Abstract
Objective:
The goal of this paper is to demonstrate and evaluate the potential efficacy of laser-generated shockwave (LGS) therapy on biofilm infected tissue.
Methods:
To demonstrate proof of concept, Staphylococcus epidermidis was allowed to proliferate on ex vivo pigskin, until mature biofilm formation was achieved, and then subjected to LGS. Bacterial load between control and treated samples was compared using the swab technique and colony counting. Scanning electron microscopy (SEM) was then used to visualize the biofilm growth and resulting reduction in biofilm coverage from treatment. Images were false colored to improve contrast of biofilm, and percent biofilm coverage was computed, along with biofilm cluster size.
Results:
LGS reduced bacterial load by 69% (p = 0.008). Imaging showed biofilm coverage reduced by 52% and significantly reduced average cluster size (p < 0.001).
Conclusion:
LGS therapy reduced the burden of bacterial biofilm on ex vivo pigskin and can be visualized using SEM imaging.
Significance:
LGS therapy is a new treatment for infected wounds, allowing rapid disruption of biofilm to 1) remove bacteria and 2) increase susceptibility of remaining biofilm to topical antibiotics. This can lead to improved wound healing times, reduced patient morbidity, and decreased healthcare costs.
Keywords: Biofilm, infected wound, laser-generated shockwaves (LGS), scanning electron microscopy (SEM)
I. Introduction
BACTERIAL biofilms have been recognized as a major contributor to healthcare-associated infections and the respective financial burdens on the healthcare industry, to both provider and patient [1]–[3]. In infected cutaneous wounds, biofilms provide a barrier to normal patient immune response and to topical antibiotic therapies [4]–[6], significantly hindering healing times and wound closure [7]. Furthermore, there has been a significant rise in bacterial strains which have adapted them-selves to commonly used antibiotic treatment, rendering those treatments ineffective [3], [8]. Several research fronts are being pursued to alleviate this rise in prolonged infections, both to increase effectiveness of current antibiotic treatment and find new methods of treatment, either chemically inactivating or mechanically disrupting the biofilm [9]. While chemical treatments are logistically easier to implement, mechanical treatments, while treating bacteria directly, have the benefit of being nonspecific to bacteria phenotype, for example gram-positive versus gram-negative bacteria, aerobic versus anaerobic bacteria, leading to a more sustainable therapy.
One such effort being explored for biofilms is nanoparticle-based antibiotic therapy, which utilizes both antimicrobial metals, particularly silver, and conjugated nanomaterials to antibiotic compounds to combat infection [10], [11]. Research groups have found promising results, however, there are issues that still need to be resolved. First, the long-term toxicity effects of nanoparticle treatment are unknown. Second, different strains of infectious agent may require a different type of nanomaterial. Third, certain bacterial strains are already showing resistant to nanoparticle treatments [12]. A second area under study is photodynamic therapy (PDT), where a photosensitive dye and a light source is used as an antibacterial treatment to target bacteria and other pathogens [13]. A limited phase II clinical experiments demonstrated minor success in improving wound healing of chronically infected wounds, observing 50% (4 out of 8) wound closures for PDT treatment group and 12% (1 out of 8) wound closures in the placebo group, after a three month period. However, median wound area of placebo arm was 3.6× greater than the PDT treatment median wound area, and after three months both groups had the same relative range of wound area [14]. The study found an initial drop in bioburden, measured as colony forming units (CFU) immediately post-PDT treatment, however the bacterial count increased to pathogenic values 24 h and 1 week posttreatment [14].
One category of mechanical treatment of biofilms is using pressure waves derived from laser energy to mechanically disrupt the bacterial biofilms. Krespi et al. demonstrated, through confocal miscroscopy, the ability of 10–20 low-energy laserinduced pressure waves to disperse bacterial cells within biofilm off of small suture, stent, and bone screw [15], [16]. Nigri et al. demonstrated no effect of laser-induced shock waves (SW) alone to biofilms, but clearly observed SW permeabilizing effects on biofilms, reporting a 102 improvement of antibiotic effectiveness when coupled with SW treatment of eight pulses at 60 MPa of pressure [17].
In this application, modified laser-generated shockwave (LGS) therapy are being developed for rapid treatment of infected wounds, rather than medical devices. In the LGS process, a short-pulsed laser ablates a thin metal film layered between an acoustic backing and a carrier substrate. This configuration causes a focused compressive shockwave to travel through a coupling medium to the biofilm layer [18]. When the pressure wave reaches the biofilm—tissue interface, acoustic impedance mismatch causes a portion of the compressive wave to reflect as a tensile wave, while the rest of the compressive wave transmits through and disperses within the tissue. If the pressure magnitude of the reflection is sufficient, axial forces from the tensile stress wave causes mechanical disruption of the biofilm matrix from the wound tissue, while the transmitted wave attenuates into the heterogeneous tissue [19], [20]. In addition to biofilm disruption on the same scale as the shockwave spatial boundaries, Navarro et al. also found that small localized delaminated areas occur outside of the shockwave boundaries, which may be attributed to traveling cavitation-induced bubbles [20]. However, analysis on that mechanism is currently under review. Further, it has been shown that these pressure waves have the ability to increase delivery of macromolecules through cutaneous tissue without damaging the stratum corneum [21], which can provide a secondary mechanism to treat biofilm by coupling LGS therapy with a topical antibiotic treatment, similar to [17].
Previous work with LGS, in the context of cutaneous wound treatment, has demonstrated bacterial bioburden reduction in vitro [19] off an agar surface and biofilm delamination, in vitro, from a polystyrene surface [20]. Clinical translation of LGS technology for biofilm-infected wounds requires an accurate tissue analogue to evaluate potential delamination in a model that is relevant to in vivo human applications. Pig skin is an ideal phantom for human skin for its physical properties and structural similarities [22]. Preliminary safety of LGS was demonstrated with minimal side effects in healthy ex vivo porcine tissue [23]. This paper describes the observed bioburden reduction after treatment of a single shockwave on ex vivo pigskin infected with Staphylococcus epidermidis, measured through CFU counting. Scanning electron microscopy (SEM) was then used to directly observe the disruptive effects of LGS on the biofilm and skin surface (see Fig. 1). These data from the SEM images are finally leveraged to calculate percent biofilm coverage and fragmented biofilm cluster size to help quantify the potential increased bacterial exposure to antibiotics in future studies.
Fig. 1.
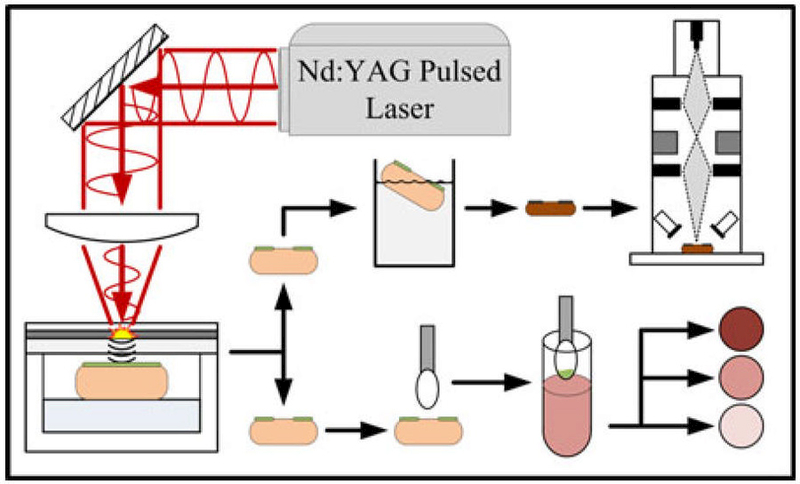
Diagram of experimental setup. Infected pigskin is treated with LGS, swabbed and plated for CFU counting. Two samples are fixed and dehydrated for SEM.
II. MATERIALS AND METHODS
A. Preparation of S. epidermidis
A 1-μL sample of S. epidermidis (ATCC 35984) was streaked onto a tryptic soy agar plate and incubated at 37 °C for 24 h to isolate bacterial colonies of the same phenotype. A single colony was then suspended into 10 mL of tryptic soy broth (TSB) and cultured in a shaker at 37 °C, 200 r/min for 18 h. The culture was centrifuged at 3000 r/min for 5 min at 4 °C. The supernatant was discarded, and the remaining pellet was resuspended in 5 mL of fresh TSB. A working S. epidermidis solution was prepared by further diluting with TSB, until an optical density of 0.2 at 600 nm was measured on a spectrophotometer, which correlates to a cell density of 4 × 107 CFU/mL, more than sufficient to initiate wound infection [24].
B. Pigskin Sample Preparation
Healthy skin samples were collected from a single pig, obtained through the University of California Los Angeles tissue-sharing protocol, excised preeuthanasia. Fat and muscle were removed to leave only the epidermal and dermal layers. The epidermal layer was removed using a razor dermatome to simulate damaged tissue and promote bacterial cellular adhesion. Eighteen samples between 0.7 and 1.0 cm2 in size were cut from the prepared tissue and washed with sterile phosphate buffered saline (PBS). Samples for treatment were placed epidermis side up in a 24 well plate and saturated with 2 mL of working S. epidermidis solution. A separate sample was placed in a Petri dish with sterile PBS for the negative control. The samples were incubated at 37 °C for 24 h for biofilm formation.
C. LGS Treatment
The shockwave-generating substrates in this experiment were 15 × 15 mm2 sections of 0.127-mm-thick polycarbonate, RF sputtered with a 0.5 μm titanium film as the ablation layer. This material was selected based on results of previous shockwave characterization experiments [18]. Confinement was created by manually applying a thin layer (200 μm) of water—glass on the titanium surface. For treatment, incubated skin samples were placed on top of a 1-cm-thick layer of ballistic gel, inside a sterile six-well plate. The gel was used to minimize any potential reflection of shockwaves from the backside of the tissue surface. For the coupling medium, sterile PBS was added to each well until the sample was submerged approximately 1 mm below the surface. A single shockwave was generated using a 1064 nm Nd:YAG laser, ablating a 3-mm-diameter spot size (1/e) with an energy fluence of 93 mJ/mm2. The shockwave travels through the polycarbonate and PBS coupling, resulting in a shockwave peak pressure on the biofilm—pigskin surface of 263.4 MPa, a value empirically determined to disrupt biofilm [20]. Samples were then removed and washed with PBS to remove any planktonic cells.
D. Bacterial Collection and Colony Counting
For CFU counting, a 2 mm tipped sterile swab was placed vertically in the center of the shocked area and twisted ten times to pick up residual bacteria and biofilm. The swab tip was then cut off, placed in 2 mL of TSB, and vortexed for 15 s to dislodge bacteria. After the tip was removed, sample solution was vortexed again for 15 s to achieve cell uniformity. Serial ten fold dilutions were made of the bacterial solution, down to 10−7, and 100 μL of each dilution was plated onto mannitol salt agar plates, in triplicate. Agar plates were then incubated at 37 °C for 24 h. Plates were removed from the incubator, and colony counting was performed by two blinded personnel. Their counts were then averaged and CFU concentrations were back calculated based on the dilutions.
E. Tissue Preparation and SEM Imaging
For SEM imaging, samples were fixed in 2.5% gluteraldehyde in 0.1 M sodium cacodylate buffer at pH 7.4 for 24 h, then washed three times with 0.1 M sodium cacodylate buffer for 5 min each. To retain bacteria and biofilm structure stability during imaging, secondary fixation was performed using three drops of osmium tetroxide on the surface of each specimen and allowed to react for 20 min. Dehydration of the skin samples was completed through submersion in 30%, 50%, 70%, 85%, 95%, and 3× 100%ethanol solutions, for 10 min each. Samples were then washed in 1.5 mL of hexamethyldisilazane for 20 min and allowed to air dry in a chemical fume hood overnight. The dried samples were then sputtered with a 5-nm-thick layer of gold particles for contrast enhancement. Each sample was imaged under SEM (JSM-6700F, JEOL, Japan) with 10.0 kV accelerating voltage and a working distance of 7.9 mm. Several representative images were taken of each sample, positive control, negative control, and treated sample, at different magnifications (200×, 600×, and 5000×) to verify biofilm confluence and capture structural changes to the biofilm.
F. Composite Image Construction
To create an image with a field of view (FOV) that spans the entire treatment spot size of 3 mm, while maintaining a resolution sufficient to distinguish biofilm and bacteria (1 μm), a composite image was constructed from 48 spatially sequential SEM images in a 6-row by 8-column format. These images were taken for both the treated sample and the positive control sample to create two composite images, visualizing the same FOV for each sample. Each set of images were stitched using Photoshop (Adobe Systems Inc., USA) to create, after processing and stitching, a 3.756 mm by 2.347 mm FOV image, with a resolution of 2130 pixels per mm (8000 × 5000 pixels image, pixel size = 0.469 μm × 0.469 μm) to resolve bacterial biofilms. Blinded personnel, with at least 20 h experience in imaging biofilm on SEM, created false-colored images by manually shading bacterial biofilm and residual cells green, and shading the underlying tissue as red. Both control sample and treated sample composite images cover the same FOV. Percentage of biofilm coverage was determined by isolating the specific hue, saturation, and brightness (HSB) value range to measure the population of green and red pixels using MATLAB (Mathworks, USA).
G. Biofilm Cluster Analysis
To investigate the fragmentation resulting from the application of LGS, and therefore its potential for increased antibiotic susceptibility, the false-colored control and LGS-treated composite images were converted to binary maps, using the same isolation of HSB values, and applied connected components analysis to tabulate the size and distributions of biofilm clusters. The connected component framework returns a histogram of the total fragmented clusters, m, detailing the number of clusters, N, for a particular cluster size bin, b. From there the distributions of clusters from each sample were compared and differences between mean cluster size, median cluster size, and skewness were noted.
H. Statistics
The comparison of mean bacterial colony counts between the positive and treated samples was analyzed using the unpaired two-way Student’s t-test, assuming equal variance (OriginPro, v8.02, OriginLab Corporation, USA). The comparison of the median biofilm cluster size between the two sample composite images was completed using a Wilcoxon rank-sum test (MATLAB, USA), due to the nonparametric dataset and nonnormal distribution of cluster sizes. Significance was established with p values ≤ 0.05 for both tests.
III. RESULTS
A. Bacterial CFU Count
Nine of the 18 infected skin samples were subjected to a single shockwave in the center of the skin. Seven samples from both treated and positive control groups were swabbed and plated for CFU counting. Results comparing the mean CFU values between control and treated samples are shown in Fig. 2. The mean CFU/mL for the control sample groups was 5.58 × 107, with a standard deviation of 3.00 × 107, and the mean for the treated was 1.71 × 107, with a standard deviation of 1.20 × 107, demonstrating a significant reduction in bioburden in treated samples (p = 0.008). This equates to a 69% reduction in bacteria on the treated pigskin.
Fig. 2.
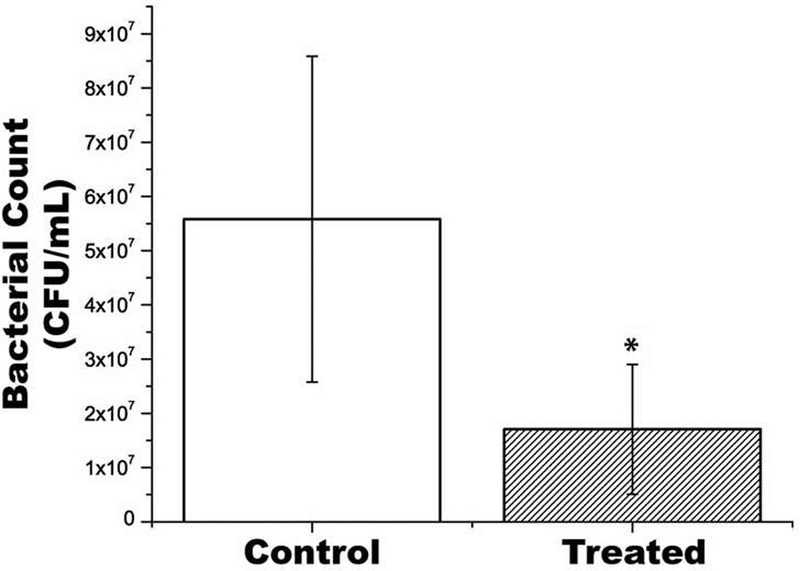
Results of bacterial CFU per mL from infected samples compared to LGS treated samples. Mean ± Standard Deviation. Asterisk indicates significant difference.
B. SEM Imaging of Biofilm
The remaining two treated and positive control samples were prepared for SEM imaging. Validation of bacterial biofilm formation and adhesion to pigskin was completed through SEM images acquired at 5000× magnification, clearly resolving the cocci shape of S. epidermidis [see Fig. 3(a), green arrows] and the presence of biofilm matrix between cell clusters [Fig. 3(a), red arrows]. Fig. 3(b–d) shows representative images of the negative control sample, positive control sample, and shockwave treated sample for 24 h biofilm growth, respectively, demonstrating clear qualitative differences between sample groups. The negative control image reveals clear skin surface, with only mild contamination of foreign, non-colonized cells [see Fig. 3(b), orange arrows]. The positive control image shows a confluent biofilm layer, and noted fractures in the biofilm along the ridge lines of the skin [see Fig. 3(c), blue arrows]. The tortuous appearance of the skin and resulting bioflim fractures are most likely caused during the tissue dehydration and drying process which results in shrinkage of the samples. In the treated sample image, significant biofilm reduction is seen with residual colonies appearing sporadically throughout the tissue [see Fig. 3(d), yellow arrows], especially in the recesses of the skin where the shockwave does not have clear line-of-sight travel.
Fig. 3.
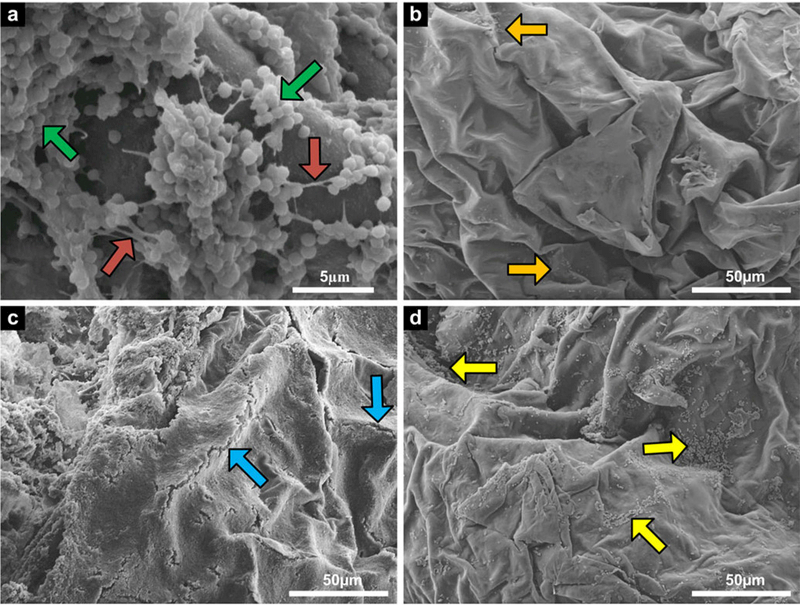
SEM images of pigskin samples: (a) 5000× magnification of S. epidermidis cells (green arrow) and biofilm matrix (red arrows) to verify biofilm formation and adhesion [scale bar = 5 μm], (b) 600× magnification of 24 h negative control sample showing very little contamination (orange arrows), (c) 600× magnification of 24 h positive control showing natural biofilm fracture along ridge lines (blue arrow), and (d) 600× magnification of 24 h treated sample showing residual bacteria and biofilm (yellow arrows) [(b)–(d) scale bar = 50 μm].
C. False-Colored Composite Images
Fig. 4 shows the complete false-colored composite image for positive control [see Fig. 4(a)], LGS treated sample [see Fig. 4(b)], and a magnified representative image from both control and treated samples [see Fig. 4(c) and (d), respectively]. Calculations on the composite images by MATLAB, by isolating the green pixel color between normalized hue values of 0.27451 and 0.39216, revealed 68.53% biofilm coverage on the positive control sample and 33.01% biofilm coverage on the treated sample, signifying a 52% reduction. This result, while lower than the observed CFU reduction, still correlates well with previously published results which showed a 55% reduction in bacterial load on a surface, measured through bacterial plating and colony counting [19]. There were observed areas on the positive control sample that were damaged, indicated by localized fracture of skin cells and total absence of biofilm, most likely from handling [see in Fig. 4(a), center, black arrows], resulting in the removal of biofilm before imaging and, therefore, from the biofilm reduction calculation.
Fig. 4.
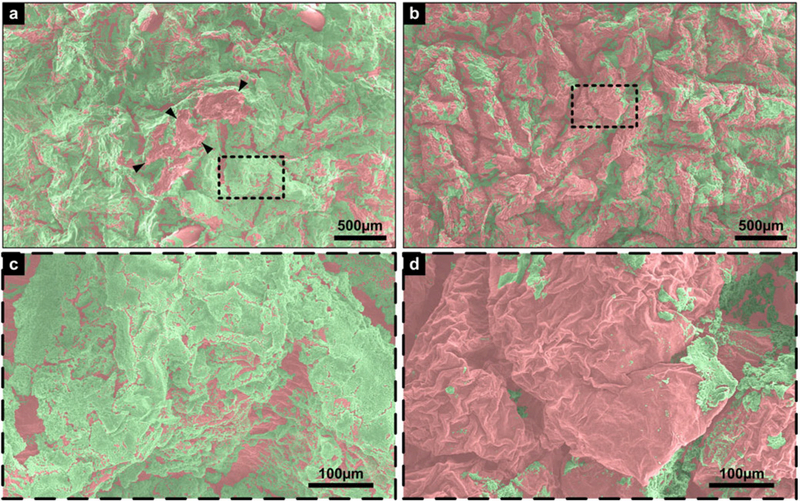
False-colored SEM images: (a) Positive control composite image with FOV of 3.756 mm by 2.347 mm, damaged area demarcated by arrowheads, (b) shockwave treated sample composite image, with same FOV as (a), (c) magnified image of boxed area in (a), with a FOV of 0.601 mm by 0.376 mm, and (d) magnified image of boxed area in (b), same FOV as (c). Green represents biofilm, red represents pigskin. Scale bar for (a) and (b) is 500 μm; scale bar for (c) and (d) is 100 μm.
D. Biofilm Fragmentation Comparison
As previously stated, biofilms act as a barrier for diffusion for topical antibiotic therapy and, therefore, much greater concentration of antibiotics are necessary to effectively kill bacteria in biofilm than in a planktonic state [5]. One of the key benefits of LGS is the potential ability to permeabilze biofilms through fragmentation, thus, rendering the remaining colonies and underlying tissue more susceptible to topical antibiotic treatments. The distribution of cluster sizes for the control sample and shockwave-treated sample composite images can be seen in Fig. 5(a) and (b), respectively, where the x-axis has been mapped to a log scale in line with standard bacterial cell population analysis [25]. Within the control sample distribution, number of clusters is well distributed across the biofilm cluster size. In contrast, the treated sample distribution shows significantly higher density of clusters below 102 μm2. This distribution shift between the two samples supports increased biofilm fragmentation from the shockwave treatment. The mean biofilm cluster areas for the control sample was calculated as 4931.2 μm2, with a standard deviation of 167 366.7 μm2, and LGS-treated sample was calculated as 474.0 μm2, with a standard deviation of 5285.7 μm2. The median biofilm cluster area for positive control sample was 34.6 μm2, while the median biofilm cluster area for shockwave treated sample was 20.5 μm2. The large discrepancies between the mean and median is due to a small number of clusters encompassing such a high percentage of total biofilm area, skewing both control and treated sample distributions to the right. The Wilcoxon rank-sum test showed significance of the medians between the distributions, and a p-value of less than 10−4 was obtained. This confirms that the remaining biofilm clusters on the shockwave-treated sample were significantly smaller in area than the biofilm clusters on the positive control. The skewness and fragmentation can also be visualized with the cumulative area distribution of biofilm coverage for the control and treated sample, shown in Fig. 5(c) and (d), respectively, and defined by where C[m] is the cumulative biofilm area in units μm2 and A is the area for each individual biofilm cluster in the series 1,..., m (total number of clusters). The distribution of the control sample trends smoothly until the large jump at the final point in the histogram, showing that this largest biofilm cluster in the image contributes to 96.5% of the total biofilm coverage. Conversely, the final point on the treated sample cumulative area distribution contributes only 11.3% of the total biofilm coverage. This distribution in Fig. 5(d) also demonstrates a significant jump at one of the first few bins again indicating that the majority of biofilm coverage is contributed by small, fragmented clusters. In fact, biofilm clusters with areas under 100 μm2 contributed to 1.92% of the total biofilm coverage for shockwave treated sample, while for the positive control samples, clusters under 1000 μm2 surface area contributed to only 0.22% of the total biofilm coverage, further characterizing the shockwave’s ability to significantly fragment biofilm clusters.
Fig. 5.
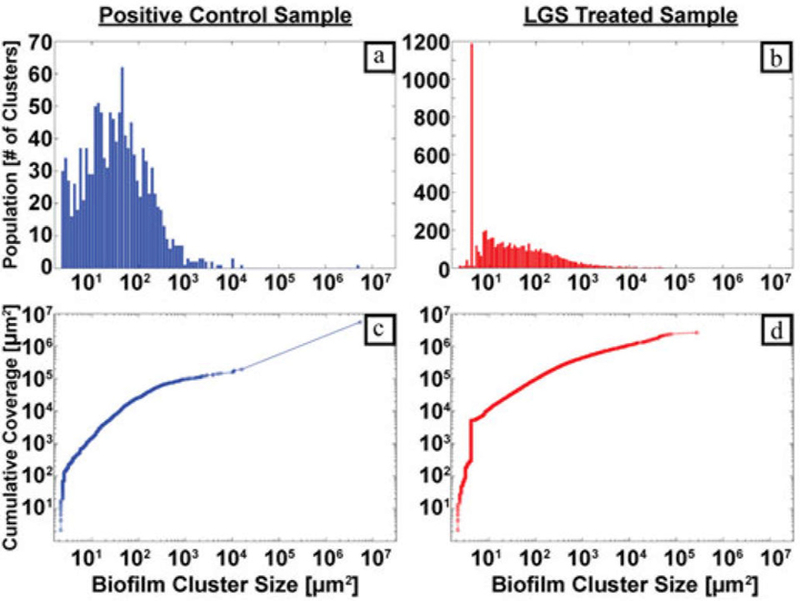
(a) Positive control biofilm cluster size distribution, (b) shock-wave treated biofilm cluster size distribution, (c) positive control cluster cumulative area distribution, and (d) shockwave treated cluster cumulative area distribution.
| (1) |
IV. DISCUSSION
The results show that LGS can disrupt appreciable amounts of bacterial biofilm, as characterized by CFU count and SEM imaging, therefore, reducing the bioburden on the tissue. While the CFU results demonstrate mathematically a significant decrease in bioburden, clinically this reduction will have limited effect on infected wounds without additional optimization. In a clinical setting, wounds are typically cleaned or debrided prior to swabbing, so that only the exudate is sampled for CFU count. This study, however, is only measuring the reduction in surface bacterial load. Future in vivo wound healing studies in an animal model will be required and sampling will follow standard clinical operating procedures. The published bacterial threshold for infection is 105 [24] and, therefore, reduction needs to fall below this value in order to benefit the wound healing process. One of the limitations of the CFU counting method in this study is that swabbing technique is required, since the sample is only subjected to a single shockwave of 3-mm diameter. Twisting of the swab consequently twists the skin surface, increasing the chance of swabbing outside the perimeter of the treateddiameter, which leads to inclusion of nontreated biofilm, and artificially increasing CFU count. Future treatments will require a scanning system to traverse the laser pulse over the entire infected area, and that development is currently underway.
As was mentioned earlier, two skin samples from positive control and LGS treatment groups were processed for SEM imaging. Only one set, however, was able to complete false-coloring due to the fact that distinction between bacterial biofilm and pigskin has to be manually discerned. Attempts at automating biofilm boundary based on morphology, boundary detection, or color differentiation were unsuccessful. Therefore, personnel trained on the SEM, able to distinguish between biofilm and underlying tissue, were required to manually add in biofilm color to create the images in Fig. 4. This time commitment to analyze both sets of stitched images equated to around 500 man-hours.
In an attempt to reduce the intensive time requirement for SEM analysis of biofilm delamination, a Monte Carlo simulation was explored to check the accuracy of coloring a subset of randomly selected images within the composite images, as a representation of the total biofilm delamination. A subset of five images, each 1000 × 833 pixels in size, were randomly selected, without repeating, from the false-colored composite images of both control and treated samples. Their mean biofilm coverage area was calculated and compared to the population mean of 68.53% for control and 33.01% for treated. This was repeated 1000× to evaluate the frequency of the mean of a random subset of images falling within 5% of the population mean of their respective composite image. This process was then repeated 1000× to find the distribution of point accuracy of a five-image subset. The same analysis was then repeated for subsets of 10, 15, and 20 images, for both the control and treated sample composite images. Two criteria of accuracy were defined: Within 5% of the population means and within 10% of the population means. The results are summarized in Table 1 and the trends are shown graphically in Fig. 6.
TABLE 1.
Simulation Results for the Accuracy of Random Image Subsets Compared to the Total Composite Image
| Within 5% accuracy | Within 10% accuracy | |
|---|---|---|
| 5 images (Control) | 37.69% ± 1.53% | 72.57% ± 1.39% |
| 10 images (Control) | 57.18% ± 1.58% | 90.31% ± 0.98% |
| 15 images (Control) | 70.85% ± 1.45% | 97.25% ± 0.52% |
| 20 images (Control) | 81.67% ± 1.22% | 99.55% ± 0.20% |
| 5 images (Treated) | 23.73% ± 1.36% | 45.56% ± 1.59% |
| 10 images (Treated) | 35.57% ± 1.51% | 64.60% ± 1.48% |
| 15 images (Treated) | 45.71% ± 1.54% | 77.96% ± 1.30% |
| 20 images (Treated) | 55.65% ± 1.54% | 87.71% ± 1.01% |
Fig. 6.
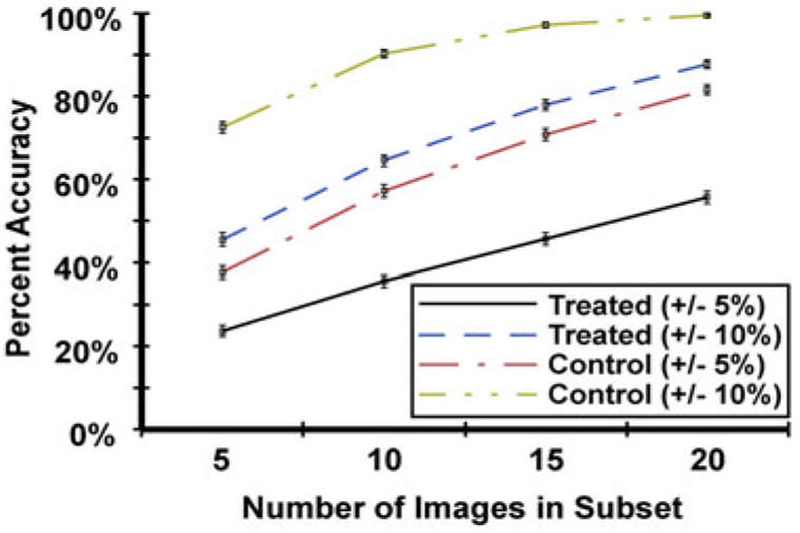
Trends in accuracy of image subset as number of images per subset is increased. Error bars are 1 standard deviation.
It is evident that a larger subset of images will be more accurate than smaller subsets, as shown in the figure. When accuracy is defined as when the subset mean is within 5% of the population mean, the point accuracy of any subset is inadequate to represent the total composite image, with the best subset of 20 images from the control sample being accurate only 81.67% of the time. If the definition of accuracy is loosened to when the subset mean is within 10% of the population mean, percentage values for all subsets, in both control and treated samples, increases significantly. For control sample, subset of ten images will be accurate 90.31% of the time, and subset of 15 images will be accurate 97.25% of the time. This would work if analysis was only required for the control sample, where biofilm population is large and spatially uniform throughout the sample surface. The treated sample, however, has a nonuniform distribution throughout the sample surface, since there are areas more delaminated than others. This, plus the fact the biofilm population is a smaller value, has a greater effect on the potential accuracy of image subsets. Even the largest subset of 20 is only accurate, to within 10% of population mean, 87.71% of the time.
The analysis is not optimum for either sample, given 1) the area of damage on the control sample decreasing biofilm percentages in that area, and 2) the treated sample includes peripheral area not exposed to LGS treatment, thereby dramatically increasing biofilm percentages in those area compared to the central treated area. The control sample analysis can easily be improved by using a sample without surface damage, however, the limiting factor in this simulation is the treated sample. For one to optimize subset image selection in order to improve accuracy, prior knowledge of the boundary between treated and nontreated areas would need to be known or discernible in preliminary SEM imaging. With morphology changes of dehydrated samples and the limited FOV for a single SEM image, neither situation can be improved. Given these limitations and the values above, SEM analysis of LGS-treated samples requires the entire surface to be false colored, which given the lack of added information gained from standard CFU count, would be a value loss to the experiment.
One important observation that was made from the SEM images and analyzed is the biofilm fracturing mechanism and resulting cluster sizes. This has important implications to increasing biofilm permeability to antibiotic treatment, giving a second mechanism to how LGS treatment can improve infected wound healing times. This fragmentation of the biofilm matrix can create more pathways for antibiotics to penetrate the biofilm, potentially increasing its effectiveness against remnant bacteria. Research is ongoing to confirm potential increases of antibiotic susceptibility of bacterial biofilms as a result of LGS treatment. However the exact mechanism, whether direct permeability of bacterial membrane, fragmentation of the biofilm, or both, is still under investigation. In the end this synergistic treatment will require in vivo studies to quantify wound healing times posttreatment. The mechanical nature of LGS therapy allows the system to be nonspecific to bacterial strains, and can be used in conjunction with any variation of standard antibiotic solutions. This property is desirable because of the polymicrobial nature of biofilms [26] and the increasing prevalence of antibiotic resistant pathogens [4]. While this demonstration is with single bacterial strain, S. epidermidis, the same procedure can accommodate any biofilm forming bacteria which hinders wound healing.
LGS reduced bacterial load on ex vivo pigskin by 69%, and reduced biofilm area in one set of samples by 52%. Biofilm disruption and fragmentation were visually confirmed through SEM imaging. LGS is shown as one mechanical method of disrupting the bacterial biofilm that hinders wound healing. It is shown that LGS can reduce appreciable amounts of biofilm and viable bacterial load on the surface of tissue with a single shockwave. These results mark an important distinction from previous work reported on shockwaves-based biofilm removal, in particular the material underlying the biofilm. In the publications reporting biofilm delamination, the candidate biofilms were grown on rigid substrates composed of either glass, polymer(s), or metals which often constitute the composite material of a medical device. In this setup there is a large acoustic impedance mismatch between the biofilm-to-surface interface, and surface-to-underlying material (e.g., air, water, ultrasound gel, etc.) interface. This large mismatch can generate significant tensile stress in either or both interfaces, resulting in substantial biofilm delamination.
The goal of this paper is to evaluate the potential biofilm delamination efficacy in an environment that mimics the acoustic properties of a cutaneous wound. Thus, great care was taken to remove materials and structures that would produce shockwave reflections and companion tensile stress components not reproducible in vivo. This warranted the growth of biofilm on ex vivo pig skin samples and the coupling of the opposite tissue surface to an acoustically thick volume of tissue-mimicking ballistic gel. The result is an in vivo material system that mimics cutaneous wounds and isolates the tensile component generation to the slight acoustic impedance mismatch between biofilm and dermal tissue. As opposed to the numerous analyses reported in the literature on delamination of biofilm from rigid growth surfaces, delamination under the conditions reported here are more difficult to obtain, but significantly more clinically relevant. There are some unknowns that remain to be explored to fully characterize the mechanical mechanism responsible for biofilm disruption. The generation of apparent traveling cavitation bubbles has been observed in high-speed microscopy of the coupling medium in the presence of the shockwaves. The implosion of the cavitation bubbles can themselves generate stress waves, and these may aid in the disruption of biofilm. More characterization work needs to be performed to understand what role bubble dynamics plays in the disruption of biofilms by LGS.
V. CONCLUSION
With the rise of bacterial infected wounds and antibioticresistant bacterial strains, new treatment strategies are required to combat chronic wound infections. LGS is one such therapy being studied to treat these infections, either as a stand-alone therapy or used synergistically with topical antibiotics. The results presented in this paper are the experimental evidence that LGS can sufficiently disrupt bacterial biofilms on dermal tissue. While the results are ex vivo these methods and materials support the feasibility of this technique in vivo. Future wound healing studies with LGS and antibiotics will be completed to evaluate efficacy of increasing time to healing, therefore, eventually reduce patient morbidity and reduce healthcare costs.
V. ACKNOWLEDGMENT
The authors would like to thank the University of California Los Angeles Materials Characterization Lab for use of their scanning electron microscope, Dr. P. Benharash for the tissue sharing protocol, and G. Kerezyte and I. Lee for their assistance in false coloring.
REFERENCES
- [1].Hassan M et al. , “Cost of hospital-acquired infection,” Hospital Topics vol. 88, no. 3, pp. 82–89, 2010. [DOI] [PubMed] [Google Scholar]
- [2].Sen CK et al. , “Human SkinWounds: A Major and Snowballing Threat to Public Health and the Economy,”Wound Repair Regener, vol. 17, no. 6, pp. 763–771, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Niederman M, “Impact of antibiotic resistance on clinical outcomes and the cost of care,” Crit. Care Med, vol. 29, no. 4, pp. N114–120, 2001. [DOI] [PubMed] [Google Scholar]
- [4].Percival SL et al. , “A review of the scientific evidence for biofilms in wounds,” Wound Repair Regener, vol. 20, pp. 647–657, 2012. [DOI] [PubMed] [Google Scholar]
- [5].Mah TF and OToole GA, “Mechanisms of biofilm resistance to antimicrobial agents,” Trends Microbiol, vol. 9, no. 1 pp. 34–39, 2001. [DOI] [PubMed] [Google Scholar]
- [6].Farber BF et al. , ‘Staphylococcus epidermidis extracted slime inhibits the antimicrobial action of glycopeptide antibiotics,” J. Infect. Dis . vol. 161, no. 1, pp. 37–40 1990. [DOI] [PubMed] [Google Scholar]
- [7].Roy S et al. , “Mixed-species biofilm compromises wound healing by disrupting epidermal barrier function,” J. Pathol, vol. 233, pp. 331–343, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Otto M, “Staphylococcus epidermidis–the “accidental” pathogen,” Nat. Rev. Microbiol, vol. 7, no. 8, pp. 555–567, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Rhoads D et al. , “Biofilms in wounds: management strategies,” J. Wound Care, vol. 17, no. 11, pp. 502–508, 2008. [DOI] [PubMed] [Google Scholar]
- [10].Huh AJ and Kwon YJ, “Nanoantibiotics: A new paradigm for treating infectious diseases using nanomaterials in the antibiotics resistant era,” J. Controlled Release, vol. 156, no. 2, pp. 128–145, 2011. [DOI] [PubMed] [Google Scholar]
- [11].Rai M, “Silver nanoparticles as a new generation of antimicrobials,” Biotechnol. Adv, vol. 27, pp. 76–83, 2008. [DOI] [PubMed] [Google Scholar]
- [12].Hajipour MJ et al. , “Antibacterial properties of nanoparticles,” Trends Biotechnol, vol. 30, no. 10, pp. 499–511, 2012. [DOI] [PubMed] [Google Scholar]
- [13].Biel MA, “Antimicrobial photodynamic therapy for treatment of biofilmbased infections,” in Biofilm-Based Healthcare-Assoc. Infections: Volume II, 1st ed. Donelli G, Ed. Switzerland: Springer, 2015, pp. 119–136. [Google Scholar]
- [14].Morley S, “Phase IIa randomized, placebo-controlled study of antimicrobial photodynamic therapy in bacterially colonized, chronic leg ulcers and diabetic foot ulcers: A new approach to antimicrobial therapy,” Brit. J. Dermatol vol. 168, no. 3, pp. 617–624, 2012. [DOI] [PubMed] [Google Scholar]
- [15].Krespi Y, Stoodley P, and Hall-Stoodley L, “Laser disruption of biofilm,” Laryngoscope vol. 118, no. 7, pp. 1168–1173, 2008. [DOI] [PubMed] [Google Scholar]
- [16].Kizhner V et al. , “Laser-generated shockwave for clearing medical device biofilms,” Photomed. Laser Surg, vol. 29, no. 4, pp. 277–282, 2011. [DOI] [PubMed] [Google Scholar]
- [17].Nigri GR et al. , “Laser-induced shock waves enhance sterilization of infected vascular prosthetic grafts,” Lasers Surg. Med, vol. 29, pp. 448–454, 2001. [DOI] [PubMed] [Google Scholar]
- [18].Francis NC et al. , “Analysis of flexible substrates for clinical translation of laser-generated shockwave therapy,” Biomed. Opt. Exp vol. 6, no. 3, pp. 827–837, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Taylor ZD et al. , “Bacterial biofilm disruption using laser generated shockwaves,” in Proc. IEEE Annu. Int. Conf. Eng.Med. Biol. Soc, Buenos Aires, Argentina, Aug. 31–Sep. 4, 2010, pp. 1028–1032. [DOI] [PubMed] [Google Scholar]
- [20].Navarro A et al. , “Bacterial biofilm disruption using laser-generated shockwaves,” Proc. SPIE, vol. 8214, 2012. [DOI] [PubMed] [Google Scholar]
- [21].Doukas A and Kollias N, “Transdermal drug delivery with a pressure wave,” Adv. Drug Del. Rev, vol. 56, pp. 559–579, 2004. [DOI] [PubMed] [Google Scholar]
- [22].Ansel DM et al. , “Animal models of wound repair: Are they cutting it?,” Exp. Dermatol, vol. 21, no. 8, pp. 581–585, 2012. [DOI] [PubMed] [Google Scholar]
- [23].Ramaprasad V et al. , “Effect of laser generated shockwaves 1 on ex-vivo pigskin,” Laser Surg. Med, vol. 46, pp. 620–627, 2014. [DOI] [PubMed] [Google Scholar]
- [24].Tan JS and File TM, “Diagnosis and treatment of diabetic foot infections,” Clin. Infectious Dis, vol. 39, pp. 885–910, 2004. [DOI] [PubMed] [Google Scholar]
- [25].Limpert E et al. , “Log-normal distributions across the sciences: Keys and clues,” BioScience, vol. 51, no. 5, pp. 341–352, 2001. [Google Scholar]
- [26].Burmølle M et al. , “Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms,” Appl. Environ. Microbiol, vol. 72, no. 6, pp. 3916–3923, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]


