Abstract
CRISPR/Cas9 is now widely used in biomedical research and has great potential for clinical applications. However, the safety and efficacy of this gene-editing technique are significant issues. Recent reports on mouse models and human cells have raised concerns that off-target mutations could hamper applying the CRISPR technology in patients. The high similarities of nonhuman primates to humans in genome content and organization, genetic diversity, physiology, and cognitive abilities have made these animals ideal experimental models for understanding human diseases and developing therapeutics. Off-target mutations of CRISPR/Cas9 have been analyzed in previous studies of nonhuman primates, but no report has investigated genome-wide off-target effects in living monkeys. Here, we used rhesus monkeys in which a genetic disorder mimicking Duchenne muscular dystrophy had previously been produced with CRISPR/Cas9. Using whole-genome sequencing to comprehensively assess on- and off-target mutations in these animals, we found that CRISPR/Cas9-based gene editing is active on the expected genomic sites without producing off-target modifications in other functional regions of the genome. These findings suggest that the CRISPR/Cas9 technique could be relatively safe and effective in modeling genetic disease in nonhuman primates and in future therapeutic research of human diseases.
Keywords: gel electrophoresis, gene knockout, gene mapping, gene therapy, gene silencing, gene transfer, genetic disease, CRISPR/Cas9, efficiency, nonhuman primate, off-target, safety
Introduction
Genome-editing technologies, especially CRISPR systems, have been growing quickly and have been extensively used in modeling of human diseases. CRISPR/Cas9 is capable of inducing precise modification of genomic sequences, which could lead to both on-target and potential off-target effects (OTs)4 because of the high sequence homology of sgRNA (1, 2). Previous reports on mouse models and human cells (3–5) have raised concern about its OTs, which could hamper the process of applying CRISPR to patients. Recent research has verified that CRISPR/Cas9 can be used to precisely modify the genome of monkeys, demonstrating that CRISPR/Cas9-mediated nonhuman primates (NHPs) have promising application potential in understanding of gene function and disease mechanisms and exploration of gene therapy (6–9). However, uncertain OTs are an inevitable concern for translational medicine research using these NHP models. We had previously generated monkey models of Duchenne muscular dystrophy (DMD) by disrupting the X-linked dystrophin gene via CRISPR/Cas9. The genotypes and predicted OTs were assessed in the placenta and umbilical cord of newborn or tissues of stillborn monkeys, and no OT was detected (8), which is the most important requirement for gene therapy research. In this study, we performed OT analysis based on whole-genome sequencing (WGS) in two CRISPR-edited monkey models. The result demonstrated that there was no off-target mutation in functional genome regions of a CRISPR/Cas9-generated monkey model of DMD.
Results
Analysis of mutations and presumptive off-target sites via DNA sequencing
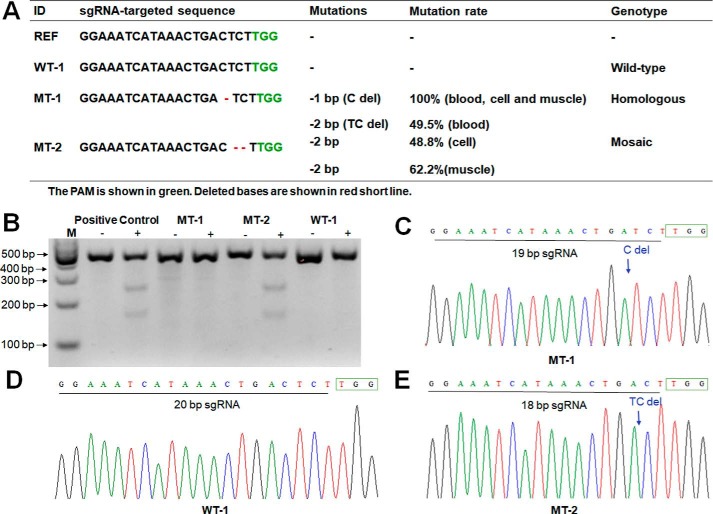
We had previously assessed the genotypes and presumptive OT sites in the placenta and umbilical cord of newborn or tissues of stillborn monkeys, and no OT was detected (8). Here, we tested two CRISPR-edited DMD monkeys (both were 4 years old) that were identified as mutant. The MT-1 is a biallelic knockout homozygous with a 1-bp deletion, and 100% of tested blood, muscle, and skin cells are mutant. The MT-2 is a mosaic mutant monkey with 2-bp deletions, containing about 50% mutant cells (Fig. 1A). OTs were analyzed in both mutant monkeys and one age-matched WT sibling control (WT-1). A total of 17 presumptive OT sites were predicted with CCTop (CRISPR/Cas9 target online predictor) software according to PAM loci and similarity of sgRNA and were identified through Sanger sequences with specific primers. Consequently, there was no OT occurring at these predicted sites (Fig. 1 (B–E) and Table S1).
Figure 1.
Identification of mutations of living DMD monkeys and SNVs and indels at WGS level. A, analysis of gene knockout in three sibling monkeys. B, T7EN1 analysis of mutations in MT-1 and MT-2 with WT-1 as control. C–E, sequence peak map of the three sibling monkeys. Framed TGG, site of PAM loci. C del and TC del, deletion of the C base and TC bases; arrows, locations of the deletions.
WGS off-target analysis
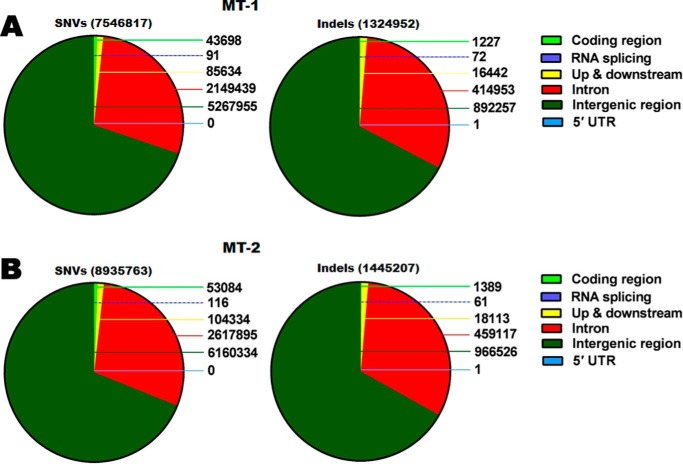
To further validate on- and off-targets at WGS level, the single-nucleotide variants (SNVs) and insertions or deletions (indels) were examined at a depth of 60× to cover variants of lower frequencies, including coding region, RNA splicing, up- and downstream, intron, intergenic region, and 5′-UTR. There are 7,546,817 SNVs and 1,324,952 indels in MT-1 (Fig. 2A) and 8,935,763 SNVs and 1,445,207 indels in MT-2 (Fig. 2B), respectively. Detailed comparisons of indels and SNVs of MT-1 versus WT-1 and MT-2 versus WT-1 are shown in Tables S2–S5.
Figure 2.
Identification of mutations of living DMD monkeys and SNVs and indels at WGS level. A, a total of 7,546,817 SNVs and 1,324,952 indels in MT-1. B, a total of 8,935,763 SNVs and 1,445,207 indels in MT-2. Detailed SNVs and indels include coding region, RNA splicing, up- and downstream, intron and intergenic region, and 5′-UTR.
Compared with the reference genome, a total of 847 homologous sequences of sgRNA were screened out with the NCBI alignment tool blastn (Table S6). There were 510 general sites in MT-1, including 442 SNVs and 68 indels, and 592 general sites in MT-2, containing 535 SNVs and 57 indels. We used the WGS sequence of the WT sibling first and then an online genomic database as reference to filter the SNVs and indels step by step. According to the identity of genomes between reference and sibling, SNVs were filtered, and there remained 208 SNVs in MT-1 and 227 SNVs in MT-2 in intronic or intergenic regions, but not in the coding region, splicing, and up- and downstream in both mutant monkeys (Table 1). In contrast with inbred mice, humans and monkeys have rich genome diversity and complex individual differences. We mainly focused on the identification of indels that contain almost all of the mutations introduced by nonhomologous end joining, because they are the hallmarks of endonuclease-induced damage (9, 10). Similarly, given the identity with reference and WT control, we filtered and excluded repetitive sequence and genomic WGS sequence assemblies (NW_xxx). The remaining three indels in MT-1 and two in MT-2 were validated using Sanger sequence, and we found that there were one intronic indel in MT-1 and two intergenic indels in MT-1 and MT-2, respectively. To investigate the potential interaction with dystrophin gene for these five indels, we searched them in the genome bank of Macaca mulatta from NCBI. The four intergenic sequences and the intronic one could not be found in known coding genes. There is a single exonic indel located at the expected locus, the same as with Sanger sequencing (Table 1). Together, these results indicated that all on-target but no off-target mutations exist in functional genome regions of DMD monkeys.
Table 1.
Summary of on- and off-target sites detected by whole-genome sequencing
| Target types | CRISPR-editing MT-1 |
CRISPR-editing MT-2 |
||
|---|---|---|---|---|
| Predicted | Filtered | Predicted | Filtered | |
| Total OT sites | 510 | 211 | 592 | 229 |
| Total SNVs | 442 | 208 | 535 | 227 |
| Coding region | 0 | 0 | 0 | 0 |
| RNA splicing | 0 | 0 | 0 | 0 |
| Upstream | 0 | 0 | 0 | 0 |
| Downstream | 3 | 0 | 4 | 0 |
| Intron | 116 | 51 | 116 | 39 |
| Intergene | 323 | 157 | 415 | 188 |
| Total indels | 68 | 3 | 57 | 2 |
| Coding region | 0 | 0 | 0 | 0 |
| RNA splicing | 0 | 0 | 0 | 0 |
| Up- and downstream | 0 | 0 | 0 | 0 |
| Intron | 15 | 1 | 12 | 0 |
| Intergene | 53 | 2 | 45 | 2 |
| On-target exon | 1 | 1 | 1 | 1 |
Discussion
The OT effect is an important issue for both animal model construction and gene therapy. Genome-modified monkey models are important for translational medicine research in safety and efficiency assessment due to their high similarity to humans in genome and genetic diversity, physiology, and cognition. Recent literature based on mouse models indicated that OT effects are still an unavoidable issue. It is essential for reliable analytical methods and in-depth sequencing to determine whether or not an OT exists. Anderson et al. (3), using next-generation sequencing, analyzed on- and off-target effects of genome-edited mice and rats and detected the existing OTs, whereas in another report, potential OTs were excluded by Sanger sequencing (11). In our work, all potential OTs in aborted tissue (8) and live monkeys in this study were excluded by T7EN1 and Sanger sequencing. To further explore the present of OTs in WGS level, Anderson's (3) but not Iyer's group (4) found the occurrence of OTs, including SNVs and indels. In view of genome diversity and complex individual differences in monkeys, we focused on indels and filtered the data with steps similar to those used in the methods for rodents. We found that no functional OTs happened in single sgRNA-induced DMD monkey models. The above OT analysis indicated that gene targets in rodents and monkeys display different consequences, case by case. Therefore, with regard to safety concerns of CRISPR/Cas9 technology, OT analysis should be a regular assessment in model construction and therapeutic research.
The result of no OT in the functional regions of the DMD monkey genome will encourage us to perform gene correction. Our present findings also imply that our previous result of presumptive OTs are faithful, demonstrating the stability in vivo and the safety of precise gene editing. With the improvement of efficacy, CRISPR/Cas9 would be better applied in clinical research and disease treatment.
Materials and methods
Animals
DMD rhesus monkey models were reported previously (8). For OT assessment, blood, ear tissues (cells), and muscle samples were taken in a minimally invasive manner from three 4-year-old healthy siblings, one of them an untreated WT control and the other two CRISPR/Cas9-mediated mutant monkeys. All animal procedures were approved in advance by the institutional animal care and use committee and were performed in accordance with the Association for Assessment and Accreditation of Laboratory Animal Care International for the ethical treatment of primates.
Identification of mutations and predicted OTs via DNA sequencing
Genomic DNA was extracted with the DNeasy blood and tissue kit (Qiagen) according to the kit instructions. The sequence of the targeting region was analyzed by PCR, and the complete process was performed by initial incubation at 95 °C for 5 min, followed by 20 cycles of 98 °C for 10 s, 60 °C for 30 s, and 72 °C for 40 s and then 10 cycles of 98 °C for 10 s, 50 °C for 30 s, and 72 °C for 40 s. The PCR products were purified, and 200 ng of DNA/sample was digested with 0.3 μl of T7 endonuclease in a 20-μl reaction volume for 30 min at 37 °C. Then the product was isolated by a 3% agarose gel. To assess mosaic status, purified PCR products were ligated with the TA cloning vector and transfected into Escherichia coli–competent cells. Single colonies were picked and sequenced, and chimeric rate was analyzed by sequencing results. A total of 17 presumptive OT sites were predicted with CCTop software (http://crispr.cos.uni-heidelberg.de/index.html)5 (14); according to PAM NGG and similarity of sgRNA, the maximum total mismatches is 4. OTs were identified through Sanger sequences with specific primers (Table S2).
DNA library preparation and WGS analysis
Blood samples of WT-1, MT-1, and MT-2 monkeys were taken, and genomic DNA was subjected to standard whole-genome DNA library preparation for high-throughput sequencing (Illumina platform) with a mean coverage of 60×. According to the Illumina sequencing features, using double-end sequencing data, we required Q30 to have an average ratio above 80% and an average error rate below 0.1%. Valid sequencing data were aligned to the reference genome of the rhesus monkey (M. mulatta) (assembly Mmul_8.0.1) by BWA (12), and the results were ranked using a comparison of SAMtools (13). Finally, we marked duplicate reads using Picard (http://sourceforge.net/projects/picard).5 All polymorphic SNVs and indel sites in the genome were extracted, and high-confidence SNVs and indel data sets were ultimately obtained and analyzed.
Prediction of potential OT sites
Obtained data sets were used to compare the sgRNA sequence with the reference genome. All potential polymorphic SNVs and indels sites in the genome were extracted by SAMtools and Picard tools. We then ultimately obtained highly confident SNVs and indel data sets along with repeatability and other factors for further filtering. These OT sites contained 1–5 base mismatches with the 20-bp target sequence and at least one mismatch in the PAM-proximal seed region via the NCBI alignment tool blastn. To find more SNVs and indel sites, sgRNA homologous regions were amplified 150 bp in their upstream and downstream, respectively. All obtained potential OT sites were differentiated as exon (coding region), RNA splicing, up- and downstream, intronic, and intergenic regions. All SNVs and indels were further filtered according to identity with reference and WT control and excluded repetitive sequence and WGS shotgun.
Author contributions
Y. C. and W. J. designed and conceived the study. W. J. and Y. N. took part in discussion; S. W., S. R., R. B., P. X., and Q. Z. performed molecular, cell, sequencing, and off-target analyses; Y. Z. and Z. Z. collected monkey materials; Y. C., S. W., and S. R. analyzed data and wrote the manuscript, which was approved by all authors.
Supplementary Material
Acknowledgments
We thank Dr. Xiao-jiang Li (Jinan University) and Yu Kang and Xinglong Chen (Kunming University of Science and Technology) for their previous efforts in establishment of animal models. We also thank Dr. Lu Zhao (University of Cambridge) for careful reading and editing of the manuscript.
This work was supported by National Key Research and Development Program of China Grants 2018YFA0107902 and 2016YFA010401, National Natural Science Foundation of China Grant U1602224, and Basic Research Project of Yunnan Province Grant 2015FA037. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1–S6.
The accession numbers for the WGS data reported in this paper are in the Sequence Read Archive (SRA) database under accession no. SRP145269 at the NCBI.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- OT
- off-target effect
- NHP
- nonhuman primate
- DMD
- Duchenne muscular dystrophy
- WGS
- whole-genome sequencing
- CCTop
- CRISPR/Cas9 target online predictor
- PAM
- protospacer-associated motif
- sgRNA
- single guide RNA
- SNV
- single-nucleotide variant
- indel
- insertion or deletion.
References
- 1. Fu Y., Foden J. A., Khayter C., Maeder M. L., Reyon D., Joung J. K., and Sander J. D. (2013) High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31, 822–826 10.1038/nbt.2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sakuma T., Nishikawa A., Kume S., Chayama K., and Yamamoto T. (2014) Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system. Sci. Rep. 4, 5400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anderson K. R., Haeussler M., Watanabe C., Janakiraman V., Lund J., Modrusan Z., Stinson J., Bei Q., Buechler A., Yu C., Thamminana S. R., Tam L., Sowick M. A., Alcantar T., O'Neil N., et al. (2018) CRISPR off-target analysis in genetically engineered rats and mice. Nat. Methods 10.1038/s41592-018-0011-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Iyer V., Shen B., Zhang W., Hodgkins A., Keane T., Huang X., and Skarnes W. C. (2015) Off-target mutations are rare in Cas9-modified mice. Nat. Methods 12, 479 10.1038/nmeth.3408 [DOI] [PubMed] [Google Scholar]
- 5. Li M., Suzuki K., Kim N. Y., Liu G. H., and Izpisua Belmonte J. C. (2014) A cut above the rest: targeted genome editing technologies in human pluripotent stem cells. J. Biol. Chem. 289, 4594–4599 10.1074/jbc.R113.488247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Niu Y., Shen B., Cui Y., Chen Y., Wang J., Wang L., Kang Y., Zhao X., Si W., Li W., Xiang A. P., Zhou J., Guo X., Bi Y., Si C., et al. (2014) Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 156, 836–843 10.1016/j.cell.2014.01.027 [DOI] [PubMed] [Google Scholar]
- 7. Cui Y., Niu Y., Zhou J., Chen Y., Cheng Y., Li S., Ai Z., Chu C., Wang H., Zheng B., Chen X., Sha J., Guo X., Huang X., and Ji W. (2018) Generation of a precise Oct4-hrGFP knockin cynomolgus monkey model via CRISPR/Cas9-assisted homologous recombination. Cell Res. 28, 383–386 10.1038/cr.2018.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen Y., Zheng Y., Kang Y., Yang W., Niu Y., Guo X., Tu Z., Si C., Wang H., Xing R., Pu X., Yang S. H., Li S., Ji W., and Li X. J. (2015) Functional disruption of the dystrophin gene in rhesus monkey using CRISPR/Cas9. Hum. Mol. Genet. 24, 3764–3774 10.1093/hmg/ddv120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhao H., Tu Z., Xu H., Yan S., Yan H., Zheng Y., Yang W., Zheng J., Li Z., Tian R., Lu Y., Guo X., Jiang Y. H., Li X. J., and Zhang Y. Q. (2017) Altered neurogenesis and disrupted expression of synaptic proteins in prefrontal cortex of SHANK3-deficient non-human primate. Cell Res. 27, 1293–1297 10.1038/cr.2017.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Veres A., Gosis B. S., Ding Q., Collins R., Ragavendran A., Brand H., Erdin S., Cowan C. A., Talkowski M. E., and Musunuru K. (2014) Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell 15, 27–30 10.1016/j.stem.2014.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shen B., Zhang W., Zhang J., Zhou J., Wang J., Chen L., Wang L., Hodgkins A., Iyer V., Huang X., and Skarnes W. C. (2014) Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat. Methods 11, 399–402 10.1038/nmeth.2857 [DOI] [PubMed] [Google Scholar]
- 12. Li H., and Durbin R. (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., and 1000 Genome Project Data Processing Subgroup (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stemmer M., Thumberger T., Del Sol Keyer M., Wittbrodt J., and Mateo J. L. (2015) CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS One 10, e0124633 10.1371/journal.pone.0124633 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.