhang FH, Wang DY, Shi JL. Long non-coding RNA MALAT1 promotes cholangiocarcinoma cell proliferation and invasion by activating PI3K/Akt pathway. Neoplasma. 2017;64:725–731. doi: 10.4149/neo_2017_510. [DOI] [PubMed] [Google Scholar]
- 12.Yang C, Wu D, Gao L, Liu X, Jin Y, Wang D, Wang T, Li X. Competing endogenous RNA networks in human cancer: Hypothesis, validation, and perspectives. Oncotarget. 2016;7:13479–13490. doi: 10.18632/oncotarget.7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo LL, Song CH, Wang P, Dai LP, Zhang JY, Wang KJ. Competing endogenous RNA networks and gastric cancer. World J Gastroenterol. 2015;21:11680–11687. doi: 10.3748/wjg.v21.i41.11680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richtig G, Ehall B, Richtig E, Aigelsreiter A, Gutschner T, Pichler M. Function and clinical implications of long non-coding RNAs in melanoma. Int J Mol Sci. 2017;18:E715. doi: 10.3390/ijms18040715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang S, Fan W, Wan B, Tu M, Jin F, Liu F, Xu H, Han P. Characterization of long noncoding RNA and messenger RNA signatures in melanoma tumorigenesis and metastasis. PLoS One. 2017;12:e0172498. doi: 10.1371/journal.pone.0172498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bian D, Gao C, Bao K, Song G. The long non-coding RNA NKILA inhibits the invasion-metastasis cascade of malignant melanoma via the regulation of NF-ĸB. Am J Cancer Res. 2017;7:28–40. [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Guide for the Care and Use of Laboratory Animals. National Academy Press; Washington (DC): 1996. [Google Scholar]
- 20.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.RDevelopment CORE TEAM . R: A Language and Environment for Statistical Computing. R foundation for statistical computing; 2014. http://www.R-project.org/ [Google Scholar]
- 22.Tan YD, Xu H. A general method for accurate estimation of false discovery rates in identification of differentially expressed genes. Bioinformatics. 2014;30:2018–2025. doi: 10.1093/bioinformatics/btu124. [DOI] [PubMed] [Google Scholar]
- 23.Talantov D, Mazumder A, Yu JX, Briggs T, Jiang Y, Backus J, Atkins D, Wang Y. Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin Cancer Res. 2005;11:7234–7242. doi: 10.1158/1078-0432.CCR-05-0683. [DOI] [PubMed] [Google Scholar]
- 24.Wang L, Shi S, Zhou Y, Mu X, Han D, Ge R, Mu K. miR-590-5p inhibits A375 cell invasion and migration in malignant melanoma by directly inhibiting YAP1 expression. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2017;33:326–330. In Chinese. [PubMed] [Google Scholar]
- 25.Shi SJ, Wang LJ, Yu B, Li YH, Jin Y, Bai XZ. LncRNA-ATB promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget. 2015;6:11652–11663. doi: 10.18632/oncotarget.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu XM, Guo W, Li N, Liu HZ, Liu J, Qiu SQ, Zhang Q, Wang LC, Li F, Li CL. The expression and function of long noncoding RNA lncRNA-ATB in papillary thyroid cancer. Eur Rev Med Pharmacol Sci. 2017;21:3239–3246. [PubMed] [Google Scholar]
- 27.Qi JJ, Liu YX, Lin L. High expression of long non-coding RNA ATB is associated with poor prognosis in patients with renal cell carcinoma. Eur Rev Med Pharmacol Sci. 2017;21:2835–2839. [PubMed] [Google Scholar]
- 28.Lei K, Liang X, Gao Y, Xu B, Xu Y, Li Y, Tao Y, Shi W, Liu J. Lnc-ATB contributes to gastric cancer growth through a MiR-141-3p/TGFβ2 feedback loop. Biochem Biophys Res Commun. 2017;484:514–521. doi: 10.1016/j.bbrc.2017.01.094. [DOI] [PubMed] [Google Scholar]
- 29.Kim HJ, Lee DW, Yim GW, Nam EJ, Kim S, Kim SW, Kim YT. Long non-coding RNA HOTAIR is associated with human cervical cancer progression. Int J Oncol. 2015;46:521–530. doi: 10.3892/ijo.2014.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y, Bian Y, Zhao S, Kong F, Li X. Suppression of PDCD4 mediated by the long non-coding RNA HOTAIR inhibits the proliferation and invasion of glioma cells. Oncol Lett. 2016;12:5170–5176. doi: 10.3892/ol.2016.5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao Y, Luo X, Ding X, Cui S, Guo C. LncRNA ATB promotes proliferation and metastasis in A549 cells by down-regulation of microRNA-494. J Cell Biochem. 2018 Apr 25; doi: 10.1002/jcb.26894. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 32.Han F, Wang C, Wang Y, Zhang L. Long noncoding RNA ATB promotes osteosarcoma cell proliferation, migration and invasion by suppressing miR-200s. Am J Cancer Res. 2017;7:770–783. [PMC free article] [PubMed] [Google Scholar]
- 33.Ma CC, Xiong Z, Zhu GN, Wang C, Zong G, Wang HL, Bian EB, Zhao B. Long non-coding RNA ATB promotes glioma malignancy by negatively regulating miR-200a. J Exp Clin Cancer Res. 2016;35:90. doi: 10.1186/s13046-016-0367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu S, Yi XM, Tang CP, Ge JP, Zhang ZY, Zhou WQ. Long non-coding RNA ATB promotes growth and epithelial-mesenchymal transition and predicts poor prognosis in human prostate carcinoma. Oncol Rep. 2016;36:10–22. doi: 10.3892/or.2016.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yue B, Qiu S, Zhao S, Liu C, Zhang D, Yu F, Peng Z, Yan D. LncRNA-ATB mediated E-cadherin repression promotes the progression of colon cancer and predicts poor prognosis. J Gastroenterol Hepatol. 2016;31:595–603. doi: 10.1111/jgh.13206. [DOI] [PubMed] [Google Scholar]
- 36.Fan YH, Ji CX, Xu B, Fan HY, Cheng ZJ, Zhu XG. Long noncoding RNA activated by TGF-beta in human cancers: A meta-analysis. Clin Chim Acta. 2017;468:10–16. doi: 10.1016/j.cca.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 37.Ke L, Xu SB, Wang J, Jiang XL, Xu MQ. High expression of long non-coding RNA ATB indicates a poor prognosis and regulates cell proliferation and metastasis in non-small cell lung cancer. Clin Transl Oncol. 2017;19:599–605. doi: 10.1007/s12094-016-1572-3. [DOI] [PubMed] [Google Scholar]
- 38.Saito T, Kurashige J, Nambara S, Komatsu H, Hirata H, Ueda M, Sakimura S, Uchi R, Takano Y, Shinden Y, et al. A long non-coding RNA activated by transforming growth factor-β is an independent prognostic marker of gastric cancer. Ann Surg Oncol. 2015;22(Suppl 3):S915–S922. doi: 10.1245/s10434-015-4554-8. [DOI] [PubMed] [Google Scholar]
- 39.Iguchi T, Uchi R, Nambara S, Saito T, Komatsu H, Hirata H, Ueda M, Sakimura S, Takano Y, Kurashige J, et al. A long noncoding RNA, lncRNA-ATB, is involved in the progression and prognosis of colorectal cancer. Anticancer Res. 2015;35:1385–1388. [PubMed] [Google Scholar]
- 40.Lu Q, Zhao N, Zha G, Wang H, Tong Q, Xin S. LncRNA HOXA11-AS exerts oncogenic functions by repressing p21 and miR-124 in uveal melanoma. DNA Cell Biol. 2017;36:837–844. doi: 10.1089/dna.2017.3808. [DOI] [PubMed] [Google Scholar]
- 41.Zhao H, Xing G, Wang Y, Luo Z, Liu G, Meng H. Long noncoding RNA HEIH promotes melanoma cell proliferation, migration and invasion via inhibition of miR-200b/a/429. Biosci Rep. 2017;37:BSR20170682. doi: 10.1042/BSR20170682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lv L, Jia JQ, Chen J. LncRNA CCAT1 upregulates proliferation and invasion in melanoma cells via suppressing miR-33a. Oncol Res. 2018;26:201–208. doi: 10.3727/096504017X14920318811749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qu J, Li M, Zhong W, Hu C. Competing endogenous RNA in cancer: A new pattern of gene expression regulation. Int J Clin Exp Med. 2015;8:17110–17116. [PMC free article] [PubMed] [Google Scholar]
- 44.Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: Possible functions and clinical implications. J Med Genet. 2015;52:710–718. doi: 10.1136/jmedgenet-2015-103334. [DOI] [PubMed] [Google Scholar]
- 45.Cui B, Li B, Liu Q, Cui Y. lncRNA CCAT1 promotes glioma tumorigenesis by sponging miR-181b. J Cell Biochem. 2017;118:4548–4557. doi: 10.1002/jcb.26116. [DOI] [PubMed] [Google Scholar]
- 46.Lv M, Zhong Z, Huang M, Tian Q, Jiang R, Chen J. lncRNA H19 regulates epithelial-mesenchymal transition and metastasis of bladder cancer by miR-29b-3p as competing endogenous RNA. Biochim Biophys Acta. 18642017:1887–1899. doi: 10.1016/j.bbamcr.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 47.Wang H, Huo X, Yang XR, He J, Cheng L, Wang N, Deng X, Jin H, Wang N, Wang C, et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017;16:136. doi: 10.1186/s12943-017-0680-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luan W, Li L, Shi Y, Bu X, Xia Y, Wang J, Djangmah HS, Liu X, You Y, Xu B. Long non-coding RNA MALAT1 acts as a competing endogenous RNA to promote malignant melanoma growth and metastasis by sponging miR-22. Oncotarget. 2016;7:63901–63912. doi: 10.18632/oncotarget.11564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen X, Dong H, Liu S, Yu L, Yan D, Yao X, Sun W, Han D, Gao G. Long noncoding RNA MHENCR promotes melanoma progression via regulating miR-425/489-mediated PI3K-Akt pathway. Am J Transl Res. 2017;9:90–102. [PMC free article] [PubMed] [Google Scholar]
- 50.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.