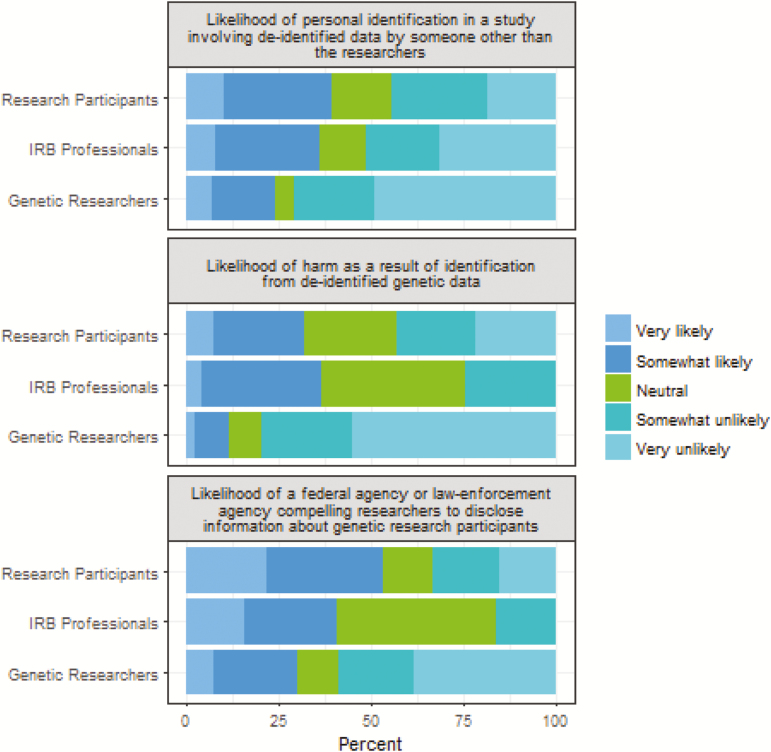
Compared to genetic researchers, institutional review board professionals and research participants felt that participants would be identified or harmed from a study involving de-identified data.
Keywords: De-identification, Genomic, Preferences, Linkage
Abstract
Data sharing of large genomic databases and biorepositories provides researchers adequately powered samples to advance the goals of precision medicine. Data sharing may also introduce, however, participant privacy concerns including possible reidentification. This study compares views of research participants, genetic researchers, and institutional review board (IRB) professionals regarding concerns about the use of de-identified data. An online survey was completed by cancer patients, their relatives, and controls from the Northwest Cancer Genetics Registry (n = 450) querying views about potential harms with the use of de-identified data. This was compared to our previous online national survey of human genetic researchers (n = 351) and IRB professionals (n = 208). Researchers were less likely to feel that participants would be personally identified or harmed from a study involving de-identified data or feel that a federal agency might compel researchers to disclose information about research participants. Compared to genetic researchers, IRB professionals and participants were significantly more likely to express that personal identification or harm was likely or that researchers might be forced to disclose information by a federal agency. An understanding of the differences in views regarding possible harm from the use of de-identified data between these three important stakeholder groups is necessary to move forward with genomic research.
Implications
Practice: Those involved in recruitment of participants into research studies need to be aware of Institutional Review Board professional and participant concerns regarding the use of their de-identified data for genomic research.
Policy: The design of research participant recruitment efforts should consider the contrasting viewpoints among stakeholders regarding the risks and use of de-identified data in genomic research.
Research: Future research is needed to understand the specific reasons for differences in perceived risks associated with the use of de- identified genomic data between stakeholder groups.
Introduction
Rapid advancement of genomic sequencing technologies has led to the ability to genotype large numbers of research participants. Linkage of large biorepositories with clinical and lifestyle data offers a new mechanism to evaluate gene–gene (G×G) and gene–environment (G×E) interactions, but large samples are needed to have adequate power to detect interactions. Data sharing now offers the ability to have adequately powered samples and advance the goals of precision medicine.
While the linkage of biological material to personal data raises challenges, including return of results, informed consent, and privacy, it is widely accepted and is required by the National Institutes of Health for certain areas (http://grants.nih.gov/grants/gwas/). While beneficial for investigators, researchers, and the public [1], consent for data sharing is often not included in the informed consent process for the original study and open data access is often at odds with participant privacy. Data points that may be used as explicit personal identifiers are often removed to safeguard the participant’s privacy and reduce the risk of identification. And although it has been shown that de-identification may be inadequate because it is possible to reverse identify participants from previously de-identified data [2, 3], genomic information is not considered identifiable data [4] and is exempt from the major US privacy law, the Health Insurance Portability and Accountability Act (HIPAA) [5].
A comparison of three stakeholder groups, including participants, researchers, and Institutional Review Board (IRB) professionals, is critical in order to find common ground, maximize study recruitment, and maintain study participation. These investigators have previously evaluated the beliefs of research participants and found that most expressed a desire for their data to be available to as many research studies as possible, with the goal of receiving personal health information, while noting the importance of protecting their privacy and information [6]. Research participants were also likely to feel that the original researcher was responsible for maintaining a link to their de-identified data, and felt that it was important to maintain a link in order to allow individual health results to be return to them and to support further research. Most research participants were not concerned about personal identification when participating in a genetic study using de-identified data.
In a study of IRB professionals, these investigators found that about two thirds felt that it is unlikely that a research participant would be personally identified or harmed from their de-identified data and about one-third believed that investigators might be compelled by a federal agency to disclose personal information about research participants [7]. Compared to a group of genetic researchers, these IRB professionals were significantly more likely to believe that a research participant would be personally identified from coded genetic data, harmed as a result of identification, or have a federal agency force disclosure about genetic research participants [8].
This study will expand on our previous work by comparing views of genetic researchers, IRB professionals, and research participants regarding concerns about the use of de-identified data.
METHODS
Eligibility and recruitment for the Genetics Research Review Issues Project (GRRIP)
Details on the GRRIP study and development of the surveys have been described previously [7–9]. Briefly, genetic researchers, recruited from the American Society of Human Genetics, and IRB professionals, recruited from The Public Responsibility in Medicine and Research, were asked to complete a web-based survey related to four areas of interest: the research study application process, the IRB review process, IRB functions, and issues in genetic research. Surveys were completed by 351 human genetic researchers and 208 IRB professionals.
Eligibility and recruitment for the Participant Issues and Expectations Project (PIP)
Details on the Northwest Cancer Genetics Registry (NWCGR), the source of research participants for this study, have been described previously [10]. Briefly, individuals currently enrolled in the NWCGR (n = 3,352) were the source of PIP participants, including people with cancer recruited from Western Washington (n = 2,027), first-degree relatives of cases (n = 451), and controls recruited from a random sample from Washington (n = 527), and people who were self-referred in response to community awareness efforts (n = 904). Up to three invitations were sent to participants at approximately 2-week intervals [11]. The online survey was completed by 450 participants.
PIP and GRRIP survey methods
GRRIP survey development was conducted among 25 genetic researchers and 31 IRB professionals using in-depth interviews and focus groups. A tailored design method was used to identify salient issues and develop survey questions. Cognitive interviews with researchers and IRB professionals were then used to assess clarity and ease of the survey. In 2009, parallel online surveys were used to anonymously collect information from human genetic researchers (n = 351) and IRB professionals (n = 208). The purpose of the PIP survey was to document the range and frequency of concerns and expectations regarding participating in genomic research studies and to compare these findings to our previous GRRIP surveys [8, 9, 12]. Detailed methods for the PIP study have been published previously [10, 11]. Briefly, administered in 2012, the confidential, online survey instrument had a total of 22 questions, including overlapping questions from the GRRIP surveys and covered six general topic areas: decision to participate in research, relationship between researchers and participants, reconsent and broad consent, return of results, use and security of de-identified data, and family communication of health issues. Three questions regarding privacy or harm from de-identified genetic data overlapped with our previous GRRIP surveys. These questions asked how likely (i) a research participant would be personally identified in a study involving de-identified data by someone other than the researchers, (ii) a research participant would be harmed as a result of identification from de-identified genetic data, and (iii) a federal agency or other law-enforcement agency might compel researchers to disclose information about genetic research participants. Wording of the overlapping questions related to privacy or harm from de-identified genetic data were either identical (“How likely is it that a federal agency or other law-enforcement agency might compel investigators to disclose information about genetic research participants”) or very similar (“How likely is it that a research participant would be harmed as a result of identification from coded genetic data” for researchers and IRB professionals vs “How likely is it that a research participant would be harmed as a result of identification from de-identified genetic data” for research participants). The response categories for these questions were 5-point Likert-scales rating level of agreement with the statement. The five categories were as follows: very likely, somewhat likely, neutral, somewhat unlikely, very unlikely, or don’t know.
A comparison of nonresponses found that of the 450 PIP participants, 15 did not answer the first question, 21 did not answer the second, and 14 did not answer the third. With regards to the GRRIP comparison group, 6 of the genetic researchers did not answer the first question, 13 did not answer the second, and 64 did not answer the third; the comparable numbers for the IRB professionals were 12, 15, and 28, respectively. All study procedures were approved by the University of Washington’s Human Subjects Division and by the University of California, Irvine Institutional Review Board. All participants provided informed consent prior to participation and were free to skip any questions that they did not wish to answer.
Statistical analysis
Responses were first summarized using frequency distributions separately for each group. We hypothesized that PIP participants would differ from researchers and IRB professionals in their concerns over the likelihood of harm. To address this, we compared questions that were asked in the same way from the three surveys. “Don’t know” responses were considered missing. Differences in frequency of responses between the three stakeholder groups were tested using ordinal logistic regression, which allows for multiple categories of the outcome variable and adjustment for potential confounders. The response categories were ordered and coded as follows: “likely” was coded as 0, “neutral” 1, and “unlikely” 2. With ordinal logistic regression, several cumulative logits were modeled using all possible cut points of the dependent variable, but a single summary odds ratio and 95% confidence interval were obtained. Comparisons within the research participant sample were adjusted for age, gender and education, and comparisons between the three groups were adjusted for gender. Because there were no differences by gender, age, or relative type within the research participant sample, all participants were combined when compared with the two other groups. R version 3.2.2 with the MASS package was used [13, 14]. A p value ≤.05 was considered statistically significant.
RESULTS
About half of the 450 research participants were cases (n = 228), one-third were controls (n = 155), and the remainder were relatives (n = 67) (Table 1). The average research participant age was 63.6 years, and most were white (94%), married or living with someone (76%), and well educated, with over 60% having a college degree. Age, gender, race/ethnicity, education, and marital status were similarly distributed in the case, control, and relative groups. Among participants with cancer at baseline, melanoma was most frequent type (29.5%), followed by thyroid cancer (18.3%), and breast cancer (15.5%). Thirty-five research participants without cancer at enrollment into a parent study reported a cancer at the time of this survey (data not shown). Researchers were less likely to be women (51.9% vs 76.0%) and to have worked in the opposite service (26.8% vs 43.8%). Compared to IRB professionals, genetic researchers were more likely to have worked long term (>5 years) in their respective area (82.3% vs. 56.7%).
Table 1.
Demographics of the research participant group
| Total (n = 450) | Cases (n = 228) | Controls (n = 155) | Relatives (n = 67) | p Value | |
|---|---|---|---|---|---|
| Age (years), Mean (SD) | 63.6 (11.8) | 64.3 (11.4) | 64.0 (11.5) | 60.5 (13.6) | .08 |
| Women | 292 (64.9%) | 145 (63.6%) | 110 (71.0%) | 37 (55.2) | .07 |
| Race | .92 | ||||
| Asian/Pacific Islander | 7 (1.6%) | 4 (1.8%) | 2 (1.3%) | 1 (1.5%) | |
| Black | 4 (0.9%) | 2 (0.9%) | 2 (1.3%) | 0 | |
| Multi-Racial/Other | 16 (3.6%) | 8 (3.5%) | 4 (2.6%) | 4 (6.0%) | |
| White | 423 (94.0%) | 214 (93.9%) | 147 (94.8%) | 62 (92.5%) | |
| Education | .61 | ||||
| High School or less | 40 (8.9%) | 19 (8.3%) | 13 (8.4%) | 8 (11.9) | |
| Some College | 107 (23.8%) | 57 (25.0%) | 37 (23.9%) | 13 (19.4%) | |
| Bachelors Degree | 276 (61.3%) | 126 (55.3%) | 105 (67.7%) | 45 (67.2%) | |
| Unknown | 27 (6.0%) | 26 (11.4) | 0 | 1 (1.5%) | |
| Marital status | .10 | ||||
| Married/living together | 343 (76.2%) | 180 (78.9%) | 110 (71.0%) | 53 (79.1%) | |
| Single | 32 (7.1%) | 14 (6.1%) | 13 (8.4%) | 5 (7.5%) | |
| Divorced/separated | 44 (9.8%) | 19 (8.3%) | 20 (12.9%) | 5 (7.5%) | |
| Widowed | 22 (4.9%) | 6 (2.6%) | 12 (7.7%) | 4 (6.0%) | |
| Unknown | 9 (2.0%) | 9 (3.9%) | 0 | 0 |
Scenario 1: Research participant would be personally identified in a study involving de-identified data by someone other than the researchers: Compared to research participants and IRB professionals, genetic researchers were less likely to feel that research participants would be personally identified. IRB professionals were twice as likely and research participants 2.6 times as likely as researchers to feel that a participant would be personally identified in a study involving de-identified data (Fig. 1).
Fig 1.
Comparison between research participants, genetic researchers, and institutional review board (IRB) professionals regarding likelihood of harm or personal identification.
Scenario 2: Research participant would be harmed as a result of identification from de-identified data: Compared to research participants and IRB professionals, genetic researchers were significantly less likely to feel that research participants would be harmed from a study involving de-identified data (Fig. 1). Participants were almost five times more likely than researchers but half as likely as IRB professionals to feel they would be harmed.
Scenario 3: A federal agency or other law-enforcement agency might compel researchers to disclose information about genetic research participants: Genetic researchers were 25% as likely as IRB professionals and participants were 3.4 times as likely as researchers to feel that a federal agency might compel researchers to disclose information about research participants. No significant difference was seen between participants and IRB professionals with this scenario (Fig. 1).
Discussion
To our knowledge, this is the first study to quantitate differences in views between research participants, IRB professionals, and genetic researchers, regarding the likelihood of harm to the participant from the use of de-identified genomic data. This study showed that research participants and IRB professionals were more similar than the views of genetic researchers. Researchers were the least likely group to believe that research participants would be personally identified or harmed from a study involving de-identified data or feel that researchers might be forced to disclose information about research participants to a federal agency.
It is possible that differences in views between these three groups may be influenced by their unique roles in research. We have previously published in this group of participants that while almost half were concerned about being personally identified when participating in a genetic study using de-identified data, most felt that the benefits outweigh the potential risks [6, 15, 16] and others have shown that knowledge of the risks would not change participants’ attitude toward joining a study [17]. While others have shown that age is directly associated with a willingness to share personal data [18], we have previously reported in this group of participants a direct association between age and the importance of protecting privacy and information when deciding to allow data for a research repository [19]. Similar to research participants’ views, about half of the IRB professionals felt that re- identification or harm was likely. This finding is somewhat higher than a previous study of IRB Chairs, which found that 27% were concerned about the sensitivity of genetic information [20]. This was based, however, on a smaller sample size (41 vs 208) and the focus of these IRB Chairs was mental health-related. It has been suggested that the IRB professionals’ role of overseeing human subject protection and compliance may foster a greater perceived risk compared with the genetic researcher, who may be more likely to minimize the likelihood of harm, especially with increased experience in managing their own protocols to protect participants [8]. Also, it is possible that researchers better understand the logistical challenge of re-identification compared to IRB professionals [8]. While gender differences in the perception of harm or re-identification are unknown, it has been shown that male research participants are 1.7 times more likely to participate in a genetic substudy compared with women [21]. In this study, the research participant and IRB groups had more female participants than the researcher group; however, the coefficient for gender was not significant for any comparisons between research participants and researchers/IRB professionals so it is unlikely that these gender differences account for the contrast in likelihood of harm or identification.
There were limitations within this participant study population. Participants were recruited from a long-standing research population and selection bias may have influenced their attitudes about data sharing and de-identified data. In addition, this participant population was highly educated and mostly white, older adults, possibly limiting generalizability of these results. As discussed previously, it should be noted that the response rate among IRB professionals who received an invitation was low, about 7.5% [7, 9]. It is possible that many of the nonresponders were not eligible to participate in this study; however, data were not available to compare responders versus nonresponders. Likewise, the response rate for genetic researchers was approximately 8%, but it is possible that the denominator is overinflated because it is unknown how many researchers received the invitation but were not involved in genomic research or how many invitations were forwarded to colleagues. Finally, the GRRIP survey of the IRB professionals and genetic researchers was completed 4 years prior to completion of the PIP research participant survey, and it is possible that outside political or cultural changes may have impacted survey responses.
This study suggests that there are differences that need to be resolved between these three stakeholder groups regarding likelihood of identification or harm in the use the de-identified genetic data. Maximizing recruitment rates and minimizing dropout rates for future genomic observational research studies can only be achieved when researchers become more aware of participants’ views of perceived risk and harm and consider these concerns when constructing policy about sharing de- identified data. Reaching a consensus between stakeholders to establish best practices and inform policy decisions regarding the protection of research participants is critical for successful genomic observational research, and the use of de-identified shared data.
Acknowledgements
The authors wish to thank the individuals enrolled in the NWCGR for their ongoing participation in and contribution to cancer research. They also acknowledge and thank Lesley Pfeiffer, Anne Renz, Joan Scott, and David Kaufmann for their work contributing to the earlier stages of this project. This research was supported by NIH grant no. R01CA149051 to K.L. Edwards (PI), “Identification of Issues and Expectations of Subjects Participating in Genetic Studies of Cancer”.
Compliance with Ethical Standards
Conflict of Interest These authors do not have actual or potential conflicts of interest.
Primary Data The authors have full control of all primary data and agree to allow the journal to review these data, if requested. This study did not involve research animals.
Authors’ Contributions: DG participated in the analysis design, interpreted the data and was a major contributor in writing the manuscript. CJ and MS performed the data analyses and contributed to the writing of the manuscript. DB participated in the study design, analysis design, data interpretation, and writing of the manuscript. LW participate in data interpretation and manuscript preparation. KE participated in the study design, analysis design, data interpretation and writing of the manuscript. All authors read and approved the final
Ethical Approval The procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000. IRB approval was obtained from all participating institutions, and all research participants provided written, informed consent.
References
- 1. Vayena E, Gasser U. Between openness and privacy in genomics. PLoS Med. 2016; 13(1): e1001937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gymrek M, McGuire AL, Golan D, Halperin E, Erlich Y. Identifying personal genomes by surname inference. Science. 2013; 339(6117): 321–324. [DOI] [PubMed] [Google Scholar]
- 3. Sweeney L, Abu A, Winn J. 2013. Identifying Participants in the Personal Genome Project by Name (a Re-Identification Experiment). Rochester, NY: Social Science Research Network (SSRN). [Google Scholar]
- 4. U.S. Department of Health and Human Services. 45 CFR, Parts 160–164. Standards for privacy of individually identifiable health information, final rule. Federal Register. 2002; 67(157): 53182–53273. [PubMed] [Google Scholar]
- 5. Wang S, Jiang X, Singh S, Marmor R, et al. Genomic privacy: challenges, technical approaches to mitigate risk, and ethical considerations in the United States. Ann. N. Y. Acad. Sci. 2017; 1387(1): 73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goodman D, Johnson CO, Bowen D, Smith M, Wenzel L, Edwards K. De-identified genomic data sharing: the research participant perspective. J. Commun. Genet. 2017; 8(3): 173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lemke AA, Trinidad SB, Edwards KL, Starks H, Wiesner GL; GRRIP Consortium Attitudes toward genetic research review: results from a national survey of professionals involved in human subjects protection. J. Empir. Res. Hum. Res. Ethics. 2010; 5(1): 83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Edwards KL, Lemke AA, Trinidad SB et al. ; GRRIP Consortium Genetics researchers’ and IRB professionals’ attitudes toward genetic research review: a comparative analysis. Genet. Med. 2012; 14(2): 236–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Edwards KL, Lemke AA, Trinidad SB et al. . Attitudes toward genetic research review: results from a survey of human genetics researchers. Public Health Genomics. 2011; 14(6): 337–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Condit CM, Korngiebel DM, Pfeifer L et al. . What should be the character of the researcher-participant relationship? Views of participants in a long-standing cancer genetic registry. IRB. 2015; 37(4): 1–10. [PubMed] [Google Scholar]
- 11. Goodman D, Johnson CO, Wenzel L, Bowen D, Condit C, Edwards KL. Consent issues in genetic research: views of research participants. Public Health Genomics. 2016; 19(4): 220–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lemke AA, Smith ME, Wolf WA, Trinidad SB; GRRIP Consortium Broad data sharing in genetic research: views of institutional review board professionals. IRB. 2011; 33(3): 1–5. [PMC free article] [PubMed] [Google Scholar]
- 13. R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: 2013. ISBN 3-900051-07-0, URL http://www.R-project.org/. [Google Scholar]
- 14. Venables WN, Ripley BD.. Modern Applied Statistics With S. 4th ed Springer, NY; 2002. [Google Scholar]
- 15. Kaufman DJ, Murphy-Bollinger J, Scott J, Hudson KL. Public opinion about the importance of privacy in biobank research. Am J Hum Genet. 2009;85(5):643–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pullman D, Etchegary H, Gallagher K et al. . Personal privacy, public benefits, and biobanks: a conjoint analysis of policy priorities and public perceptions. Genet Med. 2012; 14(2): 229–235. [DOI] [PubMed] [Google Scholar]
- 17. McCarty CA, Garber A, Reeser JC, Fost NC; Personalized Medicine Research Project Community Advisory Group and Ethics and Security Advisory Board Study newsletters, community and ethics advisory boards, and focus group discussions provide ongoing feedback for a large biobank. Am J Med Genet A. 2011; 155A(4): 737–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Trinidad SB, Fullerton SM, Bares JM, Jarvik GP, Larson EB, Burke W. Genomic research and wide data sharing: views of prospective participants. Genet Med. 2010; 12(8): 486–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goodman D, Bowen D, Johnson CO, Smith M, Wenzel L, Edwards K. Factors that motivate participation in observational genetic cancer research studies. Submitted.
- 20. Wolf LE, Catania JA, Dolcini MM, Pollack LM, Lo B. IRB Chairs’ perspectives on genomics research involving stored biological materials: ethical concerns and proposed solutions. J. Empir. Res. Hum. Res. Ethics. 2008; 3(4): 99–111. [DOI] [PubMed] [Google Scholar]
- 21. Amiri L, Cassidy-Bushrow AE, Dakki H et al. . Patient characteristics and participation in a genetic study: a type 2 diabetes cohort. J. Investig. Med. 2014; 62(1): 26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]