Summary
The history of humankind is marked by the constant adoption of new dietary habits affecting human physiology, metabolism, and even the development of nutrition-related disorders. Despite clear archaeological evidence for the shift from hunter-gatherer lifestyle to agriculture in Neolithic Europe [1], very little information exists on the daily dietary habits of our ancestors. By undertaking a complementary -omics approach combined with microscopy, we analyzed the stomach content of the Iceman, a 5,300-year-old European glacier mummy [2, 3]. He seems to have had a remarkably high proportion of fat in his diet, supplemented with fresh or dried wild meat, cereals, and traces of toxic bracken. Our multipronged approach provides unprecedented analytical depth, deciphering the nutritional habit, meal composition, and food-processing methods of this Copper Age individual.
Keywords: Iceman, stomach content, European Copper Age mummy, diet, last meal, multi-omics study, ancient DNA, lipidomics, proteomics, microscopy
Highlights
-
•
The last meal of the Iceman, a European Copper Age mummy, was reconstructed
-
•
Our multipronged approach deciphers the meal composition and food processing
-
•
His high-fat diet was supplemented with wild meat and cereals
Maixner et al. report the dietary reconstruction of the Iceman’s last meal using a combined multi-omics approach. The stomach content analysis of the 5,300-year-old glacier mummy shows that the Iceman’s diet preceding his death was a mix of carbohydrates, proteins, and lipids, well adjusted to the energetic requirements of his high-altitude trekking.
Results and Discussion
Human evolution is closely linked to dietary changes and the development of new food-processing techniques. This is clearly observed in the transition from hunter-gatherer lifestyle to agriculture, which gave rise to cultivation of crops, animal husbandry, and permanent settlements [1]. The more stable availability of food boosted the growth of Neolithic European populations [4, 5]. However, changes in diet had drawbacks for health, such as increased rates of caries [6]. Added to this, permanent large settlements combined with the adoption of agriculture promoted the spread of density-dependent infectious diseases [7]. Considering the impact of dietary habits on human evolution and health, the reconstruction of past human subsistence appears to be vital to our understanding of past societies. Stable carbon (13C/12C) and nitrogen (15N/14N) isotope analyses of bone collagen have been widely used to study the dietary adaptations of Neolithic populations [8], lipid analysis on pottery residues dated the onset of milk use in Neolithic Europe [9], and numerous archaeological studies describe the occurrences of wild and domesticated animal and plant remains as indirect measures of past diets [10]. All these studies provide insights into major dietary changes during the Neolithic period. However, specific details on past diets such as the exact meal composition or food processing are missing. Knowledge of daily dietary habits of our ancestors is pertinent as food type, intake quantity, and food-processing techniques influence human metabolism and health.
A recent radiological re-examination of the Iceman, a 5,300-year-old European natural ice mummy, identified his completely filled stomach (Figure 1A) [2]. The well-preserved stomach content still contains ancient endogenous biomolecules as demonstrated from the complete reconstruction of the Iceman’s Helicobacter pylori genome [11]. Previous isotopic analysis (15N/14N) of the Iceman’s hair suggested first a vegetarian lifestyle [12, 13] which was later, after careful re-examination of the data, changed to a omnivorous diet [14]. Further analyses of lower intestinal tract samples of the Iceman confirmed that he was omnivorous, with a diet consisting of both wild animal and plant material. Among the plant remains, there were cereals, pollen grains of hop-hornbeam, and fragments of bracken and mosses [14, 15, 16, 17, 18, 19, 20]. The detection of the Iceman’s stomach content with its pristine yet undigested food mix, provides the unique opportunity to fully reconstruct a Copper Age meal.
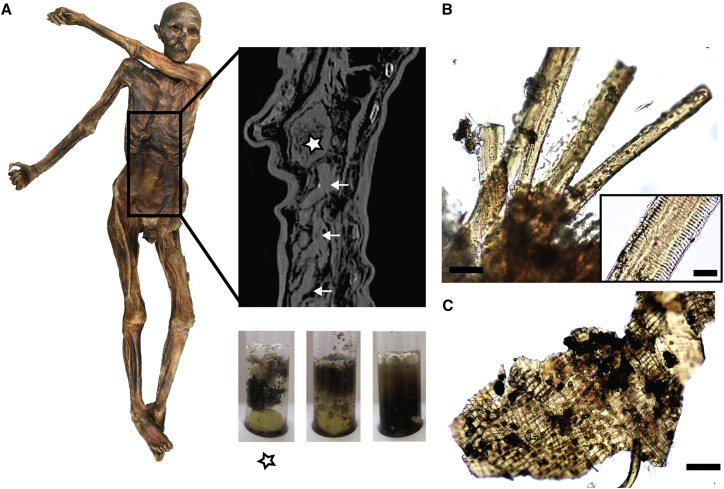
Figure 1.
The Iceman’s Gastrointestinal Tract
(A) Gastrointestinal (GI) tract preservation and content texture. The radiographic image shows the completely filled stomach (asterisk) and the intestinal loops of the lower GI tract (arrows). Content samples of the stomach (left, asterisk) and of two different sites in the lower GI tract (middle, right) that were re-hydrated in phosphate buffer are shown below the radiographic image.
(B) Animal muscle fibers detected in the stomach content using light microscopy. Scale bar, 50 μm. The black box contains a zoomed-in view of one muscle fiber (scale bar, 20 μM).
(C) Plant tissue detected in the stomach content using light microscopy. Scale bar, 50 μm. See also Figure S1 for additional plant and animal remains detected in the Iceman intestinal contents.
The study material included 11 gastrointestinal (GI) content biopsies of the stomach and small and large intestines (Figure 1A; Data S1). Using microscopy and a combined multi-omics approach targeting different biomolecules (ancient DNA, proteins, metabolites, and lipids) (see STAR Methods), we determined the exact composition of the Iceman’s diet preceding his death and present evidence how people during the Copper Age processed food.
Initial macro- and microscopic analyses showed that, compared to the small and large intestine contents, samples of the stomach content still contain compact pieces of food that display a hydrophobic “fatty-like” character (Figure 1A). The most prevalent tissue types in the stomach content samples are animal muscle fibers arranged mostly in bundles (Figures 1B and S1) and plant fragments identified as bran and glumes belonging to the Triticum/Secale type (Figures 1C and S1). The striated muscle fiber structures are characteristic for cardiac and skeletal muscle tissue (Figure S1B) [21]. Further microscopic analysis of the plant micro-remains identified cereal pollen from the Triticum-type as well as airborne arboreal pollen and an abundance of spores and sporangia from the bracken fern Pteridium aquilinum (Figure S1J).
Metabolite, glycan, stable isotope, and elemental analyses confirm the omnivorous character of the Iceman’s last meal (Data S2). Metabolic compounds indicate the presence of ruminant fat or dairy products (phytanic acid) and support the existence of whole-grain cereals (azealic acid) in the Iceman’s diet. In addition, gamma-terpinene that occurs in coriander oil, lemon oil, and other essential oils suggest the usage of herbs. The most abundant elements found in the stomach content were the nutritional minerals iron, calcium, zinc, magnesium, and sodium, consistent with the consumption of red meat or dairy products. Minor concentrations of trace nutrients of chromium, copper, manganese, selenium, molybdenum, and cobalt were also detected. These data suggest that the Iceman’s last meal was well balanced in terms of essential minerals required for good health with no evidence of toxic heavy metals such as lead, cadmium, or arsenic.
To further identify the possible food sources, we analyzed the DNA and protein traces still present in the material. Metagenomic analysis of the Iceman’s biopsy samples revealed a high number of bacterial reads in all GI tract contents (Figure 2A). At most, 39% of reads in the Iceman samples were of eukaryotic origin, the majority of which (between 94% and 42%) were identified as fungal. The Metazoa and Virdiplantae reads, important for the dietary reconstruction, comprised 0.2%–0.7% of all reads (Figures S2A–S2C). Comparative analysis of all GI shotgun datasets (n = 21) against currently available full mitochondrial and chloroplast genomes unambiguously assigned the majority of animal reads to ibex (Capra ibex) and red deer (Cervus elaphus), and the major proportion of plant reads belong to the species Pteridium aquilinum subsp. aquilinum and to members of the genus Triticum (Figures 2B and S2D). DNA traces of these four major taxa were detected in all GI content samples. A control dataset from the Iceman’s muscle tissue was negative (Figure S2D). The unambiguously assigned reads were aligned to modern reference plastid genomes, and all animal- and plant-derived reads show damage patterns indicative of ancient DNA (Figure S3) [22]. Subsequent phylogenetic assignment of the reconstructed plastid genomes confirmed and further extended the metagenomic result (Figure S3). The Triticum reads were more specifically assigned to chloroplast genomes of Triticum monococcum (einkorn) and Triticum urartu (Figure S3; Data S1).
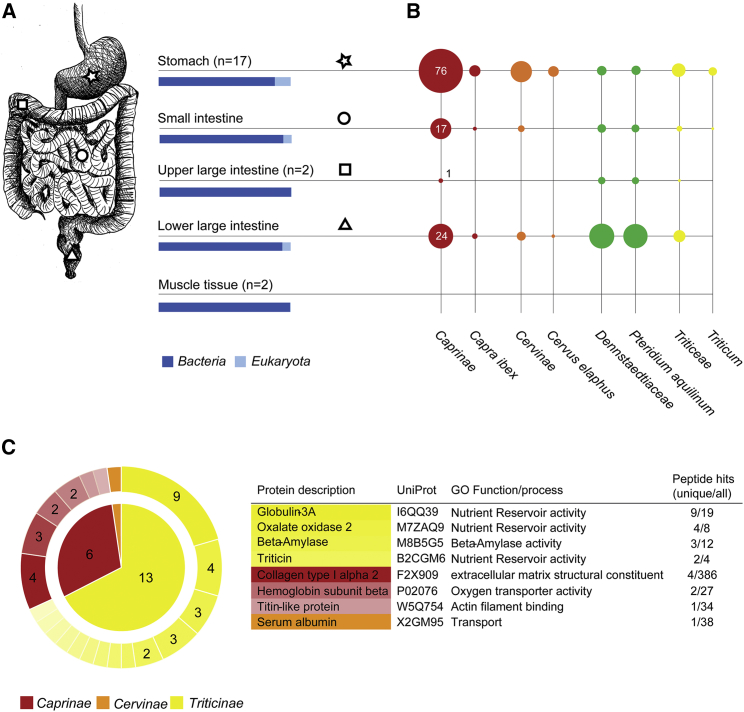
Figure 2.
Metagenomic and Proteomic Analysis
(A) GI tract samples included in the metagenomic approach. The blue-bar diagrams below the sample description display the read distribution between the two kingdoms Bacteria and Eukaryota in selected shotgun datasets (for details, please refer to STAR Methods and Data S1). The number of shotgun datasets included in all further metagenomic analyses is provided in brackets.
(B) Most abundant taxa detected in the intestinal content shotgun datasets. The circle size corresponds to the number of unambiguously assigned mitochondrial and chloroplast reads per million metagenomic reads.
(C) Plant and animal proteins detected in the stomach content. The inner circle displays the number of identified proteins for the taxa Caprinae, Cervinae, and Triticinae. The outer circle highlights the number of unique peptides present for protein identification. The table summarizes details about the proteins detected with the highest unique peptide hits. See also Figures S2, S3, and S4 for a taxonomic overview of the plastid reads, the phylogenetic assignments, damage pattern, and DNA-barcode analysis. Data S1 contains details to the datasets and the plastid mapping statistics. Data S2 provides additional information to the identified peptide hits.
Importantly, the presence of ibex and red deer in the stomach contents was independently confirmed in the Molecular Population Genetics Laboratory, Trinity College, Dublin, by sequencing of a mammalian mitochondrial capture (Data S1). Moreover, PCR-based analysis targeting animal- and plant-DNA barcodes and further assignment of the shotgun data against the DNA Barcode of Life Data dataset (www.boldsystems.org) support the first metagenomic results on a phylogenetic higher hierarchical level (genus, family) (Figure S4).
Our analysis of the ancient DNA identified the main components in the Iceman’s meal. However, the highly degraded state of this biomolecule excluded any further quantification of certain foods.
Using multistep solubilization and fractionation proteomics, we identified 167 animal and plant proteins in the stomach metaproteome, of which 13 were taxonomically unambiguously assigned to the Triticinae, 6 to the Caprinae, and 1 protein to the Cervinae subfamily (Figure 2C; Data S2). The proteomic hits not only confirm the previous metagenomic results but also provide important information on the food source. The detected Triticinae plant proteins are primarily expressed in the endosperm (Globulin 3A, Triticin) [23] and pericarp (Oxalate oxidase 2) [24] of wheat seeds. The Caprinae Titin-like protein is part of the sarcomere apparatus in muscle tissue [25]. Thus, we have combined genomic and proteomic evidence that the Iceman’s last meal contained whole seeds of the T. monococcum/T. urartu-like wheat and muscle fibers of ibex. However, due to the lack of diagnostic proteins, we cannot determine which tissues of the red deer were ingested by the Iceman.
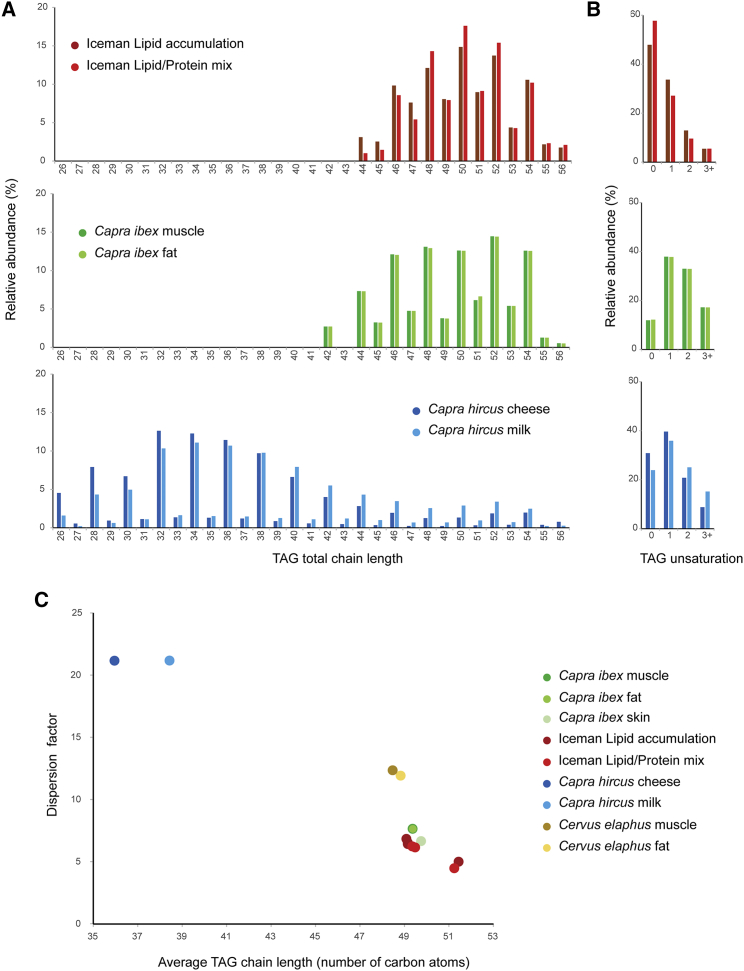
Approximately 46% ± 19% (w/w) of the stomach content is fat residues. Comparative lipid analysis of the Iceman’s stomach content to contemporary wild ruminant muscle and adipose tissue, and to goat dairy products (milk and cheese), using reverse-phase liquid chromatography separation and quadrupole time-of-flight (QToF) tandem mass spectrometry analysis (RP-LC/MSMS) identified triacylglycerols (TAGs) as a major component of the stomach content, with molecular compositions consistent with ruminant muscle/adipose fat (Figure 3A). The TAGs identified in the Iceman’s stomach contain between 44 and 56 total carbon atoms in the fatty acyl chains and resemble the TAG profile of contemporary muscle and adipose tissue. The high level (up to 20%) of odd numbered fatty acyl chains is consistent with TAG profiles from ruminant animals (Data S2) [27]. Short-chain TAG species characteristic of dairy products are absent in the Iceman TAG profile. However, the high level of saturated bonds in the TAGs of the Iceman’s stomach content compared to the fresh tissues indicates an already ongoing preferential degradation of unsaturated fatty acids in the ancient specimen (Figure 3B) [28]. Therefore, we cannot fully exclude that short-chain dairy-specific lipid species were degraded, although the identification of di- and poly-unsaturated TAG indicates a still high level of preservation. Furthermore, we have histological evidence for the presence of adipose tissue in the Iceman’s stomach content (Figure S1C). Further comparison of the dispersion factor with the average carbon number (M) of each detected TAG in the analyzed samples points to the ibex as potential source of the adipose tissue (Figure 3C). Thus, the ibex served the Iceman as food source for both meat and fat.
Figure 3.
Lipid Analysis of the Iceman’s Stomach Content
(A) Triacylglycerol (TAG) distribution in the Iceman’s stomach’s content in comparison to the TAG distribution in ibex muscle and fat tissue and goat dairy products. Displayed is the relative abundance of TAGs based on their chain length.
(B) Saturation level of the TAGs in the stomach content and the fresh tissue samples. The Iceman’s stomach TAGs contain a much higher saturated bond content than fresh tissue samples.
(C) The dispersion factor (DF) plotted against the average carbon number (M) of each TAG detected in the ancient and modern samples. The DF measures the spread of the TAG distribution. Together with M, DF can be used for discrimination between archaeological artifacts [26]. See also Data S2 for further details to the relative abundance of TAG species and to the mass spectrometry results.
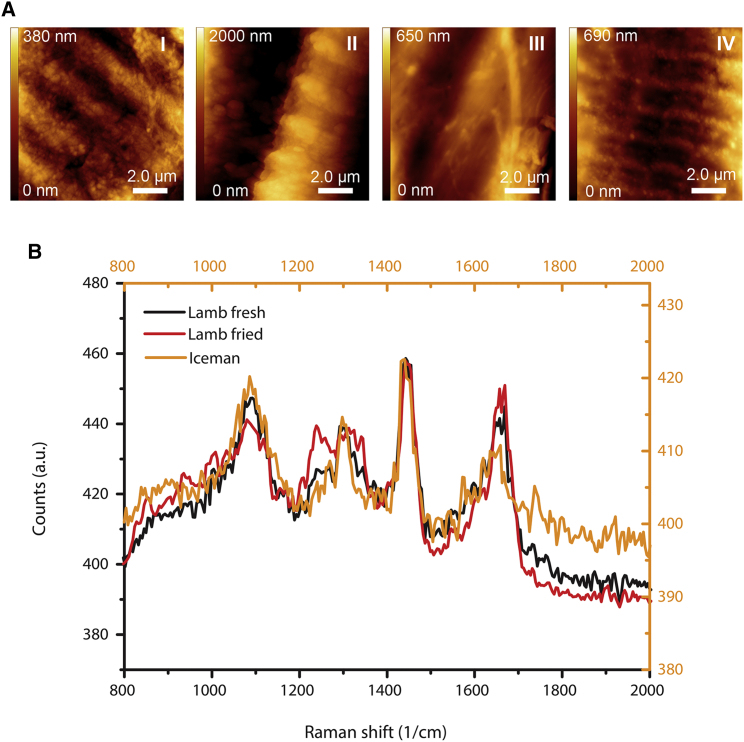
Charcoal particles detected in a previous study in the Iceman’s lower intestine content suggested the involvement of fire in the food processing [16]. We experimentally tested the effect of heat and drying on contemporary animal muscle tissue and compared their structural and chemical composition with the muscle fibers detected in the Iceman’s stomach by using atomic force microscopy (AFM) and Raman spectroscopy. High magnification AFM images revealed striated fiber structures in both ancient and fresh or dried contemporary muscle fibers while structural changes of the fibers were observed when the samples were thermally treated (Figure 4A). After frying or cooking, the regular patterns in the meat fibers disappeared, indicating the denaturation of their protein compounds. Complementary confocal Raman spectroscopy measurements support the AFM results and indicate heat-related conformational changes of the molecular composition of the meat fibers (Figure 4B). The Raman spectra of the ancient muscle fibers largely matched the spectra taken of fresh and dried contemporary meat fibers. After heat treatment, the Raman band at about 1,093 cm-1 becomes weaker, and bands at around 1,240 cm-1 appear that are indicative of heat-induced changes in the secondary structure of the myofibrils [29]. Thus, both the AFM and Raman spectroscopy results suggest that the Iceman consumed either fresh or dried wild meat. Besides, a slow drying or smoking of the meat over the fire would explain the charcoal particles detected previously in the lower intestine content.
Figure 4.
Atomic Force Microscope Images and Raman Spectra of Muscle Fibers from the Iceman’s Stomach Content
(A) Fibers obtained from the content of the Iceman’s stomach (I) show structures similar to those of recent fibers (II and VI). The typical Z-line is observed in raw (II) and air-dried lamb muscle fibers (IV). After thermal treatment such as frying or cooking of the meat, the sarcomeres disappear (III). The surface of the fibers becomes blurred, and only faint fibrillary structures can be found.
(B) Raman spectra of samples extracted from the Iceman’s stomach content show similarities to untreated (fresh) lamb meat spectra. See Data S2 for details to the modern comparative animal samples.
Our forensic multi-omics study provides important insights into the general life and nutritional habits of a Copper Age individual in the Alpine area. The microscopic and molecular data presented support the presence of three major components in the Iceman’s last meal: fat and game meat from ibex and red deer supplemented with cereals from einkorn. Intriguing is the pervasive presence of bracken particles. Despite of its toxicity, bracken is still consumed today by different indigenous people [30]. However, it is also possible that fern leaves, as it was previously suggested for the moss Neckera complanata [20], were used to wrap the food and the spores and sporangia entered the ingesta unintentionally. Unexpectedly, there is a high proportion of fat in the diet. The distribution of triglycerides and their constituting fatty acids is consistent with the consumption of ibex muscle and adipose tissues. The extreme alpine environment in which the Iceman lived and where he have been found (3,210 m above sea level) is particularly challenging for the human physiology and requires optimal nutrient supply to avoid rapid starvation and energy loss [31]. Therefore, the Iceman seemed to have been fully aware that fat displays an excellent energy source. On the other hand, the intake of animal adipose tissue fat has a strong correlation with increased risk of coronary artery disease [32]. A high saturated fats diet raises cholesterol levels in the blood, which in turn can lead to atherosclerosis. Importantly, computed tomography scans of the Iceman showed major calcifications in arteria and the aorta indicating an already advanced atherosclerotic disease state [33]. Both his high-fat diet and his genetic predisposition for cardiovascular disease [34] could have significantly contributed to the development of the arterial calcifications. Finally, we could show that the Iceman either consumed fresh or dried meat. Drying meat by smoking or in the open air are simple but highly effective methods for meat preservation that would have allowed the Iceman to store meat long term on journeys or in periods of food scarcity. In summary, the Iceman’s last meal was a well-balanced mix of carbohydrates, proteins, and lipids, perfectly adjusted to the energetic requirements of his high-altitude trekking.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological Samples | ||
| Iceman gastrointestinal tract content samples and tissues biopsies | [11] | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| EDTA disodium salt dihydrate | Carl Roth | Cat#8043.2 |
| Chloroform | SIGMA-ALDRICH | Cat#C2432 |
| Chloroform | Fisher Chemical | Cat#C297-4 |
| Chloroform -Isoamylalcohol (24:1, v/v) | SIGMA-ALDRICH | Cat#C0549 |
| Methanol | Carl Roth | Cat#4627.1 |
| Methanol | Fisher Chemical | Cat#A456-4 |
| Acetonitrile | Fisher Chemical | Cat#A955-4 |
| Isopropanol | Fisher Chemical | Cat#A461-4 |
| Cetyltrimethylammonium bromide CTAB | Carl Roth | Cat#9161.3 |
| Polyvinylpyrrolidone-K10 | SIGMA-ALDRICH | Cat#PVP10-100G |
| Tris-HCl | Carl Roth | Cat#9090.2 |
| Acrylamide | Carl Roth | Cat#0189.2 |
| Ammonium peroxydisulphate APS | Carl Roth | Cat#9592.2 |
| TEMED | Carl Roth | Cat#2367.3 |
| Sodium acetate | Carl Roth | Cat#6773.1 |
| Guanidine hydrochloride | Carl Roth | Cat#6069.2 |
| Beta-Mercaptoethanol | Carl Roth | Cat#4227.3 |
| BSA (20 mg/ml) | New England Biolabs | Cat#B9000S |
| Bst polymerase, large fragment (8 U/μl) | New England Biolabs | Cat#M0275S |
| dNTPs (2.5mM each) | Thermo Fisher Scientific | Cat#R72501 |
| T4 DNA ligase (5 U/μl) | Thermo Fisher Scientific | Cat#EL0014 |
| Critical Commercial Assays | ||
| Min Elute PCR Purification Kit | QUIAGEN | Cat#28006 |
| Amplitaq Gold mastermix 360 | Thermo Fisher Scientific | Cat#4398881 |
| NEBNext End Repair Module | New England Biolabs | Cat#E6050L |
| Accuprime Pfx Supermix | Thermo Fisher Scientific | Cat#12344040 |
| Deposited Data | ||
| Iceman gastrointestinal tract content shotgun datasets | [11] | ENA: ERP012908 |
| Iceman stomach content enriched dataset | This Study | ENA: PRJEB26465 |
| Iceman stomach content amplicon datasets | This Study | ENA: PRJEB26365 |
| Iceman stomach content proteomics data | This Study | PRIDE: PXD009565 |
| Oligonucleotides | ||
| 12419F CAAACTGGGATTAGATACCC | This Study | Thermo Fisher Scientific |
| 12564R YRGAACAGGCTCCTCTAG | This Study | Thermo Fisher Scientific |
| CBF GCGTACGCAATCTTACGATCAA | [35] | Thermo Fisher Scientific |
| CBR CTGGCCTCCAATTCATGTGAG | [35] | Thermo Fisher Scientific |
| trnL-g GGGCAATCCTGAGCCAA | [36] | Thermo Fisher Scientific |
| trnL-h CCATTGAGTCTCTGCACCTATC | [36] | Thermo Fisher Scientific |
| rbcL19 AGATTCCGCAGCCACTGCAGCCCCTGCTTC | [37] | Thermo Fisher Scientific |
| rbcLZ1 ATGTCACCACAAACAGAGACTAAAGCAAGT | [37] | Thermo Fisher Scientific |
| Software and Algorithms | ||
| Imaging Software NIS elements F 3.00 | Nikon | N/A |
| ChromaTOF software version 4.2 | LECO | N/A |
| FastQC | Babraham Bioinformatics | http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| SeqPrep | N/A | https://github.com/jstjohn/SeqPrep |
| RAPSearch2 | [38] | http://rapsearch2.sourceforge.net/ |
| Krona | [39] | https://github.com/marbl/Krona |
| MEGAN6 | [40] | https://ab.inf.uni-tuebingen.de/software/megan6 |
| Burrows-Wheeler Aligner (BWA) | [41] | http://bio-bwa.sourceforge.net/ |
| blastn | [42] | N/A |
| FastQ Screen | Babraham Bioinformatics | http://www.bioinformatics.babraham.ac.uk/projects/fastq_screen/ |
| SAMtools | [43] | http://samtools.sourceforge.net/ |
| Picard tools | N/A | http://broadinstitute.github.io/picard/ |
| ANGSD | [44] | http://www.popgen.dk/angsd/index.php/ANGSD |
| MAFFT | [45] | https://mafft.cbrc.jp/alignment/server/ |
| ARB software package | [46] | http://www.arb-home.de/ |
| AdapterRemoval | [47] | https://github.com/MikkelSchubert/adapterremoval |
| DeDup | N/A | https://github.com/apeltzer/DeDup |
| mapDamage | [21] | https://ginolhac.github.io/mapDamage/ |
| GATK | [48] | https://software.broadinstitute.org/gatk/ |
| mothur | [49] | https://www.mothur.org/ |
| Trans-Proteomic Pipeline (TPP) software tool suite | [50] | http://tools.proteomecenter.org/wiki/index.php?title=Software:TPP |
| MassHunter Qualitative Analysis Software Version B.05.01 | Agilent | https://www.agilent.com/en/products/software-informatics/masshunter-suite/masshunter/masshunter-software |
| Other | ||
| BOLD system database | [51] | http://www.boldsystems.org/ |
| Additional metadata for the Illumina shotgun datasets | [11] | http://cube.univie.ac.at/supplements/iceman |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Frank Maixner (frank.maixner@eurac.edu).
Experimental Model and Subject Details
The Iceman
The Iceman is one of the oldest human mummies ever found in human history [52]. With his body perfectly preserved throughout millennia in an Alpine glacier (3,210 m above sea level) and his secrets unlocked and shown for the first time to humanity by the melting of ice in 1991, it has been and still is a subject of great interest, fascination and speculations, since the veil of mystery enveloping the circumstances of his death has not yet fallen [3]. The Iceman was murdered when he was 40-50 years old by an arrow that lacerated the left subclavian artery, likely leading to a rapid, deadly hemorrhagic shock [53, 54]. It is not only his historical age and the bounty of prehistoric belongings that make him extremely valuable for scientists, but also how he was preserved for about 53 centuries. The Iceman is indeed an “ice mummy,” so-called for the retention of humidity in his cells [55]. The body tissues contain well preserved biomolecules such as lipids, proteins, and nucleic acids. This feature makes him extremely valuable for modern scientific investigations [11, 34, 56, 57, 58, 59, 60, 61]. Initially, the Iceman‘s stomach was not found radiologically and was generally assumed to be empty. During a recent re-examination however, the Iceman‘s stomach was not only identified radiologically, but also shown to be completely filled [2].
Method Details
Sampling
The sampling of the Iceman stomach content took place under sterile conditions at an ambient temperature of 4°C in the Iceman’s conservation chamber at the Archaeological Museum of Bolzano, Italy. The samples were immediately stored at −20°C in the ancient DNA laboratory of the Eurac Research - Institute for Mummy Studies. Furthermore, we had access to previously sampled gastrointestinal tract contents and to an Iceman muscle and lung tissue sample, which we used in addition to the extraction blanks as a negative control for our molecular analyses. See Data S1 for additional details to the samples used in this study.
Microscopic analysis
Small pieces of the stomach content (0.5cmx0.5cm) were processed for microscopy in the aDNA laboratory of the Eurac Research - Institute for Mummy Studies, Bolzano. After rehydration in 15mL solution consisting of 5 parts glycerol and 5 parts 4% formaldehyde for 48 h, the samples were fixed for 24 h in 4% formaldehyde and finally stored in 1x phosphate buffered saline solution at 4°C. For the microscopic analysis, small tissue pieces have been embedded with an aqueous mounting medium on a glass slide. In parallel, some fixed content samples were embedded in OCT cryostat embedding medium (Tissue Tek®) and further processed for histology. The embedded specimens were cut on a cryostat in 10 μm thick sections (Leica, CM1950). The cryosections were histochemically counterstained with the lipid stain Sudan III [62]. The images were recorded with a CCD camera (Nikon DS-Fi1) mounted on a light microscope (Nikon Eclipse E600), by using the imaging software NIS elements F 3.00. Additionally, ancient animal meat fibers detected in the stomach content were sent to the Center of Smart Interfaces at the TU Darmstadt, Germany, where the coarse topography of meat fibers was analyzed by laser scanning microscope (LSM) images (VK 8710, Keyence, Osaka, Japan).
Metabolite analysis
Primary metabolism was assessed using GC-TOF MS. 2 mg fresh weight of stomach contents were extracted using 1 mL of degassed and pre-cooled (−20°C) acetonitrile/isopropanol/water solvent at the volume ratio 3:3:2. After centrifugation, the supernatant was removed and evaporated to complete dryness. A clean-up step with 500 μL acetonitrile/water (1:1) removed membrane lipids and triglycerides and after centrifugation. To the dried extracts, C8-C30 fatty acid methyl esters were added as internal standards. Samples were derivatized by 10 μL methoxyamine hydrochloride in pyridine (20 mg/mL) at 30°C for 90 min, followed by the addition of 90 μL N-methyl-N-trimethylsilyltrifluoroacetamide at 37°C for 30 min. 0.5 μL of sample was injected at 50°C (ramped to 250°C by 12°C/s and held for 3 min) in splitless mode in a Gerstel cold injection system with multi-baffled glass liners. Metabolites were separated using an Agilent 6890 GC system (Santa Clara, CA, USA) equipped with a Rtx5Sil-MS column (30 m length x 0.25 mm i.d.) with a 0.25 μm 95% dimethyl/ 5% diphenyl polysiloxane film and additional 10 m integrated guard column (Restek, Bellefonte, PA, USA). Chromatography was performed at 1 mL/min helium from 50°C to 330°C with 20°C/min. Mass spectra were acquired on a Leco Pegasus IV time of flight mass spectrometer (St. Joseph, MI, USA) with electron ionization at 70 eV at 250°C. Masses were detected between m/z 85-500 at 17 spectra/s and 1800 V detector voltage. ChromaTOF software version 4.2 was used for data pre-processing including baseline subtraction and automatic mass spectral deconvolution and peak detection. The resulting chromatograms were further processed using the metabolomics BinBase database and spectra were further manually investigated by the Fiehnlib libraries and the NIST12 mass spectral library. Please refer for the list of metabolites identified in the Iceman stomach content to Data S2_Metabolomics.
Glycan analysis
Glycan analysis for the Iceman stomach content was performed at Asia-Pacific Glycomics Reference Site (AGRS) in Daejeon, Republic of Korea. Two stomach content samples of 0.021 g and 0.023 g, respectively were ground with 300 μL 70% EtOH into tissue grinder. High molecular weight materials including glycoproteins were enriched from ground samples by mini dialysis unit (10,000 MWCO). Supernatant was transferred to micro-centrifuge tube and denatured by mixture of NH4HCO3 and DTT (95°C, 2min). N-glycans were enzymatically released from denatured glycoproteins by PNGase F (37°C, overnight) and these were further purified and enriched using solid phase extraction with a porous carbon cartridge. N-glycans released from the Iceman stomach content were comprehensively characterized by nano LC PGC-chip/Q-TOF MS and tandem MS (Agilent 6530). Glycan candidates were selected from the mass-to-charge values extracted in raw LC/MS data based on monosaccharide combinations using hexose, N-acetyl hexoamine, fucose, N-acetyl neuraminic acid, and xylose. N-glycan compositions were further confirmed and determined by compositional analysis using tandem MS. In order to determine relative amounts of glycans, individual glycan compound was integrated using the ion count corresponding to individual peak and then normalized by total ion counts to obtain the relative abundance of each glycan. Additionally, isomers derived from PGC-LC analysis were combined in a single composition prior to quantitative analysis. Data S2_Glycomics lists all identified glycan structures.
Stable isotope analysis
The preparation of the stomach content (EURAC ID 1054) for the stable isotope analysis was conducted in the laboratory at the Department of Physical Anthropology, Institute of Forensic Medicine at University of Bern. Therefore, a sample of 1g was dissolved in ddH2O and freeze-dried for 47h. After homogenization by a ball triturator (Retsch), approx. 5.5mg of the lyophilized stomach content was weighed into tin capsules and shipped to IRMS at Isolab GmbH, Schweitenkirchen, Germany for measurement. Stable isotope ratios of carbon (13C/12C), nitrogen (15N/14N) and sulfur (34S/32S) were analyzed by isotope ratio mass spectrometry and the mean of six measurements was calculated to avoid measurement errors due to potential inhomogeneous material. All data are presented in δ-notation in per mil (‰) according to international standards for carbon (Vienna Pee Dee Belemnite, VPDB), nitrogen (Ambient Inhalable Reservoir, AIR) and sulfur (Canyon Diablo Troilite, CDT) [63, 64, 65]. An internal standard was used for determination of the analytical error. It amounted to ± 0.1‰ for δ13C, ± 0.2‰ for δ15N and ± 0.3‰ for δ34S. For details for the stable isotope values of the Iceman stomach content please refer to the Data S2_Stable isotope analysis.
Elemental analysis of the Iceman stomach content
An Agilent Technologies 7500cx inductively coupled plasma-mass spectrometer (ICP-MS) (Agilent Technologies, Mulgrave, Australia) was used with sample introduction via a micromist concentric nebuliser (Glass Expansion, West Melbourne, Australia) and a Scott type double pass spray chamber cooled to 2°C. The sample solution and the spray chamber waste were carried with the aid of a peristaltic pump. ICP-MS extraction lens conditions were selected to maximize the sensitivity of a 1% HNO3:HCl solution containing 1ng ml-1 of Li, Co, Y, Ce and Tl. Helium was added into the octopole reaction cell to reduce interferences. Calibration curves were constructed and the results analyzed using Agilent Technologies MassHunter software. A certified calibration standard containing Li, Be, B, Na, Mg, Al, Ca, V, Cr, Mn, Fe, Co, Ni, Cu, Zn, As, Se, Sr, Mo, Ag, Cd, Sb, Ba, La, Eu, Ho, Yb, Tl, Pb, Bi, Th, U and P as well as a Hg standard was obtained from Choice Analytical, Thornleigh, Australia. Calibration curves were constructed from seven standards at 0, 1, 10, 50, 100, 500, and 1000 ng/mL. Baseline nitric acid (HNO3) hydrochloric acid (HCl) and hydrogen peroxide (H2O2) were purchased from Choice Analytical, Thornleigh, Australia. Two stomach content samples of 0.05915 g and 0.0279 g were each digested in 2 mL HNO3, 1 mL HCl and 1 mL H2O2 in a Milestone MLS 1200 microwave digester (Milestone, Sorisole, Italy), and diluted to approximately 50 g (accurately measured on an analytical balance) before analysis. A digestion blank was subtracted from each sample. The average concentrations of the elements detected in the Iceman’s stomach is listed in the Data S2_Elemental analysis.
DNA extraction, library preparation and Illumina sequencing
The datasets used for the metagenomic dietary analysis have been published in our previous study on the Iceman’s Helicobacter pylori genome. In the following sections, all major molecular steps will be briefly outlined once more. For methodological details, however, please refer to the publication of Maixner et al. [11]. The molecular analyses were conducted at the ancient DNA Laboratory of the Eurac Research - Institute for Mummy Studies, Bolzano, Italy, and in the ancient DNA facility of the Institute of Clinical Molecular Biology, Kiel University, Germany. Sample preparation and DNA extraction was performed in pre-PCR areas dedicated to ancient DNA procedures. DNA was extracted from the gastrointestinal tract samples (250 mg) and from lung tissue (50 mg) using a chloroform-based DNA extraction method according to the protocol of Tang et al. [66]. DNA extraction from the muscle tissue (100 mg) was performed with a magnetic-bead based technology using the Biorobot®-EZ1 (QIAGEN, Hilden, Germany), following previously described procedures [67]. Negative controls from all experimental steps were included to monitor external contamination. Library preparation and sequencing were performed at the Institute of Clinical Molecular Biology, Kiel University, Germany. Libraries for the sequencing runs were generated with a modified protocol for Illumina multiplex sequencing [68, 69]. The sequencing was carried out on the Illumina HiSeq 2000 and 2500 platform by 2 × 101 cycles using the HiSeq v3 chemistry and the manufacturer’s protocol for multiplex sequencing. For details to the metagenomic shotgun datasets please refer to Data S1_Illumina shotgun datasets.
Bioinformatic analysis of the Illumina datasets
The metagenomic shotgun datasets were subjected to a bioinformatic pipeline to identify the major animal and plant dietary composition of the Iceman’s last meals. The paired-end Illumina reads (101 bp length) were first quality-checked using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and then subjected to adaptor removal and read merging using SeqPrep (https://github.com/jstjohn/SeqPrep) (“SeqPrep -f forward-read-fastqfile -r reverse-read-fastqfile −1 forward-read-output-fastqfile −2 reverse-read-output-fastqfile -L 15 –A10 “AGATCGGAAGAGCACACGTCTGAA” -B “AGATCGGAAGAGCGTCGTGTAGGG” –s merged-fastq-file). Approximately 60%–90% of reads were merged (see Data S1_Illumina shotgun datasets). To obtain a first overview of the taxonomic composition of selected samples from the different gastrointestinal tract contents and of the muscle tissue control sample, we performed a sequence similarity search of the metagenomic reads using RAPSearch2 [38] and the complete NCBI-nr database [70]. A stringent minimum bitscore filter (min bitscore 70) was applied to reduce random hits and the results were taxonomically assigned using MEGAN6 [40] with the lowest common ancestor (LCA) of hits within 10% of the bitscore of the best hits and a minimum number of 100 reads that a taxon must obtain. Finally, the data was visualized using the Krona metagenomic visualization tool [39] providing a first taxonomic overview on the bacterial and eukaryotic reads. In the subsequent step, we focused on the eukaryotic reads aiming at identifying highly abundant animal and plant species possibly resembling the Iceman’s diet. We first aligned all metagenomic reads against all currently available full mitochondrial and chloroplast genomes of the NCBI database [70] using BWA [41] with default parameters. Subsequently, to obtain a taxonomic overview of the mapped reads and to identify and filter for the most prevalent taxonomic groups in the samples, we performed a sequence similarity search using blastn [42] with default parameters and the complete NCBI-nt database as reference database [70]. Blast results were taxonomically assigned using MEGAN6 [40] with a Min percent value of 0.01 for the reads mapped against the mitochondrial genomes and with a Min percent value of 0.5 for the reads mapped against the chloroplast genomes (see Figure S2).
To further confirm our previous taxonomic assignments and to understand if the extracted reads are of ancient origin or if they display modern contamination, we reconstructed the plastid genomes of selected animal and plant species and performed phylogenetic assignments and DNA damage pattern analysis. Initially, for the plastid genome reconstruction, we extracted all reads of all intestinal content shotgun datasets (See Data S1_Illumina shotgun datasets, both UDG-treated and non-UDG treated) that were previously taxonomically assigned to the four dominant animal and plant (sub)families Caprinae Cervinae, Dennstaedtiaceae and Triticeae (see Data S1_ Mapping metagenomic reads). Next, we selected for the alignment of each (sub)family reads the modern reference sequence based on the dominant species detected in the previous taxonomic assignment: the mitochondrial genome of Capra ibex (NC_020623) for the Caprinae reads, the mitochondrial genome of Cervus elaphus (AB245427) for the Cervinae reads, and the chloroplast genome of Pteridium aquilinum subsp. aquilinum (HM535629) for the Dennstaedtiaceae reads. However, the Triticeae reads have been only taxonomically assigned down to the genus Triticum. Thus, to obtain an appropriate modern reference for the alignment, we mapped the Triticeae reads against selected chloroplast genomes of domesticated and wild species in the genus Triticum using BWA [41] (with default parameters) implemented in the program FastQ Screen [71]. Thereby obtained FastQ Screen results indicate that the Triticeae reads preferentially map to both Triticum monococcum (KC912690) and Triticum urartu (KC912693) having the most specific reads to these two Triticum species (see Data S1_ FastQScreen Triticeae reads).
The previously extracted reads (both UDG-treated and non-UDG treated) of the four dominant animal and plant (sub)families Caprinae Cervinae, Dennstaedtiaceae, and Triticeae were then re-mapped against the respective reference plastid genomes using BWA [41] and increased the mapping sensitivity parameters (-l 1000, -o 1, -n 0.03). The obtained mapping statistics are summarized in the (see Data S1_ Mapping metagenomic reads). Resulting SAM files were filtered for a minimum mapping quality of 30, converted to BAM files, sorted, and indexed with SAMtools [43]. Duplicates were removed using MarkDuplicates from Picard tools (version 115, http://broadinstitute.github.io/picard/). A consensus FASTA sequence was generated for each sample using the ANGSD tool [44] and making a majority consensus call from overlapping reads (with the arguments –doFasta2 –doCounts1).
The reconstructed plastid genome sequences were subjected to comparative sequence and phylogenetic assignment. At first, a comparative plastid genome dataset was created. Nucleic acid sequences of modern plastid genomes that are closely related to the consensus sequences were retrieved from the public database using the BLAST search tool NCBI tblastn [42]. A multiple alignment was obtained by using the MAFFT multiple sequence alignment program [45]. The automatically inferred alignment was manually refined by using the sequence editor implemented in the ARB software package [46]. Phylogenetic analyses were performed by applying the maximum-likelihood method [PhyML [72] with the JTT substitution model]. For all datasets a stringent filter approach was applied to include into the phylogenetic assignment only position present in all sequences (please refer to Figure S3 for details to the number of informative position used).
To assess the nucleotide misincorporation patterns along the DNA fragments, we performed a mapDamage analysis [22] using all non-UDG treated reads of the Iceman intestine contents mapped against the above mentioned reference genomes. Mapping and filtering has been done as described for the preparation of the consensus sequences for the phylogenetic analysis (see Data S1). Thereby generated SAM files and the corresponding reads mapped against the reference genomes were used for the mapDamage analysis (see Figure S3D).
One limitation of the previous applied taxonomic assignment approach is the currently still limited availability of full plant and animal plastid genomes. Especially the chloroplast genomes deposited in the NCBI database represent only a small percentage of the possible plant diets present in the European Alps during the Iceman’s time. Therefore, we decided to perform a comparative sequence analysis of selected shotgun read datasets against the DNA barcodes deposited in the BOLD system database (http://www.boldsystems.org/) [51]. The DNA barcodes deposited at the BOLD database comprise fragments of the cytochrome c oxidase I (COI) gene, the ribulose bisphosphate carboxylase (rbcL) gene and the maturase K (matK) gene and the number of deposited species by far exceeds the number of plastid genomes in the NCBI database. The comparative analyses were performed against the COI fragments deposited in the groups Mammalia, Actinopterygii, Astacidae, Nematoda, and Platyhelmintes and against the rbcL and matK fragments in the groups Magnoliophyta, Pinophyta, Bryophyta, and Pteridophyta. First, we performed a sequence similarity search with the reads of selected shotgun datasets using blastn [42] with default parameters against the DNA barcodes extracted from the BOLD database. In the analysis four Iceman intestinal content datasets (B0626, B0621, C1824, B0625) were included, together with the Iceman muscle tissue (C0004) and Extraction Blank (B0629) as control datasets. All hits from the previous step with minimum bitscore 70, min length 15, 80% alignment length, 95% identity were further subjected to a second sequence similarity search using blastn [42] with default parameters and the complete NCBI-nt database as reference database. Blast results were taxonomically assigned using MEGAN6 [40] with a Min percent value of 0.01 (see Figure S4A).
Animal mitochondrial genome enrichment in the Iceman’s stomach content
Ancient DNA extraction and library preparation were conducted in the dedicated ancient DNA laboratory at EURAC research, Institute for Mummy Studies, Bolzano. The DNA was extracted from stomach contents using in-solution silica based protocol for ancient DNA [73]. The extracted DNA was prepared for Illumina sequencing by blunt-end repair and ligation of universal Illumina adapters (31). The library was shipped at −20°C to Trinity College Dublin and Illumina barcoded at the 5′ end there [74]. A custom RNA capture produced from reference domesticate mitochondrial genomes by MYcroarray (Mycroarray, 5692 Plymouth Road, Ann Arbor, MI 48105) was used to enrich the indexed library for mitochondrial genomes (following the manufacturer’s protocol). The captured library was sequenced on the Illumina MiSeq platform at TrinSeq St James’s Hospital Dublin (65 bp single-end sequencing kit). Data were quality assessed with fastqc, sequencing produced 4,905,473 raw reads, of which 95.5% passed adaptor trimming by AdapterRemoval [47]. To provide an overview of which mammalian DNA sequences were captured, the recovered reads were mapped to an array of candidate mammalian genomes using FastqScreen (https://www.bioinformatics.babraham.ac.uk/projects/fastq_screen/) (see Data S1_FastQScreen enriched reads) and bwa aligner (arguments: aln –l 1000 –n 0.01). Mapping sequence data showed that the captured library contained reads specific for red deer (Cervus elaphus, reference accession code: AB245427), ibex (Capra ibex, reference accession code: NC_020623) and human (reference accession code: NC_012920.1). Mapped reads were further analyzed for these three species, SAMtools (45) filtered out reads below mapping quality 30. DeDup (https://github.com/apeltzer/DeDup) removed duplicate reads (see Data S1). mapDamage displayed the presence of lesions associated with ancient DNA [22] (see Figure S3D), showing that on 10% of fragments had deamination lesions at the 5′ end of fragments. To investigate variants in the maped mitochondrial data, GATK UnifiedCaller created vcf. files from the bam files and variants were quantified with MultiVcfAnalyser, genotypes with a quality below 30 were filtered [48]. To further reduce the potentially confounding data from cross mapping or misincorporation of deaminated positions, only SNPs with a consensus above 80% were considered with respect to the overlapping reads (see Data S1_Variants enriched reads). This filtering strategy effectively removed confounding SNPs as demonstrated by the exact match to the Iceman’s mitochondrial haplotype defining SNPs [75].
Polymerase chain reaction (PCR)-based analyses
To further confirm our metagenomic findings in the Iceman intestinal content shotgun datasets we additionally performed a PCR-based analysis with selected DNA extracts targeting small DNA fragments of animal and plant barcodes (see Key Resources Table). To assess the animal diet of the Iceman, we amplified the fragments of two different mitochondrial loci, the 12S rRNA gene (12S rRNA) and the cytochrome B gene (cytB). For the cytB gene we applied a previously published PCR assay specifically designed to target different animal species [35]. In parallel, this assay excludes the amplification of human background DNA. Furthermore, we designed for this study a second PCR assay targeting a smaller fragment of the 12S rRNA gene of different animal species, including the human 12S rRNA gene variant. To further investigate via PCR the plant diet of the Iceman we amplified in the intestinal content DNA extracts the P6 loop of the chloroplast trnL (UAA) intron [37] and a 153 bp fragment of the chloroplast gene coding for the large subunit of the ribulose bisphosphate carboxylase (rbcL) [36]. To test for possible external contaminations, we included in all PCR assays the extraction blanks and two negative PCR controls. In addition, we included in the plant PCR assays an Iceman lung tissue sample to test for possibly inhaled pollen background. All PCR assays have been performed in the ancient DNA laboratory of the Eurac research Institute for Mummy Studies conducted in 50μl volume using the Amplitaq Gold mastermix 360, 1 mM primers and 5μl DNA template. Template DNA was initially denatured at 95°C for 5 min followed by 40 cycles of denaturation (95°C, 45 s), primer annealing (55°C, 45 s) and elongation (72°C, 45 s), with 4 min of final extension at 72°C. Only for the 12S rRNA gene PCR assay 30 cycles instead of 40 cycles were applied. The first PCR was followed by a second PCR using 1:50 dilution of the first PCR product and 10 to 15 additional cycles of amplification with barcoded PCR primers [76]. The barcoded Primers consisted of the gene-specific PCR primer sequences tagged with the sequencing adapters for GS FLX Titanium chemistry, including (5′-3′): Titanium adaptor (454 Life Sciences), a sample specific ten-base multiplex identifiers (MIDs on the reverse primer), a linker sequence (“TC” for forward primer, “CA” for reverse primer), and the gene-specific PCR primer. Amplicons were sequenced form the reverse side on a GS FLX instrument using Titanium chemistry (454 Life Sciences). For details to the amplicon datasets please refer to Data S1_ 454 Amplicon datasets. Sequencing reads were de-multiplexed and filtered using the mothur software package [49]. To obtain a taxonomic overview of the reads, we performed a sequence similarity search using blastn (40) with default parameters and the complete NCBI-nt database as reference database. The results were taxonomically assigned using MEGAN6 [40] with the lowest common ancestor (LCA) of hits within 10% of the bitscore of the best hits and a minimum number of 2 reads that a taxon must obtain (see Figure S4).
Proteomic analysis of the Iceman stomach content
We studied the proteome of three distinct stomach content samples (1051L, 1051LP & 1054LP) by liquid chromatography-mass spectrometry (LC-MS) proteomics using Orbitrap and QExactive technologies. For methodological details to the sample preparation, the LC-MS approach and the mass spectrometry data analysis please refer to the publication of Maixner et al. [11]. Briefly, each stomach content sample was solubilized, tryptic digested and fractionated using 1D SDS-PAGE or OffGEL isolelectric focusing (OGE) and then analyzed by high-mass accuracy Orbitrap or QExactive mass spectrometry instruments interfaced with nanospray liquid chromatography. Peptide identities were determined using the Trans-Proteomic Pipeline (TPP) software tool suite to define proteins contained in the stomach content samples [50].
The protein database used in the comparative analysis contained amino acid sequences constructed from multiple species. 75,649 protein sequences were to Homo sapiens (UniProt reference proteome, release 2012_10 ftp.uniprot.org/pub/databases/uniprot/previous_releases/release-2012_10/relnotes.txt) supplemented with sequences translated directly from the Iceman’s genome [34, 58]. Additionally, 936 and 1,278 Cervus elaphus protein sequences were obtained from NCBI and UniProt, respectively (obtained November 17, 2015). 30,155 Capra hircus protein sequences were obtained from NCBI (October 19, 2015) as the closest surrogate for the ibex proteome. The 1,011 available Triticum monococcum protein sequences from UniProt were supplemented with 95,051 Triticum aestivum sequences from UniProt (release 2015_11) to provide a complete proteome representing Triticinae. 28,356 open reading frames of at least 30 amino acids were obtained from a previously published Pteridium aquilinum transcriptome [77]. An additional 67 protein sequences to common laboratory contaminants were also added. An equal number of shuffled decoy sequences were generated to peform database search validation, as described previously [11].
The identified proteins (167), evaluated by statistical analysis using the TPP to 1% false discovery rate, were further taxonomically screened for homologous peptides and subsequently filtered for peptide sequences that can be unambiguously assigned to certain plant and animal families. To taxonomically assign the retrieved peptides based on the lowest common ancestor we performed a BLAST search [42] against the NCBI nr database and further analyzed the BLAST hits with the MEGAN6 software package [40]. In total, 20 of the 167 proteins were unambiguously identified with peptides assigned to the families Triticeae, Caprinae, and Cervinae (see Data S2_ Proteomics).
Lipid analysis of the Iceman stomach content
During the preliminary inspection of the Iceman’s stomach content, two samples were isolated, described as “lipid accumulation” and “lipid/protein mix.” Both samples were inhomogeneous, and multiple aliquots (a few milligrams each) were taken for lipid analysis. Aliquots were extracted using chloroform-methanol/based liquid-liquid extraction and analysis using an unbiased approach with reverse phase liquid chromatography separation and quadrupole time-of-flight (QToF) tandem mass spectrometry analysis (RP-LC/MSMS). Lipid analyses of aliquots from the Iceman stomach content were undertaken independently at two laboratories in Singapore and Seattle, USA. The analytical methods were as follows:
Singapore analytical method
Sample Preparation: Portions of two Iceman stomach samples and fresh animal tissue samples were dissected with a scalpel on an aluminum foil-covered ice pack, and the weights were noted: 1) Iceman Lipid accumulation 1: 6.9 mg, 2) Iceman Lipid/protein mix 1: 4.5 mg, 3) Capra ibex muscle tissue: 17.9 mg, 4) Capra ibex adipose tissue: 8.2mg, 5) Capra ibex skin: 8.6mg, 6) Cervus elaphus muscle tissue: 18.3 mg, 7) Cervus elaphus fat tissue: 15.0 mg, 8) Capra hircus cheese: 17.8mg, 9) Capra hircus milk: 27.5mg. For details to the modern reference samples used in this study please refer to Table S12. Lipids were extracted by a modified Bligh and Dyer extraction method [78]. The tissue pieces were transferred to glass homogenization tubes, and 300 μL of methanol were added. Samples were homogenized using a benchtop homogenizer, and the homogenizer probe was rinsed with an extra 300μL methanol. 300 μL of chloroform were added to the homogenates (final volume 900 μL of chloroform/methanol 1/2 v/v). The homogenizing rotor was carefully rinsed between each sample. A tube containing methanol only was used as negative control. The bottom organic phases were recovered and transfer to clean tubes and the solvent was dried under nitrogen gas. Dried lipid extracts were kept at −80°C, until LC/MS analysis for which they were re-suspended in 200μL chloroform/methanol (1/1 v/v). To determine the approx. lipid amount (w/w) three stomach biopsies were first dried at 105°C for 24h and weighed. Then, the lipids were extracted by the modified Bligh and Dyer extraction method (described above) and the extracts were dried at 105°C for 24h and weighed.
LC/MS: Chromatography conditions were optimized to enable efficient separation of triacylglycerols (TAGs) by reverse phase separation. LC/MS experiments were undertaken using an Agilent 1200 series HPLC-Chip system connected to Agilent 6540 Q-ToF. An Agilent C18 chip was used (75μm (i.d.) x 43 mm length, 5μm particle size, 300Å pore size) with a binary solvent system at a flow rate of 0.3μL/min. Solvent A was acetonitrile/water 40/60 v/v containing 5mM ammonium acetate and 0.1% acetic acid, and solvent B was isopropanol/acetonitrile 90/10 v/v containing 5mM ammonium acetate and 0.1% acetic acid. The gradient was as follows: 0%B for 1.5min, increase to 50%B in 2.0min, increase to 100% B in 18min, stay at 100% B for 10min, decrease to 0%B in 0.1min, stay at 0%B for 3.4min, total runtime: 35min. The Agilent 6540 Q-ToF spectrometer was operated in positive ion mode; electrospray voltage was set to 1450 V (Vcap), temperature 300°C, drying gas 4L/min, fragmentor voltage 150 V. The instrument was operated with MS acquisition rate of 1 spectra/sec. Injection volume was 1 to 2 μL. To monitor sample LC carryover and cross-contamination, solvent blanks were run between samples.
Data Analyses: LC/MS data files were processed with the MassHunter Qualitative Analysis Software Version B.05.01. The data were manually curated to identify triglyceride ions ([M+NH4]+ adducts) based on retention time and accurate mass measurement with mass accuracy < 5ppm. Intensities of the [M+NH4]+ adduct mono-isotopic peaks were used for calculating the relative abundance of triglyceride molecular species. TAG have been successfully used in the identification of food remains in an archaeological context [26, 79]. Therefore, we decided to compare the relative abundance of TAG between the Iceman’s stomach samples and with contemporary animal samples (See Data S2_Modern comparative samples). Following the genomics results, C. ibex and C. elaphus meat samples were obtained from local hunters. In addition, and in view of the interest regarding the question of dairy consumption in early humans, samples of C. hircus milk and cheese were also sourced. To better compare TAG distribution between samples, Mirabaud et al. [26] made use of two parameters: the average carbon number (M) and the distribution factor (DF), which can be calculated by the following formulae:
where Pi is the relative abundance of a given TAG (in percentage of total TAG) and Ci is the total number of carbon atoms in the fatty acyl chains.
Seattle analytical method
Sample Preparation: Portions of two Iceman samples and four fresh male tissue samples were dissected with a razor on an aluminum-foil covered block on top of dry ice, and the weights were noted: 1) Iceman Lipid accumulation 5: 73 mg, 2) Iceman Lipid-protein mix 5: 70 mg, 3) 0065206-19 adipose: 182 mg, 4) 0065251-10 adipose: 135 mg, 5) 0065251-29 skin (full thickness): 209 mg, 6) 0065206-09 skin (full thickness): 184 mg. The tissue pieces were transferred to homogenization tubes containing 2.38 mm metal beads, and 1 mL methanol. To serve as negative controls, two tubes containing methanol only were carried through each step of sample preparation. Following bead beating, BCA protein estimation was performed on the soluble portion of each lysate. The fresh tissue samples each contained between 500-2000 μg of soluble protein, and, surprisingly, each of the two Iceman samples contained roughly 500 μg soluble protein. The lysates were transferred to silanized 13x100 glass vials, and total lipids were extracted with chloroform/methanol using a modified “Folch” extraction protocol [80]. The combined aqueous upper phase and interphase (containing protein precipitates) from each sample were dried and processed by multi-dimensional fractionation for proteomics analyses in Seattle (see above). The lower organic phases were dried under nitrogen and stored at −30°C. The visual appearance of the dried lipid extracts differed remarkably: Iceman extracts appeared as a dry brown powder, and the fresh tissue extracts appeared as a viscous yellow liquid. For lipid LC/MS, 8 mL chloroform/methanol (2:1 v/v) was added to each extract, and 5 μL was used for injection. The extract was also diluted 100-fold and 5 μL was again used for injection. Additionally, neutral lipids were obtained from the total lipid extract by solid-phase extraction (SPE) following a previously published method [81]. Briefly, 100 μL of the total lipid extract was evaporated under N2 gas, re-dissolved in 1 mL isooctane/ethyl acetate (75/25 v/v), sonicated, and then loaded onto 100 mg Biotage Isoelute XL silica cartridges. Neutral lipids were collected from the flow-through, and further washed and collected from the column with isooctane/ethyl acetate (75/25 v/v). Eluants were evaporated under nitrogen gas, dissolved in 1 mL chloroform/methanol (2:1 v/v), and 5 μL was used for injection. Additionally, a 10-fold dilution was also used for injection.
LC/MS: Chromatography conditions were optimized to enable efficient separation of triacylglycerols (TAGs). An Agilent 1290 Infinity LC with a 2.1 mm (i.d.) x 100 mm, 3.5 μm Agilent ZORBAX Eclipse XDB-C18 Narrow Bore Rapid Resolution column heated to 30°C was used with a binary solvent system and a flow rate of 500 μL/min. The system was equilibrated with 80% of solvent A (5 mM ammonium formate in methanol/water (99:1 v/v)) and 20% solvent B (5 mM ammonium formate in isopropanol/water (99:1 v/v)). A 5 uL injection volume of the lipid extract (in 2:1 chloroform/methanol) was applied to the column, followed by a 30.0-min linear gradient to 20% solvent A/80% solvent B, and held for 2.5 min. An Agilent 6530 Q-ToF equipped with an Agilent JetStream ESI source was used for accurate mass analysis of the LC eluent. Positive ion mass spectra data were acquired in 2 GHz Extended Dynamic Range mode at a rate of 1.02 spectra/s, and data was collected as profiled spectra over a mass range of 150 to 1500 m/z. For MS/MS analyses, a fixed collision energy of 40 V was used for fragmentation. To monitor sample LC carryover and cross-contamination, several solvent blanks were run between samples, and sample order was considered carefully.
Data Analyses: LC/MS data files were processed with the MassHunter Qualitative Analysis Software version B.03.01. Molecular features (MFs) were extracted from the raw MS1 data using the Molecular Feature Extraction (MFE) algorithm with a peak filter of ≥ 10,000 counts. The resulting MFs were then identified with MassHunter by searching a custom database containing 690 TAG ions corresponding to [M+Na]+ and [M+NH4]+ adducts, and 99 [M+NH4]+ cholesterol ester ions, using a ± 5 ppm search window. For quantitative purposes, lipid groups were compared using the [M+NH4]+ molecular ion height. See Data S2 for a summary of the lipid analysis results.
Atomic force microscopy (AFM) and Raman spectroscopy
Meat fiber assemblies within the Iceman food bolus samples were identified with an inverted optical microscope (Axiovert 135, Zeiss, Oberkochen, Germany). Subsequently, the coarse topography of the putative meat fibers was analyzed by laser scanning microscope (LSM) images (VK 8710, Keyence, Osaka, Japan) while high-resolution images of defined sample areas were taken with a NanoWizard-II AFM (JPK Instruments, Berlin, Germany). The AFM was operated in the intermittent contact mode and silicon cantilevers (BS Tap 300, Budget Sensors, Redding, USA) with nominal spring constants of 40 N/m, resonance frequencies of 300 kHz, and tip radii of 10 nm were used. All images were taken at lab conditions (22°C and about 45% relative humidity).
To assess the molecular composition of the samples, Raman spectra were taken with a confocal Raman microscope (WITec alpha 300 R, WITec GmbH, Ulm, Germany). The Raman system was operated with a laser (frequency doubled Nd:YAG-laser with an excitation wavelength of 532 nm) at 1.0 mW laser power, to avoid sample damage (photodegradation). The backscattered light from the sample was detected with a vacuum sealed, high-sensitivity, back-illuminated CCD camera cooled to −56°C. The spectrometer was equipped with a 600 g/mm grating enabling a spectral resolution of approximately 3 cm-1 per CCD-pixel. Single spectra of each sample are average spectra of 10 accumulations with up to 180 s of integration time. Due to the confocal setup of the microscope and the objectives used, Raman spectra were collected from a sample area with approximately 400 nm diameter and a focal depth of about 1 μm.
Quantification and Statistical Analysis
Phylogenetic analyses were performed by applying the maximum-likelihood method [PhyML [58] with the JTT substitution model] implemented in the ARB software package [46]. For all datasets a stringent filter approach was applied to include into the phylogenetic assignment only position present in all sequences.
Data and Software Availability
The Illumina datasets are available from the European Nucleotide Archive (ENA) under accession number ENA: ERP012908. The enriched Illumina dataset has been deposited in ENA under the accession number ENA: PRJEB26465. The 454 amplicon datasets has been deposited in ENA under the accession number ENA: PRJEB26365. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository [82] with the dataset identifier PXD009565.
Additional Resources
Additional metadata for the Illumina shotgun datasets are provided under the following link: http://cube.univie.ac.at/supplements/iceman.
Acknowledgments
We acknowledge the following funding sources: Provincia Autonoma di Bolzano, grant legge 14 agreement n. 1/40.3, 23.11.2012 (F.M., N.O., G.C., A. Paladin, M. Samadelli, A.Z.), the DFG Graduate School Human Development in Landscapes at Kiel University (B.K.-K., A.N.), and the DFG Excellence Cluster Inflammation at Interfaces at Kiel University (B.K.-K., J.F.B., A.F., A.N.). Ludwig Erhard and his team from the Provincia autonoma di Bolzano - Alto Adige, Ripartizione Foreste are highly acknowledged for providing the modern wild animal samples for this study. M.T. and V.M. were supported by European Research Council Investigator Grant 295729-CodeX to D.G.B. The National Institutes of Health from the National Institute of General Medical Sciences under Grant Nos. R01 GM087221 (R.L.M.), S10 RR027584 (R.L.M.), and the 2P50 GM076547/Center for Systems Biology (R.L.M.). The authors thank the Department of Innovation, Research and University of the Autonomous Province of Bozen/Bolzano for covering the open access publications costs. Gaia Brusco is highly acknowledged for the artwork.
Author Contributions
F.M. and A.Z. conceived the investigation. F.M., D.T., A.C.-G., M.J., M.R.H., R.L.M., A.F., A.K., M.W., R.W.S., O.F., P.D., T.R., R.G., and A.Z. designed the experiments. P.M., P.G., L.E., E.E.-V., M. Samadelli, F.M., and A.Z. were involved in the sampling campaign. F.M., A.C.-G., M.J., B.K.-K., M.R.H., U.K., M. Sartain, N.O., M.T., G.C., V.M., M.P., J.M., S.L., P.R., B.J.K., H.-J.A., and D.B. conducted the experiments. F.M., D.T., A.C.-G., M.J., M.R.H., N.O., M.T., S.L., J.F.B., B.J.K., H.-J.A., and K.O. performed analyses. F.M. wrote the manuscript with contributions from D.T., A.C.-G., M.J., M.R.H., A. Paladin, A. Putzer, G.G., U.T., A.N., D.G.B., K.O., R.G., T.R., and A.Z.
Declaration of Interests
The authors declare no competing interests.
Published: July 12, 2018
Footnotes
Supplemental Information includes four figures and two data files and can be found with this article online at https://doi.org/10.1016/j.cub.2018.05.067.
Contributor Information
Frank Maixner, Email: frank.maixner@eurac.edu.
Albert Zink, Email: albert.zink@eurac.edu.
Supplemental Information
Details to the shotgun and amplicon datasets, FastQScreen results, mapping statistics and detected variants.
Details to the comparative modern animal samples used in this study and to the metabolite, glycan, stable isotope, elemental, protein, and lipid analysis results.
References
- 1.Fowler C., Harding J., Hofmann D. Defining the ‘Neolithic in Europe’: Diverse and Contemporaneous Communities, c. 6500-2500 BC. In: Fowler C., Harding J., Hofman D., editors. The Oxford Handbook of Neolithic Europe. Oxford University press; United Kingdom: 2015. [Google Scholar]
- 2.Gostner P., Pernter P., Bonattie G., Graefen A., Zink A.R. New radiological insights into the life and death of the Tyrolean Iceman. J. Arch. Sci. 2011;38:3425–3431. [Google Scholar]
- 3.Spindler K. Harmony Books; 1994. The Man in the Ice. [Google Scholar]
- 4.Shennan S., Downey S.S., Timpson A., Edinborough K., Colledge S., Kerig T., Manning K., Thomas M.G. Regional population collapse followed initial agriculture booms in mid-Holocene Europe. Nat. Commun. 2013;4:2486. doi: 10.1038/ncomms3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richards M.P. A brief review of the archaeological evidence for Palaeolithic and Neolithic subsistence. Eur J Clin Nutr. 2002;56:1270–1278. doi: 10.1038/sj.ejcn.1601646. [DOI] [PubMed] [Google Scholar]
- 6.Nicklisch N., Ganslmeier R., Siebert A., Friederich S., Meller H., Alt K.W. Holes in teeth - Dental caries in Neolithic and Early Bronze Age populations in Central Germany. Ann. Anat. 2016;203:90–99. doi: 10.1016/j.aanat.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Diamond J. Evolution, consequences and future of plant and animal domestication. Nature. 2002;418:700–707. doi: 10.1038/nature01019. [DOI] [PubMed] [Google Scholar]
- 8.Richards M.P., Schulting R.J., Hedges R.E. Archaeology: sharp shift in diet at onset of Neolithic. Nature. 2003;425:366. doi: 10.1038/425366a. [DOI] [PubMed] [Google Scholar]
- 9.Evershed R.P., Payne S., Sherratt A.G., Copley M.S., Coolidge J., Urem-Kotsu D., Kotsakis K., Ozdoğan M., Ozdoğan A.E., Nieuwenhuyse O. Earliest date for milk use in the Near East and southeastern Europe linked to cattle herding. Nature. 2008;455:528–531. doi: 10.1038/nature07180. [DOI] [PubMed] [Google Scholar]
- 10.Fowler C., Harding J., Hofman D. Movement of Plants, Animals, Ideas and People. In: Fowler C., Harding J., Hofman D., editors. The Oxford Handbook of Neolithic Europe. Oxford University press; United Kingdom: 2015. [Google Scholar]
- 11.Maixner F., Krause-Kyora B., Turaev D., Herbig A., Hoopmann M.R., Hallows J.L., Kusebauch U., Vigl E.E., Malfertheiner P., Megraud F. The 5300-year-old Helicobacter pylori genome of the Iceman. Science. 2016;351:162–165. doi: 10.1126/science.aad2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macko S.A., Engel M.H., Andrusevich V., Lubec G., O’Connell T.C., Hedges R.E. Documenting the diet in ancient human populations through stable isotope analysis of hair. Philos Trans R Soc Lond B Biol Sci. 1999;354:65–75. doi: 10.1098/rstb.1999.0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macko S.A., Lubec G., Teschler-Nicola M., Andrusevich V., Engel M.H. The Ice Man’s diet as reflected by the stable nitrogen and carbon isotopic composition of his hair. FASEB J. 1999;13:559–562. doi: 10.1096/fasebj.13.3.559. [DOI] [PubMed] [Google Scholar]
- 14.Dickson J.H., Oeggl K., Holden T.G., Handley L.L., O’Connell T.C., Preston T. The omnivorous Tyrolean Iceman: colon contents (meat, cereals, pollen, moss and whipworm) and stable isotope analyses. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000;355:1843–1849. doi: 10.1098/rstb.2000.0739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oeggl K. The Diet of the Iceman. In: Bortenschlager S., Oeggl K., editors. The Iceman and his natural environment. Springer; Vienna: 2000. pp. 89–115. [Google Scholar]
- 16.Oeggl K., Kofler W., Schmidl A., Dickson J.H., Egarter-Vigl E., Gaber O. The reconstruction of the last itinerary of “Ötzi”, the Neolithic Iceman, by pollen analyses from sequentially sampled gut extracts. Quat. Sci. Rev. 2007;26:853–861. [Google Scholar]
- 17.Oeggl K., Kolfer W., Schmidl A. New aspects on the diet of the Tyrolean Iceman, ‘‘Ötzi”. J. Biol. Res. (Thessalon.) 2005;80:344–347. [Google Scholar]
- 18.Rollo F., Ubaldi M., Ermini L., Marota I. Otzi’s last meals: DNA analysis of the intestinal content of the Neolithic glacier mummy from the Alps. Proc. Natl. Acad. Sci. USA. 2002;99:12594–12599. doi: 10.1073/pnas.192184599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dickson J.H., Hofbauer W., Porley R., Schmidl A., Kofler W., Oeggl K. Six mosses from the Tyrolean Iceman’s alimentary tract and their significance for his ethnobotany and the events of his last days. Veg. Hist. Archaeobot. 2009;18:13–22. [Google Scholar]
- 20.Dickson J.H., Oeggl K., Handley L.L. The iceman reconsidered. Sci. Am. 2003;288:70–79. doi: 10.1038/scientificamerican0503-70. [DOI] [PubMed] [Google Scholar]
- 21.Schiaffino S., Reggiani C. Fiber types in mammalian skeletal muscles. Physiol. Rev. 2011;91:1447–1531. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- 22.Ginolhac A., Rasmussen M., Gilbert M.T., Willerslev E., Orlando L. mapDamage: testing for damage patterns in ancient DNA sequences. Bioinformatics. 2011;27:2153–2155. doi: 10.1093/bioinformatics/btr347. [DOI] [PubMed] [Google Scholar]
- 23.Guo G., Lv D., Yan X., Subburaj S., Ge P., Li X., Hu Y., Yan Y. Proteome characterization of developing grains in bread wheat cultivars (Triticum aestivum L.) BMC Plant Biol. 2012;12:147. doi: 10.1186/1471-2229-12-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jerkovic A., Kriegel A.M., Bradner J.R., Atwell B.J., Roberts T.H., Willows R.D. Strategic distribution of protective proteins within bran layers of wheat protects the nutrient-rich endosperm. Plant Physiol. 2010;152:1459–1470. doi: 10.1104/pp.109.149864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Labeit S., Kolmerer B. Titins: giant proteins in charge of muscle ultrastructure and elasticity. Science. 1995;270:293–296. doi: 10.1126/science.270.5234.293. [DOI] [PubMed] [Google Scholar]
- 26.Mirabaud S., Rolando C., Regert M. Molecular criteria for discriminating adipose fat and milk from different species by NanoESI MS and MS/MS of their triacylglycerols: application to archaeological remains. Anal. Chem. 2007;79:6182–6192. doi: 10.1021/ac070594p. [DOI] [PubMed] [Google Scholar]
- 27.Cordain L., Watkins B.A., Florant G.L., Kelher M., Rogers L., Li Y. Fatty acid analysis of wild ruminant tissues: evolutionary implications for reducing diet-related chronic disease. Eur. J. Clin. Nutr. 2002;56:181–191. doi: 10.1038/sj.ejcn.1601307. [DOI] [PubMed] [Google Scholar]
- 28.Mayer B.X., Reiter C., Bereuter T.L. Investigation of the triacylglycerol composition of iceman’s mummified tissue by high-temperature gas chromatography. J. Chromatogr. B Biomed. Sci. Appl. 1997;692:1–6. doi: 10.1016/s0378-4347(96)00501-4. [DOI] [PubMed] [Google Scholar]
- 29.Böcker U., Ofstad R., Wu Z., Bertram H.C., Sockalingum G.D., Manfait M., Egelandsdal B., Kohler A. Revealing covariance structures in fourier transform infrared and Raman microspectroscopy spectra: a study on pork muscle fiber tissue subjected to different processing parameters. Appl. Spectrosc. 2007;61:1032–1039. doi: 10.1366/000370207782217707. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y., Wujisguleng W., Long C. Food uses of ferns in China: a review. Acta Soc. Bot. Pol. 2012;81 [Google Scholar]
- 31.Wing-Gaia S.L., Wayne Askew E. Nutrition, Malnutrition, and Starvation. In: Auerbach P., Cushing T., Harris N.S., editors. Wilderness Medicine. 7th edition. Elsevier; 2016. pp. 1964–1985. [Google Scholar]
- 32.Sacks F.M., Lichtenstein A.H., Wu J.H.Y., Appel L.J., Creager M.A., Kris-Etherton P.M., Miller M., Rimm E.B., Rudel L.L., Robinson J.G., American Heart Association Dietary Fats and Cardiovascular Disease: A Presidential Advisory From the American Heart Association. Circulation. 2017;136:e1–e23. doi: 10.1161/CIR.0000000000000510. [DOI] [PubMed] [Google Scholar]
- 33.Murphy W.A., Jr., Nedden Dz Dz., Gostner P., Knapp R., Recheis W., Seidler H. The iceman: discovery and imaging. Radiology. 2003;226:614–629. doi: 10.1148/radiol.2263020338. [DOI] [PubMed] [Google Scholar]
- 34.Keller A., Graefen A., Ball M., Matzas M., Boisguerin V., Maixner F., Leidinger P., Backes C., Khairat R., Forster M. New insights into the Tyrolean Iceman’s origin and phenotype as inferred by whole-genome sequencing. Nat. Commun. 2012;3:698. doi: 10.1038/ncomms1701. [DOI] [PubMed] [Google Scholar]
- 35.Burger J., Schoon R., Zeike B., Hummel S., Hermann B. Species Determination using Species-discriminating PCR-RFLP of Ancient DNA from Prehistoric Skeletal Remains. Anc. Biomol. 2002;4:19–23. [Google Scholar]
- 36.Poinar H.N., Hofreiter M., Spaulding W.G., Martin P.S., Stankiewicz B.A., Bland H., Evershed R.P., Possnert G., Pääbo S. Molecular coproscopy: dung and diet of the extinct ground sloth Nothrotheriops shastensis. Science. 1998;281:402–406. doi: 10.1126/science.281.5375.402. [DOI] [PubMed] [Google Scholar]
- 37.Taberlet P., Coissac E., Pompanon F., Gielly L., Miquel C., Valentini A., Vermat T., Corthier G., Brochmann C., Willerslev E. Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res. 2007;35:e14. doi: 10.1093/nar/gkl938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao Y., Tang H., Ye Y. RAPSearch2: a fast and memory-efficient protein similarity search tool for next-generation sequencing data. Bioinformatics. 2012;28:125–126. doi: 10.1093/bioinformatics/btr595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ondov B.D., Bergman N.H., Phillippy A.M. Interactive metagenomic visualization in a Web browser. BMC Bioinformatics. 2011;12:385. doi: 10.1186/1471-2105-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huson D.H., Beier S., Flade I., Górska A., El-Hadidi M., Mitra S., Ruscheweyh H.J., Tappu R. MEGAN Community Edition - Interactive Exploration and Analysis of Large-Scale Microbiome Sequencing Data. PLoS Comput. Biol. 2016;12:e1004957. doi: 10.1371/journal.pcbi.1004957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 43.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Korneliussen T.S., Albrechtsen A., Nielsen R. ANGSD: Analysis of Next Generation Sequencing Data. BMC Bioinformatics. 2014;15:356. doi: 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ludwig W., Strunk O., Westram R., Richter L., Meier H., Yadhukumar, Buchner A., Lai T., Steppi S., Jobb G. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lindgreen S. AdapterRemoval: easy cleaning of next-generation sequencing reads. BMC Res. Notes. 2012;5:337. doi: 10.1186/1756-0500-5-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., Lesniewski R.A., Oakley B.B., Parks D.H., Robinson C.J. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deutsch E.W., Mendoza L., Shteynberg D., Slagel J., Sun Z., Moritz R.L. Trans-Proteomic Pipeline, a standardized data processing pipeline for large-scale reproducible proteomics informatics. Proteomics Clin. Appl. 2015;9:745–754. doi: 10.1002/prca.201400164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ratnasingham S., Hebert P.D. bold: The Barcode of Life Data System (http://www.barcodinglife.org) Mol. Ecol. Notes. 2007;7:355–364. doi: 10.1111/j.1471-8286.2007.01678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aufderheide A.C. Cambridge University Press; 2003. The Scientific Study of Mummies. [Google Scholar]
- 53.Gostner P., Vigl E.E. INSIGHT: Report of Radiological-Forensic Findings on the Iceman. J. Archaeol. Sci. 2002;29:323–326. [Google Scholar]
- 54.Pernter P., Gostner P., Egarter Vigl E., Rühli F.J. Radiologic proof for the Iceman’s cause of death (ca. 5′300 BP) J. Archaeol. Sci. 2007;34:1784–1786. [Google Scholar]
- 55.Lynnerup N. Mummies. Am. J. Phys. Anthropol. 2007;45(Suppl 45):162–190. doi: 10.1002/ajpa.20728. [DOI] [PubMed] [Google Scholar]
- 56.Coia V., Cipollini G., Anagnostou P., Maixner F., Battaggia C., Brisighelli F., Gómez-Carballa A., Destro Bisol G., Salas A., Zink A. Whole mitochondrial DNA sequencing in Alpine populations and the genetic history of the Neolithic Tyrolean Iceman. Sci. Rep. 2016;6:18932. doi: 10.1038/srep18932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Janko M., Stark R.W., Zink A. Preservation of 5300 year old red blood cells in the Iceman. J. R. Soc. Interface. 2012;9:2581–2590. doi: 10.1098/rsif.2012.0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maixner F., Overath T., Linke D., Janko M., Guerriero G., van den Berg B.H., Stade B., Leidinger P., Backes C., Jaremek M. Paleoproteomic study of the Iceman’s brain tissue. Cell. Mol. Life Sci. 2013;70:3709–3722. doi: 10.1007/s00018-013-1360-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Keller A., Kreis S., Leidinger P., Maixner F., Ludwig N., Backes C., Galata V., Guerriero G., Fehlmann T., Franke A. miRNAs in ancient tissue specimens of the Tyrolean Iceman. Mol. Biol. Evol. 2017;34:793–801. doi: 10.1093/molbev/msw291. [DOI] [PubMed] [Google Scholar]
- 60.O’Sullivan N.J., Teasdale M.D., Mattiangeli V., Maixner F., Pinhasi R., Bradley D.G., Zink A. A whole mitochondria analysis of the Tyrolean Iceman’s leather provides insights into the animal sources of Copper Age clothing. Sci. Rep. 2016;6:31279. doi: 10.1038/srep31279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Makristathis A., Schwarzmeier J., Mader R.M., Varmuza K., Simonitsch I., Chavez J.C., Platzer W., Unterdorfer H., Scheithauer R., Derevianko A., Seidler H. Fatty acid composition and preservation of the Tyrolean Iceman and other mummies. J. Lipid Res. 2002;43:2056–2061. doi: 10.1194/jlr.m100424-jlr200. [DOI] [PubMed] [Google Scholar]
- 62.Mulisch M., Welsch U. Spektrum Akademischer Verlag; 2010. Romeis - Mikroskopische Techniken. [Google Scholar]
- 63.Fry B. Springer-Verlag; New York: 2006. Stable Isotope Ecology. [Google Scholar]
- 64.Hoefs J. Sixth Edition. Springer Verlag; Berlin, Heidelberg: 2009. Stable Isotope Geochemistry. [Google Scholar]
- 65.Schoeninger M.J., DeNiro M.J. Nitrogen and carbon isotopic composition of bone collagen from marine and terrestrial animals. Geochim. Cosmochim. Acta. 1984;48:625–639. [Google Scholar]
- 66.Tang J.N., Zeng Z.G., Wang H.N., Yang T., Zhang P.J., Li Y.L., Zhang A.Y., Fan W.Q., Zhang Y., Yang X. An effective method for isolation of DNA from pig faeces and comparison of five different methods. J. Microbiol. Methods. 2008;75:432–436. doi: 10.1016/j.mimet.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 67.Lee E.J., Makarewicz C., Renneberg R., Harder M., Krause-Kyora B., Müller S., Ostritz S., Fehren-Schmitz L., Schreiber S., Müller J. Emerging genetic patterns of the European Neolithic: perspectives from a late Neolithic Bell Beaker burial site in Germany. Am. J. Phys. Anthropol. 2012;148:571–579. doi: 10.1002/ajpa.22074. [DOI] [PubMed] [Google Scholar]
- 68.Kircher M., Sawyer S., Meyer M. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 2012;40:e3. doi: 10.1093/nar/gkr771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meyer M., Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot5448. pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- 70.Coordinators N.R., NCBI Resource Coordinators Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2017;45(D1):D12–D17. doi: 10.1093/nar/gkw1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Andrew S. FastQ Screen. Available online at. 2011. http://www.bioinformatics.babraham.ac.uk/projects/fastq_screen/
- 72.Guindon S., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 73.Rohland N., Hofreiter M. Comparison and optimization of ancient DNA extraction. Biotechniques. 2007;42:343–352. doi: 10.2144/000112383. [DOI] [PubMed] [Google Scholar]
- 74.Gamba C., Jones E.R., Teasdale M.D., McLaughlin R.L., Gonzalez-Fortes G., Mattiangeli V., Domboróczki L., Kővári I., Pap I., Anders A. Genome flux and stasis in a five millennium transect of European prehistory. Nat. Commun. 2014;5:5257. doi: 10.1038/ncomms6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ermini L., Olivieri C., Rizzi E., Corti G., Bonnal R., Soares P., Luciani S., Marota I., De Bellis G., Richards M.B., Rollo F. Complete mitochondrial genome sequence of the Tyrolean Iceman. Curr. Biol. 2008;18:1687–1693. doi: 10.1016/j.cub.2008.09.028. [DOI] [PubMed] [Google Scholar]
- 76.Berry D., Ben Mahfoudh K., Wagner M., Loy A. Barcoded primers used in multiplex amplicon pyrosequencing bias amplification. Appl. Environ. Microbiol. 2011;77:7846–7849. doi: 10.1128/AEM.05220-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Der J.P., Barker M.S., Wickett N.J., dePamphilis C.W., Wolf P.G. De novo characterization of the gametophyte transcriptome in bracken fern, Pteridium aquilinum. BMC Genomics. 2011;12:99. doi: 10.1186/1471-2164-12-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bligh E.G., Dyer W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 79.Garnier N., Rolando C., Høtje J.M., Tokarski C. Analysis of archaeological triacylglycerols by high resolution nanoESI, FT-ICR MS and IRMPD MS/MS: Application to 5th century BC–4th century AD oil lamps from Olbia (Ukraine) Int. J. Mass Spectrom. 2009;284:47–56. [Google Scholar]
- 80.Folch J., Lees M., Sloane Stanley G.H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 81.Krank J., Murphy R.C., Barkley R.M., Duchoslav E., McAnoy A. Qualitative analysis and quantitative assessment of changes in neutral glycerol lipid molecular species within cells. Methods Enzymol. 2007;432:1–20. doi: 10.1016/S0076-6879(07)32001-6. [DOI] [PubMed] [Google Scholar]
- 82.Vizcaíno J.A., Csordas A., del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44(D1):D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Details to the shotgun and amplicon datasets, FastQScreen results, mapping statistics and detected variants.
Details to the comparative modern animal samples used in this study and to the metabolite, glycan, stable isotope, elemental, protein, and lipid analysis results.