Abstract
β-1,4-Mannanase degrades β-1,4-mannan polymers into manno-oligosaccharides with a low degree of polymerization. To date, only one glycoside hydrolase (GH) family 113 β-1,4-mannanase, from Alicyclobacillus acidocaldarius (AaManA), has been structurally characterized, and no complex structure of enzyme–manno-oligosaccharides from this family has been reported. Here, crystal structures of a GH family 113 β-1,4-mannanase from Amphibacillus xylanus (AxMan113A) and its complexes with mannobiose, mannotriose, mannopentaose, and mannahexaose were solved. AxMan113A had higher affinity for −1 and +1 mannoses, which explains why the enzyme can hydrolyze mannobiose. At least six subsites (−4 to +2) exist in the groove, but mannose units preferentially occupied subsites −4 to −1 because of steric hindrance formed by Lys-238 and Trp-239. Based on the structural information and bioinformatics, rational design was implemented to enhance hydrolysis activity. Enzyme activity of AxMan113A mutants V139C, N237W, K238A, and W239Y was improved by 93.7, 63.4, 112.9, and 36.4%, respectively, compared with the WT. In addition, previously unreported surface-binding sites were observed. Site-directed mutagenesis studies and kinetic data indicated that key residues near the surface sites play important roles in substrate binding and recognition. These first GH family 113 β-1,4-mannanase–manno-oligosaccharide complex structures may be useful in further studying the catalytic mechanism of GH family 113 members, and provide novel insight into protein engineering of GHs to improve their hydrolysis activity.
Keywords: enzyme structure; enzyme mechanism; enzyme catalysis; X-ray crystallography; protein design; Complex structure; endo-β-1,4-mannanase; GH family 113 catalytic mechanism; Rational design
Introduction
Mannans are major components of hemicellulose, which is widely distributed in monocotyledons, plant seeds, and beans (1, 2). They are classified as linear mannans, glucomannans, galactomannans, and galactoglucomannans. Their backbone is composed of β-1,4-linked mannose or glucose units, and galactomannan and galactoglucomannan have additional α-galactosyl branches at O-6 (2, 3).
Mannans can be hydrolyzed into soluble manno-oligosaccharides, which is a suitable candidate in functional food production (4, 5), feed manufacturing (6), and paper processes (7). However, complete degradation of mannan requires a series of enzymes, including β-1,4-mannanase (EC 3.2.1.78), β-1,4-mannosidase (EC 3.2.1.25), β-d-glucosidase (EC 3.2.1.21), and α-galactosidase (EC 3.2.1.22) (2, 8). β-1,4-Mannanases widely exist in mollusks, plants, bacteria, fungi, and actinomycetes (1, 9). According to the classification of carbohydrate-active enzymes (CAZy database, http://www.cazy.org) (29),2 β-1,4-mannanases are grouped into glycoside hydrolase (GH)3 families 5, 26, 113, and 134. Similar to GH families 5 and 26, family 113 members belong to clan GH-A, whose members share a typical (β/α)8-barrel protein-fold (TIM barrel) and a retaining GH double-displacement catalytic mechanism (10–12); on the other hand, GH family 134 β-1,4-mannanases display an inverting GH catalytic mechanism (13, 14). To date, 359 GH family 113 protein sequences have been recorded, most of them derived from bacteria. Only one β-1,4-mannanase from the prokaryote Alicyclobacillus acidocaldarius Tc-12-31 (AaManA, PDB entry 3CIV) has been structurally characterized (10). AaManA shows the (β/α)8-barrel-fold, and 2 glutamate residues that serve as a general acid/base and a nucleophile, respectively, to catalyze β-1,4-linked glucosidic bonds. However, the structure of GH family 113 enzyme–manno-oligosaccharide complexes is not available, and the precise catalytic mechanism needs to be further elucidated.
In this study, a novel GH family 113 β-1,4-mannanase gene from Amphibacillus xylanus NBRC 15112 (AxMan113A) was cloned and expressed in Escherichia coli. The three-dimensional structures of the apo form and four complexes with mannobiose (M2), mannotriose (M3), mannopentaose (M5), and mannohexaose (M6) are solved. We further performed rational design by site-directed mutagenesis of key amino acids in the substrate-binding cleft to improve the enzyme's activity. Our studies provide structural insights into the catalytic mechanism of GH family 113 β-1,4-mannanases.
Results
Enzyme characterization
A novel GH family 113 β-1,4-mannanase from A. xylanus NBRC 15112 (AxMan113A) was cloned and expressed in E. coli (Fig. S1). After purification, the denatured molecular mass of AxMan113A was 36.2 kDa on SDS-PAGE, and the native molecular mass was 38.8 kDa by gel filtration chromatography (Fig. S2), suggesting that the enzyme is a monomer. The amino acid sequence of AxMan113A showed the highest identity (43%) with the β-1,4-mannanase from A. acidocaldarius Tc-12-31 (AaManA; GenBankTM accession number ABG77968.1) (Fig. 1). AxMan113A exhibited maximal activity at pH 6.5 in 50 mm McIlvaine buffer (Fig. S3A) and was stable in a pH range of 5.0–9.0, with over 90% activity (Fig. S3B). Optimal temperature of AxMan113A was 45 °C (Fig. S3C), and the enzyme was stable at temperatures below 45 °C, retaining over 90% activity after incubation for 30 min (Fig. S3D).
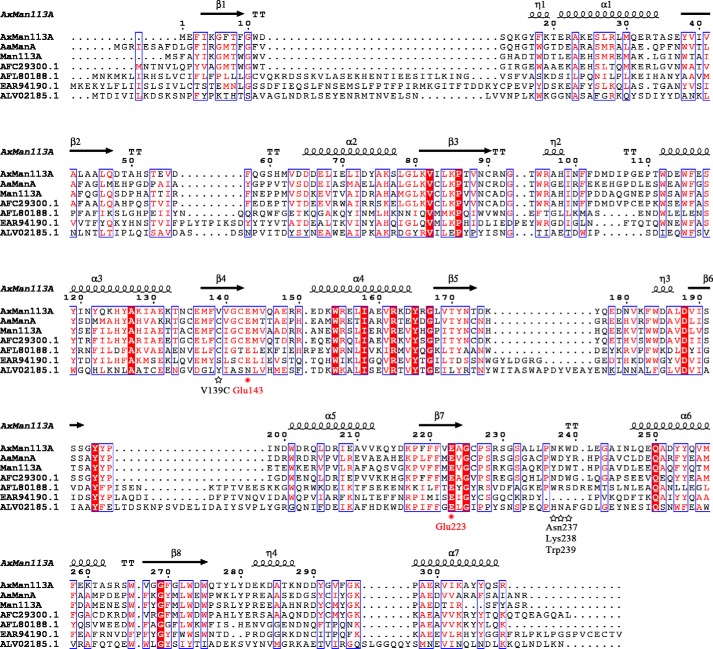
Figure 1.
Sequence alignment of AxMan113A with other GH family 113 members. Residues forming the secondary structures of AxMan113A are highlighted above the sequences. Identical residues are shown in white on red background. Two catalytic residues, Glu-143 and Glu-223, are marked by red dots. The sequences of AxMan113A (PDB entry 5YLH), A. acidocaldarius β-1,4-mannanase (AaManA; PDB entry 3CIV), Alicyclobacillus sp. strain A4 β-1,4-mannanase (Man113A; GenBankTM accession number KC460333.1), Paenibacillus mucilaginosus 3016 β-1,4-mannanase (GenBankTM accession number AFC29300.1), Aequorivita sublithincola DSM 14238 β-1,4-mannanase (GenBankTM accession number AFL80188.1) and Bacillus amyloliquefaciens (GenBankTM accession number ALV02185.1) were aligned by ClustalX2 (55) and the figure was produced in ESPript (56).
Substrate specificity and hydrolysis properties
The substrate specificity of AxMan113A was determined (Table 1). Its highest specific activity was toward M6 (8.3 × 103 units/mg), followed by M5 (7.5 × 103 units/mg), mannotetraose (M4; 6.3 × 103 units/mg), mannotriose (M3; 3.8 × 103 units/mg), and M2 (3.6 × 102 units/mg). AxMan113A showed much lower specific activity toward the mannans konjac powder (54.4 units/mg), locust bean gum (LBG; 22.4 units/mg), and guar gum (1.2 units/mg). It did not show any detectable activity toward p-nitrophenyl β-d-mannopyranoside (pNPM).
Table 1.
Substrate specificity of AxMan113A
| Substrate | Specific activitya | Relative activityb |
|---|---|---|
| units/mg | % | |
| Locust bean gum | 22.4 ± 0.21 | 100 |
| Konjac powder | 54.4 ± 0.66 | 2.4 × 102 |
| Guar gum | 1.2 ± 0.02 | 5.4 |
| Mannobiose | 3.6 × 102 ± 10.9 | 1.6 × 103 |
| Mannotriose | 3.8 × 103 ± 87.7 | 1.7 × 104 |
| Mannotetraose | 6.3 × 103 ± 1.9 × 102 | 2.8 × 104 |
| Mannopentaose | 7.5 × 103 ± 2.3 × 102 | 3.3 × 104 |
| Mannohexaose | 8.3 × 103 ± 2.9 × 102 | 3.7 × 104 |
| pNPM | NAc | 0 |
a Enzyme activity was measured at 45 °C for 10 min in 50 mm McIlvaine buffer (pH 6.5).
b Relative activity of the substrate locust bean gum was set at 100%.
c NA, no activity detected.
To investigate the hydrolysis properties of AxMan113A, various mannans and manno-oligosaccharides (Megazyme, Ireland) were hydrolyzed by the enzyme. The hydrolysate was subjected to thin-layer chromatography (TLC) (Fig. S4). AxMan113A hydrolyzed LBG and konjac powder to yield mainly mannose and a series of manno-oligosaccharides (Fig. S4A). Manno-oligosaccharides (M2–M5) were further incubated with AxMan113A. The enzyme slightly cleaved M2 to produce mannose after a 4-h incubation. AxMan113A exhibited higher hydrolysis activity toward M3, M4, and M5, mainly releasing mannose and M2, which were the end products of the hydrolysis after incubation for 4 h (Fig. S4B). Hydrolysis experiments were performed with reduced M2, M3, M4, and M5 viz. M2r, M3r, M4r, and M5r. AxMan113A could not hydrolyze M2r and M3r, even after a 4-h incubation. It exhibited very low hydrolysis ability toward M4r, yielding traces of M2 and M2r. M5r was hydrolyzed to yield M2 and M3r as the end hydrolysis products (Fig. S5).
Enzyme kinetics of manno-oligosaccharides
The kinetics constants for manno-oligosaccharides (M2–M6) by AxMan113A were determined (Table 2). The Km value order was M2 (74.3 mm) > M3 (29.1 mm) > M4 (12.7 mm) > M5 (5.3 mm) > M6 (3.9 mm), suggesting that this enzyme had higher affinity toward M6. In addition, AxMan113A had low catalytic efficiency toward M2 (0.0013 s−1 mm−1), high efficiency toward M3 (0.029 s−1 mm−1), M4 (0.077 s−1 mm−1), M5 (0.201 s−1 mm−1), and M6 (0.387 s−1 mm−1). The catalytic efficiencies (kcat/Km) of mutants Q58A and D73A were 0.0006 and 0.0007 s−1 mm−1, respectively (Table S2). Compared with WT, catalytic efficiencies of Q58A and D73A were decreased to 46.1 and 53.8%, respectively. Thus, mutants Q58A and D73A reduced substrate affinity toward M2.
Table 2.
Kinetic data of AxMan113A toward manno-oligosaccharides
The kinetic parameters were determined at 45 °C for 5 min in 50 mm McIlvaine buffer (pH 6.5).
| Substrate | Vmax | Km | kcat | kcat/Km |
|---|---|---|---|---|
| μmol min−1 mg−1 | mm | s−1 | s−1 mm−1 | |
| M2 | 162.2 ± 7.3 | 74.3 ± 4.4 | 0.098 | 0.0013 |
| M3 | 1413.8 ± 65 | 29.1 ± 2.3 | 0.853 | 0.029 |
| M4 | 1622.5 ± 86 | 12.7 ± 1.1 | 0.979 | 0.077 |
| M5 | 1764.7 ± 94 | 5.3 ± 0.4 | 1.065 | 0.201 |
| M6 | 2500 ± 152 | 3.9 ± 0.3 | 1.508 | 0.387 |
Overall structure of AxMan113A
The structure of AxMan113A was solved and the crystallographic statistics are summarized in Table 2. The orthorhombic space group of AxMan113A was P212121 with two monomers in the asymmetric unit (Fig. 2A). The monomer with approximate dimensions of 41 × 31 × 56 Å contained residues from Met-1 to Arg-309. AxMan113A displayed the typical (β/α)8-barrel-fold of GH family 113 members, containing an internal core of 8 β-strands encircled by 8 α-helices. The catalytic proton donor and nucleophile (Glu-143 and Glu-223) were determined by structural alignment to another GH family 113 β-1,4-mannanase (AaManA, PDB entry 3CIV), located in the middle of the narrow groove (Fig. 2B).
Figure 2.
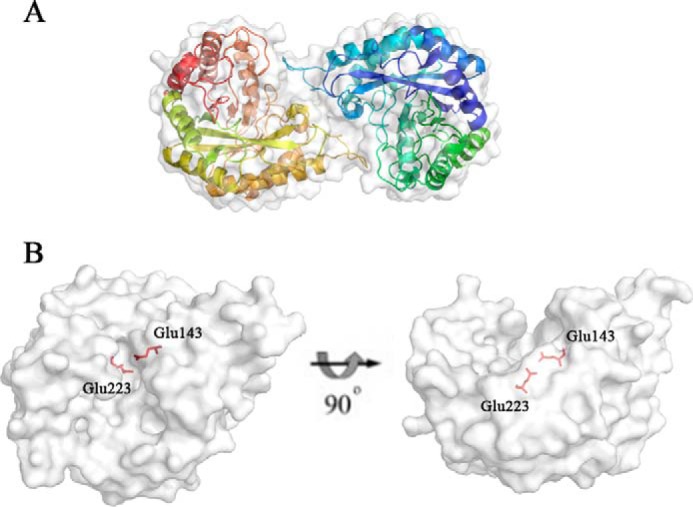
Overall structure of AxMan113A. A, cartoon form, illustrating the classical (β/α)8-barrel. Two monomer molecules are present in the asymmetric unit of AxMan113A. B, surface view. A pair of catalytic residues (Glu-143 and Glu-223) are shown as sticks in red.
Crystal structures of AxMan113A–E223A complexes
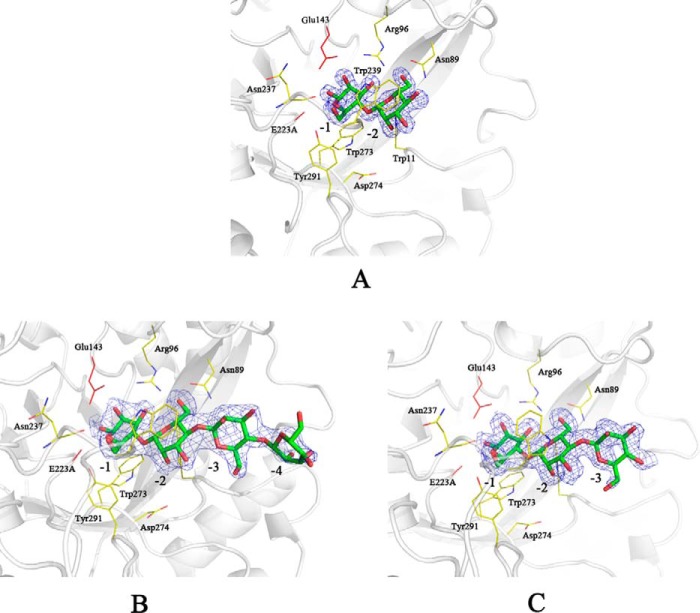
The structures of AxMan113A–E223A complexes with M2, M3, M5, and M6 were obtained by co-crystallization method. In the AxMan113A–E223A-M2 structure, one M2 molecule was embedded in the narrow groove and occupied subsites −2 and −1 in chain B. In chain A, however, three mannose units were found in subsites −2, −1, and +2, indicating the existence of two binding modes in AxMan113A–E223A-M2, occupying subsites −2 to −1 and +1 to +2. The lack of electron density for mannose moieties might have been due to the following: (i) mannoses in subsites +1 and +2 were very flexible; (ii) binding domains in subsites +1 and +2 were too narrow to completely bind one M2 molecule. In subsites −1 and −2, M2 residues formed 12 direct hydrogen bonds to Trp-11, Asn-89, Arg-96, Glu-143, Tyr-195, Asn-237, and Tyr-291. Three water-mediated hydrogen bonds were formed between Asn-237 and the mannose residues. Specifically, the amino groups of the side chains of Trp-11, Asn-89, and Arg-96 were directly hydrogen-bonded to O-2, O-5, and O-6 of the −2 mannose residue. A strong hydrogen-bond network was observed with the −1 mannose residue. The O-2, O-3, O-4, O-5, and O-6 hydroxyls of the −2 mannose residue were directly hydrogen-bonded to the side chains of Arg-96, Glu-143, Tyr-195, Asn-237, Trp-273, and Tyr-291. Furthermore, aromatic residues Trp-239 and Trp-273 were stacked against the mannose units in subsites −2 and −1, forming hydrophobic sugar-binding platforms (Fig. 3A). There were seven direct hydrogen bonds and three water-mediated interactions for the +2 mannose residue. Asn-237, Trp-239, and Asp-240 formed four interactions with the O-2, O-3, and O-4 hydroxyls of +2 mannose. The O-6 hydroxyl of +2 mannose residue was directly bound to Asn-237 and Lys-238. Trp-40 and Trp-239 provided the hydrophobic sugar-binding platform, making contact with +2 mannose residue (Fig. 3B). Superposition of AxMan113A–E223A-M2 and AxMan113A showed that most residues were in identical positions, except for Trp-239 and Asp-240. The side chain of Trp-239 was deflected in the former, forming a hydrophobic platform to bind to the mannose residues in subsites −2 and +2. The distance between the side chain of Asp-240 and +1 mannose was 3.3 to 2.7 Å, suggesting that Asp-240 was attracted during the recognition and binding with substrates. Interestingly, some additional electron-density maps representing M2 molecules were found on the surface region of the AxMan113A–E223A-M2 complex structure, which is far away from the catalytic cleft. In chain A, one M2 molecule was found in the middle of two α-helixes. Another M2 molecule was observed on the extended loop of chain B, and neither of these mannoses were structurally symmetrical (Fig. 4).
Figure 3.
Complex of mutant AxMan113A–E223A with mannobiose (M2). A, molecular surface of the complex structure. M2 residues located along the groove are shown as sticks in chains A and B. B, stereo view of the substrate interactions of AxMan113A–E223A-M2 in chains A and B. All of the mannose residues bound in the groove are shown as green/red sticks. The key protein residues are shown as yellow sticks and the catalytic residues are shown in red. The electron density of the M2 ligand is shown as an σA-weighted mFo − DFc OMIT map contoured at the 1.0 σ level.
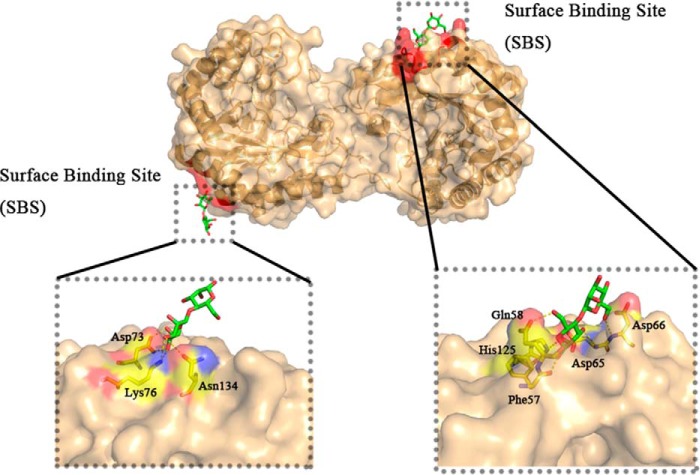
Figure 4.
Surface-binding sites of AxMan113A/E223A-M2. Structure of AxMan113A/E223A-M2 is shown as cartoon in light orange, surface-binding regions are colored in red. The key protein residues are shown in yellow sticks. Mannobiose are shown as green/red sticks.
A crystal structure of AxMan113A–E223A in complex with M3 (AxMan113A–E223A-M3) was solved at 1.68-Å resolution (Table 3). Only one M2 molecule was found to occupy to the catalytic cleft of AxMan113A (at subsites −2 to −1), suggesting that mannose occupies subsites −2 to −1 preferentially during the process of substrate binding (Fig. 5A). The complex structure of AxMan113A–E223A-M5 was determined at 2.38-Å resolution. Four mannose units (M4) linked by β-1,4-glycosidic bonds could be built into the electron-density map (at subsites −4 to −1). Mannose residues in subsites −3 and −4 extended to the terminal and lacked interactions with any surrounding key amino acids, indicating that AxMan113A can only weakly bind −3 and −4 mannoses (Fig. 5B). Superposition of AxMan113A–E223A-M5 and AxMan113A–E223A-M2 showed that the −1 and −2 mannose residues were basically in almost the same position, and all adopted the chair conformation 4C1. At least six binding sites existed in the groove, occupying subsites −4 to +2. In addition, the complex structure of AxMan113A–E223A-M6 was solved. Similarly, one M3 molecule was well-defined in the electron-density maps, occupying the catalytic cleft of AxMan113A (subsites −1 to −3) (Fig. 5C).
Table 3.
X-ray data collection and refinement statistics
| Apo-AxMan113A | AxMan113A–E223A-M2 | AxMan113A–E223A-M3 | AxMan113A–E223A-M5 | AxMan113A–E223A-M6 | |
|---|---|---|---|---|---|
| Data collection statistics | |||||
| Radiation source | SSRF-BL18U | SSRF-BL17U | SSRF-BL17U | SSRF-BL17U | SSRF-BL17U |
| Wavelength (Å) | 0.978 | 0.979 | 0.979 | 0.979 | 0.979 |
| Temperature of measurements (K) | 100 | 100 | 100 | 100 | 100 |
| Resolution (Å) | 30.38–2.29 (2.38–2.29) | 40.53–2.09 (2.17–2.09) | 32.9–1.68 (1.74–1.68) | 35.85–2.37 (2.46–2.37) | 32.75–1.81 (1.88–1.81) |
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 |
| Unit cell parameters | |||||
| a, b, c (Å) | 54.8, 69.4, 188.9 | 49.0, 72.2, 177.2 | 65.1, 69.9, 186.6 | 55.1, 69.6, 188.8 | 62.6, 70.0,186.3 |
| α, β, γ(°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Protein molecules in asymmetric unit | 2 | 2 | 2 | 2 | 2 |
| Unique reflections | 29076 (2687) | 36641 (3374) | 91072 (6371) | 29759 (2690) | 68340 (6807) |
| Completeness (%) | 88.1 (64.4) | 92.0 (96.2) | 93.0 (79.6) | 98.0 (90.4) | 91.4 (75.5) |
| Rmergea (%) | 12.2 (60.9) | 9.0 (41.9) | 19.8 (88.,1) | 8.8 (48.5) | 8.6 (45.2) |
| Mean I/sigma(I) | 4.6 | 7.3 | 6.8 | 9.1 | 9.4 |
| Wilson B-factor (Å2) | 31.67 | 22.83 | 13.82 | 35.38 | 19.04 |
| Refinement statistics | |||||
| Resolution (Å) | 2.29 | 2.09 | 1.68 | 2.37 | 1.81 |
| Rworkb (%) | 18.62 (26.67) | 15.40 (17.54) | 18.32 (22.23) | 18.04 (22.62) | 18.57 (23.76) |
| Rfreeb (%) | 23.49 (33.02) | 19.47 (25.60) | 20.68 (27.73) | 23.40 (31.28) | 21.59 (26.07) |
| No. residues | 618 | 618 | 618 | 618 | 618 |
| No. ligands | 0 | 92 | 46 | 90 | 68 |
| No. water molecules | 160 | 484 | 1035 | 216 | 734 |
| No. atoms | 5277 | 5656 | 6170 | 5418 | 5877 |
| Root mean square deviations | |||||
| Bond lengths (Å) | 0.008 | 0.009 | 0.01 | 0.009 | 0.008 |
| Bond angles (°) | 0.94 | 0.92 | 0.94 | 1.00 | 0.86 |
| Average B-factors (Å2) | 30.22 | 24.58 | 19.69 | 35.23 | 26.66 |
| Macromolecules | 29.85 | 23.66 | 17.11 | 34.94 | 24.67 |
| Ligands | - | 35.32 | 14.45 | 47.59 | 28.80 |
| Solvent | 35.29 | 32.18 | 32.62 | 36.79 | 40.24 |
| Ramachandran | |||||
| Most favored regions (%) | 97 | 98 | 98 | 98 | 98 |
| Additional allowed regions (%) | 3.1 | 2.0 | 2.1 | 2.3 | 1.8 |
| Disallowed regions (%) | 0 | 0 | 0 | 0 | 0 |
| Clashscore | 8.93 | 5.37 | 5.1 | 4.84 | 5.91 |
| PDB code | 5YLH | 5YLK | 5Z4T | 5YLI | 5YLL |
a Rmerge = Σhkl Σi|Ii(hkl) − 〈I(hkl)〉|/Σhkl ΣiIi(hkl), where Ii(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl.
b Rwork/free = Σhkl||Fobs| − k|Fcalc||/Σhkl|Fobs|; Rwork is the R value for the reflections used in the refinement, whereas Rfree is the R value for 5% of the reflections selected randomly and not included in the refinement.
Figure 5.
Complexes of mutant AxMan113A–E223A with mannotriose (M3), mannopentaose (M5), and mannohexaose (M6). A, stereo view of the substrate interactions of AxMan113A–E223A-M3. B, stereo view of the substrate interactions of AxMan113A–E223A-M5. C, stereo view of the substrate interactions of AxMan113A–E223A-M6. All of the mannose residues are shown as green/red sticks. The key protein residues are shown as yellow sticks and the catalytic residues (Glu-143 and E223A) are shown in red. Electron density of M3, M5, and M6 ligand is shown as an σA-weighted mFo − DFc OMIT map contoured at the 1.0 σ level.
Rational design of AxMan113A
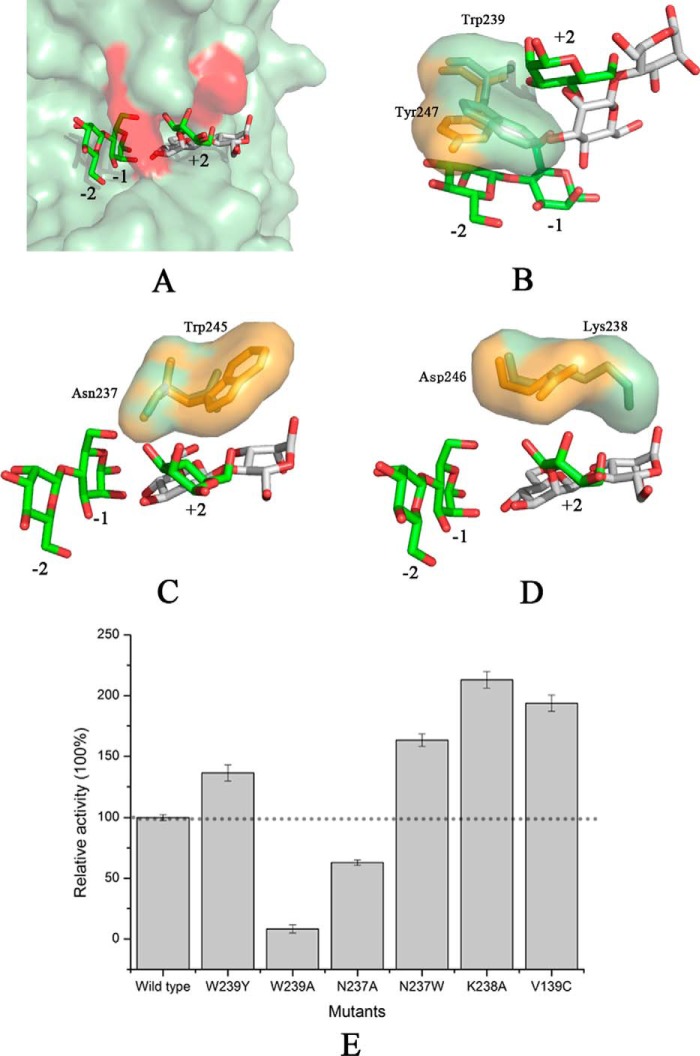
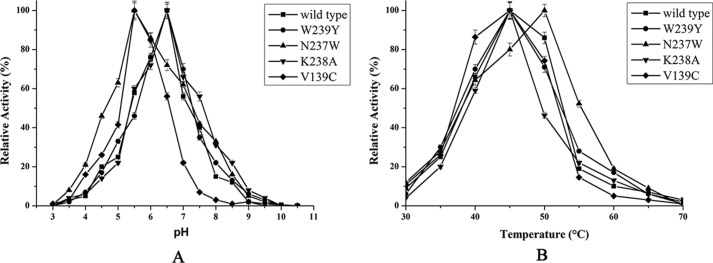
Structural alignment of AxMan113A (PDB entry 5YLH) and AaManA in complex with a synthesized inhibitor (ManIFG) and M2 (AaManA–ManIFG-M2; PDB entry 4CD7) is shown in Fig. 6A. Both of the catalytic grooves were basically identical except for several residue positions in subsites +1 and +2. Trp-239 of AxMan113A (Tyr-237 in AaManA–ManIFG-M2) showed a large aromatic ring, which formed a stacking interaction with −2 and +2 sugars. Compared with AaManA–ManIFG-M2–Tyr-247, the side chain of AxMan113A–Trp-239 rotated nearly 90°, closer to the catalytic cleft (Fig. 6B). AxMan113A–Asn-237 was located above the −1 and −2 mannoses, forming three direct hydrogen bonds with them. This amino acid was replaced by Trp-245 in AaManA–ManIFG-M2, which provided a hydrophobic sugar-binding platform that could stabilize the +2 mannose (Fig. 6C). Another proposed key amino acid residue was Lys-238 of AxMan113A, which was situated on subsite +2. The longer side chain extended into the groove, forming steric hindrance to mannose binding (Fig. 6D). Notably, AxMan113A–Lys-238 was replaced by Asp-246 in the corresponding site of AaManA–ManIFG-M2. In addition, nonpolar residue Val-139 was replaced by polar residues (Cys, Ser, and Tyr) in other GH family 113 members (Fig. 1). To investigate whether hydrolysis activity in AxMan113A could be altered by changing these residues, six mutants (W239Y, W239A, N237A, N237W, K238A, and V139C) were designed. Specific activity of W239A and N237A was decreased by 92.0 and 37.1%, respectively, whereas the specific activity of W239Y, N237W, K238A, and V139C was improved by 36.4, 63.4, 112.9, and 93.7%, respectively, relative to the WT (Fig. 6E). Compared with AxMan113A, mutants V139C and N237W shifted 1.0 pH unit toward the acidic range. However, W239Y, W239A, N237A, and K238A did not show change in optimal pH (Fig. 7A). With respect to optimal temperature, none of the mutants changed expect N237W decreased by 5 °C (Fig. 7B).
Figure 6.
Unique conformation of the amino acids near subsites +1 and +2. A, structural alignment of AxMan113A–E223A-mannobiose (M2) and AaManA–ManIFG-M2. B, a semi-enclosed space formed by Trp-239 of AxMan113A. C, Asn-237 of AxMan113A–E223A-mannobiose was replaced by Trp-245 of AaManA-ManIFG-M2. D, a steric hindrance formed by Lys-238. E, enzyme activity assay for the WT and mutants toward locust beam gum. Results are plotted as percentage of the product generated relative to the WT (100%; dashed line). All of the measurements were performed in triplicate.
Figure 7.
Determination of optimal pH (A) and optimal temperature (B) of WT and the mutants. The effect of pH on β-mannanase activity was determined in 50 mm of different buffers at optimal temperature of each enzyme. For optimal temperature, activity was measured at different temperatures in 50 mm McIlvaine buffer (optimal pH of each enzyme). The specific activities of WT (22.4 units/mg), W239Y (30.6 units/mg), N237W (36.6 units/mg), K238A (47.7 units/mg), and V139C (43.4 units/mg) were considered as 100% in determinate optimal pH and optimal temperature.
Discussion
Because of the various biochemical properties, β-1,4-mannanases are widely used in many industrial applications. In paper/pulp industries, β-1,4-mannanases are able to cleave the mannan portion of pulps to increase the brightness (2, 7). In the field of clinical applications, β-1,4-mannanases can be used to hydrolyze nondigestible oligosaccharides and polysaccharides like prebiotics and dietary fibers, producing short chain fatty acids, which can protect against inflammatory bowel disease, ulcerative colitis, and Crohn's disease (1, 15). β-1,4-Mannanases was also used to produce manno-oligosaccharides, which can effectively regulate the immune system and reduce serum cholesterol (16). In the field of functional foods, high viscosity of guar gum can be hydrolyzed by β-1,4-mannanases to yield partially hydrolyzed guar gum, which offers health benefits and nutrient digestion (17). Due to their growing industrial potential, many of β-1,4-mannanases have been studied. GH family 113 only had one structure-resolved β-1,4-mannanase (AaManA) from A. acidocaldarius Tc-12-31 (10). AaManA has been reported to share a typical (β/α)8-barrel folding motif and displays a two-step retaining catalytic mechanism, which is consistent with β-1,4-mannanases in GH families 5 and 26. The crystal structure of β-1,4-mannanase-targeted inhibitors (AaManA–ManIFG-M2) was solved after AaManA (18). However, lack of enzyme–manno-oligosaccharide complex structural information gives only limited comprehensive clarification of the catalytic mechanism of GH family 113 members. In this study, we cloned a novel GH family 113 β-1,4-mannanase gene from A. xylanus (AxMan113A) and obtained four manno-oligosaccharide complex crystals to further elucidate the catalytic mechanism of GH family 113 members.
The specific activity of AxMan113A toward konjac powder was higher than that toward LBG or guar gum, which is consistent with other GH family 113 members, such as AaManA (10) and Man113A from Alicyclobacillus sp. strain A4 (19). In contrast, β-1,4-mannanases from GH families 5, 26, and 134, such as RmMan5A from Rhizomucor miehei (20), ReTMan26 from thermophilic Bacillus subtilis (21), and AoMan134A from Aspergillus oryzae (22), exhibit high hydrolysis preference for LBG. AxMan113A and other GH family 113 β-1,4-mannanases have a similar catalytic cleft, and some key amino acids around subsites −1 and +1 form a semi-enclosed “lid” region that can only accommodate one mannose or glucose (Fig. S6A). On the other hand, structures of GH families 5, 26, and 134 β-1,4-mannanases display an “open” groove, which has enough space to accommodate the substrates with galactose side chains (13, 23, 24). Therefore, it can be deduced that enzyme activity of GH family 113 members is limited by the α-1,6-galactose residues because of that structural difference.
The catalytic mechanism of GH family 113 β-1,4-mannanases can be described as a two-step retaining catalytic mechanism (25). The catalytic residues are Glu-143 and Glu-223 in AxMan113A, as confirmed by site-directed mutagenesis and structural alignment to AaManA. Previous studies have shown that GH family 113 members hydrolyze manno-oligosaccharides with DP > 3 and exhibit considerable transglycosylation activity (10, 19). However, the smallest substrate of AxMan113A was M2 (Fig. S4B). In addition, AxMan113A had no transglycosylation activity. Thus, the catalytic mechanism of AxMan113A differs from that of other GH family 113 members. AxMan113A could hydrolyze the reduced manno-oligosaccharides with DP ≥ 4, and the hydrolysis efficiency toward M5r was higher than that of M4r (Fig. S5). It indicated that the reducing terminal substitution by mannitol could influence the recognition of mannose residue in the catalytic groove. Moreover, the hydrolysis of AxMan113A was diminished gradually with the substitutions approached to the catalytic groove. That might be one major reason that this enzyme cannot hydrolyze pNPM.
AxMan113A exhibited the different catalytic efficiency (kcat/Km) toward manno-oligosaccharides, which were decreased with DP from M6 to M2 (Table 2). Among the endo-β-mannanases, AxMan113A displayed catalytic efficiency toward M3 (0.029 s−1 mm−1), which is much lower than Man113A (6.54 s−1 mm−1) (19), AnMan5B (0.16 s−1 mm−1) (26), and PaMan5A (26.67 s−1 mm−1) (27). It had much lower catalytic efficiency toward M2 (only 0.0013 s−1 mm−1). Thus, the enzyme hydrolyzed M2 slowly after a 4-h incubation (Fig. S4B). These results could support for the main hydrolysis products being M2 and mannose.
To verify the unique catalytic mechanism of AxMan113A, crystal structures of AxMan113A and its complexes with M2, M3, M5, and M6 were solved (Table 3). Clearly, mannose residues bound to at least 6 subsites in the groove (−4 to +2) (Fig. S6B). In the structure of AxMan113A–E223A-M6, one M3 molecule was occupied in −3 to −1 subsites, and another 3 substrate-binding sites (+1 to +3) has been proved by hydrolyzing M5r (Fig. S5). Mannose moieties of M5r would preferentially bind in subsites −2 to +2. Mannitol was occupied in subsite +3 (24, 28). Thus, it could be deduced that another M3 molecule was occupied in +1 to +3 subsites. A bulk hydrogen-bond network was formed around the subsite −1, which firmly fixed the position of the −1 mannose. Three residues (Arg-96, Tyr-195, and Trp-273) of GH family 113 β-1,4-mannanases were conserved (Fig. 1). Arg-96 not only hydrogen bonded with the −1 mannose, but also formed one hydrogen bond and salt bridge with the side chain of Asp-143 at a distance of 2.9 Å, stabilizing the latter's orientation. Similarly, the presence of Tyr-195 (2.7 Å away from Glu-223) was vital to orienting catalytic residue Glu-223 (Fig. S7A). This is consistent with studies of chitosanases OU01 and N174 (30, 31). Trp-273 in AxMan113A provided a hydrophobic sugar-binding platform with the +1 sugar ring. The importance of three residues for substrate-binding was corroborated by site-directed mutagenesis. Mutants R96A, Y195A, and W273A lost most of this activity (Fig. S7C). Compared with AaManA–ManIFG-M2, one key water molecular in AxMan113A–E223A-M2 was found above −1 subsite, forming three hydrogen bonds with −1 mannose, better fixing the sugar's position. This suggests that the binding of −1 mannose in AxMan113A is stronger than in AaManA. Moreover, in subsite +1, the conformation of three residues (Trp-95, Lys-176, and Asp-240) in AxMan113A changed a great deal compare with AaManA–ManIFG-M2. Trp-95 flipped up ∼30°, which brought the aromatic nucleus close to subsite +1, strengthening the interaction with +1 sugar. The side chains of Lys-176 and Asp-240 (His-183 and Arg-248 in AaManA, respectively) extended to subsite +1, forming more hydrogen bonds with the mannose (Fig. S7B). Specific activity of W95A, K176A, and D240A was decreased to 7.1, 9.6, and 10.2% of the WT, respectively (Fig. S7C). AxMan113A exhibited higher affinity for −1 and +1 mannoses, which might be an important reason why the enzyme can hydrolyze M2. As already noted, unlike other members of GH family 113, AxMan113A had no transglycosylation activity. In the GHs with retaining catalytic mechanism, some amino acids near subsite +2 play an important role in stabilizing the transition state of transglycosylation (32). Compared with AaManA–ManIFG-M2, the conformation of mannose in the subsite +2 of AxMan113A changed significantly. This suggested that the +2 sugar is very flexible, making its binding to subsite +2 difficult. Aromatic amino acids in the binding subsite +2 can stabilize the transglycosylation receptor (33, 34). In AaManA–ManIFG-M2, Trp-245 provided a hydrophobic sugar-binding platform in subsite +2. However, AxMan113A lacked aromatic residues near this subsite. This special structural feature may result in the enzyme with no transglycosylation activity.
Two different M2 electron-density maps were observed on the surface of AxMan113A–E223A-M2, which is far away from the catalytic sites (Fig. 4). A number of structural studies have revealed that carbohydrates are not only bound on carbohydrate-binding modules, but also observed on one or more surface-binding sites (SBSs). Compared with carbohydrate-binding modules, carbohydrates binding in SBSs are often through a flexible linker (35, 36). Most of SBSs existed in α-amylases, other amylolytic enzymes, transglycosidases, xylanases, and other GHs. To date, there is no report on SBSs of GH family 113. The superposition of AxMan113A–E223A-M2 on AaManA showed a position change in some amino acids near the surface-binding site. In chain A, the side chain of AxMan113A–Asp-73 shifted to the binding site, which formed two hydrogen bonds with mannose. Similarly, AxMan113A–Lys-76 and AxMan113A–Asn-134 were also hydrogen bonded with the mannose. However, AaManA–Glu-81, AaManA–His-84, and AaManA–Gly-142 in the same position revealed no interaction with M2 (Fig. S8A). In chain B, Gln-58 and Asp-66 in a loop of AxMan113A, equivalent to Gly-66 and Asp-74 in AaManA, were flipped 90°, forming a “tweezer” binding linker that can target more mannose molecules efficiently (Fig. S8B) (37, 38). It has been reported that a β-1,4-mannanase from Cryptopygus antarcticus (PDB entry 4OOZ) and several α-amylases have surface-binding sites (37–39). Moreover, mutations of the key amino acids near surface-binding sites decreased enzyme activity. To investigate the significance of these residues, site-directed mutagenesis was performed (Fig. S8C). The specific activity of six mutants (D66A, D73A, K76A, Q58A, N134A, and D65A) decreased, especially that of Q58A and D73A, which retained 56.8 and 82.7% of the initial enzyme activity, respectively. This indicated that these six residues play key roles in binding with ligands. The catalytic efficiencies (kcat/Km) of Q58A and D73A toward M2 were decreased to 46.1 and 53.8% of WT, respectively. It could be deduced that residues around the surface-binding sites have an effect on M2 affinity.
Rational design is an important method to modulate enzyme properties, in an attempt to understand structural information. Many GHs, such as β-gal, β-glycosidase, and α-l-arabinofuranosidase, have been modified through protein engineering (40–42). However, protein engineering of GH family 113 β-1,4-mannanases has never been reported. There were some differences in the structures of AxMan113A compared with other GH family 113 members. On the basis of the structural comparison of AxMan113A–E223A-M2 to AaManA–ManIFG-M2, three residues (Trp-239, Asn-237, and Lys-238) were identified in subsites +1 and +2 of AxMan113A, which had a unique amino acid arrangement (Fig. 6). Trp-239 in subsite +1 had a larger side chain and extended into the groove, forming a semi-enclosed space, which impeded the accommodation of mannose units. However, Trp-239 formed hydrophobic interactions with −2 and +2 mannoses. Thus, mutant W239Y was designed to reduce steric hindrance and retain the hydrophobic sugar-binding platform. W239A was designed to open the catalytic cleft. N237W not only retained the hydrogen bond, but also created an indole ring. K238A eliminated the side chain of Lys and partly opened the groove of subsite +2. Mutant W239A lost most of its activity, confirming that the hydrophobic interaction is crucial for binding with ligands. However, β-1,4-mannanase activity of W239Y increased significantly to 136.4% of the WT. Mutant N237W enhanced affinity to the +2 mannose, and enzyme activity was improved to 163.4%. The enzyme activity of K238A was improved to 212.9% of the WT (Fig. 6E). It has been reported that increasing intramolecular hydrophobic interactions can improve structure stability (43, 44). Herein, hydrophilic residue Asn-237 was mutated to hydrophobic tryptophan, forming a more favorable hydrophobic region with Trp-273, which is beneficial for substrate binding. Indeed, compared with WT, mutant N237W displayed the highest activity in acidic and higher temperature conditions (Fig. 7). According to the result of sequence alignment (Fig. 1), several polar residues (Cys, Ser, and Tyr) exist in six GH family 113 β-1,4-mannanases, but were replaced by nonpolar residue Val-139 in AxMan113A. Mutant V139C showed an acidic shift of 1.0 unit of optimal pH. In general, polar group burial makes a large contribution to structure stability (45, 46). Mutating the nonpolar residue Val-139 to a polar residue Cys might be one reason for the optimal pH change. These results provided novel insights into the mechanism of GH family 113 members, which might have great significance in improving the hydrolysis efficiency of β-1,4-mannanases.
In conclusion, we performed structural and biochemical analyses to provide an in-depth description of the catalytic mechanism of GH family 113 members. Analysis of the complex structures revealed at least six binding sites (−4 to +2) in the groove, but Lys-238 and Trp-239 were located near subsites +1 and +2, blocking the catalytic cleft. This prominent structural feature can mostly accommodate mannose units at subsites −4 to −1. Site-directed mutagenesis yielded a series of mutations designed to affect the enzyme's properties. Mutants K238A and W239Y successfully opened the catalytic cleft and their enzyme activities were improved to 212.9 and 163.4% of the WT, respectively. In addition, two M2-binding sites were discovered on the surface of AxMan113A, never before reported in GH family 113 members. Kinetic analysis of key residues near the surface sites suggested these sites' important role in substrate binding. The structural information of AxMan113A not only provides further insights into its catalytic mechanism, but also a direction for protein engineering in GH family 113 members.
Experimental procedures
Cloning, expression, and purification
The β-1,4-mannanase gene (designated AxMan113A) from A. xylanus NBRC 15112 (GenBankTM accession number BAM48369.1) was amplified by PCR with primers AxMan113A-up and AxMan113A-down (Table S1). After digestion with EcoRI and NotI, the PCR products were purified and inserted into vector pET-28a (Novagen, USA). Mutants E143A, E223A, Q58A, D65A, D66A, D73A, K76A, W95A, R96A, V139C, K175A, K175H, N237A, N237W, K238A, W239A, W239Y, and W273A were created in AxMan113A using the Fast Mutagenesis System site-directed mutagenesis kit (TransGen Biotech, China) with the primers listed in Table S1. All transformants were confirmed by DNA sequencing.
The recombinant plasmid pET28a–AxMan113A was transformed into E. coli Rosetta (DE3) for expression. Seed cultures of E. coli harboring AxMan113A were incubated in Luria Bertani (LB) medium containing 50 μg/ml of kanamycin at 37 °C on a rotary shaker at 200 rpm to an A600 of 0.6–0.8. Isopropyl β-d-1-thiogalactopyranoside (1 mm) was added to induce expression at 30 °C overnight. Cells were harvested by centrifugation at 10,000 × g for 5 min. The precipitate was suspended in buffer A (20 mm imidazole, 20 mm Tris-HCl, pH 8.0, 500 mm NaCl) and disrupted by ultrasonication. The cell debris was centrifuged at 10,000 × g for 10 min and the supernatant was loaded into a Ni-IDA column (1 × 5 cm; GE Life Sciences) pre-equilibrated with buffer A. The column was washed with buffer A followed by buffer B (50 mm imidazole, 20 mm Tris-HCl, pH 8.0, 500 mm NaCl) and then eluted with buffer C (200 mm imidazole, 20 mm Tris-HCl, pH 8.0, 500 mm NaCl) at a flow rate of 1.0 ml/min. The eluted enzyme was subjected to a Sephacryl S-100 gel filtration column (1 × 100 cm; GE Life Sciences), eluted with buffer D (20 mm Tris-HCl, pH 8.0, 100 mm NaCl) and concentrated to 10 mg/ml by ultrafiltration for crystallization. The concentration of the protein was measured by the Lowry method (47). All mutants were expressed and purified by the same method.
SDS-PAGE was performed using a 12.5% (w/v) separation gel. The molecular mass of native AxMan113A was determined by gel filtration on a Sephacryl S-100 HR column (40 × 1 cm) pre-equilibrated with buffer D. The protein was eluted at 0.3 ml/min. The standard markers used for calibration were phosphorylase b (97.2 kDa), albumin (66.4 kDa), ovalbumin (44.3 kDa), chymotrypsinogen A (25.7 kDa), and lysozyme (14.3 kDa).
Enzyme characterization
Enzyme assay for AxMan113A was performed using the 3,5-dinitrosalicylic acid (DNS) method (48) with 0.5% (w/v) LBG. Briefly, 100 μl of suitably diluted enzyme was added to 900 μl of LBG solution in 50 mm McIlvaine buffer (pH 6.5). After incubation at 45 °C for 10 min, the reaction was terminated by adding 1 ml of DNS and boiling for 15 min. Enzyme activity in the reaction mixture was immediately measured at 540 nm after the addition of 1.0 ml of sodium potassium tartrate (40%, w/v). One unit of enzyme activity was defined as the amount of enzyme producing 1 μmol of reducing sugars (mannose equivalents) per min. The optimal pH of AxMan113A was determined in various buffers (50 mm): McIlvaine buffer (pH 3.0–7.0), citrate buffer (pH 3.0–6.0), phosphate buffer (pH 6.0–8.0), CHES (pH 8.0–10.0), Gly-NaOH (pH 9.0–10.5), and CAPS (pH 10–11). The pH stability of AxMan113A was examined by determining the residual activity after incubation in these buffers at 30 °C for 30 min. The optimal temperature was determined in the temperature range of 30–70 °C in 50 mm McIlvaine buffer (pH 6.5). The thermostability of AxMan113A was investigated by measuring residual activity after incubation at different temperatures for 30 min in the same buffer. All measurements were performed in triplicate.
Substrate specificity and hydrolysis properties
Substrate specificity toward 0.5% (w/v) of various mannans: konjac powder, LBG, and guar gum, was determined using the DNS method (48). HPLC with a refractive index detector was used to evaluate the substrate specificity of AxMan113A toward 0.1% (w/v) manno-oligosaccharides M2, M3, M4, M5, and M6 (49). Suitably diluted AxMan113A and manno-oligosaccharides were incubated at 45 °C in 50 mm McIlvaine buffer (pH 6.5) for 10 min. A 10-μl aliquot of the mixture was then injected into a Shodex sugar KS-802 (4.5 × 250 mm) column and eluted by mobile phase (water) at a flow rate of 0.8 ml/min. The temperatures of column and detector were 35 and 60 °C, respectively. One unit of enzyme activity was defined as the amount of enzyme that released 1 μmol of manno-oligosaccharides/min. For determination of β-mannosidase activity of AxMan113A, 50 μl of diluted enzyme was added to 200 μl of 3.75 mm pNPM in 50 mm McIlvaine buffer (pH 6.5), then incubated at 45 °C for 10 min. A total of 750 μl of saturated sodium tetraborate was added to terminate the reaction. Enzyme activity in the reaction mixture was measured at 405 nm. One unit of enzyme activity was defined as the amount of enzyme liberating 1 μmol of pNP/min under the described conditions. The degradation of different mannans or manno-oligosaccharides by AxMan113A was detected by TLC. AxMan113A (5 units) was mixed into 1% (w/v) mannans or manno-oligosaccharides in 50 mm McIlvaine buffer (pH 6.5) at 45 °C for 12 or 4 h. The hydrolysis of M2r, M3r, M4r, and M5r by AxMan113A were also analyzed. Purified AxMan113A (5 units) was incubated with 1% (w/v) M2r, M3r, M4r, and M5r under the same conditions for 4 h. Samples were withdrawn at the different time points and boiled for 10 min. The supernatants were spotted on a silica gel plate (Merck Silica Gel 60 F254, Germany) and developed in n-butyl alcohol:acetic acid:water (2:1:1, v/v). The plates were heated at 130 °C for 5 min after spraying with methanol containing 2% (v/v) H2SO4.
Kinetic parameters for manno-oligosaccharides
The Michaelis-Menten parameters of AxMan113A for various manno-oligosaccharides (M2, M3, M4, M5, and M6) were determined by HPLC. Different concentrations of manno-oligosaccharides were incubated with the appropriate amount of AxMan113A at 45 °C in 50 mm McIlvaine buffer (pH 6.5) for 5 min. The mixtures were withdrawn followed by boiling for 5 min. The injection volume was 10 μl and eluted by water at a flow rate of 0.8 ml/min. The temperatures of column and detector were 35 and 60 °C, respectively. The catalytic efficiencies of mutants (Q58A and D73A) toward M2 were determined using the same method. The Km and Vmax values were calculated by nonlinear regression using Grafit software.
Crystallization and data collection
Crystallization experiments were performed in 48-well plates by sitting drop vapor diffusion at 293 K. The crystals were obtained from a drop containing 1 μl of protein solution (10 mg/ml) and 1 μl of reservoir solution (100 mm HEPES sodium, pH 6.8, 15% (v/v) 2-propanol, and 20% (w/v) PEG 4000). Initial crystals of AxMan113A were grown in 100 mm HEPES sodium (pH 7.5), 10% 2-propanol, and 20% PEG 4000. To obtain high-quality crystals, we tried to optimize the initial screening conditions. Better crystals were obtained in the drops by mixing 2 μl of protein solution and 1 μl of reservoir solution at 293 K for 7 days. To obtain complex structures, 5% (w/v) M2, M3, M5, or M6 were added to the mutant AxMan113A–E223A. The mixtures were first incubated at 293 K for 2 h, and then screened under the same conditions as for the apo form.
For the X-ray diffraction experiments, a single crystal was fished out and immediately soaked in cryoprotectant solution (20% (v/v) glycerol under crystallization conditions), and then flash-cooled in liquid nitrogen. The X-ray diffraction data of AxMan113A were collected by beamline BL18U at the Shanghai Synchrotron Research Facility (SSRF, China). X-ray data for other complex crystals were obtained by beamline BL17U at SSRF. All data were indexed, integrated and scaled by the HKL-2000 package (50).
Structure determination and refinement
The structure of AxMan113A was determined by a molecular replacement method using AaManA (sequence identity 43%) as the search model. Phenix.autobuild was used for automatic building. The structures were completed with alternating rounds of manual model building with Coot (51) and Phenix.refine programs (52). The final model was analyzed by MolProbity (53). Structure illustrations were prepared with PyMOL. Ligplus (54) was used to analyze the protein–ligand interactions. The sequence alignments were created with ClustalX2 (55) and ESPript (56).
Author contributions
X. Y. and Y. L. investigation; X. Y. writing-original draft; Z. Q. and S. Y. data curation; Q. Y. and Z. J. supervision.
Supplementary Material
Acknowledgments
We are grateful to the staff of the National Center for Protein Science, Shanghai (NCPSS), and the Shanghai Synchrotron Radiation Facility (SSRF) for assistance with the X-ray diffraction data collection.
This work was supported in part by the Key Program of the National Natural Science Foundation of China Grant 31630096 and Program for Changjiang Scholars Grant T2014055. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S8 and Tables S1 and S2.
The atomic coordinates and structure factors (codes 5YLH, 5YLK, 5Z4T, 5YLI, and 5YLL) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- GH
- glycoside hydrolase
- M2
- mannobiose
- M3
- mannotriose
- M4
- mannotetraose
- M5
- mannopentaose
- M6
- mannohexaose
- LBG
- locust bean gum
- pNPM
- p-nitrophenyl β-d-mannopyranoside
- DP
- degree of polymerization
- DNS
- 3,5-dinitrosalicylic acid
- CHES
- 2-(cyclohexylamino)ethanesulfonic acid
- CAPS
- 3-(cyclohexylamino)-1-propanesulfonic acid
- PEG
- polyethylene glycol
- SBS
- surface-binding site.
References
- 1. Srivastava P. K., and Kapoor M. (2017) Production, properties and applications of endo-β-1,4-mannanases. Biotechnol. Adv. 35, 1–19 10.1016/j.biotechadv.2016.11.001 [DOI] [PubMed] [Google Scholar]
- 2. Moreira L. R., and Filho E. X. (2008) An overview of mannan structure and mannan-degrading enzyme systems. Appl. Microbiol. Biotechnol. 79, 165–178 10.1007/s00253-008-1423-4 [DOI] [PubMed] [Google Scholar]
- 3. Dhawan S., and Kaur J. (2007) Microbial mannanases: an overview of production and applications. Crit. Rev. Biotechnol. 27, 197–216 10.1080/07388550701775919 [DOI] [PubMed] [Google Scholar]
- 4. Yamabhai M., Sak-Ubol S., Srila W., and Haltrich D. (2016) Mannan biotechnology: from biofuels to health. Crit. Rev. Biotechnol. 36, 32–42 10.3109/07388551.2014.923372 [DOI] [PubMed] [Google Scholar]
- 5. Asano I., Hamaguchi K., Fujii S., and Iino H. (2003) In vitro digestibility and fermentation of mannooligosaccharides from coffee mannan. Food Sci. Technol. Res. 9, 62–66 10.3136/fstr.9.62 [DOI] [Google Scholar]
- 6. Daskiran M., Teeter R. G., Fodge D., and Hsiao H. Y. (2004) Evaluation of endo-β-D-mannanase (Hemicell) effects on broiler performance and energy use in diets varying in β-mannan content. Poult. Sci. 83, 662–668 10.1093/ps/83.4.662 [DOI] [PubMed] [Google Scholar]
- 7. Benech R. O., Li X. M., Patton D., Powlowski J., Storms R., Bourbonnais R., Paice M., and Tsang A. (2007) Recombinant expression, characterization, and pulp prebleaching property of a Phanerochaete chrysosporium endo-β-1,4-mannanase. Enzyme Microb. Technol. 41, 740–747 10.1016/j.enzmictec.2007.06.012 [DOI] [Google Scholar]
- 8. Puchart V., and Biely P. (2005) Glycosylation of internal sugar residues of oligosaccharides catalyzed by α-galactosidase from Aspergillus fumigatus. Biochim. Biophys. Acta 1726, 206–216 10.1016/j.bbagen.2005.07.015 [DOI] [PubMed] [Google Scholar]
- 9. Ma Y., Xue Y., Dou Y., Xu Z., Tao W., and Zhou P. (2004) Characterization and gene cloning of a novel β-1,4-mannanase from alkaliphilic Bacillus sp. N16-5. Extremophiles 8, 447–454 10.1007/s00792-004-0405-4 [DOI] [PubMed] [Google Scholar]
- 10. Zhang Y., Ju J., Peng H., Gao F., Zhou C., Zeng Y., Xue Y., Li Y., Henrissat B., Gao G. F., and Ma Y. (2008) Biochemical and structural characterization of the intracellular mannanase AaManA of Alicyclobacillus acidocaldarius reveals a novel glycoside hydrolase family belonging to Clan GH-A. J. Biol. Chem. 283, 31551–31558 10.1074/jbc.M803409200 [DOI] [PubMed] [Google Scholar]
- 11. Vocadlo D. J., and Davies G. J. (2008) Mechanistic insights into glycosidase chemistry. Curr. Opin. Chem. Biol. 12, 539–555 10.1016/j.cbpa.2008.05.010 [DOI] [PubMed] [Google Scholar]
- 12. Davies G., and Henrissat B. (1995) Structures and mechanisms of glycosyl hydrolases. Structure 3, 853–859 10.1016/S0969-2126(01)00220-9 [DOI] [PubMed] [Google Scholar]
- 13. Jin Y., Petricevic M., John A., Raich L., Jenkins H., Portela De Souza L., Cuskin F., Gilbert H. J., Rovira C., Goddard-Borger E. D., Williams S. J., and Davies G. J. (2016) A β-1,4-mannanase with a lysozyme-like fold and a novel molecular catalytic mechanism. ACS Cent. Sci. 2, 896–903 10.1021/acscentsci.6b00232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. You X., Qin Z., Li Y. X., Yan Q. J., Li B., and Jiang Z. Q. (2018) Structural and biochemical insights into the substrate-binding mechanism of a novel glycoside hydrolase family 134 β-mannanase. Biochim. Biophys. Acta 1862, 1376–1388 10.1016/j.bbagen.2018.03.016 [DOI] [PubMed] [Google Scholar]
- 15. Li J. M., and Nie S. P. (2016) The functional and nutritional aspects of hydrocolloids in foods. Food Hydrocolloids 53, 46–61 10.1016/j.foodhyd.2015.01.035 [DOI] [Google Scholar]
- 16. Guo W., Yang C., Cui L., Lin H., and Qu F. (2014) An enzyme-responsive controlled release system of mesoporous silica coated with Konjac oligosaccharide. Langmuir. 30, 243–249 10.1021/la403494q [DOI] [PubMed] [Google Scholar]
- 17. Yoon S. J., Chu D. C., and Raj Juneja L. (2008) Chemical and physical properties, safety and application of partially hydrolized guar gum as dietary fiber. J. Clin. Biochem. Nutr. 42, 1–7 10.3164/jcbn.2008001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Williams R. J., Iglesias-Fernández J., Stepper J., Jackson A., Thompson A. J., Lowe E. C., White J. M., Gilbert H. J., Rovira C., and Davies G. J. (2014) Combined inhibitor free-energy landscape and structural analysis reports on the mannosidase conformational coordinate. Angew. Chem. Int. Ed. Engl. 53, 1087–1091 10.1002/anie.201308334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xia W., Lu H., Xia M., Cui Y., Bai Y., Qian L., Shi P., Luo H., and Yao B. (2016) A novel glycoside hydrolase family 113 endo-β-1,4-mannanase from Alicyclobacillus sp. strain A4 and insight into the substrate recognition and catalytic mechanism of this family. Appl. Environ. Microbiol. 82, 2718–2727 10.1128/AEM.04071-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Katrolia P., Yan Q., Zhang P., Zhou P., Yang S., and Jiang Z. (2013) Gene cloning and enzymatic characterization of an alkali-tolerant endo-1,4-β-mannanase from Rhizomucor miehei. J. Agric. Food. Chem. 61, 394–401 10.1021/jf303319h [DOI] [PubMed] [Google Scholar]
- 21. Luo Z., Miao J., Li G., Du Y., and Yu X. (2017) A recombinant highly thermostable β-1,4-mannanase (ReTMan26) from thermophilic Bacillus subtilis (TBS2) expressed in Pichia pastoris and its pH and temperature stability. Appl. Biochem. Biotechnol. 182, 1259–1275 10.1007/s12010-017-2397-4 [DOI] [PubMed] [Google Scholar]
- 22. Sakai K., Mochizuki M., Yamada M., Shinzawa Y., Minezawa M., Kimoto S., Murata S., Kaneko Y., Ishihara S., Jindou S., Kobayashi T., Kato M., and Shimizu M. (2017) Biochemical characterization of thermostable β-1,4-mannanase belonging to the glycoside hydrolase family 134 from Aspergillus oryzae. Appl. Microbiol. Biotechnol. 101, 3237–3245 10.1007/s00253-017-8107-x [DOI] [PubMed] [Google Scholar]
- 23. Kumagai Y., Yamashita K., Tagami T., Uraji M., Wan K., Okuyama M., Yao M., Kimura A., and Hatanaka T. (2015) The loop structure of Actinomycete glycoside hydrolase family 5 mannanases governs substrate recognition. FEBS J. 282, 4001–4014 10.1111/febs.13401 [DOI] [PubMed] [Google Scholar]
- 24. Le Nours J., Anderson L., Stoll D., Stålbrand H., and Lo Leggio L. (2005) The structure and characterization of a modular endo-β-1,4-mannanase from Cellulomonas fimi. Biochemistry 44, 12700–12708 10.1021/bi050779v [DOI] [PubMed] [Google Scholar]
- 25. Vuong T. V., and Wilson D. B. (2010) Glycoside hydrolases: catalytic base/nucleophile diversity. Biotechnol. Bioeng. 107, 195–205 10.1002/bit.22838 [DOI] [PubMed] [Google Scholar]
- 26. Rosengren A., Reddy S. K., Sjöberg J. S., Aurelius O., Logan D. T., Kolenová K., and Stålbrand H. (2014) An Aspergillus nidulans β-mannanase with high transglycosylation capacity revealed through comparative studies within glycosidase family 5. Appl. Microbiol. Biotechnol. 98, 10091–10104 10.1007/s00253-014-5871-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Couturier M., Roussel A., Rosengren A., Leone P., Stålbrand H., and Berrin J. G. (2013) Structural and biochemical analyses of glycoside hydrolase families 5 and 26 β-(1,4)-mannanases from Podospora anserina reveal differences upon manno-oligosaccharide catalysis. J. Biol. Chem. 288, 14624–14635 10.1074/jbc.M113.459438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Larsson A. M., Anderson L., Xu B., Muñoz I. G., Usón I., Janson J. C., Stålbrand H., and Ståhlberg J. (2006) Three-dimensional crystal structure and enzymic characterization of β-mannanase Man5A from blue mussel Mytilus edulis. J. Mol. Biol. 357, 1500–1510 10.1016/j.jmb.2006.01.044 [DOI] [PubMed] [Google Scholar]
- 29. Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., and Henrissat B. (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495 10.1093/nar/gkt1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lyu Q., Shi Y., Wang S., Yang Y., Han B., Liu W., Jones D. N., and Liu W. (2015) Structural and biochemical insights into the degradation mechanism of chitosan by chitosanase OU01. Biochim. Biophys. Acta 1850, 1953–1961 10.1016/j.bbagen.2015.06.011 [DOI] [PubMed] [Google Scholar]
- 31. Fukamizo T., Juffer A. H., Vogel H. J., Honda Y., Tremblay H., Boucher I., Neugebauer W. A., and Brzezinski R. (2000) Theoretical calculation of pKa reveals an important role of Arg-205 in the activity and stability of Streptomyces sp. N174 chitosanase. J. Biol. Chem. 275, 25633–25640 10.1074/jbc.M002574200 [DOI] [PubMed] [Google Scholar]
- 32. Teze D., Hendrickx J., Czjzek M., Ropartz D., Sanejouand Y. H., Tran V., Tellier C., and Dion M. (2014) Semi-rational approach for converting a GH1 β-glycosidase into a β-transglycosidase. Protein Eng. Des. Sel. 27, 13–19 10.1093/protein/gzt057 [DOI] [PubMed] [Google Scholar]
- 33. Zakariassen H., Hansen M. C., Jøranli M., Eijsink V. G., and Sørlie M. (2011) Mutational effects on transglycosylating activity of family 18 chitinases and construction of a hypertransglycosylating mutant. Biochemistry 50, 5693–5703 10.1021/bi2002532 [DOI] [PubMed] [Google Scholar]
- 34. Horn S. J., Sikorski P., Cederkvist J. B., Vaaje-Kolstad G., Sørlie M., Synstad B., Vriend G., Vårum K. M., and Eijsink V. G. (2006) Costs and benefits of processivity in enzymatic degradation of recalcitrant polysaccharides. Proc. Natl. Acad. Sci. U.S.A. 103, 18089–18094 10.1073/pnas.0608909103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cuyvers S., Dornez E., Delcour J. A., and Courtin C. M. (2012) Occurrence and functional significance of secondary carbohydrate binding sites in glycoside hydrolases. Crit. Rev. Biotechnol. 32, 93–107 10.3109/07388551.2011.561537 [DOI] [PubMed] [Google Scholar]
- 36. Cockburn D., Wilkens C., Ruzanski C., Andersen S., Willum Nielsen J., Smith A. M., Field R. A., Willemoës M., Abou Hachem M., and Svensson B. (2014) Analysis of surface binding sites (SBSs) in carbohydrate active enzymes with focus on glycoside hydrolase families 13 and 77-a mini-review. Biologia 69, 705–712 10.2478/s11756-014-0373-9 [DOI] [Google Scholar]
- 37. Nielsen P. K., Bønsager B. C., Berland C. R., Sigurskjold B. W., and Svensson B. (2003) Kinetics and energetics of the binding between barley α-amylase/subtilisin inhibitor and barley α-amylase 2 analyzed by surface plasmon resonance and isothermal titration calorimetry. Biochemistry 42, 1478–1487 10.1021/bi020508+ [DOI] [PubMed] [Google Scholar]
- 38. Robert X., Haser R., Gottschalk T. E., Ratajczak F., Driguez H., Svensson B., and Aghajari N. (2003) The structure of barley α-amylase isozyme 1 reveals a novel role of domain C in substrate recognition and binding. Structure 11, 973–984 10.1016/S0969-2126(03)00151-5 [DOI] [PubMed] [Google Scholar]
- 39. Kim M. K., An Y. J., Song J. M., Jeong C. S., Kang M. H., Kwon K. K., Lee Y. H., and Cha S. S. (2014) Structure-based investigation into the functional roles of the extended loop and substrate-recognition sites in an endo-β-1,4-d-mannanase from the Antarctic springtail, Cryptopygus antarcticus. Proteins 82, 3217–3223 10.1002/prot.24655 [DOI] [PubMed] [Google Scholar]
- 40. Wu Y., Yuan S., Chen S., Wu D., Chen J., and Wu J. (2013) Enhancing the production of galacto-oligosaccharides by mutagenesis of Sulfolobus solfataricus β-galactosidase. Food Chem. 138, 1588–1595 10.1016/j.foodchem.2012.11.052 [DOI] [PubMed] [Google Scholar]
- 41. Feng H. Y., Drone J., Hoffmann L., Tran V., Tellier C., Rabiller C., and Dion M. (2005) Converting a β-glycosidase into a β-transglycosidase by directed evolution. J. Biol. Chem. 280, 37088–37097 10.1074/jbc.M502873200 [DOI] [PubMed] [Google Scholar]
- 42. Arab-Jaziri F., Bissaro B., Dion M., Saurel O., Harrison D., Ferreira F., Milon A., Tellier C., Faure R., and O'Donohue M. J. (2013) Engineering transglycosidase activity into a GH51 α-l-arabinofuranosidase. New Biotechnol. 30, 536–544 10.1016/j.nbt.2013.04.002 10.1016/j.nbt.2013.04.002 [DOI] [PubMed] [Google Scholar]
- 43. Golovanov A. P., Efremov R. G., Jaravine V. A., Vergoten G., Kirpichnikov M. P., and Arseniev A. S. (1998) A new method to characterize hydrophobic organization of proteins: application to rational protein engineering of barnase. J. Biomol. Struct. Dyn. 4, 673–687 [DOI] [PubMed] [Google Scholar]
- 44. Murphy G. S., Mills J. L., Miley M. J., Machius M., Szyperski T., and Kuhlman B. (2012) Increasing sequence diversity with flexible backbone protein design: the complete redesign of a protein hydrophobic core. Structure 20, 1086–1096 10.1016/j.str.2012.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Takano K., Yamagata Y., and Yutani K. (2001) Contribution of polar groups in the interior of a protein to the conformational stability. Biochemistry 40, 4853–4858 10.1021/bi002792f [DOI] [PubMed] [Google Scholar]
- 46. Pace C. N. (2001) Polar group burial contributes more to protein stability than nonpolar group burial. Biochemistry 40, 310–313 10.1021/bi001574j [DOI] [PubMed] [Google Scholar]
- 47. Lowry O. H., Rosebrough N. J., Farr A. L., and Randall R. J. (1951) Protein measurement with the folin phenol reagent. J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 48. Miller G. L. (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31, 426–428 10.1021/ac60147a030 [DOI] [Google Scholar]
- 49. Li Y. X., Liu Y., Yan Q. J., Yang S. Q., and Jiang Z. Q. (2015) Characterization of a novel glycoside hydrolase family 5 β-mannosidase from Absidia corymbifera with high transglycosylation activity. J. Mol. Catalysis B Enzymatic 122, 265–274 10.1016/j.molcatb.2015.09.018 [DOI] [Google Scholar]
- 50. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- 51. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., and Adams P. D. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 10.1107/S0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Laskowski R. A., and Swindells M. B. (2011) LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 51, 2778–2786 10.1021/ci200227u [DOI] [PubMed] [Google Scholar]
- 55. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., and Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 56. Robert X., and Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.