Abstract
Breast cancer cell invasion is influenced by growth factor concentration gradients in the tumor microenvironment. However, studying the influence of growth factor gradients on breast cancer cell invasion is challenging due to both the complexities of in vivo models and the difficulties in recapitulating the tumor microenvironment with defined gradients using in vitro models. A defined hyaluronic acid (HA)-based hydrogel crosslinked with matrix metalloproteinase (MMP) cleavable peptides and modified with multiphoton labile nitrodibenzofuran (NDBF) was synthesized to photochemically immobilize epidermal growth factor (EGF) gradients. We demonstrate that EGF gradients can differentially influence breast cancer cell invasion and drug response in cell lines with different EGF receptor (EGFR) expression levels. Photopatterned EGF gradients increase the invasion of moderate EGFR expressing MDA-MB-231 cells, reduce invasion of high EGFR expressing MDA-MB-468 cells, and have no effect on invasion of low EGFR-expressing MCF-7 cells. We evaluate MDA-MB-231 and MDA-MB-468 cell response to the clinically tested EGFR inhibitor, cetuximab. Interestingly, the cellular response to cetuximab is completely different on the EGF gradient hydrogels: cetuximab decreases MDA-MB-231 cell invasion but increases MDA-MB-468 cell invasion and cell number, thus demonstrating the importance of including cell-microenvironment interactions when evaluating drug targets.
Keywords: EGF, hydrogels, invasion, breast cancer, gradients
1. Introduction
Breast cancer cell invasion through the stroma is influenced by signals the breast cancer cells receive from the microenvironment, including the extracellular matrix (ECM), mechanical properties, stromal cells, and cytokines, among others. [1-5] Of particular interest are gradients of chemoattractants (often cytokines) that arise in the microenvironment and guide cell invasion. In vivo, breast cancer cells have been shown to respond to and migrate towards increasing epidermal growth factor (EGF) concentrations secreted by macrophages within the microenvironment. [6,7]
Epidermal growth factor receptor (EGFR) overexpression is associated with aggressive phenotypes, decreased patient survival, and occurs in approximately 6-19% of all breast cancers, and 30-52% of triple negative breast cancers – that is cells that are negative for estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2 (HER2). [8-12] EGF is a known mitogen and causes increased cell motility, invasion, and proliferation through the mitogen-activated protein kinase (MAPK) and phospatidylinositol-3 kinase (PI-3K) downstream signalling pathways. [13-16] However, EGF has also been shown to have an antitumorigenic effect on several EGFR overexpressing cancer cell lines including the triple negative breast cancer cell line MDA-MB-468 and the vulvar epidermoid cancer cell line A431, where exposure to high levels of EGF induces apoptosis. [17,18] It has been hypothesized that EGFR expression switches from a tumor promoting role in situ to a tumour inhibiting role in metastatic breast cancer. [19] It is therefore important to develop an in vitro model with EGF gradients to improve our understanding of the role of EGF and EGFR expression in breast cancer progression.
To study the role of EGF gradients on cancer cell invasion, a three-dimensional (3D) in vitro hydrogel model is needed: two-dimensional (2D) culture is inherently limited by a lack of matrix through which cells can invade and in vivo models are overly complex to study the role of defined gradients. Boyden chambers are widely used in invasion assays as the “gold standard”, but they produce poorly defined, transient gradients. Microfluidic devices have been used to form well-defined gradients; however, this strategy often lacks a matrix for cell invasion, forcing cells to migrate along hard plastic surfaces and poorly recapitulating invasion through native tissues. [14] Several studies have investigated the influence of EGF gradients on breast cancer cell invasion; however, migration was either along 2D surfaces or model compounds, such as dextran, were used as evidence of a relevant EGF gradient. [14,20,21] Truong et al. recently developed a microfluidic invasion platform to study breast cancer invasion through 3D matrices: SUM-159 breast cancer cells encapsulated in a hydrogel responded to a predicted EGF gradient with enhanced cell invasion and altered cellular morphology. [20] With this platform, diffusional gradients of EGF were predicted based on simulations with 10 kDa molecules – there was no direct characterization of the EGF gradients. [20] Wang et al. investigated EGF gradients with concentration slopes of 0.071, 0.143, and 0.286 ng mL−1 μm−1 to study MDA-MB-231 breast cancer cell migration, but did so along 2D surfaces that do not recapitulate invasion through ECM. [14] The current study is, to the best of our knowledge, the first to investigate the influence of well-characterized, immobilized EGF gradients on cell invasion in a 3D model.
3D hydrogels patterned with biomolecules in known concentrations and gradients have been previously designed. [22-28] For example, EGF gradients were patterned in hyaluronic acid (HA) hydrogels crosslinked with bismaleimide-poly(ethylene glycol) (PEG); however, these hydrogels did not allow cell invasion and matrix remodelling. To improve upon past hydrogel models, PEG crosslinkers were replaced by a matrix metalloproteinase (MMP) cleavable (GPQG↓IWGQ) peptide crosslinker (MMPx), which breast cancer cells can actively degrade through the expression of MMPs 1, 2, 3, 7, 8, 9. [29–31] Furthermore, bromohydroxy coumarin (Bhc) was used as a caging molecule to facilitate EGF photopatterning: two photon irradiation cleaves Bhc, exposing a thiol that can immobilize thiol-reactive EGF. [24] However, Bhc isomerizes to form a product that quenches reactive thiols when irradiated. [32,33] When Bhc is modified with a methyl group at the endocyclic 3 position (mBhc), photoisomerization is blocked, thereby improving photopatterning efficiency over Bhc. [32] Nitrodibenzofuran (NDBF), another photocaging molecule, does not undergo photoisomerization and has been shown to efficiently uncage thiol-containing peptides with one- and two-photon irradiation; [33] however, NDBF, Bhc, and mBhc have not been directly compared in terms of photo-uncaging within 3D hydrogels. Herein, we compare NDBF, Bhc, and mBhc, and demonstrate the superior photopatterning efficiency of NDBF.
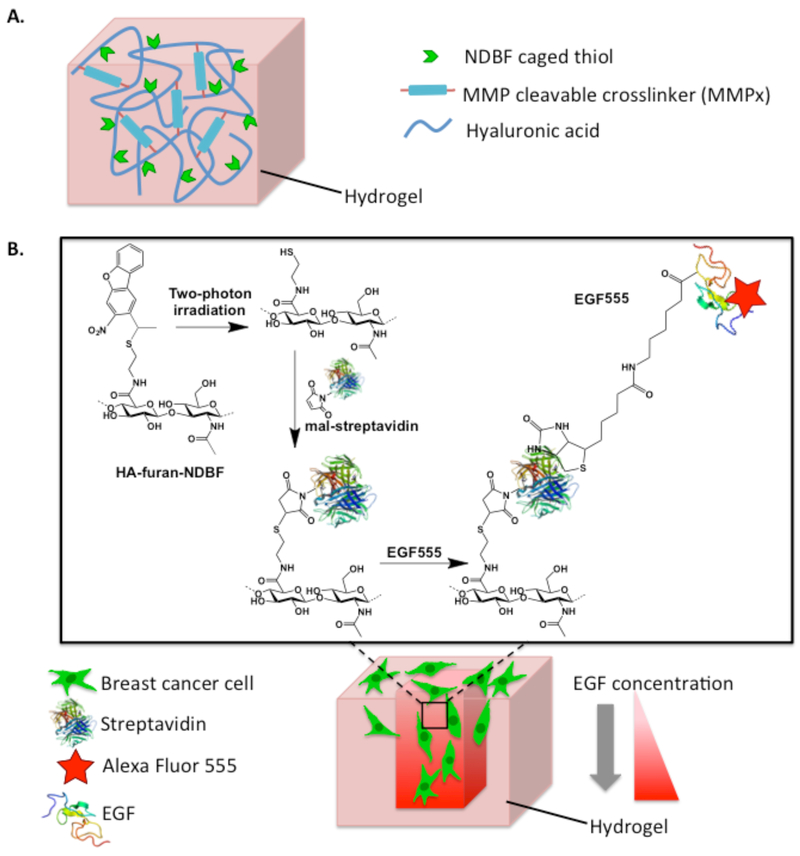
In the current study, a 3D in vitro breast cancer invasion platform was developed in NDBF-conjugated HA hydrogels crosslinked with MMPx (HANDBF/MMPx) (Figure 1). HA hydrogels are ideal in studies of breast cancer cell invasion because HA is often overexpressed in the breast cancer microenvironment and provides an inherently bioactive and degradable material. [35-40] Two-photon irradiation of HANDBF/MMPx cleaves NDBF, revealing a free thiol that subsequently reacts to immobilize maleimide-modified biomolecules into the hydrogel. To form gradients of EGF, maleimide-streptavidin (mal-streptavidin) is first patterned into HANDBF/MMPx. Adding biotinylated EGF modified with Alexa Fluor 555 (EGF555) for visualization of the patterns, results in EGF555 selectively binding to the immobilized streptavidin, forming a pattern of EGF555. Using this two-photon patterning approach, EGF gradients were formed in HANDBF/MMPx hydrogels, demonstrating the utility of NDBF for 3D photopatterning. These hydrogel platforms contain spatially defined EGF gradients that allow the role of EGF on breast cancer cell invasion to be studied.
Figure 1.
A) Furan modified hyaluronic acid (HA) is crosslinked with bis-maleimide, MMP cleavable peptide crosslinkers (MMPx) to form a hydrogel through a Diels-Alder Click reaction. The HA hydrogel backbone is also modified with nitrodibenzofuran (NDBF) caged thiols (HANDBF), which participate in the photopatterning reaction. B) Schematic diagram depicting photopatterning of HANDBF/MMPx hydrogels and subsequent breast cancer cell invasion. Two-photon irradiation of NDBF uncages a reactive thiol, with the concentration of the free thiol proportional to the number of two-photon scans. The free thiol then reacts with maleimide-streptavidin (mal-streptavidin), forming immobilized streptavidin patterns. Biotinylated EGF, modified with Alexa Fluor 555 for visualization (EGF555), binds to the immobilized streptavidin to create EGF gradients. Streptavidin structure obtained from Baugh et al. through the Protein Data Bank). [34]
The HANDBF/MMPx hydrogels containing EGF555 gradients are used to evaluate the response of breast cancer cell lines with different EGFR expression levels: MDA-MB-231, MDA-MB-468, and MCF-7 cell lines. MDA-MB-231 breast cancer cells are a highly invasive, triple negative breast cancer cell line that expresses EGFR. MDA-MB-468 breast cancer cells are an invasive, triple negative breast cancer cell line, with an intermediate invasive capacity that is lower than MDA-MB-231 cells but with very high EGFR expression. MCF-7 cells are a luminal A breast cancer cell line with low invasive potential that express low levels of EGFR. [41,42] We subsequently evaluate the response of the EGFR expressing breast cancer cells (MDA-MB-231 and MDA-MB-468) to the EGFR inhibitor, cetuximab. Cetuximab binds to the extracellular domain of EGFR at a higher affinity than EGF, preventing ligand binding and phosphorylation of the receptor. [43-45] Triple negative breast cancers tend to have the highest proportion of EGFR overexpression out of the breast cancer subtypes. [9-12,46] Since there are currently no hormone or receptor targeted treatment options for triple negative breast cancer, EGFR inhibitors have been evaluated clinically. [47] However, these EGFR inhibitor clinical trials have been unsuccessful; EGFR expression has not been predictive of patient response and EGFR activity has only been blocked in a minority of patients with EGFR expressing breast cancer. [48-50] Using breast cancer cell lines with different EGFR expression levels, we demonstrate how the same model microenvironment containing EGF gradients has dramatically different outcomes on breast cancer cell invasion and cetuximab response.
2. Materials and Methods
2.1. Materials
All reagents were used as received unless otherwise indicated. Lyophilized sodium hyaluronate (HA) was purchased from Lifecore Biomedical (2.15 × 105 g mol−1) (Chaska, MN, USA). Dimethyl sulfoxide (DMSO), triisopropyl silane (TIS), 4-(4,6-dimethoxy-1,3,5-triazin-2-yl)-4-methylmorpholinium chloride (DMTMM), N,N’-diisopropylcarbodiimide (DIC), 1-methyl-2-pyrrolidinone (NMP), furfurylamine, and Dulbecco’s phosphate buffered saline (PBS) were purchased from Sigma-Aldrich (St. Louis, MO, USA). N-(2-hydroxyethyl)maleimide was purchased from Strem Chemicals (Newburyport, MA, USA). Sodium chloride and 2-(N-mortholino)ethanesulfonic acid (MES) were purchased from BioShop Canada Inc. (Burlington, ON, Canada). Trifluoroacetic acid (TFA) and all solvents, unless otherwise specified, were purchased from Caledon Laboratory Chemicals (Georgetown, ON, Canada). Fmoc-protected amino acids and Wang resin were purchased from AnaSpec (Fremont, CA, USA). 3-Maleimidopropionic acid was purchased from Toronto Research Chemicals (Toronto, ON, Canada). Dialysis membranes were purchased from Spectrum Laboratories (Rancho Dominguez, CA, USA). Eight-well chamber slides (Nunc), optical glass bottom 96-well plates (Nunc), biocytin Alexa Fluor 546, Alexa Fluor 546 C5 maleimide (mal-546), Alexa Fluor 555 hydrazide, EZ-Link NHS-LC-biotin, maleimide streptavidin (mal-streptavidin), and sulfo-succinimidyl 4-[N-maleimidomethyl]cyclohexane-1-carboxylate (sulfo-SMCC) were purchased from Thermo Scientific (Waltham, MA, USA). EGF was purchased from Peprotech (Rocky Hill, NJ, USA). Nitrodibenzofuran (NDBF) was supplied by the Distefano laboratory. Distilled deionized water (ddH2O) was prepared using a Millipore Milli-RO 10 Plus and Milli-Q UF Plus at 18 M Ω resistance (Millipore, Bedford, MA, USA).
2.2. Synthesis of NDBF-cysteamine
NDBF-Br was synthesized as previously described. [33] NDBF-Br (0.40 g, 1.2 mmol), 2-(Boc-amino)ethanethiol (0.32 mL, 1.8 mmol), and DBU (1.24 mL, 1.6 mmol) were dissolved in THF and stirred at reflux. After 4 h the reaction was judged complete by TLC (2:1, Hexanes/EtOAc). After solvent removal via evaporation, the residue was extracted into 75 mL ethyl acetate from 50 mL of water. The organic layer was dried over Na2SO4 and evaporated under vacuum to afford Boc-protected NDBF-cysteamine. The resulting crude material was used directly for the following step, without further purification.
Crude Boc-protected NDBF-cysteamine was dissolved in 25 mL of a mixture of CH2Cl2 and trifluoroacetic acid (1:1, v/v) followed by addition of 0.5 mL of H2O. The mixture was stirred for 30 min followed by solvent removal under vacuum. The residue was dissolved in 50 mL of CH2Cl2 and washed with 25 mL H2O, 25 mL brine, and dried over Na2SO4. The CH2Cl2 was evaporated under vacuum and the resulting compound was purified using column chromatography (1:1 Hexanes:EtOAc) to afford the pure product (0.47 g, 70%). NDBF-cysteamine was characterized via 1H NMR in DMSO-d6 using an Agilent DD2-500 MHz NMR spectrometer (Santa Clara, CA, USA), as shown in Supplementary Data Figure S1. 1H NMR (500 MHz, DMSO-d6) δ 8.64 (s, 1H), 8.39 – 8.36 (m, 2H), 7.82 – 7.80 (m, 1H), 7.68 – 7.65 (m, 1H), 7.54 – 7.50 (m, 1H), 4.71 (q, J = 6.9 Hz, 1H), 2.71 – 2.67 (m, 2H), 2.60 – 2.54 (m, 2H), 1.72 (d, J = 6.9 Hz, 3H). HR-MS (ES) m/z calculated for [M+H]+ 317.0954, found 317.0976.
2.3. Synthesis of HA-Furan-NDBF
HA-furan was prepared as previously described. [24,29,40] Briefly, HA-furan was first synthesized by dissolving HA in MES buffer (100 mM, pH 5.5) to achieve 1% w/v HA. DMTMM was then added, followed by the dropwise addition of furfurylamine. The reaction was stirred at room temperature for 24 h and dialyzed (12-14 kDa MWCO) against NaCl (0.1 M) for 2 d followed by dH2O for 2 d. Furan substitution of the lyophilized HA-furan was confirmed via 1H NMR in D2O on an Agilent DD2-500 MHz NMR spectrometer (Santa Clara, CA, USA). The ratio of the area under the furan proton peaks (6.26, 6.46, and 7.65 ppm) to the area under the peak for N-acetyl glucosamine protons of HA (1.9 ppm) determined the degree of furan substitution. [40] For this current study a furan substitution of 46% was used.
To synthesize HA-furan-NDBF, HA-furan was dissolved in NMP:MES (100 mM, pH 5.5) at a ratio of 1:1 to achieve 0.5% w/v HA-furan. DMTMM was then added (2.5 molar equivalence relative to free carboxylic acids) followed by the dropwise addition of a solution of NDBF-cysteamine in DMSO (0.5 molar equivalence relative to free carboxylic acids). The reaction was stirred at room temperature in the dark for 24 h and then dialyzed against dH2O:NMP:DMSO at a volume ratio of 2:1:1 for 1 d. The organic fraction of the solution was halved every 24 h for 3 d before being replaced with only dH2O for the final 2 d. HA-furan-NDBF was lyophilized and the NDBF substitution was confirmed via 1H NMR in DMSO-d6 on an Agilent DD2-500 MHz NMR spectrometer (Santa Clara, CA, USA). NDBF substitution was determined to be 44% by comparing the integrated areas under the aromatic proton peaks of NDBF (8.68 – 8.13 ppm) to the furan peaks at 6.38 – 6.29 ppm and N-acetyl glucosamine peak of HA at 1.79 ppm (Supplementary Data Figure S2). HA-furan-NDBF is herein referred to as HANDBF.
HA-furan conjugated with either bromohydroxy coumarin (Bhc) or bromohydroxy methylcoumarin (mBhc) was prepared using the same method as HANDBF, [32] where NDBF-cysteamine was substituted with either Bhc-cysteamine or mBhc-cysteamine.
2.4. Synthesis of Maleimide Functionalized Peptides
MMP cleavable peptide crosslinkers (MMPx) were synthesized as previously described. [29] Briefly, peptides (Supplementary Data Figure S3) were synthesized using standard Fmoc chemistry on a Liberty 1 peptide synthesizer equipped with a Discover microwave reactor (CEM, Matthews, NC, USA). Following final deprotection of the N-terminus, 3-maleimidopropionic acid was coupled to the free amines with DIC, adding maleimide functionalities to the peptide. The MMPx peptides were then cleaved from the Wang resin with a solution of 80% TFA, 10% dH2O, and 10% TIS, precipitated with ice-cold ethyl ether, and purified with high-performance liquid chromatography (HPLC).
2.5. Synthesis of Functionalized EGF: EGF555
EGF was functionalized with biotin by reacting of EGF with EZ-Link NHS-LC-biotin in MES buffer (100 mM, pH 6). Every 2 h, 10 molar equivalence of EZ-Link NHS-LC-biotin (relative to free amines on EGF) was added to the reaction until 100 molar equivalence was reached. The reaction was stirred overnight at 4 °C and dialyzed (2 kDa MWCO) against PBS for 4 d at 4 °C. The biotin functionalization of EGF (biotinEGF) was confirmed via mass spectrometry (Supplementary Data Figure S4). The biotinEGF was further functionalized with Alexa Fluor 555 in order to visualize the EGF patterns within the hydrogel. BiotinEGF was reacted with Alexa Fluor 555 hydrazide (25 molar equivalence relative to EGF) in MES buffer (100 mM, pH 5.5) overnight, purified via FPLC, and concentrated to 9.89 μg mL−1. The average degree of Alexa Fluor 555 substitution was determined to be 6.756 (Alexa Fluor 555/EGF) by measuring the EGF concentration via micro BCA assay and the Alexa Fluor 555 concentration via UV absorbance. EGF modified with biotin and Alexa Fluor 555 is herein referred to as EGF555.
2.6. Preparation of HANDBF Hydrogels for Photopatterning
HANDBF was dissolved overnight in MES (100 mM, pH 5.5):DMSO (1:1) and mixed with an equal volume solution of either bis-maleimide peptide crosslinker (MMPx) or bis-maleimide-poly(ethylene glycol) (PEG) dissolved in MES buffer (100 mM, pH 5.5):DMSO (1:1). The mixture was pipetted into either 8-well chamber slides (125 μL) or glass bottom 96-well plates (40 μL) and gelled overnight at 37 °C to form hydrogels with a final concentration of 1.0% HANDBF and a 1:1 ratio of furan:maleimide. The unreacted furans in the hydrogel were quenched with 30 mM N-(2-hydroxyethyl)maleimide in MES buffer (100 mM, pH 5.5) for 24 h at room temperature. The N-(2-hydroxyethyl)maleimide was washed from the gel with PBS (pH 6.8). Solutions of either maleimide Alexa Fluor 546 (mal-546, 50 μM) or mal-streptavidin (0.63 mg mL−1) in PBS (pH 6.8) were then soaked into the hydrogel overnight at 4 °C and excess supernatant was removed prior to photopatterning.
2.7. Photopatterning HANDBF Hydrogels
HA/PEG and HA/MMPx hydrogels were photopatterned on a Zeiss LSM710 META confocal microscope equipped with a Coherent Chameleon two-photon laser using a 10× objective. For patterning experiments, the two-photon laser was set to 740 nm with 30-38% power (1679 mW max power) and scan speeds ranging between 0.009 and 0.527 μm μs−1. Following patterning, gels were washed thoroughly with PBS. Hydrogels patterned with mal-streptavidin were further reacted overnight with either EGF555 or Alexa546. Unreacted EGF555 or Alexa546 was washed from the hydrogel, leaving patterns of immobilized EGF555 or Alexa546 in the hydrogels, respectively. Patterns were imaged on an Olympus Fluoview FV1000 confocal microscope with xy two-photon laser scans every 1-5 μm in the z direction. Imaged photopatterns were quantified on ImageJ against a standard curve of Alexa Fluor 555 to determine the concentration of immobilized EGF55.
2.8. Maintenance of Cancer Cell Lines
All cell lines were purchased from ATCC (Manassas, VA, USA). Cell lines were maintained in tissue culture flasks in an incubator (37 °C, 5% CO2, 95% humidity) using their corresponding growth medium: MDA-MB-231, MDA-MB-468 in RPMI-1640, and MCF-7 in Dulbecco’s modified Eagle’s medium Nutrient Mixture F-12 Ham. All cell culture media were supplemented with 10% FBS, penicillin (10 U mL−1), and streptomycin (10 μg mL−1). MCF-7 growth media was also supplemented with insulin (10 μg mL−1).
2.9. Flow Cytometry
The EGFR expression in MCF-7, MDA-MB-231 and MDA-MB-468 breast cancer cells was determined using flow cytometry. Breast cancer cells were first detached from cell culture flasks using Accutase and resuspended in 2% FBS in PBS (FACS buffer). Cells were then incubated with a primary anti-EGFR antibody (Abcam, ab30) in FACS buffer (10 μg mL−1) at room temperature for 1 h, and washed 3 times in FACS buffer. A secondary antibody (goat anti-mouse, Alexa Fluor 488, Invitrogen, A-11001) was added to the cell suspension (10 μg mL−1 in FACS buffer) and incubated for 30 min at room temperature. Cells were then washed 3 times in FACS buffer and resuspended in PBS. Unstained (no primary or secondary antibodies) and secondary antibody only controls were also prepared. Flow cytometry was performed on an Accuri C6 Cytometer (BD Biosciences) and analyzed with FlowJo software.
2.10. Quantification of Cell Invasion
HA/MMPx hydrogels (40 μL) in glass bottom 96-well plates were photopatterned with EGF555 gradients (0.7242, 0.8624, and 1.2074 ng mL−1 μm−1) and the unbound EGF555 was thoroughly washed from the gels with PBS before seeding 3×104 cancer cells per well on the hydrogel surface. Cells were cultured on the hydrogels for 6 d before being fixed with 4% paraformaldehyde and stained with phalloidin for cell cytoskeleton and DAPI for cell nuclei. The hydrogels were imaged on an Olympus Fluoview FV1000 confocal microscope with x-y two-photon laser (740 nm) scans every 5 μm in the z direction. For quantification of invasion and cell number, the position coordinate of each nuclei was determined using Imaris Bitplane. The depth of invasion was calculated as the depth occupied between the top and bottom nuclei within the pattern and normalized to control patterns of Alexa546. Cell number was quantified as the total number of nuclei within the pattern; this number includes both invading and non-invading cells. Cell number was represents the response of cells to gradients of EGF555 - either an increase in cell number due to proliferation or a decrease in cell number due to cell death. Non-patterned regions of the hydrogel were quantified by determining the invasion and cell number in a ROI of equal volume to the patterns.
For EGF inhibitor experiments with cetuximab, HA/MMPx hydrogels were patterned with either EGF555 or Alexa546 gradients using the same procedure used in the gradient invasion studies. Prior to seeding cells, the hydrogels were equilibrated with either 0 or 100 μg/mL of the EGF inhibitor cetuximab. Cells (3×104 cancer cells per well) were then seeded on the hydrogel surface with 0 or 100 μg mL−1 cetuximab and cultured for 6 d with media changes every 2 d. Cells were then fixed, stained, and analyzed using the same procedure outlined above for the invasion studies.
2.11. Statistical Analysis
All statistical analyses were performed using GraphPad Prism version 6 (GraphPad Software, San Diego, CA, USA, www.graphpad.com). Differences among 3 or more groups were assessed using a one-way or two-way ANOVA followed by Tukey’s post hoc corrections to identify statistical significance. The two-way ANOVA results indicate the overall effect of the gradient slope (low, medium, high, and non-patterned region) and the presence of EGF555 (EGF555 vs. Alexa546). Subsequent post hoc tests are depicted graphically and show significance within groups, such as the significance between medium EGF555 and medium Alexa546 gradients. Differences between two groups were assessed using an unpaired t-test. For all statistical analysis α was set at 0.05. Data is displayed with p values represented as *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001. Cell invasion and cell number were normalized to Alexa546 controls to remove potential convoluting factors, such as changes in hydrogel physical properties following irradiation. To normalize the data, the measured values for each experimental repeat were divided by the relevant Alexa546 control value; this gave a normalized value of 1 for the Alexa546 control groups.
3. Results
3.1. NDBF Improves Photopatterning Efficiency in HA Hydrogels
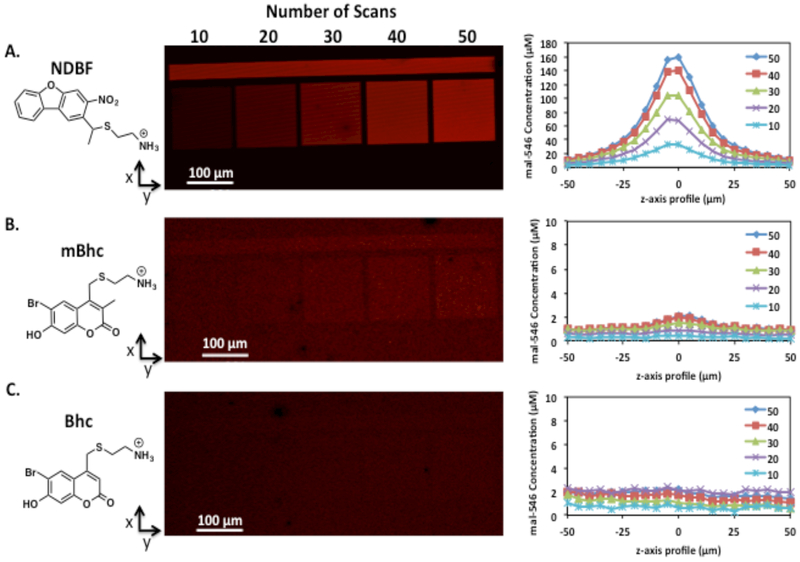
HA-furan hydrogels crosslinked with bis(maleimide)-PEG and conjugated with one of NDBF, Bhc, or mBhc (HANDBF/PEG, HABhc/PEG, and HAmBhc/PEG, respectively) were modified with maleimide-Alexa Fluor 546 (mal-546) to determine the optimal multiphoton labile molecule for photopatterning. Concentrations of NDBF, mBhc, and Bhc within the HA hydrogels were matched based on 1H NMR substitutions. Tiles were irradiated with a two-photon laser (740 nm) in an x-y plane within the hydrogels, with the number of two-photon laser scans varying between 10 and 50, as shown in Figure 2. The concentration of the immobilized mal-546 in the z-axis profile was quantified and the patterns were confirmed to be in a single x-y plane. Resolution of the patterned tiles in HANDBF hydrogels was approximately ± 25 μm from the centre of the pattern in the z-axis, while the x-axis profile of the patterned tiles revealed high resolution. Additionally, the concentration of immobilized mal-546 strongly correlates with the number of two-photon scans, and thus irradiation time of the HANDBF/PEG hydrogel (Supplementary Data Figure S5). The HA hydrogels can also be patterned with any shaped region of interest (ROI) (Supplementary Data Figure S6), but was limited to rectangular or circular planes or gradients for ease of analysis. Comparing mal-546 patterns in HANDBF, HAmBhc, and HABhc hydrogels in Figure 2, it was clear that HANDBF/PEG hydrogels had the highest patterning efficiency, where 50 scans immobilized 160 μM of mal-546. Patterns formed in HAmBhc hydrogels were far weaker than those in HANDBF hydrogels, where 50 scans immobilized 2 μM of mal-546, while patterns in HABhc were just visible above the baseline. Due to the superior photopatterning efficiency, NDBF- conjugated HA hydrogels were used for photopatterning in this study.
Figure 2.
Two-photon patterning of mal-546 using A) NDBF-, B) mBhc-, and C) Bhc- caged thiols in HA/PEG hydrogels. Regions of interest were scanned 10 to 50 times at a fixed z-dimension in HANDBF//PEG, HABhc//PEG, and HAmBhc/PEG hydrogels. Confocal images of the x-y planes are shown along with the z-axis profile of the mal-546 square patterns with the maximum intensity centred at 0 μm. Background mal-546 concentration was subtracted from the immobilized mal-546 concentration. The concentrations of NDBF, Bhc, and mBhc were matched based on 1H NMR substitution. The bar above the tiles was scanned 50 times to verify that the laser power was constant across the x-axis.
3.2. Photopatterned Gradients in HA Hydrogels
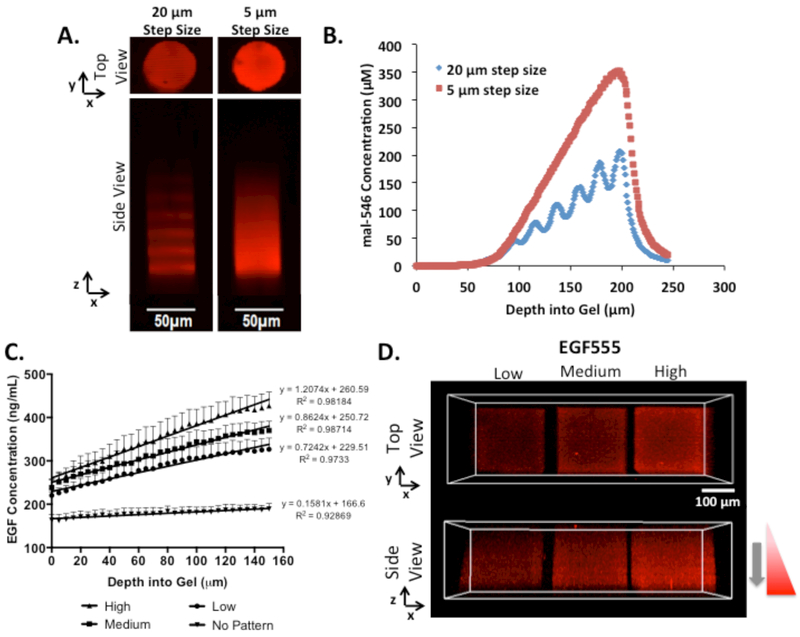
With increasing numbers of two-photon scans, there is increasing NDBF cleaved and hence more reactive thiols available for ultimate EGF immobilization. Thus, to form gradients within the HANDBF hydrogels, the number of two-photon scans increased from 1 scan at the hydrogel surface to 41 scans at a depth of 200 μm into the hydrogel, as shown in Figure 3A. To produce a smooth vs. discrete gradient, the interval step between each scanning plane was either 5 or 20 μm along the z-axis. Hence, for the 20 μm interval pattern, the number of scans was increased in increments of 4 scans per step whereas in the 5 μm interval pattern, the number of scans was increased in increments of 1 scan per step. The 5 μm interval pattern was ultimately irradiated for a greater amount of time, resulting in a steeper gradient and a higher concentration of mal-546 immobilized (Figure 3B). In the 20 μm scanning interval gradient, discrete scanning planes were visible, as shown in Figure 3A and B. Because a scanning interval step size of 5 μm resulted in a smooth gradient slope, this parameter was selected for patterning EGF gradients.
Figure 3.
A) Confocal image of mal-546 gradients in HANDBF/PEG with scanning intervals either 20 or 5 μm apart in the z-axis. Gradients were formed using a scan speed of 0.009 μm μs−1. Confocal images were enhanced for visualization. B) Quantification of mal-546 gradients in HANDBF/PEG with scanning intervals either 20 or 5 μm apart in the z-axis. Background mal-546 concentration was subtracted from the immobilized mal-546 concentration. C) Quantification of low, medium, and high (0.7242, 0.8624, and 1.2074 ng mL−1 μm−1, respectively) EGF555 gradients in HANDBF/MMPx hydrogels created at scan speeds of 0.527, 0.263, and 0.132 μm μs−1, respectively. At the surface of the hydrogel (0 μm) the hydrogel was scanned one time. The scan number was increased by one scan every 5 μm interval going into the hydrogel until a gradient of 150 μm in depth was formed. The amount of EGF555 in the non-patterned regions of the hydrogel was also quantified (0.1581 ng mL−1 μm−1). Slopes were found to be significantly different (****p<0.0001). Error bars displayed as SEM, n=4. D) Confocal images of low, medium, and high EGF555 gradients in HANDBF/MMPx hydrogels.
In order to form gradients of EGF, the free, photocleaved HA-thiols were first reacted with mal-streptavidin followed by the addition of biotinylated and Alexa Fluor 555-modified EGF (EGF555). Importantly, EGF555 showed similar bioactivity to soluble, unmodified EGF as determined by a cell proliferation assay: both EGF and EGF555 significantly increased MCF-7 cell number, with no significant difference between them (Supplementary Data Figure S7).
Three concentration gradients of immobilized EGF555 were synthesized. First HANDBF/MMPx hydrogels were exposed to varying two-photon laser scan speeds to achieve three different gradient slopes of reactive thiols that reacted immediately with mal-streptavidin already present in the gels. Slower scan speeds correspond to longer irradiation times, thus increasing the number of reactive thiols and creating steeper gradient slopes. Free mal-streptavidin that had not been immobilized into the hydrogel was then washed out, and EGF555 was added, binding selectively to the mal-streptavidin pattern. EGF555 gradients were formed in the HANDBF/MMPx hydrogels, as shown in Figure 3C and D, following the removal of unbound EGF555. The gradients of EGF555 were quantified and determined to have slopes of 0.72, 0.86, and 1.20 ng mL−1 μm−1, herein referred to as low, medium, and high, respectively. Importantly, these three gradients were determined to have significantly different slopes (****p<0.0001).
3.3. Breast Cancer Cells Respond Differently to EGF Gradients
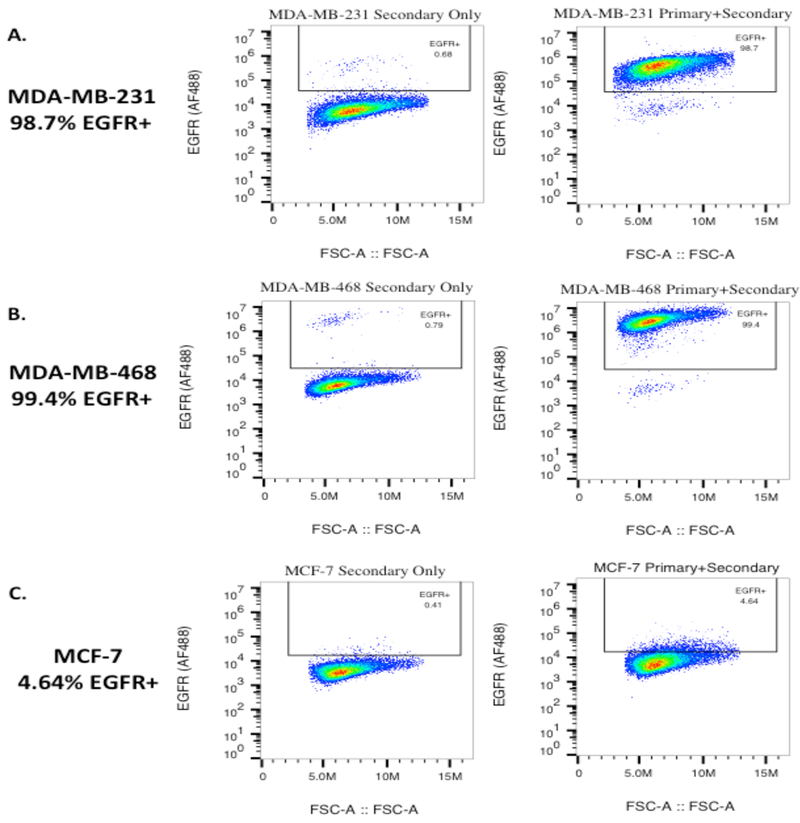
To test the response of breast cancer cells to the EGF555 gradients photopatterned into the HANDBF/MMPx hydrogels, breast cancer cells with different EGFR expression levels were used. EGFR expression in MDA-MB-231, MDA-MB-468, and MCF-7 breast cancer cell lines was determined using flow cytometry (Figure 4): 98.7% of the MDA-MB-231 population are EGFR+, expressing intermediate levels of EGFR, 99.4% of the MDA-MB-468 population are EGFR+, expressing high levels of EGFR, and 4.64% of the MCF-7 population express low levels of EGFR.
Figure 4.
Flow cytometry analysis of the expression of EGFR on: A) MDA-MB-231, B) MDA-MB-468, and C) MCF-7 breast cancer cell lines. The majority of the MDA-MB-231 and MDA-MB-468 cell populations were positive for EGFR (98.7% and 99.4%, respectively), with MDA-MB-468 cells having a greater EGFR staining intensity. MCF-7 cells were found to express EGFR in 4.64% of the population. The forward scatter is displayed on the x-axis.
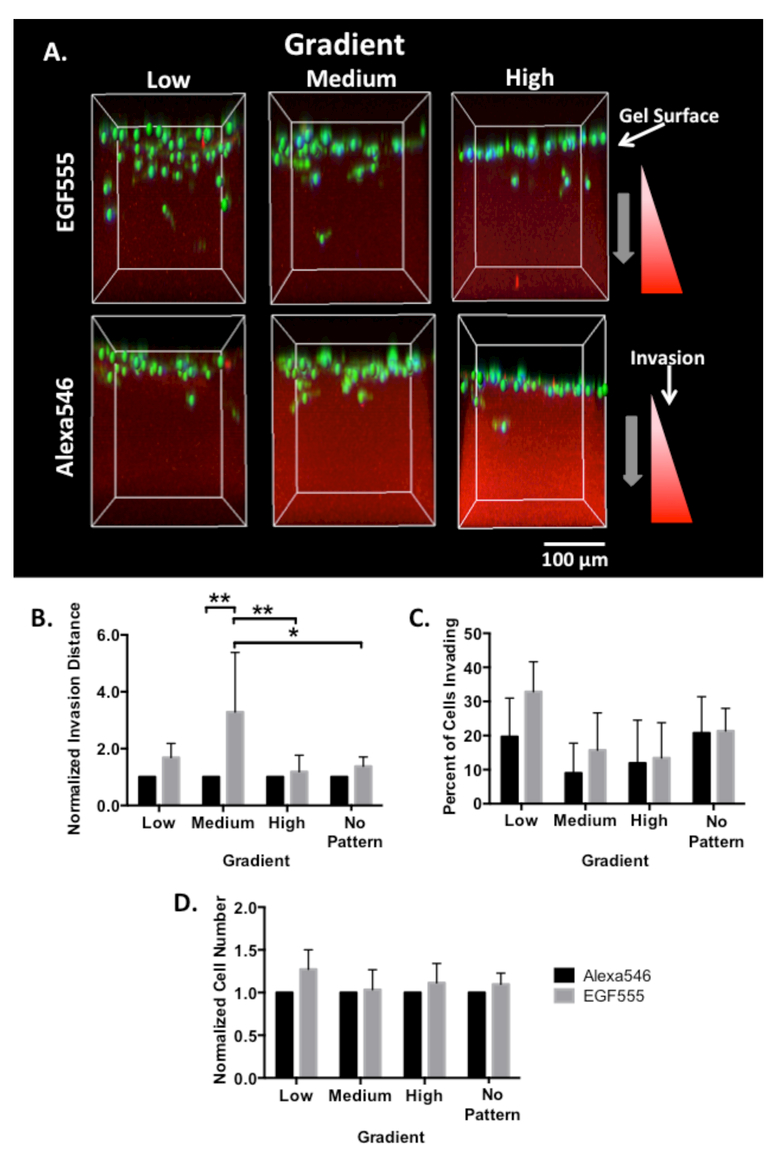
The EGF gradient invasion platform was tested with all three cell lines. The highly invasive MDA-MB-231 breast cancer cells seeded on the surface of each of the low, medium, and high EGF555 gradient hydrogels were compared to identical control gradients of simply biotinylated AlexaFluor 546 (Alexa546 – i.e., without the EGF). The Alexa546 gradients control for any changes in mechanical properties that may influence invasion. After 6 days of culture in low serum media, the cells were stained with DAPI to visualize nuclei and phalloidin to visualize the cytoskeleton. Cell invasion (the depth of position relative to the hydrogel surface), percent of invading cells, and total cell number within the gradient (including non-invading cells on the surface of the gradient) were measured using confocal z-stack images processed with IMARIS software, as shown in Figure 5A. To elucidate the influence of the EGF555 gradient on breast cancer cell invasion, the invasion distance in the EGF555 gradients was normalized to the invasion distance in the corresponding Alexa546 control gradients.
Figure 5.
A) Confocal reconstruction of MDA-MB-231 cells invading into HANDBF/MMPx hydrogels with low, medium, and high gradients of EGF555 and Alexa546. B) Normalized invasion distance of MDA-MB-231 cells in low, medium, and high gradients of Alexa546 and EGF555 and non-patterned regions of the HANDBF/MMPx hydrogel (day 6, n=5, mean + standard deviation). EGF555 significantly increased invasion compared to the control Alexa546 gradients (**p<0.01, two-way ANOVA). The slope of the gradient also significantly affected invasion (*p<0.05). Post hoc comparisons are shown graphically. C) Percent of invading MDA-MB-231 cells in low, medium, and high gradients of Alexa546 and EGF555. (day 6, n=5, mean + standard deviation). D) Normalized cell number of MDA-MB-231 cells in low, medium, and high gradients of Alexa546 and EGF555 and non-patterned regions of the HANDBF/MMPx hydrogel (day 6, n=5, mean + standard deviation).
EGF555 gradients significantly increased invasion distance relative to those of control Alexa546 using a two-way ANOVA (**p<0.001) and the slope of the gradient also significantly affected invasion (*p<0.05). Post hoc tests found that MDA-MB-231 invasion distance was significantly greater in the medium EGF555 gradients compared to: the medium Alexa546 control gradient (**p<0.001), the high EGF555 gradient (**p<0.001), and non-patterned regions of the EGF555 hydrogel (*p<0.05), as shown in Figure 5B. The slope of the gradient seemed to influence the percent of invading MDA-MB-231 cells, with fewer cells in the medium and high vs. low gradient hydrogels (Figure 5C). Since this was observed for both EGF555 and Alexa546 gradient gels, we hypothesize that this difference is due to increased mechanical constraints in the hydrogel following irradiation (Supplementary Data Figure S8). EGF555 was found to significantly increase MDA-MB-231 cell number (**p<0.001, two-way ANOVA); however, post hoc testing found no significance between groups (Figure 5D). In summary, it was observed that a greater number of MDA-MB-231 cells invaded the low EGF555 gradient compared to the medium EGF555 gradient. Additionally, there was no significant difference between the number of MDA-MB-231 cells that invaded the medium EGF555 gradient relative to the medium Alexa546 gradient; however, the cells invaded significantly further in the medium EGF555 gradient.
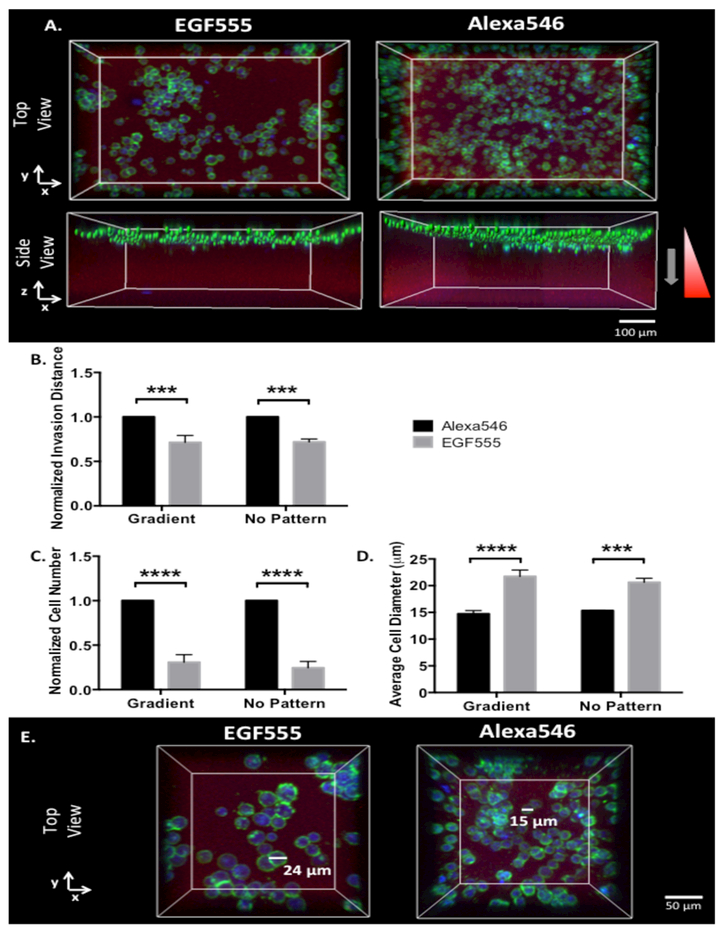
Since high concentrations of EGF can have an apoptotic effect on many EGFR overexpressing cancer cells, including MDA-MB-468 cells, [17,51,52] we studied their response to medium EGF555 gradients in the HANDBF/MMPx hydrogels as this gradient had the greatest impact on invasion distance of MDA-MB-231 cells. As shown in Figure 6A, the invasion distance of MDA-MB-468 cells in EGF555 gradient hydrogels was significantly decreased compared to their invasion distance in identical control gradients of Alexa546 (***p<0.001), as shown quantitatively in Figure 6B. Additionally, MDA-MB-468 cell number significantly decreased with EGF555 medium gradients compared to control Alexa546 gradients (****p<0.0001, Figure 6C). Interestingly, the diameter of the MDA-MB-468 cells increased when cultured on the medium gradient EGF555 hydrogels compared to the medium gradient of Alexa546 (****p<0.0001, Figure 6D, E). The invasion distance, cell number, and cell diameter of MDA-MB-468 cells had a similar response to EGF555 in non-patterned regions of the hydrogel, as well, indicating that the EGF itself, and not the gradient, had the dominant effect on MDA-MB-468 cellular response.
Figure 6.
A) Confocal reconstruction of MDA-MB-468 cells invading into HANDBF/MMPx hydrogels with medium gradients of EGF555 and Alexa546 B) Normalized invasion distance of MDA-MB-468 cells in medium gradients of Alexa546 and EGF555 and non-patterned regions of the HANDBF/MMPx hydrogel (day 6, n=3, mean + standard deviation). EGF555 significantly decreased MDA-MB-468 invasion distance compared to Alexa546 on both the gradient and non-patterned regions (****p<0.0001, two-way ANOVA). C) Normalized cell number of MDA-MB-468 cells in medium gradients of Alexa546 and EGF555 and non-patterned regions of the HANDBF/MMPx hydrogel (day 6, n=3, mean + standard deviation). EGF555 significantly decreased MDA-MB-468 cell number compared to Alexa546 on both the gradient and non-patterned regions (****p<0.0001, two-way ANOVA). D) Average cell diameter of MDA-MB-468 cells in medium gradients of Alexa546 and EGF555 and non-patterned regions of the HANDBF/MMPx hydrogel (day 6, n=3, mean + standard deviation). EGF555 significantly increased MDA-MB-468 cell diameter compared to Alexa546 on both the gradient and non-patterned regions (***p<0.001, two-way ANOVA). E) Representative images showing MDA-MB-468 cells with larger diameters when cultured on EGF555 gradients compared to the control Alexa546 gradients.
To confirm that the antitumorigenic effect of EGF is not due to the hydrogel platform, MDA-MB-468 cells were also treated with 20 ng mL−1 of unmodified, soluble EGF in 2D culture. After 5 days of culture, it was found that EGF significantly decreased MDA-MB-468 cell number (****p<0.0001) to similar levels seen in the EGF555 photopatterned HANDBF/MMPx hydrogels (Supplementary Data Figure S9).
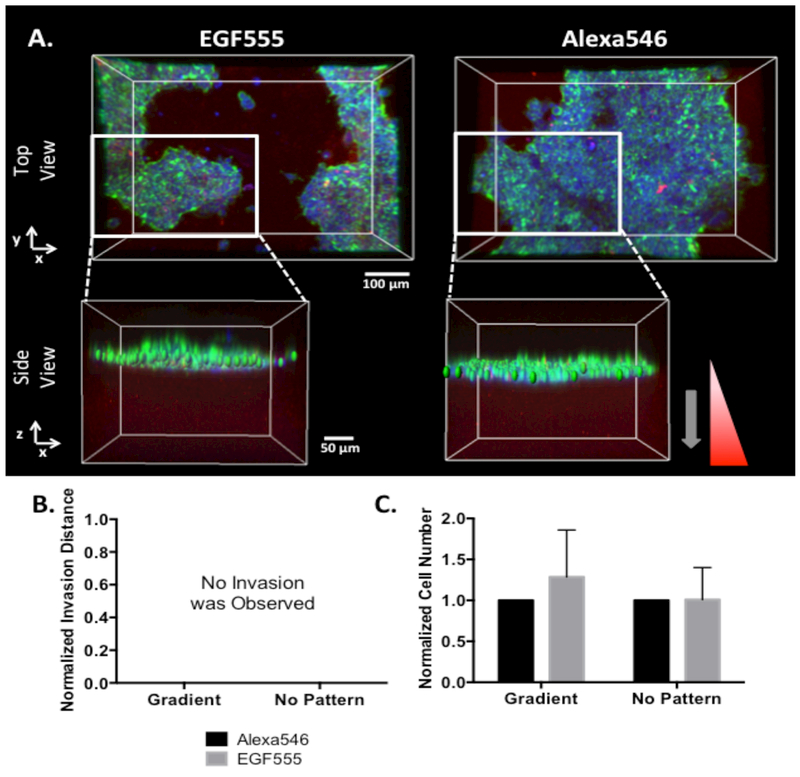
To examine the effect of EGF555 gradients on the low EGFR expressing MCF-7 breast cancer cells, MCF-7 cells were seeded on top of HANDBF/MMPx hydrogels containing medium gradients of EGF555. MCF-7 cells have low invasive potential and invaded into neither the medium EGF555 nor the Alexa546 control gradients, as shown in Figure 7A and B. No significant difference was found in MCF-7 cell number between the medium EGF555 and Alexa546 gradients (Figure 7C), demonstrating that EGF555 gradients do not influence MCF-7 cell migration. MCF-7 cell diameter was not measured since the cells grow in clusters on the hydrogel surface and it is difficult to distinguish the diameters of individual cells.
Figure 7.
A) Confocal reconstruction of MCF-7 cells cultured on HANDBF/MMPx hydrogels with medium gradients of EGF555 and Alexa546. Side views are zoomed in on a portion of the gradient so the MCF-7 cells can be clearly visualized as a cluster on the surface of the hydrogel. B) MCF-7 cells failed to invade into medium Alexa546 and EGF555 gradients after 6 days of culture (n=6). C) Normalized cell number of MCF-7 cells in medium gradients of Alexa546 and EGF555 and non-patterned regions of the HANDBF/MMPx hydrogel (day 6, n=6, mean + standard deviation). EGF555 had no significant influence on MCF-7 cell number (two-way ANOVA).
3.4. Breast Cancer Cells Respond Differently to Cetuximab when Cultured in Hydrogels with Photopatterned EGF Gradients
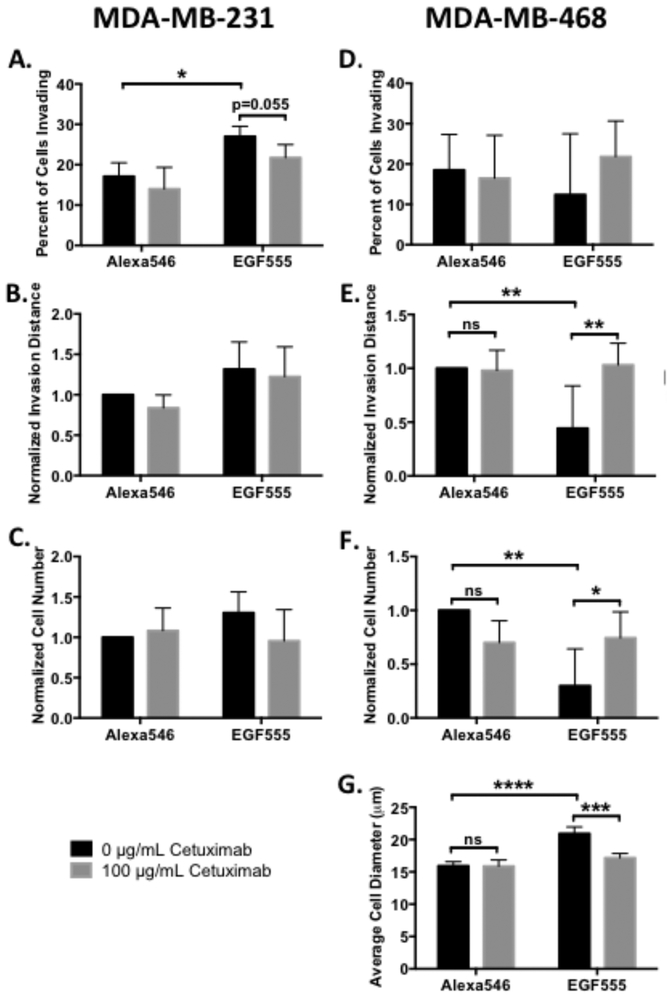
To determine whether the poor clinical outcomes of EGFR inhibitors could be better understood with in vitro 3D culture, we evaluated the response of both MDA-MB-231 and MDA-MB-468 cells to the EGFR inhibitor cetuximab on EGF555 and control Alexa546 low gradient hydrogels. HANBDF/MMPx hydrogels with low EGF concentration gradients were used: the higher percentages of invading cells in the low gradient gels relative to medium and high gradient gels increased the likelihood of distinguishing differences in cell invasion. In medium gradients, MDA-MB-231 cells invaded further, but fewer cells invaded, leading us to use the low EGF555 gradient hydrogels for the cetuximab studies. MDA-MB-231 cells were seeded on hydrogels with gradients of EGF555 vs. Alexa546 and treated with either 0 or 100 μg mL−1 of cetuximab. Significantly more MDA-MB-231 cells invaded into EGF555 than Alexa546 gradients (*p<0.05) and, cetuximab decreased the percent of invading MDA-MB-231 in EGF555 gradients (p=0.055, Figure 8A). While the percentage of cells invading decreased, neither the invasion distance (Figure 8B) nor the cell number (Figure 8C) were affected by cetuximab in either the EGF555 or Alexa546 control gradients. In contrast to the round morphology of MDA-MB-468 cells, MDA-MB-231 cells have a mesenchymal phenotype that is irregular in shape, making cell diameter difficult to measure.
Figure 8.
Cetuximab differentially affects MDA-MB-231 and MDA-MB-468 cell invasion and cell number. Post hoc comparisons are depicted graphically. A) Percent of invading MDA-MB-231 cells in low concentration gradient HANBDF/MMPx hydrogels of Alexa546 or EGF555 treated with either 0 or 100 μg mL−1 of cetuximab (day 3, n=4, mean + standard deviation). EGF555 significantly increased the percent of MDA-MB-231 cells invading into the gradients relative to Alexa546 gradients (***p<0.001, two-way ANOVA), while cetuximab significantly decreased the percent of invading MDA-MB-231 cells relative to no treatment (*p<0.05, two-way ANOVA). B) Normalized invasion distance of MDA-MB-231 cells on low gradients of Alexa546 or EGF555 treated with either 0 or 100 μg mL−1 of cetuximab (day 3, n=4, mean + standard deviation). C) Normalized cell number of MDA-MB-231 cells were similar on low gradients of Alexa546 or EGF555 treated with either 0 or 100 μg mL−1 of cetuximab (day 3, n=4, mean + standard deviation). D) Percent of invading MDA-MB-468 cells in low concentration gradient HANBDF/MMPx hydrogels of Alexa546 or EGF555 treated with either 0 or 100 μg mL−1 of cetuximab (day 6, n=5, mean + standard deviation). E) Normalized invasion distance of MDA-MB-468 cells on low gradients of Alexa546 or EGF555 treated with either 0 or 100 μg/mL of cetuximab (day 6, n=5, mean + standard deviation). Cetuximab treatment increased MDA-MB-468 cell invasion relative to no treatment (*p<0.05, two-way ANOVA). F) Normalized cell number of MDA-MB-468 cells on low gradients of Alexa546 or EGF555 treated with either 0 or 100 μg/mL of cetuximab (day 6, n=5, mean + standard deviation). Cetuximab treatment increased MDA-MB-468 cell number relative to no treatment (**p<0.01, two-way ANOVA). G) Average MDA-MB-468 cell diameter on low gradients of Alexa546 or EGF555 treated with either 0 or 100 μg/mL of cetuximab (day 6, n=4, mean + standard deviation). Cetuximab treatment decreased MDA-MB-468 cell diameter relative to no treatment (***p<0.001, two-way ANOVA).
MDA-MB-468 cells seeded on EGF555 and Alexa546 gradient hydrogels were similarly treated with 0 or 100 μg mL−1 of cetuximab. The percent of invading MDA-MB-468 cells was highly variable and was neither affected by EGF555 nor cetuximab treatment (Figure 8D). In the absence of cetuximab, there was both less invasion distance and fewer MDA-MB-468 cells on the EGF555 concentration gradient hydrogels (**p<0.01) compared to Alexa546 control gradient hydrogels (Figure 8E and F). Remarkably, cetuximab rescued the invasion and cell number of MDA-MB-468 cells on EGF555 gradients, significantly increasing both relative to no cetuximab treatment (**p<0.01 and *p<0.05, respectively). Without cetuximab, cell diameter significantly increased on EGF555 gradients (****p<0.0001) compared to control Alexa546 gradients whereas with cetuximab treatment, the MDA-MB-468 cell diameter decreased relative to no cetuximab (***p<0.001) on EGF555 gradients (Figure 8G). This suggests that cetuximab treatment prevents the antitumorigenic effects of EGF on MDA-MB-468 cells. Importantly, the negative consequence of cetuximab treatment was only observed in MDA-MB-468 cells in the EGF555 gradients. This emphasizes the need to incorporate microenvironmental factors into culture conditions, such as 3D culture conditions, when evaluating drugs that target cell invasion by specific cell-microenvironment interactions, such as blocking EGF binding to EGFR.
4. Discussion
Bhc has been used as a thiol-caging group to immobilize thiol-reactive biomolecules into hydrogels [22-24] However, the decreased photopatterning efficiency of both Bhc and mBhc relative to NDBF limits their use in photopatterning applications. To immobilize the same concentration of a given biomolecule, higher substitutions of both mBhc and Bhc on the hydrogel backbone are required relative to that of NDBF. The improved photopatterning efficiency of NDBF consequently allows reduced two-photon irradiation times (faster scan speeds) to immobilize biologically relevant concentrations of biomolecules, reducing the exposure of biomolecules to irradiation. A two-photon laser scanning speed of 0.009 μm μs−1 was used to compare the photopatterning efficiency of NDBF, Bhc, and mBhc whereas scan speeds of 0.527, 0.263, and 0.132 μm μs−1 were used to form the low, medium and high EGF555 concentration gradients, respectively.
In vivo, it is difficult to characterize biomolecule gradients, such as EGF, due to their transient nature. EGF gradients arise in breast tumors predominately due to EGF secretion by macrophages in the microenvironment, confounding their characterization. [7,53] EGF is present in serum and tumors of cancer patients [54-56] including breast tumors; [57] however, with only a handful of studies, there is a significant knowledge gap of EGF concentrations and profiles within breast tumors, and large disparities between studies.
To gain insight into the effect of EGF on breast cancer cells, we developed a hydrogel platform with defined EGF gradients. Due to challenges characterizing in vivo gradients, developing EGF gradients in invasion-permissive hydrogel models allows the influence of EGF on breast cancer cell invasion to be elucidated. To develop a model that recapitulates the in vivo breast cancer cell response to chemotactic EGF gradients, it is important to use biologically relevant concentrations. The concentration of EGF in benign breast tumors (0.13 ng mg−1 of total protein) and breast cancer (0.15 ng mg−1 of total protein) has been shown to be higher than normal breast tissue (0.04 ng mg−1 of total protein). [57] Additionally, a few studies have investigated the concentration of EGF within breast cysts and have found high EGF concentrations between 141.3 ng mL−1 and 506.2 ng mL-1. [58,59] Our EGF555 gradients contain similarly high concentrations of EGF (220 to 427 ng mL−1), recapitulating the high EGF concentration found in the breast tumor microenvironment.
Medium and high EGF555 concentration gradient hydrogels had similar numbers of invading MDA-MB-231 cells, however the cells were able to invade much further in the medium gradients likely due to the reduced mechanical constraints compared to the high gradients. Increasing the polymer concentration or increasing crosslink density can increase hydrogel stiffness. Since the hydrogel polymer concentration remained constant throughout the photopatterning process, we hypothesize that the increase in stiffness due to irradiation resulted in an increased crosslink density. It has been shown that stiffer, higher crosslinked hydrogels will reduce cell invasion distance, speed, and the percent of invading cells because cells have to degrade more crosslinks to create large enough gaps in the hydrogel network to allow for the passage of the cell body. [29,60] Additionally, breast cancer cells are known to invade through MMP independent mechanisms by squeezing their cell body through existing pores in the matrix. [61,62] Thus, smaller pore size in stiffer hydrogels will reduce both MMP-dependent and -independent invasion mechanisms. In the high gradients, the crosslinking density was likely too great for the MDA-MB-231 cells to overcome despite the higher EGF555 concentration gradient. In contrast, the MDA-MB-231 cells likely invaded farther into the medium gradient hydrogels compared to the low gradient hydrogels, due to the higher EGF555 concentration gradient. EGF is known to increase cell motility and MMP expression, both contributing to enhanced invasion in response to EGF. [63,64], [65] While triple negative MDA-MB-231 breast cancer cells have been shown to respond to chemotactic gradients of EGF, [14] EGFR expressing CHO cells have been shown to have increased directional persistence – the ratio of displacement to trajectory length - in response to EGF in the absence of chemotactic gradients. [66] Kim et al. found that U87MG human glioblastoma cells migrated faster in type I collagen matrices when cultured in the presence of EGF: cell migration in low barrier matrices was driven by increased cell speed whereas in high barrier, denser matrices, cell migration was more directional and mediated by protease degradation. [64] This is consistent with our findings: MDA-MB-231 cells invaded into hydrogels containing low EGF555 concentration gradients likely due to increased motility in response to EGF whereas they migrated into medium EGF555 concentration gradients by a chemotactic response. The latter is substantiated by the significant difference in cell invasion observed between the medium EGF555 concentration gradient hydrogels compared to the medium Alexa546 gradient controls.
Interestingly, relative to the MDA-MB-231 cells, the MDA-MB-468 cells responded completely differently to the EGF gradient hydrogels. MDA-MB-468 cells are invasive, but less invasive than MDA-MB-231 cells. [41] This was observed in our HANDBF/MMPx hydrogels, where both a higher percent (20.8 ± 10.6% vs. 10.2 ± 3.0%) of MDA-MB-231 cells invaded and to a greater distance (115 ± 46 μm vs. 82 ± 16 μm) into the gels compared to the MDA-MB-468 cells. MDA-MB-468 cells have very high EGFR expression, making them more sensitive to EGF at lower concentrations. It has been established that MDA-MB-468 cells have an apoptotic response to high concentrations of EGF. [17,51] The decreased MDA-MB-468 cell number and invasion in response to EGF555 gradients was likely due to this apoptotic effect. In response to the EGF concentration gradients, MDA-MB-468 cells showed minimal cell invasion into both the EGF555 gradient and non-patterned hydrogel likely because the residual EGF555 in the non-patterned regions (approximately 160 ng mL−1 μm−1) was sufficient to induce an apoptotic effect. An increase in MDA-MB-468 cell diameter in response to EGF555 is likely due to growth inhibition of the cells in response to cytotoxic concentrations of EGF. [67,68] A similar morphological response of MDA-MB-468 cells to EGF has been observed previously. [17,67,69] While it would be interesting to confirm that increasing concentrations of EGF555 result in MDA-MB-468 cell apoptosis, this study would be challenging on the hydrogels. The majority of MDA-MB-468 cells remained on the surface of the hydrogel and did not invade; any cells on the surface of the hydrogel that died would detach and wash away. Additionally, the non-patterned regions of the hydrogel had enough residual EGF to induce the same loss of cell number as the EGF555 gradients. Therefore, to determine any concentration dependence on MDA-MB-468 cell death, concentrations that are lower than the non-patterned EGF555 regions would have to be tested.
While MDA-MB-231 and MDA-MB-468 cells had opposite responses to EGF555 gradients, EGF555 gradients had no effect on MCF-7 cells. EGF has been shown to induce an invasive phenotype in MCF-7 cells; decreasing the expression of the cell-cell adhesion molecule E-cadherin, and increasing motility and invasion. [13,70,71] Additionally, it has been shown that MCF-7 cells will form cellular protrusions and migrate towards increasing EGF concentrations in response to gradients of EGF within a 3D matrix. [72] However, due to the low EGFR expression on only 4.64% of MCF-7 cells, any change in MCF-7 cell number and invasive potential would be limited to this population. The EGFR expression level on MCF-7 cells was likely too low and on too few cells for us to detect a response to EGF555 gradients, resulting in no detectable invasion. Although MCF-7 cells failed to invade the medium EGF555 gradients tested in this study, gradients with steeper slopes or higher EGF concentrations may induce an invasive response in low EGFR expressing MCF-7 cells. Likewise, while the medium EGF555 gradients reduced MDA-MB-468 cell number and invasion, gradients, much lower EGF concentrations may not be cytotoxic to these cells.
Micropatterned models have been developed to study cancer cell-microenvironment interactions; localized physical features or ECM deposition control the spatial organization of cancer cells and direct cell fate. [73-76] HA hydrogels containing encapsulated breast cancer cells were patterned next to fibrin hydrogels containing endothelial colony forming cells to study spatial regulation of angiogenesis in the tumor microenvironment. These micropatterned models are beneficial for elucidating the role of matrix organization on cancer cell invasion. Combining micropatterning techniques that control the distribution of ECM or cell types with photopatterning techniques that create complex gradients of biomolecules will capture the heterogeneous physical and chemical properties of the breast cancer microenvironment. This should lead to improved models for recapitulating the tumor microenvironment.
It has been established that EGFR expression may not be predictive of the clinical response to EGFR inhibitors such as cetuximab. [48-50] This may be due to some EGFR overexpressing breast cancers exhibiting apoptotic responses to EGF, which results in EGFR inhibitors reversing the antitumorigenic effect of EGF. In vivo studies have demonstrated the failure of EGFR inhibitors when treating EGFR overexpressing breast cancer. [19] Treatment with the EGFR inhibitor erlotinib only minimally delayed tumor formation following engraftment of metastatic transformed EGFR overexpressing murine NMuMg cells, and Erlotinib significantly increased tumor reoccurrence following tumor excision. [19] In addition, approximately 11% of triple negative breast cancers harbor mutations in the EGFR gene, which may influence their response to EGFR inhibitors. [9] Mutations in EGFR activated pathways may lead to EGFR inhibitor resistance. [77] Furthermore, components of the microenvironment may influence drug response; HA has been shown to alter the response of glioblastoma cells to the EGFR inhibitor erlotinib through colocalization and coimmunoprecipitation of the CD44 receptor with EGFR. [78] The majority of in vitro experiments evaluating breast cancer cell response to EGFR inhibitors use standard 2D cell culture conditions. However, in vivo, the breast microenvironment contains mechanical signals from the matrix, facilitates cell-ECM interactions and cell migration, and may contain high concentrations of EGF, especially in the diseased state. [57] Thus, standard 2D culturing models may fail to capture potential apoptotic responses of certain breast cancer cells to high EGF concentrations, resulting in false negatives where the cells are thought to be unresponsive to an EGFR inhibitor when, in fact, the drug may reverse the antitumorigenic effect of high EGF concentrations. Our model allowed us to gain insight into this phenomenon where cetuximab treatment actually increased MDA-MB-468 cell invasion yet decreased invasion of MDA-MB-231 cells. Improved models of the breast cancer microenvironment, such as the one described herein, are needed to recapitulate the breast cancer cell-matrix interactions involved in processes such as cell invasion. Models that better represent these interactions may shed light on clinical outcomes and help to identify and predict the benefit of new therapies leading to improved clinical translation.
5. Conclusions
We developed defined EGF gradients within HA/MMPx hydrogels using NDBF protecting groups coupled with multiphoton confocal patterning to study breast cancer cell invasion. Using three breast cancer cell lines, with different EGFR expression, we showed the differential cellular response to the EGF gradient hydrogels in terms of both cell invasion, cell number and cell size. With low EGFR-expressing MCF7 cells, EGF gradients had no effect on the cells whereas with high EGFR-expressing MDA-MB-231 cells, EGF gradients promoted cell invasion, which was decreased with cetuximab. In contrast, highly EGFR over-expressing MDA-MB-468 cells were arrested by EGF gradient scaffolds, which was reversed, unexpectedly, by the EGF inhibitor cetuximab. The differential responses of breast cancer cells to the EGF inhibitor cetuximab reflects the poor clinical outcome of this drug. These confounding results demonstrate the importance of pre-screening drugs in relevant, high content screening platforms, such as that developed herein, where cell invasion and cell viability are probed. The HANDBF/MMPx hydrogels developed in this study provide a system for studying breast cancer cell-microenvironment interactions and probing the effect of drugs that target EGF interactions.
Supplementary Material
Acknowledgements
We are grateful for funding from NSERC (Discovery and CHRP to MSS and CREATE in M3 to SAF), CIHR (Foundation and CHRP to MSS) and NIH (R21 CA185783 and R01 GM084152 to MDD). We acknowledge the Canada Foundation for Innovation, project number 19119, and the Ontario Research Fund for funding of the Centre for Spectroscopic Investigation of Complex Organic Molecules and Polymers. We thank members of the Shoichet lab for thoughtful review of this manuscript.
Contributor Information
Stephanie A. Fisher’, The Donnelly Centre for Cellular and Biomolecular Research, Department of Chemical Engineering and Applied Chemistry, and Institute of Biomaterials and Biomedical Engineering, University of Toronto, 160 College Street, Toronto Ontario M5S 3E1
Dr. Roger Y. Tam, The Donnelly Centre for Cellular and Biomolecular Research, Department of Chemical Engineering and Applied Chemistry, and Institute of Biomaterials and Biomedical Engineering, University of Toronto, 160 College Street, Toronto Ontario M5S 3E1
Dr. Ana Fokina, The Donnelly Centre for Cellular and Biomolecular Research, Department of Chemical Engineering and Applied Chemistry, and Institute of Biomaterials and Biomedical Engineering, University of Toronto, 160 College Street, Toronto Ontario M5S 3E1
M. Mohsen Mahmoodi, Department of Chemistry, University of Minnesota, Minneapolis, Minnesota, 55455, USA.
Prof. Mark D. Distefano, Department of Chemistry, University of Minnesota, Minneapolis, Minnesota, 55455, USA
Prof. Molly S. Shoichet, The Donnelly Centre for Cellular and Biomolecular Research, Department of Chemical Engineering and Applied Chemistry, and Institute of Biomaterials and Biomedical Engineering, University of Toronto, 160 College Street, Toronto Ontario M5S 3E1.
References
- [1].Lu P, Weaver VM, Werb Z, The extracellular matrix: A dynamic niche in cancer progression, The Journal of Cell Biology. 196 (2012) 395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wei SC, Fattet L, Tsai JH, Guo Y, Pai VH, Majeski HE, et al. , Matrix stiffness drives epithelial-mesenchymal transition and tumour metastasis through a TWIST1-G3BP2 mechanotransduction pathway, Nature Cell Biology. 17 (2015) 678–688. doi: 10.1038/ncb3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mao Y, Keller ET, Garfield DH, Shen K, Wang J, Stromal cells in tumor microenvironment and breast cancer, Cancer Metastasis Rev. 32 (2013) 303–315. doi: 10.1007/s10555-012-9415-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Muller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, et al. , Involvement of chemokine receptors in breast cancer metastasis, Nature. 410 (2001) 50–56. [DOI] [PubMed] [Google Scholar]
- [5].Infanger DW, Lynch ME, Fischbach C, Engineered Culture Models for Studies of Tumor-Microenvironment Interactions, Annu. Rev. Biomed. Eng 15 (2013) 29–53. doi : 10.1146/annurev-bioeng-071811-150028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wyckoff J, Wang W, Lin EY, Wang Y, Pixley F, Stanley ER, et al. , A Paracrine Loop between Tumor Cells and Macrophages Is Required for Tumor Cell Migration in Mammary Tumors, Cancer Research. 64 (2004) 7022–7029. [DOI] [PubMed] [Google Scholar]
- [7].Goswami S, Sahai E, Wyckoff JB, Cammer M, Cox D, Pixley FJ, et al. , Macrophages Promote the Invasion of Breast Carcinoma Cells via a Colony-Stimulating Factor-1/Epidermal Growth Factor Paracrine Loop, Cancer Research. 65 (2005)5278–5283. [DOI] [PubMed] [Google Scholar]
- [8].Bhargava R, Gerald WL, Li AR, Pan Q, Lal P, Ladanyi M, et al. , EGFR gene amplification in breast cancer: correlation with epidermal growth factor receptor mRNA and protein expression and HER-2 status and absence of EGFR-activating mutations, Mod Pathol. 18 (2005) 1027–1033. doi: 10.1038/modpathol.3800438. [DOI] [PubMed] [Google Scholar]
- [9].Teng YH-F, Tan WJ, Thike A-A, Cheok P-Y, Tse GM-K, Wong N-S, et al. , Mutations in the epidermal growth factorreceptor (EGFR) gene in triple negative breastcancer: possible implications for targeted therapy, Breast Cancer Research. 13 (2011) R35. doi: 10.1186/bcr2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Thike A-A, Cheok P-Y, Jara-Lazaro AR, Tan B, Tan P, Tan P-H, Triple-negative breast cancer: clinicopathological characteristics and relationship with basal-like breast cancer, Mod Pathol. 23 (2010) 123–133. doi : 10.1038/modpathol.2009.145. [DOI] [PubMed] [Google Scholar]
- [11].Rakha EA, El-Sayed ME, Green AR, Lee AHS, Robertson JF, Ellis IO, Prognostic markers in triple-negative breast cancer, Cancer. 109 (2007) 25–32. doi : 10.1002/cncr.22381. [DOI] [PubMed] [Google Scholar]
- [12].Pintens S, Neven P, Drijkoningen M, Van Belle V, Moerman P, Christiaens M-R, et al. , Triple negative breast cancer: a study from the point of view of basal CK5/6 and HER-1, Journal of Clinical Pathology. 62 (2009) 624–628. doi: 10.1136/jcp.2008.061358. [DOI] [PubMed] [Google Scholar]
- [13].Vergara D, Valente CM, Tinelli A, Siciliano C, Lorusso V, Acierno R, et al. , Resveratrol inhibits the epidermal growth factor-induced epithelial mesenchymal transition in MCF-7 cells, Cancer Letters. 310 (2011) 1–8. doi: 10.1016/j.canlet.2011.04.009. [DOI] [PubMed] [Google Scholar]
- [14].Wang S-J, Saadi W, Lin F, Minh-Canh Nguyen C, Li Jeon N, Differential effects of EGF gradient profiles on MDA-MB-231 breast cancer cell chemotaxis, Experimental Cell Research. 300 (2004) 180–189. doi: 10.1016/j.yexcr.2004.06.030. [DOI] [PubMed] [Google Scholar]
- [15].Oda K, Matsuoka Y, Funahashi A, Kitano H, A comprehensive pathway map of epidermal growth factor receptor signaling, Mol Syst Biol. 1 (2005) E1–E17. doi: 10.1038/msb4100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Scaltriti M, Baselga J, The Epidermal Growth Factor Receptor Pathway: A Model for Targeted Therapy, Molecular Pathways. 12 (2006) 5268–5272. doi: 10.1158/1078-0432.CCR-06-1554. [DOI] [PubMed] [Google Scholar]
- [17].Armstrong DK, Kaufmann SH, Ottaviano YL, Furuya Y, Buckley JA, Isaacs JT, et al. , Epidermal growth factor-mediated apoptosis of MDA-MB-468 human breast cancer cells, Cancer Research. 54 (1994) 5280–5283. [PubMed] [Google Scholar]
- [18].Alanazi I, Ebrahimie E, Hoffmann P, Adelson DL, Combined gene expression and proteomic analysis of EGF induced apoptosis in A431 cells suggests multiple pathways trigger apoptosis, Apoptosis. 18 (2013) 1291–1305. doi: 10.1007/s10495-013-0887-6. [DOI] [PubMed] [Google Scholar]
- [19].Wendt MK, Williams WK, Pascuzzi PE, Balanis NG, Schiemann BJ, Carlin CR, et al. , The Antitumorigenic Function of EGFR in Metastatic Breast Cancer is Regulated by Expression of Mig6, Neo. 17 (2015) 124–133. doi: 10.1016/j.neo.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Truong D, Puleo J, Llave A, Mouneimne G, Kamm RD, Nikkhah M, Breast Cancer Cell Invasion into a Three Dimensional Tumor-Stroma Microenvironment, Sci. Rep 6 (2016) 34094–18. doi: 10.1038/srep34094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kim BJ, Hannanta-anan P, Chau M, Kim YS, Swartz MA, Wu M, Cooperative Roles of SDF-1a and EGF Gradients on Tumor Cell Migration Revealed by a Robust 3D Microfluidic Model, PLoS ONE. 8 (2013) e68422–9. doi : 10.1371/journal.pone.0068422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wylie RG, Ahsan S, Aizawa Y, Maxwell KL, Morshead CM, Shoichet MS, Spatially controlled simultaneous patterning of multiple growth factors in three-dimensional hydrogels, Nat Mater. 10 (2011) 799–806. doi: 10.1038/nmat3101. [DOI] [PubMed] [Google Scholar]
- [23].Tam RY, Fisher SA, Baker AEG, Shoichet MS, Transparent Porous Polysaccharide Cryogels Provide Biochemically Defined, Biomimetic Matrices for Tunable 3D Cell Culture, Chem. Mater 28 (2016) 3762–3770. doi : 10.1021/acs.chemmater.6b00627. [DOI] [Google Scholar]
- [24].Owen SC, Fisher SA, Tam RY, Nimmo CM, Shoichet MS, Hyaluronic Acid Click Hydrogels Emulate the Extracellular Matrix, Langmuir. 29 (2013) 7393–7400. doi: 10.1021/la305000w. [DOI] [PubMed] [Google Scholar]
- [25].DeForest CA, Anseth KS, Cytocompatible click-based hydrogels with dynamically tunable properties through orthogonal photoconjugation and photocleavage reactions, Nature Chemistry. 3 (2011) 925–931. doi: 10.1038/nchem.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Guo C, Kim H, Ovadia EM, Mourafetis CM, Yang M, Chen W, et al. , Bio-orthogonal conjugation and enzymatically triggered release of proteins within multi-layered hydrogels, Acta Biomaterialia. (2017) 1–11. doi: 10.1016/j.actbio.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wade RJ, Bassin EJ, Gramlich WM, Burdick JA, Nanofibrous Hydrogels with Spatially Patterned Biochemical Signals to Control Cell Behavior, Adv. Mater 27 (2015) 1356–1362. doi: 10.1002/adma.201404993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Phillips JE, Burns KL, Le Doux JM, Guldberg RE, Garcia AJ, Engineering graded tissue interfaces, Proceedings of the National Academy of Sciences. 105 (2008)12170–12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Fisher SA, Anandakumaran PN, Owen SC, Shoichet MS, Tuning the Microenvironment: Click-Crosslinked Hyaluronic Acid-Based Hydrogels Provide a Platform for Studying Breast Cancer Cell Invasion, Adv. Funct. Mater 25 (2015) 7163–7172. doi: 10.1002/adfm.201502778. [DOI] [Google Scholar]
- [30].Nagase H, Fields GB, Human Matrix Metalloproteinase Specificity Studies Using Collagen Sequence-Based Synthetic Peptides, Biopolymers. 40 (1996) 399–416. [DOI] [PubMed] [Google Scholar]
- [31].Lutolf MP, Lauer-Fields JL, Schmoekel HG, Metters AT, Weber FE, Fields GB, et al. , Synthetic matrix metalloproteinase-sensitive hydrogels for the conduction of tissue regeneration: Engineering cell-invasion characteristics, Proceedings of the National Academy of Sciences. 100 (2003) 5413–5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Mahmoodi MM, Fisher SA, Tam RY, Goff PC, Anderson RB, Wissinger JE, et al. , 6-Bromo-7-hydroxy-3-methylcoumarin (mBhc) is an efficient multi-photon labile protecting group for thiol caging and three-dimensional chemical patterning, Org. Biomol. Chem 14 (2016) 8289–8300. doi: 10.1039/C6OB01045H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Mahmoodi MM, Abate-Pella D, Pundsack TJ, Palsuledesai CC, Goff PC, Blank DA, et al. , Nitrodibenzofuran: A One-and Two-Photon Sensitive Protecting Group That Is Superior to Brominated Hydroxycoumarin for Thiol Caging in Peptides, J. Am. Chem. Soc 138 (2016) 5848–5859. doi: 10.1021/jacs.5b11759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Baugh L, Le Trong I, Stayton PS, Stenkamp RE, Lybrand TP, A Streptavidin Binding Site Mutation Yields an Unexpected Result: An Ionized Asp128 Residue Is Not Essential for Strong Biotin Binding, Biochemistry. 55 (2016) 5201–5203. doi: 10.1021/acs.biochem.6b00698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Li P, Xiang T, Li H, Li Q, Yang B, Huang J, et al. , Hyaluronan synthase 2 overexpression is correlated with the tumorigenesis and metastasis of human breast cancer, International Journal of Clinical Experimental Pathology. 8 (2015) 12101–12114. [PMC free article] [PubMed] [Google Scholar]
- [36].Toole BP, Hyaluronan: from extracellular glue to pericellular cue, Nat Rev Cancer. 4 (2004) 528–539. doi: 10.1038/nrc1391. [DOI] [PubMed] [Google Scholar]
- [37].Auvinen P, Tammi R, Parkkinen J, Tammi M, Agren U, Johansson R, et al. , Hyaluronan in Peritumoral Stroma and Malignant Cells Associates with Breast Cancer Spreading and Predicts Survival, American Journal of Pathology. 156 (2000) 529–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Misra S, Heldin P, Hascall VC, Karamanos NK, Skandalis SS, Markwald RR, et al. , Hyaluronan-CD44 interactions as potential targets for cancer therapy, FEBS Journal. 278 (2011) 1429–1443. doi: 10.1111/j.1742-4658.2011.08071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Misra S, Hascall VC, Markwald RR, Ghatak S, Interactions between Hyaluronan and Its Receptors (CD44, RHAMM) Regulate the Activities of Inflammation and Cancer, Front. Immunol 6 (2015) 201. doi: 10.3389/fimmu.2015.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Nimmo CM, Owen SC, Shoichet MS, Diels-Alder Click Cross-Linked Hyaluronic Acid Hydrogels for Tissue Engineering, Biomacromolecules. 12 (2011) 824–830. doi: 10.1021/bm101446k. [DOI] [PubMed] [Google Scholar]
- [41].Gordon LA, Mulligan KT, Maxwell-Jones H, Adams M, Walker RA, Jones JL, Breast cell invasive potential relates to the myoepithelial phenotype, Int. J. Cancer 106 (2003) 8–16. doi: 10.1002/ijc.11172. [DOI] [PubMed] [Google Scholar]
- [42].Subik K, Lee J-F, Baxter L, Strzepek T, Costello D, Crowley P, et al. , The expression patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67 and AR by immunohistochemical analysis in breast cancer cell lines, Breast Cancer Basic and Clinical Research. 4 (2010) 35–41. [PMC free article] [PubMed] [Google Scholar]
- [43].Graham J, Muhsin M, Kirkpatrick P, Fresh from the pipeline: Cetuximab, Nat Rev Drug Discov. 3 (2004) 549–550. doi: 10.1038/nrd1445. [DOI] [PubMed] [Google Scholar]
- [44].Li S, Schmitz KR, Jeffrey PD, Wiltzius JJW, Kussie P, Ferguson KM, Structural basis for inhibition of the epidermal growth factor receptor by cetuximab, Cancer Cell. 7 (2005) 301–311. doi: 10.1016/j.ccr.2005.03.003. [DOI] [PubMed] [Google Scholar]
- [45].Martinelli E, De Palma R, Orditura M, De Vita F, Ciardiello F, Anti-epidermal growth factor receptor monoclonal antibodies in cancer therapy, Clinical & Experimental Immunology. 158 (2009) 1–9. doi: 10.1111/j.1365-2249.2009.03992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Changavi A, Shashikala A, Ramji A, Epidermal growth factor receptor expression in triple negative and nontriple negative breast carcinomas, J Lab Physicians. 7 (2015) 79–83. doi: 10.4103/0974-2727.163129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Nakai K, Hung M-C, Yamaguchi H, A perspective on anti-EGFR therapies targeting triple-negative breast cancer, American Journal of Cancer Research. 6 (2016)1609–1623. [PMC free article] [PubMed] [Google Scholar]
- [48].Dickler MN, Rugo HS, Eberle CA, Brogi E, Caravelli JF, Panageas KS, et al. , A Phase II Trial of Erlotinib in Combination with Bevacizumab in Patients with Metastatic Breast Cancer, Clinical Cancer Research. 14 (2008) 7878–7883. doi: 10.1158/1078-0432.CCR-08-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].von Minckwitz G, Jonat W, Fasching P, du Bois A, Kleeberg U, Luck H-J, et al. , A multicentre phase II study on gefitinib in taxane-and anthracycline-pretreated metastatic breast cancer, Breast Cancer Res Treat. 89 (2005) 165–172. [DOI] [PubMed] [Google Scholar]
- [50].Carey LA, Rugo HS, Marcom PK, Mayer EL, Esteva FJ, Ma CX, et al. , TBCRC 001: Randomized Phase II Study of Cetuximab in Combination With Carboplatin in Stage IV Triple-Negative Breast Cancer, Jco. 30 (2012) 2615–2623. doi: 10.1200/JC0.2010.34.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Chen K-Y, Huang L-M, Kung H-J, Ann DK, Shih H-M, The role of tyrosine kinase Etk/Bmx in EGF-induced apoptosis of MDA-MB-468 breast cancer cells, Oncogene. 23 (2003) 1854–1862. doi: 10.1038/sj.onc.1207308. [DOI] [PubMed] [Google Scholar]
- [52].Hognason T, Chatterjee S, Vartanian T, Ratan RR, Ernewein KM, Habib AA, Epidermal growth factor receptor induced apoptosis: potentiation by inhibition of Ras signaling, Federation of European Biochemical Societies Letters. 491 (2001) 9–15. [DOI] [PubMed] [Google Scholar]
- [53].Knutsdottir H, Condeelis JS, Palsson E, 3-D individual cell based computational modeling of tumor cell-macrophage paracrine signaling mediated by EGF and CSF-1 gradients, Integr. Biol 8 (2016) 104–119. doi: 10.1039/C5IB00201J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Erol-Demirbilek M, Komurcu HF, Komurcu S, Kilic N, EGF and TNF-α levels and oxidative/nitrosative stress in breast and non-small cell lung cancer patients, Hacettepe Journal of Biology and Chemistry. 41 (2013) 357–363. [Google Scholar]
- [55].Baron AT, Lafky JM, Boardman CH, Balasubramaniam S, Suman VJ, Podratz KC, et al. , Serum sErbB1 and Epidermal Growth Factor Levels As Tumor Biomarkers in Women with Stage III or IV Epithelial Ovarian Cancer, Cancer Epidemiology. 8 (1999) 129–137. [PubMed] [Google Scholar]
- [56].Lemos-Gonzalez Y, Rodriguez-Berrocal FJ, Cordero OJ, C. G0mez, M. Paez de la Cadena, Alteration of the serum levels of the epidermal growth factor receptor and its ligands in patients with non-small cell lung cancer and head and neck carcinoma, Br J Cancer. 96 (2007) 1569–1578. doi: 10.1038/sj.bjc.6603770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Bolufer P, Lluch A, Molina R, Alberola V, Vazquez C, Padilla J, et al. , Epidermal growth factor in human breast cancer, endometrial carcinoma and lung cancer. Its relationship to epidermal growth factor receptor, estradiol receptor and tumor TNM, Clinica Chimica Acta. 215 (1993) 51–61. [DOI] [PubMed] [Google Scholar]
- [58].Boccardo F, Lunardi GL, Petti AR, Rubagotti A, Enterolactone in breast cyst fluid: correlation with EGF and breast cancer risk, Breast Cancer Res Treat. 79 (2003)17–23. [DOI] [PubMed] [Google Scholar]
- [59].Smith K, Miller WR, Fennelly JA, Matthews JNS, Scott WN, Harris AL, Quantification of epidermal growth in human breast cyst fluids: Correlation with dehydroepiandrosterone-sulphate and electrolyte concentrations, Int. J. Cancer 44 (1989)229–232. [DOI] [PubMed] [Google Scholar]
- [60].Kyburz KA, Anseth KS, Three-dimensional hMSC motility within peptide-functionalized PEG-based hydrogels of varying adhesivity and crosslinking density, Acta Biomaterialia. 9 (2013) 6381–6392. doi: 10.1016/j.actbio.2013.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Friedl P, Alexander S, Cancer Invasion and the Microenvironment: Plasticity and Reciprocity, Cell. 147 (2011) 992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- [62].Sabeh F, Shimizu-Hirota R, Weiss SJ, Protease-dependent versus -independent cancer cell invasion programs: three-dimensional amoeboid movement revisited, The Journal of Cell Biology. 185 (2009) 11–19. doi: 10.1083/jcb.200807195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Lu Z, Jiang G, Blume-Jensen P, Hunter T, Epidermal Growth Factor-Induced Tumor Cell Invasion and Metastasis Initiated by Dephosphorylation and Downregulation of Focal Adhesion Kinase, Molecular and Cellular Biology. 21 (2001) 4016–4031. doi: 10.1128/MCB.21.12.4016-4031.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Kim H-D, Guo TW, Wu AP, Wells A, Gertler FB, Lauffenburger DA, Epidermal Growth Factor-induced Enhancement of Glioblastoma Cell Migration in 3D Arises from an Intrinsic Increase in Speed But an Extrinsic Matrix-and Proteolysis-dependent Increase in Persistence, Molecular Biology of the Cell. 19 (2008) 4249–4259. doi: 10.1091/mbc.E08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Dilly M, Hambruch N, Haeger JD, Pfarrer C, Epidermal growth factor (EGF) induces motility and upregulates MMP-9 and TIMP-1 in bovine trophoblast cells, Mol. Reprod. Dev 77 (2010) 622–629. doi: 10.1002/mrd.21197. [DOI] [PubMed] [Google Scholar]
- [66].Harms BD, Bassi GM, Horwitz AR, Lauffenburger DA, Directional Persistence of EGF-Induced Cell Migration Is Associated with Stabilization of Lamellipodial Protrusions, Biophysical Journal. 88 (2005) 1479–1488. doi: 10.1529/biophysj.104.047365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Prasad KAN, Church JG, EGF effects on p53 in MDA-468 human breast cancer cells: implications for G1 arrest, Cell Proliferation. 30 (1997) 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Ryu J-W, Choe SS, Ryu S-H, Park E-Y, Lee BW, Kim TK, et al. , Paradoxical induction of growth arrest and apoptosis by EGF via the up-regulation of PTEN by activating Redox factor-1/Egr-1 in human lung cancer cells, Oncotarget. 8 (2017) 4181–4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Thomas T, Balabhadrapathruni S, Gardner CR, Hong J, Faaland CA, Thomas JT, Effects of Epidermal Growth Factor on MDA-MB-468 Breast Cancer Cells: Alterations in Polyamine Biosynthesis and the Expression of p21/CIP1/WAF1, J. Cell. Physiol. 179 (1999) 257–266. [DOI] [PubMed] [Google Scholar]
- [70].Garcia R, Franklin RA, McCubrey JA, EGF Induces Cell Motility and Multi-Drug Resistance Gene Expression in Breast Cancer Cells, Cell Cycle. 5 (2006) 2820–2826. doi: 10.4161/cc.5.23.3535. [DOI] [PubMed] [Google Scholar]
- [71].Kim J, Kong J, Chang H, Kim H, Kim A, EGF induces epithelial-mesenchymal transition through phospho-Smad2/3-Snail signaling pathway in breast cancer cells, Oncotarget. 7 (2016) 85021–85032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Liu T, Li C, Li H, Zeng S, Qin J, Lin B, A microfluidic device for characterizing the invasion of cancer cells in 3-D matrix, Electrophoresis. 30 (2009) 4285–4291. doi: 10.1002/elps.200900289. [DOI] [PubMed] [Google Scholar]
- [73].Dickinson LE, Lutgebaucks C, Lewis DM, Gerecht S, Patterning microscale extracellular matrices to study endothelial and cancer cell interactions in vitro, Lab Chip. 12 (2012) 4244–10. doi: 10.1039/c2lc40819h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Peela N, Sam FS, Christenson W, Truong D, Watson AW, Mouneimne G, et al. , A three dimensional micropatterned tumor model for breast cancer cell migration studies, Biomaterials. 81 (2016) 72–83. doi: 10.1016/j.biomaterials.2015.11.039. [DOI] [PubMed] [Google Scholar]
- [75].Smulovitz C, Dickinson LE, Gerecht S, Micropatterned Surfaces for the Study of Cancer and Endothelial Cell Interactions with Hyaluronic Acid, Isr. J. Chem 14 (2013) 710–718. doi: 10.1002/ijch.201300058. [DOI] [Google Scholar]
- [76].Pedron S, Harley BAC, Impact of the biophysical features of a 3D gelatin microenvironment on glioblastoma malignancy, J. Biomed. Mater. Res 101 (2013) 3404–3415. doi: 10.1002/ibm.a.34637. [DOI] [PubMed] [Google Scholar]
- [77].Rimawi MF, Shetty PB, Weiss HL, Schiff R, Osborne CK, Chamness GC, et al. , Epidermal growth factor receptor expression in breast cancer association with biologic phenotype and clinical outcomes, Cancer. 116 (2010) 1234–1242. doi: 10.1002/cncr.24816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Pedron S, Hanselman JS, Schroeder MA, Sarkaria JN, Harley BAC, Extracellular Hyaluronic Acid Influences the Efficacy of EGFR Tyrosine Kinase Inhibitors in a Biomaterial Model of Glioblastoma, Advanced Healthcare Materials. 6 (2017) 1700529–9. doi: 10.1002/adhm.201700529. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.