Abstract
Atopic dermatitis is a chronic and recurrent inflammatory skin disease. Recently, probiotics have been shown to suppress allergic symptoms through immunomodulatory responses. In the present study, combinatorial effects on allergic symptoms were identified in BALB/c mice fed with a mixture of four species of probiotics, Bifidobacterium lactis, Lactobacillus casei, Lactobacillus rhamnosus, and Lactobacillus plantarum, and sodium butyrate. Following sensitization with whey protein, the mice were challenged and divided into two groups: (1) mice administered with phosphate-buffered saline as a control and (2) mice administered with the probiotic mixture and sodium butyrate. Allergic symptoms were assessed by measuring ear thicknesses, serum histamine and IL-10 concentrations, and the quantities of leaked Evans blue. T cell differentiation was determined by analyzing the T cells groups in the mesenteric lymph nodes (MLNs) and spleen. To examine changes in the total gut microbiota, total fecal microflora was isolated, species identification was performed by DNA sequencing using Illumina MiSeq, and changes in intestinal beneficial bacteria were analyzed using quantitative polymerase chain reaction. Treatment with the probiotic mixture and sodium butyrate reduced ear thicknesses, the quantity of leaked Evans blue, and serum histamine values, while increasing serum IL-10 values. In the mouse model, the probiotic mixture and sodium butyrate increased Th1 and Treg cell differentiation in MLN and spleen tissues; the ratio of Firmicutes/Bacteroidetes, which is associated with reduction in allergic reactions; and microorganisms that lead to cell differentiation into Treg. These results suggest that the probiotic mixture and sodium butyrate can prevent and alleviate allergic symptoms.
Keywords: : Galectin9, gut microbiota, Th1 cells/Th2 cells, Treg cells
Introduction
Atopic dermatitis is accompanied with erythema, severe itching, and hemorrhage1 and its causes include environmental changes, hyperimmune responses, stress, and genetic factors.2 It is a type 1 hypersensitivity response mediated by mast cell degranulation and a type 2 helper T cells (Th2)-mediated allergic disease.3 The imbalance between Th1 and Th2 cells leads to autoimmune disorder,4 and, in particular, increases the levels of Th2 cells associated with the onset of atopy.5 Th2 cells produce cytokines such as IL-4, IL-5, and IL-13,6 these cytokines produce antigen-specific IgE, and the resulting IgE activates mast cells to release histamine.7
Probiotics are defined as live microorganisms that are beneficial to the health of the host.8 Specifically, they have been shown to alleviate allergic reactions by modifying the structures of allergens and by inhibiting the adherence of pathogens to intestinal epithelia or mucous membranes as well as the growth of pathogens.9 In addition, probiotics fine-tune the intestinal microflora composition to induce immune-modulatory effects.10
Short chain fatty acids such as acetate, butyrate, and propionate can be produced when the gut microbiota utilizes dietary fibers as an energy source.11,12 Especially, butyrate-producing bacteria promotes the development of tight junction leading to enhancing the function of intestine barrier, while inhibits proinflammatory signaling.13 Mechanistically, butyrate has been known to bind to receptors such as G protein-coupled receptors GPR41, GPR43, and GPR109A to modulate cell activation, proliferation, and differentiation.14 In terms of host immune responses in the colon, butyrate elevates the activation and proliferation of Treg cells to repress CD4+ T cells, maintaining of human gut health.15–17
However, studies on the effects of probiotics and sodium butyrate on changes in the gut microbiota are still insufficient. In this study, we investigated the mitigation effect of probiotic mixture and sodium butyrate on atopic dermatitis in an in vivo mouse model.
Materials and Methods
Animals
Three-week-old female BALB/c mice (Samtako, Osan, Korea) and 4- and 6-week-old male Sprague–Dawley rats (Samtako) were maintained at room temperature (22°C ± 1°C), with a 12-h light–dark cycle during the experimental period and were provided ad libitum access to food (AIN-76A; Central Lab, Seoul, Korea) and water. The probiotic mixture, which was a mixture of four species of lactic acid bacteria (Lactobacillus casei, Lactobacillus rhamnosus, Lactobacillus plantarum, and Bifidobacterium lactis [Cell Biotech, Gimpo, Korea]), was provided at a concentration 2% wt:wt 2 × 109 CFU/g, and sodium butyrate (Sigma-Aldrich, CA, USA) was provided at a concentration of 100 mM/0.2 mL phosphate-buffered saline (PBS). After a preliminary experimental period of 1 week, the experimental animals were randomly divided into five groups: (1) control group (C: normal diet), (2) negative control group (N: normal diet+whey proteins), (3) probiotics mixture group (T1: normal diet+whey proteins+probiotics mixture), (4) sodium butyrate group (T2: normal diet+whey proteins+sodium butyrate), and (5) probiotics mixture+sodium butyrate group (T3: normal diet+whey proteins+probiotics mixture+sodium butyrate). The experimental protocol was approved by the Institutional Animal Care Board of Gyeongnam National University of Science and Technology (Approval No. 2016-7).
Acute hypersensitivity response
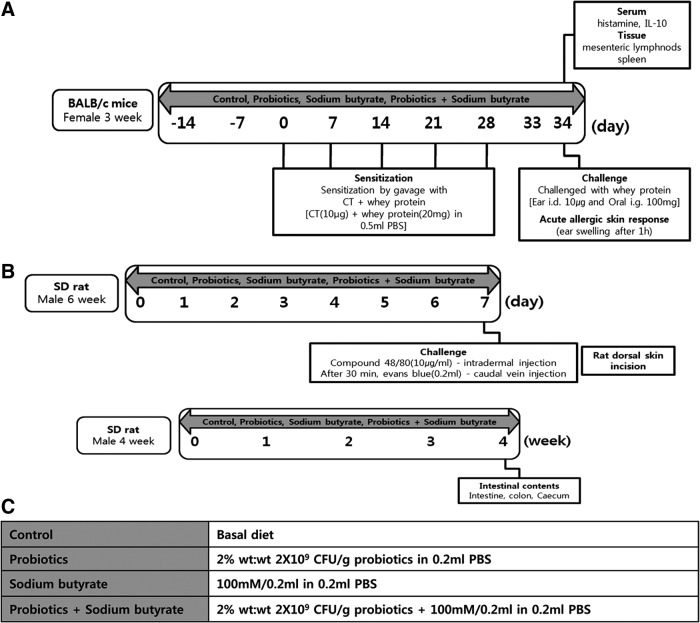
According to the method presented by de Kivit et al.,18 allergic symptoms were induced in BALB/c mice using whey proteins and cholera toxin (CT; Fig. 1A). Whey protein consists of lactalbumin and lactoglobulin, which are considered to be major cow's milk allergens.19 The probiotics mixture and sodium butyrate were dissolved in 0.2 mL PBS and orally administered once per day from 2 weeks before sensitization until the end of the experimental period. Whey proteins (20 mg) and CT (10 μg) were dissolved in 0.5 mL PBS and orally administered once per week from day 0 to 28 to sensitize the mice. At day 33, whey proteins were dissolved in PBS and were orally administered (100 mg) and intradermally injected (10 μg) to challenge the mice. Ear thickness was measured using digital calipers (Bluebird; NA500-150S) 1 h after challenging with whey proteins. The following day, the BALB/c mice were sacrificed and blood samples were collected from the vena cava; the blood samples were centrifuged for 10 min at 1300 g to separate the serum. Histamine levels in the separated serum were measured using the Histamine EIA kit (LDN, Germany), and IL-10 levels in the separated serum were measured using the Quantikine Human IL-10 Immunoassay kit (R&D, USA).
FIG. 1.
Experimental design. (A) Acute hypersensitivity response. (B) Histamine induced vasodilation. (C) Analysis of Intestinal microbiota. CT, cholera toxin; PBS, phosphate-buffered saline.
Histamine-induced vasodilation
The atopic dermatitis model was established by triggering histamine release using compound 48/80 (COM; Sigma-Aldrich) (Fig. 1B).20 In brief, 6-week-old Sprague–Dawley rats were used as atopic dermatitis models, and the probiotics mixture and sodium butyrate dissolved in 0.5 mL PBS were orally administered to the mice once per day for 7 days. Before inducing atopic dermatitis, the dorsal fur of the mice was resected using electric shavers. Next, 50 μL of COM (10 μg/mL) was intradermally injected into the dorsal dermis, and 30 min later, Evans blue (Sigma-Aldrich) was injected into the tail vein. All the animals were sacrificed 30 min there after. To measure the quantity of leaked Evans blue, the dorsal dermis of the mice was resected, maintained for 48 h in 1.0 mL of 1.0 N KOH at 37°C, followed by adding 0.6 N H3PO4 and acetone, and centrifuged for 10 min at 900 g. Next, the concentration of Evans blue was measured at 620 nm using a spectrophotometer (Biodrop Ltd., United Kingdom).
Separation and analysis of the RNA of mouse spleen and mesenteric lymph nodes
To analyze the immunomodulatory effects of the probiotics mixture and sodium butyrate, acute hypersensitivity BALB/c mice were sacrificed and mesenteric lymph node (MLN) and spleen were separated. TRIzol® was added to MLN and spleens of BALB/c mice and homogenized using SilentCrusher M (Heidolph, Germany). According to the method presented by Chomczynski and Sacchi,21 RNA was extracted and stored at −20°C until cDNA synthesis. cDNA was synthesized using reverse transcription-polymerase chain reaction (RT-PCR) kits (TaKaRa Co., Tokyo, Japan) according to the manufacturer's instructions. The list of the primer sequences is given in Table 1. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the reference gene.
Table 1.
Reverse Transcription-Polymerase Chain Reaction Primer Sequences for Immune-Modulatory Analysis
| Target gene | Primer | |
|---|---|---|
| GAPDH | Forward | 5′ CCACCCAGAAGACTGTGGAT 3′ |
| Reverse | 5′ CACATTGGGGGTAGGAACAC 3′ | |
| T-bet | Forward | 5′ TCAACCAGCACCAGACAGAG 3′ |
| Reverse | 5′ AAACATCCTGTAATGGCTTGTG 3′ | |
| Gata3 | Forward | 5′ CATTACCACCTATCCGCCCTATG 3′ |
| Reverse | 5′ CACACACTCCCTGCCTTCTGT 3′ | |
| Rorγt | Forward | 5′ TTCACCCCACCTCCACTG 3′ |
| Reverse | 5′ TGCAAGGGATCACTTCAATTT 3′ | |
| Foxp3 | Forward | 5′ CCCATCCCCAGGAGTCTTG 3′ |
| Reverse | 5′ CCATGACTAGGGGCACTGTA 3′ | |
| Galectin9 | Forward | 5′ GAGAGGAAGACACACATGCCTTTC 3′ |
| Reverse | 5′ GACCACAGCATTCTCATCAAAACG 3′ | |
Analysis of gut microbiota
Four-week-old Sprague–Dawley rats were used to examine the effects of the probiotics mixture and sodium butyrate on changes in gut microbiota (Fig. 1C). Same volumes of the probiotics mixture and sodium butyrate as the acute hypersensitivity response experiment were dissolved in PBS, and 0.5 mL of the solution was orally administered once per day for 4 weeks. Four weeks later, all the animals were sacrificed and fecal samples of the Sprague–Dawley rats were collected and kept at −80°C until genomic DNA (gDNA) extraction. DNA was extracted using Fecal DNA MiniPrep Kits (Zymo Research, CA, USA), and the sequence was analyzed using Illumina MiSeq (ChunLab, Inc., Seoul, Korea).
Quantitative real-time PCR
As mentioned above, DNA was extracted from rat fecal samples. Quantitative PCR (qPCR) was performed using Rotor-Gene SYBR® Green PCR Kits (QIAGEN GmbH, Germany). A list of the primer sequences of intestinal beneficial bacteria is presented in Table 2. Universal was used as the reference gene.
Table 2.
Quantitative Polymerase Chain Reaction Primer Sequences for Beneficial Bacteria Analysis
| Target gene | Primer | |
|---|---|---|
| Universal | Forward | 5′ GTGSTGCAYGGYYGTCGTCA 3′ |
| Reverse | 5′ ACGTCRTCCMCNCCTTCCTC 3′ | |
| Faecalibacterium prausnitzii | Forward | 5′ GGAGGAAGAAGGTCTTCGG 3′ |
| Reverse | 5′ AATTCCGCCTACCTCTGCACT 3′ | |
| Ruminococcus | Forward | 5′ GGCGGCYTRCTGGGCTTT 3′ |
| Reverse | 5′ CCAGGTGGATWACTTATTGTGTTAA 3′ | |
| Bifidobacterium | Forward | 5′ TCGCGTCYGGTGTGAAAG 3′ |
| Reverse | 5′ GGTGTTCTTCCCGATATCTACA 3′ | |
| Weissella cibaria | Forward | 5′ TTGATTGACATAGAACCTGAT 3′ |
| Reverse | 5′ TTCGGTGCTAGTTCTTCAATA 3′ | |
Statistical analysis
The variance of the results obtained through repeated experiments was analyzed using SPSS 12.0 (SPSS, Inc., Chicago, IL, USA). The significance of the results of analysis in the treatment groups were tested using Duncan's multiple range test at a level of P < .05, and intergroup significance was tested using the independent-sample t-test at levels of P < .05 and P < .01. All results are indicated as mean ± standard deviation.
Results
Allergic symptoms in mice
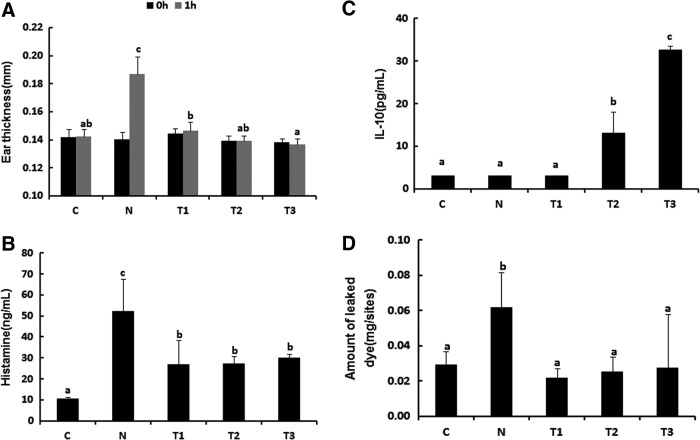
To examine the effects of administration of the probiotics mixture and sodium butyrate on the allergic symptoms, we first measured the ear thickness, which is a pathological phenotype in mice orally sensitized and challenged with whey protein. As shown in Figure 2A, ear thickness decreased significantly in the T1, T2, and T3 groups compared with that in the N group (Fig. 2A; P < .05). Next, we examined the histamine and IL-10 levels because allergic symptoms are accompanied with increases in serum histamine levels and decreases in serum IL-10 levels due to increases in the Th2 cells immune response.5 Serum histamine levels decreased significantly in all treatment groups, whereas IL-10 concentration increased in the T2 and T3 groups compared with those in the N group (Fig. 2B, C; P < .05). Similar results were also observed in mice intradermally injected with COM, an allergic reaction inducer. Reduced Evans blue staining was observed in the dorsal dermis of mice in the T1, T2, and T3 groups due to vasodilation (Fig. 2D). Therefore, these results suggest that the probiotics mixture and sodium butyrate inhibit allergic symptoms.
FIG. 2.
Probiotic mixture and sodium butyrate suppress allergic symptoms in mice. (A) One hour after intradermal challenge with whey in the ear, acute hypersensitivity response was reduced in whey-sensitized mice treated with probiotics mixture and sodium butyrate compared with whey-sensitized mice (n = 6). (B) Total amount of serum histamine was reduced and (C) serum IL-10 was increased treated with probiotics mixture and sodium butyrate at after the last challenge (n = 3). (D) After an intradermal injection with compound 48/80 in the skin, amount of leaked dye was reduced in whey-sensitized mice treated with probiotics mixture and sodium butyrate compared with whey-sensitized mice (n = 5). a–cMeans are significantly different within the same row (P < .05). C, control; N, negative control group (allergy induce); T1, probiotic mixture group (2% wt:wt 2 × 109 CFU/g-Lactobacillus casei, Lactobacillus rhamnosus, Lactobacillus plantarum, Bifidobacterium lactis+allergy inducer); T2, sodium butyrate group (100 mM/0.2 mL-sodium butyrate+allergy inducer); T3, probiotic mixture+sodium butyrate group (probiotic mixture+sodium butyrate+allergy inducer).
T cell differentiation in MLN and spleen
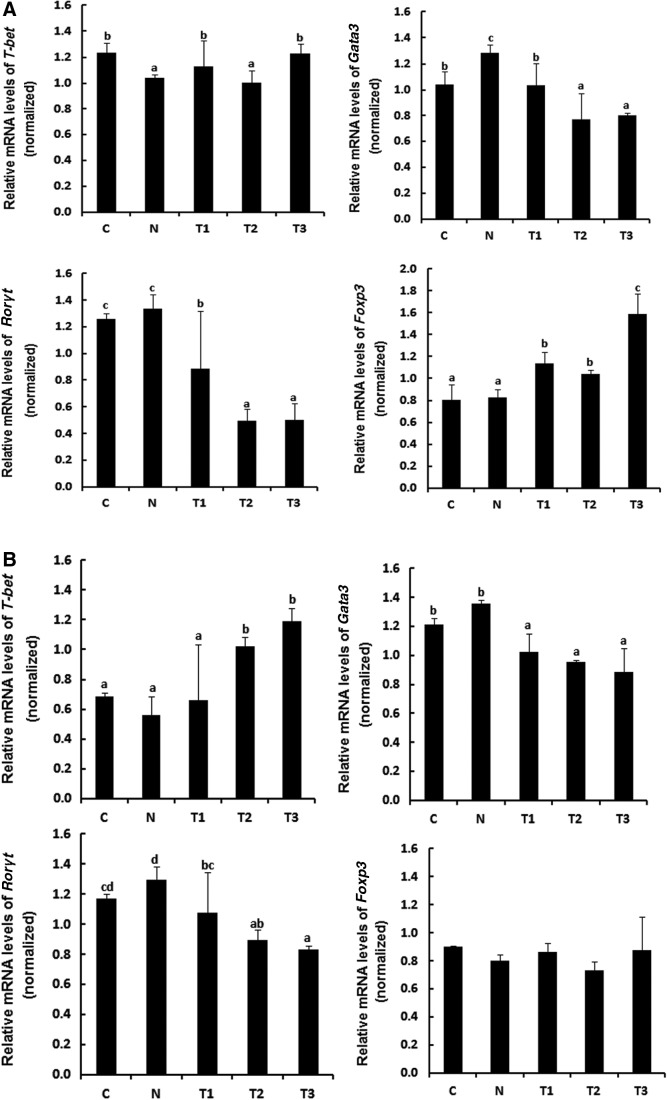
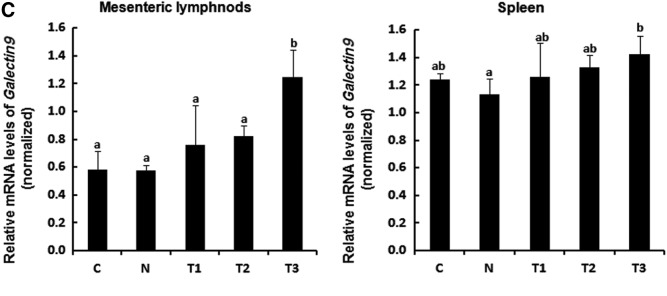
To understand the effects of the probiotics mixture and sodium butyrate on changes in T cells population, mRNA levels of T-bet, Gata3, Rorγt, and Foxp3, which are transcription factors specific to Th1, Th2, Th17, and Treg cells, respectively, were analyzed by RT-PCR in MLN and spleen tissues from whey-allergic mice. mRNA expression of T-bet and Foxp3 increased in the T1, T2, and T3-treated MLN, but the difference between T2-treated MLN and the N group was not significant (Fig. 3A). However, the treatment with T1, T2, and T3 caused a marked decrease in GATA3 and Rorγt mRNA expression. In particular, the T3 group exhibited a significant increase in the mRNA expression of Foxp3 by 1.9-fold compared with the N group, indicating that the treatment with probiotics mixture and sodium butyrate may induce immunomodulatory responses by changing the balance of Th1, Th2, and Treg cells, resulting in alleviating atopic dermatitis occurring due to immune imbalance states. mRNA expression of T-bet increased in the T2 and T3-treated spleen, but there is no statistical significant in the Foxp3 mRNA expression of experimental groups in spleen (Fig. 3B). To further determine the effect of the probiotics mixture and sodium butyrate on Galectin9 expression, Galectin9 mRNA expression in MLN and spleen was determined by RT-PCR. Galectin9 modulates mast cell degranulation and T cell differentiation.18 As shown in Figure 3C, Galectin9 expression increased significantly in the MLN of the T3 group compared with that in the MLN of the N group (P < .05). However, in the spleen, Galectin9 expression increased slightly in the T1 and T2 groups and increased significantly in the T3 group (P < .05). It is known that Gata3 gene is a specific marker during Th2 cell differentiation.22 In the Figure 3, due to the low expression level of Gata3 in T2 groups, we believe probiotic mixture and sodium butyrate group reduces Th2 cell differentiation. Our results suggest that treatment with the probiotics mixture+sodium butyrate reduces Th2 cell differentiation while increasing Th1 and Treg cell differentiation. Therefore, we propose that probiotics mixtures and sodium butyrate can alleviate atopic dermatitis through immunomodulation.
FIG. 3.
Whey-allergic mice fed probiotics mixture and sodium butyrate show enhanced Th1- and Treg-cell development in MLNs and spleen. T cell polarization in (A) MLN and (B) spleen was evaluated by analyzing the expression of T-bet (Th1), GATA3 (Th2), RORγT (Th17), and Foxp3 (Treg). The probiotics mixture and sodium butyrate increased the differentiation of Th1 and Treg cells and decreased the differentiation of Th2 and Th17 cells. (C) Galectin9 is expressed by intestinal epithelial cells and the probiotics mixture and sodium butyrate increased the Galectin9 in MLN and spleen. a–dMeans are significantly different within the same row (P < .05). C, control; N, negative control group (allergy inducer); T1, probiotic mixture group (2% wt:wt 2 × 109 CFU/g-L. casei, L. rhamnosus, L. plantarum, B. lactis+allergy inducer); T2, sodium butyrate group (100 mM/0.2 mL-sodium butyrate+allergy inducer); T3, probiotic mixture+sodium butyrate group (probiotic mixture+sodium butyrate+allergy inducer). MLN, mesenteric lymph node.
Effects of probiotics mixture and sodium butyrate on gut microbiota
Gastrointestinal microbiota influence Th1, Th2, and Treg maturation.23 As shown in Figure 4A, the ratio of Firmicutes to Bacteroides increased significantly in the T3 group compared with that in the C, T1, and T2 groups. In addition, the results of the analysis of gut microbiota indicated that Firmicutes (64.85–75.31%) existed in the highest ratio, which was more than half of the total population of gut microbiota. When the results were analyzed at the class level, Clostridia was dominant at 50.52–62.17% in the Firmicutes (Fig. 4B).
FIG. 4.
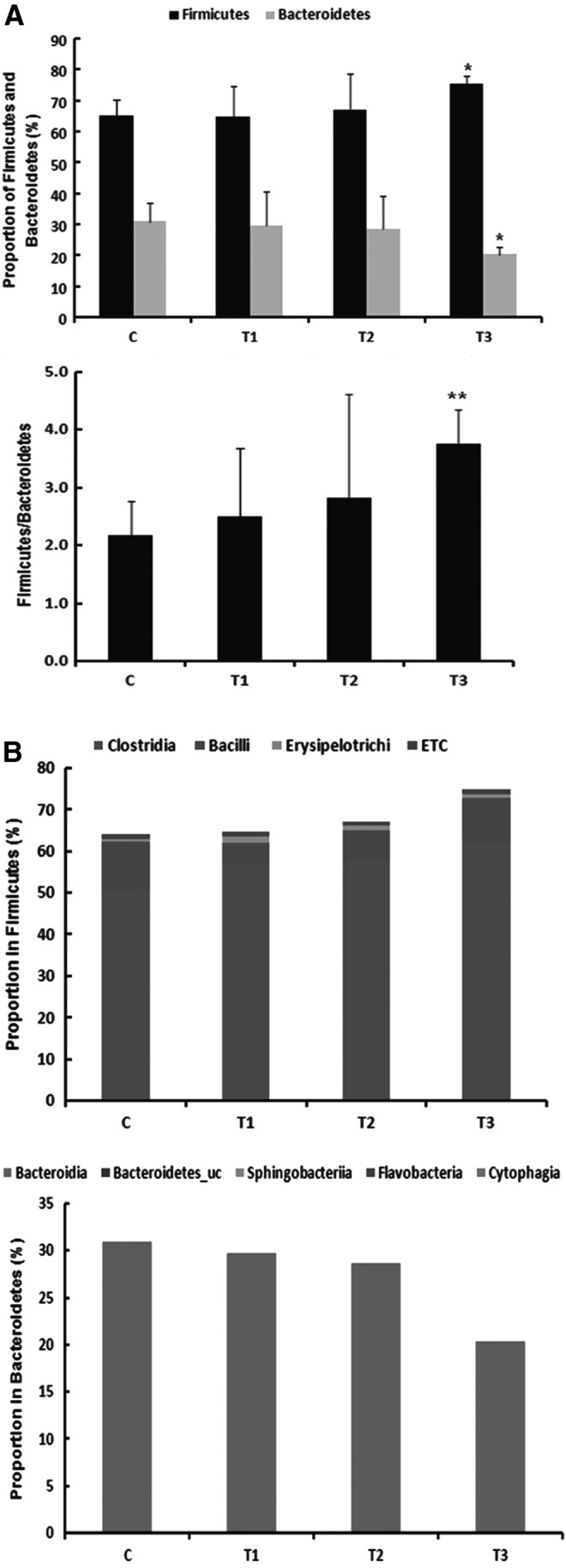
Pyrosequencing analysis of the fecal microbiota composition from mice fed the probiotics mixture and sodium butyrate. (A) Relative abundance ratio of Firmicutes and Bacteroidetes in fecal microbiota. (B) Population of Firmicutes and Bacteroidet at class level. *Means are significantly different within the same row (P < .05), **Means are significantly different within the same row (P < .01). C, control; T1, probiotic mixture group (2% wt:wt 2 × 109 CFU/g-L. casei, L. rhamnosus, L. plantarum, B. lactis+allergy inducer); T2, sodium butyrate group (100 mM/0.2 mL-sodium butyrate+allergy inducer); T3, probiotic mixture+sodium butyrate group (probiotic mixture+sodium butyrate+allergy inducer).
Effects of probiotics mixture and sodium butyrate on increases in intestinal beneficial bacteria
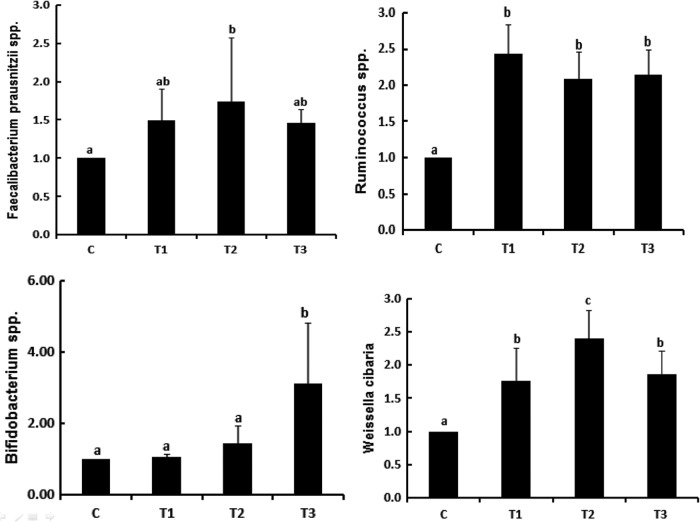
We could not observe synergistic effects of combinatorial treatment of probiotics mixtures and sodium butyrate to regulate intestinal beneficial bacteria such as Faecalibacterium prausnitzii spp. and Weissella cibaria. In Figure 5, it seems to be more effective of sodium butyrate to increase their populations. It is known that F. prausnitzii spp. promotes Treg cell differentiation by producing butyrate as a butyrate-producing bacteria in the gut.24 In addition, W. cibaria has been shown to block Th2 cell differentiation as well as IgE.25 Taken together, we concluded T2 group containing sodium butyrate repress the symptoms of atopy dermatitis.
FIG. 5.
The abundance of functional bacteria in the feces of probiotics mixture and sodium butyrate by quantitative PCR. a–cMeans are significantly different within the same row (P < .05). C, control; T1, probiotic mixture group (2% wt:wt 2 × 109 CFU/g-L. casei, L. rhamnosus, L. plantarum, B. lactis+allergy inducer); T2, sodium butyrate group (100 mM/0.2 mL-sodium butyrate+allergy inducer); T3, probiotic mixture+sodium butyrate group (probiotic mixture+sodium butyrate+allergy inducer). PCR, polymerase chain reaction.
Increase in immunomodulatory effects by probiotics mixture and sodium butyrate
Among the gut microbiota, Clostridia has been implicated in preventing diseases such as food allergies.26 As shown in Table 3, Clostridia population increased significantly to 62.17% in the T3 group compared with the C group with a ratio of 50.52%. In particular, the IV and XIVa clusters of Clostridia alleviate allergy and inflammation by inducing Treg cells.27 Therefore, we verified the immunomodulatory effects of Clostridia clusters IV and XIVa in gut microbiota. According to our results, at the genus level, Eubacterium, which belongs to Clostridia cluster XIVa, increased significantly in the T3 group to 10.54% compared with that in the C group, where its proportion was 5.55% (P < .01). In addition, Clostridium, which belongs to Clostridia clusters XIVa and IV, increased significantly in the T1 (7.00%) and T3 groups (8.49%) compared with that in the C group (5.09%; P < .05). Clostridium was also shown to exist in the highest ratio in the T3 group. Eubacterium (7.37%) and Clostridium (7.18%) increased in the T2 group; however, the differences were not significant. Therefore, the present study indicates that the probiotics mixture and sodium butyrate can induce Treg cells through increases in Clostridia clusters IV and XIVa and increase IL-10 concentrations in the blood, there by alleviating allergic reactions.
Table 3.
Pyrosequencing Analysis of the Fecal Microbiota Composition from Mice Fed the Probiotics Mixture and Sodium Butyrate
| Treatments | ||||
|---|---|---|---|---|
| C n = 4 Mean% (SD) | T1 n = 4 Mean% (SD) | T2 n = 4 Mean% (SD) | T3 n = 4 Mean% (SD) | |
| Firmicutes | 65.01 (5.15) | 64.85 (9.92) | 67.11 (11.64) | 75.31 (2.49)* |
| Clostridia | 50.52 (6.87) | 56.99 (11.81) | 57.87 (8.23) | 62.17 (4.02)* |
| Lachnospiraceae | 33.69 (3.74) | 37.57 (9.15) | 38.93 (7.52) | 41.40 (2.65)* |
| Ruminococcaceae | 13.56 (1.33) | 13.52 (3.79) | 14.99 (4.34) | 17.50 (1.82)* |
| Eubacterium | 5.55 (1.71) | 6.26 (3.61) | 7.37 (2.77) | 10.54 (1.96)** |
| Clostridium | 5.09 (0.74) | 7.00 (1.28)* | 7.18 (3.09) | 8.49 (2.48)* |
| Bacilli | 11.87 (4.38) | 5.18 (3.69) | 7.08 (2.60) | 10.65 (3.80) |
| Lactobacillaceae | 11.76 (4.36) | 5.12 (3.69) | 7.00 (2.56) | 10.55 (3.78) |
| Lactobacillales_uc | 0.03 (0.01) | 0.02 (0.01) | 0.02 (0.01) | 0.03 (0.01) |
| Streptococcaceae | 0.04 (0.02) | 0.03 (0.01) | 0.03 (0.01) | 0.04 (0.01) |
| Bacteroidetes | 30.99 (6.15) | 29.78 (10.81) | 28.65 (10.52) | 20.33 (2.53)* |
| Bacteroidia | 30.98 (6.16) | 29.77 (10.81) | 28.65 (10.51) | 22.13 (5.74)* |
| Prevotellaceae | 18.60 (12.19) | 14.84 (11.32) | 13.75 (11.66) | 2.91 (1.64)* |
| S24-7_f | 10.09 (6.51) | 10.88 (1.47) | 9.13 (1.87) | 9.86 (1.60) |
Means are significantly different within the same row (P < .05).
Means are significantly different within the same row (P < .01).
C, control; SD, standard deviation; T1, probiotics mixture group (2% wt:wt 2 × 109 CFU/g-Lactobacillus casei, Lactobacillus rhamnosus, Lactobacillus plantarum, Bifidobacterium lactis+allergy induce); T2, sodium butyrate group (100 mM/0.2 mL-sodium butyrate+allergy induce); T3, probiotics mixture+sodium butyrate group (probiotics mixture+sodium butyrate+allergy induce).
Discussion
Atopic dermatitis accompanies serum histamine release6 and IL-10 decrease by mast cell degranulation.28 In this study, we demonstrate the effects of the treatment with probiotic mixture and sodium butyrate to alleviate atopic dermatitis in vivo. Because COM induces mast cell degranulation and histamine releases in systemic anaphylaxis, it has been used for establishing vasodilation in atopic dermatitis model.29 Several studies have shown that a probiotic, Lactococcus chungangensis, could improve atopic dermatitis symptoms in a COM-induced mouse model.30 Using this model, we observed that a composite mixture of four species of probiotic bacteria and sodium butyrate mitigated atopic dermatitis symptoms due to histamine secretion.
Atopic dermatitis is characterized by Th1/Th2 cells imbalance and dominant Th2 cells-mediated responses.31,32–34 Recent studies have reported that Treg cells could trigger inflammatory responses in atopic dermatitis through Th2 cells regulation.25,35 In addition, Galectin9 is associated with the alleviation of allergy-related diseases as it induces Th1 and Treg cells immune responses and directly inhibits mast cell degranulation.18 In MLN and spleen tissues of atopic dermatitis mouse models, we demonstrated that the probiotics mixture and sodium butyrate increased the populations of Th1 and Treg cells and Galectin9 expression, while reducing the population of Th2 and Th17 cells. These data imply that probiotics mixture and sodium butyrate can induce positive effects by alleviating symptoms of atopic dermatitis.
Symbiotic microorganisms in the large intestine have significant effects on metabolism and immunity36 and cause allergic airway inflammation in response to allergens.37 Therefore, we hypothesized that administration of a probiotic mixture supplemented with sodium butyrate may be able to modulate atopic dermatitis by immunoregulation of the changes in intestinal symbiotic microorganisms. Most of the intestinal microorganisms (90–99%) are composed of Firmicutes and Bacteroidetes, and 50–80% of Firmicutes are primarily Clostridium clusters.38 In this study, we have shown that treatment with probiotics mixture and sodium butyrate increased Firmicutes while decreasing Bacteroidetes in the gut microbiota of rats. In addition, consistent with our findings, it has been shown that Firmicutes (phylum) and Clostridia (class) are present in high numbers in infants after completely treating milk allergies, whereas Bacteroidetes (phylum) are present in high numbers in infants suffering from milk allergies.39 Microorganisms belonging to the increasing Firmicutes population and the decreasing Bacteroidetes population were analyzed. Bacteroidia (class) and Prevotellaceae (family) were found to be the dominant species in Bacteroidetes. The population of intestinal Prevotellaceae (family), a pathogenic bacterium that causes chronic intestinal inflammation,40 decreased following administration of the probiotics mixture and sodium butyrate. When the families belonging to Firmicutes were analyzed, Lachnospiraceae and Ruminococcaceae were shown to be the dominant intestinal species, and their numbers increased after administration of the probiotics mixture and sodium butyrate. Lachnospiraceae and Ruminococcaceae are butyrate-producing bacteria.41 Gut microbiota utilize dietary fiber as an energy source to produce lactate and acetate, which are converted to butyrate by butyrate-producing bacteria.17 Bacteroidia probiotics exhibit a butyrogenic effect through interaction with bacteria that produce butyrate,12 and butyrate binds to receptors such as G protein-coupled receptors GPR43 and GPR109a to stimulate the differentiation of Treg cells.42 In a recent study, it was found that butyrate stimulates Treg cells to increase the production of IL-10.16 In addition, changes in the gut microbiota reduce the Th2 cell population increase caused by allergic reactions and promote Treg cell development,43 indicating that there is a correlation between gut microbiota and Treg cells during an allergic response.44 It is plausible that probiotics exert immunomodulatory effects by increasing butyrate that activates GPR43- and GPR109a-mediated signaling pathways. Clostridia is classified into 19 clusters, of which clusters IV, XIVa, and XVIII are known as Treg-inducing strains.45 In this study, Eubacterium belonging to Clostridia cluster XIVa and Clostridium belonging to Clostridia clusters XIVa and IV increased significantly following administration of the probiotics mixture and sodium butyrate. However, the increase in Ruminococcus, belonging to the Clostridia cluster XIVa, was not remarkable.
In conclusion, the intake of probiotics mixture promotes butyrate production by increasing the population of butyrate-producing bacteria. Butyrate enhances Treg cells, thereby acting as an immunomodulatory agent, to mitigate and prevent atopic dermatitis. In future studies, the pathways through which probiotics play the role of an immunomodulator between the gut microbiota and T cell differentiation should be investigated in detail.
Acknowledgments
This research was performed with the support of the Industry Core Technology Development Project (No. 10063302), Ministry of Trade, Industry, and Energy, Korea.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Inoue R, Otsuka M, Nishio A, Ushida K: Primary administration of Lactobacillus johnsonii NCC533 in weaning period suppresses the elevation of proinflammatory cytokines and CD86 gene expressions in skin lesions in NC/Nga mice. FEMS Immunol Med Microbiol 2007;50:67–76 [DOI] [PubMed] [Google Scholar]
- 2.Hwang JS, Im SH: Probiotics as an immune modulator for allergic disorders. Pediatr Allergy Respir Dis (Korea) 2012;22:325–335 [Google Scholar]
- 3.Soumelis V, Reche PA, Kanzler H, et al. : Human epithelial cell strigger dendritic cell mediated allergic inflammation by producing TSLP. Nat Immunol 2002;3:673–680 [DOI] [PubMed] [Google Scholar]
- 4.Asarat M, Apostolopoulos V, Vasiljevic T, Donkor O: Short-chain fatty acids regulate cytokines and Th17/Treg cells in human peripheral blood mononuclear cells in vitro. Immunol Invest 2016;45:205–222 [DOI] [PubMed] [Google Scholar]
- 5.Prescott SL, Macaubas C, Smallacombe T, et al. : Development of allergen-specific T-cell memory in atopic and normal children. Lancet 1999;353:196–200 [DOI] [PubMed] [Google Scholar]
- 6.Kim JY, Park BK, Park HJ, et al. : Atopic dermatitis-mitigating effects of new Lactobacillus strain, Lactobacillus sakei probio65 isolated from Kimchi. J Appl Microbiol 2013;115:517–526 [DOI] [PubMed] [Google Scholar]
- 7.Weber MB, Petry V, Weis L, Mazzotti NG, Cestari TF: Evaluating the relation between pruritus, serum IgE levels and severity of clinical manifestations in atopic dermatitis patients. An Bras Dermatol 2005;80:245–248 [Google Scholar]
- 8.Betsi GI, Papadavid E, Falagas ME: Probiotics for the treatment or prevention of atopic dermatitis: A review of the evidence from randomized controlled trials. Am J Clin Dermatol 2008;9:93–103 [DOI] [PubMed] [Google Scholar]
- 9.Yeşilova Y, Çalka Ö, Akdeniz N, Berktaş M: Effect of probiotics on the treatment of children with atopic dermatitis. Ann Dermatol 2012;24:189–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meneghin F, Fabiano V, Mameli C, Zuccotti GV: Probiotics and atopic dermatitis in children. Pharmaceuticals 2012;5:727–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bian F, Xiao Y, Zaheer M, et al. : Inhibition of NLRP3 inflammasome pathway by butyrate improves corneal wound healing in corneal alkali burn. Int J Mol Sci 2017;18:pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rivière A, Selak M, Lantin D, Leroy F, De Vuyst L: Bifidobacteria and butyrate-producing colon bacteria: Importance and strategies for their stimulation in the human gut. Front Microbiol 2016;7:979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nylund L, Nermes M, Isolauri E, et al. : Severity of atopic disease inversely correlates with intestinal microbiota diversity and butyrate-producing bacteria. Allergy 2015;70:241–244 [DOI] [PubMed] [Google Scholar]
- 14.Kim CH, Park J, Kim M: Gut microbiota-derivedshort-chain fatty acids, T cells, and inflammation. Immune Netw 2014;14:277–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim JM: Roles of enteric microbial composition and metabolismin health and diseases. Korean J Gastroenterol 2013;62:191–205 [DOI] [PubMed] [Google Scholar]
- 16.Anand S, Kaur H, Mande SS: Comparative in silico analysis of butyrate production pathways in gut commensals and pathogens. Front Microbiol 2016;7:1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holscher HD: Dietary fiber and prebiotics and the gastrointestinal microbiota. Gut Microbes 2017;8:172–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Kivit S, Saeland E, Kraneveld AD, et al. : Galectin-9 induced by dietary synbiotics is involved in suppression of allergic symptoms in mice and humans. Allergy 2012;67:343–352 [DOI] [PubMed] [Google Scholar]
- 19.Lam HY, van Hoffen E, Michelsen A, et al. : Cow's milk allergy in adults is rare but severe: Both casein and whey proteins are involved. Clin Exp Allergy 2008;38:995–1002 [DOI] [PubMed] [Google Scholar]
- 20.Ishiguro K, Oku H, Suitani A, Yamamoto Y: Effects of conjugated linoleic acid on anaphylaxis and allergic pruritus. Biol Pharm Bull 2002;25:1655–1657 [DOI] [PubMed] [Google Scholar]
- 21.Chomczynski P, Sacchi N: Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 1987;162:156–159 [DOI] [PubMed] [Google Scholar]
- 22.Kanhere A, Hertweck A, Bhatia U, et al. : T-bet and GATA3 orchestrate Th1 and Th2 differentiation through lineage-specific targeting of distal regulatory elements. Nat Commun 2012;3:1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.West CE, Jenmalm MC, Prescott SL: The gut microbiota and its role in the development of allergic disease: A wider perspective. Clin Exp Allergy 2015;45:43–53 [DOI] [PubMed] [Google Scholar]
- 24.Vernocchi P, Del Chierico F, Quagliariello A, et al. : A metagenomic and in silico functional prediction of gut microbiota profiles may concur in discovering new cystic fibrosis patient-targeted probiotics. Nutrients 2017;9:1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim SK, Kwon MS, Lee J, et al. : Weissella cibaria WIKIM28 ameliorates atopic dermatitis-like skin lesions by inducing tolerogenic dendritic cells and regulatory T cells in BALB/c mice. Sci Rep 2017;7:40040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blázquez AB, Berin MC: Microbiome and food allergy. Transl Res 2017;179:199–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Atarashi K, Tanoue T, Oshima K, et al. : Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013;500:232–236 [DOI] [PubMed] [Google Scholar]
- 28.Xie RD, Xu LZ, Yang LT, et al. : Galectin-1 inhibits oral-intestinal allergy syndrome. Oncotarget 2017;8:13214–13222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim YY, Je IG, Kim MJ, et al. : 2-Hydroxy-3-methoxybenzoic acid attenuates mast cell-mediated allergic reaction in micevia modulation of the FcɛRI signaling pathway. Acta Pharmacol Sin 2017;38:90–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi WJ, Konkit M, Kim Y, Kim MK, Kim W: Oral administration of Lactococcus chungangensis inhibits 2,4-dinitrochlorobenzene-induced atopic-like dermatitis in NC/Nga mice. J Dairy Sci 2016;99:6889–6901 [DOI] [PubMed] [Google Scholar]
- 31.Böhm I, Bauer R: Th1 cells, Th2 cells and atopic dermatitis. Hautarzt 1997;48:223–227 [DOI] [PubMed] [Google Scholar]
- 32.Muñoz FC, Cervantes MM, Cervantes-García D, et al. : Glycomacropeptide attenuates inflammation, pruritus, and Th2 response associated with atopic dermatitis induced by 2,4-dinitrochlorobenzene in rat. J Immunol Res 2017;2017:6935402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brandt EB, Sivaprasad U: Th2 cytokines and atopic dermatitis. J Clin Cell Immunol 2011;2:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grewe M, Bruijnzeel-Koomen CA, Schöpf E, et al. : A role for Th1 and Th2 cells in the immunopathogenesis of atopic dermatitis. Immunol Today 1998;19:359–361 [DOI] [PubMed] [Google Scholar]
- 35.Kim HJ, Kim YJ, Lee SH, et al. : Effects of Lactobacillus rhamnosuson allergic march model by suppressing Th2, Th17, and TSLP responses via CD4(+)CD25(+)Foxp3(+) Tregs. Clin Immunol 2014;153:178–186 [DOI] [PubMed] [Google Scholar]
- 36.Gophna U: Microbiology. The guts of dietary habits. Science 2011;334:45–46 [DOI] [PubMed] [Google Scholar]
- 37.Noverr MC, Huffnagle GB: The “microflora hypothesis” of allergic diseases. Clin Exp Allergy 2005;35:1511–1520 [DOI] [PubMed] [Google Scholar]
- 38.Biagi E, Candela M, Fairweather-Tait S, Franceschi C, Brigidi P: Aging of the human metaorganism: The microbial counterpart. Age (Dordr) 2012;34:247–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bunyavanich S, Shen N, Grishin A, et al. : Early-life gut microbiome composition and milk allergy resolution. J Allergy Clin Immunol 2016;138:1122–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang T, Ding C, Zhao M, et al. : Sodium butyrate reduces colitogenic immunoglobulinA-coated bacteria and modifies the composition of microbiota in IL-10 deficient mice. Nutrients 2016;8:pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhute SS, Suryavanshi MV, Joshi SM, et al. : Gut microbial diversity assessment of indian Type-2-diabetics reveals alterations in eubacteria, archaea, and eukaryotes. Front Microbiol 2017;8:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cho KK. and Choi IS: Allergy immunity regulation and synergism of Bifidobacteria. J Life Sci 2017;27:482–499 [Google Scholar]
- 43.Noverr MC, Falkowski NR, McDonald RA, McKenzie AN, Huffnagle GB: Development of allergic airway disease in mice following antibiotic therapy and fungal microbiota increase: Role of host genetics, antigen, and interleukin-13. Infect Immun 2005;73:30–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noverr MC, Huffnagle GB: Does the microbiota regulate immune responses outside the gut? Trends Microbiol 2004;12:562–568 [DOI] [PubMed] [Google Scholar]
- 45.Narushima S, Sugiura Y, Oshima K, et al. : Characterization of the 17 strains of regulatory T cell-inducing human-derived Clostridia. Gut Microbes 2014;5:333–339 [DOI] [PMC free article] [PubMed] [Google Scholar]