Abstract
This report describes the identification of a genetically confirmed linked heterosexual human immunodeficiency virus (HIV) superinfection (HIV-SI) in a woman with chronic HIV infection who acquired a second strain of the virus from her husband. Serum neutralizing antibody (NAb) responses against their homologous and heterologous viruses, including the superinfecting strain, in the woman and her husband were examined before and after onset of HIV-SI. The woman displayed a moderately potent and broad anti-HIV NAb response prior to superinfection but did not possess NAb activity against the superinfecting strain. This case highlights the unique potential of linked HIV-SI studies to examine natural protection from HIV infection.
Keywords: HIV Superinfection, NAb, Africa
Human immunodeficiency virus (HIV) superinfection (HIV-SI) occurs when an HIV-infected individual acquires a new HIV strain that is phylogenetically distinct from their existing viral population [1]. The majority of studies examining HIV-SI have examined high-risk populations; however, HIV-SI also occurs at significant rates in the general HIV-infected population [1–4]. Screening for cases of HIV-SI in large population cohorts has allowed for the examination of immunological characteristics that may be associated with protection against HIV-SI by comparing superinfection cases to matched HIV-infected controls who do not become superinfected. Of particular interest in these studies has been the potential role of preexisting HIV-specific neutralizing antibodies (NAb) in protecting against HIV-SI. Two matched case control studies observed that individuals who became superinfected appear to have lower NAb responses as compared to controls, but a larger study of female bar workers in Kenya found no association between preexisting NAb and protection from HIV-SI [5–9]. An alternative approach for exploring HIV-SI risk is to examine HIV-infected couples who acquired their viruses from different sources, thereby making their viral populations phylogenetically unlinked [4, 10]. These couples can then be examined at multiple time points for a linked HIV-SI event if one or both members of the couple pass their virus onto their partner, which then allows for the examination of the underlying immune response to their partner’s viral population before and after the HIV-SI event [4, 10, 11].
METHODS
Participants in this study were enrolled in a general population cohort established in 1989 by the then MRC Programme on AIDS in rural southwest Uganda (Supplementary Methods) [12]. All participants were in Monogamous (n = 15) and polygamous (n = 6) relationships, attended the Rural Clinical Cohort in southwest Uganda, were previously identified as being HIV infected, and had virus populations determined by bulk HIV sequence analysis to be unlinked. The presence of HIV-SI was determined by examining longitudinal serum samples obtained from each member of the partnership, using a previously described next-generation sequencing (NGS) assay of 3 viral genomic regions (gag, pol, and gp41; Supplementary Methods) [10, 13]. Individuals with successful NGS of 2 longitudinal samples for at least 1 genetic region whose corresponding partner also had NGS data available from the same genetic region were assessed for linked HIV-SI (Supplementary Table 1). One such event was detected.
For the linked HIV-SI case, serum samples collected before and after the time of HIV-SI from both the female participant and her husband were subjected to single-genome amplification (SGA) to generate full envelope gene sequences (Supplementary Figure 1 and Supplementary Methods). For samples collected from the husband before the female partner’s HIV-SI event, full-length SGA was unsuccessful; therefore, total RNA was amplified using universal primers and was sequenced using a shotgun sequencing method (Supplementary Methods). NGS amplicons specific for the HIV Env gene were matched to the SGA sequences from other time points to verify similarity. Full-length Env amplicons from SGA were subcloned or synthesized and used to generate Env pseudoviruses. All pseudoviruses were examined for functionality and neutralization susceptibility to known monoclonal antibodies, as well as to a variety of subtype A and A/D serum from historic serum samples and nonsuperinfected Ugandans in the same cohort. Env-pseudoviruses were tested for their neutralization susceptibility to their homologous serum, as well as their partner’s heterologous serum from before and after HIV-SI (Supplementary Methods). Viral sequences are available in Genbank (accession numbers MG722983-MG724743).
RESULTS
Ten individuals had NGS data from at least 1 genomic region for 2 time points that matched the same region from their partner’s NGS data. Of these, 1 case of linked HIV-SI was identified. The case occurred in a polygamous relationship in which an HIV-infected uncircumcised male had 4 wives who were also HIV positive (Supplementary Table 1). Longitudinal NGS data were available for both the male participant and one of his wives from the initial screen (Supplementary Figure 1). NGS data were available for the pol and gp41 region from only 1 time point for 2 of his other 3 wives, and these regions were linked to those of virus from their husband (Supplementary Figure 2). The fourth wife’s virus did not amplify at either time point examined. The male participant was initially infected approximately 4 years prior to the HIV-SI event with a recombinant virus that contained HIV subtype D in the Pol region and subtype A in the gp41 region (Figure 1A and Supplementary Figure 2). The female participant with HIV-SI was also initially infected approximately 4 years before the HIV-SI event, as well as before marrying her husband, with a pure subtype A virus in both pol and gp41 (Figure 1A). It was observed that superinfection occurred 19–22 months after her initial sample was collected, and that the superinfecting virus was phylogenetically linked to her husband’s viral strain (Supplementary Figure 1). During this 3-month period, the woman also became pregnant, and although antiretroviral therapy to prevent mother-to-child transmission was not available in this area of Uganda at this time (the early 2000s), she later gave birth to a baby that did not become infected with HIV.
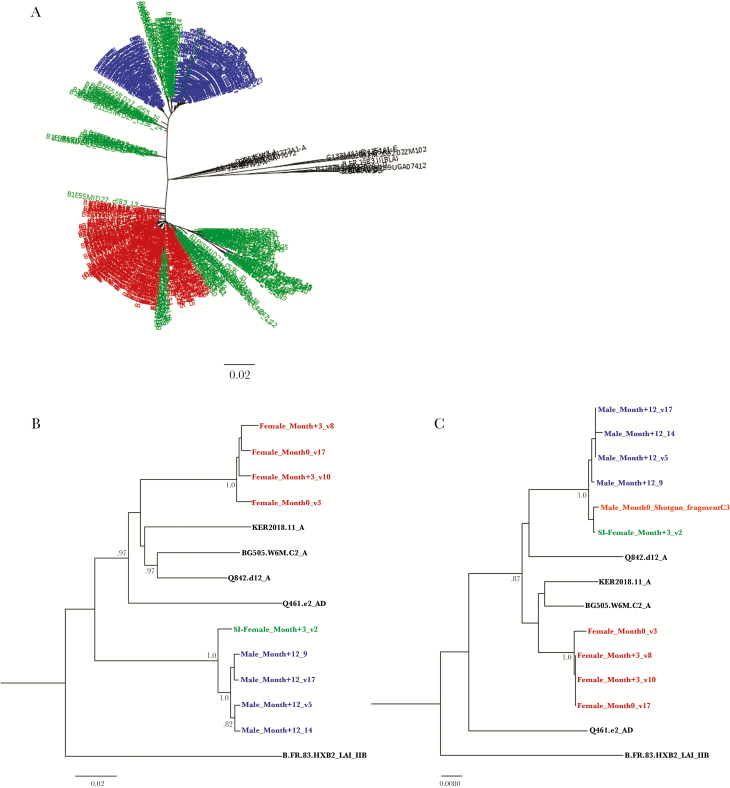
Figure 1.
Sequencing results demonstrate a linked human immunodeficiency virus superinfection (HIV-SI) event. A, Neighbor-joining phylogenetic tree of consensus gp41 viral sequences (≥10 reads) derived from next-generation sequencing (NGS) of viruses collected at the initial time point from the female participant (red) and her husband (blue; at −19 months), as well as viral sequences collected from the female participant immediately after HIV-SI (ie, during month 3; green), with the superinfecting viral strains clustering with virus from her husband. The number of repeated sequences represented by each NGS consensus sequence is shown at the end of the consensus identifier. B, Neighbor-joining tree of full SGA-derived viral envelopes used for pseudotyped viruses. C, Neighbor-joining tree of 230 base pairs of the 5′ end of the viral envelope from the pseudotyped viral isolates aligned with the NGS shotgun-sequencing fragment from husband’s sample collected before HIV-SI (orange). The fragment clusters with the superinfecting strain found in his wife immediately after HIV-SI. Distances are indicated for the tree by the scale at bottom, and samples are grouped with a selection of subtype reference sequences (black). Bootstrap values >80% are indicated (1000 replicates).
Full-length viral Env sequences were obtained from the female partner immediately before HIV-SI (at month 0; n = 21) and when HIV-SI was first detected 3 months later (at month 3; n = 10; Supplementary Figure 4). Three of the viral sequences from this later sample were phylogenetically linked to the husband’s viruses, thus representing the superinfecting strain (Figure 1B and Supplementary Figure 4). Full-length Env sequences collected from the husband 12 months after he superinfected the female participant were also generated (n = 24; Supplementary Figure 4). Full-length Env sequences collected from the husband and subjected to SGA after HIV-SI contained regions from both subtype A and D, indicating a unique A/D recombinant and corroborating the assertion that the NGS data from gag and pol came from the same virus (Supplementary Figure 3). The husband had no indication of HIV-SI between the 2 time points examined by NGS or in any of the SGA sequences examined later (Supplementary Figure 2). Repeated attempts to amplify full-length envelope genes of viruses collected from the husband at earlier time points were unsuccessful; however, shotgun NGS analysis of viral RNA recovered from his serum sample at the time of HIV-SI (ie, during month 0.3) identified 1 fragment with a 230–base pair overlap into the 5′ end of the viral envelope region. This fragment differed by only 1 nonsynonymous nucleotide mutation from the 3 superinfecting strains found in the female after HIV-SI (Figure 2C).
Figure 2.
Sera from the female subject sera did not neutralize the superinfecting viral strain. A, Values in table indicate the dilution of the heat-inactivated serum required to block 50% of a standard infectious dose (ID50): weak (green), moderate (yellow), and strong (orange) neutralization values are highlighted. Along the top of the table are indicated time points at which sera were collected from the woman and her husband, as well as a collection of sera from individuals infected with human immunodeficiency virus (HIV) subtype A and A/D. The 3 columns to the left show information on Env pseudoviruses tested, including the month and visit time point. The female SI virus (SI-Female Month+3_v2) is in green. Sera from individuals screened for linked superinfection are indicated by couple number and member identifier (Supplementary Table 1). To provide a benchmark for the varied levels of neutralization activity against autologous viruses, sera from the husband and wife were also tested against a panel of 6 previously described HIV pseudoviruses (heterologous virus panel). Serum samples collected from the woman before HIV-SI displayed a measurable neutralizing antibody response to 5 of 6 unassociated pseudoviruses, and the serum sample collected from the husband at the time of HIV-SI was weakly neutralizing against all pseudoviruses tested. B, ID50 values of the female participant’s samples over time against the corresponding heterologous and homologous pseudoviruses are shown.
Full-length Env amplicons from SGA were subcloned or synthesized and used to generate Env pseudoviruses for both the female participant (2 from month 0 and 3 from month 3, including 1 superinfecting strain) and her husband (9 from month 12, with only 4 used for subsequent assays; Figure 2C and Supplementary Figure 4) [14]. All pseudoviruses were examined for functionality and neutralization susceptibility to well-described anti-HIV monoclonal antibodies, as well as to a variety of subtype A and A/D serum from historic serum samples and nonsuperinfected Ugandans in the same cohort [15]. These pseudoviruses demonstrated varying susceptibility to the monoclonal antibodies and serum tested (Figure 2 and Supplementary Figure 5). Based on this susceptibility, none of the Env pseudoviruses from the couple were unusually sensitive to neutralization and all had a tier 2–like phenotype.
Serum samples from the female participant (collected at months −3, 0, 3, and 10) and the husband (collected at months 0.3 and 12) were tested for their neutralization activity against the couple’s Env pseudoviruses (Figure 2). The female participant’s serum samples collected before HIV-SI displayed moderate NAb activity against her homologous virus. However, serum specimens collected before and immediately after HIV-SI contained no detectable NAb activity to the superinfecting strain and weak responses to her husband’s strains collected 1 year later (ie, during month 12) that were genetically similar to the superinfecting strain (Figure 2A). Ten months after HIV-SI, the female participant had developed a moderate response to the superinfecting strain (Figure 2). In contrast, there was no increase in NAb response to the other viruses collected from her husband during month 12 (Figure 2).
The serum specimen collected from the husband at the time of HIV-SI had no detectable neutralizing activity against his wife’s strains, and his NAb responses to those strains did not improve 1 year after HIV-SI (Figure 2). However, sera collected from him 1 year after HIV-SI had high-titer NAb activity against the superinfecting strain (Figure 2).
DISCUSSION
This identification and characterization of a genetically confirmed case of a linked heterosexual HIV-SI event provides a unique opportunity to examine HIV-SI in an individual whose infecting partner is known. In this case, HIV-SI occurred in a chronically infected female who had moderately potent and broad anti-HIV NAb responses. Despite this, she possessed no detectable NAb response to the superinfecting strain during the estimated window when HIV-SI occurred, which potentially could have protected her against the superinfecting strain. This lack of response was not due to an inability to develop a NAb response to this strain, since she developed a moderate NAb response to the superinfecting virus approximately 7 months after superinfection, as well as a low response to 3 of 4 other viruses isolated from her male partner. It is interesting that the husband possessed a very limited NAb response to the viruses tested, even after being infected for >30 months at the time of HIV-SI. However, like his female partner, his NAb response to the superinfecting strain, which originally came from him, increased significantly 12 months after the superinfection occurred.
There is a large body of preclinical data indicating that NAb can confer protective immunity against animal lentiviruses. The data from this case report agree with the widely held concept that NAb are an important component of protective immunity against HIV infection, and thus that a successful HIV vaccine should aim to elicit a broadly reactive NAb response [16]. As with any single case, these data are supportive but not conclusive. Also, this study was limited by the sample types (serum only) and volumes available, as this was a secondary analysis of a previous study performed >15 years ago. The limited sample volume for this couple precluded examination of other interesting aspects of the humoral immune response that may play a role in protection against HIV-SI, and limited our ability to fully characterize the neutralization breadth of the couple before and after HIV-SI. The totality of the data were also limited by the inability to amplify full envelope sequences from the male partner prior to HIV-SI. However, the superinfecting strain’s viral envelope sequence isolated from the woman at the time of HIV-SI was almost identical to a fragment of envelope sequence collected from the man prior to HIV-SI, suggesting that this isolate is extremely similar to the superinfecting viral strain.
Notably, the husband possessed no detectable NAb response to the woman’s heterologous virus, yet he did not become superinfected. This could be influenced by the possibility that NAb have no protective role against HIV-SI, by the increased risk of male-to-female transmission as compared to female-to male transmission, or by protection due to a different immunological response not examined here [8, 17]. In summary, this case demonstrates the exciting amount of potential information that even a small number of these types of cases could provide and supports the need to further examine historical cohorts for linked HIV-SI events.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments We thank all of the study physicians and staff; all of the study participants; and Julie Overbaugh, Stephanie Rainwater, Rebecca Lynch, and Patrick Madden, for PCR protocols.
Disclaimer. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Financial support. This work was supported by the United Kingdom Medical Research Council (MRC) and the United Kingdom Department for International Development (DFID) under the MRC/DFID Concordat agreement; and by the Vaccine Research Center and Division of Intramural Research, National Institute of Allergy and Infectious Disease, National Institutes of Health. This study was approved by the Science and Ethics Committee of the Uganda Virus Research Institute and the Uganda National Council for Science and Technology (institutional review board number 00001693). Clinical data, epidemiological data, and blood samples were obtained following receipt of written informed consent.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Redd AD, Quinn TC, Tobian AA. Frequency and implications of HIV superinfection. Lancet Infect Dis 2013; 13:622–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Redd AD, Ssemwanga D, Vandepitte J et al. Rates of HIV-1 superinfection and primary HIV-1 infection are similar in female sex workers in Uganda. AIDS 2014; 28:2147–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ronen K, McCoy CO, Matsen FA et al. HIV-1 superinfection occurs less frequently than initial infection in a cohort of high-risk Kenyan women. PLoS Pathog 2013; 9:e1003593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kraft CS, Basu D, Hawkins PA et al. Timing and source of subtype-C HIV-1 superinfection in the newly infected partner of Zambian couples with disparate viruses. Retrovirology 2012; 9:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Basu D, Kraft CS, Murphy MK et al. HIV-1 subtype C superinfected individuals mount low autologous neutralizing antibody responses prior to intrasubtype superinfection. Retrovirology 2012; 9:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smith DM, Strain MC, Frost SD et al. Lack of neutralizing antibody response to HIV-1 predisposes to superinfection. Virology 2006; 355:1–5. [DOI] [PubMed] [Google Scholar]
- 7. Cortez V, Odem-Davis K, McClelland RS, Jaoko W, Overbaugh J. HIV-1 superinfection in women broadens and strengthens the neutralizing antibody response. PLoS Pathog 2012; 8:e1002611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ronen K, Dingens AS, Graham SM et al. Comprehensive characterization of humoral correlates of human immunodeficiency virus 1 superinfection acquisition in high-risk Kenyan women. EBioMedicine 2017; 18:216–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Blish CA, Dogan OC, Derby NR et al. Human immunodeficiency virus type 1 superinfection occurs despite relatively robust neutralizing antibody responses. J Virol 2008; 82:12094–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Redd AD, Collinson-Streng A, Martens C et al. ; Rakai Health Sciences Program. Identification of HIV superinfection in seroconcordant couples in Rakai, Uganda, by use of next-generation deep sequencing. J Clin Microbiol 2011; 49:2859–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Blick G, Kagan RM, Coakley E et al. The probable source of both the primary multidrug-resistant (MDR) HIV-1 strain found in a patient with rapid progression to AIDS and a second recombinant MDR strain found in a chronically HIV-1-infected patient. J Infect Dis 2007; 195:1250–9. [DOI] [PubMed] [Google Scholar]
- 12. Asiki G, Murphy G, Nakiyingi-Miiro J et al. ; GPC team. The general population cohort in rural south-western Uganda: a platform for communicable and non-communicable disease studies. Int J Epidemiol 2013; 42:129–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ssemwanga D, Lyagoba F, Ndembi N et al. Multiple HIV-1 infections with evidence of recombination in heterosexual partnerships in a low risk Rural Clinical Cohort in Uganda. Virology 2011; 411:113–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Montefiori DC. Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol Biol 2009; 485:395–405. [DOI] [PubMed] [Google Scholar]
- 15. Bonsignori M, Montefiori DC, Wu X et al. Two distinct broadly neutralizing antibody specificities of different clonal lineages in a single HIV-1-infected donor: implications for vaccine design. J Virol 2012; 86:4688–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lewis GK, Pazgier M, DeVico AL. Survivors Remorse: antibody-mediated protection against HIV-1. Immunol Rev 2017; 275:271–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hughes JP, Baeten JM, Lingappa JR et al. ; Partners in Prevention HSV/HIV Transmission Study Team. Determinants of per-coital-act HIV-1 infectivity among African HIV-1-serodiscordant couples. J Infect Dis 2012; 205:358–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.