Abstract
Epigenetic regulation is critical in normal cardiac development. We have demonstrated that the deletion of Jarid2 (Jumonji (Jmj) A/T-rich interaction domain 2) in mice results in cardiac malformations recapitulating human congenital cardiac disease and dysregulation of gene expression. However, the precise developmental and epigenetic functions of Jarid2 within the developing heart remain to be elucidated. Here, we determined the cardiac-specific functions of Jarid2 and the genetic networks regulated by Jarid2. Jarid2 was deleted using different cardiac-specific Cre mice. The deletion of Jarid2 by Nkx2.5-Cre mice (Jarid2Nkx) caused cardiac malformations including ventricular septal defects, thin myocardium, hypertrabeculation, and neonatal lethality. Jarid2Nkx mice exhibited elevated expression of neural genes, cardiac jelly, and other key factors including Isl1 and Bmp10 in the developing heart. By employing combinatorial genome-wide approaches and molecular analyses, we showed that Jarid2 in the myocardium regulates a subset of Jarid2 target gene expression and H3K27me3 enrichment during heart development. Specifically, Jarid2 was required for PRC2 occupancy and H3K27me3 at the Isl1 promoter locus, leading to the proper repression of Isl1 expression. In contrast, Jarid2 deletion in differentiated cardiomyocytes by cTnt-Cre mice caused no gross morphological defects or neonatal lethality. Thus, the early deletion of Jarid2 in cardiac progenitors, prior to the differentiation of cardiac progenitors into cardiomyocytes, results in morphogenetic defects manifested later in development. Our studies reveal that there is a critical window during early cardiac progenitor differentiation when Jarid2 is crucial to establish the epigenetic landscape at later stages of development.
Keywords: heart development, epigenetics, gene regulation, gene expression, histone modification, Congenital heart disease, Jarid2
Introduction
Human congenital cardiac defects are one of the most common forms of birth defects (1). Normal cardiovascular development requires precise control of gene expression in a spatial and temporal-dependent manner. Eukaryotic gene transcription is regulated by chromatin structure partly via modifications of histone tails. Due to a groundbreaking discovery of histone demethylases such as Jumonji (Jmj) family factors, histone methylation is now considered a reversible epigenetic mark. Methylated histone tails are recognized as a marker for transcriptional activation or repression. In general, methylation at histone H3 lysine 9 (H3K9), H3K27, or H4K20 is associated with gene repression, whereas methylation at H3K4, H3K36, or H3K79 is correlated with gene activation (2, 3). However, the regulatory roles of histone methylation status in gene expression are not fully understood. Histone lysine demethylases show exquisite substrate specificity and participate in diverse biological processes. Mutations or deregulation of histone demethylases are often linked to human diseases (4, 5).
Jarid22 (JMJ) is a nuclear factor critical for mouse embryonic development (6). Jarid2 is the founding member of the Jmj family that functions as histone lysine demethylases. However, Jarid2 is enzymatically inactive due to amino acid substitutions in the JmjC domain that is a catalytic domain (7, 8). Nonetheless, Jarid2 is essential for embryonic development in the heart, liver, and hematopoietic tissues (6). Jarid2 knockout (Jarid2 KO) mice die perinatally and exhibit cardiac defects mimicking human congenital cardiac defects including ventricular septal defect (VSD), double-outlet right ventricle, thin myocardium and hypertrabeculation (9, 10). Left ventricular noncompaction (LVNC) in humans is characterized by a spongy ventricular myocardium with excessive trabeculations and deep trabecular recesses in the left ventricle leading to a thin ventricular wall (11), which is manifested in Jarid2 KO hearts (9). The American Heart Association formally classified LVNC as a distinct cardiomyopathy (11). The genesis of LVNC has been speculated to represent an arrest of the final stage of myocardial morphogenesis, which is often referred to as “myocardial compaction” for a lack of better terminology. The etiology of LVNC remains unclear partly because the genetic causes of LVNC are heterogeneous, and there is insufficient knowledge on the molecular control of normal trabeculation and compaction during ventricular myocardial wall development.
During mammalian heart development, the ventricles undergo complex morphogenetic events (12). The initial step is the formation of a single cell layer of myocardium at an early developmental stage, followed by the formation of a trabecular and compact myocardium at early midgestation stage. The final step involves the myocardial compaction to give rise to the thickened ventricular wall with the reduced trabecular layer at late midgestation stage, but molecular events leading to the mature ventricular wall remain poorly understood. In mouse models, noncompaction cardiomyopathy has been used to describe the thin compact layer with normal or excessive trabeculations, leading to the thin ventricular myocardial wall at mid- to late stages of cardiac development (13). Notch pathways, Neuregulin1, and bone morphogenetic protein 10 (Bmp10) expression are critical for initiation and expansion of the trabecular layer, which in turn affect the ventricular wall thickness (12). Cardiac jelly between the endocardium and the underlying trabecular layer is also essential for initiation and growth of the trabecular layer (12, 14). Interestingly, all of the above signals in the heart are significantly reduced in later developmental stages, which coincides with the cessation of trabeculation as well as compaction of the ventricular myocardium.
Jarid2 functions as a major component of the transcriptional networks that balance pluripotency and differentiation in embryonic stem (ES) cells. Jarid2 is associated with the Polycomb repressive complex 2 (PRC2) in ES cells and is required for an efficient accumulation of PRC2 on the chromatin (7, 15, 16). Major components of PRC2 consist of the SET domain containing histone methylases, EZH1/EZH2 and SUZ12, and EED, which specifically methylate at H3K27. Trimethylation of H3K27 (H3K27me3) is associated with repressed chromatin states, and widely distributed among genes encoding developmental regulators. However, it is debated whether Jarid2 loss in ES cells causes increases or decreases in H3K27me3 levels. PRC2 and H3K27me3 occupy a set of genes controlling differentiation and prevent full expression of these genes until lineage commitment in ES cells (17, 18). Although loss of Jarid2 or PRC2 function results in defective ES cell differentiation, the epigenetic role of Jarid2 remains unclear during heart development. We have demonstrated that the endothelial deletion of Jarid2 partially recapitulates cardiac defects observed in Jarid2 KO mice (19). In the endocardial layer, Jarid2 represses Notch1 expression by interacting with Setdb1 via H3K9me3 enrichment at the Notch1 locus (8). This seems crucial for termination of trabeculation and initiation of compaction of the ventricular myocardial wall. As a major epigenetic marker, H3K9 methylation is known as a “histone code” for gene silencing. Conditional deletion of Ezh2, a catalytic subunit of PRC2, using Nkx2.5-Cre mice results in cardiac developmental defects such as hypertrabeculation, thinning of the compact myocardium, and VSD, which are similar to the defects observed in Jarid2 KO mice (9, 20). However, it remains unknown whether Jarid2 cooperates with PRC2 during heart development.
In this study, we demonstrate that cardiac-specific deletion of Jarid2 in cardiac progenitors and their progeny causes neonatal lethality and cardiac malformations including VSD, hypertrabeculation, and thin compact layer. In contrast, Jarid2 deletion in differentiated cardiomyocytes did not result in overt cardiac malformation. These data indicate that there is a critical window during early cardiac progenitor differentiation when Jarid2 is crucial to establish the epigenetic landscape at later stages of development. We provide evidence that Jarid2 cooperates with PRC2 for H3K27me3 accumulation on a subset of Jarid2 target genes in the developing heart, which contributes to repress differentiation of other lineages such as neural differentiation, and to guide normal myocardial development.
Results
Cardiac-specific deletions of Jarid2
Jarid2 deletion in mice causes congenital heart defects and death right after birth (9). However, the precise developmental and molecular functions of Jarid2 remain to be elucidated within the early developing heart. Thus, we set out to determine the cardiac-specific function of Jarid2 by deleting Jarid2 in cardiac progenitors and their progeny using Nkx2.5-Cre KI mice (Jarid2Nkx) (21). We first analyzed Mendelian ratios from embryonic day (E) 9.5 through postnatal day (P) 10 (Table 1). The Mendelian ratios for Jarid2Nkx mutants (Nkx2.5-Cre/+;Jarid2f/f) were normal until birth, but all mutants succumbed to death within 1 day after birth. Heterozygous mutant mice (Nkx2.5-Cre/+;Jarid2f/+) were present at the expected Mendelian ratio. We further examined roles of Jarid2 within the myocardium after cardiac cells had differentiated to cardiomyocytes. Cardiomyocyte-specific deletion of Jarid2 using αMHC-Cre mice causes neither gross cardiac malformation nor perinatal lethality (19). It is plausible that Jarid2 plays a critical role earlier than expected. cTnt-Cre mice express Cre from E7.5 onwards, whereas αMHC-Cre mice express Cre at E8.5 (22, 23). Thus Jarid2 deletion mice using cTnt-Cre mice, cTnt-Cre/+;Jarid2f/f (Jarid2cTnt), were generated. Jarid2cTnt mice were born at the expected Mendelian ratio without overt cardiac defects (data not shown) and survived to adulthood (Table 1). These results suggest that Jarid2 in differentiated cardiomyocytes is dispensable for cardiac morphogenesis, supporting the critical roles of Jarid2 early in cardiac progenitors.
Table 1.
Mendelian ratios of embryonic (E) or postnatal (P) mice
| Cre mouse | Age | No. of litters | No. of live mice | Genotype of live mice |
Dead | |||
|---|---|---|---|---|---|---|---|---|
| +/+;f/f | +/+;f/+ | C/+;f/+ | C/+;f/f | |||||
| Nkx2.5 | E9.5–14.5 | 41 | 344 | 79 | 90 | 88 | 87 | 4a |
| (23.0%) | (26.2%) | (25.6%) | (25.3%) | |||||
| E15–19.5 | 27 | 192 | 46 | 48 | 52 | 46 | 1b | |
| (24.0%) | (25.0%) | (27.1%) | (24.0%) | |||||
| P1–10 | 24 | 107 | 37 | 35 | 35 | 0 | 8c | |
| (34.6%) | (32.7%) | (32.7%) | (0%) | |||||
| cTnt | P21 | 8 | 56 | 16 | 13 | 14 | 13 | |
| (28.6%) | (23.2%) | (25.0%) | (23.2%) | |||||
a Two were not possible for genotyping because of necrosis, and the other two were +/+;f/+.
b One was C/+;f/f.
c Five dead mice were C/+;f/f , two were +/+;f/f, and one was C/+;f/+. All dead ones were observed at P1.
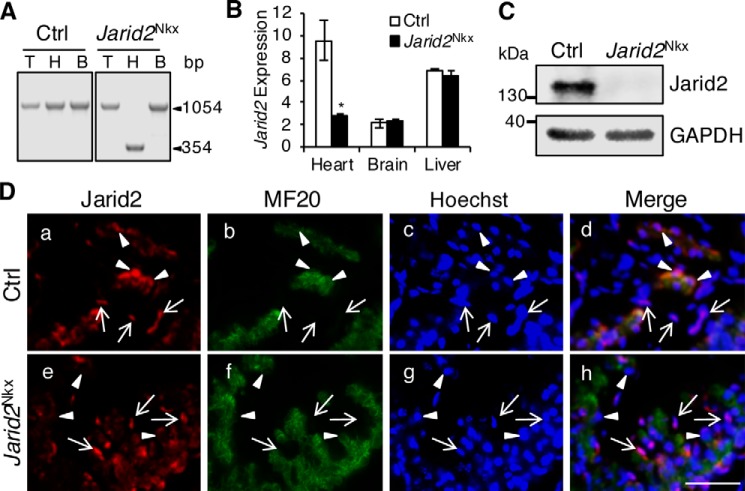
We first confirmed that Jarid2 was efficiently deleted in the heart. PCR data on isolated genomic DNAs showed that the floxed exon 3 of Jarid2 was deleted only in the heart, but not in the tail or the brain of Jarid2Nkx embryos (Fig. 1A). Jarid2 transcripts and protein levels were significantly decreased in Jarid2Nkx versus control embryonic hearts (Fig. 1, B and C). Jarid2f/f mice were used as the control (Ctrl) throughout this study.
Figure 1.
Cardiac-specific deletion of Jarid2 using Nkx2.5-Cre mice (Jarid2Nkx). A, genomic DNAs were isolated from the tail (T), heart (H), and brain (B) at E18.5. PCR was performed using primers located outside two loxP sites containing exon3 of Jarid2 (61), yielding floxed allele (1054 bp) or floxed-out allele (354 bp). B, Jarid2 mRNA levels were detected by qRT-PCR relative to 18S RNA on the control or Jarid2Nkx heart, brain, or liver at E18.5, n = 3. *, p ≤ 0.05. C, Western blotting was performed with Jarid2 antibody on control or Jarid2Nkx hearts at E13.5. GAPDH is a loading control. D, immunostaining analysis was performed on comparable transverse heart sections from E10.5 control (a–d) or Jarid2Nkx (e–h) embryos using Jarid2 (red) and MF20 (green) antibodies. Arrows indicate the endocardium and arrowheads indicate the myocardium. Scale bar, 50 μm.
Cardiac progenitors can contribute to different cardiac cell types such as endocardium. Conflicting reports exist that Nkx2.5-Cre mice delete a floxed allele only in the myocardium or both in the myocardium and endocardium due to an early expression of Cre in cardiac progenitors (20, 21, 24). To determine whether Jarid2 is deleted only in the myocardium or also in the endocardium, co-immunostaining was performed using antibodies against Jarid2 and MF20, a cardiomyocyte marker (Fig. 1D). Jarid2 was detected in the control myocardium but was not detectable in the Jarid2Nkx myocardium (arrowheads). In contrast, Jarid2 was detected in the endocardium of the Jarid2Nkx heart similar to the control endocardium as indicated by arrows. When primary cultured cells isolated from Jarid2Nkx hearts were co-immunostained using Jarid2 and MF20 or PECAM antibodies, Jarid2 expression was detected in PECAM-positive cells, whereas it was not detectable in MF20 positive cells (Fig. S1, A and B). These data suggest that Jarid2 was deleted in cardiomyocytes but not in endothelial/endocardial cells. We have previously demonstrated that Jarid2 plays important roles in the endocardium by repressing Notch1-Neuregulin1 (Nrg1) signaling pathways to the underlying myocardium (8). Because we cannot exclude a possibility that Jarid2 may be deleted in a subpopulation of endocardial cells in Jarid2Nkx hearts, we examined whether endocardial signaling pathways are altered in Jarid2Nkx hearts. qRT-PCR data showed that Notch1-Nrg1 pathways were not increased in Jarid2Nkx versus control hearts (Fig. S1C). Furthermore, none of the Jarid2 mutants generated by other cardiac Cre drivers including αMHC-, MLC2v-, and Nkx2.5-transgenic (Tg) Cre showed cardiac malformations or perinatal lethality (19). These data support that Jarid2 functions normally in the endocardial cells of Jarid2Nkx hearts.
Jarid2Nkx mice exhibit cardiac developmental defects
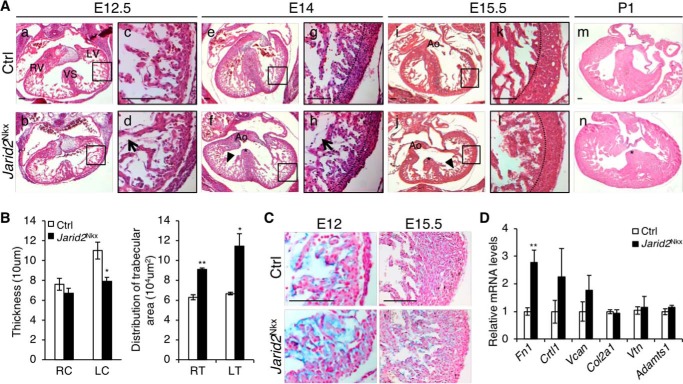
Next, we examined cardiac defects in mutant hearts during development by H&E staining of transverse sections (Fig. 2). At E12.5, Jarid2Nkx hearts showed the grossly normal trabecular and compact layers in the ventricle as compared with the control hearts (Fig. 2A, a and b). However, an increased space between the endocardium and the myocardium was observed in Jarid2Nkx versus control hearts as indicated by an arrow (Fig. 2A, c and d). Around E14, the interventricular septum in the control heart fused to the endocardial cushion and separated the right and left ventricles (Fig. 2A, e). In contrast, Jarid2Nkx hearts showed defective interventricular septation leading to VSD as indicated by a star (Fig. 2A, f). Due to decreased cardiac jelly in the normal heart around E14, the endocardium is in direct contact with the myocardium for termination of trabeculation and compaction of trabeculae into the compact layer, leading to the development of a thick ventricular wall (14). However, Jarid2Nkx ventricles at E14 showed an increased subendocardial space (arrow), a thin compact layer (Fig. 2A, h), and hypertrabeculae (Fig. 2A, f, arrowhead) as compared with controls (Fig. 2A, e and g). At E15.5, the mutants continued to show VSDs (star), thin myocardium, and hypertrabeculation (Fig. 2A, j and l, arrowhead) compared with controls (Fig. 2A, i and k), which persisted in Jarid2Nkx mutant hearts at P1 (Fig. 2A, n). Ventricular wall thickness and a distribution of trabeculae were quantitated at E15.5, indicating a decrease in compact layer thickness mainly in the left ventricle, and an increase in trabeculation in the ventricle of Jarid2Nkx hearts (Fig. 2B). Jarid2Nkx hearts exhibited partially penetrant cardiac defects as summarized in Table 2 (see details in Table S1). Some Jarid2 heterozygous mutant mice (Nkx2.5-Cre/+;Jarid2f/+) showed cardiac defects, whereas either Nkx2.5-Cre alone (Nkx2.5-Cre/+;Jarid2+/+, Table 2 and Fig. S2) or Jarid2 whole body heterozygous mutants (9) did not show any ventricular defects or lethality. Because Nkx2.5-Cre mice contain a deletion of Nkx2.5 in one allele, these results suggest that Nkx2.5 and Jarid2 may cooperate functionally or genetically during development. In summary, Jarid2Nkx mutants exhibit ventricular defects including increased subendocardial space, VSD, hypertrabeculation, and the thin compact layer of the ventricular wall.
Figure 2.
Cardiac defects were observed in Jarid2Nkx embryos. A, H&E staining was performed on transverse sections at E12.5 (a–d), E14 (e–h), E15.5 (i–l), and P1 (m and n) of Jarid2Nkx (b, d, f, h, j, l, and n) versus control (a, c, e, g, i, k, and m) mice. The boxed regions of a, b, e, f, I, and j are magnified in c, d, g, h, k, and l, respectively. Representative images of Jarid2Nkx embryos show VSD (*, f, j, and n), thin myocardium (dashed line, h and l), and disorganized hypertrabeculae (arrowheads, f and j) are shown. Arrows (d and h) indicate the increased distance between the endocardium and myocardium in Jarid2Nkx. The dotted lines separate the compact and trabecular layers. Scale bar, 100 μm. B, compact layer thickness was measured by drawing lines, and distribution of trabecular area was measured using NIH ImageJ software on the right (R) or left (L) compact layer (C) and trabecular layer (T) at E15.5. Three slides per heart were measured, n = 4. C, Alcian blue staining showed increased mucopolysaccharides in Jarid2Nkx at E12 and E15.5. The sections were counterstained with nuclear fast red. Scale bar, 100 μm. D, qRT-PCR was performed to determine the expression levels of extracellular matrix components and, a metalloproteinase, Adamts1 on control or Jarid2Nkx hearts at E13.5. The expression levels were normalized to the control; n = 3 (*, p ≤ 0.05; **, p ≤ 0.01).
Table 2.
Cardiac phenotypic defects observed in Jarid2Nkx mice
Mice at various stages were examined by H&E staining on transverse sections.
| Stages | Genotype | No. of mice | VSD | Hypertrabeculation | Thin myocardium |
|---|---|---|---|---|---|
| E15.5–E19.5 | Control (+/+;f/f) | 10 | 0 | 0 | 0 |
| Jarid2Nkx (C/+;f/f) | 11 | 10 (91%) | 9 (82%) | 8 (73%) | |
| Nkx2.5-Cre (C/+;+/+) | 7 | 0 | 0 | 0 | |
| Heterozygous (C/+;f/+) | 7 | 1 (14%) | 1 (14%) | 1 (14%) |
Increased subendocardial space in Jarid2Nkx hearts is indicative of increased cardiac jelly. The cardiac jelly in the ventricle is critical for ventricular wall development and required for the initiation and growth of trabeculation between E9.5 and 13.5 (14). Cardiac jelly components are mainly produced by the myocardium in the ventricle during the early stages of cardiac development until E12, and then the amounts of cardiac jelly begin to diminish at E12.5. The cardiac jelly is degraded by matrix metalloproteinases that are generated by the endocardium, which signals termination of trabecular growth. Thus, the heart sections were stained with Alcian blue to detect the presence of mucopolysaccharides, components of the cardiac jelly (Fig. 2C). Alcian blue staining was increased in the mutant versus control ventricle at E12. At E15.5, the mutant heart showed cardiac jelly between the endocardium and the trabecular myocardium, whereas the control heart did not show any staining. The thin compact layer is evident in the mutant ventricle compared with the control.
Because cardiac jelly was increased in Jarid2Nkx hearts, we analyzed the expression levels of cardiac jelly components that play important roles in heart development. Our previous gene expression profile showed that Fibronectin 1 (Fn1), Cartilage link protein 1 (Crtl1, Hapln1), and Versican (Vcan) are highly elevated in Jarid2 KO hearts (8). Fn1 is critical for cardiac development (25). It is produced by the myocardium and endocardium, and secreted into the cardiac jelly (26). Crtl1 is mainly expressed in the endocardial lining of the heart and in the atrioventricular junction. At later stages, it becomes restricted to endocardially derived mesenchyme. Crtl1 functions to stabilize the interaction between hyaluronan and proteoglycan such as Vcan (27). Vcan, a chondroitin sulfate proteoglycan produced by the myocardium, is normally decreased at E12.5. Vcan is proteolyzed by the ADAMTS (a disintegrin and metalloproteinase with thrombospondin motif) family for myocardial compaction (28). Our qRT-PCR data showed that only Fn1 was significantly increased in Jarid2Nkx versus control hearts (Fig. 2D). Collagen2a1 (Col2a1) was not increased, and collagen staining of heart sections using Masson's trichrome stain also indicated no increase in collagen in Jarid2Nkx versus control ventricles at E13.5 (data not shown). Vitronectin (Vtn) is a component of the extracellular matrix, and functions in cell attachment by interacting with other components or receptors (29). Vtn expression was not altered in Jarid2Nkx hearts versus controls. Adamts1 is a metalloproteinase that breaks down the cardiac jelly and contributes to the termination of trabeculation (14). Adamts1 expression was not altered, implying normal degradation of the cardiac jelly in Jarid2Nkx hearts. These data suggest that Jarid2 in the myocardium inhibits the production of the cardiac jelly but may not affect the degradation of cardiac jelly.
To determine whether the cardiac defects in Jarid2Nkx mice are due to altered cell proliferation in the ventricle, we analyzed cell proliferation rates. The number of phospho-Histone H3 or Ki67-positive cardiomyocytes in mutant heart sections was similar to control sections at E13.5 and 15.5 (Fig. S3, A and B). Both control and mutant hearts showed very low levels of cleaved caspase3 expression, indicating no significant changes in apoptosis in Jarid2Nkx versus control hearts (Fig. S3C). Our previous study also indicates no significant change in apoptosis in Jarid2 KO versus WT hearts (30).
Determination of the genetic network regulated by Jarid2
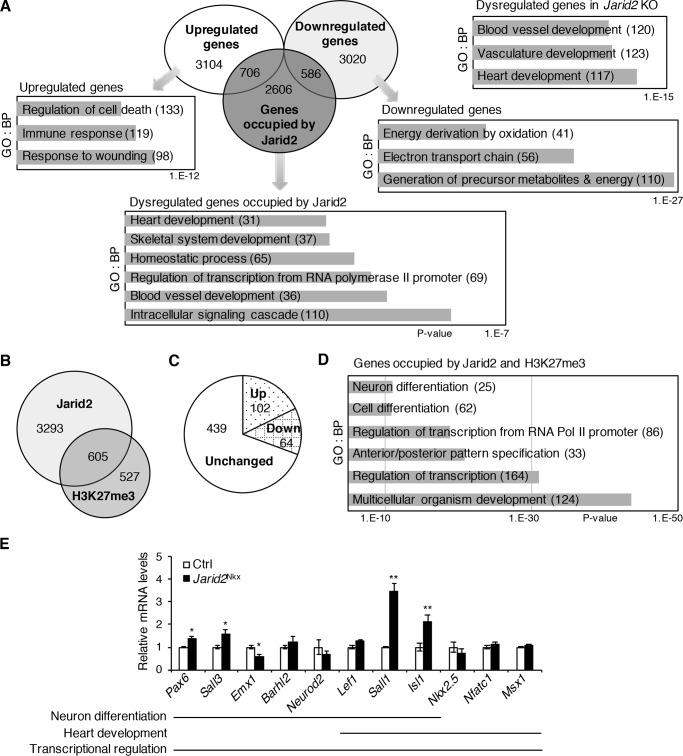
Analyses of our gene expression profile data indicated that 3606 genes were down-regulated, and 3810 genes were up-regulated in Jarid2 KO versus WT embryonic hearts (Fig. 3A, fold-change cutoff of >1.2) (8). The dysregulated genes were involved mainly in heart development and vasculature development by gene ontology (GO) analysis of biological pathways using DAVID functional analysis software (https://david.ncifcrf.gov/)3 (GO/DAVID) (Fig. 3A). Among the dysregulated genes, up-regulated genes were involved in response to wounding, whereas down-regulated genes represented a generation of precursor metabolites and energy by GO/DAVID analyses. Jarid2 has been shown to occupy the promoter regions to regulate target gene expression (31). We have identified genome-wide Jarid2 occupancy on the promoters in the developing heart using the RefSeq promoter arrays (8). To investigate the genetic network regulated by Jarid2, we overlapped the dysregulated genes in Jarid2 KO hearts with the genes whose promoters were occupied by Jarid2. Our results revealed that of 3898 promoters that were occupied by Jarid2, 1292 genes were dysregulated in Jarid2 KO hearts, of which 706 genes were up-regulated, and 586 genes were down-regulated. The overlapping genes were mainly involved in intracellular signaling cascade, blood vessel development, transcription, and skeletal system and heart development by GO/DAVID analyses.
Figure 3.
Gene expression profile analyses on the promoter occupancy by Jarid2 or H3K27me3. A, Venn diagram demonstrated the overlap of up- or down-regulated genes in Jarid2 KO hearts by microarray analyses and the genes occupied by Jarid2 from ChIP-chip (8). Highly significant biological pathways (BPs) for dysregulated genes, up- or down-regulated genes, or overlapping genes with Jarid2 ChIP-chip data were determined by GO/DAVID analyses. Numbers indicate the number of genes in each category. The x axis represents the p values. B, Venn diagram demonstrates the overlap of genome-wide occupancy of Jarid2 or H3K27me3 by ChIP-chip. C, 605 overlapping genes in B were overlaid with the microarray data. The pie chart shows the number of dysregulated genes in Jarid2 KO. D, highly significant BPs for the 605 overlapping genes were determined by GO/DAVID. E, expression levels of dysregulated genes, occupied by Jarid2, Ezh2, and H3K27me3 (Fig. S4), were examined by qRT-PCR on control or Jarid2Nkx hearts at E13.5 and normalized to the control, n = 3–4 (*, p ≤ 0.05; **, p ≤ 0.01).
Although Jarid2 can regulate H3K27 methylation (7), it remains unknown whether Jarid2 is involved in H3K27 methylation during heart development. To identify the promoters where H3K27me3 is enriched, we performed ChIP-chip on embryonic hearts using H3K27me3 antibodies, yielding 1,132 promoters (Fig. 3B). When Jarid2 ChIP data were overlapped with H3K27me3, 605 promoters were identified, which mainly represented multicellular organism development and transcription pathways by GO/DAVID analyses (Fig. 3D). Of those, 102 genes were up-regulated, and 64 genes were down-regulated in Jarid2 KO hearts (Fig. 3C), indicating a subset of Jarid2 target promoters show H3K27me3 accumulation and simultaneously dysregulated in the absence of Jarid2. These analyses also indicate that Jarid2 regulates target genes by other pathways.
To determine potential targets of Jarid2 that are co-occupied by the PRC2 complex, we overlapped Jarid2 targets with the published PRC2 targets in embryonic hearts (20), yielding 263 potential target genes (Fig. S4). To determine targets of both Jarid2 and PRC2, which concomitantly show H3K27me3 accumulation, we overlapped the three ChIP data sets. A total of 224 genes among 263 genes showed H3K27me3 accumulation, indicating that a remarkably high percentage (85.2%) of the targets occupied by both Jarid2 and Ezh2 show H3K27me3 accumulation, whereas only 15.5% (605/3898) of Jarid2 targets showed H3K27me3 accumulation. These results strongly suggest that Jarid2 forms a functional complex with PRC2 to increase or maintain H3K27me3 levels on the specific promoters in the developing heart. Next, we investigated the transcriptional network regulated by the Jarid2–PRC2–H3K27me3 axis. We performed GO/DAVID analyses with 224 genes. The top two significant pathways were regulation of RNA metabolic process and regulation of transcription. Of the 224 genes, 67 genes (29.9%) were dysregulated in Jarid2 KO hearts. Among those dysregulated genes, 39 genes were up-regulated, whereas 28 genes were down-regulated (Tables S2 and S3, respectively). Interestingly, among the up-regulated genes, 12 genes were involved in neuron differentiation (Ngfr, Sall1, Lhx1, Sall3, Emx1, Barhl2, Neurod2, and Isl1) or function (Npas3, Syt6, Lef1, and Drd4). These results imply that Jarid2 together with PRC2 inhibits neuronal pathways via H3K27me3 enrichment on specific promoter loci within the developing heart.
Among the down-regulated genes in Jarid2 KO hearts, 586 genes were occupied by Jarid2 (Fig. 3A), which represent generation of precursor metabolites and energy, and regulation of transcription pathways by GO/DAVID. Among the 586 genes, 36 genes were co-occupied by PRC2, and of the 36 genes, 28 genes also showed H3K27me3 accumulation (Table S3). These 36 or 28 genes represent embryonic organ development, and multicellular organismal processes, respectively. It is unknown how these genes are down-regulated in Jarid2 KO hearts. Because only the promoter occupancy was analyzed in this study, it is plausible that other enhancer regions of these genes may elicit dominant roles, which warrants further investigation.
We then analyzed gene expression levels in Jarid2Nkx hearts, which are selected among the 67 dysregulated genes (Fig. 3E). Neural developmental genes, including Isl1, Sall1, Sall3, and Pax6 were up-regulated in Jarid2Nkx hearts, suggesting that Jarid2 with PRC2 represses these genes via H3K27me3 accumulation. In contrast, expression levels of some genes involved in neural differentiation, including Barhl2, Neurod2, and Lef1 showed no significant difference, implying that certain potential targets are not actively repressed by myocardial Jarid2 at E13.5. The genes involved in heart development (Nkx2.5, Nfatc1, and Msx1) were not significantly changed in Jarid2Nkx hearts versus controls. Interestingly, Isl1 was significantly up-regulated. Isl1 is a critical transcription factor for the development of the secondary heart field and is highly expressed until E10 in the ventricle (32). Lef1 and Sall1 were not included in heart developmental genes by GO/DAVID analyses. However, Tcf/Lef1-mediated Wnt signaling regulates the transcription of cardiac factors, such as Isl1, Mesp1, and Anf, and cardiac development (33–35). In addition, mutations of Sall1 cause Townes-Brocks syndrome in humans with heart anomalies, and Sall1 expresses in the undifferentiated cardiac progenitor cell (36, 37). Sall1 expression was significantly increased in Jarid2Nkx hearts. Our data suggest that neural developmental pathways are suppressed by myocardial Jarid2 in the developing heart. Jarid2 elicits fine transcriptional regulatory function during ventricular myocardial differentiation partly by depositing H3K27me3 via PRC2 on specific promoter loci.
The transcriptional network regulated by Jarid2
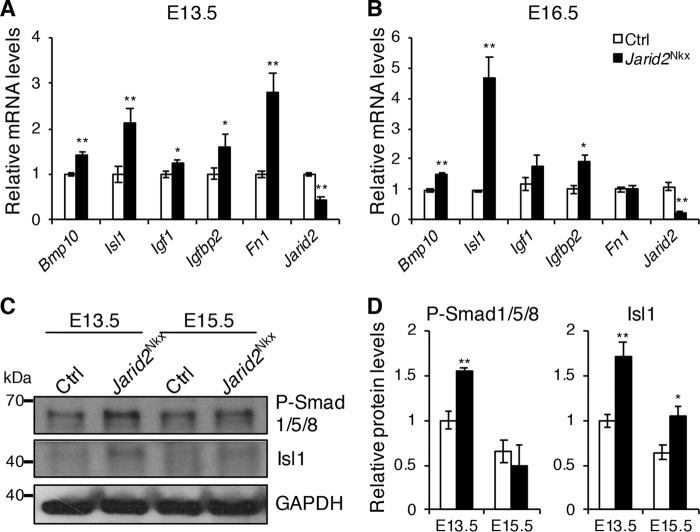
To identify transcriptional networks governed by Jarid2 in cardiac development, we have selected up-regulated genes that show more than an 1.7-fold increase in Jarid2 KO hearts (8), yielding 586 genes. An average fold-increase of all the up-regulated genes was 1.7 in our gene expression profile data. Among these, we analyzed expression levels of important genes in cardiac development, such as Bmp10, Isl1, Igf1, Igfbp2, and Fn1 in Jarid2Nkx hearts. qRT-PCR indicated that Bmp10, Isl1, and Igfbp2 were significantly elevated in Jarid2Nkx versus control hearts at E13.5 and E16.5 (Fig. 4, A and B). Igf1 and Fn1 were transiently up-regulated in Jarid2Nkx at E13.5. To confirm the protein levels, Western blotting was performed using antibodies against phosphorylated Smad1/5/8 and Isl1. P-Smad1/5/8 levels were significantly elevated in Jarid2Nkx hearts at E13.5, and Isl1 expression was increased at E13.5 and 15.5 compared with control hearts (Fig. 4, C and D). P-Smad1/5/8 is used as a marker for Bmp10 signaling. When a Bmp10 signal is activated, Smad1/5/8 is phosphorylated and activated (38). Bmp10 expression and P-Smad1/5/8 were elevated in Jarid2Nkx hearts, which correlates well with hyper-trabecular defects or ventricular myocardial maturation defects. Isl1 is expressed in conduction cells during heart development, and its expression persists in a subset of cardiac cells after birth such as in nodal cells or cardiac progenitors (39). Our gene expression profile data showed up-regulation of Bmp2, Tbx2, and Gjd3 in Jarid2 KO hearts, which are expressed in conduction cells (8, 40). Hcn4 is a marker of the sinoatrial node with co-expression of Isl1 (41). All four genes seem increased in Jarid2Nkx hearts at E13.5 although only Gjd3 showed statistical significance (Fig. S5). Thus, up-regulation of Isl1 appears to correlate with an increased expression of the conduction system genes in Jarid2Nkx hearts. Ventricular wall maturation requires the precise regulation of gene expression in the trabecular and compact layer. We examined whether the trabecular versus compact layer fate decision is defective in Jarid2Nkx hearts. Bmp10 is critical for trabecular formation and expressed only in the trabecular myocardium between E9 and E13.5 (38). In situ hybridization analyses showed that Bmp10 expression was expanded deep into the compact layer in Jarid2Nkx ventricles at E13.5 (Fig. S6). Anf is a trabecular-specific gene in the embryonic ventricle, and its expression decreases as the heart develops (9). Anf expressing cardiomyocytes are detected deep into the compact layer in Jarid2 KO embryonic hearts compared with controls. In contrast, the Hey2 expression level, a compact layer-specific gene, is not altered in Jarid2 KO hearts by qRT-PCR (19), and it was detected only in the compact layer of the Jarid2Nkx ventricle by in situ hybridization (data not shown). Thus, in Jarid2Nkx hearts, the trabecular layer appears to be expanded into the compact layer, but the expression pattern of the compact layer-specific gene has not been altered. These data imply that the fate determination of the trabecular versus compact layer is normal.
Figure 4.
Identification of dysregulated genes in Jarid2Nkx mice. qRT-PCR was performed on control or Jarid2Nkx hearts at E13.5 (A) and E16.5 (B). The expression levels were normalized to the control, n = 3–5. C, Western blotting was performed on E13.5 and E15.5 control or Jarid2Nkx hearts with phospho-Smad1/5/8 or Isl1 antibody. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a loading control. D, the graph shows the protein levels of phospho-Smad1/5/8 and Isl1 that were standardized to GAPDH and normalized to the control heart at E13.5, n = 4 (*, p ≤ 0.05; **, p ≤ 0.01).
Epigenetic mechanisms of Jarid2 in regulation of Isl1 expression
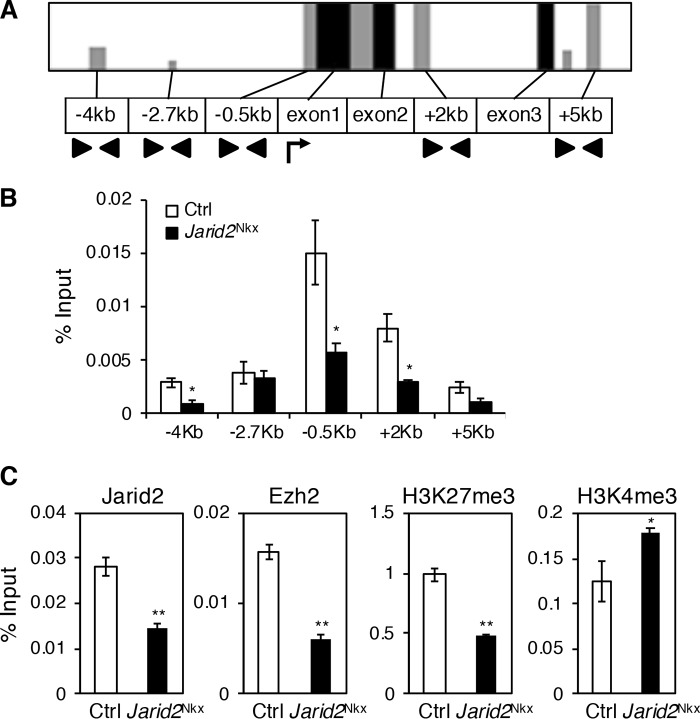
Our data indicate that Isl1 is a putative direct target of Jarid2, and one of the most up-regulated genes in the absence of Jarid2 during heart development. To determine the precise regulatory mechanism of Isl1 expression by Jarid2, we analyzed genomic occupancy of the Isl1 locus by qChIP assays. We performed VISTA alignments to identify conserved regions at the Isl1 locus (Fig. 5A). The conserved regions from 5 kilobases (kb) upstream and downstream of the Isl1 transcription start site were selected for q-ChIP assays using a Jarid2 antibody on E14.5 hearts when Isl1 is normally reduced (32). In control hearts, the high level of Jarid2 occupancy was detected at −0.5 kb relative to the transcriptional start site, which was significantly reduced in Jarid2Nkx hearts (Fig. 5B). Our data indicate that Jarid2 accumulates at the Isl1 promoter region in the developing myocardium.
Figure 5.
Jarid2 occupies a specific region at the Isl1 locus. A, VISTA alignment was performed at the Isl1 genomic locus spanning about 70 kb for mouse, monkey, human, and rat to determine conserved regions. Here, the promoter region around the 10–kb region near the transcription start site (arrow) was analyzed. Gray bars indicate regions with greater than 75% conservation, and black bars indicate exons. Arrowheads indicate primer sites. B, Jarid2 occupancy at the conserved regions was measured by qChIP assays on Jarid2Nkx or control hearts using Jarid2 antibody. The bars show enrichment compared with each input. C, qChIP assays were performed on control or Jarid2Nkx hearts at the −0.5–kb region of Isl1 using Jarid2, Ezh2, H3K27me3, or H3K4me3 antibody, n = 3 (*, p ≤ 0.05; **, p ≤ 0.01).
PRC2 is necessary for normal cardiac development, and Isl1 expression is also elevated in PRC2 KO hearts (20). However, it remains unknown whether Jarid2 cooperates with PRC2 to regulate Isl1 expression in the developing heart. We hypothesize that Jarid2 is essential for recruiting PRC2 to the Isl1 genomic locus and for histone modifications of H3K27. To test this, q-ChIP assays were performed to analyze Ezh2, a PRC2 component, and H3K27me3 accumulation in Jarid2Nkx versus control hearts at E14.5. Both Ezh2 and H3K27me3 were enriched at the −0.5–kb promoter region in control hearts, but significantly reduced in Jarid2Nkx hearts (Fig. 5C). These results suggest that the Isl1 promoter locus is occupied by Jarid2, and the same region is enriched with Ezh2 and H3K27me3 in a Jarid2-dependent manner. Although the physical interaction between Jarid2 and Ezh2 in the PRC2 complex has been well-demonstrated (7, 15), their interaction has not been shown in the heart. Our co-immunoprecipitation showed the physical interaction between Jarid2 and Ezh2 in embryonic heart extracts in vivo (Fig. S7A). To determine whether H3K4 methylation, a transcriptional activation marker, is altered, we measured H3K4me3 enrichment at the Isl1 promoter locus. H3K4me3 was increased at the −0.5–kb region of the Isl1 locus in Jarid2Nkx versus control hearts, which correlates well with the active transcriptional status of Isl1 in Jarid2Nkx hearts. Thus, early cardiac-specific deletion of Jarid2 by Nkx2.5-Cre causes a decreased PRC2 and H3K27me3 accumulation on the Isl1 promoter locus, which correlates well with a failure of Isl1 suppression in Jarid2Nkx hearts during development.
Jarid2 suppresses transcriptional activity of Isl1
To investigate whether Jarid2 represses Isl1 transcription, we constructed two reporter plasmids containing the Isl1 promoter with or without the −0.5–kb region, which is the site of Jarid2 occupancy. The reporter containing the −0.5–kb region showed a decrease in luciferase levels when co-transfected with Jarid2 in a dose-dependent manner (Fig. 6A). However, the reporter without the −0.5–kb region or pGL3 basal vector did not show any changes in luciferase levels by Jarid2. These results indicate that the −0.5–kb region is critical for repression of Isl1 transcription by Jarid2.
Figure 6.
Jarid2 represses the Isl1 reporter gene. A, pGL3, pGL3-Isl1 including the −0.5–kb region (−0.9/+0.15 kb) or without the −0.5–kb region (−0.12/+0.15 kb) was transfected into 10T1/2 cells with increasing amounts of Jarid2 (μg). B, a schematic diagram shows Jarid2, and Jarid2 mutants (N-term, 1–528 aa; TR, 1–222 aa; C-term, 529–1234 aa; NLS/C-term, 1–130/529-1234 aa). Jarid2 (0.2 μg) was transfected into 10T1/2 cells with the pGL3-Isl1 (−0.9/+0.15 kb) reporter. TR, transcription repression domain; JN, Jumonji N domain; JC, Jumonji C domain; ARID, AT-rich interaction domain. C, pGL3-Isl1 (−0.9/+0.15 kb) reporter was transfected into 10T1/2 cells with Jarid2, EED, and/or EZH2 at a low dose (50 ng). D, the TR domain of Jarid2 (1–222 aa, 25 ng) was transfected into 10T1/2 cells with or without EED and/or EZH2 (50 ng) for luciferase activity assays of pGL3-Isl1 (−0.9/+0.15 kb). Luciferase activity was normalized to the reporter gene alone. Asterisks indicate a significant difference compared with the reporter gene alone (*, p ≤ 0.05; **, p ≤ 0.01, n = 3). A number sign indicates a significant difference between a combination of any two factors and all three factors together.
To determine which domain of Jarid2 is required to repress Isl1 transcriptional activity, various Jarid2 mutant plasmids (42) were co-transfected with the Isl1 reporter (Fig. 6B). Jarid2 mutants, the N-term (aa 1–528) or TR (aa 1–222), showed about 30–40% decreases in Isl1 reporter activity. These constructs also contain the EZH2 interaction site (31). In contrast, the C-term (aa 529–1234, cytoplasmic protein) or NLS/C-term (aa 1–130/529-1234, nuclear protein) displayed no repressive activity. Because Jarid2 contains NLS in the N-terminal region (aa 1–130), the C-term protein without NLS does not localize in the nucleus. The mutant containing NLS fused to the C-term localizes in the nucleus (42). These data indicate that the TR and EZH2 interacting domain of Jarid2 is required to repress Isl1 reporter activities. Next, we determined whether PRC2 components, EZH2 and EED, regulate Isl1 transcriptional activity in cooperation with Jarid2. Jarid2 or EZH2 alone at a low dose did not repress Isl1 reporter, but EED showed a 10% reduction (Fig. 6C). Co-transfection of Jarid2 with either EZH2 or EED showed about a 30% reduction, whereas Jarid2 together with both EZH2 and EED resulted in a 50% reduction. Therefore, we tested whether the TR domain of Jarid2 is sufficient to repress Isl1 by interacting with PRC2 (Fig. 6D). Although a low dose of TR domain did not display a significant repression of Isl1, it showed a synergistic repression of about 30–40% with EED or EZH2. Additionally, a combination of EED and EZH2 with the TR domain showed about a 50% repression similar to the full-length Jarid2. Jarid2 together with EED and EZH2 significantly repressed the reporter activity as compared with EED and EZH2 without Jarid2 (Fig. 6, C and D). Altogether, our data indicate that Jarid2 and PRC2 cooperate to significantly inhibit Isl1 transcription, and that the TR domain of Jarid2 mediates PRC2-dependent inhibition of Isl1 transcription.
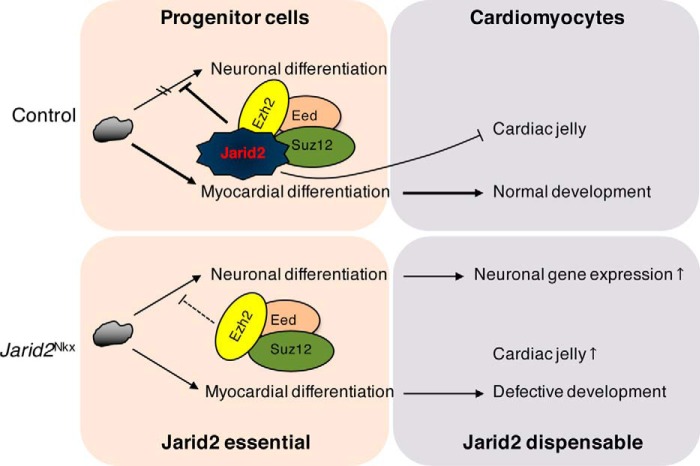
In summary, our working model depicts the mechanism of Jarid2 function within the developing myocardium (Fig. 7). There is a critical window during early cardiac progenitor differentiation when Jarid2 is required for normal cardiac development. In contrast, Jarid2 in differentiated cardiomyocytes is dispensable for cardiac morphogenesis. Our finding indicates that Jarid2 is necessary to recruit PRC2 to the promoter region of a subset of Jarid2 target genes and to establish a proper histone methyl code such as methylation of H3K27, which leads to transcriptional repression of the target genes in the developing heart. This process is instrumental for normal myocardial differentiation, in part by repressing noncardiac lineage developmental pathways and regulating cardiac jelly components.
Figure 7.
Jarid2-mediated gene repression is required for normal cardiomyocyte differentiation. Jarid2 and PRC2 complex repress neuronal gene expression by deposition of H3K27me3 epigenetic marks in early cardiac cells. However, Jarid2 deficiency in cardiac cells relieves the suppressive function of PRC2 complex on neuronal genes, and increases cardiac jelly production, all of which contribute to abnormal cardiac differentiation.
Discussion
Jarid2 is essential for embryonic development and is a recognized component of pluripotency networks (5–7). The present study demonstrates that cardiac-specific deletion of Jarid2 using Nkx2.5-Cre mice causes neonatal lethality and ventricular defects including VSD, hypertrabeculation, and the thin compact layer leading to the thin ventricular myocardial wall. The genes whose promoters are occupied by Jarid2, PRC2, and H2K27me3 showed dysregulated biological pathways in Jarid2-deficient hearts such as neuronal differentiation pathways. Of note, cardiac-specific deletion of PRC2 results in cardiac developmental defects, which are remarkably similar to those observed in Jarid2 KO mice (9, 20). In contrast, cardiomyocyte-specific deletion of Jarid2 by cTnT-Cre mice did not cause gross cardiac malformations or perinatal lethality, indicating that Jarid2 in differentiated cardiomyocytes is dispensable for cardiac morphogenesis.
To determine roles of Jarid2 in differentiated cardiomyocytes, Jarid2-floxed mice have been crossed with different myocardial-specific Cre mice, including αMHC-Cre, Nkx2.5-Cre Tg (19). These mutant mice show grossly normal hearts and survive to adulthood, which might be due to the late expression of Cre. Nkx2.5-Cre Tg mice show Cre activity at E8.0 in the myocardium and a subset of endocardial cells (43). Because αMHC-Cre mice activate Cre from E8.5 in the myocardium (23), we employed cTnT-Cre mice that express Cre early from E7.5 (22). However, Jarid2 deletion by cTnT-Cre mice did not cause cardiac developmental defects or perinatal lethality. Although Nkx2.5-Cre and cTnt-Cre mice start to express Cre recombinase from E7.5 in the heart, a complete recombination in the cardiogenic fields is observed at E8.0 and E8.5, respectively (44). In addition, cTnt-Cre expression starts later in the secondary heart field. These may have caused a lack of cardiac developmental defects in Jarid2cTnt. Thus, our results indicate that Jarid2 is necessary in early cardiac progenitors for normal heart development at later stages. Perturbation of this early process may contribute to cardiac malformations manifested later in development. It would be interesting to investigate the impact of Jarid2 deletion on adult heart pathophysiology using αMHC-Cre mice.
Nkx2.5 is a cardiac-specific transcription factor and expressed in the cardiac mesoderm from E7.5 to adult cardiomyocytes. It is believed that endocardial cells are also derived from Nkx2.5-positive cardiac progenitors in the cardiac mesoderm (21, 24). When the progenitor cells differentiate into endocardial cells around E7.5–8.0, they lose Nkx2.5 expression, whereas myocardial cells continue to express Nkx2.5. Because the Nkx2.5-Cre mice employed in this study express Cre from E7.5 in cardiac progenitors, other lineages such as endocardial cells may have expressed Cre (21). However, our analyses indicate a myocardial-specific deletion of Jarid2 in Jarid2Nkx hearts (Fig. 1, and Fig. S1). Thus, the cardiac defects observed in Jarid2Nkx hearts are mainly caused by myocardial deletion of Jarid2, although we cannot exclude a possibility that Jarid2 may be deleted in a subset of endocardial cells. Nkx2.5-Cre mice are haploinsufficient for Nkx2.5, and Nkx2.5 haploinsufficiency is associated with atrial septal defects, VSD, and conduction system abnormalities albeit at a low rate in mouse models (45, 46). Thus, we examined Nkx2.5-Cre (Nkx2.5-Cre/+;Jarid2+/+) and heterozygous (Nkx2.5-Cre/+;Jarid2f/+) mice for possible cardiac defects. All Nkx2.5-Cre embryos (Table 2, Fig. S2) or Jarid2 heterozygous KO embryos (9) showed normal ventricular structure in our genetic background. Nkx2.5-Cre/+;Jarid2f/+ compound heterozygous mice showed cardiac defects with low penetrance, suggesting putative functional and/or genetic interactions between Jarid2 and Nkx2.5. Additional mice with Nkx2.5-Cre/+;Jarid2f/+ need to be examined to determine possible interactions between Jarid2 and Nkx2.5. However, the Mendelian ratio of Nkx2.5-Cre/+;Jarid2f/+ mice was normal (Table 1). Moreover, Nkx2.5 transcript levels appear unchanged in Jarid2Nkx hearts versus controls (Fig. 3E), implying that Nkx2.5 haploinsufficiency may not have contributed significantly to cardiac defects or lethality observed in Jarid2Nkx mice. Jarid2Nkx embryos exhibited partial penetrance (Table 2). It may be due to a limitation of Cre-loxP technology, an incomplete Cre-mediated deletion of the Jarid2 floxed allele in a subset of cells. However, it should be noted that each mutant showed at least one or more cardiac defects (Table S1).
Regulation of the cardiac jelly is crucial for normal trabeculation in the ventricle wall, which requires cross-talk between the endocardium and myocardium (14). Increased cardiac jelly in the Jarid2Nkx ventricle may have contributed to hypertrabeculation and thin myocardium, likely due to the failure of repressing trabeculation and compacting the ventricular wall. Intriguingly, both Jarid2 KO and endothelial deletion cause increased subendocardial space (9, 19), which indicate that complex regulatory mechanisms exist to regulate the cardiac jelly expression involving both the myocardium and endocardium. In our data, Fn1 was significantly increased at E13.5 in Jarid2Nkx hearts. Deletion of the Fn1 gene in mice causes embryonic lethality due to severe cardiac defects (25). Thus, it would be interesting to determine the mechanism by which Jarid2 regulates Fn1 expression during heart development.
Various mouse models have been generated and studied for ventricular wall morphogenesis or noncompaction cardiomyopathy. Some models exhibit reduced cell proliferation in the developing heart. These include cardiac-specific PRC2 deletion mice exhibiting hypertrabeculation and thin compact layer (20), and Bmp10 deletion mice showing decreased trabeculation and thin myocardium leading to the hypoplastic ventricle (47). To the contrary, some mouse models show increased cell proliferation mainly in the trabecular layer. For example, Jarid2 KO, Fkbp1a deletion, or cardiac-specific deletion of Mib1 in mice result in hypertrabeculation and thin compact layer leading to the thin ventricular myocardium (9, 48, 49). In some mouse models, cell proliferation was not altered. For example, Daam1-deficient hearts show hypertrabeculation and noncompaction phenotype (50), and the endothelial deletion of Brg1 causes reduced cardiac jelly, reduced trabeculae, and thin compact layer (14). Our Jarid2Nkx mice also showed no significant changes in cell proliferation at E13.5 and E15 when ventricular defects were obvious. Thus, although cell proliferation is critically involved in the formation of trabecular and compact layers, other processes such as planar cell polarity, cell adhesion and alignment, and proper myocardial differentiation are also important. There has been no direct evidence that the trabecular layer contributes to the thickened ventricular wall at late stages. Interestingly, recent studies using lineage-tracing experiments indicate that both trabecular and compact myocardium contribute to generating the middle hybrid myocardial zone of the ventricular myocardium, although the myocytes from the compact layer contribute more than the trabecular cardiomyocytes (51). Some coronary vessels in the myocardium seem to arise from an endocardial lineage, suggesting endocardial cells are trapped during trabecular coalescence (52).
Isl1, an important early cardiac transcription factor, was identified as one of the direct target genes of Jarid2 and up-regulated in Jarid2Nkx hearts. Isl1 expression is also up-regulated in Nkx2.5-Cre mediated PRC2 KO hearts (20), supporting functional cooperation between Jarid2 with PRC2 within the developing heart. Isl1-null mice are embryonic lethal at E10.5 with severe abnormalities in the heart (32). However, the effect of Isl1 overexpression within the heart remains unknown although Isl1 overexpression enhances differentiation of ES cells into cardiac progenitors (53). Isl1 is a marker of the secondary heart field, but recent studies indicate that Isl1 also expresses in the primary heart field (24, 53). Although Isl1 expression is reduced around E10.5 during normal heart development, Isl1-positive cells have been reported as cardiac progenitors or nodal cells in embryonic and adult hearts (39). Therefore, overexpression of Isl1 in Jarid2Nkx hearts may be indicative of myocardial differentiation defects or increased progenitor populations in the Jarid2Nkx heart. Persistent expression of Isl1 may contribute to a failure of neuronal gene repression and/or an increase in conduction system-specific gene expression, rendering defective ventricular maturation. Although Bmp10 expression was increased in Jarid2Nkx hearts, Bmp10 promoter loci were not identified as Jarid2 targets by ChIP-chip, suggesting indirect regulation of Bmp10 expression by Jarid2. Interestingly, Bmp10 is also induced in PRC2 KO hearts, but not directly regulated by PRC2 (20). Because Bmp10 expression is restricted to the trabecular layer in the normal heart and critical for trabecular formation (38), it would be interesting to identify the regulatory mechanism of Bmp10 expression.
In this study, we used whole heart extracts. The heart contains heterogeneous cell populations including fibroblasts and endothelial cells, but cardiomyocytes are a major cell type in the embryonic heart. Jarid2 expresses at a higher level in cardiomyocytes compared with other cell types in the embryonic heart (30), and the fibroblasts express very low levels of Jarid2 (54). As shown in Fig. 1, Jarid2 expression levels are significantly reduced in Jarid2Nkx versus control hearts. Thus, major molecular changes in Jarid2Nkx hearts are likely to be detected using whole heart extracts.
In undifferentiated ES cells, many genes that are required for subsequent states of development are enriched with histones modified simultaneously for active transcription (H3K4me2/3) and PRC2-mediated repression (H3K27me3), which is referred to as being “bivalent” (17). This serves to prime undifferentiated cells to respond rapidly to lineage-dependent induction.
Histone methylation is tightly regulated in part by balancing functions of Jmj histone demethylases and SET domain containing histone methylases (3). Moreover, the methylation status of H3K27 impacts on H3K4 methylation and vice versa (55, 56). These cross-talks are important for fine regulation of the histone methyl code, and developmental gene expression. Jarid2 may mediate H3K4 methylation as shown in Fig. 5C. It would be interesting to determine whether Jarid2 facilitates demethylation of H3K4 or inhibits methylation of H3K4. Jarid1B, a H3K4 demethylase, regulates mouse development by protecting developmental genes from inappropriate H3K4me3 accumulation such as neural master regulators (57). Recently, de novo mutations identified in congenital heart disease patients are mainly in histone modifying genes (4). In particular, five genes encode proteins that regulate H3K4me3 including Jarid1B, highlighting the importance of H3K4 methylation status during heart development.
Deletion of Ezh2 in the secondary heart field causes postnatal myocardial pathology and destabilizes cardiac gene expression with the activation of Six1 (58). This work suggests that epigenetic dysregulation in embryonic progenitor cells is a predisposing factor for adult disease and dysregulated stress responses. Because our data indicate that Jarid2 regulates only a subset of targets through PRC2 in the developing heart, other target genes of Jarid2 should be regulated by different mechanisms. Indeed, Jarid2 regulates other target gene expression via interaction with Setdb1 by depositing H3K9me3 epigenetic marks during heart and immune cell development (8, 59). Jarid2 also interacts with long noncoding RNAs, such as MEG3 for proper recruitment of PRC2 at target genes in pluripotent stem cells or with Xist long noncoding RNA for X chromosome inactivation (60). Thus, complex epigenetic regulatory mechanisms exist to confer distinct roles of Jarid2 in different developmental processes. Together, our results indicate that Jarid2 is necessary during a narrow developmental window to establish correct epigenetics on the target genomic loci, which is prior to differentiation of cardiac progenitors into cardiomyocytes. Once cardiac progenitors are differentiated to cardiomyocytes, Jarid2 appears dispensable for cardiac morphogenesis. It would be interesting to determine whether the cardiac progenitors at early stages around E7.5 already show an elevated neuronal profile in the Jarid2Nkx mice.
Experimental procedures
Animal husbandry and genotyping
All the mice were housed at the animal facility in accordance with University of Wisconsin Research Animal Resource Center policies and the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals. All animal research has been reviewed and approved by an Institutional Animal Care and Use Committee (protocol M005971). All mice were littermate or age-matched control and mutants. Studies were not blinded. Herein, Jarid2 conditional deletion mice using Nkx2.5-Cre knock-in mice (21), Nkx2.5-Cre/+;Jarid2f/f, are designated as Jarid2Nkx. To generate Jarid2Nkx mice, females with floxed Jarid2 alleles (Jarid2f/f) (61) were mated with Nkx2.5-Cre/+;Jarid2f/+ males. cTnt-Cre mice (Jackson Laboratories) were employed to delete Jarid2 in differentiated cardiomyocytes (cTnt-Cre/+;Jarid2f/f), designated as Jarid2cTnt. Embryos were isolated from timed-mated females at E9.5–19.5 days postcoitum. All mice employed in this study were bred to a mixed 129/Svj and C57BL/6 genetic background, and genotyping was performed as described previously (61).
Western blotting, coimmunoprecipitation, and primary cultures of cardiomyocytes
To determine the protein levels, Western blotting was performed using embryonic heart extracts, as described previously (19). The primary antibodies used were anti-Jarid2 peptide antibodies (19), anti-Isl1 (DSHB, Developmental Studies Hybridoma Bank), anti-Phospho-Smad 1/5/8 (CST), or anti-GAPDH (EMD) followed by HRP-conjugated secondary antibodies (Santa Cruz). Protein bands were detected by chemiluminescence (Thermo Fisher) and quantitated with NIH ImageJ. Coimmunoprecipitation was performed as described (8). Briefly, precleared nuclear extracts from E15.5 hearts were immunoprecipitated with nonspecific rabbit IgG or Jarid2 antibody, followed by incubation with protein A/G-agarose beads (Santa Cruz), SDS-PAGE, and Western blotting with Ezh2 antibody (CST). Primary cultures of embryonic hearts at E15.5 were prepared as described (62), yielding about 70% cardiomyocytes under our culture conditions. The cells on coverslips were subjected to co-immunostaining using Jarid2 with PECAM (BD Biosciences) or MF20 (DSHB) antibodies.
In situ hybridization, histology, and immunohistochemistry
In situ hybridization was performed to examine the expression pattern of Bmp10 mRNA in mouse embryonic hearts. Section in situ at E13.5 was carried out using digoxigenin/UTP-labeled antisense cRNA probes (Roche) as described (19). Bmp10-C1/pSK(+) plasmid was obtained from Dr. W. Shou (20).
Hematoxylin and eosin (H&E) staining was performed as described (9). Immunohistochemistry was performed on paraffin-embedded sections as described (19). Briefly, tissue sections were incubated with primary antibodies, anti-Jarid2, anti-MF20, anti-Ki67 (Abcam), or anti-P-H3 (EMD). Alexa dye-conjugated secondary antibodies (Thermo Fisher) or Biotin (Sigma)/streptavidin-HRP (Thermo Fisher) systems with diaminobenzidine substrate kit (Vector Laboratories) were used for visualization. Hoechst dye was used for the counterstaining of nuclei. Images were taken using a Zeiss Axiovert 200 microscope and an AxoiCam HRc camera. Alcian blue staining for cardiac jelly and Masson's trichrome staining for collagen were performed as described (9, 63).
Quantitative ChIP (q-ChIP) and ChIP-chip assays
qChIP experiments were performed as described previously (8). All the experiments were repeated in duplicate at least three times on E14.5 control and Jaird2Nkx hearts with preimmune serum, Jarid2, Ezh2 (CST), H3K27me3 (EMD), or H3K4me3 (EMD) antibody. For the amplification of the Isl1 locus, the following primers were used: −4–kb forward, 5′-caaagattccggagaaaggaatg-3′; −4–kb reverse, 5′-gagttcaggtggttgtttctgtcat-3′; −2.7–kb forward, 5′-gaagtccaattttgacaggagagtgt-3′; −2.7–kb reverse, 5′-cctcttgtgttcaatgagggatt-3′; −0.5–kb forward, 5′-gttccaagtgccccccttt-3′; −0.5–kb reverse, 5′-agtagctggtgggtaggtccttc-3′; +2 kb forward, 5′-gaattagacagagcagatcaaattgc-3′; +2–kb reverse, 5′-ccaattgttcgcagacagatga-3′; +5 kb forward, 5′-ttttaaaaaggagcctgcctctt-3′; and +5–b reverse, 5′-caccaaatcacgtagaatgaatgg-3′.
ChIP-chip for H3K27me3 was performed as we described (8, 64). Briefly, sonicated chromatin from 20 pooled E17.5-fixed hearts was immunoprecipitated using H3K27me3 antibody, followed by the reversal of cross-linking and DNA purification. Immunoenriched DNA targets were amplified by whole genome amplification and fluorescently labeled, which were then hybridized onto the Roche NimbleGen 3X720K RefSeq promoter arrays and scanned with an Axon 4000B. After the arrays were extracted using Nimblescan (Roche), global and local normalization and data smoothing in R was performed, and peaks were detected using ChIPOlte (64) and in-house algorithms. Peaks with a p value less than 10−14 were used for analyses.
Reporter gene assays and qRT-PCR
Reporter gene assays were performed as described previously (19). An Isl1 reporter plasmid containing the Jarid2 occupied region (−0.5–kb region) was constructed by subcloning the Isl1 locus from −0.9 to +0.15 kb of the transcriptional start site into the pGL3 basic vector (Promega). A reporter plasmid lacking the Jarid2-occupied region was constructed by subcloning a region from −0.12 to +0.15 kb into the pGL3 basic vector. The reporter vector (100 ng) was transfected into 10T1/2 cells in a 24-well plate. Jarid2 or Jarid2 mutants in pcDNA3.1-HisB-Xpress (42) were co-transfected with or without EED/pCDH, or EZH2/pCDH (from Dr. P. Lewis) using Lipofectamine 2000 (Thermo Fisher). Luciferase assays were performed 2 days after transfection using the luciferase assay system (Promega). A β-gal–cytomegalovirus vector was used for normalizing the luciferase activity. The Jarid2 mutant constructs have been characterized in detail (42, 65), and were expressed equally well when transfected as previously reported (Fig. S7B). Thus, differences in their transcriptional activities are not caused by different expression levels of the mutants.
qRT-PCR was performed as we described (19). Briefly, mRNAs extracted from embryonic hearts were reverse transcribed to cDNA followed by qRT-PCR using FastStart SYBR Green Master (Roche) on a Bio-Rad iCycler. The appropriate primers for each gene are listed in Table S4. All primers were thoroughly evaluated by melt curve analysis to ensure the amplification of a single, desired amplicon. All samples were assayed in duplicate with nearly identical replicate values. Data were generated using the standard curve method and normalized to 18S expression. qRT-PCR data were analyzed by the RQ analysis algorithm (Bio-Rad).
Statistical analysis
Data represent the average of 3 to 5 replicates and mean ± S.E. The replicate numbers are indicated in the text. Significance was tested by the Student's t test for 2 groups: *, p ≤ 0.05; **, p ≤ 0.01.
Author contributions
E. C. and Y. L. conceptualization; E. C. and Y. L. data curation; E. C. and Y. L. formal analysis; E. C. visualization; E. C. and Y. L. methodology; E. C. and Y. L. writing-original draft; M. R. M., C. D. C., A. Z. A., and R. J. S. resources; M. R. M. and Y. L. writing-review and editing; C. D. C. and A. Z. A. software; Y. L. funding acquisition; Y. L. investigation; Y. L. project administration.
Supplementary Material
Acknowledgments
We thank Drs. Weinian Shou and Peter Lewis for generously providing Bmp10 in situ probe, and EZH2 and EED in expression vector, respectively. We thank Dr. Chad Vezina for technical assistance for in situ hybridization.
This work was supported in part by National Institutes of Health Grant HL067050 and American Heart Association Grant 12GRNT12070021 (to Y. L.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S7 and Tables S1–S4.
The ChIP-chip data reported in this paper have been submitted to the Gene Expression Omnibus (GEO) under GEO accession no. GSE113895.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- Jarid2
- Jumonji A/T-rich interaction domain-2
- PRC2
- polycomb repressive complex-2
- H3K27me3
- trimethylation of histone H3 lysine 27
- H3K4me3
- trimethylation of H3K4
- Jarid2Nkx
- Nkx2.5-Cre/+;Jarid2f/f genotype
- Jarid2cTnt
- cTnt-Cre/+;Jarid2f/f genotype
- VSD
- ventricular septal defect
- LVNC
- left ventricular noncompaction
- ES
- embryonic stem
- Vcan
- Versican
- qRT
- quantitative-real time
- GO
- gene ontology
- aa
- amino acid(s)
- NLS
- nuclear localization signal
- C-term
- C-terminal.
References
- 1. Benjamin E. J., Blaha M. J., Chiuve S. E., Cushman M., Das S. R., Deo R., de Ferranti S. D., Floyd J., Fornage M., Gillespie C., Isasi C. R., Jiménez M. C., Jordan L. C., Judd S. E., Lackland D., et al. (2017) Heart disease and stroke statistics-2017 update: a report from the American Heart Association. Circulation 135, e146–e603 10.1161/CIR.0000000000000485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Greer E. L., and Shi Y. (2012) Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 13, 343–357 10.1038/nrg3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dimitrova E., Turberfield A. H., and Klose R. J. (2015) Histone demethylases in chromatin biology and beyond. EMBO Rep. 16, 1620–1639 10.15252/embr.201541113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zaidi S., Choi M., Wakimoto H., Ma L., Jiang J., Overton J. D., Romano-Adesman A., Bjornson R. D., Breitbart R. E., Brown K. K., Carriero N. J., Cheung Y. H., Deanfield J., DePalma S., Fakhro K. A., et al. (2013) De novo mutations in histone-modifying genes in congenital heart disease. Nature 498, 220–223 10.1038/nature12141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Johansson C., Tumber A., Che K., Cain P., Nowak R., Gileadi C., and Oppermann U. (2014) The roles of Jumonji-type oxygenases in human disease. Epigenomics 6, 89–120 10.2217/epi.13.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jung J., Mysliwiec M. R., and Lee Y. (2005) Roles of JUMONJI in mouse embryonic development. Dev. Dyn. 232, 21–32 10.1002/dvdy.20204 [DOI] [PubMed] [Google Scholar]
- 7. Shen X., Kim W., Fujiwara Y., Simon M. D., Liu Y., Mysliwiec M. R., Yuan G. C., Lee Y., and Orkin S. H. (2009) Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell 139, 1303–1314 10.1016/j.cell.2009.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mysliwiec M. R., Carlson C. D., Tietjen J., Hung H., Ansari A. Z., and Lee Y. (2012) Jarid2 (Jumonji, AT-rich interactive domain 2) regulates NOTCH1 expression via histone modification in the developing heart. J. Biol. Chem. 287, 1235–1241 10.1074/jbc.M111.315945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee Y., Song A. J., Baker R., Micales B., Conway S. J., and Lyons G. E. (2000) Jumonji, a nuclear protein that is necessary for normal heart development. Circ. Res. 86, 932–938 10.1161/01.RES.86.9.932 [DOI] [PubMed] [Google Scholar]
- 10. Takeuchi T., Kojima M., Nakajima K., and Kondo S. (1999) jumonji gene is essential for the neurulation and cardiac development of mouse embryos with a C3H/He background. Mech. Dev. 86, 29–38 10.1016/S0925-4773(99)00100-8 [DOI] [PubMed] [Google Scholar]
- 11. Towbin J. A., Lorts A., and Jefferies J. L. (2015) Left ventricular non-compaction cardiomyopathy. Lancet 386, 813–825 10.1016/S0140-6736(14)61282-4 [DOI] [PubMed] [Google Scholar]
- 12. Zhang W., Chen H., Qu X., Chang C. P., and Shou W. (2013) Molecular mechanism of ventricular trabeculation/compaction and the pathogenesis of the left ventricular noncompaction cardiomyopathy (LVNC). Am. J. Med. Genet. C Semin. Med. Genet. 163C, 144–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen H., Zhang W., Li D., Cordes T. M., Mark Payne R., and Shou W. (2009) Analysis of ventricular hypertrabeculation and noncompaction using genetically engineered mouse models. Pediatr. Cardiol. 30, 626–634 10.1007/s00246-009-9406-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stankunas K., Hang C. T., Tsun Z. Y., Chen H., Lee N. V., Wu J. I., Shang C., Bayle J. H., Shou W., Iruela-Arispe M. L., and Chang C. P. (2008) Endocardial Brg1 represses ADAMTS1 to maintain the microenvironment for myocardial morphogenesis. Dev. Cell 14, 298–311 10.1016/j.devcel.2007.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Peng J. C., Valouev A., Swigut T., Zhang J., Zhao Y., Sidow A., and Wysocka J. (2009) Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 139, 1290–1302 10.1016/j.cell.2009.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Landeira D., Sauer S., Poot R., Dvorkina M., Mazzarella L., Jørgensen H. F., Pereira C. F., Leleu M., Piccolo F. M., Spivakov M., Brookes E., Pombo A., Fisher C., Skarnes W. C., Snoek T., et al. (2010) Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA polymerase II to developmental regulators. Nat. Cell Biol. 12, 618–624 10.1038/ncb2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jones A., and Wang H. (2010) Polycomb repressive complex 2 in embryonic stem cells: an overview. Protein Cell 1, 1056–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vizán P., Beringer M., Ballaré C., and Di Croce L. (2015) Role of PRC2-associated factors in stem cells and disease. FEBS J. 282, 1723–1735 10.1111/febs.13083 [DOI] [PubMed] [Google Scholar]
- 19. Mysliwiec M. R., Bresnick E. H., and Lee Y. (2011) Endothelial Jarid2/Jumonji is required for normal cardiac development and proper Notch1 expression. J. Biol. Chem. 286, 17193–17204 10.1074/jbc.M110.205146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. He A., Ma Q., Cao J., von Gise A., Zhou P., Xie H., Zhang B., Hsing M., Christodoulou D. C., Cahan P., Daley G. Q., Kong S. W., Orkin S. H., Seidman C. E., Seidman J. G., and Pu W. T. (2012) Polycomb repressive complex 2 regulates normal development of the mouse heart. Circ. Res. 110, 406–415 10.1161/CIRCRESAHA.111.252205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moses K. A., DeMayo F., Braun R. M., Reecy J. L., and Schwartz R. J. (2001) Embryonic expression of an Nkx2–5/Cre gene using ROSA26 reporter mice. Genesis 31, 176–180 10.1002/gene.10022 [DOI] [PubMed] [Google Scholar]
- 22. Jiao K., Kulessa H., Tompkins K., Zhou Y., Batts L., Baldwin H. S., and Hogan B. L. (2003) An essential role of Bmp4 in the atrioventricular septation of the mouse heart. Genes Dev. 17, 2362–2367 10.1101/gad.1124803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Agah R., Frenkel P. A., French B. A., Michael L. H., Overbeek P. A., and Schneider M. D. (1997) Gene recombination in postmitotic cells: targeted expression of Cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J. Clin. Invest. 100, 169–179 10.1172/JCI119509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ma Q., Zhou B., and Pu W. T. (2008) Reassessment of Isl1 and Nkx2–5 cardiac fate maps using a Gata4-based reporter of Cre activity. Dev. Biol. 323, 98–104 10.1016/j.ydbio.2008.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen D., Wang X., Liang D., Gordon J., Mittal A., Manley N., Degenhardt K., and Astrof S. (2015) Fibronectin signals through integrin α5β1 to regulate cardiovascular development in a cell type-specific manner. Dev. Biol. 407, 195–210 10.1016/j.ydbio.2015.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ffrench-Constant C., and Hynes R. O. (1988) Patterns of fibronectin gene expression and splicing during cell migration in chicken embryos. Development 104, 369–382 [DOI] [PubMed] [Google Scholar]
- 27. Lockhart M., Wirrig E., Phelps A., and Wessels A. (2011) Extracellular matrix and heart development. Birth Defects Res. Part A Clin. Mol. Teratol. 91, 535–550 10.1002/bdra.20810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nandadasa S., Foulcer S., and Apte S. S. (2014) The multiple, complex roles of versican and its proteolytic turnover by ADAMTS proteases during embryogenesis. Matrix Biol. 35, 34–41 10.1016/j.matbio.2014.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leavesley D. I., Kashyap A. S., Croll T., Sivaramakrishnan M., Shokoohmand A., Hollier B. G., and Upton Z. (2013) Vitronectin: master controller or micromanager? IUBMB Life 65, 807–818 [DOI] [PubMed] [Google Scholar]
- 30. Jung J., Kim T. G., Lyons G. E., Kim H. R., and Lee Y. (2005) Jumonji regulates cardiomyocyte proliferation via interaction with retinoblastoma protein. J. Biol. Chem. 280, 30916–30923 10.1074/jbc.M414482200 [DOI] [PubMed] [Google Scholar]
- 31. Pasini D., Cloos P. A., Walfridsson J., Olsson L., Bukowski J. P., Johansen J. V., Bak M., Tommerup N., Rappsilber J., and Helin K. (2010) JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature 464, 306–310 10.1038/nature08788 [DOI] [PubMed] [Google Scholar]
- 32. Cai C. L., Liang X., Shi Y., Chu P. H., Pfaff S. L., Chen J., and Evans S. (2003) Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev. Cell 5, 877–889 10.1016/S1534-5807(03)00363-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lu H., Li Y., Wang Y., Liu Y., Wang W., Jia Z., Chen P., Ma K., and Zhou C. (2014) Wnt-promoted Isl1 expression through a novel TCF/LEF1 binding site and H3K9 acetylation in early stages of cardiomyocyte differentiation of P19CL6 cells. Mol. Cell. Biochem. 391, 183–192 10.1007/s11010-014-2001-y [DOI] [PubMed] [Google Scholar]
- 34. Li Y., Yu W., Cooney A. J., Schwartz R. J., and Liu Y. (2013) Brief report: Oct4 and canonical Wnt signaling regulate the cardiac lineage factor Mesp1 through a Tcf/Lef-Oct4 composite element. Stem Cells 31, 1213–1217 10.1002/stem.1362 [DOI] [PubMed] [Google Scholar]
- 35. Zhang C. G., Jia Z. Q., Li B. H., Zhang H., Liu Y. N., Chen P., Ma K. T., and Zhou C. Y. (2009) β-Catenin/TCF/LEF1 can directly regulate phenylephrine-induced cell hypertrophy and Anf transcription in cardiomyocytes. Biochem. Biophys. Res. Commun. 390, 258–262 10.1016/j.bbrc.2009.09.101 [DOI] [PubMed] [Google Scholar]
- 36. Morita Y., Andersen P., Hotta A., Tsukahara Y., Sasagawa N., Hayashida N., Koga C., Nishikawa M., Saga Y., Evans S. M., Koshiba-Takeuchi K., Nishinakamura R., Yoshida Y., Kwon C., and Takeuchi J. K. (2016) Sall1 transiently marks undifferentiated heart precursors and regulates their fate. J. Mol. Cell. Cardiol. 92, 158–162 10.1016/j.yjmcc.2016.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Miller E. M., Hopkin R., Bao L., and Ware S. M. (2012) Implications for genotype-phenotype predictions in Townes-Brocks syndrome: case report of a novel SALL1 deletion and review of the literature. Am. J. Med. Genet. A. 158A, 533–540 [DOI] [PubMed] [Google Scholar]
- 38. Huang J., Elicker J., Bowens N., Liu X., Cheng L., Cappola T. P., Zhu X., and Parmacek M. S. (2012) Myocardin regulates BMP10 expression and is required for heart development. J. Clin. Invest. 122, 3678–3691 10.1172/JCI63635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Laugwitz K. L., Moretti A., Caron L., Nakano A., and Chien K. R. (2008) Islet1 cardiovascular progenitors: a single source for heart lineages? Development 135, 193–205 [DOI] [PubMed] [Google Scholar]
- 40. Christoffels V. M., Smits G. J., Kispert A., and Moorman A. F. (2010) Development of the pacemaker tissues of the heart. Circ. Res. 106, 240–254 10.1161/CIRCRESAHA.109.205419 [DOI] [PubMed] [Google Scholar]
- 41. Liang X., Zhang Q., Cattaneo P., Zhuang S., Gong X., Spann N. J., Jiang C., Cao X., Zhao X., Zhang X., Bu L., Wang G., Chen H. S., Zhuang T., et al. (2015) Transcription factor ISL1 is essential for pacemaker development and function. J. Clin. Invest. 125, 3256–3268 10.1172/JCI68257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim T. G., Kraus J. C., Chen J., and Lee Y. (2003) JUMONJI, a critical factor for cardiac development, functions as a transcriptional repressor. J. Biol. Chem. 278, 42247–42255 10.1074/jbc.M307386200 [DOI] [PubMed] [Google Scholar]
- 43. McFadden D. G., Barbosa A. C., Richardson J. A., Schneider M. D., Srivastava D., and Olson E. N. (2005) The Hand1 and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner. Development 132, 189–201 [DOI] [PubMed] [Google Scholar]
- 44. Ilagan R., Abu-Issa R., Brown D., Yang Y. P., Jiao K., Schwartz R. J., Klingensmith J., and Meyers E. N. (2006) Fgf8 is required for anterior heart field development. Development 133, 2435–2445 10.1242/dev.02408 [DOI] [PubMed] [Google Scholar]
- 45. Panzer A. A., Regmi S. D., Cormier D., Danzo M. T., Chen I. D., Winston J. B., Hutchinson A. K., Salm D., Schulkey C. E., Cochran R. S., Wilson D. B., and Jay P. Y. (2017) Nkx2–5 and Sarcospan genetically interact in the development of the muscular ventricular septum of the heart. Sci. Rep. 7, 46438 10.1038/srep46438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Terada R., Warren S., Lu J. T., Chien K. R., Wessels A., and Kasahara H. (2011) Ablation of Nkx2–5 at mid-embryonic stage results in premature lethality and cardiac malformation. Cardiovasc. Res. 91, 289–299 10.1093/cvr/cvr037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen H., Shi S., Acosta L., Li W., Lu J., Bao S., Chen Z., Yang Z., Schneider M. D., Chien K. R., Conway S. J., Yoder M. C., Haneline L. S., Franco D., and Shou W. (2004) BMP10 is essential for maintaining cardiac growth during murine cardiogenesis. Development 131, 2219–2231 10.1242/dev.01094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen H., Zhang W., Sun X., Yoshimoto M., Chen Z., Zhu W., Liu J., Shen Y., Yong W., Li D., Zhang J., Lin Y., Li B., VanDusen N. J., Snider P., et al. (2013) Fkbp1a controls ventricular myocardium trabeculation and compaction by regulating endocardial Notch1 activity. Development 140, 1946–1957 10.1242/dev.089920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Luxán G., Casanova J. C., Martínez-Poveda B., Prados B., D'Amato G., MacGrogan D., Gonzalez-Rajal A., Dobarro D., Torroja C., Martinez F., Izquierdo-García J. L., Fernández-Friera L., Sabater-Molina M., Kong Y. Y., et al. (2013) Mutations in the NOTCH pathway regulator MIB1 cause left ventricular noncompaction cardiomyopathy. Nat. Med. 19, 193–201 10.1038/nm.3046 [DOI] [PubMed] [Google Scholar]
- 50. Li D., Hallett M. A., Zhu W., Rubart M., Liu Y., Yang Z., Chen H., Haneline L. S., Chan R. J., Schwartz R. J., Field L. J., Atkinson S. J., and Shou W. (2011) Dishevelled-associated activator of morphogenesis 1 (Daam1) is required for heart morphogenesis. Development 138, 303–315 10.1242/dev.055566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tian X., Li Y., He L., Zhang H., Huang X., Liu Q., Pu W., Zhang L., Zhao H., Wang Z., Zhu J., Nie Y., Hu S., Sedmera D., Zhong T. P., et al. (2017) Identification of a hybrid myocardial zone in the mammalian heart after birth. Nat. Commun. 8, 87 10.1038/s41467-017-00118-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tian X., Hu T., Zhang H., He L., Huang X., Liu Q., Yu W., Yang Z., Yan Y., Yang X., Zhong T. P., Pu W. T., and Zhou B. (2014) Vessel formation: de novo formation of a distinct coronary vascular population in neonatal heart. Science 345, 90–94 10.1126/science.1251487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dorn T., Goedel A., Lam J. T., Haas J., Tian Q., Herrmann F., Bundschu K., Dobreva G., Schiemann M., Dirschinger R., Guo Y., Kühl S. J., Sinnecker D., Lipp P., Laugwitz K. L., Kühl M., and Moretti A. (2015) Direct nkx2–5 transcriptional repression of isl1 controls cardiomyocyte subtype identity. Stem Cells 33, 1113–1129 10.1002/stem.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang Z., Jones A., Sun C. W., Li C., Chang C. W., Joo H. Y., Dai Q., Mysliwiec M. R., Wu L. C., Guo Y., Yang W., Liu K., Pawlik K. M., Erdjument-Bromage H., et al. (2011) PRC2 complexes with JARID2, MTF2, and esPRC2p48 in ES cells to modulate ES cell pluripotency and somatic cell reprogramming. Stem Cells 29, 229–240 10.1002/stem.578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schmitges F. W., Prusty A. B., Faty M., Stützer A., Lingaraju G. M., Aiwazian J., Sack R., Hess D., Li L., Zhou S., Bunker R. D., Wirth U., Bouwmeester T., Bauer A., Ly-Hartig N., et al. (2011) Histone methylation by PRC2 is inhibited by active chromatin marks. Mol. Cell 42, 330–341 10.1016/j.molcel.2011.03.025 [DOI] [PubMed] [Google Scholar]
- 56. Kim D. H., Tang Z., Shimada M., Fierz B., Houck-Loomis B., Bar-Dagen M., Lee S., Lee S. K., Muir T. W., Roeder R. G., and Lee J. W. (2013) Histone H3K27 trimethylation inhibits H3 binding and function of SET1-like H3K4 methyltransferase complexes. Mol. Cell. Biol. 33, 4936–4946 10.1128/MCB.00601-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Albert M., Schmitz S. U., Kooistra S. M., Malatesta M., Morales Torres C., Rekling J. C., Johansen J. V., Abarrategui I., and Helin K. (2013) The histone demethylase Jarid1b ensures faithful mouse development by protecting developmental genes from aberrant H3K4me3. PLoS Genet. 9, e1003461 10.1371/journal.pgen.1003461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Delgado-Olguín P., Huang Y., Li X., Christodoulou D., Seidman C. E., Seidman J. G., Tarakhovsky A., and Bruneau B. G. (2012) Epigenetic repression of cardiac progenitor gene expression by Ezh2 is required for postnatal cardiac homeostasis. Nat. Genet. 44, 343–347 10.1038/ng.1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pereira R. M., Martinez G. J., Engel I., Cruz-Guilloty F., Barboza B. A., Tsagaratou A., Lio C. W., Berg L. J., Lee Y., Kronenberg M., Bandukwala H. S., and Rao A. (2014) Jarid2 is induced by TCR signalling and controls iNKT cell maturation. Nat. Commun 5, 4540 10.1038/ncomms5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. da Rocha S. T., Boeva V., Escamilla-Del-Arenal M., Ancelin K., Granier C., Matias N. R., Sanulli S., Chow J., Schulz E., Picard C., Kaneko S., Helin K., Reinberg D., Stewart A. F., Wutz A., Margueron R., and Heard E. (2014) Jarid2 is implicated in the initial Xist-induced targeting of PRC2 to the inactive X chromosome. Mol. Cell 53, 301–316 10.1016/j.molcel.2014.01.002 [DOI] [PubMed] [Google Scholar]
- 61. Mysliwiec M. R., Chen J., Powers P. A., Bartley C. R., Schneider M. D., and Lee Y. (2006) Generation of a conditional null allele of jumonji. Genesis 44, 407–411 10.1002/dvg.20221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Brody M. J., Cho E., Mysliwiec M. R., Kim T. G., Carlson C. D., Lee K. H., and Lee Y. (2013) Lrrc10 is a novel cardiac-specific target gene of Nkx2–5 and GATA4. J. Mol. Cell. Cardiol. 62, 237–246 10.1016/j.yjmcc.2013.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Brody M. J., Hacker T. A., Patel J. R., Feng L., Sadoshima J., Tevosian S. G., Balijepalli R. C., Moss R. L., and Lee Y. (2012) Ablation of the cardiac-specific gene leucine-rich repeat containing 10 (lrrc10) results in dilated cardiomyopathy. PloS One 7, e51621 10.1371/journal.pone.0051621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tietjen J. R., Zhang D. W., Rodriguez-Molina J. B., White B. E., Akhtar M. S., Heidemann M., Li X., Chapman R. D., Shokat K., Keles S., Eick D., and Ansari A. Z. (2010) Chemical-genomic dissection of the CTD code. Nat. Struct. Mol. Biol. 17, 1154–1161 10.1038/nsmb.1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mysliwiec M. R., Kim T. G., and Lee Y. (2007) Characterization of zinc finger protein 496 that interacts with Jumonji/Jarid2. FEBS Lett. 581, 2633–2640 10.1016/j.febslet.2007.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.