Abstract
The antiviral cGMP–AMP (cGAMP)–stimulator of interferon genes (STING) pathway is well characterized in mammalian cells. However, whether this pathway also plays a role in insect antiviral immunity is unknown. In this study, we found that cGAMP is produced in silkworm (Bombyx mori) cells infected with nucleopolyhedrovirus (NPV). In searches for STING-related sequences, we identified BmSTING, a potential cGAMP sensor in B. mori. We observed that BmSTING overexpression effectively inhibits NPV replication in silkworm larvae, whereas dsRNA-mediated BmSTING knockdown resulted in higher viral load. Cleavage and nuclear translocation of BmRelish, a NF-κB–related transcription factor, was also observed when BmSTING was overexpressed and was enhanced by cGAMP stimulation or viral infection of B. mori larvae. Moreover, we identified a caspase-8–like protein (BmCasp8L) as a BmSTING-interacting molecule and as a suppressor of BmSTING-mediated BmRelish activation. Interestingly, cGAMP stimulation decreased BmCasp8L binding to BmSTING and increased BmRelish activity. Of note, an interaction between death-related ced-3/Nedd2-like caspase (BmDredd) and BmSTING promoted BmRelish cleavage for efficient antiviral signaling and protection of insect cells from viral infection. Our findings have uncovered BmSTING as a critical mediator of antiviral immunity in the model insect B. mori and have identified several BmSTING-interacting proteins that control antiviral defenses.
Keywords: immunosuppressor, immunology, infection, silkworm, signal transduction, virus, stimulator of interferon genes (STING), cyclic GMP-AMP (cGAMP), caspase-8-like (Casp8L), NF-kappaB, antiviral
Introduction
Over recent decades, the innate immune responses elicited by nucleic acids derived from viruses, bacteria, or protozoan parasites have been intensively studied in mammals (1, 2). An endoplasmic reticulum protein, stimulator of interferon genes (STING;4 also known as MITA, MPYS, or ERIS), was identified as a critical mediator for inducing the production of type I interferons (IFNs) and other proinflammatory cytokines in response to viral DNA (3–7). Later studies demonstrated that cGMP–AMP (cGAMP) synthesized in DNA virus–infected cells functions as a second messenger to stimulate STING (8). Activated STING then recruits the protein kinases IκB kinase (IKK) and TANK-binding kinase 1 (TBK1), which in turn activate NF-κB and interferon regulatory factor 3 (IRF3) to induce potent IFN production (9). In addition to the antiviral immune response to DNA viruses, STING is also involved in RNA virus–elicited responses and can directly bind cyclic dinucleotides produced by certain bacteria (10). The essential role of the STING pathway has been widely recognized in antiviral innate immunity; however, whether it exists or functions in invertebrates is unknown.
Recent studies in insects have indicated that in addition to RNAi-mediated antiviral mechanisms, Toll and immune deficiency (IMD) pathways, which are canonical NF-κB–dependent signaling pathways and well characterized in response to bacteria or fungi infection, also confer antiviral activity (11–14), although the mechanism is not clear. CG1667, which encodes a homolog of mammalian STING in Drosophila, has been reported to be up-regulated 48 h after Drosophila C virus infection (15). Its expression level was also increased in WT flies compared with microbiota-treated ones and decreased in mutants bearing deficiency in Relish, the central transcriptional factor in the IMD pathway (16), suggesting that the insect STING homolog may be involved in responses to viral or bacterial infections. Considering the prominent role of STING in antiviral immunity to DNA viruses, a DNA virus, nucleopolyhedrovirus (NPV), and its natural host, Bombyx mori, were used in this study to investigate insect STING functions. BmNPV belongs to the Baculoviridae family, which has been proven to evoke type I IFN production through STING in mammalian cells (10). In addition, a genome-wide study of silkworm transcription factors revealed that an NF-κB–like protein, BmRel, was up-regulated 6 h after BmNPV infection (17). Expression of several antimicrobial peptides, including gloverin-1, gloverin-2, gloverin-3, gloverin-4, and lysozyme, was also induced in silkworm fat body and hemocytes by BmNPV infection (18). Gloverin has been identified as an antiviral protein combating baculovirus infection and highly expressed in BmNPV-resistant silkworm strains (19, 20). Therefore, it is of interest to investigate whether activation of the NF-κB signaling pathway by viral infection in insects is mediated by certain unknown molecules, such as putative STING.
In this study, we found that cGAMP was produced in cells after BmNPV infection, and it promoted the expression of antimicrobial peptides, such as BmCecropinA (BmCecA) and BmCecropinB (BmCecB). We then identified BmSTING, the potential sensor of cGAMP in silkworm. Knockdown of BmSTING in BmE and BmN4-SID1 cells resulted in less activation of BmRelish and rendered cells more susceptible to BmNPV infection. Furthermore, we identified BmCaspase-8–like (BmCasp8L), a paralogue of BmDredd that is responsible for proteolytic activation of BmRelish, as an interacting protein of BmSTING through immunoprecipitation coupled with LC-MS/MS analysis. Interestingly, BmCasp8L lacks the caspase domain, and it attenuated immune responses by inhibiting BmDredd-mediated cleavage of BmRelish. Our findings reveal a molecular mechanism of the regulation of antiviral immunity in insects.
Results
cGAMP is produced in BmNPV-infected cells and induces antiviral immunity
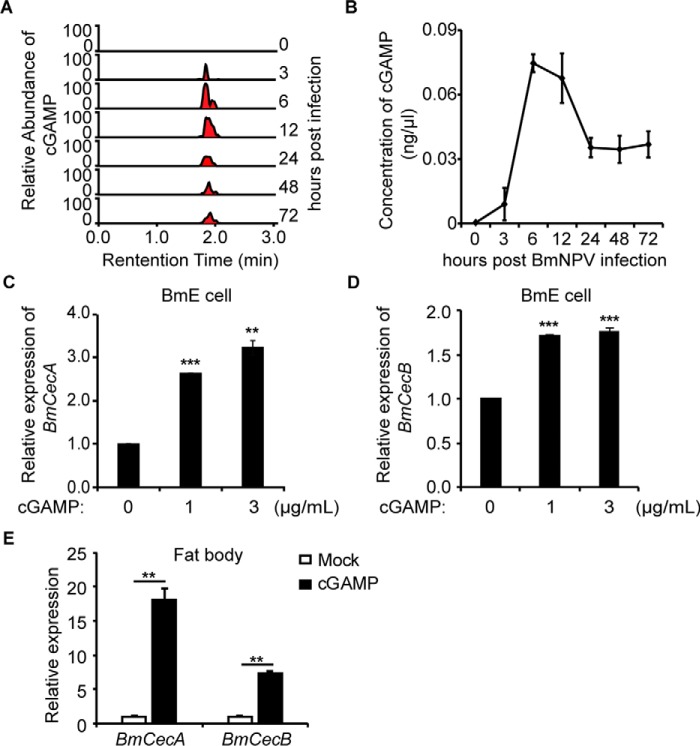
An endogenous second messenger, cGAMP, was reported to be induced by DNA and DNA viruses (21, 22). To determine whether DNA viral infection leads to the production of an activator in insect cells that triggers the innate immunity, we infected BmE cells with BmNPV-GFP and then performed a LC coupled with tandem MS (LC-MS/MS) analysis of the cytosolic extract. In-depth examination of the MS spectra revealed one ion with an m/z of 675.11 (Z = 1+), and collision-induced dissociation fragmentation of this ion (Fig. S1A) revealed several prominent ions with m/z values consistent with those expected of cGAMP (Fig. S1B). Moreover, quantitative MS showed that the abundance of ions representing cGAMP varied at different times after BmNPV-GFP infection. Based on the calibration curves of chemical-synthesized cGAMP, we found that the concentrations of cytosolic cGAMP increased upon viral infection and reached a maximal level at ∼6 h postinfection (hpi). The concentrations of cGAMP then decreased and sustained from 24 to 72 hpi (Fig. 1, A and B). These results demonstrate the production of cGAMP in BmE cells after BmNPV infection.
Figure 1.
Identification of cGAMP in BmNPV-infected cells. A and B, relative abundance of cGAMP in BmNPV-infected cells at different times. Fractions of cell extracts from BmNPV-infected cells were analyzed for the presence of cGAMP by selective reaction monitoring of the expected ions (n = 5). C and D, the levels of BmCecA and BmCecB mRNA were increased in BmE cells after cGAMP stimulation. BmE cells were transfected with cGAMP (3 μg/ml) for 9 h, followed by qRT-PCR analysis (n = 3). E, relative levels of BmCecA and BmCecB mRNA in the fat body of larvae injected with X-treme/cGAMP complexes and X-treme. The relative mRNA levels in the treated fat body of larvae were increased compared with the mRNA levels in untreated larvae (n = 6). **, p < 0.01; ***, p < 0.001. Error bars, S.D.
To determine whether cGAMP activates innate immunity in insects, we examined the mRNA levels of the antibacterial peptides BmCecA and BmCecB. Quantitative RT-PCR analysis showed that chemically synthesized cGAMP induced BmCecA and BmCecB in BmE cells (Fig. 1, C and D) as well as in the fat body of silkworm larvae (Fig. 1E).
BmSTING mediates antiviral effects against BmNPV
Because STING has been proven to be a direct immune sensor of cGAMP (23), we suspected that there exists a STING-homologous molecule in silkworm, which might play a similar role in antiviral immune response as in mammals. Using human STING sequence as the query to BLAST against the silkworm genome database, we identified a sequence (XP_004923946) encoding a putative protein comprising 329 amino acids with an estimated molecular mass of 37.9 kDa. Full-length BmSTING was subsequently cloned by 5′-RACE. The C-terminal domain (amino acids 126–318) of BmSTING, which consists of five α helices and six β sheets, presumably is highly conserved with human and mouse STINGs, whereas the N-terminal sequence is more variable (Fig. S2A). Residues that are thought to be critical for binding its ligands and activation, such as Ser-133, Tyr-138, and Asn-224, which correspond to Ser-162, Tyr-167, and Asn-241, respectively, in human STING are also conserved in BmSTING.
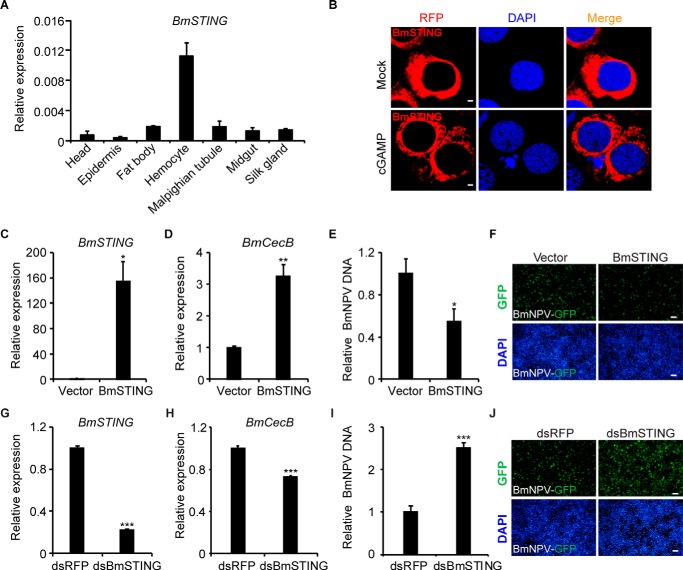
To analyze sequence homology, STING sequences from B. mori, Drosophila melanogaster, Danio rerio, Xenopus tropicalis, Gallus gallus, Rattus norvegicus, Mus musculus, Bos taurus, Canis lupus, Macaca mulatta, Pan troglodytes, and Homo sapiens were used for phylogenetic analysis (Fig. S2B). The sequences were primarily clustered into two groups; all of the sequences from vertebrates were clustered together, whereas BmSTING and DmSTING (CG1667) formed a separate branch. Immunoblotting confirmed the molecular weight of BmSTING (Fig. S2C). Although BmSTING was ubiquitously expressed in a variety of tissues of silkworm larvae, prominent expression was noticed in hemocytes (Fig. 2A). Interestingly, we also found that under cGAMP induction, BmSTING predominantly congregated to the perinucleus from the cytoplasm (Fig. 2B), which was similarly observed for the localization of STING upon viral infection or interferon stimulatory DNA (ISD) transfection in mammalian cells (4). Taken together, these results suggest that BmSTING may respond to cGAMP and thus be involved in antiviral immunity.
Figure 2.
BmSTING expression affects the host defense against BmNPV virus. A, expression profile of BmSTING in B. mori tissues. BmSTING was found to be ubiquitously expressed in a variety of tissues, as determined by qRT-PCR analysis. B, BmSTING tagged with RFP was primarily found in the cytoplasm. cGAMP induced predominant congregation of BmSTING to the perinucleus in cells. Cells were fixed and imaged by confocal microscopy (scale bar, 2 μm). C and D, overexpression of BmSTING induced BmCecB expression. E and F, BmSTING is a strong antiviral protein. BmE cells were treated with BmNPV at an MOI of 1, total genomes were extracted, and relative viral DNA level was determined by qPCR at 72 hpi. Fluorescence microscopy (GFP) of BmSTING-overexpressing cells or controls after infection with BmNPV (MOI 1). G and H, BmCecB was down-regulated by the RNAi of BmSTING. qRT-PCR analysis of BmSTING and BmCecB mRNA in dsBmSTING-treated or control BmE cells (scale bar, 100 μm). I and J, cells treated with dsBmSTING were more susceptible to BmNPV infection. BmE cells were treated with dsRNA and infected with BmNPV at an MOI of 1, total genomes were extracted, and relative viral DNA level was determined by qPCR at 72 hpi. Fluorescence microscopy (GFP) of BmSTING RNAi or control cells (scale bar, 100 μm). *, p < 0.05; **, p < 0.01; ***, p < 0.001. Error bars, S.D.
To determine whether BmSTING is involved in regulating innate immunity, we first examined whether overexpression of BmSTING contributes to the up-regulation of antimicrobial peptides, which are downstream effector molecules of insect immune pathways. As shown in Fig. 2 (C and D), overexpression of BmSTING significantly induced the expression of BmCecB. We then evaluated whether overexpression of BmSTING protects cells from the viral infection. BmE cells were infected with BmNPV-GFP at a multiplicity of infection (MOI) of 1. Viral titer measurements showed that overexpression of BmSTING resulted in a decrease of viral DNA imported to cells by 2-fold compared with that in control cells at 72 hpi (Fig. 2E). Fluorescence microscopy also revealed that virus production was decreased in BmSTING-expressing cells compared with the control cells (Fig. 2F).
To further characterize the function of BmSTING, RNAi was performed to down-regulate the expression of endogenous BmSTING (Fig. 2G). BmE cells from WT and BmSTING-knockdown cells were then infected with BmNPV-GFP at an MOI of 1 for up to 72 hpi. Our result indicated that knockdown of BmSTING in BmE cells modestly reduced the expression of BmCecB (Fig. 2H). However, those cells were extremely vulnerable to viral infection, as more progeny viruses were produced in dsBmSTING-treated cells than in dsRFP-treated cells (Fig. 2, I and J). The same result was also observed in BmN4-SID1 cells (Fig. S3).
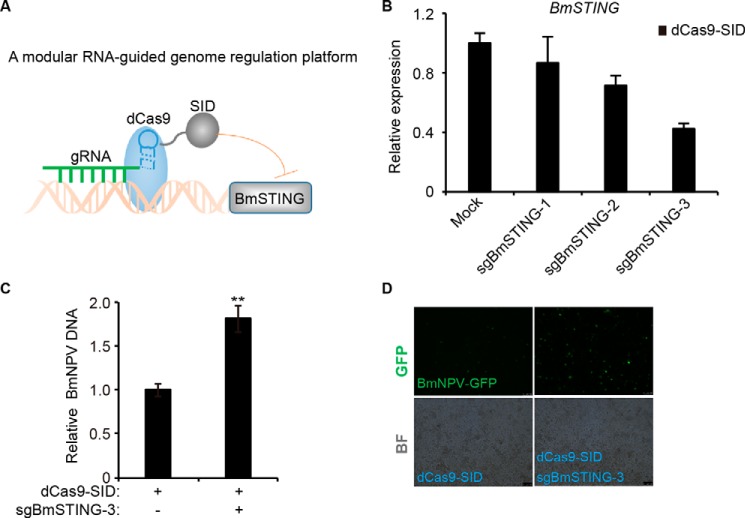
To exclude off-target effects of RNAi, we used the CRISPRi system (24), in which dCas9 was fused to effector domains to serve as an RNA-guided DNA-binding protein to target gene expression (Fig. 3A). Three gRNAs were constructed to target the BmSTING promoter. After co-transfection with dCas9, the gRNA that showed the highest degree of repression achieved by CRISPRi (sgBmSTING-3) was selected for further study (Fig. 3B). The expression of BmSTING was reduced more than 50% (Fig. 3C), and more progeny viruses were produced in dCas9-SID and sgBmSTING-3–coexpressing cells than in dCas9-SID–expressing cells (Fig. 3, D and E). These findings suggested that BmSTING promoted antiviral immune response.
Figure 3.
Targeting BmSTING by CRISPR fusion system resulted in more susceptibility to BmNPV infection. A, the modular CRISPR fusion system by which dCas9 was fused with SID domains serves as an RNA-guided DNA-binding protein to target BmSTING DNA sequence. B, the dCas9-SID fusion protein efficiently silenced BmSTING expression. Three sgRNAs targeting BmSTING were transfected into BmE cells with dCas9-SID, respectively. The expression of BmSTING was quantified by qRT-PCR. C, CRISPRi resulted in more susceptibility to BmNPV infection. BmE cells were treated with sgBmSTING-3 together with dCas9-SID and infected with BmNPV at an MOI of 0.5. Relative viral DNA level was determined by qPCR at 72 hpi. D, fluorescence microscopy (GFP) of STING-knocked-down or control cells (scale bar, 100 μm). The data are displayed as mean S.D. (error bars) for three independent experiments. **, p < 0.01.
BmSTING is essential for antiviral defense in vivo
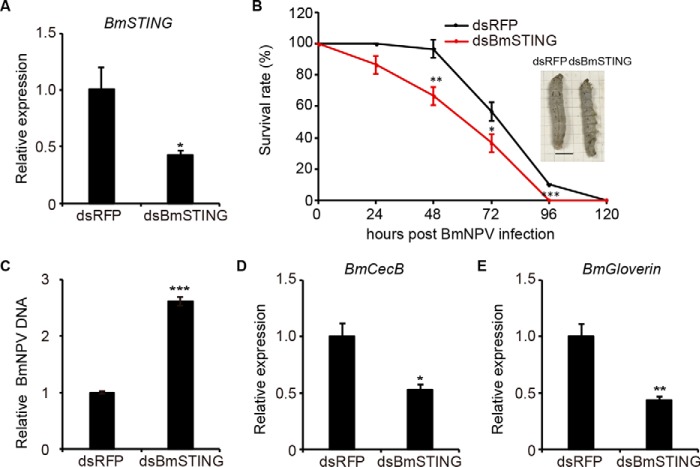
To investigate whether BmSTING plays a role in antiviral immunity in vivo, we performed RNAi to down-regulate the expression of endogenous BmSTING in the silkworms. The expression level of BmSTING was reduced by 58%, confirmed by qPCR (Fig. 4A). Then we injected BmNPV to RNAi-treated larvae and analyzed viral DNA replication with qPCR for up to 120 hpi. We found that the survival rate of animals with decreased BmSTING expression was lower than the controls at each time point sampled (Fig. 4B). Additionally, more progeny viruses were produced in dsBmSTING-treated silkworms compared with dsRFP-treated ones (Fig. 4C). The expression of BmCecB (Fig. 4D) and BmGloverin (Fig. 4E) was also reduced. Overall, these results showed that the BmSTING knockdown resulted in higher susceptibility to BmNPV infection in silkworm larvae, suggesting that BmSTING plays an important antiviral role in vivo.
Figure 4.
BmSTING expression affects the host defense against BmNPV virus in vivo. A, BmSTING was down-regulated by dsRNA injected into silkworm larvae (n = 10). B, the survival rate of silkworm injected with dsBmSTING or dsRFP after BmNPV infection (n = 30). Nm DZ silkworms treated with dsRNA against BmSTING were infected with BmNPV (106 pfu/ml) by injection. C–E, qRT-PCR analysis of viral DNA (C), BmCecB (D), and BmGloverin mRNA (E) in BmSTING RNAi or control silkworm. The data are displayed as mean ± S.D. (error bars) for three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
BmSTING promotes BmRelish cleavage
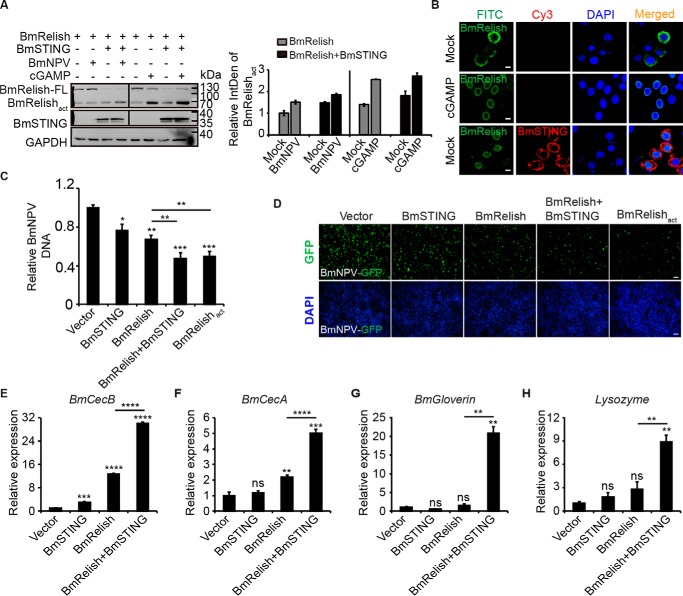
Because both cGAMP treatment and BmSTING overexpression led to induction of antimicrobial peptides, which is usually tightly controlled by NF-κB-like transcriptional factors, we next investigated whether BmRelish was properly cleaved from its full length (BmRelish-FL) into its active form BmRelishact under the same condition. We found that both BmNPV infection and cGAMP stimulation resulted in the generation of BmRelishact. When BmSTING was overexpressed, the cleavage appeared to be more prominent, even in the absence of stimuli, as indicated by quantitative analysis of relative BmRelishact level (Fig. 5A). It was also noted that more BmRelish was translocated from the cytoplasm to the nucleus (Fig. 5B). Treatment with c-di-GMP, ISD, poly(dA-dT), or poly(I:C) generated a similar result (Fig. S4).
Figure 5.
BmNPV infection activates BmRelish through the cGAMP-BmSTING pathway. A, cGAMP (3 μg/ml) and BmNPV virus were delivered to BmE cells overexpressing BmRelish together with BmSTING or BmRelish alone, and BmRelish protein was then measured by Western blotting. All Western blotting signals were analyzed, and densitometric analysis of the BmRelishact protein bands from the Western blots was performed using ImageJ software. B, confocal analysis of BmE cells overexpressing BmRelish alone or together with BmSTING and treated with cGAMP (cGAMP) or only the same transfection reagent (Mock) (scale bar, 10 μm). C, relative viral DNA level in BmE cells overexpressing both BmRelish and BmSTING, BmRelish, or BmRelishact alone and infected with BmNPV at an MOI of 1 was determined by qPCR at 72 hpi. D, fluorescence microscopy (GFP) of BmE cells overexpressing both BmRelish and BmSTING, BmRelish, or BmRelishact alone and infected with BmNPV at 72 hpi (scale bar, 100 μm). E–H, expression of BmCecB (E), BmCecA (F), BmGloverin (G), and BmLysozyme (H) in BmE cells transfected with BmSTING and BmRelish separately or together. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; ns, not significant. Error bars, S.D.
To examine whether the processing of BmRelish promoted by BmSTING affects viral infections, we evaluated the viral DNA levels in cells overexpressing BmRelish or its active form BmRelishact alone or BmRelish together with BmSTING in BmE cells (Fig. 5C). Cells co-expressing BmSTING and BmRelish as well as cells expressing BmRelishact were much more refractory to the BmNPV infection than cells expressing BmRelish alone, as determined by qRT-PCR (Fig. 5D). Fluorescence images of BmNPV-GFP–positive cells also confirmed that viral production in BmRelishact-expressing cells or BmSTING and BmRelish–co-expressing cells was lower than in BmRelish-expressing cells (Fig. 5E). Consistently, the expression levels of BmCecB, BmCecA, BmGloverin, and BmLysozyme were dramatically increased when BmSTING and BmRelish were co-expressed compared with overexpression of BmRelish alone (Fig. 5, F–I). Collectively, our data indicated that BmSTING promoted the processing of BmRelish to activate antiviral immune responses.
Identification of BmCasp8L as an BmSTING-interacting protein and an attenuator of NF-κB signaling pathway
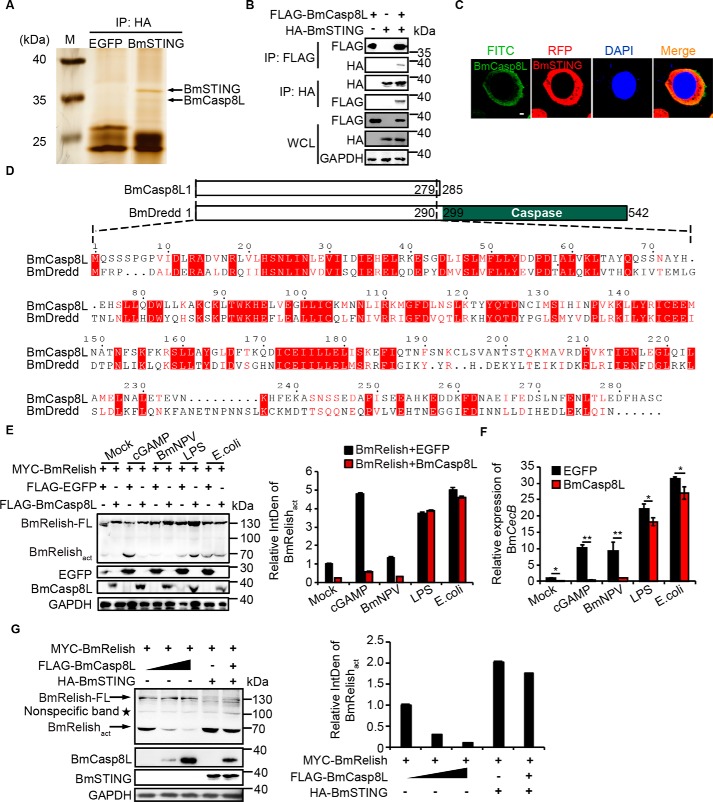
To further reveal the mechanism of STING-mediated NF-κB activation, we employed immunoprecipitation to screen molecules that interact with BmSTING by using HA-tagged BmSTING as a bait. LC-MS/MS analysis identified BmCaspase-8–like (BmCasp8L, XP_012552745) from a protein band with molecular mass about 30 kDa (Fig. 6A and Table 1). To verify the interaction between BmCasp8L and BmSTING, we generated FLAG-tagged BmCasp8L and confirmed that it was co-precipitated with HA-tagged BmSTING in BmE cells (Fig. 6B). BmCasp8l and BmSTING were also found co-localized in the cytoplasm (Fig. 6C).
Figure 6.
Identification of BmCasp8L as a suppressor of the BmSTING-NF-κB pathway. A, BmCasp8L was identified from BmSTING immunoprecipitates by LC-MS/MS. B, association of BmCasp8L and BmSTING in BmE cells. BmCasp8L–BmSTING association was detected by bidirectional co-immunoprecipitation using an anti-HA antibody and immunoblotting with the reciprocal anti-FLAG-HRP antibody or anti-FLAG antibody and immunoblotting with the reciprocal anti-HA-HRP antibody. C, subcellular localization of BmSTING and BmCasp8L in BmE cells (scale bar = 2 μm). D, sequence alignment between BmCasp8L and BmDredd. E, immunoblot of the BmRelishact in BmE cells overexpressing BmRelish alone or together with BmCasp8L and treated with cGAMP, BmNPV, LPS, or heat-killed E. coli. Densitometry analysis of the BmRelishact protein level induced by cGAMP or BmNPV was abolished by BmCasp8L expression, but not by LPS or E. coli stimulation. F, BmCecB expression level in BmE cells overexpressing BmCasp8L at 9 h after stimulation with cGAMP, BmNPV, LPS, or E. coli. G, immunoblot of the cleavage of BmRelish in BmE cells overexpressing BmRelish alone or with different amounts of BmCasp8L. In the presence of increasing amounts of BmCasp8L, less BmRelishact was detected by densitometry analysis. WCL, whole-cell lysates; LPS, lipopolysaccharide; EGFP, enhanced green fluorescent protein; *, p < 0.05; **, p < 0.01. Error bars, S.D.
Table 1.
Proteins co-immunoprecipitated with BmSTING identified with LC-MS/MS
| Protein IDs | Accession | Protein annotation | Molecular mass | iBAQa |
|---|---|---|---|---|
| kDa | ||||
| BGIBMGA008319 | XP_004928441.1 | Uncharacterized protein | 30.081 | 33,324 |
| BGIBMGA009057 | XP_004924417.1 | Mitochondrial carrier protein | 31.458 | 1,152,800 |
| BGIBMGA013965 | NP_001040428.1 | H+-transporting ATP synthase | 32.349 | 1,235,600 |
| BGIBMGA008021 | XP_004922896.1 | BmCaspase-8 like (BmCasp8l) | 29.754 | 1,449,400 |
| BGIBMGA006158 | NP_001040289.1 | Prohibitin protein WPH | 30.081 | 34,207,000 |
| BGIBMGA003829 | NP_001037072.1 | ADP/ATP translocase | 32.891 | 38,661,000 |
a Intensity-based absolute quantification.
BmDredd is an endoprotease that mediates Relish cleavage (25, 26). Interestingly, BmCasp8L shares 70% sequence similarity with the N-terminal domain of BmDredd but lacks the C-terminal caspase domain of BmDredd (Fig. 6D), implying that BmCasp8L may be involved in BmRelish processing but possesses different function. A homolog of BmCasp8L was also identified in Drosophila (Fig. S5). To further evaluate the role of BmCasp8L in BmSTING-mediated signaling, BmRelish alone or together with BmCasp8L was overexpressed in BmE cells, which were then treated with different stimuli. Generation of BmRelishact induced by cGAMP or BmNPV was abolished by BmCasp8L expression; however, this suppression was not observed when cells were treated with LPS or Escherichia coli, suggesting that BmCasp8L may mainly affect the antiviral immune signaling (Fig. 6E). The translocation of BmRelish induced by cGAMP was also attenuated by BmCasp8L (Fig. S6). Expression of BmCecB induced by viral infection or cGAMP stimulation was also remarkably reduced when BmCasp8L was co-expressed (Fig. 6F). In fact, in the presence of increasing amounts of BmCasp8L, less BmRelishact was detected, implying that BmSTING-mediated BmRelish cleavage was inhibited by BmCasp8L (Fig. 6G). These data suggested that BmCasp8L is a suppressor of the BmSTING-mediated antiviral pathway.
BmDredd interacts with BmSTING to enhance antiviral signaling
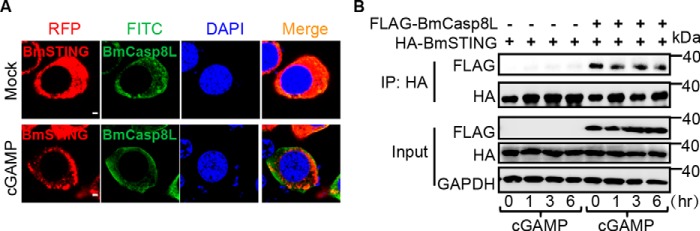
To investigate how BmSTING-mediated signaling is suppressed by BmCasp8L, we first determined the location of BmCasp8L and BmSTING after cGAMP stimulation. Confocal microscopy revealed that BmSTING and BmCasp8L were co-localized in the cytoplasm in untreated cells. However, after cGAMP treatment, BmSTING aggregated to the perinucleus space, whereas BmCasp8L did not, suggesting that less BmSTING was co-localized with BmCasp8L in the cytoplasm (Fig. 7A). Moreover, we performed a time-course study of BmSTING co-immunoprecipitation in the presence of immune stimulation to examine the interaction between BmSTING and BmCasp8L (Fig. 7B). After cGAMP stimulation, as time increased, less BmCasp8L was detected in the immunoprecipitates of BmSTING, suggesting that immune stimulation altered the interaction between BmSTING and BmCasp8L, thus reversing BmCasp8L inhibition. Interestingly, the expression of BmCasp8L after viral infection was dramatically decreased (Fig. S7), which may represent a positive feedback mechanism of lowering BmCasp8L in order to fully activate the innate immune response. Taken together, these data indicate that immune stimulation results in the release of BmSTING from the BmSTING–BmCasp8L complex.
Figure 7.
cGAMP stimulation abates the inhibition of BmCasp8L. A, subcellular localization of BmSTING and BmCasp8L in BmE cells treated with or without cGAMP (scale bar, 2 μm). B, co-immunoprecipitation assay of BmSTING and BmCasp8L in BmE cells treated with cGAMP for different times; immunoprecipitation was performed using anti-HA antibody, and the presence of BmCasp8L in BmSTING immunoprecipitates was determined with anti-FLAG-HRP antibody.
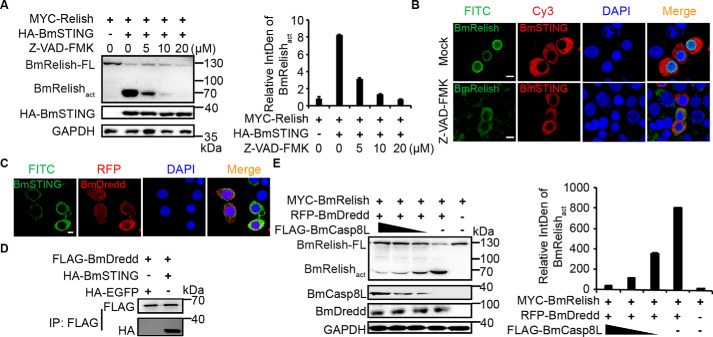
Considering the critical role of BmDredd in BmRelish processing and its high homology with BmCasp8L, we suspected that it also interacts with BmSTING. Therefore, we pretreated BmE cells co-expressing BmRelish and BmSTING with different amounts of Z-VAD-fmk, a caspase inhibitor, and then examined the cleavage and translocation of BmRelish. The level of BmRelishact decreased dramatically with increasing Z-VAD-fmk in a dose-dependent manner (Fig. 8A). Translocation of BmRelish from the cytoplasm into the nucleus promoted by BmSTING was also abrogated when cells were treated with Z-VAD-fmk (Fig. 8B). We then co-expressed BmDredd and BmSTING and found that they were co-localized in the cytoplasm (Fig. 8C). A co-immunoprecipitation assay also confirmed the interaction between these two proteins (Fig. 8D). Furthermore, when BmDredd and BmRelish were co-expressed with decreasing amounts of BmCasp8L, we found that more BmRelish was activated (Fig. 8E). It may be possible that the translocation of BmSTING following cGAMP stimulation is prone to the interaction with BmDredd rather than with BmCasp8L in favor of activating antiviral immune response.
Figure 8.
BmDredd competes with BmCasp8L to regulate the activation of Relish. A, BmE cells were treated with the indicated amounts of caspase inhibitor Z-VAD-fmk for 12 h before transfection with BmRelish and BmSTING-expressing plasmid; the cleavage of BmRelish was decreased dramatically with increasing Z-VAD-fmk by immunoblot analysis. B, subcellular localization of BmE cells treated with 1 μl of the caspase inhibitor Z-VAD-fmk, which was dissolved in DMSO at a final concentration of 20 μmol/liter (Z-VAD-fmk) or 1 μl of DMSO only (Mock) and then transfected with MYC-tagged BmRelish and BmSTING in cells (scale bar, 10 μm). C, subcellular localization of BmSTING and BmDredd in BmE cells (scale bar, 10 μm). D, co-immunoprecipitation assay of BmSTING and BmDredd. Immunoprecipitation was performed using anti-FLAG antibody, and the presence of BmSTING in BmDredd immunoprecipitates was determined with anti-HA-HRP antibody. E, the activation of BmRelish mediated by BmDredd was inhibited by BmCasp8L in a dose-dependent manner. BmRelish and BmDredd were co-expressed in BmE cells with decreasing doses of BmCasp8L. Error bars, S.D.
Discussion
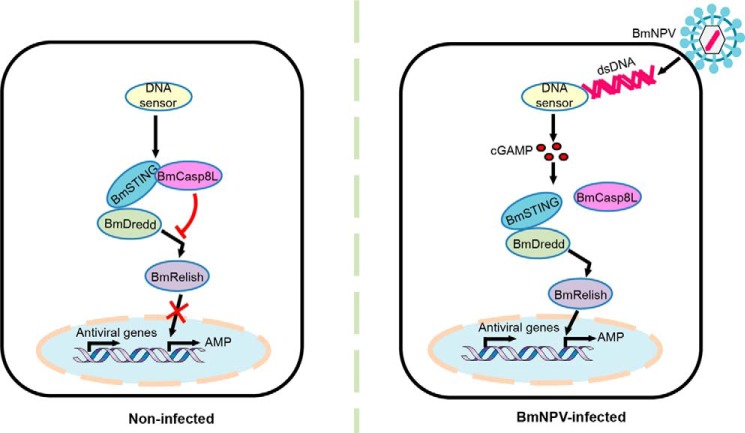
STING is a critical signaling molecule required for the immune response to cytosolic nucleic acids, particularly dsDNAs derived from pathogens and viruses as well as endogenous second messengers, such as c-di-GMP, cyclic-di-AMP (10), and cGAMP (22), in mammalian cells. These events lead to the production of innate immune response genes through the IRF3 and NF-κB pathways (1). In this study, we found that BmNPV infection led to cGAMP production in insects and that BmSTING responded to cGAMP to activate insect immune signaling pathways. The antiviral immune response that mediated by cGAMP-BmSTING in insects was attenuated by BmCasp8L. These findings provide novel insight into the mechanism of the cGAMP-mediated BmSTING-dependent NF-κB activation pathway after virus infection (Fig. 9).
Figure 9.
Schematic model representation of BmSTING-dependent activation of BmRelish/NF-κB in response to NPV infection. In noninfected cells, BmSTING interacts with BmCasp8L, which is an inhibitor of the BmSTING–BmRelish/NF-κB pathway and suppresses the activation of innate immunity. Upon viral infection, production of cGAMP is triggered by signals from some unknown DNA sensors that recognize viral DNA. cGAMP then induces the translocation of BmSTING from the cytoplasm to the prenuclear region. As a result, the interaction between BmSTING and BmCasp8L decreases, and BmSTING tends to interact with BmDredd, promoting the processing of BmRelish/NF-κB. Consequently, activated BmRelish is translocated to the nucleus to initiate the transcription of a plethora of target genes, including genes encoding antiviral factors and antimicrobial peptides.
Binding to cyclic dinucleotides is a deeply evolutionarily conserved function of STING that can be traced back to sea anemone, although co-factors are possibly required for insect STING proteins interacting with cGAMP in vitro (27). In this study, we found that BmSTING aggregated to the perinuclear region in response to cGAMP stimulation (Fig. 2E), and the expression of antimicrobial peptides induced by cGAMP was significantly down-regulated once BmSTING was knocked down by RNAi, implying that BmSTING functions as a potential sensor of cGAMP. Further study is needed to elucidate how BmSTING responds to cGAMP stimulation.
Relish, a transcription factor of the NF-κB family, is similar to mammalian p100 and p105. Its activation is dependent on signal-induced endoproteolysis, which requires the activity of the IκB kinase complex and the caspase Dredd (26). In this study, we discovered that BmCasp8L, which lacks the caspase domain, functions as an inhibitor of Relish cleavage. Based on the high similarity between BmCasp8L and the N-terminal domain of BmDredd, we speculated that BmCasp8L may compete with BmDredd in binding with DIAP2, an E3 ligase that binds to the N-terminal death effector domain of Dredd and targets it for polyubiquitylation, which is required for the full activation of Dredd. As a result, less ubiquitinated Dredd would be available to process Relish (28).
In mammals, activated STING by viral nucleic acids significantly up-regulates the production of IFN (29). However, no IFN homolog has been identified in insects. Recent studies reported that an antiviral cytokine, Vago, was produced upon viral infection in fruit fly and mosquito. Induction of Vago depends on the helicase Dicer-2, which belongs to the same DExD/H-box helicase family as RIG-I–like receptors, which sense viral infections and mediate interferon induction in mammals (30, 31). The Relish ortholog, Rel2, was also reported to be required for Vago production in Culex mosquitos infected with West Nile virus (32). Whether there exist any antiviral factors that are regulated by the BmSTING-NF-κB remain to be elucidated.
A number of DNA sensors, such as interferon γ–inducible protein 16 (IFI16) (33), DEAD box protein 41 (DDX41) (34), and cGMP–AMP synthase (cGAS) (21), have been characterized to induce STING-dependent IFN responses in mammals; nevertheless, their homologs in insects were not characterized to be intracellular DNA receptors involved in immune response. For instance, Abstrakt, the homolog of DDX41 in Drosophila, is well known for its role in the regulation of cell division (35). We also identified a homologous gene that might encode cGAS in silkworm. Further studies would be needed to uncover their possible functions in DNA-triggered immune response.
Our study demonstrated that the cGAMP–BmSTING–NF-κB–mediated antiviral signaling pathway is conserved in silkworm, B. mori, a model organism of lepidopteran insects. Identifying the mechanism of BmRelish processing promoted by BmSTING in B. mori will provide new insight into the antiviral response regulation in insects. Because BmCasp8L serves as a switch of BmSTING-mediated activation of BmRelish, targeting BmCasp8L may represent an approach to enhance early host immunity mediated by BmSTING to protect the host from viruses.
Experimental procedures
Insects, cells, and virus
Silkworm strain DaZao P50 was obtained from our laboratory. Larvae were reared following standard procedures. BmE cells (36) and BmN4-SID1 cells (37) were maintained at 27 °C in Grace medium or IPL-41 medium supplemented with 10% (v/v) fetal bovine serum, penicillin, and streptomycin. BmNPV-GFP viruses were propagated in BmE cells, and BmNPV titers were determined via a TCID50 end-point dilution assay described by O'Reilly et al. (38).
Plasmids
BmSTING (XP_0049239 46) was cloned by PCR using cDNA of BmE cells as the template. 5′-RACE was performed by using a 5′-full RACE kit with TAP (TaKaRa Bio Inc., Tokyo, Japan) to obtain the full-length sequence. HA-tagged BmSTING (HA-Bm-STING), FLAG-tagged BmCasp8L (FLAG-BmCasp8L), BmDredd (FLAG-Bm-Dredd), and MYC-tagged BmRelishact were constructed following the same procedure for construction of the MYC-BmRelish expression vector. RFP-tagged BmSTING (RFP-BmSTING), BmRelish (RFP-BmRelish), and BmDredd (RFP-BmDredd) were constructed by the same methods as described previously by Tatsuke et al. (39). The entry clones for BmSTING, BmRelish, and BmDredd were constructed on pENTR11 from Invitrogen. The primer sets were designed based on the registered sequence data and listed in Table S1. The amplified BmSTING, BmRelish, and BmDredd cDNA were digested with NcoI and XhoI and subcloned into the pENTR11 and named BmSTING-pENTR11, BmRelish-pENTR11, and BmDredd-pENTR11, respectively. pRFP-BmSTING, pRFP-BmRelish, and pRFP-BmDredd were constructed by the Gateway LR reaction between the entry vectors, BmSTING-pENTR11, BmRelish-pENTR1, and BmDredd-pENTR11, and the DEST vectors pie2RW using LR ClonaseTM Enzyme Mix (Invitrogen) according to the manufacturer's manual.
Cell stimulations
cGAMP was purchased from Invivogen. All transfections in cells were carried out using X-treme GENE Transfection Reagent according to the manufacturer's instructions (Roche, Basel, Switzerland). cGAMP, poly(dA:dT), poly(I:C), c-di-GMP, and ISD were transfected at a final concentration of 3 μg/ml as described previously (40). 10 μl of cGAMP/X-treme (0.1 μg/ml) transfection complex was injected into silkworm larvae on the second day of fifth instar through the second-to-last stoma of the abdomen with a fine needle. After 24 h, the fat body was extracted for further study.
For detection of immunostimulated activation of BmRelish, cells were first transfected with BmRelish-expressing plasmid alone or along with BmSTING-expressing plasmid using X-treme GENE Transfection Reagent (Roche). 48 h later, those cells were then transfected with cGAMP at a final concentration of 3 μg/ml using X-treme GENE Transfection Reagent or treated only with the same transfection reagent without cGAMP. After 10 h, cells were stained for immunofluorescence analysis (Fig. 5B).
Caspase inhibitor Z-VAD-fmk was purchased from Beyotime (Shanghai, China). To evaluate the effect of caspase inhibitor on BmRelish activation, cells were pretreated with 1 μl of DMSO only or 1 μl of Z-VAD-fmk (Beyotime), which was dissolved in DMSO at a final concentration of 20 μmol/liter. Then the cells were co-transfected with BmRelish- and BmSTING-expressing plasmid using X-treme GENE Transfection Reagent. 48 h later, cells were stained for immunofluorescence analysis (Fig. 8B).
Measurement of viral DNA load
At the indicated time points, BmNPV-infected BmE cells were harvested and suspended in PBS. Total DNA from each sample was prepared with a TaKaRa MiniBEST universal genomic DNA extraction kit (TaKaRa Bio) according to the protocol in the manual. The viral DNA abundance of BmNPV was examined by the expression of the GP64 gene. The silkworm GAPDH gene was used for normalization. Sequences of primers are listed in Table S1.
LC-MS/MS
BmE cells (3 × 106) were collected in 100 μl of 40% (v/v) acetonitrile, 40% (v/v) methanol, 0.1 n formic acid methanol. This slurry was incubated for 30 min at −20 °C and concentrated at 4 °C for 15 min (41). The supernatant was moved to a clean centrifuged tube, dried for 3 h in a vacuum concentrator, and then dissolved in 100 μl of water. Next, 10 μl of each sample was injected into a Thermo Fisher Scientific Ultimate 3000 system (Agilent) equipped with a Pursuit 3 PFP column (3 μm, 2 × 150 mm). The ionization source parameters were set as follows: positive mode; capillary temperature, 250 °C; spray voltage, 2.3 kV. Full-scan MS spectra were acquired in a mass range from m/z 100 to 1000. The flow phases used were as follows: A, aqueous 0.1% (v/v) formic acid; B, 0.1% formic acid + 100% acetonitrile. The gradient used was 2% B for 3.5 min, 2–30% B for 3.5 min, 30–65% B for 1.5 min, 65–95% B for 1.5 min, 95 to 2% B for 1.5 min, and re-equilibration for 1 min. The flow rate was 0.2 ml/min, and the column oven temperature was maintained at 40 °C. Targeted selected ion-monitoring (SIM)/ddMS2 mode was used in the Q-Exactive mass spectrometer with an electrospray ion source. The precursor ion used to monitor cGAMP in the samples was 675.107 [M + H]1+, and the production of MS/MS was confirmed using Mass Frontier version 7.0 software. Chemically synthesized cGAMP was dissolved in extraction buffer at concentrations of 0.245, 0.123, 0.062, 0.031, 0.016, and 0.008 nm to generate a standard curve or by calculating the cGAMP concentration in each extract. Quantifier SIM peak areas were compared with the calibration curve of external standard peak areas to determine the cGAMP concentration.
RNAi
dsRNA was synthesized using RiboMAXTM large-scale RNA production systems SP6 and T7 (Promega). The primer of T7-BmSTING is listed in Table S1. BmE cells were seeded on 12-well plates at 1 × 105 cells/well and transfected with 5 μg of dsRNA using X-treme GENE Transfection Reagent.
BmN4-SID1 cells were used for RNAi by adding 5 μg of dsRNA directly into 1 ml of medium supplemented with 10% FBS (37). After 5 days of RNAi, the rest of the process was as described above for the BmE cells.
The fifth instar larvae of Nm DZ were injected was 20 μg of dsRNA. Two days after RNAi, the larvae was injected with 5 μl of virus (106 pfu/ml). Total DNA was obtained at 3 days after injection. Each sample was extracted from 10 treated larvae, and PCR analysis of BmNPV replication was performed as described (43).
CRISPRi
The expression vector of T-hr3/A4-dCas9-SID was kept in our laboratory. The sgRNAs of BmSTING promoter were synthesized by Invitrogen and inserted into T-U6-gRNA2 vector. All of the gRNA sequences are listed in Table S1. All of the plasmids were purified with a Plasmid Mini Kit (Qiagen, Hilden, Germany). T-hr3/A4-dCas9-SID was co-transfected with T-U6-sgBmSTING into BmE cells for 60 h. Then the cells infected with BmNPV at an MOI of 0.5 were collected 3 days later for further study.
Mortality analysis
The survival rates of dsRFP-treated and dsBmSTING-treated larvae after injection with virus were investigated. Each line was infected three times, and each repetition included 30 larvae. Cumulative survival rates were calculated from the indicated time of infection up to 120 hpi. Each assay was performed three times.
Real-time PCR
Total RNA was isolated from cells using a Total RNA Kit (Omega) and reverse-transcribed by a GoScript reverse transcription system (Promega). Fluorescence real-time PCR was performed using Ex TaqII (TaKaRa) on a 7500 fast real-time PCR system (Applied Biosystems) with a program consisting of an initial denaturing step of 30 s at 95 °C and 40 amplification cycles consisting of 5 s at 95 °C followed by 30 s at 60 °C. The expression level of BmSTING or GP64 was normalized to GAPDH (XP_012549898). The primers used in qRT-PCR are available in Table S1.
Immunoprecipitation and silver stain
BmE cells (3 × 107) transfected with HA-BmSTING or HA-EGFP were separately collected and lysed in 1.5 ml of cell lysis buffer (Beyotime) per well on ice for 30 min. After centrifugation at 12,000 rpm for 15 min, the clarified supernatants were collected.
For immunoprecipitation experiments, total proteins were mixed with 50 μg of recombinant anti-HA mouse mAb (HA-7) (catalogue no. H3663-200 μl, Sigma) and incubated at 4 °C with gentle agitation for 4 h. 100 μl of protein-agarose A/G beads (Santa Cruz Biotechnology, Inc.) was added and incubated at 4 °C overnight. After washing five times with PBS containing 0.05% Tween 20, protein was eluted with 2× SDS-loading buffer by boiling for 15 min. Eluted samples (40 μl) were separated on 12% SDS-PAGE and then visualized by staining with silver nitrate (AMRESCO 0377-100G) and analyzed by LC-MS/MS, which was done by our laboratory as described previously (44).
Co-immunoprecipitation and immunoblot analysis
Cells transfected with expression vectors of FLAG-BmCasp8L or HA-BmSTING alone or together were collected after transfection and lysed in 150 μl of cell lysis buffer (Beyotime) per well on ice for 30 min. After centrifugation at 12,000 rpm for 15 min, the clarified supernatants were collected. The 500 μl of supernatants was then incubated with 5 μg of anti-FLAG antibody (catalogue no. F1804-200μg, Sigma) or 5 μg of anti-HA mouse mAb (HA-7) (catalogue no. H3663-200μl, Sigma) at 4 °C for 2–4 h separately. 20 μl of protein-agarose A/G beads (Santa Cruz Biotechnology) was added and incubated overnight. The immune complexes were washed with PBS containing 0.05% Tween 20 six times and eluted with 40 μl of 2× SDS-loading buffer by boiling for 15 min. Protein concentration of whole-cell lysate was estimated by a BCA assay (Beyotime). Sample from the co-immunoprecipitation assay and whole-cell lysate was separately resolved on 12% SDS-PAGE. After transfer to a polyvinylidene difluoride membrane (GE Healthcare), samples were immunoblotted with anti-HA-HRP antibody (catalogue no. A00169-40, Genscript, Nanjing, China), anti-FLAG-HRP antibody (catalogue no. A8592-2MG, Sigma), or GAPDH antibody (catalogue no. abs830030, Shanghai, China) following the standard procedure and then developed using SuperSignalTM West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific) as described previously (45).
ImageJ-based quantitative analysis of Western blotting data
Quantitative analysis of the visible bands in the immunoblot was performed by using the ImageJ program (46) and a procedure described previously (42). The integrated density values of all of the measured bands for the BmRelishact protein levels were normalized to amounts of loading control GAPDH.
Confocal microscopy
BmE cells transfected with HA-BmSTING were first permeabilized with 0.5% Triton X-100 and blocked with 5% BSA in PBS containing 0.1% Tween 20 (PBST) for 1 h at room temperature. Then cells were incubated with anti-HA mouse mAb (HA-7) (catalogue no. H3663-200μl, Sigma) and anti-MYC rabbit mAb (catalogue no. AM933, Beyotime) for 1 h at room temperature, followed by incubation with anti-mouse IgG conjugated with Cy3 (catalogue no. A0521, Beyotime) or anti-rabbit IgG conjugated with FITC (catalogue no. A0562, Beyotime), respectively. The stained cells were visualized under an Olympus FV-1000 confocal microscope (Olympus, Tokyo, Japan).
Statistics
The results are expressed as the means ± S.E. Statistically significant differences between the mean values were determined by Student's t test (*, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001). Silkworm experiments were performed in biological triplicate with the indicated number of silkworms per group. Cell culture experiments were conducted with three or five independent cultures for each sample.
Author contributions
X. H., H. H., and F. W. designed the experiments; B. L., L. S., C. H., X. L., D. W., Y. X., and P. Z. assisted with the experiments and provided critical reagents and intellectual input; H. H., F. W., and Q. X. provided critical reagents and intellectual input; H. H., Q. X., and F. W. supervised the study; X. H. and F. W. interpreted the data and wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Prof. Xiaofeng Wu (Zhejiang University, China) for kindly providing the BmNPV-GFP virus. We also thank Xiaogang Wang and Dr. Sanyuan Ma for help in constructing the CRISPRi system.
This work was supported by State Key Program of National Natural Science of China Grant 31530071, Natural Science Foundation of China Grants 31672495 and 31572465, and Chongqing Research Program of Basic Research and Frontier Technology CSTC2014JCYJA80010 and CSTC2015JCYJBX0035. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1 and Figs. S1–S7.
- STING
- stimulator of interferon genes
- cGAMP
- cyclic GMP–AMP
- RFP
- red fluorescent protein
- IFI16
- interferon γ–inducible protein 16
- cGAS
- cyclic GMP–AMP synthase
- DDX41
- DEAD box protein 41
- Dredd
- death-related ced-3/Nedd2-like caspase
- BmCasp8L
- BmCaspase-8–like
- RACE
- rapid amplification of cDNA ends
- SIM
- selected ion-monitoring
- IFN
- interferon
- IRF3
- interferon regulatory factor 3
- IMD
- immune deficiency
- ISD
- interferon stimulatory DNA
- NPV
- nucleopolyhedrovirus
- hpi
- hours postinfection
- MOI
- multiplicity of infection
- gRNA
- guide RNA
- sgRNA
- single guide RNA
- qPCR and qRT-PCR
- quantitative PCR and RT-PCR, respectively
- c-di-GMP
- cyclic di-GMP
- Z
- benzyloxycarbonyl
- fmk
- fluoromethyl ketone
- HA
- hemagglutinin
- HRP
- horseradish peroxidase
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- DAPI
- 4′,6-diamidino-2-phenylindole
- IP
- immunoprecipitation.
References
- 1. Barber G. N. (2014) STING-dependent cytosolic DNA sensing pathways. Trends Immunol. 35, 88–93 10.1016/j.it.2013.10.010 [DOI] [PubMed] [Google Scholar]
- 2. Roers A., Hiller B., and Hornung V. (2016) Recognition of endogenous nucleic acids by the innate immune system. Immunity 44, 739–754 10.1016/j.immuni.2016.04.002 [DOI] [PubMed] [Google Scholar]
- 3. Ishikawa H., and Barber G. N. (2008) STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678 10.1038/nature07317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ishikawa H., Ma Z., and Barber G. N. (2009) STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461, 788–792 10.1038/nature08476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jin L., Waterman P. M., Jonscher K. R., Short C. M., Reisdorph N. A., and Cambier J. C. (2008) MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol. Cell. Biol. 28, 5014–5026 10.1128/MCB.00640-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sun W., Li Y., Chen L., Chen H., You F., Zhou X., Zhou Y., Zhai Z., Chen D., and Jiang Z. (2009) ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. U.S.A. 106, 8653–8658 10.1073/pnas.0900850106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhong B., Yang Y., Li S., Wang Y.-Y., Li Y., Diao F., Lei C., He X., Zhang L., Tien P., and Shu H. B. (2008) The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29, 538–550 10.1016/j.immuni.2008.09.003 [DOI] [PubMed] [Google Scholar]
- 8. Li X.-D., Wu J., Gao D., Wang H., Sun L., and Chen Z. J. (2013) Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science 341, 1390–1394 10.1126/science.1244040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Paludan S. R., and Bowie A. G. (2013) Immune sensing of DNA. Immunity 38, 870–880 10.1016/j.immuni.2013.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burdette D. L., Monroe K. M., Sotelo-Troha K., Iwig J. S., Eckert B., Hyodo M., Hayakawa Y., and Vance R. E. (2011) STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518 10.1038/nature10429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Costa A., Jan E., Sarnow P., and Schneider D. (2009) The Imd pathway is involved in antiviral immune responses in Drosophila. PLoS One 4, e7436 10.1371/journal.pone.0007436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zambon R. A., Nandakumar M., Vakharia V. N., and Wu L. P. (2005) The Toll pathway is important for an antiviral response in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 102, 7257–7262 10.1073/pnas.0409181102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Avadhanula V., Weasner B. P., Hardy G. G., Kumar J. P., and Hardy R. W. (2009) A novel system for the launch of Alphavirus RNA synthesis reveals a role for the Imd pathway in arthropod antiviral response. PLoS Pathog. 5, e1000582 10.1371/journal.ppat.1000582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xi Z., Ramirez J. L., and Dimopoulos G. (2008) The Aedes aegypti Toll pathway controls Dengue virus infection. PLoS Pathog. 4, e1000098 10.1371/journal.ppat.1000098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dostert C., Jouanguy E., Irving P., Troxler L., Galiana-Arnoux D., Hetru C., Hoffmann J. A., and Imler J.-L. (2005) The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of Drosophila. Nat. Immunol. 6, 946–953 10.1038/ni1237 [DOI] [PubMed] [Google Scholar]
- 16. Broderick N. A., Buchon N., and Lemaitre B. (2014) Microbiota-induced changes in Drosophila melanogaster host gene expression and gut morphology. MBio 5, e01117–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang L., Cheng T., Xu P., Fang T., and Xia Q. (2012) Bombyx mori transcription factors: genome-wide identification, expression profiles and response to pathogens by microarray analysis. J. Insect Sci. 12, 40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bao Y.-Y., Tang X.-D., Lv Z.-Y., Wang X.-Y., Tian C.-H., Xu Y. P., and Zhang C.-X. (2009) Gene expression profiling of resistant and susceptible Bombyx mori strains reveals nucleopolyhedrovirus-associated variations in host gene transcript levels. Genomics 94, 138–145 10.1016/j.ygeno.2009.04.003 [DOI] [PubMed] [Google Scholar]
- 19. Moreno-Habel D. A., Biglang-awa I. M., Dulce A., Luu D. D., Garcia P., Weers P. M. M., and Haas-Stapleton E. J. (2012) Inactivation of the budded virus of Autographa californica M nucleopolyhedrovirus by gloverin. J. Invertebr. Pathol. 110, 92–101 10.1016/j.jip.2012.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guo R., Wang S., Xue R., Cao G., Hu X., Huang M., Zhang Y., Lu Y., Zhu L., Chen F., Liang Z., Kuang S., and Gong C. (2015) The gene expression profile of resistant and susceptible Bombyx mori strains reveals cypovirus-associated variations in host gene transcript levels. Appl. Microbiol. Biotechnol. 99, 5175–5187 10.1007/s00253-015-6634-x [DOI] [PubMed] [Google Scholar]
- 21. Sun L., Wu J., Du F., Chen X., and Chen Z. J. (2013) Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 10.1126/science.1232458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu J., Sun L., Chen X., Du F., Shi H., Chen C., and Chen Z. J. (2013) Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830 10.1126/science.1229963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Abe T., and Barber G. N. (2014) Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-κB activation through TBK1. J. Virol. 88, 5328–5341 10.1128/JVI.00037-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gilbert L. A., Larson M. H., Morsut L., Liu Z., Brar G. A., Torres S. E., Stern-Ginossar N., Brandman O., Whitehead E. H., Doudna J. A., Lim W. A., Weissman J. S., and Qi L. S. (2013) CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 10.1016/j.cell.2013.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ma X., Li X., Dong S., Xia Q., and Wang F. (2015) A Fas associated factor negatively regulates anti-bacterial immunity by promoting Relish degradation in Bombyx mori. Insect Biochem. Mol. Biol. 63, 144–151 10.1016/j.ibmb.2015.06.009 [DOI] [PubMed] [Google Scholar]
- 26. Stoven S., Silverman N., Junell A., Hedengren-Olcott M., Erturk D., Engstrom Y., Maniatis T., and Hultmark D. (2003) Caspase-mediated processing of the Drosophila NF-κB factor Relish. Proc. Natl. Acad. Sci. U.S.A. 100, 5991–5996 10.1073/pnas.1035902100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kranzusch P. J., Wilson S. C., Lee A. S., Berger J. M., Doudna J. A., and Vance R. E. (2015) Ancient origin of cGAS-STING reveals mechanism of universal 2′,3′ cGAMP signaling. Mol. Cell 59, 891–903 10.1016/j.molcel.2015.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meinander A., Runchel C., Tenev T., Chen L., Kim C. H., Ribeiro P. S., Broemer M., Leulier F., Zvelebil M., and Silverman N., and Meier P. (2012) Ubiquitylation of the initiator caspase DREDD is required for innate immune signalling. EMBO J. 31, 2770–2783 10.1038/emboj.2012.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barber G. N. (2011) Innate immune DNA sensing pathways: STING, AIMII and the regulation of interferon production and inflammatory responses. Curr. Opin. Immunol. 23, 10–20 10.1016/j.coi.2010.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Deddouche S., Matt N., Budd A., Mueller S., Kemp C., Galiana-Arnoux D., Dostert C., Antoniewski C., Hoffmann J. A., and Imler J.-L. (2008) The DExD/H-box helicase Dicer-2 mediates the induction of antiviral activity in Drosophila. Nat. Immunol. 9, 1425–1432 10.1038/ni.1664 [DOI] [PubMed] [Google Scholar]
- 31. Paradkar P. N., Trinidad L., Voysey R., Duchemin J.-B., and Walker P. J. (2012) Secreted Vago restricts West Nile virus infection in Culex mosquito cells by activating the Jak-STAT pathway. Proc. Natl. Acad. Sci. U.S.A. 109, 18915–18920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Paradkar P. N., Duchemin J.-B., Voysey R., and Walker P. J. (2014) Dicer-2-dependent activation of Culex Vago occurs via the TRAF-Rel2 signaling pathway. PLoS Negl. Trop. Dis. 8, e2823 10.1371/journal.pntd.0002823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Unterholzner L., Keating S. E., Baran M., Horan K. A., Jensen S. B., Sharma S., Sirois C. M., Jin T., Latz E., Xiao T. S., Fitzgerald K. A., Paludan S. R., and Bowie A. G. (2010) IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 11, 997–1004 10.1038/ni.1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang Z., Yuan B., Bao M., Lu N., Kim T., and Liu Y.-J. (2011) The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 12, 959–965 10.1038/ni.2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jiang Y., Zhu Y., Liu Z.-J., and Ouyang S. (2017) The emerging roles of the DDX41 protein in immunity and diseases. Protein Cell 8, 83–89 10.1007/s13238-016-0303-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pan M.-H., Xiao S.-Q., Chen M., Hong X.-J., and Lu C. (2007) Establishment and characterization of two embryonic cell lines of Bombyx mori. In Vitro Cell. Dev. Biol. Anim. 43, 101–104 10.1007/s11626-006-9009-x,10.1007/s11627-007-9030-0 [DOI] [PubMed] [Google Scholar]
- 37. Mon H., Kobayashi I., Ohkubo S., Tomita S., Lee J., Sezutsu H., Tamura T., and Kusakabe T. (2012) Effective RNA interference in cultured silkworm cells mediated by overexpression of Caenorhabditis elegans SID-1. RNA Biol. 9, 40–46 10.4161/rna.9.1.18084 [DOI] [PubMed] [Google Scholar]
- 38. O'Reilly D., Miller L., and Luckov V. (1992) Baculovirus Expression Vectors: A Laboratory Manual, pp. 69–77, Freeman, New York [Google Scholar]
- 39. Tatsuke T., Zhu L., Li Z., Mitsunobu H., Yoshimura K., Mon H., Lee J. M., and Kusakabe T. (2014) Roles of Piwi proteins in transcriptional regulation mediated by HP1s in cultured silkworm cells. PLoS One 9, e92313 10.1371/journal.pone.0092313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ablasser A., Goldeck M., Cavlar T., Deimling T., Witte G., Röhl I., Hopfner K.-P., Ludwig J., and Hornung V. (2013) cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 498, 380–384 10.1038/nature12306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Massie J. P., Reynolds E. L., Koestler B. J., Cong J.-P., Agostoni M., and Waters C. M. (2012) Quantification of high-specificity cyclic diguanylate signaling. Proc. Natl. Acad. Sci. U.S.A. 109, 12746–12751 10.1073/pnas.1115663109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Qian W., Gang X., Zhang T., Wei L., Yang X., Li Z., Yang Y., Song L., Wang P., Peng J., Cheng D., and Xia Q. (2017) Protein kinase A-mediated phosphorylation of the Broad-Complex transcription factor in silkworm suppresses its transcriptional activity. J. Biol. Chem. 292, 12460–12470 10.1074/jbc.M117.775130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jin S., Cheng T., Jiang L., Lin P., Yang Q., Xiao Y., Kusakabe T., and Xia Q. (2014) Identification of a new Sprouty protein responsible for the inhibition of the Bombyx mori nucleopolyhedrovirus reproduction. PLoS One 9, e99200 10.1371/journal.pone.0099200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dong Z., Zhang W., Zhang Y., Zhang X., Zhao P., and Xia Q. (2016) Identification and characterization of novel chitin-binding proteins from the larval cuticle of silkworm, Bombyx mori. J. Proteome Res. 15, 1435–1445 10.1021/acs.jproteome.5b00943 [DOI] [PubMed] [Google Scholar]
- 45. Hua X.-T., Ma X.-J., Xue R.-J., Cheng T.-C., Wang F., and Xia Q.-Y. (2016) Characterization of the Bombyx mori Cecropin A1 promoter regulated by IMD pathway. Insect Sci. 23, 297–304 10.1111/1744-7917.12210 [DOI] [PubMed] [Google Scholar]
- 46. Schneider C. A., Rasband W. S., and Eliceiri K. W. (2012) NIH image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.