Abstract
Noncoding RNAs (ncRNAs) can be divided into microRNAs (miRNAs), long noncoding RNAs (lncRNAs), circular RNAs (circRNAs), pRNAs, and tRNAs. Traditionally, miRNAs exert their biological function mainly through the inhibition of translation via the induction of target RNA transcript degradation. lncRNAs and circRNAs were once considered to have no potential to code proteins. Here, we will review the current knowledge on ncRNAs in relation to their origins, characteristics, and functions. We will also review how ncRNAs work as competitive endogenous RNA, gene transcription and expression regulators, and RNA-binding protein sponges in colorectal cancer (CRC). Notably, except for the abovementioned mechanisms, recent advances revealed that lncRNAs can also act as the precursor of miRNAs, and a small portion of lncRNAs and circRNAs was verified to have the potential to code proteins, providing new evidence for the significance of ncRNAs in CRC tumorigenesis and development.
Keywords: biomarker, competitive endogenous RNA, tumorigenesis, epithelial-to-mesenchymal transition, invasion, metastasis, chemoresistance
Introduction
Noncoding RNAs (ncRNAs) are major components of the human transcriptome.1 Recently, ncRNAs were demonstrated to play important roles in multiple biological processes by directly or indirectly interfering with gene expression in various cancers. The regulatory role of ncRNAs in multiple cancers has been summarized, such as lncRNAs and microRNAs (miRNAs) in endocrine-related cancers,2 lncRNAs in hepatocarcinogenesis,3 and circRNAs in multiple types of cancer.4 However, a comprehensive and in-depth analysis of ncRNAs in CRC has not been reported to date.
ncRNAs account for the majority of RNA transcribed by human genes, including miRNA, long noncoding RNA (lncRNA), circular RNA (circRNA), pRNA, and tRNA. With the development of RNA sequencing technologies and bioinformatics, numerous ncRNAs have been discovered that influence gene expression levels via chromatin modification, transcription, and posttranscriptional processing.5 Moreover, the abnormal expression of ncRNAs is associated with invasion, metastasis, chemoresistance, and radioresistance of colorectal cancer (CRC).6 For instance, TUG1 regulates the expression of growth-related genes, activates the expression of epithelial-to-mesenchymal transition (EMT)-associated genes, and plays important roles in signal transduction, cell morphology, migration, proliferation, and apoptosis in CRC. Overexpression of TUG1 is thought to be an independent poor prognostic factor for CRC patients.7,8 The circRNA circ_001569 is upregulated in CRC tissues and promotes CRC proliferation and invasion. This circRNA acts as a sponge to directly inhibit miR-145 transcription, which subsequently affects the functions of miR-145 targets E2F5, BAG4, and FMNL2 in CRC cells.9
Herein, we performed a systematic literature review analysis of ncRNAs in CRC and the dysregulation of ncRNAs in CRC tissues or cells. Then, we discussed how these ncRNAs work as miRNA sponges, gene transcription and expression regulators, and RNA-binding protein (RBP) sponges in CRC, providing evidence for the significance of ncRNAs in CRC.
Classification of ncRNAs
Generally, according to their product size, ncRNAs can be divided into two groups: small ncRNAs and long ncRNAs.10 The size of small ncRNAs, such as miRNA, is typically less than 200 nucleotides (nt).11,12 By contrast, long ncRNAs are typically greater than 200 nt, including long intergenic ncRNAs, long intronic ncRNAs, and pseudogene RNAs.13 Except for these linear ncRNAs, circRNAs, which are formed through the ligation of the 5′ and 3′ ends of linear RNA, have been investigated recently.14,15 Accumulating evidence has revealed that aberrant ncRNA expression is correlated with various cancers, especially CRC.
Mechanisms of ncRNAs in regulating CRC progression
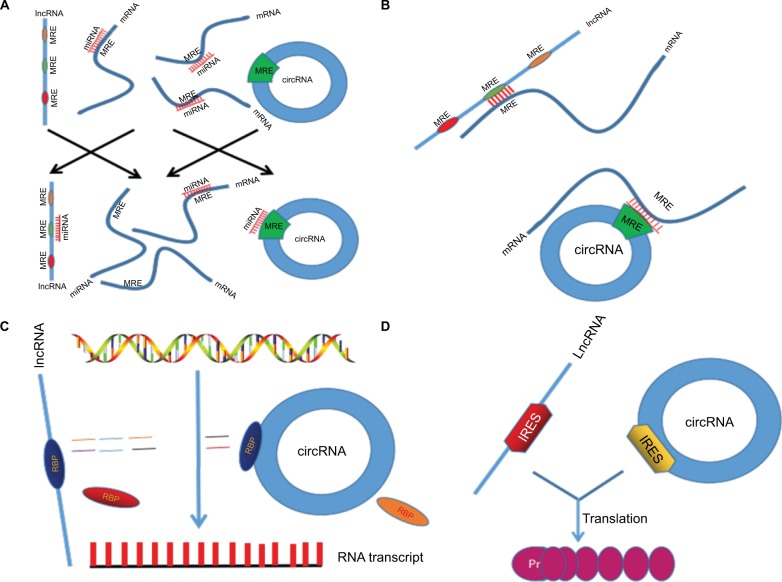
ncRNAs control individual genes and gene expression programs through changing the fundamental transcriptional mechanism or via epigenetic regulation at multiple levels, such as transcription, translation, and protein function (Figure 1).
Figure 1.
Mechanism of lncRNAs/circRNAs regulating CRC biological activities. (A) lncRNAs and circRNAs act as miRNA sponge or ceRNA. (B) Directly targeting mRNA by partial base pairing. (C) Binding RBP to regulate protein expression. (D) A small portion of lncRNAs/circRNAs can be translated to proteins.
Abbreviations: lncRNAs, long noncoding RNAs; circRNAs, circular RNAs; CRC, colorectal cancer; miRNA, microRNA; ceRNA, competitive endogenous RNA; IRES, internal ribosome entry site; RBP, RNA-binding protein.
As a competitive endogenous RNA (ceRNA) or miRNA sponge
miRNA is a small ncRNA that is 19–24 nt in length. miRNA binds to miRNA response elements (MREs) in RNA sequences and negatively regulates gene expression through the inhibition of translation via the induction of target RNA transcript degradation.16,17 ceRNA contains an MRE, which competitively binds to miRNA. Therefore, ceRNA affects the regulatory functions of miRNAs in gene expression and reduces the inhibitory effect of miRNAs on target molecules18–21 (Figure 1A). For instance, UCC may act as an endogenous sponge by competing for miR-143, thereby regulating the targets of this miRNA. UCC and miR-143 may be promising molecular targets for CRC therapy.22 The lncRNA CRNDE regulates the progression and chemoresistance of CRC via modulating the expression levels of miR-181a-5p and the activity of Wnt/β-catenin signaling.23 HOXA11-AS promotes liver metastasis in CRC by functioning as a miR-125a-5p sponge, and the novel HOXA11-AS-miR-125a-5p-PADI2 regulatory network is involved in CRC liver metastasis.24 At present, the main function of some lncRNAs and circRNAs involves acting as a miRNA sponge.
Regulating gene transcription
In addition to the abovementioned mechanisms, ncRNAs, as the product of transcription, are major regulators of the transcriptional process.25 Researchers reported that ncRNAs can function as positive regulators of their parental gene transcription, targeting mRNA by partially base pairing26 (Figure 1B). Accumulating evidence indicates that ncRNAs play a pivotal role in posttranscriptional and gene expression regulation. For example, the circRNAs circ-EIF3J and circ-PAIP2 combine with the U1 snRNP to further interact with RNA Pol II and enhance the expression of their parental genes in HeLa and HEK293 cells.27
Regulating RBPs
miRNA biogenesis is modulated by a variety of RBPs28 (Figure 1C), and the adsorption of protein factors by linear lncRNA has been reported.29–31 For example, sno-lncRNAs regulate alternative splicing of downstream genes by adsorbing the alternative splicing factor Fox2.32 NEAT1 also has a profound effect on global pri-miRNA processing. Mechanistic dissection reveals that NEAT1 broadly interacts with the NONO-PSF heterodimer and numerous other RBPs. In addition, multiple RNA segments in NEAT1, including a pseudo pri-miRNA near its 3′ end, help attract the microprocessor.33
Protein translation
A small portion of ncRNAs can also be translated to proteins similar to mRNAs. circRNA is efficiently translated in living human cells to produce abundant protein product via the RCA mechanism.34 Long-repeating polypeptide chains were synthesized from RNA circles with continuous open reading frames, and an internal ribosome entry site and the initiation codon ATG in a specific circRNA allow the circRNA translation template to function as mRNA (Figure 1D).35
Biological functions of miRNAs in CRC
miRNAs are 19–24 nt in length that regulate gene expression at the posttranscriptional level by binding to the 3′-untranslated regions or the open reading frames of target genes, leading to the degradation of target mRNAs or repression of mRNA translation.36 Numerous investigations have been performed to analyze the biological functions of miRNAs in CRC.37,38
miRNAs promote CRC cell proliferation and invasion
miRNAs exhibit a close relationship with the initiation and development of various human malignancies.39 Abundant miRNAs play oncogenic roles in CRC tumorigenesis via diverse mechanisms.40,41 On one hand, miRNAs exert their tumor oncogenic functions primarily by binding to the 3′-untranslated region of the mRNA of target genes. For instance, miR19b-3p is overexpressed in CRC tissues compared with normal tissues. Furthermore, miR19b-3p plays an oncogenic role in CRC via directly targeting ITGB8.42 Similarly, TIA1 is an important tumor suppressor in CRC. In addition, miR-19a is highly expressed in tumor tissues compared with normal tissues and exerts its oncomiR function by targeting TIA1.43 On the other hand, miRNAs promote CRC cell proliferation and invasion via other multiple mechanisms. For example, the EMT plays important roles in tumor metastasis. For example, miR-19a is upregulated in CRC tissues, and high expression of miR-19a is signifi-cantly associated with lymph node metastasis. Interestingly, miR-19a is upregulated by TNF-α and is required for TNF-α-induced EMT and metastasis in CRC cells.44
miRNAs suppress CRC cell proliferation and invasion
By contrast, miRNA may function as a tumor-repressive gene to inhibit cell proliferation in CRC. Some miRNAs suppress CRC cell proliferation and invasion by regulating tumor angiogenesis, tumor metabolism, and cancer stemness features.45,46 For instance, miR-590-5p overexpression inhibits CRC angiogenesis and metastasis by targeting nuclear factor 90, which acts as a positive regulator of vascular endothelial growth factor mRNA stability and protein synthesis. Notably, knockdown of miR-590-5p promotes the progression of CRC in vitro.47 AEG-1 is a key oncogenic factor in various tumors.48 In addition, miR-217 suppresses CRC cell proliferation and invasiveness by inhibiting AEG-1 expression with modulation of MMP2 or AMPK signaling.49,50
Biological functions of lncRNAs in CRC
lncRNA is defined as an RNA transcript of greater than 200 nt located in nuclear or cytosolic fractions.51 The overexpression, deficiency, or mutation of lncRNAs could be involved in CRC progression via a variety of mechanisms.52,53 Abundantly expressed lncRNAs play crucial roles in CRC, such as HOTAIR, CCAT1, and MALAT1 (Table 1).
Table 1.
lncRNAs reported in CRC
| lncRNA | Expression | Biofunctions in tumor | Potential mechanisms | PMID |
|---|---|---|---|---|
| H19 | Up | Oncogene | Modifies the EMT pathway; acts as a ceRNA for miR-138 and miR-200a; mediates methotrexate resistance by activating the WNT/β-catenin pathway | 26989025; 26068968; 19926638 |
| HOTAIR | Up | Oncogene | Acts as a miR-197 sponge; suppresses miR-218; activates NF-κB/TS signaling; modifies the EMT pathway | 29137688; 28918035; 27069543 |
| CCAT1 | Up | Oncogene | Activated by the super-enhancer cMYC | 25185650; 24662484 |
| MALAT1 | Up | Oncogene | Increases AKAP-9 expression by promoting SRPK1-catalyzed SRSF1 phosphorylation; regulating PRKA kinase anchor protein 9 | 26887056; 25446987 |
| XIST | Upregulated in 5FU-resistant patients | Promote 5FU resistance | Promotes the expression of thymidylate synthase | 29137332 |
| HEIH | Up | Oncogene | Counteracting miR-939-mediated transcriptional repression of Bcl-xL | 29081216 |
| lncRNA-422 | Down | Tumor suppressor | By PI3K/AKT/mTOR pathway in CRC | 29050940 |
| MIR100HG | Overexpressed in cetuximab-resistant patients | Cetuximab resistance | Host gene of cetuximab resistance, which mediates cetuximab resistance via Wnt/β-catenin signaling | 29035371 |
| CPS1-IT1 | Down | Tumor suppressor | Suppresses metastasis and EMT by inhibiting hypoxia-induced autophagy through inactivation of HIF-1α | 29017924 |
| NONHSAT062994 | Down | Tumor suppressor | By inactivating Akt signaling | 28978149 |
Abbreviations: lncRNAs, long noncoding RNAs; CRC, colorectal cancer; EMT, epithelial-to-mesenchymal transition; ceRNA, competitive endogenous RNA.
lncRNAs interact with miRNAs in CRC
The ceRNA hypothesis demonstrated that lncRNAs with shared miRNA binding sites compete for posttranscriptional control.54 In early studies, it was found that PTENP1 shares conserved miRNA seed target sites with PTEN for the miR-17, miR-21, miR-214, miR-19, and miR-26 miRNA families. The role of lncRNAs as a miRNA sponge is the main mechanism of lncRNAs in cancer. For example, H19 is overexpressed in colon cancer tissues and cell lines, whereas miR-138 expression in tumor tissues is reduced compared with normal tissues. Interestingly, the silencing of H19 strongly increases the expression of miR-138 and suppresses the expression of high-mobility group A (HMGA1) protein, indicating that H19 promotes the migration and invasion of colon cancer by sponging miR-138 to upregulate HMGA1 expression.55
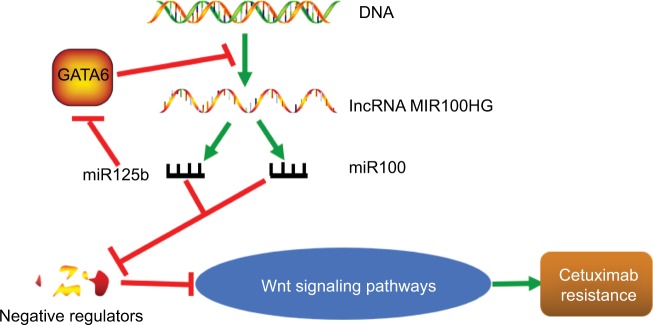
Notably, lncRNAs act as the precursor of miRNAs. For example, the lncRNA MIR100HG is the precursor of miR-100 and miR-125b, which coordinately represses five Wnt/β-catenin negative regulators, leading to cetuximab resistance. Furthermore, the transcription factor GATA6 represses MIR100HG. By contrast, the repression is relieved by miR-125b targeting of GATA6, revealing a double-negative feedback loop between MIR100HG and the transcription factor GATA6 (Figure 2). In addition, miR-17-5p expression is increased in tumors compared with paired non-tumorous tissues, and high miR-17-5p expression is significantly associated with TNM staging and lymph node metastasis. Furthermore, miR-17-5p expression is upregu-lated by CCAT2 through TCF7 L2-mediated transcriptional regulation, enhancing WNT activity.56
Figure 2.
lncRNAs function as precursors of miRNAs in CRC.
Note: Red lines represent inhibition effect; green arrows indicate promotion effect.
Abbreviations: lncRNAs, long noncoding RNAs; miRNA, microRNA; CRC, colorectal cancer.
lncRNAs function through other mechanisms in CRC
Tumor metabolism is responsible for rapid recurrence and poor survival of CRC. The relationship between ncRNAs and tumor metabolism was investigated recently. For instance, lncRNA NONHSAT062994 expression was negatively correlated with the Akt downstream targets c-Myc and cyclin D1 in clinical CRC samples, indicating that NONHSAT062994 functioned as a tumor suppressor to inhibit CRC cell growth by inactivating Akt signaling.57 Autophagy is a crucial intracellular process associated with CRC tumorigenesis and progression. lncRNAs play crucial roles in CRC by regulating tumor autophagy by targeting various proteins, such as CPS1-IT1, AC023115.3, and MALAT1.58,59 For example, the lncRNA CPS1-IT suppresses metastasis and EMT by inhibiting hypoxia-induced autophagy through inactivation of HIF-1α in CRC.60
Biological functions of circRNAs in CRC
circRNAs are a special type of endogenous ncRNA molecule. We have witnessed an explosion in published studies on all aspects of circRNA biology leading to the common understanding that these molecules are important players in cancers, especially in CRC. As previously mentioned, circRNAs differ structurally from other lncRNAs given that their 3′ and 5′ ends are covalently joined. Certain circRNAs act as highly stable sponges for specific miRNAs, such as CiRS-7 and SRY for miR-7 and miR-138, respectively, and are involved in competing endogenous RNA networks.61 Thousands of circRNAs have been identified and annotated in the circRNA repository (circBase). However, the biogenesis of circRNAs remains elusive.62
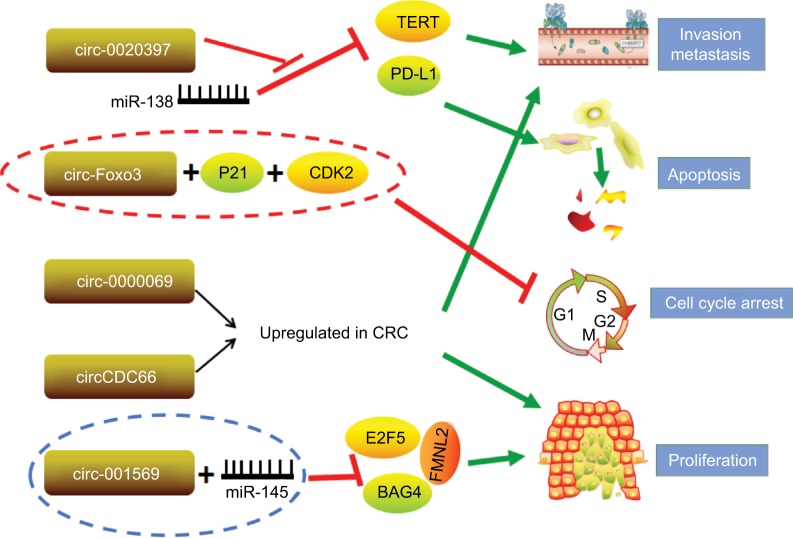
miRNAs are extremely well studied as tumor suppressors or oncogenes. It has been hypothesized that circRNAs form a class of posttranscriptional regulators, acting as epigenetic, highly stable miRNA sponges to compete with the endogenous RNA network and directly affecting the expression of any related gene.63 circRNAs sequester specific miRNA complexes and release them after cleavage.64 Circ_001569 was upregulated in CRC tissues compared with adjacent normal tissues, and high circ_001569 expression was closely correlated with differentiation and TNM classification. Interestingly, circ_001569 is negatively correlated with miR-145, and miR-145 is negatively correlated with E2F5, BAG4, and FMNL2 expression. These data indicate that circ_001569 promotes CRC cell proliferation and invasion by regulating miR-145 and its targets E2F5, BAG4, and FMNL2.65 In addition, circ_0020397 promotes CRC cell viability and invasion and inhibits their apoptosis by promoting the expression of the miR-138 target genes TERT and PD-L1.66 circCCDC66 possesses an oncogenic capacity through protecting multiple oncogenes from being attacked by a group of miRNAs, and overexpression of circCCDC66 potentiates multiple tumor characteristics, including proliferation, migration, and invasion (Figure 3).67
Figure 3.
circRNAs investigated in CRC.
Note: Red lines represent inhibition effect; green arrows indicate promotion effect.
Abbreviations: circRNAs, circular RNAs; CRC, colorectal cancer.
In addition to the abovementioned circRNAs, numerous other circRNAs govern fundamental biological process and cancer progression through multiple mechanisms instead of acting as a special class of endogenous RNAs. circ-Foxo3 functions as a tumor suppressor in CRC by forming circ-Foxo3-p21-CDK2 ternary complexes.68 CDK2 interacts with cyclin A and cyclin E to facilitate cell cycle entry, whereas p21 inhibits these interactions and arrests cell cycle progression.69
Therefore, circRNAs can also exert biological function in CRC by binding to partners.
ncRNAs as new potential tumor biomarkers and targets
An increasing number of ncRNAs are being reported to be aberrantly expressed in CRC, and differential expression of ncRNAs can be detected in the circulation of CRC patients. Importantly, ncRNAs are abundant and their expression is both step and location specific. The characteristics of ncRNAs afford them the potential to become novel diagnostic and prognostic markers for cancer. For instance, lncRNA SPRY4-IT1 is upregulated in CRC tissues and promotes proliferation and invasion by targeting miR-101-3p. These data indicate that knockdown of SPRY4-IT1 represents a rational therapeutic strategy for colorectal carcinoma.70 CircHIPK3 was significantly highly expressed in CRC tissues and positively correlated with metastasis and TNM stage, revealing that circHIPK3 may be a potential prognostic biomarker in CRC.71 On the basis of the results acquired from latest papers, circulating ncRNA profiles are emerging as potential biomarkers of diagnosis, prognosis, and therapeutic response in CRC.
Conclusion
With the development of high-throughput sequencing technologies and bioinformatics methods, ncRNAs are increasingly investigated.72 Traditionally, ncRNA has been viewed as an unstable molecule because ribonucleases are ubiquitous and extremely stable biomarkers for CRC. Emerging evidence have revealed the biological roles and relevant mechanisms of diverse ncRNAs in CRC tumorigenesis.73,74 For example, miR-17 acts as an oncogenic miRNA that promotes CRC development by activating the Wnt/β-catenin pathway and by targeting P130.75 We also found that miR-149 methylation contributes to CRC growth and invasion by targeting the transcription factor Sp1.76
Notably, the molecular mechanisms of these dysregulated ncRNAs have been investigated recent years. EMT is an important step in cancer development that involves the cooperation of a variety of signaling pathways, including the transformation growth factor-β, Sonic Hedgehog, and WNT pathways.77,78 HOTAIR, H19, AFAP1-AS1, TUG1, BANCR, lncRNA-ATB, SPRY4-IT1, SLC25A25-AS1, LINC01133, PANDAR, lncRNA-ATB, and Sox2ot are involved in the EMT pathways.
circRNAs as a new class of ncRNAs contributing to the regulatory network governing protein coding gene expression by acting as miRNA target decoys, RBP sponges and transcriptional regulators exhibit great biological potential for circRNA-related functionalities. Uncovering these additional functions (if any) and understanding these functionalities represent key research topics in the circRNA field in the future.
Via a systematic literature review, we have discussed the origins, characteristics, and main functions of ncRNAs in CRC. Continuous exploration and research in this field will provide an important molecular basis for understanding the complex regulation of CRC.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (81560385); the Medical Scientific and Technological Research Project of Henan Province (201702027); Youth Innovation Fund Project of The First Affiliated Hospital of Zhengzhou University (YNQN2017035); and the China Postdoctoral Science Foundation (2017M610462). Shuaixi Yang, Zhenqiang Sun, and Quanbo Zhou are co-first authors.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Luo J, Qu J, Wu DK, Lu ZL, Sun YS, Qu Q. Long non-coding RNAs: a rising biotarget in colorectal cancer. Oncotarget. 2017;8(13):22187–22202. doi: 10.18632/oncotarget.14728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klinge CM. Non-coding RNAs: long non-coding RNAs and microRNAs in endocrine-related cancers. Endocr Relat Cancer. 2018;25(4):R259–R282. doi: 10.1530/ERC-17-0548. [DOI] [PubMed] [Google Scholar]
- 3.Lanzafame M, Bianco G, Terracciano LM, Ng CKY, Piscuoglio S. The role of long non-coding RNAs in hepatocarcinogenesis. Int J Mol Sci. 2018;19(3) doi: 10.3390/ijms19030682. pii: E682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ji Q, Zhang C, Sun X, Li Q. Circular RNAs function as competing endogenous RNAs in multiple types of cancer. Oncol Lett. 2018;15(1):23–30. doi: 10.3892/ol.2017.7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7835):339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meng S, Zhou H, Feng Z, et al. circRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94. doi: 10.1186/s12943-017-0663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L, Zhao Z, Feng W, et al. Long non-coding RNA TUG1 promotes colorectal cancer metastasis via EMT pathway. Oncotarget. 2016;7(32):51713–51719. doi: 10.18632/oncotarget.10563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhai HY, Sui MH, Yu X, et al. Overexpression of long non-coding RNA TUG1 promotes colon cancer progression. Med Sci Monit. 2016;22:3281–3287. doi: 10.12659/MSM.897072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie H, Ren X, Xin S, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7(18):26680–26691. doi: 10.18632/oncotarget.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei Z, Batagov AO, Schinelli S, et al. Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat Commun. 2017;8(1):1145. doi: 10.1038/s41467-017-01196-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong Q, Su G. Roles of miRNAs and long noncoding RNAs in the progression of diabetic retinopathy. Biosci Rep. 2017;37(6):BSR20171157. doi: 10.1042/BSR20171157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Long M, Zhan M, Xu S, et al. miR-92b-3p acts as a tumor suppressor by targeting Gabra3 in pancreatic cancer. Mol Cancer. 2017;16(1):167. doi: 10.1186/s12943-017-0723-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu X, Sood AK, Dang CV, Zhang L. The role of long noncoding RNAs in cancer: the dark matter matters. Curr Opin Genet Dev. 2018;48:8–15. doi: 10.1016/j.gde.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia W, Qiu M, Chen R, et al. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep. 2016;6:35576. doi: 10.1038/srep35576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu W, Ma W, Yuan Y, Zhang Y, Sun S. Circular RNA hsa_cir-cRNA_103809 promotes lung cancer progression via facilitating ZNF121-dependent MYC expression by sequestering miR-4302. Biochem Biophys Res Commun. 2018;500(4):846–851. doi: 10.1016/j.bbrc.2018.04.172. [DOI] [PubMed] [Google Scholar]
- 16.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 17.Ou C, Sun Z, Li X, et al. MiR-590-5p, a density-sensitive microRNA, inhibits tumorigenesis by targeting YAP1 in colorectal cancer. Cancer Lett. 2017;399:53–63. doi: 10.1016/j.canlet.2017.04.011. [DOI] [PubMed] [Google Scholar]
- 18.Li D, Yang M, Liao A, Zeng B, et al. Linc00483 as ceRNA regulates proliferation and apoptosis through activating MAPKs in gastric cancer. J Cell Mol Med. 2018 doi: 10.1111/jcmm.13661. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu WH, Tsai ZT, Tsai HK. Comparative genomic analyses highlight the contribution of pseudogenized protein-coding genes to human lincRNAs. BMC Genomics. 2017;18(1):786. doi: 10.1186/s12864-017-4156-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Q, Jiang S, Song A, et al. HOXD-AS1 functions as an oncogenic ceRNA to promote NSCLC cell progression by sequestering miR-147a. Onco Targets Ther. 2017;10:4753–4763. doi: 10.2147/OTT.S143787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu T, Chi H, Chen J, et al. Curcumin suppresses proliferation and in vitro invasion of human prostate cancer stem cells by ceRNA effect of miR-145 and lncRNA-ROR. Gene. 2017;631:29–38. doi: 10.1016/j.gene.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 22.Huang FT, Chen WY, Gu ZQ, et al. The novel long intergenic noncoding RNA UCC promotes colorectal cancer progression by sponging miR-143. Cell Death Dis. 2017;8(5):e2778. doi: 10.1038/cddis.2017.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han P, Li JW, Zhang BM, et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol Cancer. 2017;16(1):9. doi: 10.1186/s12943-017-0583-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen D, Sun Q, Zhang L, et al. The lncRNA HOXA11-AS functions as a competing endogenous RNA to regulate PADI2 expression by sponging miR-125a-5p in liver metastasis of colorectal cancer. Oncotarget. 2017;8(41):70642–70652. doi: 10.18632/oncotarget.19956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Long Y, Wang X, Youmans DT, Cech TR. How do lncRNAs regulate transcription? Sci Adv. 2017;3(9):eaao2110. doi: 10.1126/sciadv.aao2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 27.Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 28.G Hendrickson D, Kelley DR, Tenen D, Bernstein B, Rinn JL. Widespread RNA binding by chromatin-associated proteins. Genome Biol. 2016;17:28. doi: 10.1186/s13059-016-0878-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Schwartz JC, Cech TR. Nucleic acid-binding specificity of human FUS protein. Nucleic Acids Res. 2015;43(15):7535–7543. doi: 10.1093/nar/gkv679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davidovich C, Zheng L, Goodrich KJ, Cech TR. Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol. 2013;20(11):1250–1257. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hudson WH, Ortlund EA. The structure, function and evolution of proteins that bind DNA and RNA. Nat Rev Mol Cell Biol. 2014;15(11):749–760. doi: 10.1038/nrm3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin QF, Yang L, Zhang Y, et al. Long noncoding RNAs with snoRNA ends. Mol Cell. 2012;48(2):219–230. doi: 10.1016/j.molcel.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 33.Jiang L, Shao C, Wu QJ, et al. NEAT1 scaffolds RNA-binding proteins and the microprocessor to globally enhance pri-miRNA processing. Nat Struct Mol Biol. 2017;24(10):816–824. doi: 10.1038/nsmb.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abe N, Matsumoto K, Nishihara M, et al. Rolling circle translation of circular RNA in living human cells. Sci Rep. 2015;5:16435. doi: 10.1038/srep16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268(5209):415–417. doi: 10.1126/science.7536344. [DOI] [PubMed] [Google Scholar]
- 36.Xi XP, Zhuang J, Teng MJ, et al. MicroRNA-17 induces epithelial-mesenchymal transition consistent with the cancer stem cell phenotype by regulating CYP7B1 expression in colon cancer. Int J Mol Med. 2016;38(2):499–506. doi: 10.3892/ijmm.2016.2624. [DOI] [PubMed] [Google Scholar]
- 37.Ba S, Xuan Y, Long ZW, Chen HY, Zheng SS. MicroRNA-27a promotes the proliferation and invasiveness of colon cancer cells by targeting SFRP1 through the Wnt/β-catenin signaling pathway. Cell Physiol Biochem. 2017;42(5):1920–1933. doi: 10.1159/000479610. [DOI] [PubMed] [Google Scholar]
- 38.Basati G, Razavi AE, Pakzad I, Malayeri FA. Circulating levels of the miRNAs, miR-194, and miR-29b, as clinically useful biomarkers for colorectal cancer. Tumour Biol. 2016;37(2):1781–1788. doi: 10.1007/s13277-015-3967-0. [DOI] [PubMed] [Google Scholar]
- 39.Svoronos AA, Engelman DM, Slack FJ. OncomiR or tumor suppressor? The duplicity of microRNAs in cancer. Cancer Res. 2016;76(13):3666–3670. doi: 10.1158/0008-5472.CAN-16-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu S, Qu D, Li W, et al. MiR-647 and miR-1914 promote cancer progression equivalently by downregulating nuclear factor IX in colorectal cancer. Mol Med Rep. 2017;16(6):8189–8199. doi: 10.3892/mmr.2017.7675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang L, Feng G, Zhang X, Ding Y, Wang X. MicroRNA-630 promotes cell proliferation and inhibits apoptosis in the HCT116 human colorectal cancer cell line. Mol Med Rep. 2017;16(4):4843–4848. doi: 10.3892/mmr.2017.7159. [DOI] [PubMed] [Google Scholar]
- 42.Huang L, Cai JL, Huang PZ, et al. MiR19b-3p promotes the growth and metastasis of colorectal cancer via directly targeting ITGB8. Am J Cancer Res. 2017;7(10):1996–2008. eCollection 2017. [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Y, Liu R, Yang F, et al. miR-19a promotes colorectal cancer proliferation and migration by targeting TIA1. Mol Cancer. 2017;16(1):53. doi: 10.1186/s12943-017-0625-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang L, Wang X, Wen C, et al. Hsa-miR-19a is associated with lymph metastasis and mediates the TNF-α induced epithelial-to-mesenchymal transition in colorectal cancer. Sci Rep. 2015;5:13350. doi: 10.1038/srep13350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang R, Liu R, Liu C, et al. A novel role for MiR-520a-3p in regulating EGFR expression in colorectal cancer. Cell Physiol Biochem. 2017;42(4):1559–1574. doi: 10.1159/000479397. [DOI] [PubMed] [Google Scholar]
- 46.Zhai H, Song B, Xu X, Zhu W, Ju J. Inhibition of autophagy and tumor growth in colon cancer by miR-502. Oncogene. 2013;32(12):1570–1579. doi: 10.1038/onc.2012.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou Q, Zhu Y, Wei X, et al. MiR-590-5p inhibits colorectal cancer angiogenesis and metastasis by regulating nuclear factor 90/vascular endothelial growth factor A axis. Cell Death Dis. 2016;7(10):e2413. doi: 10.1038/cddis.2016.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu X, Wang D, Liu H, et al. Knockdown of astrocyte elevated gene-1 (AEG-1) in cervical cancer cells decreases their invasiveness, epithelial to mesenchymal transition, and chemoresistance. Cell Cycle. 2014;13(11):1702–1707. doi: 10.4161/cc.28607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song H, Tian Z, Qin Y, Yao G, Fu S, Geng J. Astrocyte elevated gene-1 activates MMP9 to increase invasiveness of colorectal cancer. Tumour Biol. 2014;35(7):6679–6685. doi: 10.1007/s13277-014-1883-3. [DOI] [PubMed] [Google Scholar]
- 50.Song HT, Qin Y, Yao GD, Tian ZN, Fu SB, Geng JS. Astrocyte elevated gene-1 mediates glycolysis and tumorigenesis in colorectal carcinoma cells via AMPK signaling. Mediators Inflamm. 2014;2014:287381. doi: 10.1155/2014/287381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han D, Wang M, Ma N, Xu Y, Jiang Y, Gao X. Long noncoding RNAs: novel players in colorectal cancer. Cancer Lett. 2015;361(1):13–21. doi: 10.1016/j.canlet.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 52.Chen N, Guo D, Xu Q, et al. Long non-coding RNA FEZF1-AS1 facilitates cell proliferation and migration in colorectal carcinoma. Oncotarget. 2016;7(10):11271–11283. doi: 10.18632/oncotarget.7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Z, Zhou C, Chang Y, et al. Long non-coding RNA CASC11 interacts with hnRNP-K and activates the WNT/β-catenin pathway to promote growth and metastasis in colorectal cancer. Cancer Lett. 2016;376(1):62–73. doi: 10.1016/j.canlet.2016.03.022. [DOI] [PubMed] [Google Scholar]
- 54.Li Y, Lv M, Song Z, Lou Z, Wang R, Zhuang M. Long non-coding RNA NNT-AS1 affects progression of breast cancer through miR-142-3p/ZEB1 axis. Biomed Pharmacother. 2018;103:939–946. doi: 10.1016/j.biopha.2018.04.087. [DOI] [PubMed] [Google Scholar]
- 55.Yang Q, Wang X, Tang C, Chen X, He J. H19 promotes the migration and invasion of colon cancer by sponging miR-138 to upregulate the expression of HMGA1. Int J Oncol. 2017;50(5):1801–1809. doi: 10.3892/ijo.2017.3941. [DOI] [PubMed] [Google Scholar]
- 56.Petrov N, Zhidkova O, Serikov V, Zenin V, Popov B. Induction of Wnt/β-catenin signaling in mouse mesenchymal stem cells is associated with activation of the p130 and E2f4 and formation of the p130/Gsk3β/β-catenin complex. Stem Cells Dev. 2012;21(4):589–597. doi: 10.1089/scd.2011.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He XS, Guo LC, Du MZ, et al. The long non-coding RNA NON-HSAT062994 inhibits colorectal cancer by inactivating Akt signaling. Oncotarget. 2017;8(40):68696–68706. doi: 10.18632/oncotarget.19827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma B, Yuan Z, Zhang L, et al. Long non-coding RNA AC023115.3 suppresses chemoresistance of glioblastoma by reducing autophagy. Biochim Biophys Acta. 2017;1864(8):1393–1404. doi: 10.1016/j.bbamcr.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 59.Yuan P, Cao W, Zang Q, Li G, Guo X, Fan J. The HIF-2α-MALAT1-miR-216b axis regulates multi-drug resistance of hepatocellular carcinoma cells via modulating autophagy. Biochem Biophys Res Commun. 2016;478(3):1067–1073. doi: 10.1016/j.bbrc.2016.08.065. [DOI] [PubMed] [Google Scholar]
- 60.Zhang W, Yuan W, Song J, Wang S, Gu X. lncRNA CPS1-IT1 suppresses EMT and metastasis of colorectal cancer by inhibiting hypoxia-induced autophagy through inactivation of HIF-1α. Biochimie. 2018;144:21–27. doi: 10.1016/j.biochi.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 61.Zhao ZJ, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 2017;14(5):514–521. doi: 10.1080/15476286.2015.1122162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20(11):1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bachmayr-Heyda A, Reiner AT, Auer K, et al. Correlation of circular RNA abundance with proliferation – exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057. doi: 10.1038/srep08057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 65.Cristóbal I, Sanz-Alvarez M, Torrejón B, et al. Potential therapeutic impact of miR-145 deregulation in colorectal cancer. Mol Ther. 2018:30211–9. doi: 10.1016/j.ymthe.2018.05.008. S1525-0016(18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang XL, Xu LL, Wang F. Hsa_circ_0020397 regulates colorectal cancer cell viability, apoptosis and invasion by promoting the expression of the miR-138 targets TERT and PD-L1. Cell Biol Int. 2017;41(9):1056–1064. doi: 10.1002/cbin.10826. [DOI] [PubMed] [Google Scholar]
- 67.Hsiao KY, Lin YC, Gupta SK, et al. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77(9):2339–2350. doi: 10.1158/0008-5472.CAN-16-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Beale G, Haagensen EJ, Thomas HD, et al. Combined PI3K and CDK2 inhibition induces cell death and enhances in vivo antitumour activity in colorectal cancer. Br J Cancer. 2016;115(6):682–690. doi: 10.1038/bjc.2016.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jin J, Chu Z, Ma P, Meng Y, Yang Y. Long non-coding RNA SPRY4-IT1 promotes proliferation and invasion by acting as a ceRNA of miR-101-3p in colorectal cancer cells. Tumour Biol. 2017;39(7) doi: 10.1177/1010428317716250. 1010428317716250. [DOI] [PubMed] [Google Scholar]
- 71.Zeng K, Chen X, Xu M, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9(4):417. doi: 10.1038/s41419-018-0454-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 72.Weng M, Wu D, Yang C, et al. Noncoding RNAs in the development, diagnosis, and prognosis of colorectal cancer. Transl Res. 2017;181:108–120. doi: 10.1016/j.trsl.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 73.Fang Z, Tang J, Bai Y, et al. Plasma levels of microRNA-24, microRNA-320a, and microRNA-423-5p are potential biomarkers for colorectal carcinoma. J Exp Clin Cancer Res. 2015;34:86. doi: 10.1186/s13046-015-0198-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sun Z, Ou C, Ren W, Xie X, Li X, Li G. Downregulation of long non-coding RNA ANRIL suppresses lymphangiogenesis and lymphatic metastasis in colorectal cancer. Oncotarget. 2016;7(30):47536–47555. doi: 10.18632/oncotarget.9868. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 75.Ma Y, Zhang P, Wang F, et al. Elevated oncofoetal miR-17-5p expression regulates colorectal cancer progression by repressing its target gene P130. Nat Commun. 2012;3:1291. doi: 10.1038/ncomms2276. [DOI] [PubMed] [Google Scholar]
- 76.Wang F, Ma YL, Zhang P, et al. SP1 mediates the link between methylation of the tumour suppressor miR-149 and outcome in colorectal cancer. J Pathol. 2013;229(1):12–24. doi: 10.1002/path.4078. [DOI] [PubMed] [Google Scholar]
- 77.Zhang J, Tian XJ, Xing J. Signal transduction pathways of EMT induced by TGF-β, SHH, and WNT and their crosstalks. J Clin Med. 2016;5(4) doi: 10.3390/jcm5040041. pii: E41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]