Abstract
The evolution of cell-adhesion mechanisms in animals facilitated the assembly of organized multicellular tissues. Studies in traditional animal models have revealed two predominant adhesion structures, the adherens junction (AJ) and focal adhesions (FAs), which are involved in the attachment of neighboring cells to each other and to the secreted extracellular matrix (ECM), respectively. The AJ (containing cadherins and catenins) and FAs (comprising integrins, talin, and paxillin) differ in protein composition, but both junctions contain the actin-binding protein vinculin. The near ubiquity of these structures in animals suggests that AJ and FAs evolved early, possibly coincident with multicellularity. However, a challenge to this perspective is that previous studies of sponges—a divergent animal lineage—indicate that their tissues are organized primarily by an alternative, sponge-specific cell-adhesion mechanism called “aggregation factor.” In this study, we examined the structure, biochemical properties, and tissue localization of a vinculin ortholog in the sponge Oscarella pearsei (Op). Our results indicate that Op vinculin localizes to both cell–cell and cell–ECM contacts and has biochemical and structural properties similar to those of vertebrate vinculin. We propose that Op vinculin played a role in cell adhesion and tissue organization in the last common ancestor of sponges and other animals. These findings provide compelling evidence that sponge tissues are indeed organized like epithelia in other animals and support the notion that AJ- and FA-like structures extend to the earliest periods of animal evolution.
Keywords: adherens junction, focal adhesion, epithelium, protein evolution, evolution, Oscarella, Porifera, Sponge, Vinculin
Introduction
Cell adhesion is widely considered to be a requirement for the evolution of multicellularity (1–3). Cell–cell adhesion molecules connect adjacent cells within tissues (4), and other adhesion molecules anchor cells and tissues to the secreted extracellular matrix (ECM)4 (5). In addition, cell-adhesion mechanisms have evolved to play important roles in many other aspects of normal tissue physiology and development, including spatial landmarks that help determine cell polarity (6, 7), spindle orientation during mammalian mitotic cell division (8), signaling functions in response to mechanical forces on cells and tissues (9), and dynamic tissue remodeling during cell motility (10, 11). The critical importance of cell adhesion in multicellularity is underscored by the observation that cell adhesion is typically misregulated in cancer (12), a phenomenon unique to multicellular organisms.
Some of the earliest experimental studies of cell adhesion in animals were conducted on sponges (phylum Porifera). In 1907, Wilson (13) famously observed that cells derived from dissociated tissues of sponges could reaggregate and develop into a functional organism. This process relies on a secreted proteoglycan complex termed the aggregation factor (AF) that functions both in cell adhesion and self/nonself recognition (14–18). Core proteins of the AF have been identified and are structurally composed of calx-β domains and a sponge-specific “wreath” domain (19). Sulfated polysaccharide components of the AF undergo calcium-dependent, homophilic interactions that are considered to be the predominant adhesion mechanism in sponges (20).
The AF model of sponge cell adhesion is different from adhesion mechanisms in other animals. Two of the most important animal adhesion structures are the adherens junction (AJ) and focal adhesions (FAs). The AJ anchors adjacent cells to each other, and FAs anchor cells to the secreted ECM. These adhesive structures utilize entirely unrelated transmembrane adhesion proteins: the AJ is predominantly composed of cadherins (21, 22), whereas FAs contain integrins (23). Cadherins anchor to the actin cytoskeleton through a cytoplasmic protein complex of β-catenin, α-catenin, vinculin, and other proteins; integrins anchor to the actin cytoskeleton through interactions with a large number of cytoplasmic proteins including talin, vinculin, and paxillin (24–28).
Differences between the AF model in sponge cell adhesion and AJ and FA adhesions in other animals have contributed to the view that sponge tissues fall short of the organization level found in epithelia of other animals (29–31). Although this perspective has not gone unchallenged (31–33), there is limited ultrastructural evidence for cell junctions in sponge tissues, and where known they are not defined molecularly (32, 34). Furthermore, most sponges lack an obvious basal lamina (29), the specialized ECM to which epithelial FAs attach. Nevertheless, comparative genomic studies have established that sponges share homologs of core AJ and FA genes with other animals (35–37), and there is some evidence for their conserved biochemical interactions (38), suggesting a much earlier origin for these structures. Indeed, the apusozoan Amastigomonas sp. and the close animal outgroup Capsaspora owczarzaki have a complete integrin adhesome (39), and homologs of the vinculin/α-catenin family are widespread in eukaryotes including the social amoeba Dictyostelium discoideum (40–42). These observations raise questions about whether some form of transient multicellularity might be more ancient within eukaryotes than previously thought (42) and whether AJ and FA proteins have adhesive functions in the immediate animal stem lineage.
Here, we describe the structure and biochemical properties of a vinculin ortholog (Op vinculin) from the sponge Oscarella pearsei. O. pearsei belongs to the clade homoscleromorpha, which may have the most epithelial-like tissues of any sponge group (29, 34, 43–45). Junction-like structures have been detected in the homoscleromorph larval epithelium (46), and adult tissues have a well-developed basal lamina-like structure that contains type IV collagen (43). Additionally, a yeast two-hybrid screen revealed an interaction between Op β-catenin and Op cadherin (47). To further evaluate the molecular organization of sponge tissues, we examined: 1) the subcellular and tissue localization of vinculin in tissues of O. pearsei; 2) a structural comparison between Op vinculin and vertebrate α-catenin and vinculin; and 3) the interaction of Op vinculin with F-actin, β-catenin and talin, as a test for a role for vinculin in cadherin or integrin-based cell-adhesion mechanisms. Our results indicate that Op vinculin is an ortholog of vertebrate vinculin and that its protein-binding partners and subcellular distribution are consistent with potential roles in both AJ- and FA-mediated cell adhesions.
Results
Identification of O. pearsei VIN proteins and possible complex components
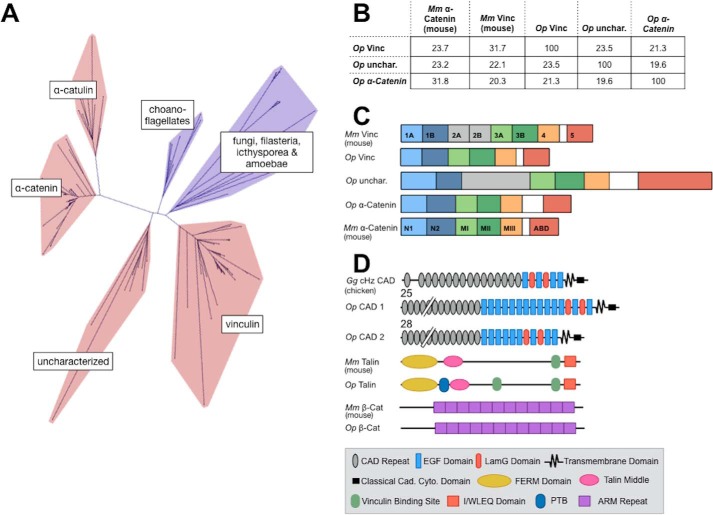
The O. pearsei genome (47) and transcriptome (48) was found to encode three predicted sequences with homology to the protein superfamily of αE-catenin, vinculin, and α-catulin (VIN-family proteins; Fig. 1). The three Op VIN-family proteins clustered with other animal sequences (Fig. 1A), consistent with previous studies (40, 71). Op vinculin grouped with other animal vinculin sequences, and Op α-catenin grouped with other animal α-catenin sequences. A third Op VIN-family protein clustered within a clade of functionally uncharacterized VIN-family proteins that appears to be unique to animals and broadly conserved across the major animal groups. It is present in sponges and cnidarians, as well as multiple protostome and deuterostome phyla (e.g. molluscs, echinoderms, and invertebrate chordates), but is absent in nonanimal eukaryotes and appears to have been lost secondarily in vertebrates and the common research models D. melanogaster and Caenorhabditis elegans (40, 71). In O. pearsei, this sequence has a vinculin-like D2 domain (Fig. 1C), and the clade to which it clustered lacks close affinity with vinculin, α-catenin, and α-catulin, so we refer to this protein as “Op uncharacterized.”
Figure 1.
VIN homology proteins of the sponge, O. pearsei. A, unrooted network depicting the phylogeny of VIN-family proteins (red, sequences from animal species; blue, sequences from nonanimal species). Three VIN-family proteins from O. pearsei were found to fall within the animal-exclusive clades corresponding to vinculin, α-catenin, and a functionally uncharacterized clade. A detailed phylogeny is provided in Figs. S1–S7. B, the percentage of identity of amino acid sequences of Mus musculus αE-catenin (Mm α-cat), Mm vinculin (Mm Vinc), and Op VIN proteins. C, domain schematics of Mm αE-catenin, Mm vinculin, and Op VIN proteins. Mm vinculin is composed of seven 4-helix bundle domains (D1–D4), a proline-rich hinge region (white), and a C-terminal 5-helix bundle (D5, the actin-binding domain). Domains 1, 2, and 3 each comprise two 4-helix bundles that share a central, long helix and are therefore subdivided into subdomains A and B. Mm αE-catenin shares a similar structure to Mm vinculin but lacks domain D2 (gray); the N-terminal domain is homologous to vinculin D1, and the M domain comprises three 4-helix bundles equivalent to vinculin D3A, D3B, and D4. The homologous domains in Op vinculin, Op uncharacterized, and Op α-catenin are color-coded. The length of the domain schematics represents the length of the corresponding amino acid sequences. D, Pfam-predicted domain composition of potential binding partners of Op VIN proteins examined in this study, shown relative to their vertebrate counterparts. Predicted Pfam domains of Op CAD1, Op CAD2, Op Talin, and Op β-catenin are annotated in the figure legend. The numbers above Op CAD1 and Op CAD2 indicate the number of extracellular cadherin repeats.
As an additional measure of similarity, we examined the predicted domain organization between Op VIN proteins and their putative orthologs in other animals. Vertebrate vinculins comprise a series of four helix bundles that form domains 1A, 1B, 2A, 2B, 3A, 3B, and 4, followed by a flexible proline rich linker and a C-terminal five-helix bundle that binds F-actin (72–74) (Fig. 1, B–D). α-Catenin family proteins have a similar structure but lack domains 2A and 2B (75). In α-catenin, domain 1A (also called NI) binds to β-catenin (69, 76, 77) and in mammals is the site for homodimerization, which occludes β-catenin binding (69, 75). In vinculin, domain 1A binds talin and mediates autoinhibition of F-actin binding. Op vinculin is 31.7% identical to Mm vinculin and 23.7% identical to Mm αE-catenin (Fig. 1B). However, Op vinculin lacks domain 2, which typically distinguishes vinculin from α-catenin in other animals and may contribute to the autoinhibition of F-actin binding (Fig. 1C). Op uncharacterized has a similar sequence identity to both α-catenin and vinculin (23.2 and 22.1% identity, respectively) and has domain 2 (Fig. 1C). Op α-catenin is most similar to Mm αE-catenin and least similar to Mm vinculin by sequence (31.8 and 20.3% identity, respectively, for α-catenin) (Fig. 1B) and also lacks domain 2.
Next, we tested the possibility of complete, vertebrate-like cell adhesion complexes in O. pearsei by searching for homologs of other AJ and FA proteins. We identified the previously described Op cadherin homolog (CAD1) and β-catenin (47), a second uncharacterized classical cadherin Op CAD2, and Op talin (Fig. 1D). Op CAD1 and 2 have large extracellular regions that contains 25 and 28 CAD repeats, respectively, and both have laminin G and epidermal growth factor-like domains (Fig. 1D). In general, cadherins in nonbilaterian animals contain a longer extracellular region with a greater variety of domains compared with vertebrate classical cadherins, which have five CAD repeats and lack laminin G and epidermal growth factor-like domains (40). Like vertebrate classical cadherins, Op CAD1 and 2 are single pass transmembrane proteins with a cytosolic catenin-binding region. The domain organizations of Op talin and β-catenin are more highly conserved than in VIN proteins or cadherins. As in their mammalian counterparts, Op talin has an N-terminal FERM domain followed by a talin middle domain, vinculin-binding sites, and an I/WLEQ domain. Just C-terminal to the FERM domain, Op talin also has a predicted phosphotyrosine-binding domain that is not present in mouse talin. Like β-catenin in most vertebrates and invertebrates, Op β-catenin is comprised of 12 armadillo repeats flanked by N- and C-terminal regions (47).
Op vinculin co-localizes with F-actin at cell boundaries, at filopodia, and at the base of microvilli in choanocytes
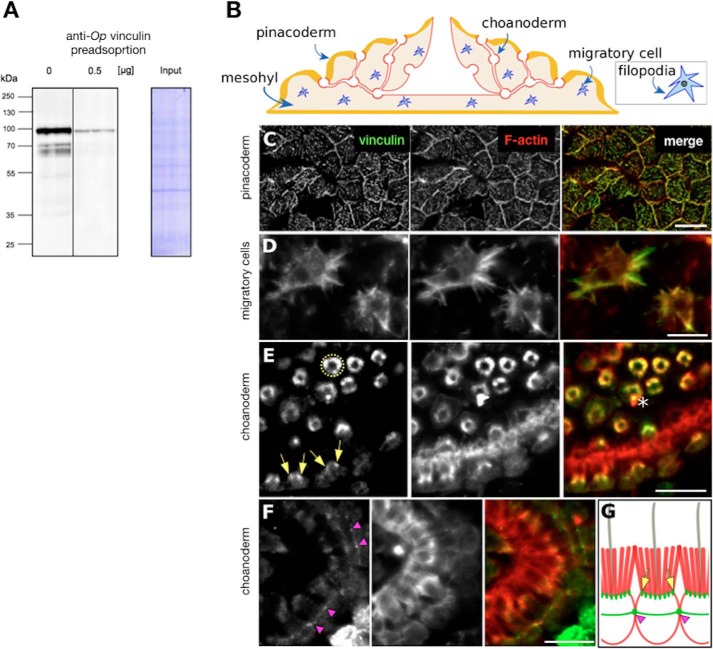
In other animals, VIN family proteins localize to the cell–cell adhesion complex and the FA cell–ECM complex. To characterize tissue-specific and subcellular localization patterns of Op vinculin, we isolated a polyclonal antibody against full-length recombinant protein. The affinity-purified antibody predominantly recognized a single band of the predicted size of Op vinculin (92 kDa) by Western blotting of whole-cell lysates, and preincubation of the antibody with recombinant protein depleted this signal (Fig. 2A). The specificity of the antibody was further validated by immunoprecipitation of endogenous Op vinculin from whole cell lysates (Fig. S8).
Figure 2.
Specificity of Op vinculin antibody and tissue localization of vinculin in O. pearsei. A, O. pearsei cell lysates probed with antibodies against His-Op vinculin in the absence and presence of a competing protein. In the absence of competing protein, the antibody recognizes a single band of ∼92 kDa, and faster migrating bands corresponding to degraded Op vinculin. Preincubation of the antibody with competing protein 0.5 μg of His-tagged Op vinculin reduced binding to Op vinculin by >90%. B, simplified illustration of the O. pearsei body plan. Op vinculin localized to the pinacoderm (yellow), the choanoderm (red), and to migratory cells in the mesohyl (blue). C–F, tissue and cellular localization of Op vinculin by immunostaining at cell– boundaries in the pinacoderm (C), filopodia in migratory in the mesohyl (D), an F-actin population at the base of microvilli in choanocytes (cross-section views of staining are visible as rings (hatched yellow circle) in the top half of the panel and longitudinal-section views of staining visible in the bottom half of the panel (yellow arrows)) (E), and at points of contact between individual choanocytes that form a continuous belt around adjacent cells (pink arrowheads) (F). Asterisk shows a foreign particle (staining artifact). G, cartoon summary of choanocyte staining (green, vinculin; red, F-actin). Scale bars, 10 μm.
O. pearsei comprises two types of epithelia: an outer epithelium termed the pinacoderm and a feeding epithelium termed the choanoderm that contains choanocytes (Fig. 2B). Immunostaining revealed four distinct Op vinculin subcellular distributions within these two epithelia: 1) a belt-like zone at points of cell–cell contact in the pinacoderm (Fig. 2C); 2) in filopodial/lamellopodial extensions of motile cells within the mesohyl (Fig. 2D); 3) at the base of the microvillar collar of choanocytes within the choanoderm (Fig. 2E), a belt-like zone of cell–cell contact in the choanoderm (Fig. 2F), and 4) within the cytoplasm of cells in the pinacoderm (Fig. 2C) (the pinacoderm is so thin that details of structures associated with vinculin were difficult to resolve in the z axis and may correspond to abundant filopodia in this tissue, as shown by transmission EM (48)). In each cellular context, Op vinculin appeared to co-localize with actin filaments, indicating an evolutionarily conserved interaction with the actin cytoskeleton. The localization of Op vinculin to cell–cell contacts in the pinacoderm and choanoderm is consistent with a possible AJ in these tissues, despite the lack of ultrastructural evidence (35).
Op vinculin is a stable monomer
Despite the phylogenetic placement of Op vinculin within the animal vinculin clade and the higher homology of individual Op vinculin domains to those of vertebrate vinculin versus α-catenin, the absence of a D2 domain and its localization to cell–cell contacts suggested that Op vinculin could be functionally closer to α-catenin. Mammalian αE-catenin, but not vinculin, exists as a monomer, homodimer, and heterodimer with β-catenin in epithelial cells, and its oligomerization state regulates its biochemical properties and function in the cell (78, 79). To determine whether Op vinculin is exclusively monomeric in solution or whether it also dimerized like mammalian αE-catenin, purified full-length Op vinculin was concentrated to 120 μm, incubated at room temperature for 1 h, and analyzed by size-exclusion chromatography and multiangle light scattering (SEC–MALS). Op vinculin eluted as a discrete peak with an estimated molecular mass of 87.0 kDa by MALS (Fig. S10), which is within 5 kDa of the predicted monomer molecular mass of 91.7 kDa, indicating that it is a monomer in solution, a property of mammalian vinculin but not αE-catenin. It should be noted, however, that some nonmammalian αE-catenins, such as those from Danio rerio, C. elegans, and Nematostella vectensis, are monomeric (71, 80, 81).
High-resolution crystal structure of full-length Op vinculin
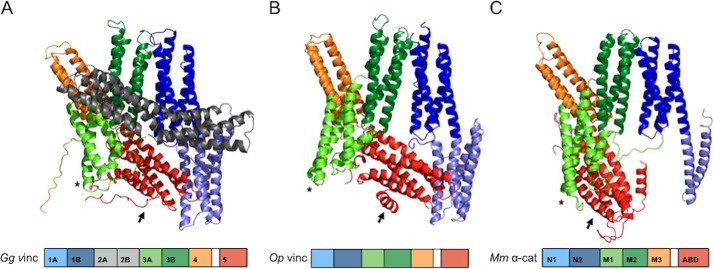
To investigate further the relationship between Op vinculin, vertebrate vinculin, and αE-catenin, we determined the X-ray crystal structure of Op vinculin at 2.3 Å resolution (Fig. 3; Table S2). The overall structure of Op vinculin (Fig. 3B) resembles the structures of Hs and Gg vinculin (Fig. 3A) (64, 73) and a single subunit of Mm αE-catenin homodimer (Fig. 3C) (75), as expected for proteins in the vinculin/α-catenin superfamily. Differences among these proteins arise in part from the relative positions of the N-terminal and central domains, which appears to be due to a somewhat flexible interface between the D1b/NII and D3b/MII helix bundles.
Figure 3.
High resolution crystal structure of full-length Op vinculin. A–C, comparison of Op vinculin structure (B) with previously solved structures of chicken vinculin (A) (73) and subunit B of human αE-catenin dimer (C) (75). Domain schematic of each protein is below the structure, and the domains of each protein are color-coded: 1A in light blue, 1B in dark blue, 2A and 2B in gray, 3A in light green, 3B in dark green, 4 in orange, and domain 5/actin-binding domain in red. Asterisks and arrows indicate conformational shifts of domains 3A and 5/actin-binding domain, respectively.
Inspection of individual structural regions reveals that Op vinculin is more similar to vertebrate vinculin than to Mm α-catenin. In particular, Op vinculin D1 closely matches vertebrate vinculin D1: subdomains D1a and b are in virtually identical positions, and the long helix that is shared between the subdomains is similarly kinked. Moreover, the fourth helix of subdomain D1a is noticeably curved in vertebrate vinculin and Op vinculin. These features are absent in the equivalent region of Mm α-catenins, known as domain N. In the α-catenin homodimer, the first two helices of domain N are disordered, and the second two form the homodimer interface with those in the partner molecule (75). A monomeric structure of domain N from αN-catenin, an isoform of αE-catenin that is expressed predominantly in neuronal cells, shows that subdomain NI forms a four-helix bundle (82), but its position differs slightly from that observed in vertebrate vinculin or Op vinculin D1a. Thus, whereas the overall structure of D1 is conserved in VIN proteins, the relative positions of the two subdomains differ from vertebrate vinculin and α-catenin.
The central portion of all VIN-family proteins contains three four-helix bundles, designated D3a, D3b, and D4 in vertebrate vinculin and MI, MII, and MIII in α-catenin. In most animals, vinculin orthologs also contain domain D2, which precedes D3. As expected from sequence analysis, D2 is missing in Op vinculin (Figs. 1 and 4, A and B). Domains D3b and D4 of vertebrate vinculin superimpose very closely with MII and MIII of α-catenin, and the Op vinculin is also very similar in these regions. In contrast to D3b and D4, the position of the Op vinculin domain D3a/MI differs among all three proteins (Fig. 3), although Op vinculin domain D3a is closer to that of α-catenin than to vinculin. This is likely due to the absence of domain D2 in Op vinculin and α-catenin; vinculin domain D2 interacts extensively with and helps to fix the position of domain D3a (Fig. 3, asterisks).
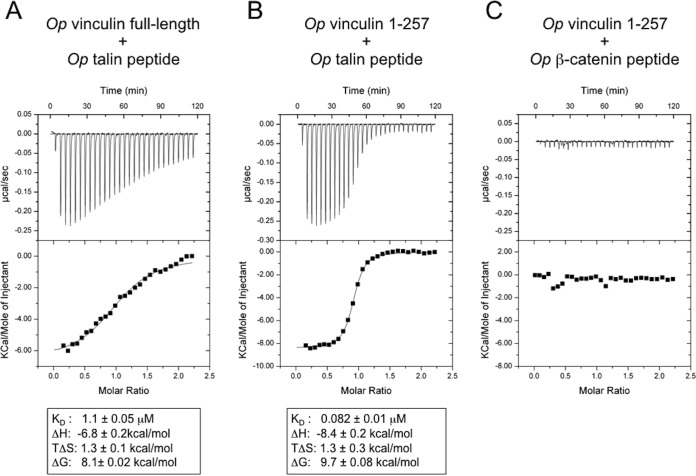
Figure 4.
Op vinculin interacts with Op talin, but not Op β-catenin. The dissociation constants and thermodynamic parameters determined by ITC of Op vinculin and different putative binding partners. A, full-length Op vinculin and a synthetic peptide comprising the vinculin-binding site 1 of Op talin (Op talin peptide). B, Op vinculin domain 1 (residues 1–257) and the Op talin peptide. C, Op vinculin domain 1 and a synthetic peptide comprising the α-catenin-binding site on Op β-catenin (Op β-cat peptide). The data shown represent the averages and the standard deviations of three independent measurements.
Vertebrate vinculin domain D2 forms a bridge that interacts with domains 1 and 3. This interaction has been proposed to lock vinculin into a closed conformation around the F-actin-binding domain D5 (Fig. 3A), which prevents constitutive binding to actin except upon activation by binding to a partner such as talin (83). α-Catenins lack domain D2, and the position of the F-actin-binding domain in the αE-catenin dimer structure is different in the two promoters (75) and is disordered in another crystal structure (83). These observations are consistent with αE-catenin binding to F-actin without prior activation. Despite the absence of domain D2, Op vinculin domain D5 is positioned similarly to vertebrate vinculin domain D5. However, the shifted position of D3a produces few contacts with D5 and thereby makes D5 more exposed, with a surface accessibility intermediate between that of the comparable Mm αE-catenin and vertebrate vinculin domains (Fig. 3).
Op vinculin binds talin, but not β-catenin
The vinculin-like position of the Op vinculin F-actin-binding domain indicates that Op vinculin might be autoinhibited with respect to F-actin binding. On the other hand, the relative exposure of this domain might enable it to bind F-actin without prior activation, which is a property of α-catenins. Therefore, we tested whether purified Op vinculin bound to talin and/or to β-catenin, and whether binding to F-actin is constitutive or requires activation.
Binding of talin to vinculin is regulated by the intramolecular vinculin head–tail interaction (73, 83, 84), whereas α-catenins bind β-catenin constitutively (85–87). Using isothermal titration calorimetry (ITC), we measured binding of full-length Op vinculin and Op vinculin D1 to synthetic peptides of Op talin and Op β-catenin that corresponded to their binding sites on vertebrate vinculin or αE-catenin, respectively (see “Experimental procedures”). Full-length Op vinculin and Op vinculin D1 bound Op talin peptide (Fig. 4, A and B), but not Op β-catenin peptide (Fig. 4C). Op vinculin D1 bound Op talin peptide with KD = 82 nm (Fig. 4B), comparable with mammalian vinculin–talin peptide binding (KD = 39 nm) (70). In mammalian vinculin, talin binding involves the rearrangement of domain 1A into a five-helix bundle (74), and interactions of D1a with the F-actin-binding domain inhibit detectable talin binding (73). Full-length Op vinculin bound to the Op talin peptide with KD = 1.1 μm, i.e. > 10-fold reduced affinity for the full-length protein (Fig. 4A). These data suggest that Op vinculin functions as vinculin, including autoinhibitory behavior likely caused by the D1–D5 interaction, although the autoinhibition is not as strong as that found in vertebrate vinculin.
Op vinculin binding to F-actin is activated by talin
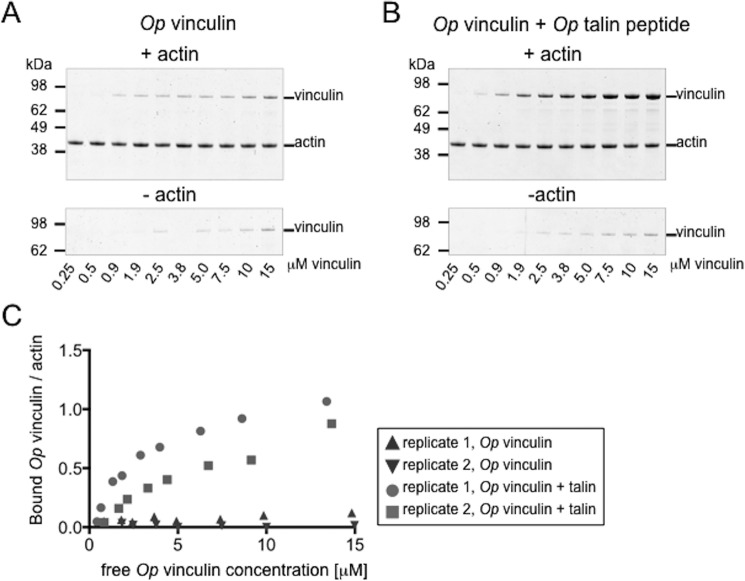
The similarity in the tertiary structures of Op vinculin and vertebrate vinculin, as well as the partial autoinhibition of full-length Op vinculin binding to talin, led us to examine the F-actin-binding properties of Op vinculin using a co-sedimentation assay (81, 85, 88). Like vertebrate vinculins, Op vinculin did not bind F-actin significantly above background at concentrations up to 15 μm (Fig. 5, A and C).
Figure 5.
Activation of Op vinculin binding to F-actin by Op talin. A and B, Op vinculin co-sedimentation with F-actin in the absence (A) or presence (B) of the Op talin peptide. Op vinculin was added at increasing concentrations to 1.5 μm F-actin. Background Op vinculin pelleting in the absence of F-actin is shown in the lower panels. Op talin peptide was used in 5-fold excess to Op vinculin. The assay was performed in duplicate. C, bound Op vinculin/actin plotted against the concentration of free Op vinculin concentration is shown for binding without (triangles) or with (circles and squares) the Op talin peptide.
The interaction between the head domain and the ABD domain in mammalian vinculin is disrupted upon binding to talin, which results in activation of vinculin binding to F-actin (74, 83). In the presence of Op talin peptide, full-length Op vinculin bound to F-actin filaments (Fig. 5, B and C). Thus, binding of talin to Op vinculin D1 releases autoinhibition for F-actin binding, similar to the behavior of vertebrate vinculin.
Discussion
VIN proteins link actin filaments to membrane proteins at the plasma membrane and are found in all animals and their close outgroups (choanoflagellates, chytridomycetes, apusozoa, amoebozoa) (40, 71). Animal VIN proteins are subdivided into four protein families based on sequence homology: vinculin, α-catenin, α-catulin, and a functionally uncharacterized clade previously called “α-catenin-like” (Fig. 1A) (40, 71).
Vinculin and α-catenin have been studied structurally and biochemically in vitro and in vivo. Mammalian vinculin predominantly localizes to cell–ECM contacts, where it anchors FA complexes to the actin cytoskeleton (23). The N-terminal domain D1 of vinculin binds talin, and the C-terminal domain binds F-actin (83); these binding sites, however, are masked by strong head–tail interactions that are stabilized by domain 2, which locks vinculin in an autoinhibited conformation (73). Vinculin binding to F-actin is activated by binding to talin or another protein (89). In contrast, homodimeric Mm αE-catenin binds F-actin constitutively, but F-actin binding by αE-catenin monomer in the cadherin complex is significant only when the complex is subjected to mechanical force (88, 90). Vinculin also associates with cell–cell adhesion AJ complexes, which also requires mechanical force (91, 92).
In invertebrate animals, vinculin has been studied in C. elegans and in D. melanogaster. In C. elegans, the vinculin ortholog DEB-1 is involved in integrin-based cell–ECM adhesions and is a component of the dense body, an anchoring structure in muscles similar to the vertebrate Z-disc. Disruption of DEB-1 function leads to disorganized muscles and paralyzed embryos and is lethal (93–96). In contrast, D. melanogaster vinculin is nonessential (97), but constitutive activation caused the formation of abundant cytoplasmic adhesion complexes with talin, producing morphological defects and death (97, 98). Biochemical differences between vertebrate and invertebrate α-catenin have been detailed elsewhere (40, 99), but all α-catenins characterized to date bind β-catenin and F-actin at the AJ and developmental disruption of α-catenin leads to morphological abnormalities and death (80, 100–102).
The O. pearsei genome encodes three VIN-family proteins: vinculin, α-catenin, and a member of the uncharacterized animal VIN-family clade (Fig. 1, A–C). Op vinculin is more similar to bilaterian vinculin than αE-catenin by sequence (32 and 24%, respectively) and would be classified as vinculin using phylogeny-based orthology assignment. However, it lacks domain 2, which is a characteristic of vinculin orthologs in other animals (Fig. 1C). Op uncharacterized is similar to α-catenin and vinculin by sequence (23 and 22%, respectively) and contains domain 2. The biochemical properties and cellular functions of these proteins have not been investigated in any organism. Op α-catenin is significantly more similar to α-catenin than vinculin (32 and 20%, respectively). However, given the uncertainty surrounding the function of Op vinculin and Op uncharacterized, we are unable to make assumptions about the specific cellular functions of any VIN proteins in O. pearsei. We focused our studies on Op vinculin to begin to dissect the functional relationship between VIN-family proteins in O. pearsei.
Immunofluorescence of O. pearsei tissue using an affinity-purified antibody raised against Op vinculin showed co-localization with cortical belts of actin filaments at cell–cell contacts in the pinacoderm and choanoderm tissues (Fig. 2, C–F) and in filopodial extensions in migratory cells within the mesohyl. Localization to cortical actin at the AJ is a function of αE-catenin in cell–cell adhesion, whereas localization to FAs in filopodia supports cell–ECM adhesion during migration. However, this strict interpretation is confounded by the contractile nature of sponge tissues (103), and the recruitment of vinculin to mammalian AJ under mechanical tension, which may stabilize the cadherin–catenin complex (104). Thus, we examined Op vinculin in vitro to test whether it might have properties of an α-catenin or vinculin homolog.
Purified recombinant Op vinculin was a stable, monomeric protein with an overall structure more similar to vertebrate vinculin than α-catenin (Figs. 3 and 4). Moreover, Op vinculin bound talin but not β-catenin (Fig. 4) and displayed a significant level of autoinhibition with respect to talin binding. Finally, Op vinculin was constitutively autoinhibited in F-actin binding, but this autoinhibition was relieved by binding talin. Collectively, these characteristics strongly suggest that Op vinculin is functionally more similar to vinculin rather than α-catenin, despite the absence of domain D2 that is characteristic of vinculin in other animals.
The presence of Op vinculin at both cell–cell and cell–ECM contacts supports the hypothesis that Op vinculin may function at both the cadherin-based AJ and integrin-based FAs by reinforcing anchorage to the underlying actin cytoskeleton. Given the tension-dependent association of mammalian vinculin with α-catenin at cell–cell junctions, we cannot exclude the possibility that Op vinculin associates with another VIN protein family member under tension at the cadherin–catenin complex. In light of these findings, future investigation of Op uncharacterized and α-catenin will be of particular interest. In the O. pearsei transcriptome, all three VIN proteins are highly expressed in adult tissues (48). Biochemical characterization will determine whether they have functional redundancy and compete with each other for association with binding partners at FA or AJ or have been co-opted for other functions.
Our results have important implications for the evolution of cell-adhesion mechanisms and the organization of sponge tissues. First, they support the hypothesis that a simple epithelium was part of the shared inheritance of all animal lineages. Second, the AF model of sponge cell adhesion appears to be incomplete, and sponge tissues are organized more like the epithelia in other animals than previously thought. One caveat to this interpretation is that O. pearsei belongs to the clade homoscleromorpha, which lacks clear orthologs of the core AF protein family characterized from demosponges (Ref. 19); but see Ref. 105); adhesion mechanisms may differ between modern sponge lineages, which underwent very ancient divergences (106). Alternatively, the AJ and FAs may regulate sponge epithelial organization, whereas AF predominantly functions during reaggregation of dissociated tissues and in histocompatibility reactions, which are the contexts in which the AF has been studied predominantly. Finally, the AF and AJ/FA adhesion systems may also interact. The putative AF receptor (107) has an RGD motif typical of ECM proteins that could facilitate interactions with integrins, thereby coupling these otherwise distinct adhesion mechanisms (30). Nevertheless, based on our studies of Op vinculin, we conclude that a complex toolkit of cell adhesion proteins and the rudiments of epithelial organization were already established in the last common ancestor of sponges and bilaterians.
Experimental procedures
Identification of O. pearsei VIN proteins and potential binding proteins
Annotated sequences of α- and β-catenin, vinculin, talin, and E-cadherin orthologs from representative bilaterian species were retrieved from Uniprot databases and used to conduct a BLAST search against the O. pearsei genome (47). The predicted sequences were then verified by BLAST search for predicted peptides against the O. pearsei transcriptome (48) to recover full-length coding sequences (Table S1). Domain composition of putative O. pearsei sequences was predicted using the HMMER and SMART web servers to annotate conserved domain motifs from the Pfam database (49–51). Protein characteristics (molecular mass, theoretical pI, extinction coefficient) were predicted using the ProtParam tool on the ExPASy server (52). An annotated alignment of vinculins is provided in Supplement 2.
Molecular phylogenetics
The NCBI PSI-BLAST server (53) was used to create a position-specific scoring matrix from an alignment of α-catenin, vinculin, and α-catulin sequences (54) in the model organism (landmark) database at NCBI. The position-specific scoring matrix was then used to search the proteome of representative animal and nonanimal species using the standalone version PSI-BLAST as implemented by the NCBI BLAST+ package (version 2.7.1) (55). Specifically, the predicted proteome (derived from either genome of transcriptome data) was searched for D. discoideum, C. owczarzaki, Monosiga brevicollis, Salpingoeca rosetta, Salpingoeca urceolata, Codosiga hollandica, Salpingoeca dolichothecata, Savilla parva, Diaphanoeca grandis, Trichoplax adhaerens, O. pearsei, Ephydatia muelleri, N. vectensis, C. elegans, D. melanogaster, Strongylocentrotus purpuratus, Branchiostoma floridae, Lottia gigantea, and Capitella teleta. Significant hits were combined with sequences from Miller et al. (40), aligned with ClustalW (40, 54) and trimmed with TrimAl (−gappyout) (56) using the phylogenetics server Phylemon-2 (57). Maximum likelihood model selection was performed using the ModelFinder (AIC selection criterion) (58) as implemented on the IQ-TREE web server (59). The best model (LG+G4+F) was implemented locally using IQ-TREE (v1.6.3) (60) with 1000 ultrafast bootstrap replicates (61), and the resulting trees were visualized using CLC Main Workbench 7 (Qiagen). Initial analyses revealed that some representative sequences from Mnemiopsis leidyi, Allomyces macrogynus, Acanthomoeba castellanii, Schistosoma mansoni, and Clonorchis sinensis were reconstructed as being exceptionally long branches. Thus, these sequences were removed from the data set, and all analyses were repeated.
Op vinculin antibody production and Western blotting analysis
Anti-Op vinculin polyclonal antibodies were generated in rabbits. The coding region from O. pearsei cDNA was amplified by PCR, and the product was cloned into the pET28a+ (Novagen) expression vector to generate an in-frame fusion between Op vinculin (MG852025) and an N-terminal hexahistidine tag (His-Op vinculin). Op vinculin was expressed in the Rosetta(DE3) strain of Escherichia coli (EMD Millipore). Recombinant protein was passed over HisPur cobalt resin, and the His-tagged protein was eluted with 150 mm imidazole in 1× PBS, pH 8.5. Imidazole was removed by buffer exchange in a PD-10 column (GE Healthcare) before immunization. Polyclonal antibodies were raised in rabbits against His-Op vinculin (Syd Labs) and affinity-purified from serum using E. coli lysates to remove nonspecific antibodies and then His-Op vinculin recombinant protein to select for antigen-specific antibodies; both steps were conducted using AminoLink Plus Coupling Columns (Thermo Scientific) per the manufacturer's instructions.
For Western blotting analysis, frozen O. pearsei tissue was dissolved in Pierce IP lysis buffer containing protease inhibitor mixture (Roche). Cell debris was removed by centrifugation at 13,000 × g for 10 min. Lysate was mixed with 4× Laemmli sample buffer, and proteins were separated by SDS-PAGE. Proteins were transferred to a PVDF membrane, and membranes were blocked for 1 h at room temperature in 5% nonfat dry milk in 1× PBST (0.1% Tween 20), incubated with anti-Op vinculin antibody (1:10,000) in blocking solution for 1 h at room temperature, and washed twice in 1× PBST. After 45 min of incubation with secondary antibody (Alexa 488 goat anti-rabbit IgG antibody, from Life Technologies; 1:500) at room temperature, the membranes were washed in 1× PBST and developed using Molecular Imager FX ProPlus (Bio-Rad). Antibodies against His-Op vinculin recognized a single band of ∼100 kDa and partially degraded protein. Preincubation of the antibody with competitor antigen (0.5 μg of His-Op vinculin) was used to confirm specificity.
Immunostaining of O. pearsei tissues
O. pearsei tissues were collected from aquaria at the University of California Santa Cruz where they grow naturally in the circulating sea water. Different fixation conditions were used for each adult tissue type. Choanoderm was fixed in 4% paraformaldehyde for 30 min at room temperature; the pinacoderm was fixed in 4% paraformaldehyde, 0.1 m phosphate buffer (EMS) for 1 h at room temperature. The samples were washed twice in 1× PBS and blocked for 1 h at room temperature with 3% BSA, 0.1% Triton in 1× PBS. Fixed tissues were incubated with primary anti-Op vinculin antibody (1:1000 to 1:2500 dilutions) in blocking buffer for 2 h at room temperature. The samples were washed three times with blocking buffer and then incubated for 45 min with Alexa 488 goat anti-rabbit IgG secondary antibody (1:500, from Life Technologies), with Alexa Fluor 568 phalloidin (1:100, Life Technologies), and Hoechst 33342 (1 μg/ml) at room temperature. The samples were washed three times in PBS and mounted on a FluoroDish plate for imaging using anti-quenching mounting medium.
Recombinant Op vinculin expression and purification
For SEC–MALS and crystallographic experiments, DNA encoding full-length (amino acids 1–846) O. pearsei vinculin was inserted into a pET His6 Sumo TEV LIC vector (Addgene plasmid 29659, a gift from Scott Gradia). Fusion proteins of Op vinculin full-length, Op vinculin 1–257, and an N-terminal TEV-cleavable GSH-S-transferase tag were used for ITC and actin-binding experiments. Recombinant fusion proteins were expressed in BL21 (DE3) codon plus E. coli (Agilent Technologies). The cells were grown at 37 °C to an A600 of 0.8 and then induced with 0.1 m isopropyl-1-thio-β-d-galactopyranoside at 16 °C. After overnight growth, the cells were harvested by centrifugation and resuspended in 20 mm Tris, pH 8.0, 150 mm NaCl, 5 mm β-mercaptoethanol (His6-tagged protein) or 1 mm DTT (GST-tagged protein) before storage at −80 °C. The cells were lysed in EmulsiFlex (Avastin) after addition of protease inhibitor mixture (Mixture Set V, Calbiochem), and lysates were cleared by centrifugation at 38,500 × g for 30 min. For purification of His6–Sumo fusion protein, supernatant was incubated with nickel–nitrilotriacetic acid–agarose beads for 30 min at 4 °C. Beads were washed with 20 mm HEPES, pH 8.0, 150 mm NaCl, 1 mm β-mercaptoethanol and equilibrated for TEV cleavage in 20 mm HEPES, pH 8.0, 150 mm NaCl, 1 mm β-mercaptoethanol, 10% glycerol. Recombinant protein was incubated with TEV protease for 4 h at 4 °C to remove the His6-Sumo tag. For purification of the GST-tagged proteins, supernatant after centrifugation was incubated with GSH-agarose beads for 30 min at 4 °C. The beads were washed with PBS containing 500 mm NaCl and 1 mm DTT and subsequently equilibrated with 20 mm HEPES, pH 8.0, 150 mm NaCl, 1 mm EDTA, 1 mm DTT, 10% glycerol. The protein was cleaved on the beads by overnight incubation with TEV at 4 °C. Cleaved proteins were eluted from beads and purified over a MonoQ anion exchange column with a 0–1 m NaCl gradient in 20 mm Tris, pH 8.0, 1 mm DTT. Op vinculin and vinculin 1–257 were further purified by Superdex 200 SEC in 20 mm HEPES, pH 8.0, 150 mm NaCl, and 1 mm DTT. Pure protein eluted as a monomer. The identity of purified vinculin 1–257 was further verified by MS after purification. Purified proteins were kept at 4 °C and used immediately for experiments.
Size-exclusion chromatography coupled to multiangle light scattering
The oligomeric state of full-length Op vinculin was assessed by SEC–MALS using a Superdex 200 column attached to a UV detector, followed by a DAWN EOS MALS detector and a refractive index detector (Wyatt Technology). The system was equilibrated with 20 mm HEPES, pH 8.0, 150 mm NaCl, and 2 mm DTT at 25 °C. Detectors were calibrated by measuring the signal of monomeric BSA. Molecular weights were calculated with ASTRA software (Wyatt Technology) using the signals from the MALS and the refractive index detectors.
Op vinculin crystallization and data collection
Purified full-length Op vinculin was concentrated to 60 mg/ml and crystallized in 0.2 m sodium/potassium tartrate, pH 8.0, 20% PEG 3350 using the sitting drop method and iterative streak seeding. Plate-shaped crystals of approximate dimensions 350 × 60 × 5 μm grew in 4–5 days at 16 °C, were transferred to crystallization solution supplemented with 10% PEG 400 for cryoprotection, and frozen in liquid nitrogen. The diffraction data were measured in 0.2° rotation frames for 0.4 s/frame with a Pilatus 6M detector on Beamline 12-2 at the Stanford Synchrotron Radiation Laboratory. An initial data set used for molecular replacement extended to 2.83 Å, in the space group C2. Data for the final model refinement extended to 2.33 Å and were in space group P1. The data from the P1 crystal were measured from four different positions of a single long crystal, with 270, 280, 360, and 360° of rotation collected respectively at the four positions. The data were integrated with XDS (62) and scaled with AIMLESS in the CCP4 package (63). The data collection statistics are shown in Table S2.
Op vinculin structure determination and refinement
The structure of full-length Op vinculin was determined by molecular replacement in the C2 crystal form using a polyalanine model of the human vinculin structure (Protein Data Bank code 1TR2) (64) with residues 250–600 deleted, which was produced by MrBUMP (65). The structure was partially refined to Rwork/Rfree of 30%/35% through iterative cycles of manual rebuilding with the program Coot and positional, individual temperature factor and translation–libration–screw refinement in Phenix (66). Two copies of the partially refined structure in C2 were then found in the P1 form using the program Phaser (67) and further refined in Phenix. The final refinement statistics are provided in Table S2. The coordinates and structure factors have been deposited in the Protein Data Bank with accession code 6BFI.
High-speed actin co-sedimentation assays
Actin-binding assays were performed with 1.5 μm F-actin and increasing amounts of full-length Op vinculin (0.25–15 μm). Rabbit skeletal muscle G-actin (prepared as described in Ref. 68) was polymerized by addition of 10× polymerization buffer (100 mm Tris, pH 7.5, 20 mm MgCl2, 500 mm KCl, 10 mm ATP) and incubated at room temperature for 1 h. Different vinculin concentrations were prepared by serial dilution starting from 20 and 30 μm vinculin, without or with 100 and 150 μm Op talin peptide, respectively (i.e. talin peptide at 5-fold molar excess) into assay buffer (20 mm HEPES, pH 8.0, 150 mm NaCl, 1 mm DTT, and 2 mm MgCl2). After the addition of an equal volume of 3 μm F-actin or assay buffer, the mixture was incubated for 30 min at room temperature. An aliquot of each concentration point was removed as a standard. The samples were centrifuged at 60,000 rpm for 20 min at room temperature in a Beckman TLA 100 rotor. The pellet and the standard were separated by SDS-PAGE. Intensities of Coomassie-stained bands were quantified with an Odyssey imaging system (LI-COR Biotechnology). After background subtraction and normalization to the actin band, the concentration of pelleted vinculin was interpolated from the standard curve. The amount of bound vinculin per actin was plotted against the concentration of free vinculin using Prism software (GraphPad Software).
Synthetic peptide design
β-Catenin and talin peptides comprising the binding regions for α-catenin and vinculin, respectively, were designed based on homology to mammalian orthologs. Homology between Mm and Op β-catenin does not extend throughout the entire α-catenin–binding region but is limited to the minimal binding region of Mm αE-catenin spanning residues 118–149 (Kd = 377 nm) (69), which correspond to residues 233–266 in Op β-catenin (AIIGSPALIGGLVHVLGTTNDPDAMRSISGTLHN). The vinculin binding site of Mm talin comprises residues 605–628 (]Kd = 39 nm) (70). In a MUSCLE alignment Mm talin 605–628 correspond to residues 598–621 in Op talin (GEKLLEAARGLAGAVRHLLKSAEP). Both peptides were commercially synthesized with a N-terminal biotin tag and a five glycine linker (Genscript).
Isothermal titration calorimetry
ITC experiments were performed in a VP-ITC calorimeter (Microcal; GE Healthcare). 80–150 μm Op β-catenin or Op talin peptide was titrated into a solution of 6–10 μm Op vinculin 1–257 or full-length vinculin. One 2-μl injection was followed by 28 10-μl injections at 240-s intervals. Titrations were performed in 20 mm HEPES, pH 8.0, 150 mm NaCl, 1 mm DTT at 25 °C. For baseline correction, the heat change signals at saturation were averaged and subtracted from all data points. A one-site specific binding model was used to fit a binding curve (Microcal software; GE Healthcare). For the interaction of full-length Op vinculin and Op vinculin 1–257 with the Op-talin peptide, binding data from three individual measurements were averaged to calculate the KD, ΔH, and TΔS. Two measurements were performed for the Op vinculin 1–257–Op β-catenin interaction.
Author contributions
P. W. M., S. P., J. M. M., D. N. C., W. N., W. I. W., and S. A. N. conceptualization; P. W. M., J. V. C., and W. I. W. data curation; P. W. M., S. P., J. V. C., D. N. C., and W. I. W. formal analysis; P. W. M., S. P., J. M. M., D. N. C., and S. A. N. investigation; P. W. M., S. P., J. M. M., D. N. C., W. N., W. I. W., and S. A. N. visualization; P. W. M., S. P., J. M. M., and J. V. C. methodology; P. W. M., S. P., J. M. M., J. V. C., D. N. C., W. N., W. I. W., and S. A. N. writing-original draft; P. W. M., S. P., W. N., W. I. W., and S. A. N. writing-review and editing; S. P. and J. M. M. validation; W. N., W. I. W., and S. A. N. resources; W. N., W. I. W., and S. A. N. supervision; W. N. and W. I. W. funding acquisition; W. N., W. I. W., and S. A. N. project administration.
Supplementary Material
Acknowledgments
We thank Hadar Feinberg for assistance in the crystallographic analysis of Op vinculin, Betsy Steele for access to the Joseph Long Marine Laboratory (University of California Santa Cruz), and Christopher Lowe for use of confocal microscopy facilities at Hopkins Marine Station (Stanford University). Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the Department of Energy Office of Biological and Environmental Research, and by NIGMS, National Institutes of Health Grant P41GM103393).
This work was supported by National Institutes of Health Grants R35GM118064 (to W. J. N.) and R01GM114462 (to W. I. W.) and NASA Grant 15-EXO15_2-0027 (to W. I. W. and S. A. N.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Tables S1 and S2, Figs. S1–S10, and sequences.
The atomic coordinates and structure factors (code 6BFI) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- ECM
- extracellular matrix
- AJ
- adherens junction
- FA
- focal adhesion
- AF
- aggregation factor
- VIN
- protein superfamily of α-catenin, α-catulin, and vinculin
- Op
- O. pearsei
- CAD
- cadherin
- SEC–MALS
- size-exclusion chromatography and multiangle light scattering
- ITC
- isothermal titration calorimetry.
References
- 1. Abedin M., and King N. (2010) Diverse evolutionary paths to cell adhesion. Trends Cell Biol. 20, 734–742 10.1016/j.tcb.2010.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Niklas K. J., and Newman S. A. (2013) The origins of multicellular organisms. Evol. Dev. 15, 41–52 10.1111/ede.12013 [DOI] [PubMed] [Google Scholar]
- 3. Knoll A. H. (2011) The multiple origins of complex multicellularity. Annu. Rev. Earth Planet. Sci. 39, 217–239 10.1146/annurev.earth.031208.100209 [DOI] [Google Scholar]
- 4. Harris T. J., and Tepass U. (2010) Adherens junctions: from molecules to morphogenesis. Nat. Rev. Mol. Cell Biol. 11, 502–514 10.1038/nrm2927 [DOI] [PubMed] [Google Scholar]
- 5. Dumbauld D. W., Lee T. T., Singh A., Scrimgeour J., Gersbach C. A., Zamir E. A., Fu J., Chen C. S., Curtis J. E., Craig S. W., and García A. J. (2013) How vinculin regulates force transmission. Proc. Natl. Acad. Sci. U.S.A. 110, 9788–9793 10.1073/pnas.1216209110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harris T. J., and Peifer M. (2005) Decisions, decisions: β-catenin chooses between adhesion and transcription. Trends Cell Biol. 15, 234–237 10.1016/j.tcb.2005.03.002 [DOI] [PubMed] [Google Scholar]
- 7. Bryant D. M., and Mostov K. E. (2008) From cells to organs: building polarized tissue. Nat. Rev. Mol. Cell Biol. 9, 887–901 10.1038/nrm2523,10.1038/nrn2534,10.1038/nrn2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. den Elzen N., Buttery C. V., Maddugoda M. P., Ren G., and Yap A. S. (2009) Cadherin adhesion receptors orient the mitotic spindle during symmetric cell division in mammalian epithelia. Mol. Biol. Cell. 20, 3740–3750 10.1091/mbc.e09-01-0023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Benham-Pyle B. W., Pruitt B. L., and Nelson W. J. (2015) Cell adhesion: mechanical strain induces E-cadherin-dependent Yap1 and β-catenin activation to drive cell cycle entry. Science. 348, 1024–1027 10.1126/science.aaa4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Collins C., and Nelson W. J. (2015) Running with neighbors: coordinating cell migration and cell–cell adhesion. Curr. Opin. Cell Biol. 36, 62–70 10.1016/j.ceb.2015.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mui K. L., Chen C. S., and Assoian R. K. (2016) The mechanical regulation of integrin–cadherin crosstalk organizes cells, signaling and forces. J. Cell Sci. 129, 1093–1100 10.1242/jcs.183699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Benjamin J. M., and Nelson W. J. (2008) Bench to bedside and back again: molecular mechanisms of α-catenin function and roles in tumorigenesis. Semin. Cancer Biol. 18, 53–64 10.1016/j.semcancer.2007.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wilson H. V. (1907) On some phenomena of coalescence and regeneration in sponges. J. Exp. Zool. 5, 245–258 10.1002/jez.1400050204 [DOI] [Google Scholar]
- 14. Humphreys T. (1963) Chemical dissolution and in vitro reconstruction of sponge cell adhesions: I. isolation and functional demonstration of the components involved. Dev. Biol. 8, 27–47 10.1016/0012-1606(63)90024-1 [DOI] [PubMed] [Google Scholar]
- 15. Cauldwell C. B., Henkart P., and Humphreys T. (1973) Physical properties of sponge aggregation factor: a unique proteoglycan complex. Biochemistry 12, 3051–3055 10.1021/bi00740a017 [DOI] [PubMed] [Google Scholar]
- 16. Henkart P., Humphreys S., and Humphreys T. (1973) Characterization of sponge aggregation factor: a unique proteoglycan complex. Biochemistry 12, 3045–3050 10.1021/bi00740a016 [DOI] [PubMed] [Google Scholar]
- 17. Haseley S. R., Vermeer H. J., Kamerling J. P., and Vliegenthart J. F. (2001) Carbohydrate self-recognition mediates marine sponge cellular adhesion. Proc. Natl. Acad. Sci. U.S.A. 98, 9419–9424 10.1073/pnas.151111298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moscona A. A. (1968) Cell aggregation: properties of specific cell-ligands and their role in the formation of multicellular systems. Dev. Biol. 18, 250–277 10.1016/0012-1606(68)90035-3 [DOI] [PubMed] [Google Scholar]
- 19. Grice L. F., Gauthier M. E. A., Roper K. E., Fernàndez-Busquets X., Degnan S. M., and Degnan B. M. (2017) Origin and evolution of the sponge aggregation factor gene family. Mol. Biol. Evol. 34, 1083–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vilanova E., Santos G. R., Aquino R. S., Valle-Delgado J. J., Anselmetti D., Fernàndez-Busquets X., and Mourão P. A. (2016) Carbohydrate–carbohydrate interactions mediated by sulfate esters and calcium provide the cell adhesion required for the emergence of early metazoans. J. Biol. Chem. 291, 9425–9437 10.1074/jbc.M115.708958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Halbleib J. M., and Nelson W. J. (2006) Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 20, 3199–3214 10.1101/gad.1486806 [DOI] [PubMed] [Google Scholar]
- 22. Nelson W. J. (2003) Adaptation of core mechanisms to generate cell polarity. Nature 422, 766–774 10.1038/nature01602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Plotnikov S. V., Pasapera A. M., Sabass B., and Waterman C. M. (2012) Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell 151, 1513–1527 10.1016/j.cell.2012.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kanchanawong P., Shtengel G., Pasapera A. M., Ramko E. B., Davidson M. W., Hess H. F., and Waterman C. M. (2010) Nanoscale architecture of integrin-based cell adhesions. Nature 468, 580–584 10.1038/nature09621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Buck C. A., and Horwitz A. F. (1987) Integrin, a transmembrane glycoprotein complex mediating cell–substratum adhesion. J. Cell Sci. 1987, 231–250 10.1242/jcs.1987.Supplement_8.13,10.1242/jcs.1987.Supplement_7.17 [DOI] [PubMed] [Google Scholar]
- 26. Horwitz A., Duggan K., Buck C., Beckerle M. C., and Burridge K. (1986) Interaction of plasma membrane fibronectin receptor with talin: a transmembrane linkage. Nature. 320, 531–533 10.1038/320531a0 [DOI] [PubMed] [Google Scholar]
- 27. Burridge K., and Mangeat P. (1984) An interaction between vinculin and talin. Nature. 308, 744–746 10.1038/308744a0 [DOI] [PubMed] [Google Scholar]
- 28. Turner C. E., Glenney J. R. Jr, and Burridge K. (1990) Paxillin: a new vinculin-binding protein present in focal adhesions. J. Cell Biol. 111, 1059–1068 10.1083/jcb.111.3.1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tyler S. (2003) Epithelium: the primary building block for metazoan complexity. Integr. Comp. Biol. 43, 55–63 10.1093/icb/43.1.55 [DOI] [PubMed] [Google Scholar]
- 30. Harwood A., and Coates J. C. (2004) A prehistory of cell adhesion. Curr. Opin. Cell Biol. 16, 470–476 10.1016/j.ceb.2004.07.011 [DOI] [PubMed] [Google Scholar]
- 31. Nickel M., Scheer C., Hammel J. U., Herzen J., and Beckmann F. (2011) The contractile sponge epithelium sensu lato–body contraction of the demosponge Tethya wilhelma is mediated by the pinacoderm. J. Exp. Biol. 214, 1692–1698 10.1242/jeb.049148 [DOI] [PubMed] [Google Scholar]
- 32. Leys S. P., and Hill A. (2012) The physiology and molecular biology of sponge tissues. Adv. Mar. Biol. 62, 1–56 10.1016/B978-0-12-394283-8.00001-1 [DOI] [PubMed] [Google Scholar]
- 33. Adams E. D., Goss G. G., and Leys S. P. (2010) Freshwater sponges have functional, sealing epithelia with high transepithelial resistance and negative transepithelial potential. PLoS One 5, e15040 10.1371/journal.pone.0015040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Leys S. P., Nichols S. A., and Adams E. D. (2009) Epithelia and integration in sponges. Integr. Comp. Biol. 49, 167–177 10.1093/icb/icp038 [DOI] [PubMed] [Google Scholar]
- 35. Nichols S. A., Dirks W., Pearse J. S., and King N. (2006) Early evolution of animal cell signaling and adhesion genes. Proc. Natl. Acad. Sci. U.S.A. 103, 12451–12456 10.1073/pnas.0604065103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Srivastava M., Simakov O., Chapman J., Fahey B., Gauthier M. E., Mitros T., Richards G. S., Conaco C., Dacre M., Hellsten U., Larroux C., Putnam N. H., Stanke M., Adamska M., Darling A., et al. (2010) The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466, 720–726 10.1038/nature09201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fahey B., and Degnan B. M. (2010) Origin of animal epithelia: insights from the sponge genome. Evol. Dev. 12, 601–617 10.1111/j.1525-142X.2010.00445.x [DOI] [PubMed] [Google Scholar]
- 38. Schippers K. J., and Nichols S. A. (2018) Evidence of signaling and adhesion roles for β-catenin in the sponge Ephydatia muelleri. Mol. Biol. Evol. 35, 1407–1421 10.1093/molbev/msy033 [DOI] [PubMed] [Google Scholar]
- 39. Sebé-Pedrós A., Roger A. J., Lang F. B., King N., and Ruiz-Trillo I. (2010) Ancient origin of the integrin-mediated adhesion and signaling machinery. Proc. Natl. Acad. Sci. U.S.A. 107, 10142–10147 10.1073/pnas.1002257107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Miller P. W., Clarke D. N., Weis W. I., Lowe C. J., and Nelson W. J. (2013) The evolutionary origin of epithelial cell–cell adhesion mechanisms. Curr. Top. Membr. 72, 267–311 10.1016/B978-0-12-417027-8.00008-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dickinson D. J., Nelson W. J., and Weis W. I. (2011) A polarized epithelium organized by β- and α-catenin predates cadherin and metazoan origins. Science 331, 1336–1339 10.1126/science.1199633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dickinson D. J., Nelson W. J., and Weis W. I. (2012) An epithelial tissue in Dictyostelium challenges the traditional origin of metazoan multicellularity. Bioessays 34, 833–840 10.1002/bies.201100187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Boute N., Exposito J. Y., Boury-Esnault N., Vacelet J., Noro N., Miyazaki K., Yoshizato K., and Garrone R. (1996) Type IV collagen in sponges, the missing link in basement membrane ubiquity. Biol. Cell. 88, 37–44 10.1016/S0248-4900(97)86829-3 [DOI] [PubMed] [Google Scholar]
- 44. Ereskovsky A. V., Borchiellini C., Gazave E., Ivanisevic J., Lapébie P., Perez T., Renard E., and Vacelet J. (2009) The homoscleromorph sponge Oscarella lobularis, a promising sponge model in evolutionary and developmental biology. Bioessays 31, 89–97 10.1002/bies.080058 [DOI] [PubMed] [Google Scholar]
- 45. Nielsen C. (2008) Six major steps in animal evolution: are we derived sponge larvae? Evol. Dev. 10, 241–257 10.1111/j.1525-142X.2008.00231.x [DOI] [PubMed] [Google Scholar]
- 46. Boury-Esnault N., Ereskovsky A., Bézac C., and Tokina D. (2003) Larval development in the Homoscleromorpha (Porifera, Demospongiae). Invertebr. Biol. 122, 187–202 10.1111/j.1744-7410.2003.tb00084.x [DOI] [Google Scholar]
- 47. Nichols S. A., Roberts B. W., Richter D. J., Fairclough S. R., and King N. (2012) Origin of metazoan cadherin diversity and the antiquity of the classical cadherin/β-catenin complex. Proc. Natl. Acad. Sci. U.S.A. 109, 13046–13051 10.1073/pnas.1120685109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ereskovsky A. V., Richter D. J., Lavrov D. V., Schippers K. J., and Nichols S. A. (2017) Transcriptome sequencing and delimitation of sympatric Oscarella species (O. carmela and O. pearsei sp. nov) from California, U.S.A. PLoS One 12, e0183002 10.1371/journal.pone.0183002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bateman A., Coin L., Durbin R., Finn R. D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E. L., Studholme D. J., Yeats C., and Eddy S. R. (2004) The Pfam protein families database. Nucleic Acids Res. 32, D138–D141 10.1093/nar/gkh121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Finn R. D., Clements J., and Eddy S. R. (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37 10.1093/nar/gkr367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Letunic I., Doerks T., and Bork P. (2012) SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40, D302–D305 10.1093/nar/gkr931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gasteiger E., Hoogland C., Gattiker A., Duvaud S'everine Wilkins M. R., Appel R. D., and Bairoch A. (2005) Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook (Walker J. M., ed.) pp. 571–607, Humana Press, Totowa, NJ [Google Scholar]
- 53. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., and Lipman D. J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., and Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 55. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., and Madden T. L. (2009) BLAST+: architecture and applications. BMC Bioinformatics 10, 421 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Capella-Gutiérrez S., Silla-Martíinez J. M., and Gabaldón T. (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sánchez R., Serra F., Tárraga J., Medina I., Carbonell J., Pulido L., de María A., Capella-Gutíerrez S., Huerta-Cepas J., Gabaldón T., Dopazo J., and Dopazo H. (2011) Phylemon 2.0: a suite of web-tools for molecular evolution, phylogenetics, phylogenomics and hypotheses testing. Nucleic Acids Res. 39, W470–W474 10.1093/nar/gkr408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kalyaanamoorthy S., Minh B. Q., Wong T. K. F., von Haeseler A., and Jermiin L. S. (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 14, 587–589 10.1038/nmeth.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Trifinopoulos J., Nguyen L.-T., von Haeseler A., and Minh B. Q. (2016) W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235 10.1093/nar/gkw256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nguyen L.-T., Schmidt H. A., von Haeseler A., and Minh B. Q. (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hoang D. T., Chernomor O., von Haeseler A., Minh B. Q., and Vinh L. S. (2018) UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522 10.1093/molbev/msx281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Borgon R. A., Vonrhein C., Bricogne G., Bois P. R., and Izard T. (2004) Crystal structure of human vinculin. Structure 12, 1189–1197 10.1016/j.str.2004.05.009 [DOI] [PubMed] [Google Scholar]
- 65. Keegan R. M., and Winn M. D. (2008) MrBUMP: an automated pipeline for molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 64, 119–124 10.1107/S0907444907037195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Spudich J. A., and Watt S. (1971) The regulation of rabbit skeletal muscle contraction I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246, 4866–4871 [PubMed] [Google Scholar]
- 69. Pokutta S., and Weis W. I. (2000) Structure of the dimerization and β-catenin-binding region of α-catenin. Mol. Cell 5, 533–543 10.1016/S1097-2765(00)80447-5 [DOI] [PubMed] [Google Scholar]
- 70. Bass M. D., Patel B., Barsukov I. G., Fillingham I. J., Mason R., Smith B. J., Bagshaw C. R., and Critchley D. R. (2002) Further characterization of the interaction between the cytoskeletal proteins talin and vinculin. Biochem. J. 362, 761–768 10.1042/bj3620761,10.1042/0264-6021:3620761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Clarke D. N., Miller P. W., Lowe C. J., Weis W. I., and Nelson W. J. (2016) Characterization of the cadherin–catenin complex of the sea anemone Nematostella vectensis and implications for the evolution of metazoan cell–cell adhesion. Mol. Biol. Evol. 33, 2016–2029 10.1093/molbev/msw084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bakolitsa C., de Pereda J. M., Bagshaw C. R., Critchley D. R., and Liddington R. C. (1999) Crystal structure of the vinculin tail suggests a pathway for activation. Cell 99, 603–613 10.1016/S0092-8674(00)81549-4 [DOI] [PubMed] [Google Scholar]
- 73. Bakolitsa C., Cohen D. M., Bankston L. A., Bobkov A. A., Cadwell G. W., Jennings L., Critchley D. R., Craig S. W., and Liddington R. C. (2004) Structural basis for vinculin activation at sites of cell adhesion. Nature 430, 583–586 10.1038/nature02610 [DOI] [PubMed] [Google Scholar]
- 74. Izard T., Evans G., Borgon R. A., Rush C. L., Bricogne G., and Bois P. R. (2004) Vinculin activation by talin through helical bundle conversion. Nature. 427, 171–175 10.1038/nature02281 [DOI] [PubMed] [Google Scholar]
- 75. Rangarajan E. S., and Izard T. (2013) Dimer asymmetry defines α-catenin interactions. Nat. Struct. Mol. Biol. 20, 188–193 10.1038/nsmb.2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Pokutta S., Choi H.-J., Ahlsen G., Hansen S. D., and Weis W. I. (2014) Structural and thermodynamic characterization of cadherin·β-catenin·α-catenin complex formation. J. Biol. Chem. 289, 13589–13601 10.1074/jbc.M114.554709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Shao X., Kang H., Loveless T., Lee G. R., Seok C., Weis W. I., Choi H.-J., and Hardin J. (2017) Cell–cell adhesion in metazoans relies on evolutionarily conserved features of the α-catenin·β-catenin–binding interface. J. Biol. Chem. 292, 16477–16490 10.1074/jbc.M117.795567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yamada S., Pokutta S., Drees F., Weis W. I., and Nelson W. J. (2005) Deconstructing the cadherin–catenin–actin complex. Cell 123, 889–901 10.1016/j.cell.2005.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Benjamin J. M., Kwiatkowski A. V., Yang C., Korobova F., Pokutta S., Svitkina T., Weis W. I., and James Nelson W. J. (2010) αE-catenin regulates actin dynamics independently of cadherin-mediated cell–cell adhesion. J. Cell Biol. 189, 339–352 10.1083/jcb.200910041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kwiatkowski A. V., Maiden S. L., Pokutta S., Choi H.-J., Benjamin J. M., Lynch A. M., Nelson W. J., Weis W. I., and Hardin J. (2010) In vitro and in vivo reconstitution of the cadherin–catenin–actin complex from Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 107, 14591–14596 10.1073/pnas.1007349107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Miller P. W., Pokutta S., Ghosh A., Almo S. C., Weis W. I., Nelson W. J., and Kwiatkowski A. V. (2013) Danio rerio αE-catenin is a monomeric F-actin binding protein with distinct properties from Mus musculus αE-catenin. J. Biol. Chem. 288, 22324–22332 10.1074/jbc.M113.458406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ishiyama N., Tanaka N., Abe K., Yang Y. J., Abbas Y. M., Umitsu M., Nagar B., Bueler S. A., Rubinstein J. L., Takeichi M., and Ikura M. (2013) An autoinhibited structure of α-catenin and its implications for vinculin recruitment to adherens junctions. J. Biol. Chem. 288, 15913–15925 10.1074/jbc.M113.453928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ziegler W. H., Liddington R. C., and Critchley D. R. (2006) The structure and regulation of vinculin. Trends Cell Biol. 16, 453–460 10.1016/j.tcb.2006.07.004 [DOI] [PubMed] [Google Scholar]
- 84. Patel B., Gingras A. R., Bobkov A. A., Fujimoto L. M., Zhang M., Liddington R. C., Mazzeo D., Emsley J., Roberts G. C., Barsukov I. L., and Critchley D. R. (2006) The activity of the vinculin binding sites in talin is influenced by the stability of the helical bundles that make up the talin rod. J. Biol. Chem. 281, 7458–7467 10.1074/jbc.M508058200 [DOI] [PubMed] [Google Scholar]
- 85. Rimm D. L., Koslov E. R., Kebriaei P., Cianci C. D., and Morrow J. S. (1995) α1(E)-catenin is an actin-binding and -bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc. Natl. Acad. Sci. U.S.A. 92, 8813–8817 10.1073/pnas.92.19.8813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Koslov E. R., Maupin P., Pradhan D., Morrow J. S., and Rimm D. L. (1997) α-Catenin can form asymmetric homodimeric complexes and/or heterodimeric complexes with β-catenin. J. Biol. Chem. 272, 27301–27306 10.1074/jbc.272.43.27301 [DOI] [PubMed] [Google Scholar]
- 87. Pokutta S., Drees F., Takai Y., Nelson W. J., and Weis W. I. (2002) Biochemical and structural definition of the l-afadin- and actin-binding sites of α-catenin. J. Biol. Chem. 277, 18868–18874 10.1074/jbc.M201463200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Drees F., Pokutta S., Yamada S., Nelson W. J., and Weis W. I. (2005) α-Catenin is a molecular switch that binds E-cadherin-β-catenin and regulates actin-filament assembly. Cell. 123, 903–915 10.1016/j.cell.2005.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Choi H.-J., Pokutta S., Cadwell G. W., Bobkov A. A., Bankston L. A., Liddington R. C., and Weis W. I. (2012) αE-catenin is an autoinhibited molecule that coactivates vinculin. Proc. Natl. Acad. Sci. U.S.A. 109, 8576–8581 10.1073/pnas.1203906109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Buckley C. D., Tan J., Anderson K. L., Hanein D., Volkmann N., Weis W. I., Nelson W. J., and Dunn A. R. (2014) Cell adhesion: the minimal cadherin–catenin complex binds to actin filaments under force. Science 346, 1254211 10.1126/science.1254211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Bays J. L., and DeMali K. A. (2017) Vinculin in cell–cell and cell–matrix adhesions. Cell. Mol. Life Sci. 74, 2999–3009 10.1007/s00018-017-2511-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Han M. K., and de Rooij J. (2016) Converging and unique mechanisms of mechanotransduction at adhesion sites. Trends Cell Biol. 26, 612–623 10.1016/j.tcb.2016.03.005 [DOI] [PubMed] [Google Scholar]
- 93. Carisey A., and Ballestrem C. (2011) Vinculin, an adapter protein in control of cell adhesion signalling. Eur. J. Cell Biol. 90, 157–163 10.1016/j.ejcb.2010.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Barstead R. J., and Waterston R. H. (1991) Vinculin is essential for muscle function in the nematode. J. Cell Biol. 114, 715–724 10.1083/jcb.114.4.715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Barstead R. J., and Waterston R. H. (1989) The basal component of the nematode dense-body is vinculin. J. Biol. Chem. 264, 10177–10185 [PubMed] [Google Scholar]
- 96. Lecroisey C., Ségalat L., and Gieseler K. (2007) The C. elegans dense body: anchoring and signaling structure of the muscle. J. Muscle Res. Cell Motil. 28, 79–87 10.1007/s10974-007-9104-y [DOI] [PubMed] [Google Scholar]
- 97. Alatortsev V. E., Kramerova I. A., Frolov M. V., Lavrov S. A., and Westphal E. D. (1997) Vinculin gene is non-essential in Drosophila melanogaster. FEBS Lett. 413, 197–201 10.1016/S0014-5793(97)00901-0 [DOI] [PubMed] [Google Scholar]
- 98. Maartens A. P., Wellmann J., Wictome E., Klapholz B., Green H., and Brown N. H. (2016) Drosophila vinculin is more harmful when hyperactive than absent, and can circumvent integrin to form adhesion complexes. J. Cell Sci. 129, 4354–4365 10.1242/jcs.189878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Kang H., Bang I., Jin K. S., Lee B., Lee J., Shao X., Heier J. A., Kwiatkowski A. V., Nelson W. J., Hardin J., Weis W. I., and Choi H.-J. (2017) Structural and functional characterization of Caenorhabditis elegans α-catenin reveals constitutive binding to β-catenin and F-actin. J. Biol. Chem. 292, 7077–7086 10.1074/jbc.M116.769778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Sarpal R., Pellikka M., Patel R. R., Hui F. Y., Godt D., and Tepass U. (2012) Mutational analysis supports a core role for Drosophila α-catenin in adherens junction function. J. Cell Sci. 125, 233–245 10.1242/jcs.096644 [DOI] [PubMed] [Google Scholar]
- 101. Costa M., Raich W., Agbunag C., Leung B., Hardin J., and Priess J. R. (1998) A putative catenin-cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. J. Cell Biol. 141, 297–308 10.1083/jcb.141.1.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Magie C. R., Pinto-Santini D., and Parkhurst S. M. (2002) Rho1 interacts with p120ctn and α-catenin, and regulates cadherin-based adherens junction components in Drosophila. Development 129, 3771–3782 [DOI] [PubMed] [Google Scholar]
- 103. Elliott G. R., and Leys S. P. (2007) Coordinated contractions effectively expel water from the aquiferous system of a freshwater sponge. J. Exp. Biol. 210, 3736–3748 10.1242/jeb.003392 [DOI] [PubMed] [Google Scholar]
- 104. Yonemura S., Wada Y., Watanabe T., Nagafuchi A., and Shibata M. (2010) α-Catenin as a tension transducer that induces adherens junction development. Nat. Cell Biol. 12, 533–542 10.1038/ncb2055 [DOI] [PubMed] [Google Scholar]
- 105. Humbert-David N., and Garrone R. (1993) A six-armed, tenascin-like protein extracted from the Porifera Oscarella tuberculata (Homosclerophorida). Eur. J. Biochem. 216, 255–260 10.1111/j.1432-1033.1993.tb18140.x [DOI] [PubMed] [Google Scholar]
- 106. Schuster A., Vargas S., Knapp I., Pomponi S. A., Toonen R. J., Erpenbeck D., and Woerheide G. (2017) Divergence times in demosponges (Porifera): first insights from new mitogenomes and the inclusion of fossils in a birth-death clock model. bioRxiv 10.1101/159806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Blumbach B., Pancer Z., Diehl-Seifert B., Steffen R., Münkner J., Müller I., and Müller W. E. (1998) The putative sponge aggregation receptor: isolation and characterization of a molecule composed of scavenger receptor cysteine-rich domains and short consensus repeats. J. Cell Sci. 111, 2635–2644 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.