Abstract
Bacterial communities are essential for the functioning of the Earth's ecosystems1. A key challenge is to quantify the functional roles of bacterial taxa in nature to understand how the properties of ecosystems change over time or under different environmental conditions2. Such knowledge could be used, for example, to understand how bacteria modulate biogeochemical cycles3, and to engineer bacterial communities to optimise desirable functional processes4. Communities of bacteria are, however, extraordinarily complex with hundreds of interacting taxa in every gram of soil and every millilitre of pond water5. Little is known about how the tangled interactions within natural bacterial communities mediate ecosystem functioning, but high levels of bacterial diversity have led to the assumption that many taxa are functionally redundant6. Here, we pinpointed the bacterial taxa associated with keystone functional roles, and show that rare and common bacteria are implicated in fundamentally different types of ecosystem functioning. By growing hundreds of bacterial communities collected from a natural aquatic environment (rainwater-filled tree holes) under the same environmental conditions, we show that negative statistical interactions among abundant phylotypes drove variation in broad functional measures (respiration, metabolic potential, cell yield), while positive interactions between rare phylotypes influenced narrow functional measures (the capacity of the communities to degrade specific substrates). The results alter our understanding of bacterial ecology by demonstrating that unique components of complex communities are associated with different types of ecosystem functioning.
Keywords: functional interactions, microbiome, biodiversity, ecosystem functioning
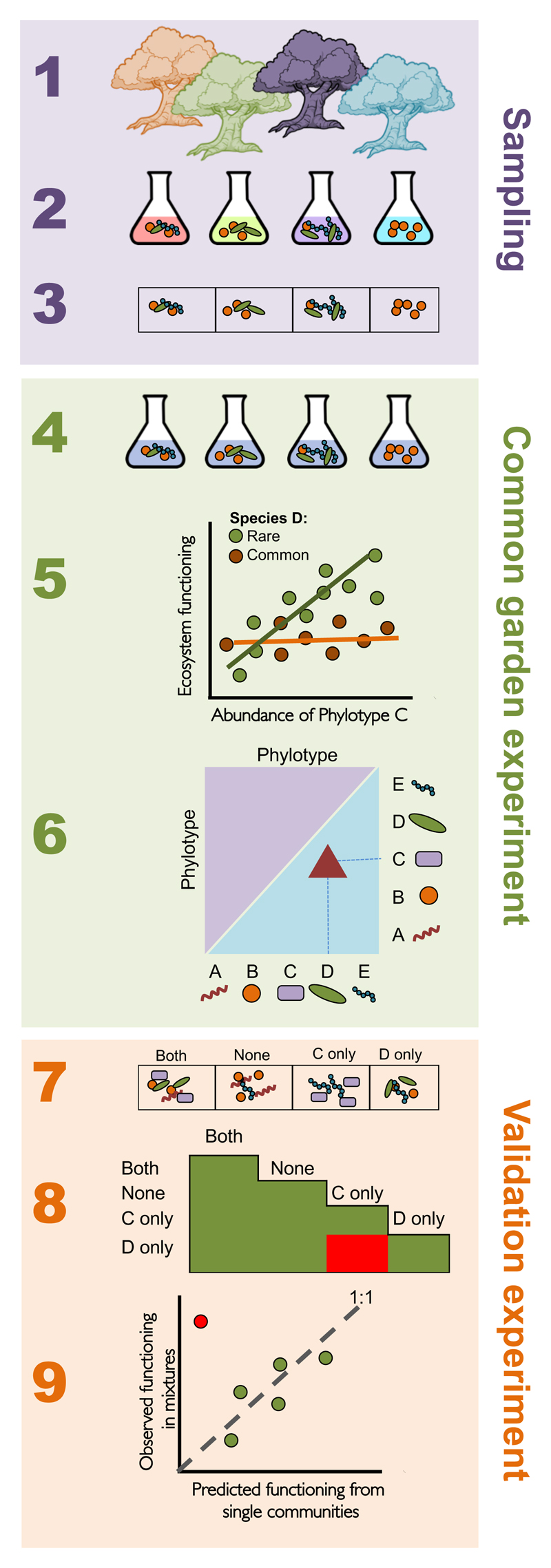
The functional roles of bacterial taxa within communities can be estimated using manipulative experiments that build communities from pure cultures or that remove taxa from intact communities7. There are many difficulties with experiments using pure cultures: most bacteria cannot be isolated in pure culture, synthetic communities constructed from pure cultures might not represent any natural community, and there are no general methods for knocking-out specific taxa from natural communities. The alternative has been to use observational methods to look for correlated changes in taxa abundance and ecosystem functioning in nature, or to infer which functional processes are important from metagenomic or metatranscriptomic data8–10. Observational approaches also have significant weaknesses because abiotic conditions (e.g. pH, temperature) can impact both ecosystem functioning and community structure, making it difficult to infer causal relationships. We developed an alternative experimental approach that exploits the natural variation in bacterial community composition, comparable to the microbiome association studies that have been proposed for inferring causal relationships between human health and microbiome community composition11 (Fig. 1, steps 1 to 6).
Figure 1. Illustration of the workflow for microbiome association studies in common gardens.
(Step 1) Samples were collected from rainwater-filled tree holes, and (Step 2) returned to the lab, where (Step 3) they were archived at -80C and a subsample was sequenced to identify community composition. Sequencing revealed 5 phylotypes (A to E) were present in the communities. (Step 4) Frozen microbiomes were resuscitated and grown in a standardised beech leaf medium while measuring ecosystem functioning. (Step 5, 6) A systematic search for associations between the abundance of each phylotype (A to E, from Step 3) and the functional measurements (from Step 4) identified a significant positive association between functioning and the abundance of Phylotype C, as well as a significant functional interaction between Phylotype C and Phylotype D. Broad functional interactions are above the diagonal (purple), while narrow functional interactions are below the diagonal (blue). (Step 7) To validate the interaction, we searched for communities where Phylotype C and D did not co-occur (C only, D only), or where they co-occurred (Both) or were both absent (None). (Step 8) The positive functional interaction between Phylotype C and D (identified in step 6) was tested by looking for an increase in functioning in mixtures that combined C-only and D-only communities (red box). (Step 9) The large increase in ecosystem functioning when mixing C-only and D-only communities (red point), and the lack of similar result for the other mixtures (green points), confirmed the functional interaction identified in step 6.
We sampled 753 aquatic microbial communities, which were taken from a natural micro-ecosystem (rainwater pools in the buttressing of beech trees)12,13. The bacterial cells were separated from the surrounding environmental matrix by filtration and then stored (frozen) so that they could be revived (thawed) for repeatable experiments. Each bacterial community was revived and placed in laboratory microcosms containing a sterile beech leaf medium, which simulated some of the environmental conditions in the natural system. We quantified 7 measures of ecosystem functioning associated with leaf litter degradation. Since the communities were assayed in a common environment, variation in ecosystem functioning was due to the initial differences in community composition. Correlations between the initial absolute abundance of each phylotype and ecosystem functioning allowed us to obtain community-wide estimates of the phylotypes that were associated with changes in functioning14, which could reflect their impact on functioning in nature. While the approach requires growing the communities in simplified microcosms, there is no need to isolate individual taxa. The approach therefore lies between the complexity of natural ecosystems and the artificiality of synthetic communities.
We searched for associations between the abundance of each phylotype and the functional measurements, analogous to a genome-wide association scan in genetics15. We found 182 significant associations between phylotype abundance and the functional measurements, involving 112 of the 522 phylotypes used in the analysis. The associations were approximately equally balanced between negative (96 significant associations) and positive (86 significant associations), but differed substantially among the functional measurements (Supplementary Figure 1). We therefore divided the functional measurements into two categories16,17. First: respiration, cell yield, and metabolic potential (ATP; adenosine triphosphate concentration) of the community were categorised as 'broad' ecosystem functions because they amalgamate many activities and are therefore performed by most community members. We expected widespread functional redundancy of taxa within the broad functional measures, which should result in few significant associations. Second: the cleavage rates of 4 substrates added to the microcosms were categorised as 'narrow' ecosystem functions because they encompass fewer activities and were therefore likely to be impacted by a relatively limited set of phylotypes. In contrast with the broad functional measures, we expected the rise and fall in abundance of particular niche specialists would be associated with the rise and fall of the narrow functional measurements18.
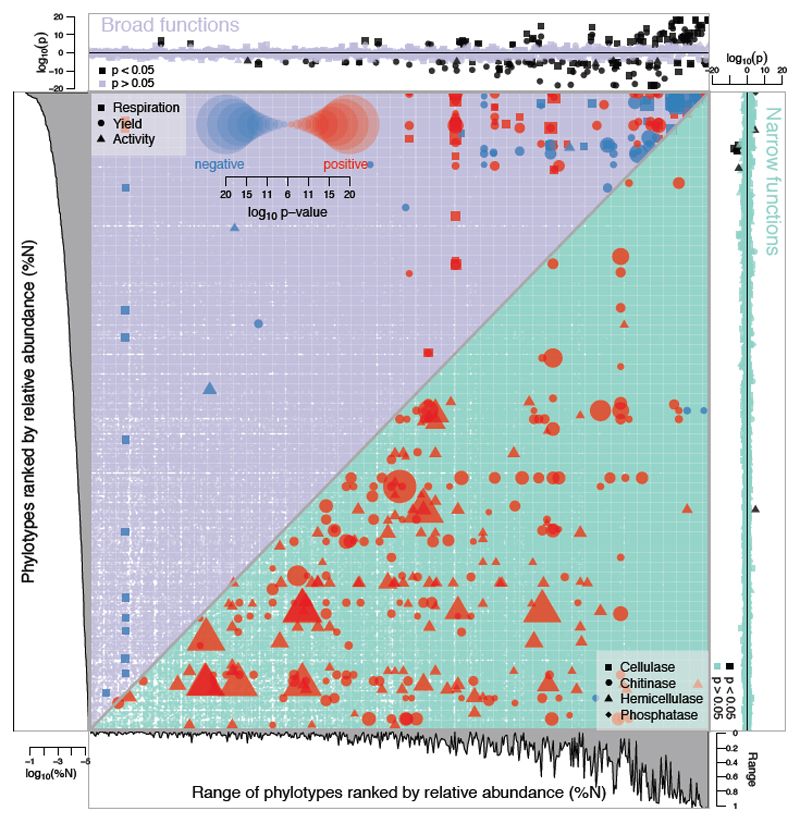
Contrasting strongly with our predictions, significant associations were largely confined to the broad functional measures. For the broad functional measures, 90% of the 174 significant associations were 'common' phylotypes (phylotypes that exceeded the median phylotype abundance) (Fig. 2, top panel). For example, the two phylotypes with the highest overall abundance across all communities (Serratia fonticola and Klebsiella pneumonia) also had among the strongest positive associations with respiration and cell yield. By contrast, only 8 significant associations were found between phylotype abundances and the narrow functional measurements (Fig. 2, right panel). The associations imply that the abundance of individual phylotypes play a role in the broad functional measures in ways that are not compensated by fully-redundant phylotypes. There were linear increases in the number of significant associations with increasing sampling effort (number of communities) (Supplementary Figure 2), showing that there are opportunities to uncover many more significant associations.
Figure 2. Associations between bacterial phylotypes and the functional measurements.
Left panel: each of the phylotypes were ranked according to their relative abundance (%N) across all 753 communities. Bottom panel: the range of each phylotype, which is the proportion of samples where each phylotype was present. Top panel: association between every phylotype and each of the broad functional measurements. Each datapoint is the log10(p-value) of the slope of the linear regression between the abundance of each phylotype and the functional measurements. Grey datapoints are non-significant, black datapoints are significant following a Bonferonni correction. Negative values indicate a negative association (the functional measurement declines with increased phylotype abundance) while positive values indicate a positive association. Symbols correspond to the broad functional measurements shown in the upper triangle of the central panel. Right panel: association between each of the phylotypes and the narrow functional measurements. Pale blue datapoints are non-significant, and black datapoints are significant following a Bonferonni correction. Negative values indicate a negative association (the functional measurement declines with increased phylotype abundance) while positive values indicate a positive association. Symbols correspond to the narrow functional measurements in the lower triangle of the central panel. Central panel: Functional interactions between all pairwise combinations of phylotypes for the broad (above diagonal) and narrow (below diagonal) functional measurements. Significant positive (red) and negative (blue) functional interactions are shown as symbols, with larger symbols being more significant (see key). Symbol definitions are given for broad functions (above diagonal, top left) and narrow functions (below diagonal, bottom right). The violet (above diagonal) or teal (below diagonal) background indicates non-significant interactions, while a white background indicates an interaction that cannot be estimated (e.g. because the phylotypes never co-occur). Phylotypes are increasingly abundant from bottom-to-top and from left-to-right within the central panel. Relationships were analysed using simple- or multiple regression. All datapoints indicated as significant are at a Bonferreonni-adjusted p-value threshold. P-values for the top and left panels are given in Supplementary Table 1, and p-values for the central panel are given in Supplementary Table 2.
We expected the impact of each bacterial phylotype on the functional measurements to be mediated by the hundreds of phylotypes that surrounded them. We used the same experiment to characterise 'functional interactions': whether the associations between the abundance of each phylotype and the functional measurements were altered by the abundance of any of the other phylotypes in the community (Fig. 1, steps 5 to 6). A positive functional interaction indicated that ecosystem functioning tended to be elevated when both phylotypes had high abundance, whereas a negative functional interaction indicated that ecosystem functioning was lower when both phylotypes had high abundance. Functional interactions might result from direct (i.e. biological) interactions among taxa, but such direct interaction would need to be verified (see Validation Experiment). The method allowed us to reconstruct a complete portrait of functional interactions within a diverse assemblage (Fig. 2, central panel).
Theoretical studies have predicted that direct inter-specific interactions (e.g. competition, mutualism) should be negative19 and weak20, but it is unclear whether these predictions extend to functional interactions. We found that negative and positive functional interactions tended to occur between common phylotypes for the broad functional measurements. In principle, all of the phylotypes contributed to the broad functional measurements, so it was unsurprising that interactions among the most common phylotypes had the largest influence. For the narrow functional measurements, functional interactions were between rare phylotypes, consistent with the idea that narrow functional measurements were driven by niche specialists (Fig. 2, central panel). The approach therefore generated predictions of which phylotypes in the 'rare biosphere' facilitate specific pathways21.
The large number of significant functional interactions among phylotypes (364 of 368 significant interactions) suggested that specific metabolic pathways were maintained by collaborations among rare phylotypes18. Positive interactions (e.g. cross-feeding) have been known to emerge over short evolutionary timescales in simplified communities22. The narrow functions used here measure the capacity of the communities to produce secreted exo-enzymes that are publicly available, and which might provide a similar mechanism for positive interactions23. Positive functional interactions were less common for the broad functional measurements (131 of 265 significant interactions), with negative interactions concentrated among the most common phylotypes (Fig. 2, central panel). This result is compatible with culture-based studies using isolated bacteria: isolates obtained from the same study system showed a strong tendency for negative functional interactions, and were also common phylotypes in our 16S gene libraries24,25. Within the limitations of the approach, the large number of positive interactions among rare phylotypes have not previously been documented, and could play a key role in generating hypotheses about how rare taxa impact functional processes.
The functional interactions uncovered here are correlations, and should be viewed as hypotheses that require independent validation using experiments that isolate each pairwise interaction. In practice, validation experiments face significant challenges because many of the functional interactions are between phylotypes that have not previously been isolated in pure culture, and because the functional interactions might be contingent on the surrounding community. An advantage of using frozen, archived communities is that it is possible to perform post hoc 'community mixture' experiments to validate the correlational results. We mixed communities in microcosms in order to place potentially interacting phylotypes in contact. We determined whether the functional interactions we observed (Figure 2) had a biological basis by mixing communities that each contained one member of the interacting pair. Mixtures of communities that placed interacting phylotypes together should increase (positive interaction) or decrease (negative interaction) the functional measurement relative to the mean functional measurement associated with each of the individual communities.
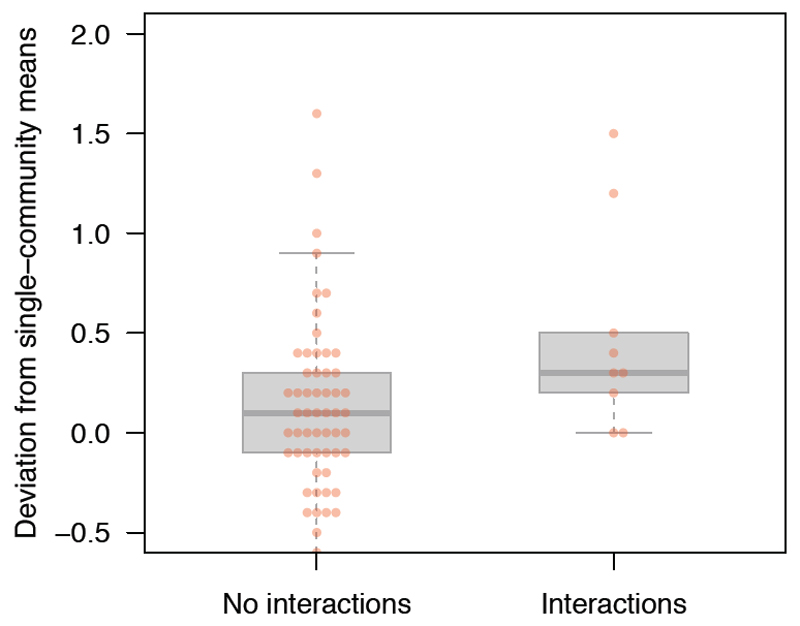
We resuscitated 12 communities to validate the positive functional interactions that we observed in the production of hemicellulase. We measured hemicellulase production in microcosms containing each of the communities on its own and all pairwise mixtures of the communities. Under a scenario of no interactions, the hemicellulase production would simply be the average of the constituent communities. We found that the results were consistent with the functional interactions we observed in Figure 2: community mixtures that placed positively-interacting phylotypes together showed a significantly elevated hemicellulase activity (Figure 3). The result implies that the functional interactions (Figure 2) resulted from direct (causal) interactions, but further work would be needed to identify mechanisms.
Figure 3. Validation of the functional interactions using community mixture experiments.
When communities were mixed together, hemicellulase activity was expected to be the mean activity of the two communities in the mixture. The y-axis is the deviation from this expectation, with positive values indicating that hemicellulase activity in the mixed communities exceeded the mean of the two constituent communities. Hemicellulase activity was elevated in pairwise mixtures of communities that placed interacting phylotypes together (“Interactions”, n = 9 mixtures) but not in communities that did not place interacting phylotypes together (“No interactions”, n = 56 mixtures) (analysis-of-variance, F1,63 = 5.1, p = 0.027). The boxes show the data quartiles, with the dark grey line indicating the mean. Data points are individual mixtures average across n = 4 replicates. Mixture were created from 12 communities.
There are important constraints to the approach described here. Notably, all communities must be tested in a common environment. Since the associations and functional interactions likely depend on environmental conditions (e.g. the abundance and complexity of the available resources), the associations between the phylotypes and the functional measurements in the microcosms might differ from those in nature. Although we found a good correspondence between community composition in the microcosms and in the natural community (Supplementary Figure 3), important changes to community structure also arose due to the experimental manipulations (freezing, fungicide addition). The simple analyses presented here could also be improved and extended. For example, we used linear relationships, but non-linear relationships are common. We also applied a conservative p-value correction, likely resulting in many false negatives. We also tested associations for every phylotype, whereas dimensionality reduction (e.g. by clustering phylotypes with similar abundance profiles across the communities) followed by multivariate analysis could yield models that better predict functioning. Here we have presented the simplest possible analysis, and expect that further studies using more complex analyses may uncover further interesting biology.
The results here are among the first to document strong relationships between structure and function in complex, non-synthetic communities under controlled conditions. Natural bacterial communities have previously been painted as world of vast functional redundancy, where high levels of niche overlap among phylotypes buffer ecosystem functioning against extinction26. It is clear from our results that the abundance of phylotypes is significantly associated with a range of functional measurements, both through their direct effects and through their interactions with other phylotypes. In both environmental microbiology and medical microbiology, there is a recognition that even those diseases or phenotypes that are caused by individual bacterial strains are mediated by complex interactions with many other taxa27. Application of the common garden method can provide an important window on the functional role phylotypes, and could in the future provide a method for unravelling the complex interactions among the thousands of phylotypes that inhabit natural environments.
Methods
Microbiome collection
We sampled 753 beech (Fagus sp.) rainwater-filled tree-holes during August 2013 to April 2014 from locations across the south of England, primarily located in the Chilterns west of London. These miniature aquatic habitats have been used extensively as 'natural microcosms'28 that house diverse and accessible microbial communities. The water from each tree-hole was homogenised by stirring, after which we collected a 1 ml sample, which was kept at ambient temperature until the samples were returned to the laboratory (<24 hours). Each sample was diluted 1:4 in sterile phosphate buffered saline (pH 7.0, Sigma-Aldrich) prior to filtration (pore size 20-22 µm, Whatman 4 filter paper) to remove debris and larIncreased ge organisms. The filtrate containing the communities was used to inoculate 5 ml into a sterile beech leaf medium supplemented with 200 µg ml-1 cyclohexamide (Sigma-Aldrich) to inhibit fungi. Although fungi were excluded here in order to simplify the communities, fungi are likely to be important decomposers in this ecosystem, and would therefore be useful additions to future studies. Beech leaf medium was created by autoclaving 50 g of dried beech leaves in 500 ml of PBS29, which gave a concentrated stock after filtration of coarse particles. Beech leaf medium is composed of a complex array of carbon sources, which are exploited to differing degrees by bacteria isolated from the tree holes29. Each microcosm (polypropylene centrifuge tube) was incubated at 22°C under static conditions for one week to allow communities to reach stationary phase (Supplementary Figure S4). Each regrown community was stored at -80°C after addition of freezing solution (final concentration 30% v/v glycerol and 0.85% w/v NaCl). Communities were stored frozen so as to allow repeatable experiments using the same starting community compositions. Community composition of the frozen communities was assessed using Illumina MiSeq (250bp-paired end) sequencing performed by Molecular Research DNA (www.mrdnalab.com). The V4 region of the 16S rRNA gene was amplified, using primers 515f/806r with the forward primer barcoded. Sequences were curated using a propriety analysis pipeline by Molecular Research DNA; any sequences <150bp and those with ambiguous base calls removed, prior to denoising and editing for chimeras. Operational taxonomic units were specified at a 97% similarity cut-off, which we refer to as 'phylotypes' in the text30. We randomly sampled 15,000 sequences per sample to normalise sequencing effort. We used the number of reads per phylotype as a measure of their relative abundance in the community. Although many biases are introduced during the DNA extraction and PCR steps, these biases would be applied equally across the experiment. Rarefaction curves indicated that we had sampled most of the diversity going into the microcosms (Supplementary Figure S5), and extrapolating to 20,000 sequencing reads31 (number of cells inoculated into each microcosm) indicated that we captured on average 95% of the total phylotype richness in the samples. We used a microbial community standard (Zymo Research) as a positive control to assess the degree to which our 16S rRNA gene amplicon sequencing produced biased estimates of abundance. We found good correspondence between observed and expected abundances for most classes of bacteria, but Enterococcaceae and Listeriaceae were considerably under-represented, while over-Enterobacteriaceae was over-represented (Supplementary Figure S6).
Common garden experiment
Microcosms were established in 1.2 ml deep-96-well plates containing 840 µl sterile beech leaf medium and inoculated with 40 µl of each revived (thawed) community (4 replicates per community, 3172 microcosms in total). Cell densities within the communities were consistent across samples (mean 4.9×105 cells ml-1 ± 2.1×104 s.e.m), thus each microcosm was initiated with on average 19,600 cells. The sequencing effort (15,000 reads per sample) was similar to the number of cells used to initiate the microcosms, so we assumed the communities were almost fully characterised. We multiplied the relative abundances obtained from the sequencing by the initial cell numbers to obtain an estimate of absolute abundance of each phylotype at the beginning of the experiment. Although there are known biases in using amplicon sequencing data to measure relative abundance (e.g. due to PCR conditions or DNA extraction methods), we expected those biases to be the same across the microcosms. The microcosms were incubated under static conditions at 22°C for 7 days, after which we quantified each of the measurements of ecosystem functioning. Our intention was not to mimic the precise conditions of their native environment since those conditions differed among the communities. However, the microcosms successfully re-created an environment that produced both a standing density (mean 1.7 ×105 cells ml-1 ± 6.9×103 s.e.m) and communities (Supplementary Figure S4) that were similar to the native environment. We also tracked cell densities over 12 days in 32 randomly chosen communities (Supplementary Figure S4). These data showed that all of the communities had reached carrying capacity well before 7 days, implying that there would have been competition both for labile substrates (typically used during growth phase) and for more recalcitrant substrates (typically important later in succession).
We categorised community respiration, cell yield, and metabolic potential as 'broad' functional measurements. Bacterial respiration was measured using the MicroResp CO2 detection system (www.microresp.com) according to the manufacturer instructions with absorbance readings converted to weight of CO2 using a linear log-log relationship (R2 = 0.965; Supplementary Figure S7). Respiration measurements were taken as the cumulative respiration of the whole community over the 7-day incubation period. Yield was the final abundance of all bacterial cells in the community, which was quantified by staining the cells with thiazole orange (42 nM, Sigma-Aldrich) followed by obtaining absolute counts using a C6 Accuri flow cytometer (size threshold of 8000 FSC-H), with cells gated on the SSC-A and FL1-A (533/30) channels. We used a threshold of 800 fluorescence units to distinguish cells from detritus. Potential metabolic activity was measured as the adenosine triphosphate (ATP) concentration within the community, measured using a Biotek Synergy 2 multimode plate reader and the BacTitr-Glo Cell Viability assay (Promega). A linear relationship was observed between concentration and luminescence (R2 = 0.998), therefore values were converted to nM ATP.
We categorised the degradation of specific substrates as 'narrow' functional measurements. We measured the breakdown of substrates labelled with 4-methylumbelliferone (MUB)32. Production of the appropriate exo-enzyme within the community induces a fluorescent signal that was quantified on a Biotek Synergy 2 multimode reader (Ex/Em: 365 nm/ 445 nm). Samples were incubated with 40 µM of the substrates (100 μl total volume) and incubated in the dark under the same conditions as the microcosms (static, 22°C) for 60 minutes. After the incubation, 10 µl of 1M NaOH was added and the fluorescence measured over four minutes with the maximum value recorded. Fluorescent values were converted to nM MUB after establishing a linear relationship between MUB concentration and fluorescence (R2 = 0.996) and using negative controls to account for any auto-fluorescence in the medium. The assays quantified the capacity of the communities to degrade the substrates within the microcosms rather than the actual rates at which the substrates were being cleaved in situ. Since the medium was derived from beech leaf litter, we selected substrates that were targeted by enzymes associated with organic matter degradation, including xylosidase (cleaves the labile substrate xylose, a monomer prevalent in hemicellulose), β-chitinase (breaks down chitin, which is the main component of arthropod exoskeletons and fungal cell walls), β-glucosidase (break down cellulose, the structural component of plants), and phosphatase (break down organic monoesters for the mineralisation and acquisition of phosphorus).
Common garden analysis
We used linear regressions to relate the functional measurements to phylotype abundance across the sites. For simplicity, we averaged the functional measurements across the 4 replicates, which yielded functional measurements from 753 communities. We excluded phylotypes from the analysis that were rare (<100 individuals across all samples) or that only occurred (abundance > 0) in <10 samples, which reduced the number of phylotypes in the analysis from 1341 to 522. These rarest phylotypes were excluded because individual data points frequently had high leverage in the regressions, and because pairwise interactions were undefined because there was no covariation in abundances between rare phylotypes. One-way associations: We performed linear regressions between the initial absolute abundance of every phylotype and every functional measurement: y = b0 + b1log10(s1+1), where y is the functional measurement, b0 is the intercept, b1 is the slope associated with phylotype 1, and s1 is the absolute abundance (number of cells per microcosm) of phylotype 1 at the start of the experiment.We used the significance of the slope as an indication of whether the phylotype was associated with the functional measurement. P-values were corrected for multiple testing across all of the one-way-analyses using a Bonferronni correction, yielding a threshold p-value of 1.4 ×10-5. Two-way functional interactions: We performed linear regressions that related the functional measurements to every pair of phylotypes: y = b0 + b1s1 + b2s2 + b3(s1×s2), where y is the functional measurement, b0 is the intercept, b1 and b2 are the slopes (coefficients) associated with phylotype 1 (s1) and phylotype 2 (s2), and b3 is the coefficient associated with the interaction between phylotype 1 and phylotype 2 (s1×s2). We used b3 as an estimate of the 'functional interaction' between each pair of phylotypes. P-values associated with b3 were corrected for multiple testing using a Bonferroni correction, yielding a significance threshold of 5.3 × 10-8. Significant functional interactions imply that correlated changes in pairs of phylotype abundance are directly linked to changes in functioning, but such causal links would need to be verified with experiments.
Validation experiment
We revived (thawed) 12 communities to validate the functional interactions. We replicated the common garden experiment (above), by dispensing 40 μl of community in to 840 μl of sterile beech leaf medium. The 40 μl of community contained either a single community, or a 50:50 mixture of two communities. When mixing communities, many phylotypes would be simultaneously put into contact. This was particularly problematic for the broad functions, since our results indicated that community mixtures would bring together both positively- and negatively-interacting phylotypes. We therefore focused on hemicellulase production because our analysis indicated that all of the significant functional interactions were positive, making qualitative predictions straightforward. Communities were incubated at 22°C for 7 days after which the hemicellulase activity was recorded as described above. We categorised the communities according to whether they contained none, one, or both interacting phylotypes from among any of the significantly interacting phylotype pairs (Fig. 2, central panel). Mixed communities that placed two interacting phylotypes together that were not found together in either of the constituent communities would be expected to 'realise' their functional interaction. For the communities we examined, the following significantly-interacting phylotypes pairs were combined: Deinococcus hohokamensis x Acidimicrobiales spp., Solibacter spp. x Aurantimonas manganoxydans, Solibacter spp. x Legionella spp., Deinococcus hohokamensis x Leptospirillum spp., Anaerosporobacter spp. x Pseudoclavibacter spp, Epilithonimonas lactis x Pseudoclavibacter spp. The phylotype pairs have not previously been implicated in hemicellulose degradation so far as we are aware.
There were 9 of 65 community mixtures that placed interacting phylotypes into contact (“Interactions”) whereas the remainder did not place interacting phylotypes into contact (“No interactions”). We tested the hypothesis that hemicellulase production exceeded what would be expected under a null model. We assumed that, in the absence of any new interactions, mixed communities should simply be the mean of the communities in the mixture. For all of the community mixtures, we therefore subtracted the mean hemicellulase production in each constituent single phylotype community from the observed hemicellulase production in the mixture. We conducted a t-test to determine whether the deviation of hemicellulase production from this null expectation was higher in mixed communities with “Interactions” than communities with “No interactions”.
Supplementary Material
Acknowledgements
The research was funded by a European Research Council starting grant (311399-Redundancy) awarded to TB. TB was also funded by a Royal Society University Research Fellowship. We are grateful for comments from T Barraclough and A Pascual Garcia.
Footnotes
Data availability
The data used in the analysis are available in the FigShare repository with digital object identifiers 10.6084/m9.figshare.6100181 (phylotype table) and 10.6084/m9.figshare.6100340 (functional data). Sequence data that support the findings of this study have been deposited in NCBI Short Read Archive with accession number SRP145037.
Author contributions. Research conceived by TB. Experimental procedures were undertaken by DWR. Analysis and writing by TB and DWR.
Competing interests. The authors declare no competing interests.
Additional information. Supplementary information is available for this paper.
References
- 1.Falkowski P, Fenchel T, Delong E. The microbial engines that drive Earth’s biogeochemical cycles. Science (80-. ) 2008;320:1034–1039. doi: 10.1126/science.1153213. [DOI] [PubMed] [Google Scholar]
- 2.Allison S, Martiny J. Resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci U S A. 2008;105 doi: 10.1073/pnas.0801925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strickland MS, Lauber C, Fierer N, Bradford MA. Testing the functional significance of microbial community composition. Ecology. 2009;90:441–451. doi: 10.1890/08-0296.1. [DOI] [PubMed] [Google Scholar]
- 4.Graham DW, Smith VH. Designed ecosystem services: Application of ecological principles in wastewater treatment engineering. Front Ecol Environ. 2004;2:199–206. [Google Scholar]
- 5.Quince C, Curtis TP, Sloan WT. The rational exploration of microbial diversity. ISME J. 2008;2:997–1006. doi: 10.1038/ismej.2008.69. [DOI] [PubMed] [Google Scholar]
- 6.Finlay BJ, Maberly SC, Cooper JI. Microbial diversity and ecosystem function. Oikos. 1997;80:209. [Google Scholar]
- 7.Bell T, et al. Biodiversity, Ecosystem Functioning, and Human Wellbeing: An Ecological and Economic Perspective. 2009:121–133. doi: 10.1093/acprof:oso/9780199547951.003.0009. [DOI] [Google Scholar]
- 8.Embree M, Liu JK, Al-Bassam MM, Zengler K. Networks of energetic and metabolic interactions define dynamics in microbial communities. Proc Natl Acad Sci. 2015;112 doi: 10.1073/pnas.1506034112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding J, et al. Integrated metagenomics and network analysis of soil microbial community of the forest timberline. Sci Rep. 2015;5:7994. doi: 10.1038/srep07994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morales SE, Holben WE. Linking bacterial identities and ecosystem processes: can ‘omic’ analyses be more than the sum of their parts? FEMS Microbiol Ecol. 2011;75 doi: 10.1111/j.1574-6941.2010.00938.x. [DOI] [PubMed] [Google Scholar]
- 11.Gilbert JA, et al. Microbiome-wide association studies link dynamic microbial consortia to disease. Nature. 2016;535:94–103. doi: 10.1038/nature18850. [DOI] [PubMed] [Google Scholar]
- 12.Bell T, Newman JA, Silverman BW, Turner SL, Lilley AK. The contribution of species richness and composition to bacterial services. Nature. 2005;436:1157–60. doi: 10.1038/nature03891. [DOI] [PubMed] [Google Scholar]
- 13.Kitching RL. An ecological study of water-filled tree-holes and their position in the woodland ecosystem. J Anim Ecol. 1971;40:281. [Google Scholar]
- 14.Reed HE, Martiny JBH. Testing the functional significance of microbial composition in natural communities. FEMS Microbiol Ecol. 2007;62 doi: 10.1111/j.1574-6941.2007.00386.x. [DOI] [PubMed] [Google Scholar]
- 15.Bergelson J, Roux F. Towards identifying genes underlying ecologically relevant traits in Arabidopsis thaliana. Nat Rev Genet. 2010;11:867–879. doi: 10.1038/nrg2896. [DOI] [PubMed] [Google Scholar]
- 16.Schimel JP, Schaeffer SM. Microbial control over carbon cycling in soil. Front Microbiol. 2012;3:348. doi: 10.3389/fmicb.2012.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schimel J. Arctic and Alpine Biodiversity: patterns, causes, and ecosystem consequences. Springer; Berlin, Heidelberg: 1995. pp. 239–254. [DOI] [Google Scholar]
- 18.Peter H, et al. Function-specific response to depletion of microbial diversity. ISME J. 2011;5:351–361. doi: 10.1038/ismej.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coyte KZ, Schluter J, Foster KR. The ecology of the microbiome: Networks, competition, and stability. Science (80-. ) 2015;350 doi: 10.1126/science.aad2602. [DOI] [PubMed] [Google Scholar]
- 20.McCann K, Hastings A, Huxel GR. Weak trophic interactions and the balance of nature. Nature. 1998;395:794–798. [Google Scholar]
- 21.Lynch MDJ, Neufeld JD. Ecology and exploration of the rare biosphere. Nat Rev Microbiol. 2015;13:217–229. doi: 10.1038/nrmicro3400. [DOI] [PubMed] [Google Scholar]
- 22.Lawrence D, et al. Species interactions alter evolutionary responses to a novel environment. PLoS Biol. 2012;10:e1001330. doi: 10.1371/journal.pbio.1001330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allison SD. Cheaters, diffusion and nutrients constrain decomposition by microbial enzymes in spatially structured environments. Ecol Lett. 2005;8:626–635. [Google Scholar]
- 24.Rivett DW, et al. Resource-dependent attenuation of species interactions during bacterial succession. ISME J. 2016 doi: 10.1038/ismej.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foster KR, Bell T. Competition, not cooperation, dominates interactions among culturable microbial species. Curr Biol. 2012;22:1845–50. doi: 10.1016/j.cub.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 26.Fernández A, et al. How stable is stable? Function versus community composition. Appl Environ Microbiol. 1999;65:3697–704. doi: 10.1128/aem.65.8.3697-3704.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Byrd AL, Segre JA. Adapting Koch’s postulates. Science (80-. ) 2016;351 doi: 10.1126/science.aad6753. [DOI] [PubMed] [Google Scholar]
- 28.Srivastava DS, et al. Are natural microcosms useful model systems for ecology? Trends Ecol Evol. 2004;19:379–84. doi: 10.1016/j.tree.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 29.Lawrence D, et al. Species Interactions Alter Evolutionary Responses to a Novel Environment. PLoS Biol. 2012;10:e1001330. doi: 10.1371/journal.pbio.1001330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prosser JI. Ecosystem processes and interactions in a morass of diversity. FEMS Microbiol Ecol. 2012;81:507–519. doi: 10.1111/j.1574-6941.2012.01435.x. [DOI] [PubMed] [Google Scholar]
- 31.Hsieh TC, Ma KH, Chao A. iNEXT: an R package for rarefaction and extrapolation of species diversity (Hill numbers) Methods Ecol Evol. 2016;7:1451–1456. [Google Scholar]
- 32.Baldrian P. (A. V.). Microbial enzyme-catalyzed processes in soils and their analysis. A review. Plant, Soil Environ - UZEI (Czech Republic) 2009;55:370–378. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.