Abstract
Streptococcus agalactiae is an important human opportunistic pathogen that can cause serious health problems, particularly among newborns and older individuals. S. agalactiae contains the CAMP factor, a pore-forming toxin first identified in this bacterium. The CAMP reaction is based on the co-hemolytic activity of the CAMP factor and is commonly used to identify S. agalactiae in the clinic. Closely related proteins are present also in other Gram-positive pathogens. Although the CAMP toxin was discovered more than a half century ago, no structure from this toxin family has been reported, and the mechanism of action of this toxin remains unclear. Here, we report the first structure of this toxin family, revealing a structural fold composed of 5 + 3-helix bundles. Further analysis by protein truncation and site-directed mutagenesis indicated that the N-terminal 5-helix bundle is responsible for membrane permeabilization, whereas the C-terminal 3-helix bundle is likely responsible for host receptor binding. Interestingly, the C-terminal domain inhibited the activity of both full-length toxin and its N-terminal domain. Moreover, we observed that the linker region is highly conserved and has a conserved DLXXXDXAT sequence motif. Structurally, this linker region extensively interacted with both terminal CAMP factor domains, and mutagenesis disclosed that the conserved sequence motif is required for CAMP factor's co-hemolytic activity. In conclusion, our results reveal a unique structure of this bacterial toxin and help clarify the molecular mechanism of its co-hemolytic activity.
Keywords: bacterial toxin, crystal structure, Streptococcus, Gram-positive bacteria, glycosylphosphatidylinositol (GPI anchor), CAMP factor, CAMP test, Group B streptococcus, novel structural fold, pore-forming toxin, bacterial virulence, virulence factor, neonatal sepsis, opportunistic pathogen
Introduction
Streptococcus agalactiae (group B streptococcus; GBS)5 can cause serious illness, particularly among newborn infants, in whom it is the leading cause of sepsis (1), and in immune-compromised or elderly patients. Asymptomatic GBS colonization is common; the bacterium resides in the vagina or rectum of about 25% of all healthy adult women in the United States. GBS also occurs in animals; it is an important cause of contagious mastitis in cattle (2) and also a fish pathogen that may compromise food safety and represents a zoonotic hazard (3, 4).
Virtually all S. agalactiae strains (5–7) express CAMP factor, a pore-forming (8) protein toxin of ∼24 kDa (9). CAMP factor gives rise to the so-called CAMP reaction, which consists of a zone of strong hemolysis that is observed when S. agalactiae is streaked next to Staphylococcus aureus on sheep blood agar. S. aureus secretes sphingomyelinase. Sheep red blood cells are rich in sphingomyelin and, upon exposure to sphingomyelinase, become greatly sensitized to CAMP factor, which then effects hemolysis. Accordingly, hemolysis is most pronounced in the zone between the colonies of the two bacterial species.
This co-hemolytic phenomenon is useful for the presumptive identification of S. agalactiae in diagnostic microbiology laboratories. It was first described by Christie, Atkins, and Munch-Petersen (10), and the “CAMP” acronym represents these investigators' last names. More recently, the cfb gene that encodes CAMP factor has also been used to identify GBS by PCR (5, 6). The high abundance of the gene among GBS strains suggests a role of CAMP factor in pathogenesis, even though such a role was not substantiated in cell culture and mouse experiments (11).
Paralogs of S. agalactiae CAMP factor genes have been sequenced in a wide range of Gram-positive pathogens (Figs. S1 and S2), including Streptococcus uberis (12) and Propionibacterium acnes (13). In addition to S. agalactiae, Streptococcus porcinus and Streptococcus iniae produce positive results in the CAMP co-hemolysis test (14). Although group A streptococci (GAS; Streptococcus pyogenes) are phenotypically CAMP test–negative, most strains nevertheless harbor a CAMP factor (cfa) gene. The cfa gene product, when recombinantly expressed, also has co-hemolytic activity, which suggests that the CAMP-negative phenotype of GAS is due to strict control over the expression of the cfa genes under conditions of in vitro culture (15).
In the membranes of sphingomyelinase-pretreated sheep red blood cells, CAMP factor forms circular pores of somewhat variable diameter that have been visualized by EM (8). CAMP factor attaches to the cell membrane by binding to the glycosyl moieties of GPI-anchored proteins (16). Recently, through the synthesis of biotin-labeled core glycans of GPI anchors, Gao et al. showed that the phosphate group at the GPI inositol 1-O-position is most critical for GPI–CAMP binding (17). However, the mechanism of CAMP's co-hemolytic activity at the molecular level and the structural details of this toxin are unknown.
CAMP factor does not share obvious sequence homology to any proteins that have structures available, which precludes homology-based assignment of structure and function. Therefore, we set out to determine the structure of this protein to establish its structure/function relationships. We here report the first three-dimensional structure of CAMP factor, which consists of two well-separated domains. We also describe biochemical experiments that assign functional roles to each domain.
Results
GBS CAMP factor defines a novel structure
To gain insight into the structure/function relationships of CAMP family proteins, we solved the X-ray crystal structure of the GBS CAMP factor at a resolution of 2.45 Å (Table 1 and Fig. 1). There are three copies of CAMP factor molecules in one asymmetric unit (Fig. S3). The all-atom alignment with PyMOL produced an RMSD of 2.95 Å between chain B and chain A and 2.68 Å between chain C and chain A (Fig. S4). Whereas the overall folding architecture of three chains is similar, the biggest variation occurs in the loop between helices α2 and α3, which is herein referred to as L23. The L23 of chain A and chain B are in different conformation, whereas the density of that of chain C (residues Ala93–Asp100) is missing, and no model was built (Fig. S4). This suggests that the L23 region is highly flexible.
Table 1.
X-ray data collection and refinement parameters for GBS CAMP factor
Values in parentheses correspond to the highest-resolution shell.
| Data collection | |
| Space group | P21212 |
| Unit cell parameters | |
| a, b, c (Å) | 90.9, 116.0, 72.8 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 50–2.45 (2.60–2.45) |
| No. of reflections (total/unique) | 152,348/28,921 |
| Redundancy | 5.2 (5.2) |
| Completeness (%) | 99.6 (99.0) |
| I/σ(I) | 20.4 (2.3) |
| Rmeasa | 0.060 (0.841) |
| CC(1/2) | 0.999 (0.784) |
| Refinement | |
| Resolution (Å) | 50–2.45 |
| No. of protein atoms | 4956 |
| No. of solvent/hetero-atoms | 35/28 |
| RMSD bond lengths (Å) | 0.002 |
| RMSD bond angles (degrees) | 0.44 |
| Rwork (%)b | 23.4 |
| Rfree (%)c | 28.5 |
| Wilson B-factor | 52.5 |
| Ramachandran plot (favored/disallowed)d | 95.6/0 |
| PDB code | 5H6I |
a Rmeas = Σh(n/n − 1)½ Σi |Ii(h) − 〈I(h)〉|/ΣhΣiIi(h), where Ii(h) and 〈I(h)〉 are the ith and the mean measurement, respectively, of the intensity of reflection h.
b Rwork = Σh‖Fo (h)| − |Fc (h)‖/Σh|Fo (h) , where Fo (h) and Fc (h) are the observed and calculated structure factors, respectively. No I/σ cutoff was applied.
c Rfree is the R value obtained for a test set of reflections consisting of a randomly selected 10% subset of the data set excluded from refinement.
d Values from the MolProbity server at http://molprobity.biochem.duke.edu/ (49). Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
Figure 1.
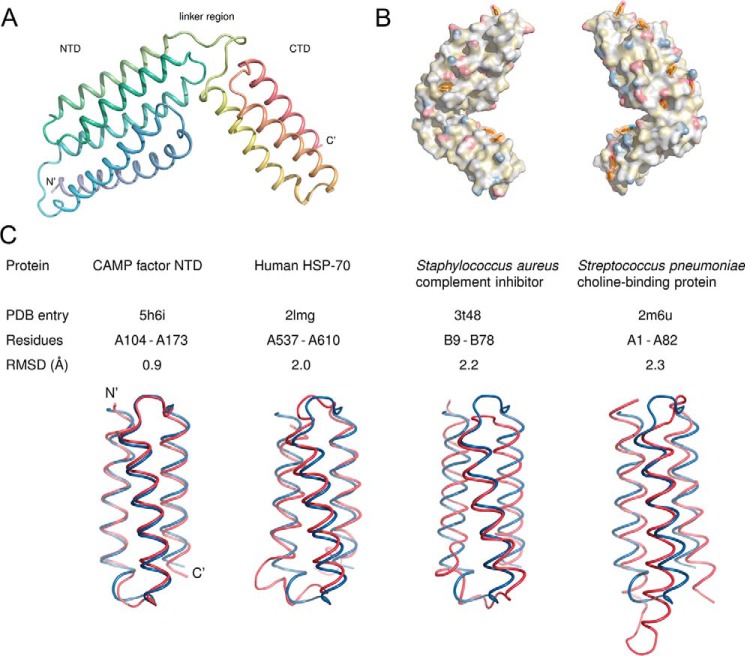
Structure of GBS CAMP factor. A, overall structure. The protein consists of two helical domains connected by a linker. The N terminus in the structure is formed by residue Val41; the preceding residues were not resolved. B, aromatic residues and polar moieties on the surface of the CAMP factor molecule. Aromatic residues are shown in a stick representation and with semitransparent surfaces; the remainder of the molecule is shown as an opaque surface. Polarity is highlighted using the YRB scheme (56), which shows the side-chain nitrogens of lysine and arginine in blue, the side-chain oxygens of aspartate and glutamate in red, carbons not linked to oxygen or nitrogen atoms in yellow, and everything else without color. C, structural alignment of the CAMP factor CTD (blue) with helices 3–5 of the NTD (red) and with domains of three other proteins that belong to group 46996 of the SCOP database (24). The alignments were carried out with PyMOL.
The recombinant mature form of CAMP protein, which starts from Ala29 of the sequence, was used for crystallization in this study. The N-terminal loop regions (Ala29–His40 of chain A, Ala29–Asn42 of chain B, and Ala29–Gln45 of chain C, respectively) are not visible in the electron density map, which suggests that they are disordered, although we have not rigorously eliminated the possibility that the loop was proteolytically cleaved during crystallization. A flexible structure is in agreement with secondary structure prediction by PSIPRED (18), which indicated that this region has predominantly coil structure. Furthermore, this loop region is not part of the conserved consensus CAMP factor core region, judging from multiple-sequence alignment of CAMP factor proteins from different bacteria (Fig. S1).
For clarity, we focus on chain A for the description of CAMP factor structure. The final model of chain A contains 215 residues from Val41 to Lys255, which defines a core structure of the CAMP family. CAMP factor is an all-helix protein composed of two well-separated structural modules (Fig. 1A): an N-terminal 5-helix bundle (Asn42–Thr173) and C-terminal 3-helix bundle (Ile185–Leu254). The two domains are linked by a loop that comprises a DLXXXDXAT sequence motif that is conserved among CAMP factor genes from different species (Fig. S1).
Because a BLAST search did not identify any homologous structural model by sequence similarity, we performed a search of the Protein Data Bank (PDB) using the DALI server (DaliLite version 3) (19), pdbefold (20), and VAST (21) to identify similar structural folds in the structural fold databases, including SCOP (22) and CATH (23). We did not find any other protein with the same folding architecture as CAMP factor. Considering the separate domains, the 3-helix bundle of the C-terminal domain (CTD) shares high similarity in terms of architecture and topology to the immunoglobulin/albumin binding domain-like fold (group 46996 in the SCOP database (24); see Fig. 1C). Notably, the 5-helix bundle of the N-terminal domain (NTD) has a left-handed twisted superhelical topology. We could not assign the NTD of CAMP factor to any known protein structural fold in the SCOP/CATH database, which strongly suggests that the CAMP structure defines a novel structural fold. However, a close inspection showed that the 3-helix bundle formed by its helices 3–5 is nearly superimposable to the CTD (Fig. 1C), suggesting that an event of gene duplication gave rise to this fold.
CAMP factor lacks large surface hydrophobic patches
Because of its capability of forming membrane-inserted pores (8), one might expect the surface of the molecule to contain continuous hydrophobic patches, as commonly found in typical transmembrane proteins. However, the surface is decorated with both hydrophilic and hydrophobic residues (Fig. 1B). In this respect, CAMP factor resembles other pore-forming toxins that are initially secreted as monomeric, water-soluble molecules (25). Fig. 1 also shows several surface-exposed large aromatic residues, rarely seen on the surface of globular proteins.
Functional roles of the two domains
The novelty of CAMP factor's structure implies that its functional assignments cannot be based on precedent. For example, unlike other predominantly helical toxins (or membrane-binding domains of such toxins) (e.g. diphtheria toxin (26, 27) or colicin Ia (28)), CAMP factor does not contain an obvious “hydrophobic hairpin” (Fig. S5). Moreover, neither sequence-based (29) nor structure-based (30, 31) bioinformatics tools were able to locate any carbohydrate binding motifs in CAMP factor that would account for its experimentally observed binding to GPI anchors (16, 32). Therefore, CAMP factor must contain an as yet unknown carbohydrate binding motif.
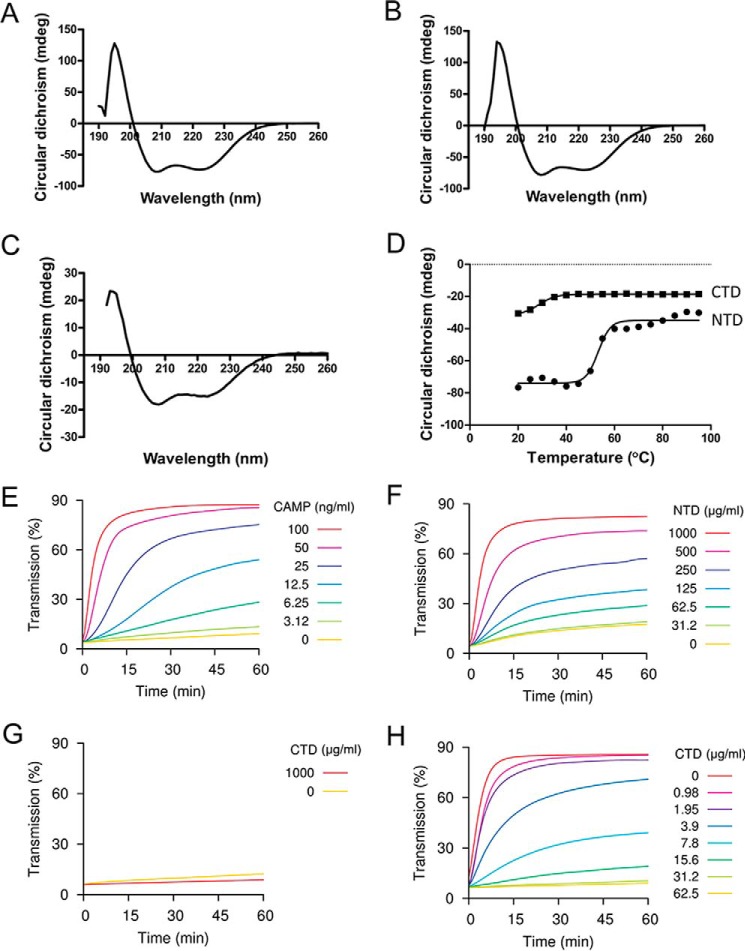
To examine the respective functions of the N- and the C-terminal domains, we expressed them as separate fragments. Gel filtration analysis showed that that both the NTD and CTD are monomeric in solution (data not shown). Furthermore, CD spectroscopic analysis showed that both full-length CAMP factor and NTD contain a high percentage of helical structure, whereas CTD has a lower negative peak at the same protein concentration (0.3 mg/ml) (Fig. 2, A–C), suggesting that NTD, but not CTD, is well-folded when they are expressed alone (Fig. 2C). Notably, the CTD is also less thermally stable compared with NTD, as evaluated by CD spectrum (Fig. 2D). The fitted Tm for NTD is 53.3 ± 0.9 °C, whereas the Tm for CTD is 26.7 ± 0.3 °C.
Figure 2.
CD spectrum analysis and co-hemolytic activity assay. A, CD spectrum analysis of intact CAMP factor. B, CD spectrum analysis of NTD. C, CD spectrum analysis of CTD is shown to the right. D, thermal titration of NTD and CTD by CD spectrum. The fitted Tm for NTD is 53.3 ± 0.9 °C, whereas the Tm for CTD is 26.7 ± 0.3 °C. E, co-hemolytic activity assay of intact CAMP factor. F, co-hemolytic activity assay of the NTD. G, co-hemolytic activity assay of the CTD. H, inhibition of intact CAMP factor by CTD. Sheep red blood cells were pretreated with sphingomyelinase and then incubated with CAMP factor or its fragments at the concentrations indicated. The transmission at 650 nm of the cell suspension was monitored over time. An increase in transmission indicates reduced turbidity (i.e. hemolysis). In H, intact CAMP factor was used at 100 ng/ml, and CTD was used at the concentrations indicated.
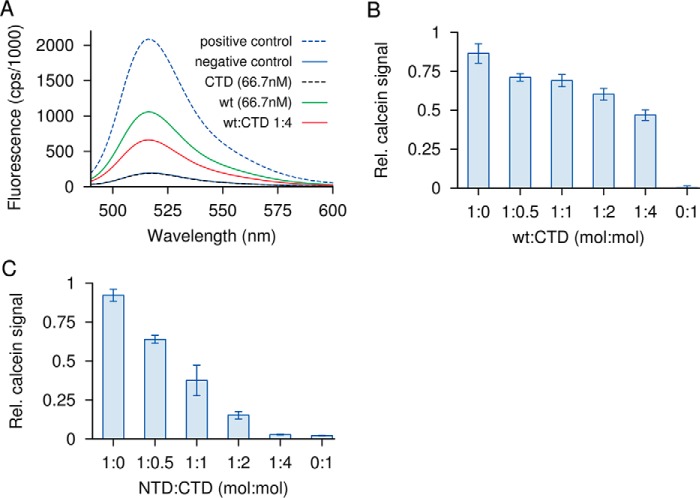
We further compared activities of individual domains with that of WT toxin on red blood cells (RBCs), as well as on liposomes composed of phosphatidylcholine, phosphatidylglycerol, and cholesterol (Figs. 2 and 3). The CTD alone permeabilized neither RBCs nor liposomes. The NTD retained the ability to lyse both RBCs and liposomes, suggesting that the membrane insertion and pore-forming activities reside chiefly within this domain. NTD also permeabilized liposomes. It is interesting to compare the specific activity of NTD with that of WT between the two membranes. On liposomes, NTD was about 15 times less active than WT, whereas on RBCs, its activity was ∼104 times lower. Thus, the CTD enhances the activity of NTD much more on RBCs, which contain GPI anchors, than on liposomes, which do not. This observation suggests that CTD is important for binding to GPI anchors.
Figure 3.
Permeabilization of liposomes by CAMP factor or its fragments and inhibitory activity of the C-terminal domain. Liposomes containing phosphatidylcholine, cholesterol, and phosphatidylglycerol were loaded with self-quenching concentrations of calcein. Membrane permeabilization releases and dilutes calcein, which increases its fluorescence. Detergent permeabilization is used as a positive control. A, intact CAMP factor (wt) at 66.7 nm causes half-maximal permeabilization. The CTD alone has no effect; its addition to WT at a 4-fold molar excess partially inhibits the latter. B, the same experiment plotted using a relative scale for calcein release, with additional molar ratios for CTD/WT included. C, NTD at 1 μm causes a similar extent of membrane permeabilization as a 15-fold lower amount of WT. NTD too is subject to inhibition by CTD. Error bars, S.E.
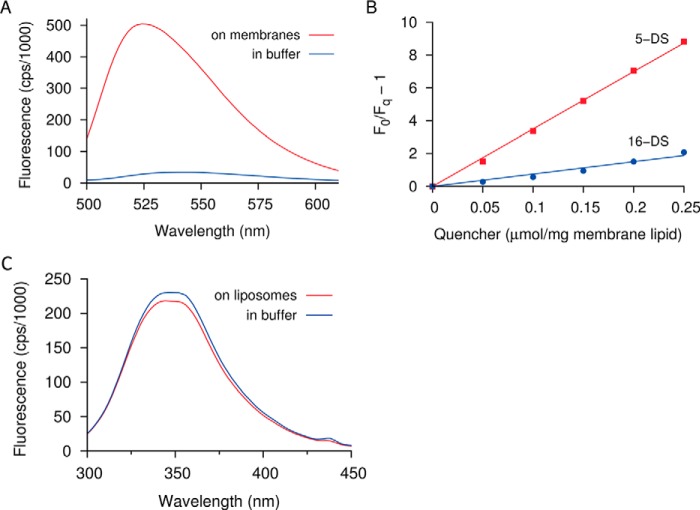
Membrane insertion of the NTD was monitored with residue Phe138, which is located approximately in the middle of the fourth helix of this domain. We replaced this residue with cysteine and then labeled it with the polarity-sensitive fluorescent dye NBD (33). Upon incubation with RBCs or liposome membranes, the fluorescence emission was greatly increased, and the emission maximum was blue-shifted by ∼10 nm (Fig. 4A). These changes are often observed when NBD enters the hydrophobic interior of a lipid membrane (34), and the fluorescence was indeed susceptible to quenching by the membrane-affine probe 5-doxylstearic acid (Fig. 4B). Conversely, when CTD was incubated with liposome membranes, no change in the fluorescence of its single intrinsic tryptophan residue was observed (Fig. 4C). It must be noted, however, that other residues within the CTD may still participate in membrane insertion; this possibility is suggested by the observation that hydrophobic regions are similarly prevalent in both domains (Figs. 1B and Fig. S5).
Figure 4.
Fluorescence effects of membrane interaction. A, residue Phe138 in the NTD was replaced with cysteine and labeled with the polarity-sensitive probe NBD. Incubation with cell membranes causes a large increase and a blue shift of the fluorescence, consistent with an insertion of the label into the hydrophobic membrane interior. B, quenching of NBD fluorescence by lipophilic quenchers 5- and 16-doxylstearate (5-DS and 16-DS, respectively; Stern–Volmer plot). The fluorescence is quenched more strongly by 5-doxylstearate, consistent with a shallow insertion of the labeled residue into the membrane. C, the tryptophan fluorescence of the CTD does not change significantly upon incubation with liposomes.
When WT toxin was mixed with CTD before the addition to red blood cells, its hemolytic activity was inhibited in a dose-dependent manner (Fig. 2H). CTD also inhibited the hemolytic activity of NTD (not shown). Whereas this might conceivably be due to competition for binding to GPI-anchored proteins, inhibition was also observed on liposomes (Fig. 3). Because liposomes are devoid of GPI anchors, the inhibition observed on liposomes is likely due to a direct interaction between the isolated CTD on one hand and WT or NTD on the other. This suggests that CTD also participates in oligomerization and that this function involves some interaction in trans with NTD. In summary, we tentatively assign membrane permeabilization to NTD, carbohydrate binding to CTD, and toxin oligomerization to both domains. All of these assignments await further experimental validation.
Functional significance of conformational flexibility within the linker and NTD regions
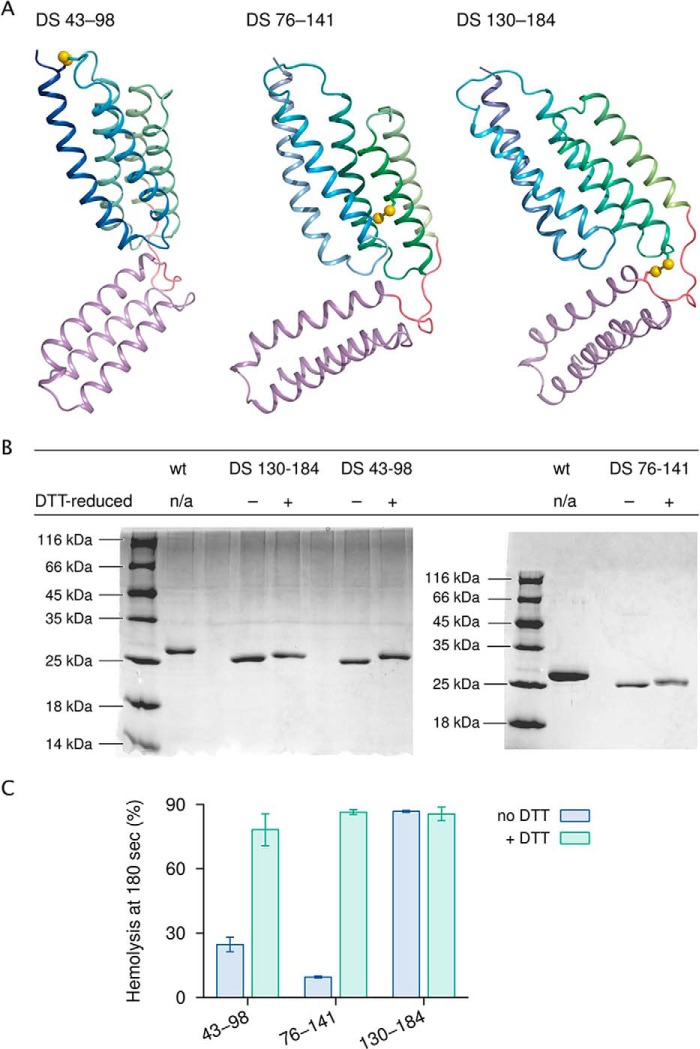
Membrane-permeabilizing proteins usually undergo conformational changes, sometimes extensive ones (25), when inserting into membranes. One method to probe such conformational changes is to restrict them by introducing engineered disulfide bonds and then determining the effect on function. Fig. 5 shows three such experiments.
Figure 5.
Engineered disulfide bonds and their effects on the hemolytic activity of CAMP factor. A, locations of mutant cysteine pairs. The NTD points upward. B, SDS-PAGE of disulfide mutants in the absence and presence of the disulfide-reducing agent DTT. Accelerated migration indicates presence of disulfide bonds. C, hemolytic activity of each mutant with and without DTT. n/a, not applicable. Error bars, S.E.
Successful disulfide bond formation was verified by accelerated migration on SDS-PAGE under nonreducing conditions and by electrospray ionization MS. Cross-linking two different pairs of helices within the NTD, namely Ser43 of α1 and Val98 of L23, and Pro76 of α2 and Thr141 of α4, caused measurable decreases in hemolytic activity, although hemolysis was not fully suppressed. Somewhat surprisingly, no major effect on activity was seen with a disulfide that connects the Ser130 residue in the NTD with the Thr184 residue in the CTD, which should restrict motions of the two domains relative to one another.
Overall, these findings indicate that the N-terminal domain contribute mostly to the membrane-permeabilizing activity, whereas mobility in the linker region may not be as significant as the appearance of the structure might suggest. In this context, it is noteworthy that the linker region is connected internally as well as to both domains by a considerable number of hydrogen bonds that should limit conformational freedom (Fig. 6A).
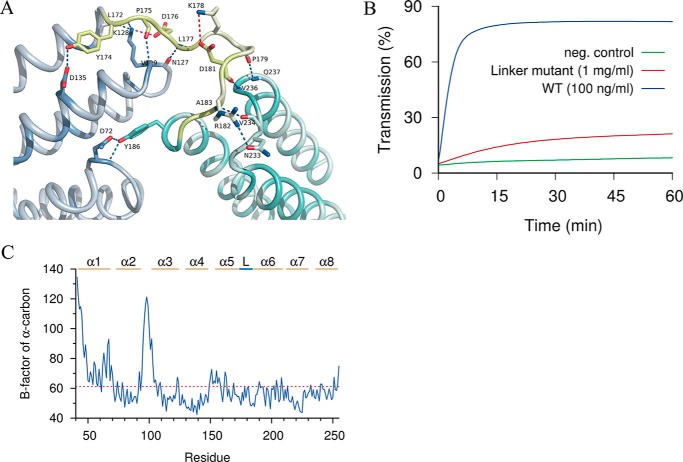
Figure 6.
Structural details of conserved linker region. A, hydrogen bonds (blue dotted lines) and ionic bonds (red dotted lines) between residues within the linker region (lime) as well as adjacent residues in the NTD (blue-gray) and CTD (blue-green) are shown. B, mutation of the conserved linker region abolished the hemolytic activity of intact CAMP. C, B-factor analysis of the conserved linker region revealed low flexibility. The B-factors of the α-carbons of all residues of the A chain are shown. Helical segments are indicated as α1–α8. L, the linker region between the N-terminal and C-terminal domains.
Based on the sequence alignment of the CAMP family proteins, we noticed that the linker region has a conserved DLXXXDXAT motif between NTD and CTD (Fig. S1). After replacing each of the 5 conserved residues in this motif with glycine, the hemolytic activity decreased 1000 times as compared with that of the WT CAMP factor of GBS (Fig. 6B). Of note, of all of the proteinogenic amino acids, glycine constrains conformational flexibility the least. This suggests that the mutated motif contributes to activity in a manner that is more specific than facilitating conformational flexibility. Indeed, the linker region has a well-defined electron density, and its B-factors are comparable with the adjacent helical regions of the structure (Fig. 6C), which proves that this region has low flexibility in the crystal.
Discussion
Overall, we determined the first structure of the CAMP family bacterial pore-forming toxin. Our study shows that the structure of CAMP factor is unique among pore-forming toxins, and it reports initial assignments of function to elements of this novel structure. Knowledge of the structure will facilitate more detailed biophysical studies to better understand the membrane interaction and pore-formation by this unique family of bacterial toxins.
GPI anchors have a complex composition. The core of GPI consists of phosphatidylinositol, a glycan moiety that comprises one glucosamine and three mannose residues, and a terminal phosphoethanolamine, which is amide-bonded to the C terminus of the protein. The GPI backbone can be further modified with phosphoethanolamine and/or various glycan side branches, depending on the organism, cell type, and protein. There can be various forms in its lipid moiety as well (35, 36). After determination of the first structure of CAMP factor, it will be interesting to determine the exact interaction mode between CAMP factor and GPI anchors. More systematic biochemical as well as structural studies are needed to determine whether the NTD, CTD, or both are required for interaction with GPI and which functional parts of GPI anchors are mostly involved in the recognition by CAMP family toxins.
Pore-forming toxins are a unique group of proteins that exist as soluble forms after being synthesized, while undergoing significant structural changes concomitantly with host membrane insertion. Thus, elucidating the molecular details of membrane interaction and structural rearrangement of pore-forming toxins is critical in understanding their function. The structures of soluble as well as membrane-inserted conformations have been determined for several types of bacterial toxins, including the α-hemolysin of S. aureus (37), cytolysin A of Escherichia coli (38), TcdA1 from Photorhabdus luminescens (39), and pneumolysin of Streptococcus pneumoniae (40). Each type of these toxins has their unique way of structural reorganization upon membrane insertion. Thus, the structural determination of the soluble form of CAMP factor described in this study is only the first step toward understanding its mechanism of action; the next challenge will be to determine the structure of the membrane-inserted pore complex.
Experimental procedures
Protein expression, purification, and crystallization
The WT CAMP factor protein Ala29–Lys255 (NCBI accession code ADA13253.1) from S. agalactiae was expressed with pGEX vector in bacteria with a thrombin-cleavable GST tag as described previously (8). Truncated CAMP proteins (i.e. NTD (Val41–Asp181) and CTD (Arg182–Lys255)) were expressed with an MBP tag. After obtaining the fusion proteins, tobacco etch virus protease was used to remove the MBP tag, and tagless proteins were further purified by a second nickel affinity step and then gel filtration chromatography. Purified proteins were analyzed by SDS-PAGE.
Purified WT CAMP factor was concentrated by Amicon centrifugal concentrators (Millipore, Billerica, MA) to 20–30 mg/ml before setting up the hanging-drop vapor diffusion method for crystallization. WT native CAMP factor (Ala29–Lys255) was crystallized in a well solution containing 0.2 m NH4NO3, 20% PEG 3350, 0.1 m MES, pH 6.0. Thin plate crystals appeared after 1 week at room temperature. The same solution, with 20% ethylene glycol added to a final concentration of 20%, was used as the cryoprotectant for quick washing and flash-cooling the crystals in liquid nitrogen before collecting X-ray diffraction data.
X-ray diffraction, structure determination, and refinement
X-ray diffraction data were collected at GM/CA-CAT at the Advanced Photon Source, Argonne National Laboratory. Data were processed with the HKL2000 program suite (41) and with XDS (42). The best native CAMP crystal diffracted to 2.45 Å. The space group is P21212 with unit cell parameters a = 90.98 Å, b = 116.21 Å, and c = 73.93 Å (also see Table 1). The solvent content is 48.4% for three CAMP molecules in each asymmetric unit.
To solve the phase problem, a series of methods were tried, including selenomethionine labeling and heavy element soaking, but none of them succeeded. Inspired by the recent successes of using MBP as a crystallization chaperone in the structural determination of a variety of challenging proteins (43–45), we fused GBS CAMP to the C terminus of MBP and purified the fusion protein for crystallization screening. Because the linker sequence between MBP and target protein is the most critical factor for the crystallization of such fusion proteins, we tested different N-terminal residues starting from Ala29 to Met50 of CAMP, with a two-residue increment each time. Finally, we obtained crystals for several fusion proteins, but only the fusion protein starting from Asn42 of CAMP produced crystals that diffracted to 3.1 Å. Molecular replacement was calculated with Phaser (46) in CCP4 GUI (47), using MBP as a search model to solve the structure of fusion protein. The backbones of visible helices corresponding to CAMP were manually built in Coot (48). Due to the low resolution and lack of homology model, the side chains were not confidently assigned. The structure of native CAMP was determined by molecular replacement using a C-α model of CAMP obtained from the MBP-CAMP fusion crystal as a search model. Structure was refined by Phenix.refine alternated with manual model adjustment in Coot. The native CAMP crystal structure was validated by the MolProbity server (49) and RCSB ADIT validation server (50) before deposition. Molecular graphics were prepared with PyMOL (Schrödinger, LLC).
Mutagenesis
Mutations of the CAMP gene were done by a jointing PCR–based method with the Phusion DNA polymerase (New England Biolabs). CAMP mutants were expressed and purified in similar protocol as the WT protein. Cu(II)-1,10-phenanthroline was used to induce disulfide bridge formation between introduced adjacent cysteines (51, 52). 0.1 volume of 3 mm Cu(II)-(1,10-phenanthroline)3 was added to purified protein, followed by incubation at 30 °C for 30 min. The reaction was quenched by the addition of 65 mm EDTA from a stock solution of 0.5 m NaEDTA, pH 8. The formation of disulfide bonds was confirmed by SDS-PAGE (see Fig. 5B) and LC-MS.
Hemolysis assay
Sheep red blood cells (Cedarlane, Burlington, Canada) were pretreated with staphylococcal sphingomyelinase (Sigma; 50 milliunits/ml) in HEPES-buffered saline (HBS) with 10 mm MgCl2, washed in the same buffer by centrifugation, and resuspended to 1% (v/v). An equal volume of the cell suspension was added to CAMP factor (WT, NTD, or CTD alone or in combination (see “Results”)) in the wells of a 96-well microtiter plate. Progress of hemolysis was monitored using the optical density at 650 nm, a wavelength that lies outside the absorption band of hemoglobin and thus reflects cell turbidity alone. These measurements were performed using a SpectroMax Plus 384 microplate spectrophotometer (Molecular Devices, Sunnyvale, CA).
Liposome permeabilization
1,2-Dimyristoyl-sn-glycero-3-phosphocholine, cholesterol, and 1,2-dimyristoyl-sn-glycero-3-phosphoglycerol were purchased from Avanti Polar Lipids (Alabaster, AL). To prepare liposomes, the desired lipids were dissolved in chloroform and mixed at a molar ratio of 6.5:2.5:1. The mixture was dried down under a stream of nitrogen in a round-bottom flask for 5 min and then dried under vacuum for an additional 3 h to remove any residual chloroform. The dried lipids were resuspended by vortexing for 30 min with 3.0 ml of HBS (10 mm HEPES, 150 mm NaCl, pH 7.4) containing 50 mm calcein (Sigma-Aldrich) at room temperature to a final total lipid concentration of 3 mg/ml. The resulting suspension of multilamellar liposomes was converted to unilamellar liposomes by extruding 15 times through a 100-nm polycarbonate membrane filter (Whatman). Subsequently, nonencapsulated calcein was removed by chromatography using a Bio-Gel P6-DG (Bio-Rad) column pre-equilibrated with HBS.
The calcein-loaded liposomes were diluted to 45 μg/ml total lipid with HBS; mixed with CAMP factor, NTD, or CTD alone or in combination (compositions and final concentrations as indicated under “Results”); and then incubated for 30 min at room temperature. The calcein fluorescence intensity was then measured (excitation, 478 nm; emission, 516 nm) on a QuantaMaster 4 spectrofluorometer (PTI, London, Canada). The extent of membrane permeabilization P was then calculated using the formula, P = (Fsample − F0)/(FTriton − F0), where F0 is the fluorescence of a liposome control without toxin, and FTriton is that of a liposome sample solubilized with Triton X-100 at a final concentration of 0.01%.
Fluorescent labeling and spectrofluorimetry
The cysteine mutant F138C was transferred to labeling buffer consisting of 100 mm sodium phosphate buffer with 1 mm tris(2-carboxyethyl)phosphine, pH 7.3, using gel filtration. The samples were supplemented with 0.5 mm IANBD amide (Invitrogen, Burlington, Canada). The samples were incubated at room temperature for 120 min, and the excess label was removed by gel filtration. To determine the labeling efficiency, the molar ratio of fluorophore to protein was calculated from UV-visible spectra using an extinction coefficient of 23,500 liters/mol × cm for IANBD amide at 478 nm. The extinction coefficient of CAMP factor was determined to be 25,040 liters/mol × cm at 280 nm (53). The measured absorbance values at 280 nm of the labeled proteins were corrected for absorbance of the dye at 280 nm. Labeling efficiencies ranged from 85 to 95%.
Fluorescence measurements were performed using red cell ghost membranes. Sheep red blood cells were pretreated with sphingomyelinase as for hemolysis assays. They were then lysed osmotically with 5 mm sodium phosphate buffer and washed repeatedly by centrifugation. The membranes were suspended in HBS and incubated with the NBD-labeled mutant for 60 min at room temperature. They were then collected by centrifugation; the supernatant was retained, and the membrane pellet was resuspended in HBS buffer. Fluorescence emission spectra were acquired in a PTI QuantaMaster spectrofluorometer, using an excitation wavelength of 478 nm. The emission intensity was corrected for incomplete membrane binding; the extent of binding was determined by comparing the fluorescence of the saved supernatant with that of an equivalent amount of labeled mutant that had not been incubated with membranes.
CD spectroscopic study of CAMP protein
To study the secondary structure and stability of CAMP proteins, CD spectra were acquired on a Chirascan spectrometer (Applied Photophysics, Leatherhead, UK). Two CD spectra measurements were collected for each sample from 180 to 260 nm in a 1-nm step with a 4-s averaging time at 20 °C, using a 1-mm path length cell. Before CD spectrum analysis, sample buffer was changed to PBS, and protein concentration was adjusted to 0.3 mg/ml, as determined by its absorbance at 280 nm. For thermal titration, two CD spectra measurements were also collected between 20 and 95 °C with temperature steps of 5 °C and wavelengths between 180 and 260 nm. CD signals at 220 nm were adopted to characterize the conformational change during thermal titration (54). The data (Θ220) were fitted by Prism to calculate the Tm values (55).
Author contributions
T. J., W. F., Y. Z., and M. P. conceptualization; T. J., W. F., Y. Z., and M. P. resources; T. J., E. B.-M., W. Z., and Y. L. data curation; T. J. software; T. J., E. B.-M., W. Z., Y. L., and M. P. formal analysis; T. J. supervision; T. J. and Y. L. funding acquisition; T. J., E. B.-M., and M. P. investigation; T. J. methodology; T. J., E. B.-M., and M. P. writing-original draft; T. J., Y. Z., and M. P. project administration; T. J. and M. P. writing-review and editing.
Supplementary Material
Acknowledgments
We thank Tsan Sam Xiao (Case Western Reserve University) and Jiansheng Jiang (National Institutes of Health) for support, critical reading, and comments on the manuscript. We thank the staff at GM/CA-CAT at the Advanced Photon Source and the BL17U1, BL18U, and BL19U1 beamline at the Shanghai Synchrotron Radiation Facility for assistance during data collection.
The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S5.
The atomic coordinates and structure factors (code 5H6I) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- GBS
- group B streptococcus
- GAS
- group A streptococcus
- GPI
- glycosylphosphatidylinositol
- RMSD
- root mean square deviation
- CTD
- C-terminal domain
- NTD
- N-terminal domain
- RBC
- red blood cell
- NBD
- nitrobenzoxadiazole
- IANBD amide
- N,N′-dimethyl-N-(iodoacetyl)-N′-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)ethylenediamine
- MBP
- maltose-binding protein
- HBS
- HEPES-buffered saline.
References
- 1. Verani J. R., McGee L., and Schrag S. J. (2010) Prevention of perinatal group B streptococcal disease: Revised guidelines. Centers for Disease Control and Prevention, Atlanta: [PubMed] [Google Scholar]
- 2. Mweu M. M., Nielsen S. S., Halasa T., and Toft N. (2014) Spatiotemporal patterns, annual baseline and movement-related incidence of Streptococcus agalactiae infection in Danish dairy herds: 2000–2009. Prev. Vet. Med. 113, 219–230 10.1016/j.prevetmed.2013.10.020 [DOI] [PubMed] [Google Scholar]
- 3. Liu G., Zhang W., and Lu C. (2013) Comparative genomics analysis of Streptococcus agalactiae reveals that isolates from cultured tilapia in China are closely related to the human strain A909. BMC Genomics 14, 775 10.1186/1471-2164-14-775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu L., Li Y. W., He R. Z., Xiao X. X., Zhang X., Su Y. L., Wang J., and Li A. X. (2014) Outbreak of Streptococcus agalactiae infection in barcoo grunter, Scortum barcoo (McCulloch & Waite), in an intensive fish farm in China. J. Fish Dis. 37, 1067–1072 10.1111/jfd.12187 [DOI] [PubMed] [Google Scholar]
- 5. Grandjean F., Goffinet P., and Hougardy N. (2007) Detection of colonization by Streptococcus agalactiae: prospective study comparing real-time gene amplification with a new chromogenic medium Strepto B ID. Pathol. Biol. 55, 407–411 10.1016/j.patbio.2007.07.008 [DOI] [PubMed] [Google Scholar]
- 6. Shabayek S., Abdalla S., and Abouzeid A. M. (2010) Comparison of scpB gene and cfb gene polymerase chain reaction assays with culture on Islam medium to detect Group B Streptococcus in pregnancy. Ind. J. Med. Microbiol. 28, 320–325 10.4103/0255-0857.71821 [DOI] [PubMed] [Google Scholar]
- 7. Shome B. R., Bhuvana M., Mitra S. D., Krithiga N., Shome R., Velu D., Banerjee A., Barbuddhe S. B., Prabhudas K., and Rahman H. (2012) Molecular characterization of Streptococcus agalactiae and Streptococcus uberis isolates from bovine milk. Trop. Anim. Health Prod. 44, 1981–1992 10.1007/s11250-012-0167-4 [DOI] [PubMed] [Google Scholar]
- 8. Lang S., and Palmer M. (2003) Characterization of Streptococcus agalactiae CAMP factor as a pore-forming toxin. J. Biol. Chem. 278, 38167–38173 10.1074/jbc.M303544200 [DOI] [PubMed] [Google Scholar]
- 9. Schneewind O., Friedrich K., and Lütticken R. (1988) Cloning and expression of the CAMP factor of group B streptococci in Escherichia coli. Infect. Immun. 56, 2174–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Christie R., Atkins N. E., and Munch-Petersen E. (1944) A note on a lytic phenomenon shown by group B streptococci. Aust. J. Exp. Biol. Med. Sci. 22, 197–200 10.1038/icb.1944.26 [DOI] [PubMed] [Google Scholar]
- 11. Hensler M. E., Quach D., Hsieh C.-J., Doran K. S., and Nizet V. (2008) CAMP factor is not essential for systemic virulence of Group B Streptococcus. Microb. Pathog. 44, 84–88 10.1016/j.micpath.2007.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jiang M., Babiuk L. A., and Potter A. A. (1996) Cloning, sequencing and expression of the CAMP factor gene of Streptococcus uberis. Microb. Pathog. 20, 297–307 10.1006/mpat.1996.0028 [DOI] [PubMed] [Google Scholar]
- 13. Valanne S., McDowell A., Ramage G., Tunney M. M., Einarsson G. G., O'Hagan S., Wisdom G. B., Fairley D., Bhatia A., Maisonneuve J.-F., Lodes M., Persing D. H., and Patrick S. (2005) CAMP factor homologues in Propionibacterium acnes: a new protein family differentially expressed by types I and II. Microbiology 151, 1369–1379 10.1099/mic.0.27788-0 [DOI] [PubMed] [Google Scholar]
- 14. Collins M. D., Hutson R. A., Falsen E., Nikolaitchouk N., LaClaire L., and Facklam R. R. (2000) An unusual Streptococcus from human urine, Streptococcus urinalis sp. nov. Int. J. Syst. Evol. Microbiol. 50, 1173–1178 10.1099/00207713-50-3-1173 [DOI] [PubMed] [Google Scholar]
- 15. Gase K., Ferretti J. J., Primeaux C., and McShan W. M. (1999) Identification, cloning, and expression of the CAMP factor gene (cfa) of group A streptococci. Infect. Immun. 67, 4725–4731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lang S., Xue J., Guo Z., and Palmer M. (2007) Streptococcus agalactiae CAMP factor binds to GPI-anchored proteins. Med. Microbiol. Immunol. 196, 1–10 10.1007/s00430-006-0021-2 [DOI] [PubMed] [Google Scholar]
- 17. Gao J., Zhou Z., Guo J., and Guo Z. (2017) Synthesis of biotin-labelled core glycans of GPI anchors and their application in the study of GPI interaction with pore-forming bacterial toxins. Chem. Commun. 53, 6227–6230 10.1039/C7CC03056H [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Buchan D. W., Minneci F., Nugent T. C., Bryson K., and Jones D. T. (2013) Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 41, W349–W357 10.1093/nar/gkt381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Holm L., and Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 10.1093/nar/gkq366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Krissinel E., and Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 10.1107/S0907444904026460 [DOI] [PubMed] [Google Scholar]
- 21. Gibrat J.-F., Madej T., and Bryant S. H. (1996) Surprising similarities in structure comparison. Curr. Opin. Struct. Biol. 6, 377–385 10.1016/S0959-440X(96)80058-3 [DOI] [PubMed] [Google Scholar]
- 22. Andreeva A., Howorth D., Chothia C., Kulesha E., and Murzin A. G. (2014) SCOP2 prototype: a new approach to protein structure mining. Nucleic Acids Res. 42, D310–D314 10.1093/nar/gkt1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sillitoe I., Lewis T. E., Cuff A., Das S., Ashford P., Dawson N. L., Furnham N., Laskowski R. A., Lee D., Lees J. G., Lehtinen S., Studer R. A., Thornton J., and Orengo C. A. (2015) CATH: comprehensive structural and functional annotations for genome sequences. Nucleic Acids Res. 43, D376–DD381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Andreeva A., Howorth D., Chandonia J.-M., Brenner S. E., Hubbard T. J., Chothia C., and Murzin A. G. (2008) Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 36, D419–D425 10.1093/nar/gkm993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dal Peraro M., and van der Goot F. G. (2016) Pore-forming toxins: ancient, but never really out of fashion. Nat. Rev. Microbiol. 14, 77–92 10.1038/nrmicro.2015.3 [DOI] [PubMed] [Google Scholar]
- 26. Kaul P., Silverman J., Shen W. H., Blanke S. R., Huynh P. D., Finkelstein A., and Collier R. J. (1996) Roles of Glu 349 and Asp 352 in membrane insertion and translocation by diphtheria toxin. Protein Sci. 5, 687–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ren J., Sharpe J. C., Collier R. J., and London E. (1999) Membrane translocation of charged residues at the tips of hydrophobic helices in the T domain of diphtheria toxin. Biochemistry 38, 976–984 10.1021/bi981576s [DOI] [PubMed] [Google Scholar]
- 28. Kienker P. K., Qiu X.-Q., Slatin S. L., Finkelstein A., and Jakes K. S. (1997) Transmembrane insertion of the colicin Ia hydrophobic hairpin. J. Membr. Biol. 157, 27–37 10.1007/s002329900213 [DOI] [PubMed] [Google Scholar]
- 29. Malik A., Firoz A., Jha V., and Ahmad S. (2010) PROCARB: a database of known and modelled carbohydrate-binding protein structures with sequence-based prediction tools. Adv. Bioinformatics 2010, 436036 10.1155/2010/436036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Doxey A. C., Cheng Z., Moffatt B. A., and McConkey B. J. (2010) Structural motif screening reveals a novel, conserved carbohydrate-binding surface in the pathogenesis-related protein PR-5d. BMC Struct. Biol. 10, 23 10.1186/1472-6807-10-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhao H., Yang Y., von Itzstein M., and Zhou Y. (2014) Carbohydrate-binding protein identification by coupling structural similarity searching with binding affinity prediction. J. Comput. Chem. 35, 2177–2183 10.1002/jcc.23730 [DOI] [PubMed] [Google Scholar]
- 32. Wu X., Shen Z., Zeng X., Lang S., Palmer M., and Guo Z. (2008) Synthesis and biological evaluation of sperm CD52 GPI anchor and related derivatives as binding receptors of pore-forming CAMP factor. Carbohydr. Res. 343, 1718–1729 10.1016/j.carres.2008.03.033 [DOI] [PubMed] [Google Scholar]
- 33. Fager R. S., Kutina C. B., and Abrahamson E. (1973) The use of NBD chloride (7-chloro-4-nitrobenzo-2-oxa-1,3-diazole) in detecting amino acids and as an N-terminal reagent. Anal. Biochem. 53, 290–294 10.1016/0003-2697(73)90433-8 [DOI] [PubMed] [Google Scholar]
- 34. Shepard L. A., Heuck A. P., Hamman B. D., Rossjohn J., Parker M. W., Ryan K. R., Johnson A. E., and Tweten R. K. (1998) Identification of a membrane-spanning domain of the thiol-activated pore-forming toxin Clostridium perfringens perfringolysin O: an α-helical to β-sheet transition identified by fluorescence spectroscopy. Biochemistry 37, 14563–14574 10.1021/bi981452f [DOI] [PubMed] [Google Scholar]
- 35. Ferguson M. A. J., Hart G. W., and Kinoshita T. (2015) Glycosylphosphatidylinositol anchors. in Essentials of Glycobiology (Varki A., Cummings R. D., Esko J. D., Stanley P., Hart G. W., Aebi M., Darvill A. G., Kinoshita T., Packer N. H., Prestegard J. H., Schnaar R. L., and Seeberger P. H. eds) pp. 137–150, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 36. Kinoshita T. (2016) Glycosylphosphatidylinositol (GPI) anchors: biochemistry and cell biology: introduction to a thematic review series. J. Lipid Res. 57, 4–5 10.1194/jlr.E065417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Song L., Hobaugh M. R., Shustak C., Cheley S., Bayley H., and Gouaux J. E. (1996) Structure of staphylococcal α-hemolysin, a heptameric transmembrane pore. Science 274, 1859–1866 10.1126/science.274.5294.1859 [DOI] [PubMed] [Google Scholar]
- 38. Mueller M., Grauschopf U., Maier T., Glockshuber R., and Ban N. (2009) The structure of a cytolytic α-helical toxin pore reveals its assembly mechanism. Nature 459, 726–730 10.1038/nature08026 [DOI] [PubMed] [Google Scholar]
- 39. Gatsogiannis C., Merino F., Prumbaum D., Roderer D., Leidreiter F., Meusch D., and Raunser S. (2016) Membrane insertion of a Tc toxin in near-atomic detail. Nat. Struct. Mol. Biol. 23, 884–890 10.1038/nsmb.3281 [DOI] [PubMed] [Google Scholar]
- 40. Van Pee K., Neuhaus A., D'Imprima E., Mills D. J., Kuhlbrandt W., and Yildiz O. (2017) CryoEM structures of membrane pore and prepore complex reveal cytolytic mechanism of Pneumolysin. Elife 6, e23644 10.7554/eLife.23644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Macromol. Crystallogr. A 276, 307–326 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- 42. Kabsch W. (2010) Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 10.1107/S0907444909047374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moon A. F., Mueller G. A., Zhong X., and Pedersen L. C. (2010) A synergistic approach to protein crystallization: combination of a fixed-arm carrier with surface entropy reduction. Protein Sci. 19, 901–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jin T., Perry A., Smith P., Jiang J., and Xiao T. S. (2013) Structure of the absent in melanoma 2 (AIM2) pyrin domain provides insights into the mechanisms of AIM2 autoinhibition and inflammasome assembly. J. Biol. Chem. 288, 13225–13235 10.1074/jbc.M113.468033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jin T., Chuenchor W., Jiang J., Cheng J., Li Y., Fang K., Huang M., Smith P., and Xiao T. S. (2017) Design of an expression system to enhance MBP-mediated crystallization. Sci. Rep. 7, 40991 10.1038/srep40991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Potterton E., Briggs P., Turkenburg M., and Dodson E. (2003) A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 59, 1131–1137 10.1107/S0907444903008126 [DOI] [PubMed] [Google Scholar]
- 48. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 49. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yang H., Guranovic V., Dutta S., Feng Z., Berman H. M., and Westbrook J. D. (2004) Automated and accurate deposition of structures solved by X-ray diffraction to the Protein Data Bank. Acta Crystallogr. D Biol. Crystallogr. 60, 1833–1839 10.1107/S0907444904019419 [DOI] [PubMed] [Google Scholar]
- 51. Czabotar P. E., Westphal D., Dewson G., Ma S., Hockings C., Fairlie W. D., Lee E. F., Yao S., Robin A. Y., Smith B. J., Huang D. C., Kluck R. M., Adams J. M., and Colman P. M. (2013) Bax crystal structures reveal how BH3 domains activate Bax and nucleate its oligomerization to induce apoptosis. Cell 152, 519–531 10.1016/j.cell.2012.12.031 [DOI] [PubMed] [Google Scholar]
- 52. van Montfort B. A., Schuurman-Wolters G. K., Duurkens R. H., Mensen R., Poolman B., and Robillard G. T. (2001) Cysteine cross-linking defines part of the dimer and B/C domain interface of the Escherichia coli mannitol permease. J. Biol. Chem. 276, 12756–12763 10.1074/jbc.M010728200 [DOI] [PubMed] [Google Scholar]
- 53. Gill S. C., and von Hippel P. H. (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182, 319–326 10.1016/0003-2697(89)90602-7 [DOI] [PubMed] [Google Scholar]
- 54. Jin T., Albillos S. M., Chen Y. W., Kothary M. H., Fu T. J., and Zhang Y. Z. (2008) Purification and characterization of the 7S vicilin from Korean pine (Pinus koraiensis). J. Agric. Food Chem. 56, 8159–8165 10.1021/jf801138q [DOI] [PubMed] [Google Scholar]
- 55. Herrmann L. M., and Bowler B. E. (1997) Thermal denaturation of iso-1-cytochrome c variants: comparison with solvent denaturation. Protein Sci. 6, 657–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hagemans D., van Belzen I. A., Morán Luengo T., and Rüdiger S. G. (2015) A script to highlight hydrophobicity and charge on protein surfaces. Front. Mol. Biosci. 2, 56 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.