Abstract
Introduction
Prenylated Rab acceptor 1 domain family member 2 (PRAF2), a novel oncogene, has been shown to be essential for the development of several human cancers; however, its role in hepatocellular carcinoma (HCC) remains unclear.
Materials and methods
PRAF2 mRNA and protein expressions were examined in fresh tissues by quantitative reverse transcription-polymerase chain reaction and Western blot, respectively, and in 518 paraffin-embedded HCC samples by immunohistochemistry. The correlation of PRAF2 expression and clinical outcomes was determined by the Student’s t-test, Kaplan–Meier test, and multivariate Cox regression analysis. The role of PRAF2 in HCC was investigated by cell viability, colony formation, and migration assays in vitro and with a nude mouse model in vivo.
Results
In our study, the PRAF2 expression was noticeably increased in HCC tissues at both the mRNA and protein levels compared with that of the nontumorous tissues. Kaplan–Meier analysis indicated that high PRAF2 expression was correlated with worse overall survival in a cohort of 518 patients with HCC. The prognostic implication of PRAF2 was verified by stratified survival analysis. The multivariate Cox regression model revealed PRAF2 as an independent poor prognostic factor for overall survival (hazard ratio = 1.244, 95% CI: 1.039–1.498, P<0.017) in HCC. The in vitro data demonstrated that PRAF2 overexpression markedly enhanced cell viability, colony formation, and cell migration. Moreover, ectopic expression of PRAF2 promoted tumor growth and metastasis in vivo.
Conclusion
Collectively, we conclude that PRAF2 is increased in HCC and is a novel unfavorable biomarker for prognostic prediction for patients with HCC.
Keywords: PRAF2, prognosis, proliferation, migration, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) represents the fifth most common cancer in the world and the third-ranked tumor type in terms of lethality.1,2 Progress has been made in the clinical treatment; however, patients with HCC have a very low 5-year survival rate (15%–30%) due to late diagnosis, impaired liver function, and tumor metastasis.3–6 Therefore, identification of potential biomarkers and therapeutic targets in HCC to provide personalized therapy would be of great value.7–9
Prenylated Rab acceptor 1 domain family member 2 (PRAF2, also known as JM4) is a 19-kDa protein with a prenylated Rab acceptor motif and four transmembrane domains.10 PRAF2, as an endoplasmic reticulum (ER)-resident domain protein, has been identified as a major GABAB gatekeeper11–13 and a mediator in ER-to-Golgi transport.14,15 Human PRAF2 is highly expressed in many normal tissues such as the lung and brain.10 Unlike other members of the PRA family (such as PRAF3) that induce cell apoptosis and inhibit metastasis, thus functioning as tumor suppressors,16,17 PRAF2 and PRAF1 are considered as oncogenes since they are frequently overexpressed.18–20 Increased expression of PRAF2 was significantly correlated with malignant clinical features that predict unfavorable outcome in malignant gliomas and neuroblastoma, by either blocking cell apoptosis21 or stimulating cell growth and migration.22 However, Vento et al23 reported that PRAF2 overexpression induces apoptotic cell death in U2OS (Human Osteosarcoma cell line) and may exert as a tumor suppressor gene. These data suggest that PRAF2 is involved in tumor progression in most tumor types.
The aim of the present study was to determine the expression of PRAF2 and its correlation with clinical outcomes in HCC. The role of PRAF2 in HCC development was also investigated.
Materials and methods
Patients, tissue specimens, and follow-up
A total of 518 paraffin-embedded HCC specimens were obtained from the archives of the Department of Pathology of the Sun Yat-sen University Cancer Center (SYSUCC) between January 2000 and December 2010. Another 41 cases of paired fresh HCC and adjacent nontumorous liver tissues were collected from patients at the time of surgical resection for determination of PRAF2 mRNA and protein expressions. None of the patients received any chemotherapy or radiotherapy before the surgery. The follow-up period was defined as the interval from the date of surgery to the date of death or the last follow-up.
Ethics approval and consent to participate
This study has been approved by the Institutional Review Board and Human Ethics Committee of SYSUCC. Written informed consent for using the samples for research purposes was obtained from all patients prior to surgery. This is a retrospective study and the specimens from all patients were anonymous, so informed consent has been exempted by Human Ethics Committee of SYSUCC. The project has been examined by Animal Ethical and Welfare Committee of SYSUCC and is in compliance with animal protection, animal welfare, and ethical principles and is in compliance with the relevant provisions of the National Laboratory Animal Welfare Ethics.
Tissue microarray construction and immunohistochemistry (IHC)
Using a tissue array instrument (MinicoreExcilone, Minicore, UK), a 0.6 mm diameter tissue core was punched out of the marked area and embedded again. All specimens were fixed with 4% paraformaldehyde in 0.1 M phosphate buffer for 24 hours and embedded in paraffin. Paraffin-embedded HCC sections were cut into 4 µm sections and mounted on glass slides. After dewaxing, the slides were treated with a 3% methanolic hydrogen peroxide solution and blocked with a biotin blocking kit (DAKO, Hamburg, Germany). After blocking, the slides were incubated with PRAF2 polyclonal antibody (1:50, PAB11686, Abnova, Walnut, CA, USA) in a humidified chamber at 4°C overnight. After three washes in PBS, the slides were incubated with biotinylated goat anti-rabbit antibody for 1 hour. The slides were stained with DAKO liquid 3,3′-diaminobenzidine tetrahydrochloride. Finally, the slides were stained with Mayer hematoxylin and observed under a microscope.
Protein levels were determined using a semiquantitative IHC assay. Positive staining samples: 0, positive staining cells <5%; 1, 6%–24% of positive staining cells; 2, 25%–49% of positive cells; 3, 50%–74% of positive staining cells; 4, 75%−100% of positive staining of cells. Intensity was scored according to the following criteria: 0, negative staining; 1, weak staining; 2, moderate staining; and 3, strong staining. The final score is calculated by multiplying the fractional score by the intensity score of the staining. These scores were independently determined by two pathologists (Dr Jing-Ping Yun and Dr Hui-Zhong Zhang). The median IHC value was chosen as the cutoff to define high and low expressions.
Cell lines and cell culture
The human hepatic carcinoma cell lines Bel-7402, QGY-7703, HepG2, and Bel-7404 were obtained from the Chinese Academy of Sciences (Shanghai, China). These cell lines were cultured in DMEM supplemented with 10% heat-inactivated fetal bovine serum (HyClone, Logan, UT, USA) at 37°C in a humidified atmosphere of 5% CO2.
Migration assay
The cells (2–4×104) were plated in a transwell chamber (US Millipore 8 µm pore size) in serum-free medium. After 24 hours of incubation, migrating cell counts were performed on the lower membrane after staining with 0.1% crystal violet and 20% methanol. Experiments were done in triplicate.
Proliferation and colony formation assays
Transfected cells were seeded in 96-well plates (2–4 × 104 cells/mL) with 100 µL medium in each culture for 5 days. The diffusion test was performed by adding 20 µL of MTT reagent (5 mg/mL, AMRESCO, Solon, OH, USA) for 4 hours at 37°C. Then, the formazan crystals were dissolved in dimethyl sulfoxide (150 µL/well). The absorbance values (OD 590 nm) were measured by a multilabel plate reader (PerkinElmer). For the colony formation assay, 500 cells were seeded onto six-well plates with 2 mL DMEM per well. After 10 days of culture, cell colonies were fixed with methanol and then stained with 0.1% crystal violet, and the number of colonies was counted by microscope.
Plasmid construction and transfection
The plasmid expressing PRAF2 (pcDNA 3.1/hygro+) was confirmed by sequencing. We transduced PRAF2 plasmids into QGY-7703, SMMC-7721, and Bel-7402 cells by Lipofectamine™ 2000 (Invitrogen, Carlsbad, CA, USA). After antibiotic selection with 800 µg/mL G418 (Clontech, Mountain View, CA, USA), G418-resistant colonies were isolated, expanded into cell lines, and PRAF2 expression was confirmed by Western blot.
Small interfering RNA
The duplex small interfering RNA (siRNA) targeting PRAF2 (siRNA#1: 5′-CCAGGUCAAGACAUUGCCAAA-3′; siRNA#2: 5′-GUGUUGCUGCAACAAUAAA-3′) and a negative control (NC) siRNA duplex (forward: 5′-UUCUC-CGAACGUGUCACGUTT-3′; reverse: 5′′-ACGUGACAC-GUUCGGAGAATT-3′) were designed and synthesized by Shanghai GenePharma Co. Ltd. (Shanghai, China).
In vivo tumor growth and metastasis
Male athymic nude mice (4 weeks, 18–22 g) were bred at the animal facility of the Center of Experimental Animals, Sun Yat-Sen University (Guangzhou, China). The right flanks of mice (5 weeks of age) were subcutaneously injected with Bel-7402 cells (5×106) that stably expressed PCDNA3.1-PRAF2 or the NC that stably expressed PCDNA3.1 vector (n=6 per group). Tumor growth was monitored every 3 days. Tumor volumes were calculated using the following formula: volume = length × width2 × 0.5. Thirty days later, the mice were sacrificed and tumors were collected for further measurement. For in vivo tumor metastasis, 5×105 cells were injected into the mice from the caudal vein. Seven weeks later, the mice were sacrificed and the lung tissue was obtained. Lung metastases were detected using H&E staining and were quantified by counting the metastatic lesions in each section (n=6 per group, BALB/c-nu, male, 3–4 weeks old). All animal studies were conducted with the approval of the Medical Experimental Animal Care Commission of Sun Yat-sen University Cancer Center.
Statistical analysis
Statistical analysis was conducted using SPSS software (version 16.0, Chicago, IL, USA). Differences were analyzed by Student’s t-test. The correlation between the expression level of PRAF2 and the clinical and pathological variables was examined by Pearson’s chi-squared test or Fisher’s exact test. Survival curves were evaluated with the Kaplan–Meier method (log-rank test). A multivariate Cox proportional hazards regression model was constructed to evaluate the independent influence of PRAF2 in prognosis. P<0.05 was considered significant.
Results
Expression of PRAF2 in HCC
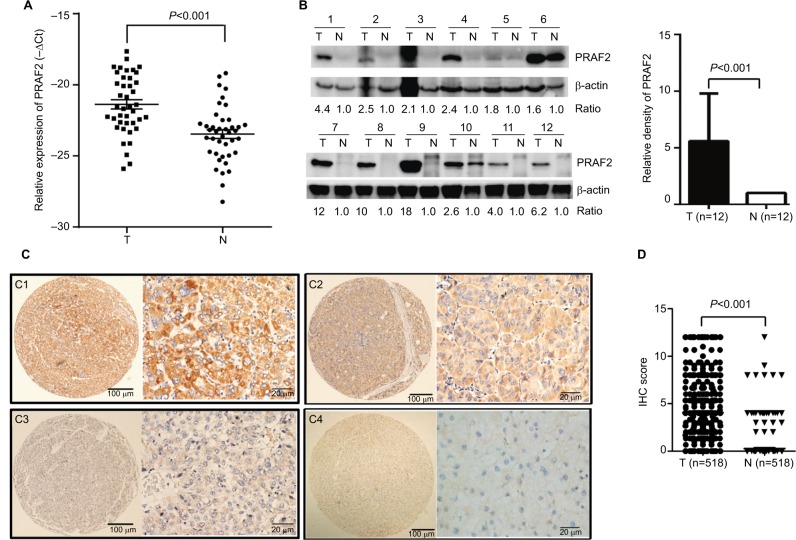
The expression of PRAF2 was detected in fresh HCC tissues by quantitative reverse transcription-polymerase chain reaction (qRT-PCR) and Western blot. The results showed that the PRAF2 mRNA expression levels in the 41 HCC fresh samples were markedly increased compared with those in the adjacent nontumorous tissues (Figure 1A). Consistently, an increase of the PRAF2 protein level was observed in the 12 pairs of fresh HCC tissues (Figure 1B). The tissue microarray-based IHC analysis showed that the cellular localization of PRAF2 mainly existed in the cytoplasm in most of the HCC cells (Figure 1C1–C3). Positive expression of PRAF2 was exhibited in 66.8% (346/518) of HCC tissues, but rarely in the nontumorous tissue (Figure 1C4). In 66.2% (343/518) of the samples, PRAF2 expression was higher in HCC than in the nontumorous tissue. The alteration of PRAF2 expression was statistically significant (Figure 1D).
Figure 1.
PRAF2 gene and protein expression levels are increased in HCC.
Notes: (A) The mRNA levels of PRAF2 in HCC (T) and the corresponding adjacent liver tissue (N) were determined in 41 patients with qRT-PCR. (B) Expression of PRAF2 protein in 12 paired HCC and adjacent liver tissues was examined by Western blot. (C) PRAF2 expression is determined in HCC by immunohistochemistry. PRAF2 was presented predominantly in cytoplasm within tumor and adjacent nontumorous liver cells. The micrographs showed strong C1, moderate C2, and weak C3 staining in HCC, along with C4 staining in nontumorous liver tissues (left panel: magnification ×100; right panel: magnification ×400). (D) Reproducibility of the measurement in 518 patients was calculated using the Wilcoxon matched paired test.
Abbreviations: HCC, hepatocellular carcinoma; IHC, immunohistochemistry; PRAF2, prenylated Rab acceptor 1 domain family member 2; qRT-PCR, quantitative reverse transcription-polymerase chain reaction.
Association of PRAF2 expression and clinical outcomes in HCC
Kaplan–Meier survival analysis was conducted to determine the prognostic impact of PRAF2 in HCC patients. Patients were separated into high or low expression groups according to the median IHC score (4.00). High PRAF2 expression was identified in 53.3% (276/518) of cases. HCC cases with high PRAF2 expression were often associated with a worse prognosis in terms of overall survival (log-rank test; P=0.011; Figure 2). Furthermore, according to the mRNA levels of PRAF2 in 370 HCC patients of The Cancer Genome Atlas dataset, high expression of PRAF2 suggests a trend of poor prognosis in terms of overall survival (log-rank test; P=0.084; Figure S5). In our HCC cohort, high PRAF2 expression was more likely to present in older age (P=0.024), but failed to detect correlation with other clinical features (Table S1).
Figure 2.
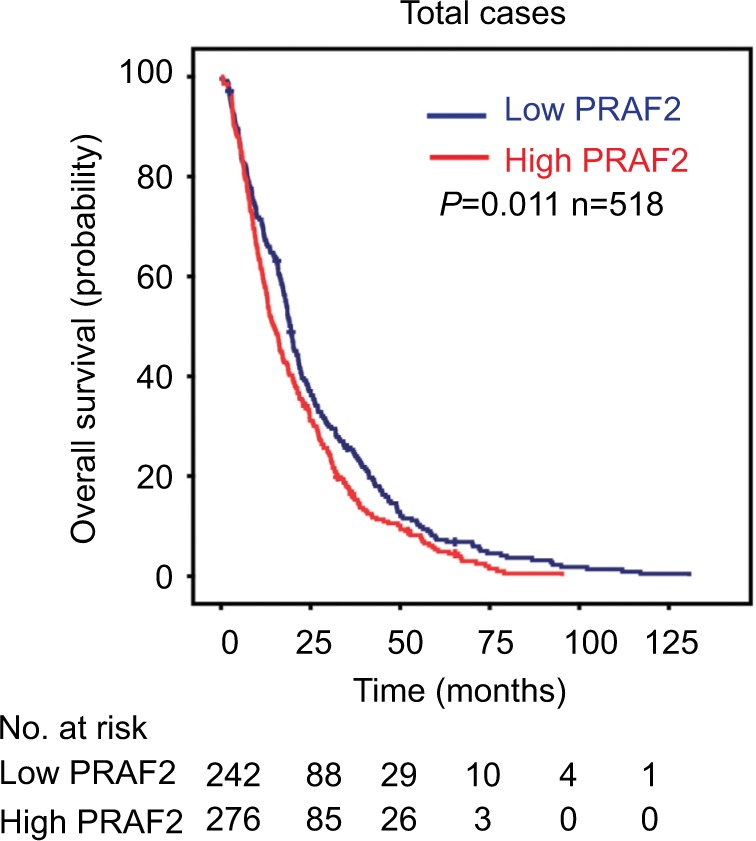
PRAF2 expression is correlated with poor prognosis.
Notes: The patients with HCC were divided into high and low PRAF2 expression groups according to the median IHC score. Kaplan–Meier analysis was conducted to evaluate the relationship between PRAF2 expression and overall survival.
Abbreviations: HCC, hepatocellular carcinoma; IHC, immunohistochemistry; PRAF2, prenylated Rab acceptor 1 domain family member 2.
Univariate and multivariate analyses of prognostic variables in HCC
To evaluate the representativeness of our samples, univariate analysis was performed. Along with PRAF2, serum alpha-fetoprotein level, tumor size, tumor multiplicity, tumor capsule, liver cirrhosis, vascular invasion, tumor differentiation, clinical stage, and lymph node metastasis were found to significantly contribute to the outcome of overall survival (Table 1). After adjusting for the prognostic factors established in the univariate analysis, multivariate analysis indicated that PRAF2 expression was an independent prognostic factor for overall survival (hazard ratio = 1.244, 95% CI: 1.039–1.498, P=0.017).
Table 1.
Univariate and multivariate analyses of clinicopathological and PRAF2 expression for overall survival in HCC patients (n=518)
| Variables | Univariate analysis
|
Multivariate analysis
|
||
|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
| Age (<49 vs ≥49 years) | 0.911 (0.765–1.086) | 0.298 | N/A | N/A |
| Gender (female vs male) | 1.019 (0.756–1.447) | 0.900 | N/A | N/A |
| HBV (positive vs negative) | 1.124 (0.878–1.438) | 0.353 | N/A | N/A |
| Tumor size (≥5 vs <5 cm) | 1.531 (1.235–1.897) | 0.000 | 1.328 (1.014–1.694) | 0.023 |
| Tumor multiplicity (multiple vs single) | 1.259 (1.049–1.510) | 0.013 | 1.006 (0.814–1.242) | 0.959 |
| Tumor capsule (absent vs present) | 0.694 (0.579–0.832) | 0.000 | 0.813 (0.670–0.987) | 0.037 |
| Liver cirrhosis (no vs yes) | 0.740 (0.586–0.935) | 0.012 | 0.842 (0.658–1.078) | 0.173 |
| AFP (≥20 vs <20 ng/mL) | 1.438 (1.169–1.769) | 0.001 | 1.293 (1.044–1.602) | 0.019 |
| Vascular invasion (yes vs no) | 1.836 (1.478–2.280) | 0.000 | 1.279 (0.998–1.637) | 0.052 |
| Tumor differentiation | 1.440 (1.184–1.752) | 0.000 | 1.293 (1.056–1.584) | 0.013 |
| TNM (III–IV vs I–II) | 1.630 (1.364–1.949) | 0.000 | 1.208 (0.951–1.535) | 0.121 |
| LNM (yes vs no) | 1.594 (1.124–2.261) | 0.009 | 1.509 (1.049–2.171) | 0.027 |
| PRAF2 expression (high vs low) | 1.256 (1.052–1.499) | 0.011 | 1.244 (1.039–1.498) | 0.017 |
Notes: Bold values indicate statistically significant values (p<0.05)
Abbreviations: AFP, alpha-fetoprotein; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; HR, hazard ratio; LNM, Lymph node metastasis; N/A, not applicable; PRAF2, prenylated Rab acceptor 1 domain family member 2.
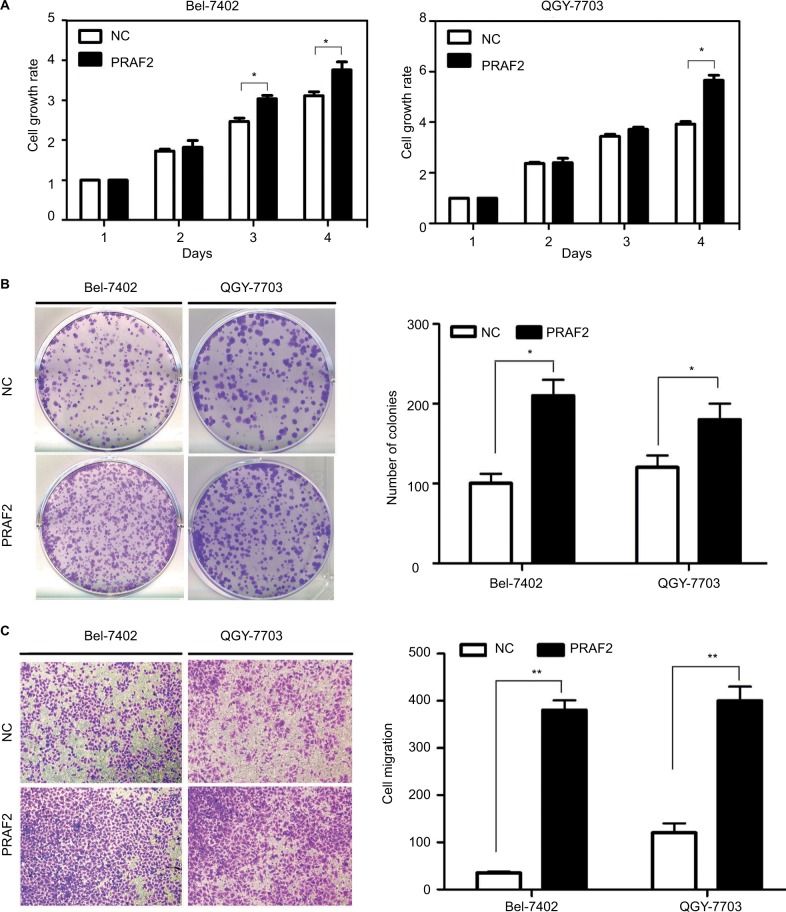
Overexpression of PRAF2 improves cell proliferation and migration in vitro
To further investigate the potential of PRAF2 in HCC progression, QGY-7703 and Bel-7402 cells were stably transfected with PRAF2 plasmid, and the expression of PRAF2 in both HCC cell lines was confirmed by western blot (Figure S1). The MTT assay revealed that overexpression of PRAF2 significantly enhanced cell viability (Figure 3A). The colony formation assay confirmed that PRAF2 promoted cell proliferation in HCC, by showing that the number of colonies formed by cells expressing PRAF2 was much higher than that of the control groups (Figure 3B). Moreover, the transwell assays demonstrated that PRAF2 reexpression significantly accelerated the cell migration (Figure 3C). Furthermore, the knockdown of PRAF2 protein in Bel-7404 and HepG2 cell lines was confirmed by Western blot analysis (Figure S2A, D), and the transwell assays demonstrated that PRAF2 knockdown virtually attenuated the numbers of migrating cells (Figure S2B, C).
Figure 3.
PRAF2 promotes cell proliferation and migration in vitro.
Notes: (A) Cells cultured in 96-well plates were transfected with a PRAF2-expressing vector for 4 days. Cell viabilities were determined using MTT assays. (B) Cells stably expressing PRAF2 were cultured with 400 µg/mL G418 for 2 weeks. The number of colonies was calculated and depicted in the histogram. Data are represented as the mean + SEM of three independent experiments (magnification, ×100). (C) The effects of PRAF2 on cell migration were determined by transwell assays, using cells with PRAF2 overexpression. The quantification of three randomly selected fields is shown. *P<0.05; **P<0.01 (magnification, ×100).
Abbreviations: NC, Negative control; PRAF2, prenylated Rab acceptor 1 domain family member 2.
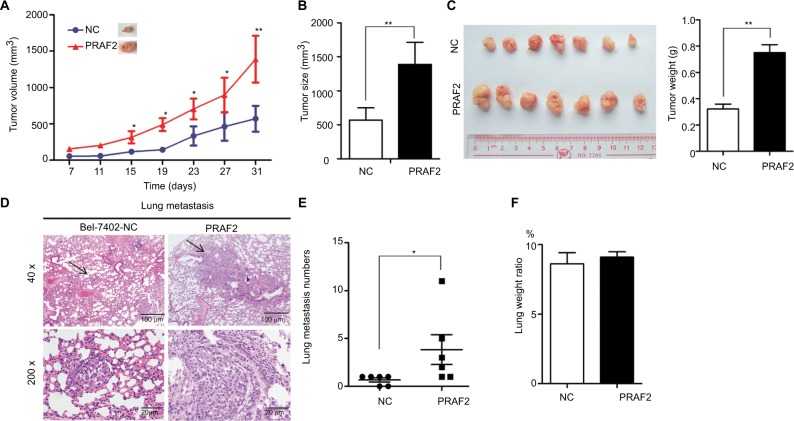
Ectopic PRAF2 expression enhances tumor growth and metastasis in vivo
In vivo experiments were performed to verify the roles of PRAF2 on cell growth and metastasis. As shown in Figure 4A, tumors with PRAF2 overexpression grew faster than those without PRAF2. Bel-7402 cells stably expressing PRAF2 generated tumors with greater volumes and weights (Figure 4B, C). The HCC metastatic model was used to confirm the effect of PRAF2 on HCC metastasis. As shown in Figure 4D, overexpression of PRAF2 promoted the metastasis of HCC xenografts. Compared with the control groups, lung metastatic nodules were significantly increased in the PRAF2-overexpressing groups (Figure 4E), although the lung weight ratio showed no difference between the two groups (Figure 4F).
Figure 4.
PRAF2 promotes tumor growth and metastasis in vivo.
Notes: (A–C) Bel-7402 cells (1×107) stably expressing PRAF2 were injected into the right flank of nude mice. Tumor volume was measured every 4 days (A). After 30 days, the tumors were measured (B) and weighed (C right); the representative images of the xenograft are shown (C left). (D) Cells expressing PRAF2 were injected into the mice through the tail vein. The lungs were fixed in 4% paraformaldehyde and sectioned for H&E staining. (E) The number of metastases was counted. Values are presented as mean ± SEM. (F) Ratio of lung weight vs body weight. *P<0.05, **P<0.01.
Abbreviations: H&E, Hematoxylin Eosin; NC, Negative control; PRAF2, prenylated Rab acceptor 1 domain family member 2.
Discussion
PRAF family proteins have been implicated in the cell cycle, extracellular matrix, and cell migration/invasion.24,25 Recent studies have shown that PRAF2 is closely associated with metastasis and poor prognosis in several cancer types, largely owing to its effects on enhancing cell proliferation, migration, and metastasis in malignant glioma or neuroblastoma.18,21,22 In our study, PRAF2 expression was frequently increased in HCC tissues compared with adjacent nontumorous tissues. PRAF2 expression was also correlated with worse patient outcomes. In vitro and in vivo data demonstrated that PRAF2 was capable of promoting cell proliferation, migration, and tumor metastasis.
Dysregulation of PRAF2 has been reported in several human cancers. However, its prognostic implication has been rarely studied. In this study, PRAF2 was identified as an independent factor affecting the overall survival in a large cohort of 518 patients with HCC. Patients with high PRAF2 expression usually have a shorter life span. Furthermore, according to the mRNA levels of PRAF2 in 370 HCC patients of TCGA dataset, high expression of PRAF2 suggests a trend of poor prognosis in terms of overall survival. Similar to our data, Yco et al22 reported that high PRAF2 expression was significantly correlated with unfavorable prognosis in neuroblastoma. These data suggest that PRAF2 is of clinical significance for monitoring the surgical outcomes of HCC patients.
The imbalance between cell survival and cell death is known to be an early and critical event in tumorigenesis. In the present study, we found that PRAF2 overexpression promoted the proliferation and mobility of HCC cells in vitro, whereas PRAF2 depletion attenuated the cell migration potential. A mechanistic study showed that PRAF2 overexpression did not alter that expression levels of biomarkers related to the epithelial–mesenchymal transition, such as N-cadherin, vimentin, and Twist1 (Figure S3). Ectopic expression of PRAF2 promoted tumor growth and metastasis in vivo. These data suggest that PRAF2 might play an oncogenic role in HCC progression. PRAF2 was reported to be involved in cell apoptosis in neuroblastoma and U2OS cells.21,23 However, in the present study, overexpression of PRAF2 did not influence the expression of apoptotic biomarkers, including Bim, Bad, and PRAP1 (Figures S4 and S6). Except for apoptotic pathways, we hope to investigate the potential mechanism through which PRAF2 is acting to drive tumorigenesis in HCC in other pathway.
As a result, the underlying mechanism by which PRAF2 participates in HCC progression and the triggers for PRAF2 expression require further investigation.
Conclusion
In summary, we determined the clinical implication of PRAF2 expression in a cohort of 518 HCC patients. High PRAF2 expression unfavorably affected the survival of HCC patients and served as an independent factor for worse outcomes. In vitro and in vivo studies involving the overexpression and silencing of PRAF2 in HCC cells confirmed the effect of PRAF2 on HCC progression. Taken together, our data suggest PRAF2 as a promising biomarker for prognosis of patients with HCC.
Acknowledgments
The study was supported by grants from the National Natural Science Foundation of China (Nos 81372572, 81572406, 81572405, 81602135, 81702755) and the National Natural Science Foundation of Guangdong Province (No. 2016A030310215). The authenticity of this article has been validated by uploading the key raw data onto the Research Data Deposit public platform (www.researchdata.org.cn), with the approval RDD number RDDB2018000342.
Footnotes
Author contributions
Conceived and designed the experiments: Wang CH, Liu LL, Liao DZ, Zhang HZ and Yun JP; performed the experiments: W-CH and Fu J; analyzed the data: Wang CH, Chen SL and Wang H; contributed reagents/materials/analysis tools: Wang CH, Zhang MF, Lu SX, Cai SH; and wrote the paper: Wang CH, Zhang CZ, Yun JP. All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365(12):1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 2.Han ZG. Functional genomic studies: insights into the pathogenesis of liver cancer. Annu Rev Genomics Hum Genet. 2012;13:171–205. doi: 10.1146/annurev-genom-090711-163752. [DOI] [PubMed] [Google Scholar]
- 3.Villanueva A, Hernandez-Gea V, Llovet JM. Medical therapies for hepatocellular carcinoma: a critical view of the evidence. Nat Rev Gastroenterol Hepatol. 2013;10(1):34–42. doi: 10.1038/nrgastro.2012.199. [DOI] [PubMed] [Google Scholar]
- 4.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379(9822):1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 5.Visvader JE. Cells of origin in cancer. Nature. 2011;469(7330):314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 6.Wei KR, Yu X, Zheng RS, et al. Incidence and mortality of liver cancer in China, 2010. Chin J Cancer. 2014;33(8):388–394. doi: 10.5732/cjc.014.10088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer. 2005;5(11):845–856. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 8.Xie B, Lin W, Ye J, et al. DDR2 facilitates hepatocellular carcinoma invasion and metastasis via activating ERK signaling and stabilizing SNAIL1. J Exp Clin Cancer Res. 2015;34:101. doi: 10.1186/s13046-015-0218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu H, Liu Y, Liu W, Zhang W, Xu J. EZH2-mediated loss of miR-622 determines CXCR4 activation in hepatocellular carcinoma. Nat Commun. 2015;6:8494. doi: 10.1038/ncomms9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fo CS, Coleman CS, Wallick CJ, Vine AL, Bachmann AS. Genomic organization, expression profile, and characterization of the new protein PRA1 domain family, member 2 (PRAF2) Gene. 2006;371(1):154–165. doi: 10.1016/j.gene.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 11.Doly S, Marullo S. PRAF2, an endoplasmic reticulum gatekeeper, controls the cell-surface export of the GABA(B) receptor in neurons. Med Sci. 2015;31(10):834–836. doi: 10.1051/medsci/20153110008. [DOI] [PubMed] [Google Scholar]
- 12.Cifuentes-Diaz C, Marullo S, Doly S. Anatomical and ultrastructural study of PRAF2 expression in the mouse central nervous system. Brain Struct Funct. 2016;221(8):4169–4185. doi: 10.1007/s00429-015-1159-8. [DOI] [PubMed] [Google Scholar]
- 13.Doly S, Marullo S. Gatekeepers controlling GPCR export and function. Trends Pharmacol Sci. 2015;36(10):636–644. doi: 10.1016/j.tips.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 14.Schweneker M, Bachmann AS, Moelling K. JM4 is a four-transmembrane protein binding to the CCR5 receptor. FEBS Lett. 2005;579(7):1751–1758. doi: 10.1016/j.febslet.2005.02.037. [DOI] [PubMed] [Google Scholar]
- 15.Ruggiero AM, Liu Y, Vidensky S, et al. The endoplasmic reticulum exit of glutamate transporter is regulated by the inducible mammalian Yip6b/GTRAP3-18 protein. J Biol Chem. 2008;283(10):6175–6183. doi: 10.1074/jbc.M701008200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen H, Bai J, Ye J, et al. JWA as a functional molecule to regulate cancer cells migration via MAPK cascades and F-actin cytoskeleton. Cell Signal. 2007;19(6):1315–1327. doi: 10.1016/j.cellsig.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 17.Wang GL, Shi X, Salisbury E, Timchenko NA. Regulation of apoptotic and growth inhibitory activities of C/EBP alpha in different cell lines. Exp Cell Res. 2008;314(7):1626–1639. doi: 10.1016/j.yexcr.2008.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borsics T, Lundberg E, Geerts D, et al. Subcellular distribution and expression of prenylated Rab acceptor 1 domain family, member 2 (PRAF2) in malignant glioma: Influence on cell survival and migration. Cancer Sci. 2010;101(7):1624–1631. doi: 10.1111/j.1349-7006.2010.01570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koomoa DL, Go RC, Wester K, Bachmann AS. Expression profile of PRAF2 in the human brain and enrichment in synaptic vesicles. Neurosci Lett. 2008;436(2):171–176. doi: 10.1016/j.neulet.2008.03.030. [DOI] [PubMed] [Google Scholar]
- 20.Kim JT, Cho MY, Choi SC, et al. Prenylated Rab acceptor 1 (PRA1) inhibits TCF/beta-catenin signaling by binding to beta-catenin. Biochem Biophys Res Commun. 2006;349(1):200–208. doi: 10.1016/j.bbrc.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 21.Geerts D, Wallick CJ, Koomoa DL, et al. Expression of prenylated Rab acceptor 1 domain family, member 2 (PRAF2) in neuroblastoma: correlation with clinical features, cellular localization, and cerulenin-mediated apoptosis regulation. Clin Cancer Res. 2007;13(21):6312–6319. doi: 10.1158/1078-0432.CCR-07-0829. [DOI] [PubMed] [Google Scholar]
- 22.Yco LP, Geerts D, Koster J, Bachmann AS. PRAF2 stimulates cell proliferation and migration and predicts poor prognosis in neuroblastoma. Int J Oncol. 2013;42(4):1408–1416. doi: 10.3892/ijo.2013.1836. [DOI] [PubMed] [Google Scholar]
- 23.Vento MT, Zazzu V, Loffreda A, et al. Praf2 is a novel Bcl-xL/Bcl-2 interacting protein with the ability to modulate survival of cancer cells. PLoS One. 2010;5(12):e15636. doi: 10.1371/journal.pone.0015636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hendrix A, Westbroek W, Bracke M, de Wever O. An ex(o)citing machinery for invasive tumor growth. Cancer Res. 2010;70(23):9533–9537. doi: 10.1158/0008-5472.CAN-10-3248. [DOI] [PubMed] [Google Scholar]
- 25.Peinado F, Lavotshkin S, Lyden D. The secreted factors responsible for pre-metastatic niche formation: old sayings and new thoughts. Semin Cancer Biol. 2011;21(2):139–146. doi: 10.1016/j.semcancer.2011.01.002. [DOI] [PubMed] [Google Scholar]