SUMMARY
FOG-3 is a master regulator of sperm fate in Caenorhabditis elegans and homologous to Tob/BTG proteins, which in mammals are monomeric adaptors that recruit enzymes to RNA binding proteins. Here, we determine the FOG-3 crystal structure and in vitro demonstrate that FOG-3 forms dimers that can multi-merize. The FOG-3 multimeric structure has a basic surface potential, suggestive of binding nucleic acid. Consistent with that prediction, FOG-3 binds directly to nearly 1,000 RNAs in nematode spermatogenic germ cells. Most binding is to the 3′ UTR, and most targets (94%) are oogenic mRNAs, even though assayed in spermatogenic cells. When tethered to a reporter mRNA, FOG-3 represses its expression. Together these findings elucidate the molecular mechanism of sperm fate specification and reveal the evolution of a protein from monomeric to multimeric form with acquisition of a distinct mode of mRNA repression.
Graphical Abstract
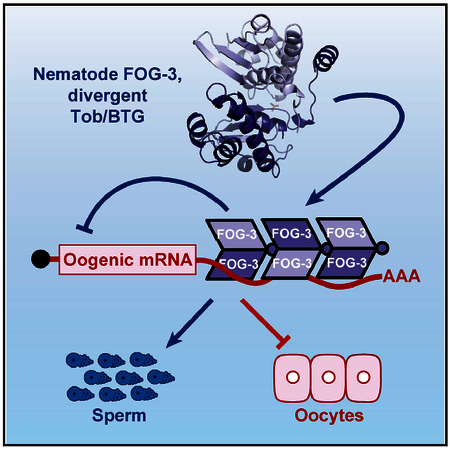
In Brief
The mechanism of the sperm or oocyte fate decision has been elusive. Aoki et al. report that nematode FOG-3, a Tob/BTG protein driving sperm fate, has evolved from monomeric to multimeric form with acquisition of a divergent Tob/BTG mechanism for mRNA repression.
INTRODUCTION
Gene discovery often paves the way to a molecular understanding of mysterious biological phenomena. But how do we learn molecular functions of newly identified genes? A common method takes advantage of amino acid comparisons to find homologs or at least protein domains that can provide clues. The major challenge is to test the predictions of those clues and tease out the function biochemically. Our focus is molecular regulation of the sperm or oocyte cell fate decision, which has remained elusive despite analyses over decades. Here, we report studies of a protein harboring a broadly conserved domain whose biochemical analysis reveals a striking case of protein evolution that could not have been predicted from sequence comparison.
Metazoan germ cells differentiate as either sperm or oocyte, depending on organismal sex. Molecular regulation of germline sexual fate relies in part on sex-determining signals from somatic tissues and in part on regulators responding to those signals in germ cells (Ellis and Schedl, 2007; Murray et al., 2010). In the nematode Caenorhabditis elegans, a network of intrinsic germline fate regulators drives expression of two key proteins that execute the sperm or oocyte fate decision (Ellis and Schedl, 2007; Kimble and Crittenden, 2007). Those two key terminal regulators are FOG-1 and FOG-3 (Figure 1A), named for their loss-of-function Fog (feminization of germline) phenotype (Barton and Kimble, 1990; Ellis and Kimble, 1995). FOG-1 is a C. elegans ortholog of the cytoplasmic polyadenylation element RNA binding protein (CPEB) (Jin et al., 2001; Luitjens et al., 2000), and FOG-3 harbors a protein domain that places it in the Tob/BTG family (Chen et al., 2000; Ellis and Kimble, 1995). Mammalian Tob/BTGs interact with RNA binding proteins and recruit enzymes to modify mRNAs and repress their expression (Hosoda et al., 2011; Ogami et al., 2014; Yu et al., 2016). Most relevant to this work, mammalian Tob/BTG binds CPEB proteins, and they recruit the CCR4-Not complex to shorten poly(A) tails and repress mRNA translation of CPEB target mRNAs (Hosoda et al., 2011). Similar to its mammalian counterparts, nematode FOG-3/Tob binds FOG-1/CPEB, and both proteins associate with a common set of 76 mRNAs, with 90% belonging to the oogenesis program (Noble et al., 2016). Because FOG-1 and FOG-3 specify the sperm fate, the inferred function was repression of oogenic RNAs.
Figure 1. FOG-3 Is a Divergent Tob/BTG Protein.
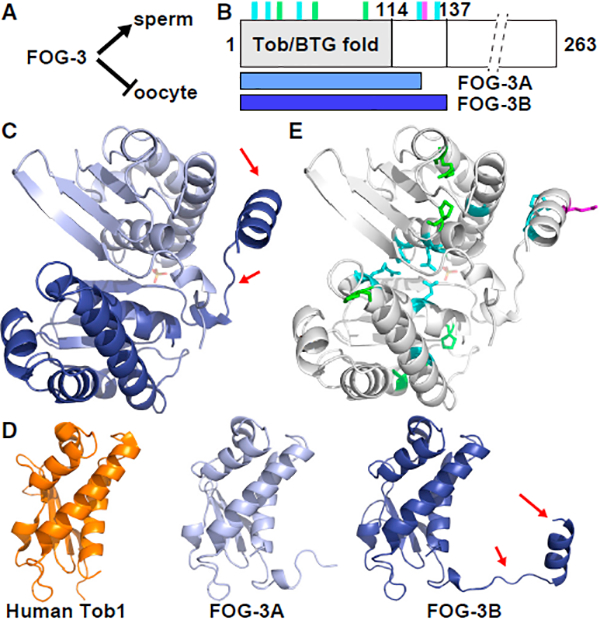
(A) FOG-3 is a terminal regulator of the sperm fate, and hence essential for sperm fate specification in both hermaphrodite larvae and males.
(B) Linear diagram of FOG-3 protein. Predicted Tob/BTG fold is N-terminal (gray). C terminus is abbreviated (dashed lines). Vertical lines mark missense mutation sites (also see Figure S1): green, conserved in all orthologs; cyan, conserved in nematode orthologs; magenta, missense mutation generated in this study. Horizontal bars below show extents of subunits in crystal dimer, termed FOG-3A (light blue) and FOG-3B (dark blue).
(C) Asymmetric unit of the FOG-3 crystal structure (PDB: 5TD6). FOG-3A (chain A, light blue) and FOG-3B (chain B, dark blue). Arrows highlight the nematode-specific linker-helix extension.
(D) FOG-3 and human Tob/BTG structures. RMSD of human Tob1 (orange, PDB: 2Z15) compared with FOG-3A and FOG-3B was 1.062Å and 1.086Å, respectively. Arrows highlight the linker-helix extension.
(E) Location of FOG-3 missense mutants in the crystal structure. Color scheme matches that in (B).
We began this work with the idea that the FOG-3 mechanism of post-transcriptional control would echo that of its mammalian relatives, but we instead found a distinct mechanism that advances understanding of sperm fate specification and highlights the potential for undiscovered protein multimers in biology.
RESULTS
FOG-3 Is a Divergent Member of the Tob/BTG Family
Like canonical Tob/BTG family members, the FOG-3 primary sequence possesses a predicted N-terminal Tob/BTG domain and a disordered C-terminal region (Figures 1B and S1) (Chen et al., 2000). Comparison of FOG-3 amino acid sequences from several Caenorhabditis species revealed a further nematode-specific conservation that extends ~20 amino acids past the predicted Tob/BTG fold. We used recombinant C. elegans FOG-3 to identify a single domain (amino acids 1–137) that spans the canonical Tob/BTG fold plus this nematode-specific extension (see Supplemental Experimental Procedures and Figures S2A and S2B).
We pursued the FOG-3 crystal structure to gain insight into this putative Tob/BTG protein. FOG-3 crystals were obtained with C-terminally truncated recombinant protein that had changes to two non-conserved amino acids (1–137; C117A). These crystals provided a full dataset to 2.03 Å (Research Collaboratory for Structural Bioinformatics [RCSB] PDB: 5TD6; Table S1). Phase information was acquired using a human Tob structure (see Supplemental Experimental Procedures). Each asymmetric unit contained two copies of FOG-3; we dub chain A as FOG-3A and chain B as FOG-3B (Figures 1B and 1C). For FOG-3A, we could model residues 1–123, and for FOG-3B we could model nearly the entire peptide chain (1–136) (Figures 1B and 1C). The structural alignment was excellent between FOG-3A, FOG-3B, and a previously determined human Tob1 structure (root-mean-square deviation [RMSD] 1.062–1.084Å; Figure 1D) (Horiuchi et al., 2009), confirming the predicted Tob/BTG fold in FOG-3. However, unlike other Tob/BTG structures, the FOG-3 structure included a linker-helix extension past the classic Tob/BTG fold (Figures 1C and 1D).
Details of the FOG-3 crystal structure supported the idea that the two FOG-3 subunits in the asymmetric unit (ASU) represent a bona fide dimer. The buried surface area between the two FOG-3 subunits was large (1,034.2Å), favorable for assembly (DG = −7.2 kcal/mol), and the interface intricate (Figures S2D and S2E). The linker-helix extension of FOG-3B folded around FOG-3A (Figure 1C), making several hydrogen bonds (Figures S2D and S2E). The Tob/BTG folds of FOG-3A and FOG-3B also contacted each other at their N-terminal helices, with hydrogen bonds and arginine planar stacking between conserved residues (Figures S2G and S2H). To ask whether this potential FOG-3 dimer may have biological significance, we analyzed the sites of the missense mutations (Chen et al., 2000). All mutations abolish sperm fate specification and hence render the protein non-functional (Ellis and Kimble, 1995). The FOG-3 structure included all eight missense sites (Figures 1B, 1E, S1, and S2F). Three mutations (P21L, R56Q, P94S) alter residues conserved across Tob/BTG folds, and five others change residues conserved only in FOG-3 and nematode paralogs (Figure S1). The three Tob/BTG fold mutations include two prolines located between helices and an arginine making a hydrogen bond characteristic of Tob/BTG folds (Figures S2G and S2I) (Horiuchi et al., 2009; Yang et al., 2008). Because of their locations, contacts, and conservation, we speculate that these residues facilitate protein folding. Five other mutations change residues conserved only in nematodes. These include three (E7K, R14K, G33K) in the Tob/BTG fold (Figures S2G and S2H) and two (P125L, A132T) in the nematode-specific linker-helix extension (Figures S2G and S2J). At the potential dimer interface of the Tob/BTG folds, two missense residues (E7K, R14K) map to the interface itself (Figures S2G and S2H) and the third (G33K) maps to a central helix, where a bulky lysine residue could disrupt dimerization via steric hindrance (Figures S2G and S2I). The two mutations outside the Tob fold (P125L, A132T) map to the internal face of the link-helix extension in FOG-3B (Figures S2G and S2J). Thus, the FOG-3 crystal structure and sites of fog-3 mutations provide evidence that FOG-3 has a Tob/BTG fold but is divergent with potential to dimerize.
FOG-3 Dimerization and Higher-Order Assembly
The mutations in the linker-helix suggest that this extension is crucial for FOG-3 function. The asymmetrical interaction between the linker-helix of one subunit and the Tob/BTG fold of the other seemed an unusual strategy for dimerization (Figure 1C). We wondered whether the linker-helix might also mediate an interaction between dimers. To explore this idea, we extended the crystal symmetry to visualize FOG-3 dimerdimer interactions in the structure and found the linker-helix extension tucked neatly into a cleft of the adjacent FOG-3 dimer (Figure 2A). Each dimer was rotated 180° relative to its neighbor in a continuous pattern to form a polymeric-like assembly within the crystal (Figure 2B). Moreover, features of the structure suggested that the dimer-dimer interface may be authentic: its surface area is 1,112.5Å (Figure S3A), a value similar to that between subunits in the dimer, and the predicted interface included 10 hydrogen bonds and four salt bridges (Figures S3A–S3C), with many residues conserved among FOG-3 orthologs (Figure S1).
Figure 2. FOG-3 Forms Higher-Order, Multi-dimer Assemblies.
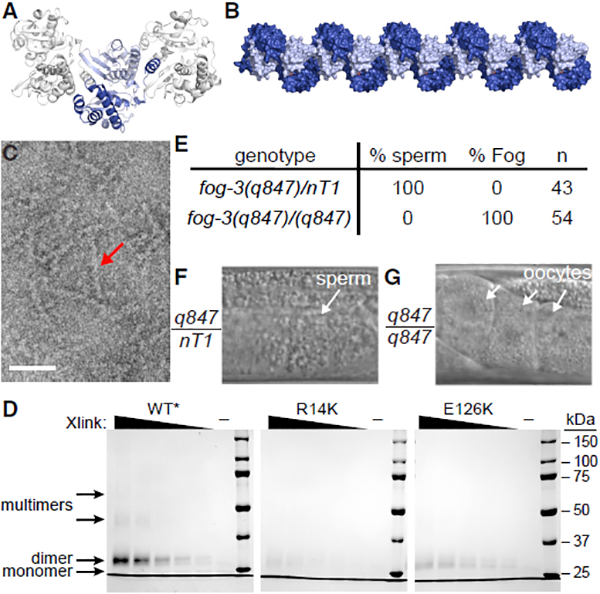
(A) Crystal packing of the FOG-3 dimer. FOG-3A and FOG-3B are represented in light and dark blue, respectively.
(B) Model for multimerization of FOG-3 dimers, as observed in the crystal. Image was generated by extending the crystal symmetry. Subunits are colored as in (A).
(C) Negative-stain electron microscopy of recombinant FOG-3 1–137. Note the presence of long rods (arrow). Scale bar, 50 nm.
(D) Coomassie-stained gel of modified FOG-3 recombinant protein incubated with increasing amounts of BS3 crosslinker. ‘‘–’’ represents no BS3 included. Different exposure shown in Figure S3I.
(E–G) Mutation of a key residue, E126K, transforms the germline from spermatogenic to oogenic. The E126K allele, fog-3(q847), was maintained over a balancer (nT1). (E) Heterozygous or homozygous adult males were analyzed for sperm or oocytes. (F and G) Representative DIC images of adult males, heterozygous (F) or homozygous (G) for the E126K mutation.
See Figures S3L–S3N for similar results in hermaphrodites.
We sought to test FOG-3 dimerization and higher-order multimerization in vitro. On a sizing column, recombinant FOG-3 eluted distinctly at high versus low concentrations (Figure S2A), suggesting dimerization at higher protein concentration. The column gave no hint of larger multimers (Figure S2A), but they might be unstable. We turned next to negative-stain electron microscopy (EM), which revealed rods when recombinant FOG-3 was assayed at high concentration (Figure 2C). The FOG-3 rods were consistent with multimerization, but their formation was sporadic, making EM an unreliable assay to compare wild-type versus mutant protein. We finally turned to biochemistry and used the crystal structure to insert lysine substitutions at nonconserved residues to facilitate chemical crosslinking (Figure S3D–S3F). The lysine variant could be purified (Figures S3G and S3H) and robustly crosslinked as a dimer when incubated (Figures 2D and S3I). At a higher crosslinker concentration, larger species formed, which we attribute to multi-dimer assemblies (Figures 2D and S3I). We next made a lysine variant that also harbored a missense mutation (R14K) (Chen et al., 2000), which is predicted from the structure to impede dimerization (Figures S2F–S2H). Indeed, dimerization was greatly reduced with this R14K mutant and multimerization was abolished (Figures 2D and S3I). Therefore, R14K disrupts dimerization, as predicted, and dimerization is required for higher-ordered assembly.
Classical genetics failed to generate a mutation that would disrupt the dimer-dimer interaction. To make our own, we identified a glutamate (E126) on the solvent-exposed surface of the linker-helix that contacts the neighboring dimer (Figures S3J and S3K). We reasoned that changing E126 to a positively charged amino acid (E126K) should disrupt the dimer-dimer interface specifically. Indeed, an E126K lysine variant still formed dimers, but not higher-order, multimeric species (Figures 2D and S3I). To ask whether dimer-dimer contact was critical for sperm fate specification in vivo, we introduced E126K in the endogenous fog-3 gene (see Supplemental Experimental Procedures). Animals homozygous for either of two independently generated E126K alleles failed to make sperm and instead had a fully penetrant Fog phenotype in both males (Figures 2E–2G) and hermaphrodites (Figures S3L–S3N). The E126K mutation may affect FOG-3 in ways other than multimerization, but the simplest explanation is that FOG-3 functions in vivo as a multimer of dimers to promote sperm fate.
FOG-3 Binds RNA Directly to 3′ UTRs of Oogenic-Associated Transcripts
The electrostatic surface potential of the FOG-3 multimer is highly basic (Figures S4A and S4B) and prompted us to ask whether FOG-3 binds directly to RNA. In nematode spermatogenic germ cells, FOG-3 immunoprecipitated radiolabeled RNA after UV crosslinking (Figures S4C–S4F), a treatment creating covalent bonds between protein and RNA (Huppertz et al., 2014). Using in vivo crosslinking and immunoprecipitation (iCLIP) (Huppertz et al., 2014), we identified RNAs crosslinked to FOG-3 (Figures S4G–S4I; Tables S2 and S3) and sites of FOG-3 binding within those RNAs. After normalization to a negative control (see Supplemental Experimental Procedures), FOG-3 enriched for 955 mRNA targets and 38 non-coding RNAs (Figure 3A). Remarkably, ~94% of FOG-3-bound mRNAs belonged to the oogenesis program (Figures 3B and S4J), despite immunoprecipitation from sperm-fated germ cells. The FOG-3 targets identified by iCLIP overlapped with those identified by microarray (Figures S4J and S4K; p < 1 × 10‒208), but iCLIP significantly increased both number of targets as well as their enrichment for oogenic RNAs (Figures S4J and S4K). Because iCLIP is more stringent than microarray methods (Wang et al., 2009), we suggest that iCLIP improved the signal-to-noise ratio of RNAs immunoprecipitating with FOG-3. We conclude that FOG-3 binds directly to RNAs that belong largely to the oogenesis program.
Figure 3. FOG-3 in Spermatogenic Germ Cells Binds Directly to 3′ UTRs of Oogenic mRNAs.
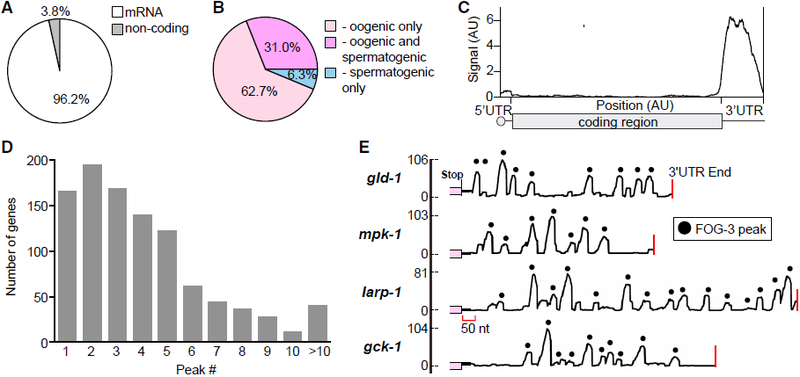
(A) FOG-3 iCLIP enriches for mRNAs.
(B) Most FOG-3-bound mRNAs belong to the oogenesis program, which includes RNA expressed only in oogenic germlines (light pink) and in both oogenic and spermatogenic germlines (dark pink), as categorized previously (Noble et al., 2016).
(C) Distribution of FOG-3 iCLIP sequence reads within mRNAs. Transcript lengths are normalized so that 5′ UTRs, coding sequences (CDS), and 3′ UTRs (50, 1,000, and 200 nt, respectively) are reported as arbitrary units (AU).
(D) Many FOG-3 targets possess multiple binding peaks in their 3′ UTRs, as shown in (E).
(E) Examples of FOG-3 binding peaks across 3′ UTRs. x axis, 3′ UTR with coding region in pink and 3′ end marked by red line; y axis, number of mapped reads. Peaks are marked by black dots; their heights correspond to number of mapped reads.
The vast majority of FOG-3 binding sites mapped to 3′ UTRs (Figures 3C and S4L–S4N), implying that FOG-3 binding is largely restricted. A multimer is expected to leave an extensive footprint. Consistent with this idea, 624 of 955 protein-coding genes (65.3%) had two or more sequence peaks in their 3′ UTRs (Figures 3D, 3E, and S4O). This pattern is reminiscent of multi-site RNA binding proteins, like HuR (Lebedeva et al., 2011). Gaps between peaks might signify authentic binding absences from breaks in FOG-3 multimerization or binding by independent dimers. These gaps may also be sites of preferred enzymatic digestion during iCLIP (see Supplemental Experimental Procedures). Regardless, we conclude that FOG-3 binds the 3′ UTRs of its target mRNAs and that most of its targets belong to the oogenic program, despite being immunoprecipitated from spermatogenic cells.
Tethered FOG-3 Represses Expression of a Reporter mRNA
Genetically, FOG-3 is a master regulator of sperm fate (Ellis and Kimble, 1995) and FOG-3 binds many oogenic mRNAs directly in spermatogenic germ cells (this work). The simplest model is that FOG-3 specifies the sperm fate by repression of oogenic mRNAs. One cannot remove FOG-3 to test this idea because germ cells make oocytes rather than sperm without FOG-3. As an alternative approach, we turned to a protein-mRNA tethering reporter assay used in other systems (Coller and Wickens, 2002) and C. elegans (Wedeles et al., 2013), which takes advantage of lN22 peptide binding to boxB RNA hairpins (Baron-Benhamou et al., 2004) (Figure 4A). Endogenous FOG-3 was engineered to include a C-terminal 3xFLAG epitope tag, with or without lN22 (see Supplemental Experimental Procedures). The reporter mRNA expressed GFP-tagged histone and carried three boxB hairpins in its 3′ UTR (Figure 4A). Both engineered FOG-3 proteins promoted the sperm fate and were expressed as expected (Figures 4B and 4F). Thus, our modifications did not affect FOG-3 function or expression.
Figure 4. FOG-3 Represses mRNA Reporter Expression When Tethered in Nematodes.
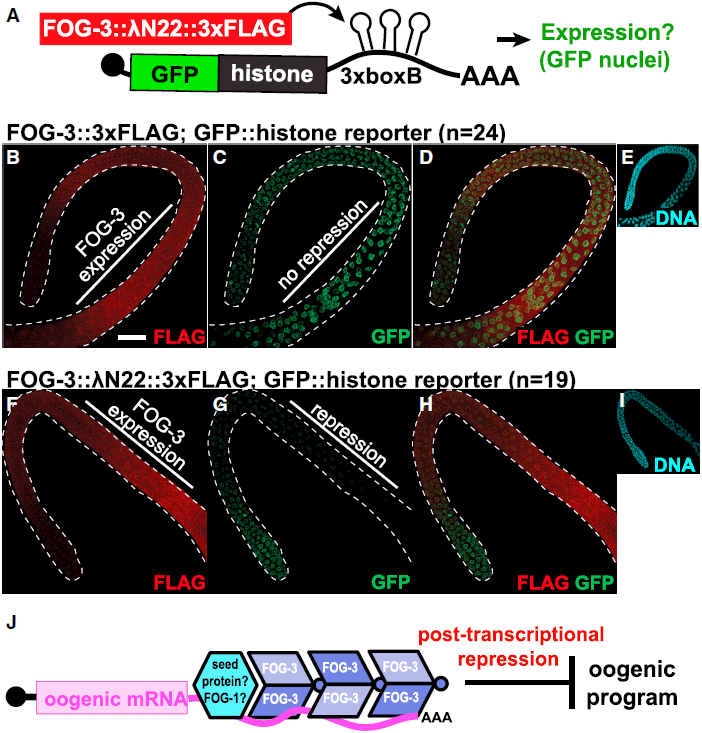
(A) Summary of the protein-mRNA tethering assay. With lN22, FOG-3 can bind and regulate the protein expression of a GFP::histone reporter transcript containing 3xboxB hairpins in its 3′ UTR.
(B–I) Maximum intensity projections from confocal images of representative male adult germlines expressing both modified FOG-3 and the GFP reporter. FOG-3::3xFLAG (B–E) or FOG-3::lN22::3xFLAG (F–I) with the GFP::histone reporter were imaged by fluorescent confocal microscopy for (B and F) FLAG, (C and G) GFP, (D and H) FLAG and GFP overlay, and (E and I) DNA (DAPI). DNA images reduced in size 2.5-fold. Dashed white line outlines male germlines. Scale bar, 20 μm.
(J) Model of the FOG-3 molecular mechanism. Multimers of FOG-3 dimers bind 3′ UTRs and promote the sperm fate by repressing mRNAs in the oogenic program. FOG-3 may find its targets by interacting with a distinct sequence-specific seed protein, one candidate being FOG-1 (see Discussion)..
We compared fluorescence in germ cells expressing engineered FOG-3 and the reporter. GFP was easily detected throughout the male germline when FOG-3::3xFLAG was not fused to lN22 (Figures 4B–4E). However, when FOG-3::3xFLAG was fused to λN22, GFP fluorescence decreased in the region of FOG-3 expression (Figures 4F–4I). Therefore, FOG-3 represses GFP expression when tethered to a reporter mRNA.
DISCUSSION
Our findings support a model for the molecular mechanism of sperm fate regulation (Figure 4J). We propose that the functional form of FOG-3 is multimeric, that FOG-3 multimers bind directly to the 3′ UTRs of target mRNAs, and that FOG-3 is a broad-spectrum repressor of the oogenesis program. Because FOG-3 and its orthologs specify sperm fate in both C. elegans sexes and related dioecious species (Chen et al., 2001), this mechanism is likely fundamental to nematode sperm fate specification. Protein expression appears sufficient for such function; FOG-3 is readily observed by immunoblot and its staining is granular (Noble et al., 2016), implying that FOG-3 may be further concentrated within the cytoplasm for multimerization. Our results challenge the idea that FOG-3 regulates RNAs via the mechanism elucidated for mammalian Tob/BTG proteins, which function as monomeric adapters. Although FOG-3 might also function as a monomeric adaptor, such a canonical Tob/BTG mechanism is unlikely for its role in sperm fate. Terminal regulators of mammalian germ cell fate are not yet known, so we cannot exclude the possibility that the biological role of Tob/BTG proteins may have been conserved in germ cells.
The restricted footprint of FOG-3 to 3′ UTRs raises questions about the regulation of FOG-3 binding. FOG-3 may itself provide RNA binding specificity, although no sequence element could be detected beneath its iCLIP peaks. More likely, another sequence-specific RNA-binding protein seeds FOG-3 assembly (Figure 4J). FOG-1/CPEB is a strong candidate for such a seed protein, because it is also drives sperm fate specification (Barton and Kimble, 1990), binds to FOG-3, and associates with oogenic mRNAs (Noble et al., 2016). The mechanism of repression remains a challenge for the future. The FOG-3 multimer might recruit effector complexes to modify the mRNA, similar to mammalian Tob/BTG. If true, details are likely different because residues corresponding to the deadenylase binding interface of human Tob (Horiuchi et al., 2009) are not conserved in FOG-3 and no missense mutations mapped to that potential interface. Other possibilities include competition with an activator or moving mRNAs to sites of repression within the cell.
Fundamental aspects of nematode FOG-3 function diverge from mammalian Tob/BTG. Although mRNA repression is shared, FOG-3 functions as a multimer and binds RNA directly, whereas mammalian Tob/BTG proteins function as monomeric adaptors that link sequence-specific RNA binding proteins to enzymes (Hosoda et al., 2011; Ogami et al., 2014; Yu et al., 2016). Consistent with this divergence, FOG-3 possesses a nematode-specific linker-helix extension that mediates dimer-dimer interactions, and those dimer-dimer interactions are integral to sperm fate specification. This strategy of FOG-3 assembly is reminiscent of certain viral RNA binding proteins that package RNA viral genomes using a core domain plus a C-terminal linker-helix or linker-b sheet extension to drive multimerization (Harrison, 2017). In an analogous but distinct strategy, yeast RNA-binding protein Rim4 assembles into amyloid-like fibers to repress translation of mRNAs required for gametogenesis (Berchowitz et al., 2015). Collaborative assembly with RNA is common among viral assembly proteins and other nucleic acid binding polymers (Ghosal and Löwe, 2015). FOG-3 may similarly require an mRNA scaffold for higher-order assembly, given that FOG-3 dimers could not multimerize into higher-order assemblies at low protein concentration.
Our work on FOG-3 highlights the concept that evolution can usurp a well-conserved domain to form multimers with only modest changes to its primary sequence and structural fold. Multimerization appears to be a unique feature of FOG-3-related Tob/BTG proteins. The critical residues at the dimer interface are conserved in nematodes, but not in mammalian Tob/BTG proteins, and mammalian homologs show no evidence of multimerization (Horiuchi et al., 2009; Yang et al., 2008). Because only a few Tob/BTG proteins have been characterized biochemically, FOG-3 might yet have a vertebrate counterpart. However, we favor instead the idea that an existing protein fold was adapted during evolution to transform a protein that acts as a monomer into a multimeric repressor with RNA binding properties. The transformation from monomer to multimer required evolution of inter-subunit interacting surfaces to form dimers and evolution of inter-dimer interacting surfaces to permit multimerization. In the case of FOG-3, the inter-dimer interface was created by adding a linker-helix extension that fits into the cleft of its neighbor, adding a binding surface to one side of the dimer. This extension provides directionality for multimer assembly. Therefore, FOG-3 provides an elegant example of protein evolution, in which a broadly conserved protein domain is redeployed to acquire a distinct mode of mRNA repression.
EXPERIMENTAL PROCEDURES
Crystallization and Structure Determination
Crystallization conditions were screened with sitting drop trays set up using the Mosquito (TTP Labtech, Cambridge, MA, USA). We obtained crystals using recombinant FOG-3 (1–137 H48N C117A) with an intact histidine tag and incubating our trays at 4° C. Coordinates, reflection data, and further experimental details are available at RCSB (PDB: 5TD6) and in the Supplemental Experimental Procedures.
Negative-Stain Electron Microscopy
EM was performed with recombinant FOG-3 (1–137 H48N C117A) as previously described (Bozzola and Russell, 1999). See Supplemental Experimental Procedures.
Molecular Genetics
Worm Maintenance
C. elegans were maintained as described previously (Brenner, 1974). Strains used were JA1515: weSi2 II; unc-119 III; JK2739: hT2[qIs48](I;III)/lin-6(e1466) dpy-5(e61)I; JK4871: fog-3(q520) I; qSi41[fog-3::3xFLAG] II; JK5437: fog-3(q847) I/hT2[qIs48](I;III); JK5439: fog-3(q849) I/hT2[qIs48](I;III); JK5921: qSi375 [mex-5 promoter::EGFP::linker::his-58::3xboxB::tbb-2 3′ UTR] II; JK5942: fog-3(q873[fog-3::3xFLAG]) I; qSi375 II; JK5943: fog-3(q874[fog-3::lN22::3xFLAG]) I; qSi375 II; and N2 Bristol. Strains are available at the Caenorhabditis Genetics Center (https://cbs.umn.edu/cgc/home) or upon request. See Supplemental Experimental Procedures for details on generation of the fog-3 alleles and in vivo fluorescent reporter.
Fertility and Fog Phenotype
Heterozygous and homozygous fog-3(q847), or fog-3(q849), were singled onto plates as L4 larvae. After 3 and 4 days, worms were scored for the presence of L1 larvae and Fog phenotype.
Imaging
Live worms were imaged by differential interference contrast (DIC) microscopy with 0.1 mM levamisole (Sigma, St. Louis, MO, USA). For fluorescent imaging, germlines were extruded, fixed, permeabilized with Triton-X (0.5%), and stained as previously described (Crittenden et al., 2017). See Supplemental Experimental Procedures for further details.
iCLIP
In vivo crosslinking and immunoprecipitation (iCLIP) was carried out essentially as described (Huppertz et al., 2014), with modifications to worm growth, crosslinking, lysis, RNase digestion, and data analysis described in the Supplemental Experimental Procedures. All reads within a gene had their position randomized to empirically determine a cluster p value. A Benjamini-Hochberg (BH) correction was applied (1% false discovery rate [FDR]). Only overlapping clusters called independently as significant in at least two replicates were retained. Fisher’s exact test was used to compare our results with previous FOG-3 RIP results (Noble et al., 2016).
Supplementary Material
Highlights.
FOG-3 crystal structure reveals sites of missense mutations
FOG-3 assembles into dimers that can multimerize
FOG-3 binds directly to mRNAs in the oogenic program
FOG-3 recruited to a reporter mRNA represses its expression
ACKNOWLEDGMENTS
The authors thank J. Claycomb and J. Ahringer for worm strains; R. Massey for electron microscopy image acquisition; M. Cox for equipment; A. Helsley-Marchbanks for help preparing the manuscript; L. Vanderploeg for help with the figures; and members of the Kimble and Wickens labs for helpful discussions. Use of the Advanced Photon Source was supported by the U.S. DOE under Contract No. DE-AC02–06CH11357. GM/CA@APS has been funded in whole or in part with federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). The Eiger 16M detector was funded by an NIH–Office of Research Infrastructure Programs, High-End Instrumentation Grant (1S10OD012289–01A1). S.T.A. was supported by NIH grants F32HD071692 and K99HD081208. C.A.B. was supported by NIH grants GM094584, GM094622, and GM098248. M.W. was supported by NIH grant GM50942. J.K. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
DATA AND SOFTWARE AVAILABILITY
The accession number for the structure reported in this paper is PDB: 5TD6. The accession number for the raw sequence files of all replicates reported in this paper is GEO: GSE76521 (https://www.ncbi.nlm.nih.gov/geo/).
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, four figures, and three tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.05.095.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Baron-Benhamou J, Gehring NH, Kulozik AE, and Hentze MW (2004). Using the lambdaN peptide to tether proteins to RNAs. Methods Mol. Biol 257, 135–154. [DOI] [PubMed] [Google Scholar]
- Barton MK, and Kimble J (1990). fog-1, a regulatory gene required for specification of spermatogenesis in the germ line of Caenorhabditis elegans. Genetics 125, 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz LE, Kabachinski G, Walker MR, Carlile TM, Gilbert WV, Schwartz TU, and Amon A (2015). Regulated formation of an amyloid-like translational repressor governs gametogenesis. Cell 163, 406–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzola JJ, and Russell LD (1999). Electron Microscopy: Principles and Techniques for Biologists (Jones and Bartlett Publishers; ). [Google Scholar]
- Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P-J, Singal A, Kimble J, and Ellis RE (2000). A novel member of the tob family of proteins controls sexual fate in Caenorhabditis elegans germ cells. Dev. Biol 217, 77–90. [DOI] [PubMed] [Google Scholar]
- Chen P-J, Cho S, Jin S-W, and Ellis RE (2001). Specification of germ cell fates by FOG-3 has been conserved during nematode evolution. Genetics 158, 1513–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J, and Wickens M (2002). Tethered function assays using 3′ untranslated regions. Methods 26, 142–150. [DOI] [PubMed] [Google Scholar]
- Crittenden SL, Seidel HS, and Kimble J (2017). Analysis of the C. elegans germline stem cell pool. Methods Mol. Biol 1463, 1–33. [DOI] [PubMed] [Google Scholar]
- Ellis RE, and Kimble J (1995). The fog-3 gene and regulation of cell fate in the germ line of Caenorhabditis elegans. Genetics 139, 561–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R, and Schedl T (2007). Sex determination in the germ line. WormBook, The C. elegans Research Community. http://www.wormbook.org/chapters/www_sexgermline.2/sexgermline.html. [DOI] [PMC free article] [PubMed]
- Ghosal D, and Löwe J (2015). Collaborative protein filaments. EMBO J. 34, 2312–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SC (2017). Protein tentacles. J. Struct. Biol 200, 244–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi M, Takeuchi K, Noda N, Muroya N, Suzuki T, Nakamura T, Kawamura-Tsuzuku J, Takahasi K, Yamamoto T, and Inagaki F (2009). Structural basis for the antiproliferative activity of the Tob-hCaf1 complex. J. Biol. Chem 284, 13244–13255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoda N, Funakoshi Y, Hirasawa M, Yamagishi R, Asano Y, Miyagawa R, Ogami K, Tsujimoto M, and Hoshino S (2011). Anti-proliferative protein Tob negatively regulates CPEB3 target by recruiting Caf1 deadenylase. EMBOJ. 30, 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppertz I, Attig J, D’Ambrogio A, Easton LE, Sibley CR, Sugimoto Y, Tajnik M, König J, and Ule J (2014). iCLIP: protein-RNA interactions at nucleotide resolution. Methods 65, 274–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S-W, Kimble J, and Ellis RE (2001). Regulation of cell fate in Caenorhabditis elegans by a novel cytoplasmic polyadenylation element binding protein. Dev. Biol 229, 537–553. [DOI] [PubMed] [Google Scholar]
- Kimble J, and Crittenden SL (2007). Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu. Rev. Cell Dev. Biol 23, 405–433. [DOI] [PubMed] [Google Scholar]
- Lebedeva S, Jens M, Theil K, Schwanhäusser B, Selbach M, Landthaler M, and Rajewsky N (2011). Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol. Cell 43, 340–352. [DOI] [PubMed] [Google Scholar]
- Luitjens C, Gallegos M, Kraemer B, Kimble J, and Wickens M (2000). CPEB proteins control two key steps in spermatogenesis in C. elegans. Genes Dev. 14, 2596–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray SM, Yang SY, and Van Doren M (2010). Germ cell sex determination: a collaboration between soma and germline. Curr. Opin. Cell Biol 22, 722–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble DC, Aoki ST, Ortiz MA, Kim KW, Verheyden JM, and Kimble J (2016). Genomic analyses of sperm fate regulator targets reveal a common set of oogenic mRNAs in Caenorhabditis elegans. Genetics 202, 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogami K, Hosoda N, Funakoshi Y, and Hoshino S (2014). Antiproliferative protein Tob directly regulates c-myc proto-oncogene expression through cytoplasmic polyadenylation element-binding protein CPEB. Oncogene 33, 55–64. [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, and Snyder M (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet 10, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedeles CJ, Wu MZ, and Claycomb JM (2013). Protection of germline gene expression by the C. elegans Argonaute CSR-1. Dev. Cell 27, 664–671. [DOI] [PubMed] [Google Scholar]
- Yang X, Morita M, Wang H, Suzuki T, Yang W, Luo Y, Zhao C, Yu Y, Bartlam M, Yamamoto T, and Rao Z (2008). Crystal structures of human BTG2 and mouse TIS21 involved in suppression of CAF1 deadenylase activity. Nucleic Acids Res. 36, 6872–6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Ji SY, Sha QQ, Dang Y, Zhou JJ, Zhang YL, Liu Y, Wang ZW, Hu B, Sun QY, et al. (2016). BTG4 is a meiotic cell cycle-coupled maternal-zygotic-transition licensing factor in oocytes. Nat. Struct. Mol. Biol 23, 387–394. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


