Abstract
Breast cancer (BC) is the second leading cause of death among women in the US, and its subtype triple-negative BC (TNBC) is the most aggressive BC with poor prognosis. In the current study, we investigated the anticancer effects of the natural product plumbagin (PL) on racially different TNBC cells. The PL effects were examined in two TNBC cell lines: MDA-MB-231 (MM-231) and MDA-MB-468 (MM-468), representing Caucasian Americans and African Americans, respectively. The results obtained indicate that PL inhibited cell viability and cell proliferation and induced apoptosis in both cell lines. Notably, MM-468 cells were 5-fold more sensitive to PL than MM-231 cells were. Testing PL and Taxol® showed the superiority of PL over Taxol® as an antiproliferative agent in MM-468 cells. PL treatment resulted in an approximately 20-fold increase in caspase-3 activity with 3 μM PL in MM-468 cells compared with an approximately 3-fold activity increase in MM-231 cells with 8 μM PL. Moreover, the results indicate a higher sensitivity to PL in MM-468 cells than in MM-231 cells. The results also show that PL downregulated CCL2 cytokine expression in MM-468 cells by 30% compared to a 90% downregulation in MM-231 cells. The ELISA results confirmed the array data (35% vs. 75% downregulation in MM-468 and MM-231 cells, respectively). Moreover, PL significantly downregulated IL-6 and GM-CSF in the MM-231 cells. Indeed, PL repressed many NF-қB-regulated genes involved in the regulation of apoptosis, proliferation, invasion, and metastasis. The compound significantly downregulated the same genes (BIRC3, CCL2, TLR2, and TNF) in both types of cells. However, PL impacted five more genes in MM-231 cells, including BCL2A1, ICAM1, IKBKE, IL1β, and LTA. In conclusion, the data obtained in this study indicate that the quinone compound PL could be a novel cancer treatment for TNBC in African American women.
Introduction
Breast cancer (BC) contributes to 23% of all cancer cases diagnosed in the US, making it the most common type and the second leading cause of cancer death among women [1]. In the United States, female BC cases are expected to rise and reach over 1.7 million with more than 600,000 death cases in 2018 [2]. The subtype TNBC is the most aggressive and metastatic BC that represents approximately 10–15% of all BC cases [3]. For BC treatments, hormone-based agents are targeting three characteristic receptors: estrogen (ER), progesterone (PR) or human epidermal growth factor (Her2/neu) [4, 5]. Although TNBC has initial higher response rates to a variety of chemotherapy agents, repeated exposure to chemotherapy can develop resistance to these agents [6], leading to poor prognosis and treatment failure. Moreover, TNBC aggressiveness is known to be more profound among African American patients (AA) than Caucasian American patients (CA) [7].
On the other hand, it is known that 15 to 20% of all cancer-related deaths worldwide are linked to inflammation [8]. The aggressiveness of TNBC is associated with the increased activity of nuclear factor-kappa B (NF-қB) transcription factor [9], which is thought to orchestrate the link between inflammation and cancer [10, 11]. NF-қB regulates genes involved in inflammation, cell- survival, apoptosis and proliferation in many solid tumors, including BC [12]. While NF-қB activation is immediate in normal cells, various oncogenic molecular modifications can lead to NF-қB pathway activation [13] that impacts the expression of different proinflammatory cytokines [14]. Moreover, cytokines such as IL-1β and TNF-α stimulated by NF-қB directly activate the NF-қB pathway to establish NF-қB autostimulation [15]. Indeed, inhibition of the NF-қB pathway can lead to significant downregulation of the inflammatory, procancerous events in several malignant tumors [16].
Tumors that arise at sites of chronic inflammation are characterized by the presence of macrophages, cytokines and chemokines [17]. In invasive BC, tumor-associated macrophages (TAMs) produce many proinflammatory cytokines, mainly, tumor-necrosis factor (TNF)-α [18, 19]. In BC patients, highly expressed TNF-α [18] stimulates proliferation of T47D cells of the human mammary gland [20] and upregulates several genes involved in cancer cell proliferation, invasion, and metastasis [21]. Additionally, an increase in TNF-α stimulates a release of the chemokine C-C Motif Ligand 2 (CCL2), which is also known as monocyte chemoattractant protein-1 (MCP-1) [22]. CCL2 is the most cancer-involved member of the CC chemokine family [23], and its high expression in the tumor tissue promotes tumorigenesis and metastasis [24]. Moreover, CCL2 suppression is associated with reduced tumor aggressiveness in BC [25].
Many studies show the anticancer and anti-inflammatory properties of plumbagin (PL), the main active constituent of Plumbago zeylanica roots. The plant roots have been used in India for many centuries in treating skin diseases, diarrhea, dyspepsia, piles, anasarca, plague, leprosy, urinary tract infections, scabies, and ulcers [26]. Moreover, the plant was found to have neuroprotective, hepatoprotective, antiatherogenic, and cardiotonic properties [27]. PL is found in other medicinal plants belonging to the Plumbaginaceae, Droseraceae, and Ebenaceae families [28]. Recent reports indicate the use of PL in treating diseases that are associated with inflammation, such as rheumatoid arthritis [29]. Our previous study indicates that PL has a potent anti-inflammatory effect in BV-2 microglia cells [30]. The PL anticancer properties have been studied in many cancers including breast [31], prostate [32, 33], and ovarian [34] cancers. The anticancer property of PL was also reported in pancreatic [35], lung [36], cervical [37, 38], and brain [28] cancers.
Therefore, we selected two human TNBC cell lines, MDA-MB-231 (MM-231) and MDA-MB-468 (MM-468), as associated with CA and AA races, respectively [39]. We hypothesized that the NF-қB pathway is involved in the PL-repressing effect of CCL2 and may also impact NF-қB-regulated genes which orchestrate the intra- and inter-cellular anticancer action.
Results
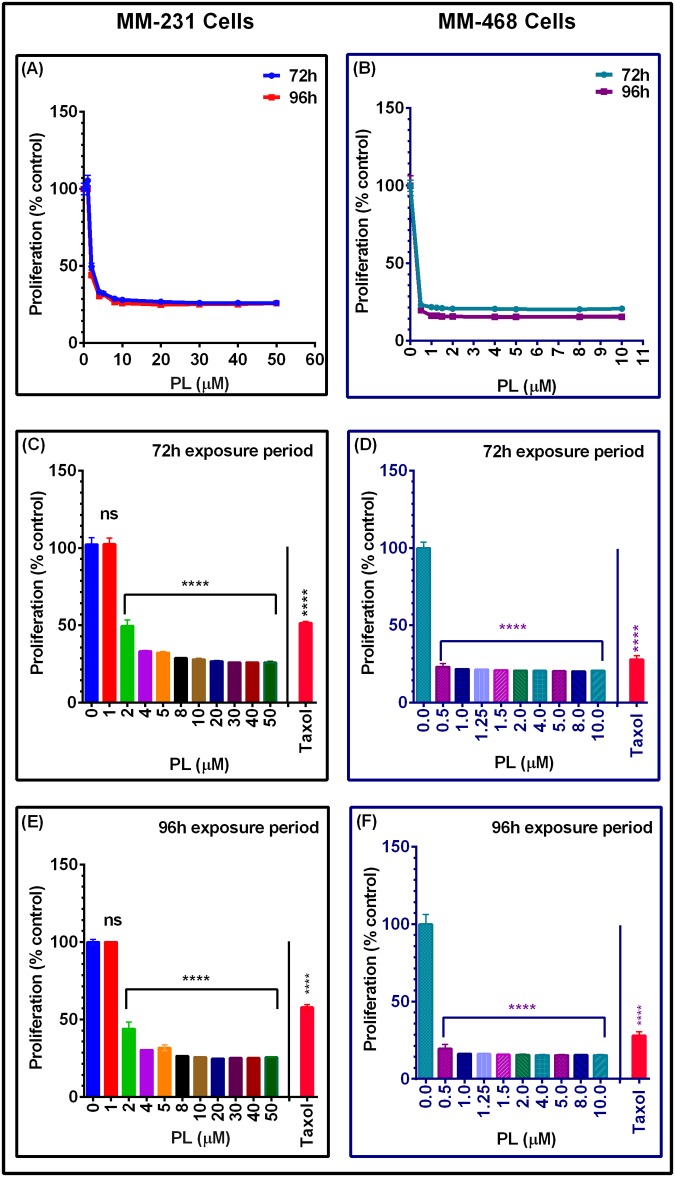
To determine the anticancer effects of PL on TNBC cells, we first examined the cytotoxicity of PL in both MM-231 and MM-468 cell lines. As shown in Fig 1A and 1B, a highly significant effect (p < 0.0001) was found in different PL concentration ranges tested in MM-231 and MM-468 cells. The obtained data indicate that PL was 5-fold more effective in MM-468 cells (IC50 = 2.03 ± 0.09 μM) than in the MM-231 cell line (IC50 = 9.91 ± 0.18 μM). Additionally, we identified the optimum concentration of the proinflammatory cytokine TNF-α to stimulate inflammatory cytokines in both cell lines. Fig 1C and 1D show that increasing concentrations (1–100 ng/mL) of TNF-α had no significant effect on the cell lines examined compared to the control. From these results, as well as from previous reports [40], we selected 50 ng/mL TNF-α as a working concentration in the study.
Fig 1. The effect of plumbagin (PL) and TNF-α on the viability of MM-231 and MM-468 cell lines.
Cells were treated for 24 h with PL in concentration ranges of 1–50 μM (A) and 0.5–10 μM (B) in MM-231 and MM-468 cells, respectively. Both cell lines, MM-231 (C) and MM-468 (D), were treated with TNF-α in a 0–100 ng/mL concentration range. On the x-axis, the circles represent the working concentrations to be used in the study. The percentages of cell survival compared to the control were calculated. The data points are expressed as the mean ± SEM of three independent studies, n = 4. The significance of the difference between the control and treated groups was determined using the one-way ANOVA followed by the Bonferroni’s multiple comparisons. ***p <0.001, ****p <0.0001, and nonsignificant (ns).
The antiproliferative assay was used to evaluate the inhibitory effect of PL on the proliferation of MM-231 and MM-468 cells in comparison with the antiproliferative effect of the standard anticancer drug Taxol®. Inhibition of TNBC cell proliferation was verified by measuring the metabolic activity and the ability to reduce resazurin after a 72-h or 96-h exposure period. In both cell lines, measurements of the proliferation rate at 72 or 96 h of PL exposure showed significant inhibition compared to the rate in the control cells, but there was no significant difference in cell proliferation inhibition between the two periods of incubation (Fig 2A and 2B) for MM-231 and MM-468 cells, respectively. The data also show that PL significantly inhibited MM-231 and MM-468 cell proliferation (p< 0.0001) at ≥2 μM concentrations in MM-231 cells (Fig 2C and 2E) and ≥0.5 μM concentrations in MM-468 cells (Fig 2D and 2F). Additionally, the viability study data indicate that PL was highly potent when tested in MM-468 cells. During the 96-h exposure period, 0.5 μM PL decreased the proliferation rate by 80% as compared with a 70% decrease in cells treated with 1 μM of Taxol® (Fig 2F). Considering these data together with the nontoxic effect of 0.5 μM PL on MM-468 cells (Fig 1B), we excluded the probability that the decrease in proliferation was due to cytotoxicity.
Fig 2. The effect of PL as an antiproliferative agent in MM-231 and MM-468 TNBC cells.
Cells were incubated for 72 or 96 h with PL in concentration ranges of 0–50 or 0–10 μM, respectively (A and B). For the 72-h (C and D) or 96-h exposure periods (E and F), Taxol® (1 μM) was applied to both cell lines. Each data point represents the mean ± SEM of three independent experiments, n = 4. The significance of the difference between 72-h vs. 96-h exposure period was calculated using two-way ANOVA, while that between the control vs. treatment was established with one-way ANOVA, and both were followed by Bonferroni’s multiple comparisons. **** p< 0.0001 indicates the statistically significant difference between the control and treated groups after the 72-h or 96-h exposure periods.
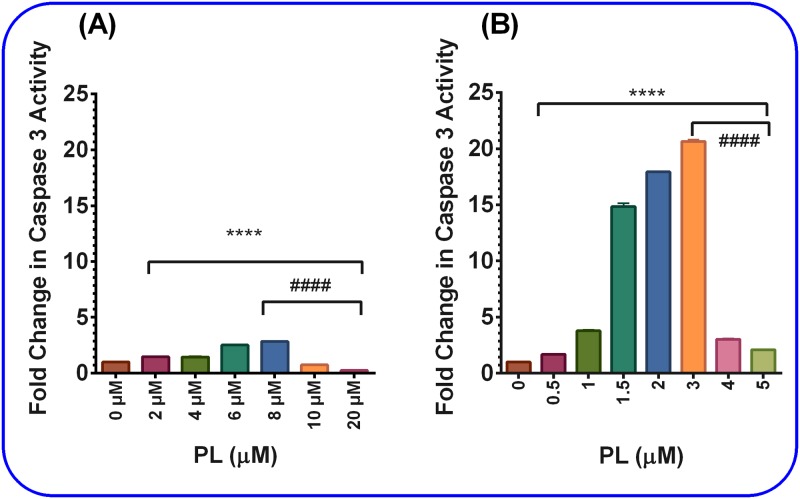
The PL effect on different TNBC cell lines was tested in both MM-231 and MM-468 cells to determine whether apoptosis mediates the antiproliferative effect of PL. The change in caspase-3 activity, as a marker for apoptosis and subsequent cell death, was assessed in both cell lines relative to that in the control cells. After the 24-h treatment period, a significant gradual increase in caspase-3 level (p< 0.0001) in a dose-dependent manner was detected in both cell lysates, reaching its maxima at 8 μM in MM-231 and 3 μM in MM-468 cells. After that, a dramatically significant decrease in caspase-3 activities (p< 0.0001) with increasing PL concentrations was detected in both cell lines. Noticeably, 8 μM PL caused a three-fold increase in caspase-3 activity in MM-231 cells (Fig 3A), while 3 μM PL caused a 20-fold increase in caspase-3 activity in MM-468 cells (Fig 3B). These results confirmed that apoptosis was taking place in both TNBC cell lines and indicated a higher sensitivity of MM-468 cells to PL.
Fig 3. The effect of PL on caspase-3 activation.
PL caused caspase-3 activation in both MM-231 and MM-468 cell lysates after cell treatment with PL for 24 h in concentration ranges of 0–20 μM for MM-231 cells (A) or 0–5 μM for MM-468 cells (B). The data represent two independent studies with n = 3 and are expressed as a fold-increase compared to the control. **** p < 0.0001 indicate the highly significant difference between the control vs. treated cells. Likewise, #### p < 0.0001 between the PL concentration-induced highest caspase level and caspase levels at further increasing PL concentrations in both cell lines.
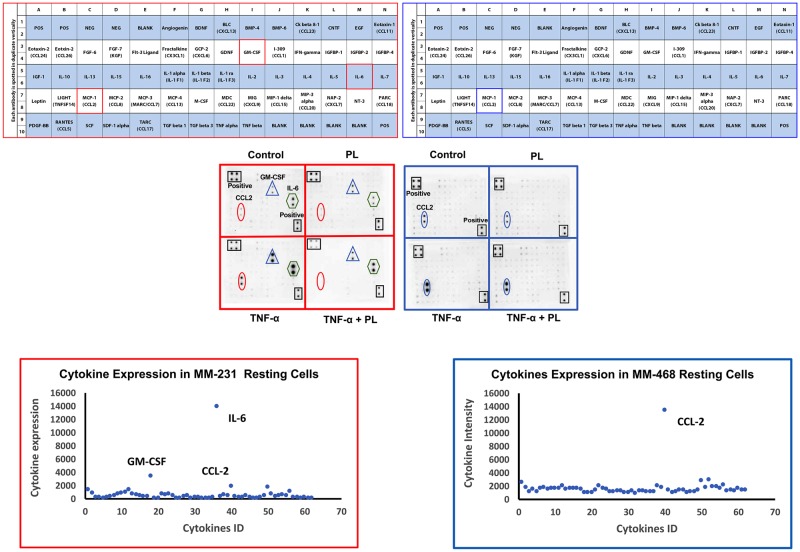
To further assess the PL effects on TNBC cells, we measured the expression of cytokines secreted into both MM-231 and MM-468 cell-free supernatants in the presence and the absence of PL and TNF-α. Optimum concentrations of PL and TNF-α were established for cytokine expression assays according to the viability study results and previous studies [40, 41]. Both cell lines were stimulated with 50 ng/mL TNF-α, in addition to the low doses of PL that slightly affected cell viability (4 μM in MM-231 cells or 1 μM in MM-468 cells). Two different sets of AAH-CYT blots, AAH-CYT-6 and AAH-CYT-7, were used. However, the data obtained from AAH-CYT-7 blot were not significant (not presented). Three cytokines were highly expressed in the resting MM-231 cells: interleukin 6 (IL-6), granulocyte-colony stimulating factor (GM-CSF), and CCL2 (Fig 4A1–4A3). In comparison, CCL2 was the only dominant and highly expressed cytokine in MM-468 cells (Fig 4B1–4B3). Indeed, the data show that CCL2 was highly expressed in both cell lines, although its expression in MM-468 cells was 7-fold higher than in MM-231 cells, as shown in Fig 4A3 and 4B3. High expression of the cytokines was visually observed in the arrays as higher spot intensities upon treatment of both cell lines with 50 ng/mL TNF-α (Fig 4A2 and 4B2). Moreover, PL (4 and 1 μM in MM-231 and MM-468 cells, respectively) attenuated spot intensities.
Fig 4. The effect of PL on different cytokine expression in TNF-α-activated MM-231 and MM-468 TNBC models.
Microarray map AAH-CYT-6 was used to assess chemokine/cytokine expression in the cell-free supernatants (A1 and B1). For each cell line, four blots represent the supernatants of the following cells: control, 4 μM PL (MM-231 cells) or 1 μM PL (MM-468 cells), TNF-α (50 ng/mL)-treated and, lastly, PL and TNF-α-treated. Microarray chemiluminescence detected the changes in cytokine expression in MM-231 and MM-468 cells (A2 and B2). Panels A3 and B3 represent highly expressed cytokines in resting cells. For MM-231 cells, the most-attenuated cytokines are CCL2, GM-CSF, and IL-6. However, CCL2 was the only impacted cytokine in MM-468 cells.
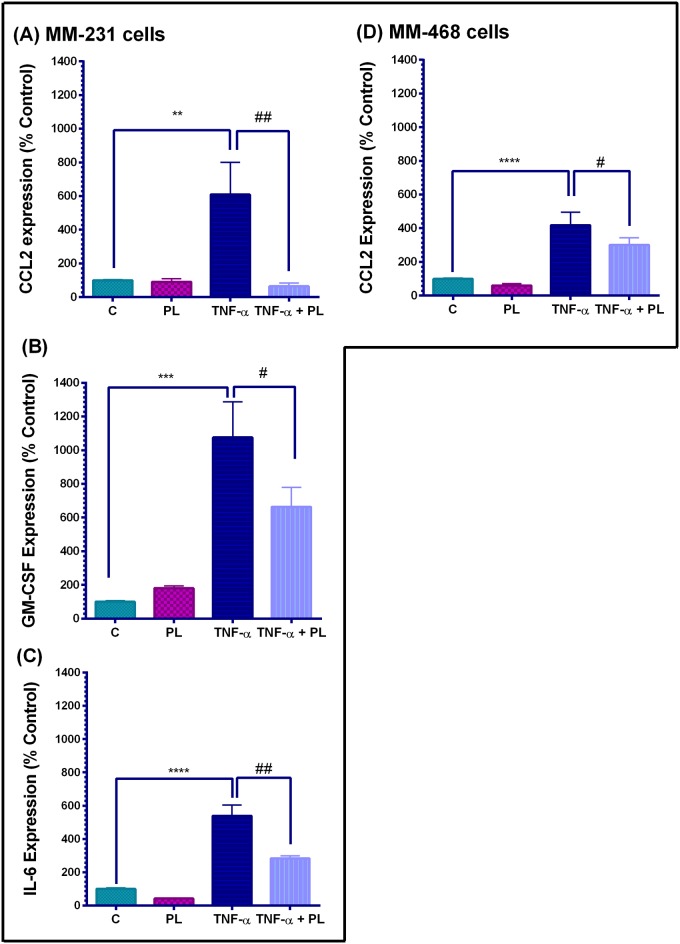
Cytokine array quantification in TNF-α-stimulated TNBC cell lines is presented in Fig 5. Indeed, TNF-α significantly increased the expression of specific cytokines to varying degrees (p<0.01—p<0.0001). In the meantime, no significant difference was found between resting vs. PL-treated cells in both cell lines. In MM-231 cells, the simultaneous presence of PL and TNF-α significantly attenuated three highly TNFα-induced proinflammatory cytokines (Fig 5A–5C), and the CCL2 expression was inhibited by as much as 90% (p < 0.01) (Fig 5A). On the other hand, treatment of TNF-α-stimulated MM-468 cells with PL attenuated the level of highly expressed chemokine CCL2 by 30% (p < 0.05) (Fig 5D). Therefore, these data show different effects of PL on the same CCL2 cytokine in the TNBC cells different by racial origin. Moreover, cytokine GM-CSF exceeded the 10-fold increase compared with the resting MM-231 cells, thus being the most-upregulated cytokine by TNF-α. PL significantly inhibited this cytokine by 40% (p < 0.05) (Fig 5B) and attenuated the highly expressed IL-6 by 50% (p < 0.01) (Fig 5C).
Fig 5. Cytokine array quantification in TNF-α-stimulated TNBC cell lines.
The effect of PL on the expression of CCL2, GM-CSF, and IL-6 in MM-231 cells (A-C), and CCL2 expression in MM-468 cells (D). The normalized data for each cell line show cytokine expression in four sets of experimental cell supernatants; control cells, PL-treated cells (4 or 1 μM PL, in MM-231 and MM-468 cells, respectively), TNF-α-stimulated cells (50 ng/mL), and co-treated cells (TNF-α +PL). The data points are expressed as the mean ± SEM of two independent studies, and the intensities are expressed as percent relative to control. The significant difference between the resting and TNF-α-activated cell groups (*), was determined by an unpaired t-test; the same for TNF-α stimulated vs. TNF-α+PL-treated cells (#). Significance is considered at **p < 0.01, ***p < 0.001, ****p < 0.0001, #p < 0.05 and ##p < 0.01.
To further validate the data, two independent ELISA studies were conducted for MM-231 and MM-468 cells to quantify CCL2 attenuation by PL (Fig 6A and 6B). In both cell lines, a highly significant increase was found in resting vs. TNF-α-treated cells (p < 0.0001 and p < 0.001 for MM-231 and MM-468, respectively) as well as in TNF-α vs. TNF-α + PL-treated cells (p < 0.0001 and p < 0.001 for MM-231 and MM-468, respectively). Overall, the data obtained were consistent with the array findings, and CCL2 was attenuated by 75% and 35% in MM-231 and MM-468 cells, respectively.
Fig 6. The effect of PL on CCL2 release in TNF-α-treated MM-231 (A) and MM-468 (B) cells.
For each cell line, chemokine quantification (pg/mL) was done in four sets of samples. The data points are expressed as the mean ± SEM of two independent studies. The significant difference between the resting vs. TNF-α -activated groups was determined by an unpaired t-test (*), as the same for TNF-α -treated vs. TNF-α + PL-treated cells (#). Significance is considered at ****p < 0.0001, ***p < 0.001, ####p < 0.0001 and ###p < 0.001.
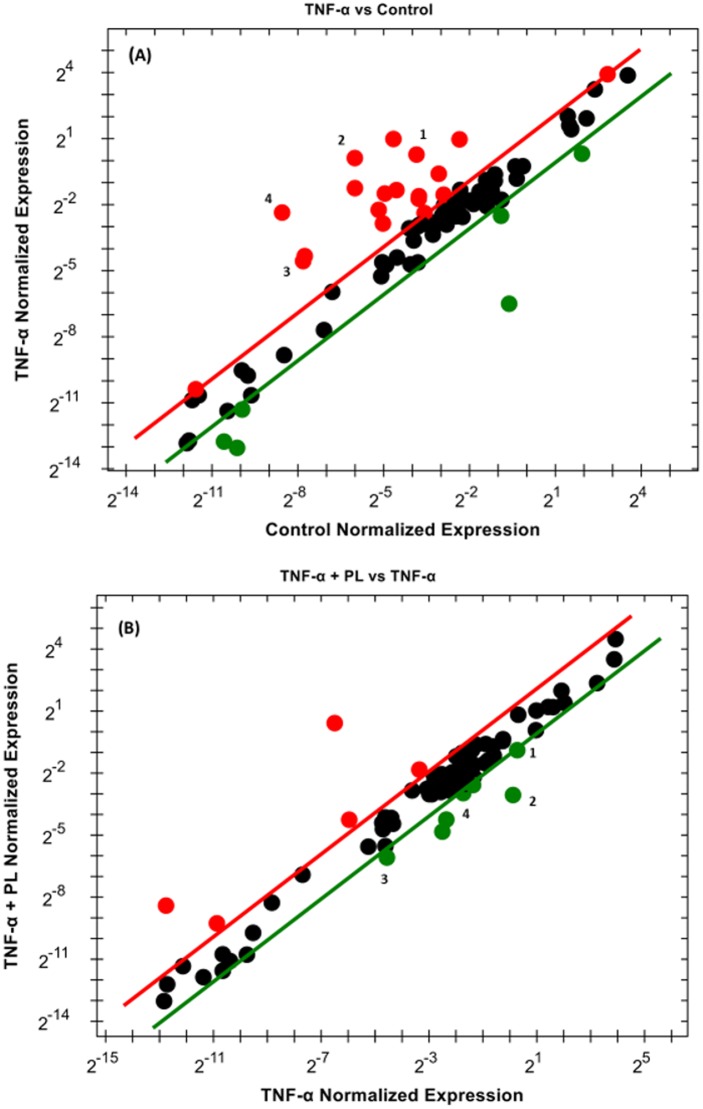
To elucidate the mechanism by which PL attenuates CCL2 release in TNF-α-treated TNBC cell models, RT-PCR was performed to investigate the NF-қB signaling pathway. In both cell lines, the normalized mRNA expression in the NF-қB signaling pathway indicates the cells’ response to either TNF-α or TNF-α + PL. Here, we present only the genes that are significantly repressed by PL. The data show that PL significantly downregulated four genes in both cell lines (BIRC3, CCL2, TLR2, and TNF). However, in MM-231 cells five more genes were downregulated (BCL2A1, ICAM1, IKBKE, IL1β, and LTA) (Table 1). The elevated expression of these genes by TNF-α is visually recognized as red dots in MM-231 and MM-468 cells (Figs 7A and 8A, respectively). In the presence of TNF-α + PL, the same genes appear in green as an indication of PL potency to downregulate their expression (Figs 7B and 8B) in MM-231 and MM-468 cells, respectively.
Table 1. mRNA gene expression changes in MM-231 TNBC.
The left side of the table presents the NF-қB-controlled genes that are upregulated (+ fold-changes) by 50 ng/mL TNF-α. In contrast, the right side presents expression of genes that are downregulated (- fold-changes) by 4 μM PL. p < 0.05 is considered statistically significant.
| Control vs. TNF-α | TNF-α vs. TNF-α + PL | ||||
|---|---|---|---|---|---|
| Target gene | Fold changes | p-value | Target gene | Fold changes | p-value |
| BCL2A1 | 60.29 | < 0.0001 | BCL2A1 | -5.36 | 0.001 |
| BIRC3 | 3.05 | 0.011 | BIRC3 | -1.80 | 0.002 |
| CCL2 | 2.90 | 0.039 | CCL2 | -25.58 | 0.007 |
| ICAM1 | 6.21 | 0.006 | ICAM1 | -5.47 | 0.007 |
| IKBKE | 1.57 | 0.378 | IKBKE | -6.05 | 0.010 |
| IL1β | 11.79 | 0.003 | IL1B | -9.50 | 0.004 |
| LTA | 5.67 | 0.005 | LTA | -6.98 | 0.005 |
| TLR2 | 2.43 | 0.116 | TLR2 | -2.76 | 0.040 |
| TNF | 56.13 | < 0.0001 | TNF | -9.25 | 0.0002 |
Fig 7. Scatter plot for MM-231 cells.
Normalized expression of mRNA target genes for the control vs. TNF-α-stimulated cells (A) and TNF-α-stimulated vs. cotreated cells (B). The plot image shows the following changes in target gene expression based on the threshold set: upregulation, red dots; downregulation, green dots; and no change, black dots. The most downregulated genes are enumerated from 1–9 as follows: BCL2A1, BIRC3, CCL2, ICAM1, IKBKE, IL1β, LTA, TLR2, and TNF.
Fig 8. Scatter plot for MM-468 cells.
Normalized mRNA expression of target genes for the control vs. TNF-α stimulated cells (A) and TNF-α vs. TNF-α + PL (B). The plot image shows the following changes in target gene expression based on the threshold set: upregulation, red dots, downregulation, green dots, and no change, black dots. The most-downregulated genes are enumerated from 1–4 as follows: BIRC3, CCL2, TLR2, and TNF.
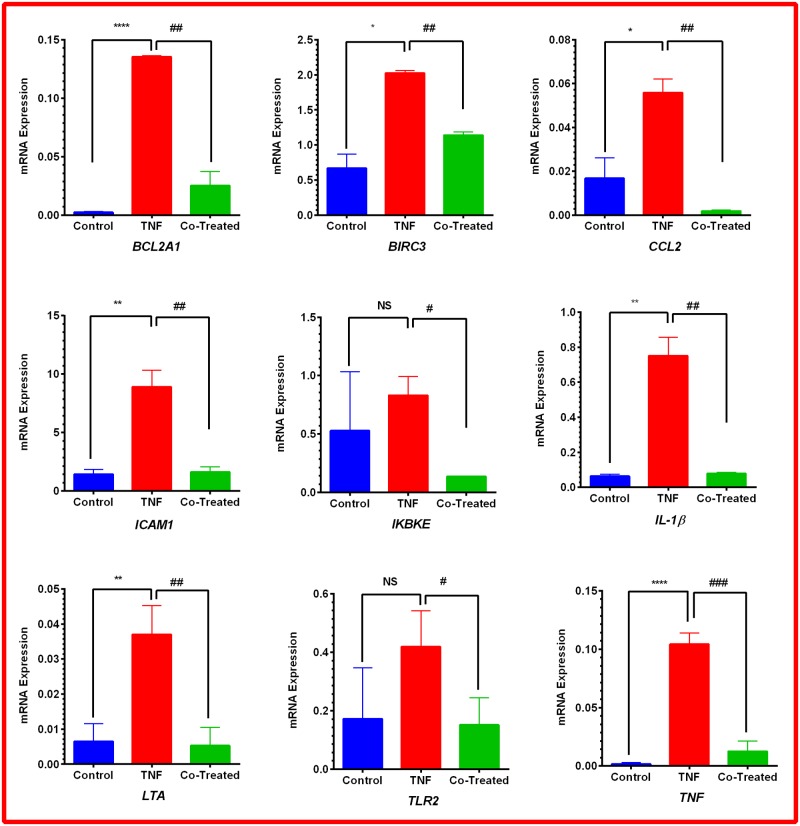
Notably, among these nine upregulated genes in MM-231 cells, seven genes were significantly upregulated by TNF-α, giving the highest significant (p < 0.0001) fold-increases in BCL2A1 (60.29) and TNF (56.13) expression (Table 1 and Fig 9). On the other hand, in MM-468 cells, TNF and CCL2 genes were significantly upregulated (p < 0.05) with 70.09 and 53.05-fold increases, respectively (Table 2 and Fig 10). Interestingly, in the TNF-α-treated cells, the expression of TNF and CCL2 genes was higher in MM-468 cells than in MM-231 cells; however, the fold-decrease was always higher in MM-231 cells. Noticeably, the fold-increase in TNF gene in MM-468 cells was 70.09 vs. 56.13 in MM-231 cells. However, MM-231 cells were more affected by PL, with the expression of TNF decreasing 9.25-fold compared with 3.44-fold in MM-468 cells. Similarly, the fold-increase in CCL2 was 2.90 vs. 53.05 in MM-231 and MM-468 cells, respectively. Nevertheless, a dramatic decrease in expression of this gene was found in MM-231 cells compared to MM-468 cells (25.58 vs. 8.75, respectively). Nevertheless, whereas a nonsignificant TNF-α-induced upregulation was observed for two genes, IKBKE and TLR2 (Table 1 and Fig 9), PL significantly inhibited their mRNA expression (p < 0.05).
Fig 9. Gene expression quantification in MM-231 cells.
Normalized data show a significant increase in seven genes in the TNF-α-stimulated MM-231 cells vs. resting cells. On the other hand, nine genes were significantly repressed in cotreated cells. The data points are expressed as the mean ± SEM of three independent studies. The significance of the difference was determined by an unpaired t-test between the resting vs. TNF-α-activated cells (*), as well as TNF-α-treated vs. TNF-α + PL-treated cells (#). Significance is considered at *p < 0.05, **p < 0.01, ****p < 0.0001, #p < 0.05, ##p < 0.01, and ###p < 0.001 or the difference was nonsignificant (ns).
Table 2. mRNA gene expression changes in MM-468 TNBC.
The left side of the table presents the NF-қB-controlled genes that are upregulated (+ fold-changes) by 50 ng/mL TNF-α. In contrast, the right side presents expression of genes that are downregulated (- fold-changes) by 1 μM PL in MM-468 cells. p < 0.05 is considered statistically significant.
| Control vs TNF-α | TNF-α vs TNF-α + PL | ||||
|---|---|---|---|---|---|
| Target gene | Fold changes | p-value | Target gene | Fold changes | p-value |
| BIRC3 | 14.92 | 0.008 | BIRC3 | -2.211 | 0.040 |
| CCL2 | 53.05 | 0.030 | CCL2 | -8.750 | 0.038 |
| TLR2 | 9.28 | 0.013 | TLR2 | -2.870 | 0.024 |
| TNF | 70.09 | 0.012 | TNF | -3.441 | 0.044 |
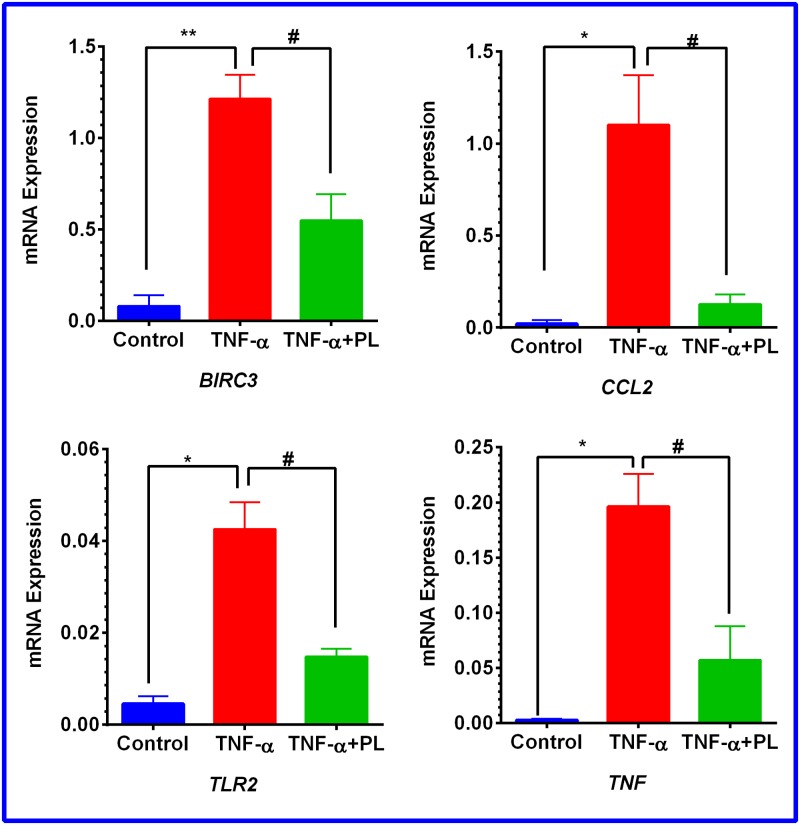
Fig 10. Gene expression quantification in MM-468 cells.
Normalized data show a significant increase in four genes in TNF-α-stimulated cells. On the other side, significant gene repression was observed in cotreated cells. The data points are expressed as the mean ± SEM of two independent experiments. The significant difference between resting and TNF-α-activated groups was determined by an unpaired t-test (*), as the same for TNF-α-treated vs. TNF-α + PL-treated cells (#). Significance is considered at *p < 0.05, **p < 0.01, and #p < 0.05.
Discussion
In the cancer cells, targeting inter- and intra-cellular signaling pathways involved in proliferation, inflammation, and apoptosis is a well-suited approach to the development of TNBC anticancer agents. NF-қB is a critical component of cancer proliferation and metastasis [14]. It regulates the genes linked to tumor proliferation [42], plays a proapoptotic role [14] and mediates the expression of antiapoptotic genes [43].
The present study elucidated the anticancer mechanism of the natural quinone PL. This compound attenuated the TNF-α-mediated increase in expression of CCL2 and other cytokines through the downregulation of the mRNA expression of specific genes involved in the NF-қB pathway (Figs 4–10). The data obtained show the differences in the antiproliferative, apoptotic, and cytotoxic effects of PL with a higher response in MM-468 cells than in MM-231 TNBC cells (Figs 1–3).
Under normal conditions, the secretion and release of signals for maintaining homeostasis are precisely controlled. However, deregulation of these signals by triggering distinctive gene expression is the major key to uncontrolled cell proliferation of cancer tissue [44]. Furthermore, the products of these genes, including caspases, are critical in the regulation of apoptosis. Caspase-3, a downstream caspase has been shown to play a critical role in the terminal execution segment of apoptosis induced by diverse stimuli [45, 46]. Previous studies have reported that PL can affect cellular proliferation, carcinogenesis, and radio-resistance, and all are regulated by activation of the transcription factor NF-қB. Consistent with the current data, PL exhibited significant antitumor effects by inducing apoptosis and decreasing proliferation in both MM-231 and MM-468 TNBC cells, yet MM-468 was more sensitive to PL in the proliferation and apoptosis experiments (Figs 2 and 3, respectively).
It is known that chemokine CCL2 is released into the tumor microenvironment by endothelial cells and fibroblasts [47] to activate lymphocytes and macrophages [48]. Additionally, CCL2 is overexpressed in several malignant tumors, including MM-231-tumor xenograft for TNBC models [49, 50]. Indeed, the reported overexpression of CCL2 was linked to the decreased survival of TNBC patients [51]. Additionally, the reduction of CCL2 expression after antibody treatment and genetic modification [52] led to reduced metastasis and improved survival [53]. Moreover, targeting CCL2 decreased the recruitment of both tumor-associated macrophages (TAMs) and myeloid-derived suppressor cells (MDSCs) to the tumor site and reduced primary tumor growth [54]. Downregulation of CCL2 was also reported to enhance tumor cell apoptosis [55] and block the recruitment of inflammatory monocytes, inhibit lung metastasis of BC cells and prolong the survival of tumor-bearing mice [52] and TNBC patients [56]. Thus, the reported wide distribution and increase in the expression of this cytokine in different cancers, particularly TNBC, indicate its significant role in cancer aggressiveness.
The mechanism by which CCL2 impacts on cancer is reported to be widely diverse. The interaction between CCL2 and its primary receptor CCR2 is crucial for recruiting large numbers of monocytes to the tumor tissue [51]. This interaction can lead to the enhancement of tumor growth and survival [57], initiation of tumor angiogenesis [56], inhibition of antitumor immunity [58], and finally, promotion of tumor invasion and metastasis [59]. Moreover, CCL2 regulates cancer stem cells through an abnormal Notch activation in various tissues [60], which is also fundamental to BC [61, 62].
Upon exposure to TNF-α, overexpressed extracellular CCL2 was dramatically and significantly amplified in both cell lines (Figs 4–6), an observation consistent with previous reports [63, 64]. It has been reported that TNF-α is highly expressed in BC patients [18]. However, its function as tumor necrosis or tumor promoting factor is related to whether it is endogenous or is administered in a high dose, respectively [65]. When adding the quinone PL to our TNF-α-activated TNBC cell models, CCL2 expression was significantly and differentially reduced. At nontoxic concentrations of PL, despite a highly aggressive nature of TNBC in African-Americans, the MM-468 model showed lower CCL2 inhibition compared to MM-231 (Figs 5 and 6). The presence of over-expressed and conceivably stimulated CCL2 in both TNBC racially disparate models emphasizes its crucial role in TNBC aggressiveness and, hence, highlights involvement of the PL suppression mechanism in TNBC treatment, particularly, of CCL2 in the MM-231 model that was dramatically suppressed (Figs 5A and 6A).
In BC, NF-қB activation increases the expression of CCL2 [66] and other inflammatory cytokines/chemokines/interleukins, including IL-1, IL-6, IL-8, TNF-α, and CXCL8, as well as cyclooxygenase 2 (COX2) and nitric oxide synthase (NOS) [67, 68]. Moreover, NF-қB influences cancer radio- and chemo-resistance [69]. Our results show that upon exposure to TNF-α, the number of genes significantly upregulated by NF-қB was higher in MM-231 than MM-468 cells, even with the previously reported poor sensitivity of MM-231 cells at the same TNF-α concentration as in our study [40].
The profile of NF-қB-regulated genes in our data indicates the racial differences in BC (Table 1 and Figs 7–10). Consistently, in TNF-α + PL-treated MM-468 cells, the reduction in the TNFα-induced increase in gene expression was always lower than in their counterpart MM-231 cells (Table 1). In the presence of TNF-α, PL was able to significantly downregulate nine genes in MM-231 cells (BCL2A1, BIRC3, CCL2, ICAM1, IKBKE, IL1β, LTA, TLR2, and TNF) (Table 1). These genes are involved in other inter/intracellular mechanisms controlling apoptosis, proliferation, invasion, and metastasis. The MM-468 cell line had only four genes downregulated (BIRC3, CCL2, TLR2, and TNF) (Table 2). These data indicate that PL is more potent in affecting the CA MM-231 cells through the NF-қB-downregulated genes than the AA MM-468 cell model of TNBC.
PL dramatically decreased the TNF-α-stimulated TNF and LTA gene expression only in MM-231 cells (Table 1). Together, the TAM-derived TNF and LTA are involved in breast carcinogenesis [70, 71]. The LTA gene is a member of the TNF family that activates NF-қB [72] and facilitates inflammation [73]. Mainly, LTA was considered as an important biomarker of BC predisposition, and its gene polymorphism was linked to an elevated risk of BC in AA [74], and CA [75].
In both cell lines, TNF-α upregulated BIRC3 expression, which was significantly suppressed by PL. BIRC3 is a member of the family of inhibitors of apoptosis (IAP), and NF-қB is involved in its gene transcription [76]. The IAP genes can, in turn, drive and potentiate NF-қB activity [77] and consequently increase CCL2 expression. Particularly, the BIRC3 gene was reported to regulate the caspase activity [78], apoptosis, cellular differentiation, and proliferation [79]. Rationally, its repressor PL showed an antiproliferative mechanism by inducing G2-M arrest and autophagy in BC cells MCF-7 and MM-231 [80]. Thus, our data highlight the importance of PL in targeting BIRC3 to indirectly attenuate CCL2 expression and inhibit proliferation in both TNBC cell lines.
Additionally, TLR2 was equally downregulated by PL in both cell lines (~3-fold decrease). Our results are consistent with a previous study that linked TLR2 inhibition to the reduction of constitutively active NF-қB, inhibition of the cytokine IL-6 production and decrease in cell proliferation [14]. TLR2/4 and their coactivator CD14 are highly expressed in ER- tumors [81] and are involved in NF-қB activation and an increase in metastasis [82, 83]. Thus, our data support recently proposed TLR2 inhibition in fighting BC [84].
Notably, in MM-231 cells, more genes were significantly downregulated by PL: BCL2A1, ICAM1, IKBKE, and IL-1β. These genes are overexpressed in TNBC cells and involved in the NF-қB pathway activation and CCL2 release. The BCL2A1 gene is critical for drug resistance [85, 86]. Therefore, targeting BCL2A1 has shown preclinical promise, either by itself or in combination with other anticancer agents [87], similarly to the ICAM1 gene [88]. Downregulation of the IKBKE gene was previously reported to attenuate CCL2 release [89] that led to cancer cell death [90]. Additionally, the IL-1β gene is linked to the invasiveness of TNBC cells [91], and certainly, downregulation of this gene will reduce cancer cell proliferation, invasion, and metastasis [21].
In summary, the current study sheds light on a novel compound targeting TNBC, among African Americans in particular. This compound impacts MM-231 and MM-468 cells differently. PL attenuated the expression of three cytokines (CCL2, IL-6, and GM-CSF) in MM-231 cells and cytokine CCL2 in MM-468 cells. The data obtained showed the potency of PL to attenuate many regulatory genes involved in the NF-қB pathway, the major factor in releasing CCL2. PL downregulated the expression of four genes in MM-468 cell model shared with MM-231 cells (BIRC3, CCL2, TLR2, and TNF), and five more distinct genes were downregulated in the MM-231 cells (BCL2A1, ICAM1, IKBKE, IL1β, and LTA). This finding explains the dramatic downregulation of CCL2 in MM-231 cells compared to their counterpart in MM-468 cells. However, PL was more potent in the MM-468 than in MM-231 cells in decreasing cell viability and proliferation and inducing apoptosis by increasing caspase-3. These findings might indicate the existence of two different anticancer mechanisms of PL in TNBC cells, a mechanism that tends to have a more effective apoptotic action in MM-468 cells, and another that has a more NF-қB-induced CCL2 inhibitory action in MM-231 cells. In conclusion, the data obtained in this study indicate that the quinone compound PL could be an attractive targeting agent for cancer therapy for African American women with TNBC.
Experimental section
Materials and reagents
PL (purity 99%), dimethyl sulfoxide (DMSO), 0.25% Trypsin-EDTA solution, Alamar Blue® (solution of resazurin fluorescence dye), Taxol® (paclitaxel), and lipopolysaccharides from Escherichia coli O111: B4 (LPS) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Cell culture flasks and plates, Dulbecco’s Modified Eagle Medium (DMEM) and fetal bovine serum (FBS) were purchased from VWR International (Radnor, PA, USA). Penicillin/streptomycin and DPBS were obtained from Atlanta Biologicals (Atlanta, GA, USA). Human Cytokine Antibody Array Kit (Cat # AAH-CYT-1000), Human MCP-1 ELISA kit (Cat # ELH-MCP1) and TNF-α were purchased from RayBiotech (Norcross, GA, USA). SsoAdvancedTM Universal SYBR® Green Supermix and NF-қB Signaling Pathway (SAB Target List) H96 were purchased from Bio-Rad (Bio-Rad, Hercules, CA). DNA-free™ Kit (Cat # AM1907) and EnzChek® Caspase-3 Assay were purchased from Life Technologies Inc. (Grand Island, NY, USA).
Cell culture
TNBC cell models, MM-231 and MM-468, were purchased from American Type Culture Collection (ATCC). Both cell lines were grown in 75-cm TC flasks at 37°C in humidified 5% CO2 incubator and were subcultured, as needed, with trypsin/EDTA. The DMEM growth medium contained 4 mM L-glutamine and was supplemented with 10% heat-inactivated FBS (v/v), and 1% penicillin/streptomycin salt solution (100 U/mL and 0.1 mg/mL, respectively). The DMEM experimental medium was phenol-free and supplemented with 2.5% heat-inactivated FBS.
Cell viability assay
In this experiment, cells were incubated overnight in the experimental media at a density of 5×104 cell/well in 96-well plates. Both types of cells were treated for 24 h with TNF-α (0–100 ng/mL) or PL (concentration ranges of 0–50 μM in MM-231 or 0–10 μM in MM-468 cells). PL was solubilized in DMSO, while TNF-α was dissolved in cell culture water. Control wells were treated with DMSO at the highest used concentration (< 0.1%). Equivalently treated wells without cells were used as blanks. In this assay, Alamar Blue® was used to determine cell viability at a concentration level of 10% v/v and an incubation time up to 6 h. The fluorescent fuchsia—reduced resazurin dye was measured at an excitation/emission of 530/590 nm using a Synergy HTX Multi-Reader (BioTek, USA).
Cell proliferation study
The effects of PL on cell proliferation were determined for MM-231 and MM-468 TNBC cells based on the dose-response viability study concentrations [92] using Alamar Blue®. Briefly, cells were plated at an initial density of 5×103 cell/well in 96-well plates and incubated overnight. The cells were treated for 72 or 96 h with PL at concentrations ranging between 0–50 μM or 0–10 μM in MM-231 or MM-468 cells, respectively, in a final volume of 200 μL/well. Control cells were exposed to DMSO at a concentration < 0.1%. Taxol® was used as a positive control only at 1 μM concentration level [93], and equivalent wells without cells were used as a blank. Proliferation was measured after the predesigned experimental period by adding Alamar Blue® to each well at 10% v/v and incubating for an additional 4 h. The plates were read at an excitation/emission of 530/590 nm using a Synergy HTX Multi-Reader (BioTek, USA).
Caspase-3 apoptosis study
The apoptotic effect of PL was determined in MM-231 and MM-468 cells using the EnzChek® Caspase-3 Assay kit. The assay detects apoptosis by assessing the increase in caspase-3 activity. Briefly, each cell line was seeded at an initial concentration of 1×106 cell/well in 6-well plates and incubated overnight. The tested concentrations were determined based on the IC50 values of the dose-response viability curve for each cell line. After 24 h, cells were treated with PL at concentrations ranging between 0–20 or 0–5 μM in MM-231 or MM-468 cells, respectively, in a final volume of 3 mL/well experimental media. Control cells were exposed to DMSO at a concentration < 0.1%. After the 24 h incubation period, treated cells from each well were harvested, pelleted, and washed in PBS. Cell pellets were resuspended in 50 μL lysis buffer (200 mM Tris, pH 7.5, 2 M NaCl, 20 mM EDTA, 0.2% Triton™ X-100) for 30 min on ice followed by centrifugation for 5 min at 5,000 × g to pellet the debris. Lastly, 50 μL of each of the sample supernatant and the reaction buffer-substrate working solution (50 mM PIPES, pH 7.4, 10 mM EDTA, 0.5% CHAPS, 1 M DTT/5 mM Z-DEVD-R110), were combined in another microplate well for 30 min at RT, and the fluorescence background was determined by using 50 μL of the cell lysis buffer. Fluorescence intensity for each sample was measured (excitation/emission ~342/441 nm) using a Synergy HTX Multi-Reader (BioTek, USA).
Human cytokine/chemokine protein microarray
For cytokine microarray analysis, four flasks for each cell line were seeded at a density of 10×106 cells/75-cm2 TC flask using the same cell culture media. On the day of the experimentation, the media were discarded, and the cells were immediately washed with phenol- free experimental media. Based on our cytotoxicity assay, the MM-231 cells were treated as follows: TNF-α (50 ng/mL), PL (4 μM), or TNF-α + PL (50 ng/mL + 4 μM, respectively). Similarly, MM-468 cells were treated with TNF-α (50 ng/mL), PL (1 μM), and TNF-α + PL (50 ng/mL + 1 μM, respectively). In both experimental sets, control samples were exposed to only DMSO at a concentration < 0.1%. After a 24-h exposure period, the cell-free supernatant of each sample was collected, aliquoted, and stored at -80°C for later use. At the same time, the cells from each sample were pelleted and similarly stored at -80°C for RT-PCR study. For each cell line, a semiquantitative method was established to evaluate chemokine/cytokine expression in the cell culture supernatants using antibody-coated array membranes. The assay was conducted following the protocols supplied with the kits from RayBiotech. Briefly, membranes were carefully placed in the incubation tray and blocked with the provided buffer on a shaker for 30 min at RT. Thereafter, the blocking buffer was decanted, and replaced with 1 mL cell-free supernatant from resting, PL-treated, TNF-α-stimulated or cotreated cells, then placed overnight on a low-speed shaker at 4°C. After 24 h, the media were decanted from each chamber, and the membranes were washed with the kit buffers. Next, 1 mL of freshly constituted biotinylated antibody cocktail was pipetted to each membrane and incubated at RT for 2 h followed by washing with the same wash buffer. The membranes were incubated again for another 2 h with 2 mL of diluted horseradish peroxidase-conjugated streptavidin (HRP-Streptavidin) followed by the final washes. Spot intensities on the blots were evaluated using chemiluminescence detection. The blot images were captured using a Flour-S Max Multiimager (Bio-Rad Laboratories, Hercules, CA, USA), and the spot intensities were obtained with the Quantity-One Software (Bio-Rad Laboratories, Hercules, CA). The Excel-based data analysis was established, using the Human Cytokine Array software C1000 (CODE: S02-AAH-CYT-1000) from RayBiotech.
Human MCP-1 cytokine ELISA study
Enzyme-Linked Immunosorbent Assay (ELISA) kits were used for both cell lines to confirm the effect of PL on CCL2 chemokine expression detected by the microarray. Briefly, the standard curves, samples, and reagents were prepared at RT. Standards and samples of 100 μL were incubated with the antibody precoated 96-well ELISA plates for 2.5 h. The supernatant was replaced by 100 μL of freshly constituted biotinylated antibody for another hour, then decanted. Streptavidin solution (100 μL) was added for 45 min followed by an addition of 100 μL of the substrate reagent for 30-min incubation. Washes were always performed after each step according to the manufacturer’s protocol. The reaction was terminated by the addition of 50 μL of a stop-solution, and the intensity was measured at 450 nm using a Synergy HTX Multi-Reader (BioTek, USA).
Reverse transcription-polymerase chain reaction (RT- RCR) array
The current study was performed using the previously -80°C-stored cells as mentioned previously. The total RNA was isolated from each sample using 1 mL Trizol reagent. Thereafter, samples were treated with chloroform (0.2 mL), vortexed, incubated at RT for 2–3 min and centrifuged at 10,000 x g for 15 min at 2–8°C. The aqueous phase was collected, and the RNAs were precipitated by mixing with 0.5 mL of isopropyl alcohol. RNA pellets were then dissolved in nuclease-free water to measure the RNA quantity and quality using a NanoDrop spectrophotometer, and cDNA was synthesized using the iScript™ cDNA Synthesis Kit. Following the manufacturer’s protocol, the obtained cDNA was reconstituted in nuclease-free water, and the NF-қB Signaling Pathway 96-well plate was loaded with 10 μL each of the reconstituted cDNA (2.3 ng) and SsoAdvanced™ Universal SYBR® Green Supermix, then placed for 5 min in a shaker and centrifuged at 1000 x g for 1 min. The PCR run was established with 39 cycles of denaturation as follows: 30-sec activation at 95°C, 10-sec denaturation at 95°C; 20-sec annealing at 60°C; and 31-sec extension at 65°C using a Bio-Rad CFX96 Real-Time System (Hercules, Ca, USA). All real-time PCR reactions were performed in triplicate for each cell line.
Statistical analysis
The data were analyzed using the Graph Pad Prism 6.2 Software (San Diego, CA, USA). All data points were obtained from the average of at least two independent studies and expressed as the mean ± SEM. For viability studies, IC50 values were determined by nonlinear regression with the R2 best fit and lowest 95% confidence interval. The significance of the difference between each control and its related treated groups was determined using the one-way ANOVA followed by Bonferroni’s multiple comparison tests. For the proliferation assay, the statistical analysis was performed with a two-way ANOVA between the 72-h and 96-h exposure periods and followed with Bonferroni’s multiple comparison tests. The significance of the difference was considered at *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. For blots and ELISA studies, Student’s t-test was used to verify the significance of the difference between the control and TNF-α groups, or between the TNF-α and TNF-α +PL groups. Gene expression was analyzed using the CFX 3.1 Manager software (Bio-Rad, Hercules, CA) and verified similarly with Student’s t-test.
Data Availability
All relevant data are within the manuscript.
Funding Statement
The work was supported by US National Institute on Minority Health and Health Disparities (NIH) grants G12 MD 007582 and P20 MD 006738 to KFAS.
References
- 1.DeSantis CE, Fedewa SA, Goding Sauer A, Kramer JL, Smith RA, Jemal A. Breast cancer statistics, 2015: Convergence of incidence rates between black and white women. CA: a cancer journal for clinicians. 2016;66(1):31–42. Epub 2015/10/30. 10.3322/caac.21320 . [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA: a cancer journal for clinicians. 2018;68(1):7–30. Epub 2018/01/10. 10.3322/caac.21442 . [DOI] [PubMed] [Google Scholar]
- 3.Anders CK, Carey LA. Biology, metastatic patterns, and treatment of patients with triple-negative breast cancer. Clinical breast cancer. 2009;9 Suppl 2: S73–81. Epub 2009/07/15. 10.3816/CBC.2009.s.008 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beaumont T, Leadbeater M. Treatment, and care of patients with metastatic breast cancer. Nursing standard (Royal College of Nursing (Great Britain): 1987). 2011;25(40):49–56. Epub 2011/07/16. 10.7748/ns2011.06.25.40.49.c8566 . [DOI] [PubMed] [Google Scholar]
- 5.Fernandez Y, Cueva J, Palomo AG, Ramos M, de Juan A, Calvo L, et al. Novel therapeutic approaches to the treatment of metastatic breast cancer. Cancer treatment reviews. 2010;36(1):33–42. Epub 2009/11/04. 10.1016/j.ctrv.2009.10.001 . [DOI] [PubMed] [Google Scholar]
- 6.Liu P, Kumar IS, Brown S, Kannappan V, Tawari PE, Tang JZ, et al. Disulfiram targets cancer stem-like cells and reverses resistance and cross-resistance in acquired paclitaxel-resistant triple-negative breast cancer cells. British journal of cancer. 2013;109(7):1876–85. Epub 2013/09/07. 10.1038/bjc.2013.534 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albain KS, Unger JM, Crowley JJ, Coltman CA Jr., Hershman DL. Racial disparities in cancer survival among randomized clinical trials patients of the Southwest Oncology Group. Journal of the National Cancer Institute. 2009;101(14):984–92. Epub 2009/07/09. 10.1093/jnci/djp175 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuper H, Adami HO, Trichopoulos D. Infections as a major preventable cause of human cancer. Journal of internal medicine. 2000;248(3):171–83. Epub 2000/09/06. . [DOI] [PubMed] [Google Scholar]
- 9.Wang X, Belguise K, Kersual N, Kirsch KH, Mineva ND, Galtier F, et al. Oestrogen 3signaling inhibits invasive phenotype by repressing RelB and its target BCL2. Nature cell biology. 2007;9(4):470–8. Epub 2007/03/21. 10.1038/ncb1559 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pierce BL, Ballard-Barbash R, Bernstein L, Baumgartner RN, Neuhouser ML, Wener MH, et al. Elevated biomarkers of inflammation are associated with reduced survival among breast cancer patients. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2009;27(21):3437–44. Epub 2009/05/28. 10.1200/jco.2008.18.9068 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karin M. NF-kappaB as a critical link between inflammation and cancer. Cold Spring Harbor perspectives in biology. 2009;1(5):a000141 Epub 2010/01/13. 10.1101/cshperspect.a000141 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zubair A, Frieri M. Role of nuclear factor-kB in breast and colorectal cancer. Current allergy and asthma reports. 2013;13(1):44–9. Epub 2012/09/08. 10.1007/s11882-012-0300-5 . [DOI] [PubMed] [Google Scholar]
- 13.Karin M, Cao Y, Greten FR, Li ZW. NF-kappaB in cancer: from innocent bystander to major culprit. Nature Reviews Cancer. 2002;2(4):301–10. Epub 2002/05/11. 10.1038/nrc780 . [DOI] [PubMed] [Google Scholar]
- 14.Prasad S, Ravindran J, Aggarwal BB. NF-kappaB and cancer: how intimate is this relationship. Molecular and cellular biochemistry. 2010;336(1–2):25–37. Epub 2009/10/14. 10.1007/s11010-009-0267-2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamamoto Y, Gaynor RB. Therapeutic potential of inhibition of the NF-kappaB pathway in the treatment of inflammation and cancer. The Journal of clinical investigation. 2001;107(2):135–42. Epub 2001/02/13. 10.1172/JCI11914 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katanov C, Lerrer S, Liubomirski Y, Leider-Trejo L, Meshel T, Bar J, et al. Regulation of the inflammatory profile of stromal cells in human breast cancer: prominent roles for TNF-alpha and the NF-kappaB pathway. Stem cell research & therapy. 2015;6:87 Epub 2015/05/01. 10.1186/s13287-015-0080-7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allavena P, Garlanda C, Borrello MG, Sica A, Mantovani A. Pathways connecting inflammation and cancer. Current opinion in genetics & development. 2008;18(1):3–10. Epub 2008/03/08. 10.1016/j.gde.2008.01.003 . [DOI] [PubMed] [Google Scholar]
- 18.Leek RD, Landers R, Fox SB, Ng F, Harris AL, Lewis CE. Association of tumour necrosis factor alpha and its receptors with thymidine phosphorylase expression in invasive breast carcinoma. British journal of cancer. 1998;77(12):2246–51. Epub 1998/07/02. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang N, Liang H, Zen K. Molecular mechanisms that influence the macrophage m1-m2 polarization balance. Frontiers in immunology. 2014;5:614 Epub 2014/12/17. 10.3389/fimmu.2014.00614 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baumgarten SC, Frasor J. Minireview: Inflammation: an instigator of more aggressive estrogen receptor (ER) positive breast cancers. Molecular endocrinology (Baltimore, Md). 2012;26(3):360–71. Epub 2012/02/04. 10.1210/me.2011-1302 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin Y, Chen X, Shu Y. Gene expression of the invasive phenotype of TNF-alpha-treated MCF-7 cells. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2009;63(6):421–8. Epub 2009/07/01. 10.1016/j.biopha.2009.04.032 . [DOI] [PubMed] [Google Scholar]
- 22.Balkwill F. TNF-alpha in promotion and progression of cancer. Cancer metastasis reviews. 2006;25(3):409–16. Epub 2006/09/05. 10.1007/s10555-006-9005-3 . [DOI] [PubMed] [Google Scholar]
- 23.Matsushima K, Larsen CG, DuBois GC, Oppenheim JJ. Purification and characterization of a novel monocyte chemotactic and activating factor produced by a human myelomonocytic cell line. The Journal of experimental medicine. 1989;169(4):1485–90. Epub 1989/04/01. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J, Patel L, Pienta KJ. CC chemokine ligand 2 (CCL2) promotes prostate cancer tumorigenesis and metastasis. Cytokine & growth factor reviews. 2010;21(1):41–8. Epub 2009/12/17. 10.1016/j.cytogfr.2009.11.009 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nam JS, Kang MJ, Suchar AM, Shimamura T, Kohn EA, Michalowska AM, et al. Chemokine (C-C motif) ligand 2 mediates the prometastatic effect of dysadherin in human breast cancer cells. Cancer research. 2006;66(14):7176–84. Epub 2006/07/20. 10.1158/0008-5472.CAN-06-0825 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jetty A, Subhakar C, Rajagopal D, Jetty M, Subramanyam M, Marthanda Murthy M. Antimicrobial activities of neo- and 1-epineo-isoshinanolones from Plumbago zeylanica roots. Pharmaceutical biology. 2010;48(9):1007–11. Epub 2010/08/25. 10.3109/13880200903433760 . [DOI] [PubMed] [Google Scholar]
- 27.Tilak JC, Adhikari S, Devasagayam TP. Antioxidant properties of Plumbago zeylanica, an Indian medicinal plant and its active ingredient, plumbagin. Redox report: communications in free radical research. 2004;9(4):219–27. Epub 2004/10/14. 10.1179/135100004225005976 . [DOI] [PubMed] [Google Scholar]
- 28.Khaw AK, Sameni S, Venkatesan S, Kalthur G, Hande MP. Plumbagin alters telomere dynamics, induces DNA damage and cell death in human brain tumour cells. Mutation research Genetic toxicology and environmental mutagenesis. 2015;793:86–95. Epub 2015/11/02. 10.1016/j.mrgentox.2015.06.004 . [DOI] [PubMed] [Google Scholar]
- 29.Poosarla A, D NR, Athota RR, Sunkara VG. Modulation of T cell proliferation and cytokine response by Plumbagin, extracted from Plumbago zeylanica in collagen induced arthritis. BMC complementary and alternative medicine. 2011;11:114 Epub 2011/11/17. 10.1186/1472-6882-11-114 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Messeha SS, Zarmouh NO, Mendonca P, Kolta MG, Soliman KFA. The attenuating effects of plumbagin on pro-inflammatory cytokine expression in LPS-activated BV-2 microglial cells. Journal of neuroimmunology. 2017. Epub 2017/09/28. 10.1016/j.jneuroim.2017.09.007 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang XQ, Yang CY, Rao XF, Xiong JP. Plumbagin shows anti-cancer activity in human breast cancer cells by the upregulation of p53 and p21 and suppression of G1 cell cycle regulators. European journal of gynaecological oncology. 2016;37(1):30–5. Epub 2016/04/07. . [PubMed] [Google Scholar]
- 32.Aziz MH, Dreckschmidt NE, Verma AK. Plumbagin, a medicinal plant-derived naphthoquinone, is a novel inhibitor of the growth and invasion of hormone-refractory prostate cancer. Cancer research. 2008;68(21):9024–32. Epub 2008/11/01. 10.1158/0008-5472.CAN-08-2494 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nair HA, Snima KS, Kamath RC, Nair SV, Lakshmanan VK. Plumbagin Nanoparticles Induce Dose and pH Dependent Toxicity on Prostate Cancer Cells. Current drug delivery. 2015;12(6):709–16. Epub 2015/03/17. . [DOI] [PubMed] [Google Scholar]
- 34.Thasni KA, Rakesh S, Rojini G, Ratheeshkumar T, Srinivas G, Priya S. Estrogen-dependent cell signaling and apoptosis in BRCA1-blocked BG1 ovarian cancer cells in response to plumbagin and other chemotherapeutic agents. Annals of oncology: official journal of the European Society for Medical Oncology. 2008;19(4):696–705. Epub 2008/01/12. 10.1093/annonc/mdm557 . [DOI] [PubMed] [Google Scholar]
- 35.Wang F, Wang Q, Zhou ZW, Yu SN, Pan ST, He ZX, et al. Plumbagin induces cell cycle arrest and autophagy and suppresses epithelial to mesenchymal transition involving PI3K/Akt/mTOR-mediated pathway in human pancreatic cancer cells. Drug design, development and therapy. 2015;9:537–60. Epub 2015/01/30. 10.2147/DDDT.S73689 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gomathinayagam R, Sowmyalakshmi S, Mardhatillah F, Kumar R, Akbarsha MA, Damodaran C. Anticancer mechanism of plumbagin, a natural compound, on non-small cell lung cancer cells. Anticancer research. 2008;28(2a):785–92. Epub 2008/05/30. . [PubMed] [Google Scholar]
- 37.Appadurai P, Rathinasamy K. Plumbagin-silver nanoparticle formulations enhance the cellular uptake of plumbagin and its antiproliferative activities. IET nanobiotechnology. 2015;9(5):264–72. Epub 2015/10/06. 10.1049/iet-nbt.2015.0008 . [DOI] [PubMed] [Google Scholar]
- 38.Srinivas P, Gopinath G, Banerji A, Dinakar A, Srinivas G. Plumbagin induces reactive oxygen species, which mediate apoptosis in human cervical cancer cells. Molecular carcinogenesis. 2004;40(4):201–11. Epub 2004/07/21. 10.1002/mc.20031 . [DOI] [PubMed] [Google Scholar]
- 39.Tate CR, Rhodes LV, Segar HC, Driver JL, Pounder FN, Burow ME, et al. Targeting triple-negative breast cancer cells with the histone deacetylase inhibitor panobinostat. Breast cancer research: BCR. 2012;14(3):R79 Epub 2012/05/23. 10.1186/bcr3192 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blot E, Chen W, Vasse M, Paysant J, Denoyelle C, Pille JY, et al. Cooperation between monocytes and breast cancer cells promotes factors involved in cancer aggressiveness. British journal of cancer. 2003;88(8):1207–12. Epub 2003/04/17. 10.1038/sj.bjc.6600872 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gallelli L, Falcone D, Cannataro R, Perri M, Serra R, Pelaia G, et al. Theophylline action on primary human bronchial epithelial cells under proinflammatory stimuli and steroidal drugs: a therapeutic rationale approach. Drug design, development and therapy. 2017;11:265–72. Epub 2017/02/09. 10.2147/DDDT.S118485 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Senftleben U, Cao Y, Xiao G, Greten FR, Krahn G, Bonizzi G, et al. Activation by IKKalpha of a second, evolutionary conserved, NF-kappa B signaling pathway. Science (New York, NY). 2001;293(5534):1495–9. Epub 2001/08/25. 10.1126/science.1062677 . [DOI] [PubMed] [Google Scholar]
- 43.Xiao G, Harhaj EW, Sun SC. NF-kappaB-inducing kinase regulates the processing of NF-kappaB2 p100. Molecular cell. 2001;7(2):401–9. Epub 2001/03/10. . [DOI] [PubMed] [Google Scholar]
- 44.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. Epub 2011/03/08. 10.1016/j.cell.2011.02.013 . [DOI] [PubMed] [Google Scholar]
- 45.Thornberry NA, Lazebnik Y. Caspases: enemies within. Science (New York, NY). 1998;281(5381):1312–6. Epub 1998/08/28. . [DOI] [PubMed] [Google Scholar]
- 46.Jin Z, El-Deiry WS. Overview of cell death signaling pathways. Cancer biology & therapy. 2005;4(2):139–63. Epub 2005/02/24. . [DOI] [PubMed] [Google Scholar]
- 47.Strieter RM, Wiggins R, Phan SH, Wharram BL, Showell HJ, Remick DG, et al. Monocyte chemotactic protein gene expression by cytokine-treated human fibroblasts and endothelial cells. Biochemical and biophysical research communications. 1989;162(2):694–700. Epub 1989/07/31. . [DOI] [PubMed] [Google Scholar]
- 48.Zachariae CO, Anderson AO, Thompson HL, Appella E, Mantovani A, Oppenheim JJ, et al. Properties of monocyte chemotactic and activating factor (MCAF) purified from a human fibrosarcoma cell line. The Journal of experimental medicine. 1990;171(6):2177–82. Epub 1990/06/01. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dwyer RM, Potter-Beirne SM, Harrington KA, Lowery AJ, Hennessy E, Murphy JM, et al. Monocyte chemotactic protein-1 secreted by primary breast tumors stimulates migration of mesenchymal stem cells. Clinical cancer research: an official journal of the American Association for Cancer Research. 2007;13(17):5020–7. Epub 2007/09/06. 10.1158/1078-0432.ccr-07-0731 . [DOI] [PubMed] [Google Scholar]
- 50.Fang WB, Jokar I, Zou A, Lambert D, Dendukuri P, Cheng N. CCL2/CCR2 chemokine signaling coordinates survival and motility of breast cancer cells through Smad3 protein- and p42/44 mitogen-activated protein kinase (MAPK)-dependent mechanisms. The Journal of biological chemistry. 2012;287(43):36593–608. Epub 2012/08/29. 10.1074/jbc.M112.365999 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J, Zhuang ZG, Xu SF, He Q, Shao YG, Ji M, et al. Expression of CCL2 is significantly different in five breast cancer genotypes and predicts patient outcome. International journal of clinical and experimental medicine. 2015;8(9):15684–91. Epub 2015/12/03. . [PMC free article] [PubMed] [Google Scholar]
- 52.Qian BZ, Li J, Zhang H, Kitamura T, Zhang J, Campion LR, et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475(7355):222–5. Epub 2011/06/10. 10.1038/nature10138 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bonapace L, Coissieux MM, Wyckoff J, Mertz KD, Varga Z, Junt T, et al. Cessation of CCL2 inhibition accelerates breast cancer metastasis by promoting angiogenesis. Nature. 2014;515(7525):130–3. Epub 2014/10/23. 10.1038/nature13862 . [DOI] [PubMed] [Google Scholar]
- 54.Zhu X, Fujita M, Snyder LA, Okada H. Systemic delivery of neutralizing antibody targeting CCL2 for glioma therapy. Journal of neuro-oncology. 2011;104(1):83–92. Epub 2010/12/01. 10.1007/s11060-010-0473-5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hembruff SL, Jokar I, Yang L, Cheng N. Loss of transforming growth factor-beta signaling in mammary fibroblasts enhances CCL2 secretion to promote mammary tumor progression through macrophage-dependent and -independent mechanisms. Neoplasia (New York, NY). 2010;12(5):425–33. Epub 2010/05/11. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salcedo R, Ponce ML, Young HA, Wasserman K, Ward JM, Kleinman HK, et al. Human endothelial cells express CCR2 and respond to MCP-1: direct role of MCP-1 in angiogenesis and tumor progression. Blood. 2000;96(1):34–40. Epub 2000/07/13. . [PubMed] [Google Scholar]
- 57.Mizutani K, Sud S, McGregor NA, Martinovski G, Rice BT, Craig MJ, et al. The chemokine CCL2 increases prostate tumor growth and bone metastasis through macrophage and osteoclast recruitment. Neoplasia (New York, NY). 2009;11(11):1235–42. Epub 2009/11/03. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vitiello PF, Shainheit MG, Allison EM, Adler EP, Kurt RA. Impact of tumor-derived CCL2 on T cell effector function. Immunology letters. 2004;91(2–3):239–45. Epub 2004/03/17. 10.1016/j.imlet.2003.12.009 . [DOI] [PubMed] [Google Scholar]
- 59.Mantovani A, Schioppa T, Porta C, Allavena P, Sica A. Role of tumor-associated macrophages in tumor progression and invasion. Cancer metastasis reviews. 2006;25(3):315–22. Epub 2006/09/13. 10.1007/s10555-006-9001-7 . [DOI] [PubMed] [Google Scholar]
- 60.Tsuyada A, Chow A, Wu J, Somlo G, Chu P, Loera S, et al. CCL2 mediates cross-talk between cancer cells and stromal fibroblasts that regulates breast cancer stem cells. Cancer research. 2012;72(11):2768–79. Epub 2012/04/05. 10.1158/0008-5472.CAN-11-3567 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koury J, Zhong L, Hao J. Targeting Signaling Pathways in Cancer Stem Cells for Cancer Treatment. Stem cells international. 2017;2017:2925869 Epub 2017/03/31. 10.1155/2017/2925869 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pires BR, DEA IS, Souza LD, Rodrigues JA, Mencalha AL. Targeting Cellular Signaling Pathways in Breast Cancer Stem Cells and its Implication for Cancer Treatment. Anticancer research. 2016;36(11):5681–91. Epub 2016/10/30. 10.21873/anticanres.11151 . [DOI] [PubMed] [Google Scholar]
- 63.Saji H, Koike M, Yamori T, Saji S, Seiki M, Matsushima K, et al. Significant correlation of monocyte chemoattractant protein-1 expression with neovascularization and progression of breast carcinoma. Cancer. 2001;92(5):1085–91. Epub 2001/09/26. . [DOI] [PubMed] [Google Scholar]
- 64.Ueno T, Toi M, Saji H, Muta M, Bando H, Kuroi K, et al. Significance of macrophage chemoattractant protein-1 in macrophage recruitment, angiogenesis, and survival in human breast cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2000;6(8):3282–9. Epub 2000/08/24. . [PubMed] [Google Scholar]
- 65.Anderson GM, Nakada MT, DeWitte M. Tumor necrosis factor-alpha in the pathogenesis and treatment of cancer. Current opinion in pharmacology. 2004;4(4):314–20. Epub 2004/07/15. 10.1016/j.coph.2004.04.004 . [DOI] [PubMed] [Google Scholar]
- 66.Zollo M, Di Dato V, Spano D, De Martino D, Liguori L, Marino N, et al. Targeting monocyte chemotactic protein-1 synthesis with bindarit induces tumor regression in prostate and breast cancer animal models. Clinical & experimental metastasis. 2012;29(6):585–601. Epub 2012/04/10. 10.1007/s10585-012-9473-5 . [DOI] [PubMed] [Google Scholar]
- 67.Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis. 2009;30(7):1073–81. Epub 2009/05/27. 10.1093/carcin/bgp127 . [DOI] [PubMed] [Google Scholar]
- 68.Mantovani A. Molecular pathways linking inflammation and cancer. Current molecular medicine. 2010;10(4):369–73. Epub 2010/05/12. . [DOI] [PubMed] [Google Scholar]
- 69.Gupta SC, Sundaram C, Reuter S, Aggarwal BB. Inhibiting NF-kappaB activation by small molecules as a therapeutic strategy. Biochimica et biophysica acta. 2010;1799(10–12):775–87. Epub 2010/05/25. 10.1016/j.bbagrm.2010.05.004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gaudet MM, Egan KM, Lissowska J, Newcomb PA, Brinton LA, Titus-Ernstoff L, et al. Genetic variation in tumor necrosis factor and lymphotoxin-alpha (TNF-LTA) and breast cancer risk. Human genetics. 2007;121(3–4):483–90. Epub 2007/01/12. 10.1007/s00439-006-0315-x . [DOI] [PubMed] [Google Scholar]
- 71.Miles DW, Happerfield LC, Naylor MS, Bobrow LG, Rubens RD, Balkwill FR. Expression of tumour necrosis factor (TNF alpha) and its receptors in benign and malignant breast tissue. International journal of cancer. 1994;56(6):777–82. Epub 1994/03/15. . [DOI] [PubMed] [Google Scholar]
- 72.Wolf MJ, Seleznik GM, Zeller N, Heikenwalder M. The unexpected role of lymphotoxin beta receptor signaling in carcinogenesis: from lymphoid tissue formation to liver and prostate cancer development. Oncogene. 2010;29(36):5006–18. Epub 2010/07/07. 10.1038/onc.2010.260 . [DOI] [PubMed] [Google Scholar]
- 73.Aggarwal BB. Signalling pathways of the TNF superfamily: a double-edged sword. Nature reviews Immunology. 2003;3(9):745–56. Epub 2003/09/02. 10.1038/nri1184 . [DOI] [PubMed] [Google Scholar]
- 74.Quan L, Gong Z, Yao S, Bandera EV, Zirpoli G, Hwang H, et al. Cytokine and cytokine receptor genes of the adaptive immune response are differentially associated with breast cancer risk in American women of African and European ancestry. International journal of cancer. 2014;134(6):1408–21. Epub 2013/09/03. 10.1002/ijc.28458 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang Y, Yu X, Wang L, Zhou S, Sun J, Feng N, et al. Four genetic polymorphisms of lymphotoxin-alpha gene and cancer risk: a systematic review and meta-analysis. PloS one. 2013;8(12):e82519 Epub 2013/12/19. 10.1371/journal.pone.0082519 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chu ZL, McKinsey TA, Liu L, Gentry JJ, Malim MH, Ballard DW. Suppression of tumor necrosis factor-induced cell death by inhibitor of apoptosis c-IAP2 is under NF-kappaB control. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(19):10057–62. Epub 1997/09/18. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Levkau B, Garton KJ, Ferri N, Kloke K, Nofer JR, Baba HA, et al. xIAP induces cell-cycle arrest and activates nuclear factor-kappaB: new survival pathways disabled by caspase-mediated cleavage during apoptosis of human endothelial cells. Circulation research. 2001;88(3):282–90. Epub 2001/02/17. . [DOI] [PubMed] [Google Scholar]
- 78.Liston P, Fong WG, Korneluk RG. The inhibitors of apoptosis: there is more to life than Bcl2. Oncogene. 2003;22(53):8568–80. Epub 2003/11/25. 10.1038/sj.onc.1207101 . [DOI] [PubMed] [Google Scholar]
- 79.Los M, Stroh C, Janicke RU, Engels IH, Schulze-Osthoff K. Caspases: more than just killers? Trends in immunology. 2001;22(1):31–4. Epub 2001/04/05. . [DOI] [PubMed] [Google Scholar]
- 80.Kuo PL, Hsu YL, Cho CY. Plumbagin induces G2-M arrest and autophagy by inhibiting the AKT/mammalian target of rapamycin pathway in breast cancer cells. Molecular cancer therapeutics. 2006;5(12):3209–21. Epub 2006/12/19. 10.1158/1535-7163.MCT-06-0478 . [DOI] [PubMed] [Google Scholar]
- 81.Mehmeti M, Allaoui R, Bergenfelz C, Saal LH, Ethier SP, Johansson ME, et al. Expression of functional toll like receptor 4 in estrogen receptor/progesterone receptor-negative breast cancer. Breast cancer research: BCR. 2015;17:130 Epub 2015/09/24. 10.1186/s13058-015-0640-x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(31):E2110–6. Epub 2012/07/04. 10.1073/pnas.1209414109 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Green TL, Santos MF, Ejaeidi AA, Craft BS, Lewis RE, Cruse JM. Toll-like receptor (TLR) expression of immune system cells from metastatic breast cancer patients with circulating tumor cells. Experimental and molecular pathology. 2014;97(1):44–8. Epub 2014/05/20. 10.1016/j.yexmp.2014.05.003 . [DOI] [PubMed] [Google Scholar]
- 84.Kuo AH, Scheeren FA. Cell-intrinsic TLR2/MyD88 pathway in breast and colon cancer. Cell cycle (Georgetown, Tex). 2014;13(24):3785–6. Epub 2014/12/03. 10.4161/15384101.2014.989947 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vogler M. BCL2A1: the underdog in the BCL2 family. Cell death and differentiation. 2012;19(1):67–74. Epub 2011/11/15. 10.1038/cdd.2011.158 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Haq R, Yokoyama S, Hawryluk EB, Jonsson GB, Frederick DT, McHenry K, et al. BCL2A1 is a lineage-specific antiapoptotic melanoma oncogene that confers resistance to BRAF inhibition. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(11):4321–6. Epub 2013/03/01. 10.1073/pnas.1205575110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kang MH, Reynolds CP. Bcl-2 inhibitors: targeting mitochondrial apoptotic pathways in cancer therapy. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009;15(4):1126–32. Epub 2009/02/21. 10.1158/1078-0432.ccr-08-0144 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guo P, Huang J, Wang L, Jia D, Yang J, Dillon DA, et al. ICAM-1 as a molecular target for triple negative breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(41):14710–5. Epub 2014/10/01. 10.1073/pnas.1408556111 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bauer D, Redmon N, Mazzio E, Soliman KF. Apigenin inhibits TNFalpha/IL-1alpha-induced CCL2 release through IKBK-epsilon signaling in MDA-MB-231 human breast cancer cells. PloS one. 2017;12(4):e0175558 Epub 2017/04/26. 10.1371/journal.pone.0175558 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Boehm JS, Zhao JJ, Yao J, Kim SY, Firestein R, Dunn IF, et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129(6):1065–79. Epub 2007/06/19. 10.1016/j.cell.2007.03.052 . [DOI] [PubMed] [Google Scholar]
- 91.Jeon M, Han J, Nam SJ, Lee JE, Kim S. Elevated IL-1beta expression induces invasiveness of triple negative breast cancer cells and is suppressed by zerumbone. Chemico-biological interactions. 2016;258:126–33. Epub 2016/08/29. 10.1016/j.cbi.2016.08.021 . [DOI] [PubMed] [Google Scholar]
- 92.Citalingam K, Abas F, Lajis NH, Othman I, Naidu R. Anti-proliferative effect and induction of apoptosis in androgen-independent human prostate cancer cells by 1,5-bis(2-hydroxyphenyl)-1,4-pentadiene-3-one. Molecules (Basel, Switzerland). 2015;20(2):3406–30. Epub 2015/02/19. 10.3390/molecules20023406 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Deiab S, Mazzio E, Eyunni S, McTier O, Mateeva N, Elshami F, et al. 1,2,3,4,6-Penta-O-galloylglucose within Galla Chinensis Inhibits Human LDH-A and Attenuates Cell Proliferation in MDA-MB-231 Breast Cancer Cells. Evidence-based complementary and alternative medicine: eCAM. 2015;2015:276946 Epub 2015/04/29. 10.1155/2015/276946 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the manuscript.