Graphical abstract
Abbreviations: ACh, acetylcholine; mAChR, muscarinic acetylcholine receptor; GPCR, G protein-coupled receptor; AD, Alzheimer’s disease; PAM, positive allosteric modulator; NAM, negative allosteric modulator; TM, transmembrane domain; ECL, extracellular loop; CHO, Chinese hamster ovary; WT, wild type; DMEM, Dulbecco modified eagle medium; FBS, fetal bovine serum; BSA, bovine serum albumin; PBS, phosphate-buffered saline; HBSS, Hanks’ balanced salt solution; IP1, inositol monophosphate; NMS, N-methylscopolamine; PBZ, phenoxybenzamine; BQCA, benzylquinolone carboxylic acid, 1-(4-methoxybenzyl)-4-oxo-1,4-dihydroquinoline-3-carboxylic acid; PF-06767832, N-((3R,4S)-3-hydroxytetrahydro-2H-pyran-4-yl)-5-methyl-4-(4-(thiazol-4-yl)benzyl)pyridine-2-carboxamide; VU6004256 (4, 6-difluoro-N-(1S,2S)-2-hydroxycyclohexyl-1-((6-(1-methyl-1H-pyrazol-4-yl)pyridine-3-yl)methyl)-1H-indole-3-carboxamide; MIPS1780, 3-(2-hydroxycyclohexyl)-6-(2-((4-(1-methyl-1H-pyrazol-4-yl)- benzyl)oxy)phenyl)pyrimidin-4(3H)-one; BQZ-12, 3-((1S,2S)-2-hydroxycyclohexyl)-6-((6-(1-methyl-1H-pyrazol-4-yl)pyridin-3-yl)methyl)benzo [h]-quinazolin-4(3H)-one; ANOVA, analysis of variance
Keywords: Allosteric modulation, Muscarinic acetylcholine receptor, BQCA, Mutagenesis, Drug discovery
Abstract
Subtype-selective allosteric modulation of the M1 muscarinic acetylcholine (ACh) receptor (M1 mAChR) is an attractive approach for the treatment of numerous disorders, including cognitive deficits. The discovery of benzyl quinolone carboxylic acid, BQCA, a selective M1 mAChR positive allosteric modulator (PAM), spurred the subsequent development of newer generation M1 PAMs representing diverse chemical scaffolds, different pharmacodynamic properties and, in some instances, improved pharmacokinetics. Key exemplar molecules from such efforts include PF-06767832 (N-((3R,4S)-3-hydroxytetrahydro-2H-pyran-4-yl)-5-methyl-4-(4-(thiazol-4-yl)benzyl)pyridine-2-carboxamide), VU6004256 (4,6-difluoro-N-(1S,2S)-2-hydroxycyclohexyl-1-((6-(1-methyl-1H-pyrazol-4-yl)pyridine-3-yl)methyl)-1H-indole-3-carboxamide) and MIPS1780 (3-(2-hydroxycyclohexyl)-6-(2-((4-(1-methyl-1H-pyrazol-4-yl)-benzyl)oxy)phenyl)pyrimidin-4(3H)-one). Given these diverse scaffolds and pharmacodynamics, the current study combined pharmacological analysis and site-directed mutagenesis to explore the potential binding site and function of newer M1 mAChR PAMs relative to BQCA. Interestingly, the mechanism of action of the novel PAMs was consistent with a common model of allostery, as previously described for BQCA. Key residues involved in the activity of BQCA, including Y179 in the second extracellular loop (ECL) and W4007.35 in transmembrane domain (TM) 7, were critical for the activity of all PAMs tested. Overall, our data indicate that structurally distinct PAMs share a similar binding site with BQCA, specifically, an extracellular allosteric site defined by residues in TM2, TM7 and ECL2. These findings provide valuable insights into the structural basis underlying modulator binding, cooperativity and signaling at the M1 mAChR, which is essential for the rational design of PAMs with tailored pharmacological properties.
1. Introduction
Muscarinic acetylcholine receptors (mAChRs) are members of the Class A G protein-coupled receptor (GPCR) family [1] involved in central and peripheral biology [2]. Five mAChR subtypes (M1-M5), have been identified; M1, M3 and M5 mAChRs preferentially couple to Gq/11 proteins; M2 and M4 mAChRs preferentially couple to Gi/o proteins [3]. Of note, the M1 mAChRs are highly expressed in forebrain regions, including the cerebral cortex, hippocampus and striatum [4]. Transgenic M1 mAChR studies implicated roles for this receptor in neuronal excitability, locomotor activity and learning and memory [Reviewed in [5]]. Therefore, selective activation of the M1 mAChR has emerged as an approach for the treatment of cognitive deficits associated with disorders such as Alzheimer’s disease (AD) and schizophrenia [6], [7]. This is of particular relevance due to limitations associated with current cognition-enhancing agents [8]. For instance, loss of cholinergic neurons is a hallmark of AD [9], [10] and inhibitors of acetylcholinesterase remain the primary treatment for disease symptoms [11] yet are associated with substantial adverse effects [12]. Unfortunately, drug discovery efforts aimed at developing directly acting M1 mAChR agonists have been unsuccessful. The best clinical example is the M1/M4-preferring agonist xanomeline, which proved beneficial in improving cognitive function and psychotic symptoms in AD and schizophrenia [13], but was not further developed due to off-target gastrointestinal adverse effects from interaction with other mAChRs [14].
An alternative approach to developing subtype-selective drugs is through targeting topographically distinct allosteric sites [15], [16]. In this regard, the discovery of benzyl quinolone carboxylic acid (BQCA; 1-(4-methoxybenzyl)-4-oxo-1,4-dihydroquinoline-3-carboxylic acid), a selective M1 positive allosteric modulator (PAM) was a major development for the field [17], leading to additional M1 mAChR PAMs. However, BQCA has a low affinity for the M1 mAChR [18], and was associated with liabilities that precluded further development, such as poor brain penetration and solubility, and high plasma protein binding [19]. The work with BQCA also highlighted complexities associated with design of allosteric modulators as potential therapeutics. In general, such challenges are two-fold. The first relates to understanding of the molecular properties associated with allosteric drugs, including affinity, cooperativity with the orthosteric agonist (positive or negative), and whether the allosteric ligand possesses intrinsic signaling efficacy; the interplay between these properties is only starting to be appreciated, and varies depending on the target and disease [20]. The second major challenge for discovery programs is optimizing physicochemical/pharmacokinetic properties of candidate molecules to ensure appropriate target coverage when/where required, while minimizing off-target activity. A key development in this regard is the identification of new allosteric scaffolds with which to target GPCRs. For instance, Pfizer recently disclosed a novel M1 mAChR ‘PAM-agonist’, PF-06767832, with promising physiochemical and pharmacokinetic properties. However, PF-06767832 exhibited seizure liability, and cardiovascular and gastrointestinal side-effects, indicating that M1 mAChR over-activation may directly contribute to adverse events [21]. A more recent study using PAMs with reduced intrinsic allosteric agonist activity also noted adverse effects [22]. This may reflect deficiencies in our understanding of the ‘optimal’ degree of positive cooperativity required for specific disease intervention [18], [23]. An alternative selective M1 PAM-agonist, VU6004256, reversed cognitive deficits in a mouse model, indicative of preclinical efficacy [24]. The chemotype and molecular pharmacology of this M1 PAM-agonist is similar to PF-06764427, yet they display different in vivo activities [25]. Our recent structure-activity studies also led to discovery of another series of M1 mAChR PAMs [26], with a key exemplar being MIPS1780 (compound 29 in [26]).
However, despite the increasing availability of a range of novel M1 mAChR PAM scaffolds with different molecular and functional properties, the extent to which their differential activities are driven from interaction with a common allosteric binding pocket, or from alternative regions is not known. Previously, we utilized mutagenesis to reveal that BQCA binds to a “common” allosteric mAChR binding site, located in an extracellular vestibule defined by residues predominantly in TM2, TM7 and ECL2 [27]. However, there is also pharmacological evidence that the mAChRs possess at least a second allosteric site [28], [29], [30]. Thus, the aim of the current study was to apply a combination of analytical pharmacology and site-directed mutagenesis (Fig. 1A) to explore the potential binding site and function of novel selective M1 mAChR PAMs with diverse scaffolds, PF-06767832, VU6004256 and MIPS1780, in comparison to the first-generation BQCA (Fig. 1B). Our results provide evidence that these ligands act solely as modulators of ACh affinity, not efficacy, and may bind to a similar binding pocket at the M1 mAChR and exert their effects with subtle differences, but largely consistent, with those of BQCA. This provides valuable insight into the structural basis underlying allosteric ligand binding and function at the M1 mAChR.
Fig. 1.
Mutations and ligands investigated in the current study. (A) A snake diagram of the hM1 mAChR highlighting the mutated residues and (B) chemical structures of the M1 positive allosteric modulators used in the current study.
2. Materials and methods
2.1. Materials
Dulbecco’s modified Eagle’s medium (DMEM) and fetal bovine serum (FBS) were purchased from Invitrogen (Carlsbad, CA) and ThermoTrace (Melbourne, Australia), respectively. Hygromycin B was purchased from Roche (Mannheim, Germany). IP-One assay kit and reagents were purchased from Cisbio (Codolet, France). [3H]N-methylscopolamine ([3H]NMS); specific activity, 75 Ci/mmol) and Ultima gold and MicroScint scintillation liquid were purchased from PerkinElmer Life Sciences (Boston, MA). BQCA and MIPS1780 were synthesized in-house at the Monash Institute of Pharmaceutical Sciences as described previously [26], [31]. VU6004256 was synthesized in-house at Vanderbilt University as described previously [25]. All other chemicals were purchased from Sigma Aldrich (St. Louis, MO), or as otherwise stated below. FlpIn Chinese Hamster Ovary (CHO) cells stably expressing the wild type (WT) or mutant c-myc hM1 mAChRs (passage numbers 8–25) were generated as described previously [32], and maintained in DMEM supplemented with 5% FBS, 16 mM HEPES and 600 μg/ml hygromycin B at 37 °C in humidified atmosphere containing 5% CO2.
2.2. Whole cell radioligand binding assays
Saturation binding assays were performed to estimate the expression levels and equilibrium dissociation constant of the radioligand (KD). FlpIn CHO cells stably expressing the WT or mutant c-myc hM1 mAChRs were plated at the density of 25,000 per well of 96-well white clear bottom Isoplates (PerkinElmer Life Sciences, Boston, MA), and grown overnight at 37 °C. The following day, cells were washed twice with Phosphate Buffer Saline (PBS), and incubated with 0.03–10 nM [3H]NMS in a final volume of 100 µl buffer (20 mM HEPES, 100 mM NaCl, 10 mM MgCl2, pH 7.4) for 4 h at room temperature. For binding interaction assays, cells were incubated with increasing concentrations of ACh in the presence or absence of increasing concentrations of PAMs, and [3H]NMS (0.3 nM for the WT, F772.56I, Y179ECL2A, W4007.35A, and 0.6 nM for Y822.61A M1 mAChRs) in a final volume of 100 µl. Atropine at the final concentration of 100 µM was used to determine non-specific binding. The assays were terminated by rapid removal of the radioligand, and two washes with 100 µl/well ice-cold 0.9% NaCl buffer. Radioactivity was determined by addition of 100 µl/well MicroScint scintillation liquid (PerkinElmer Life Sciences, Boston, MA), and counting in a MicroBeta plate reader (PerkinElmer Life Sciences, Boston, MA).
2.3. IP-one accumulation assays
The IP-One assay kit (Cisbio) was used for the direct quantitative measurement of myo-Inositol 1 phosphate (IP1). Cells were seeded at 25,000 per well into 96-well transparent cell culture plates and incubated overnight at 37 °C. The following day, cells were pre-incubated with IP1 stimulation buffer (1 mM CaCl2, 0.5 mM MgCl2, 4.2 mM KCl, 146 mM NaCl, 5.5 mM D-Glucose, 10 mM HEPES and 50 mM LiCl, pH 7.4) for 1 h before stimulation with ACh in the presence or absence of increasing concentrations of PAMs in IP1 stimulation buffer for 1 h at 37 °C, 5% CO2. Cells were then lysed with IP1 lysis buffer (50 mM HEPES pH 7.0, 15 mM KF, 1.5% V/V Triton-X-100, 3% V/V FBS, 0.2% W/V BSA), and IP1 levels were measured by incubation of cell lysates with FRET reagents (the cryptate-labeled anti-IP1 antibody and the d2-labeled IP1 analogue) for 1 h at 37 °C. The emission signals were measured at 590 and 665 nm after excitation at 340 nm, using the Envision plate reader (PerkinElmer Life Sciences, Boston, MA). Signals were expressed as the FRET ratio: F = (fluorescence665 nm/fluorescence590 nm) × 104, and normalized to the response to maximal ACh concentration (100 µM).
2.4. Receptor alkylation assays
Cells were pre-treated with vehicle or the irreversible orthosteric alkylating agent, phenoxybenzamine (PBZ) for 30 min at 37 °C, 5% CO2 to reduce functional receptor availability, followed by three extensive washes with PBS. IP1 accumulation assays were then performed as described above.
2.5. Flow cytometric detection of cell surface receptor expression
Cells were harvested with PBS supplemented with 2 mM EDTA, and transferred to a 96-well v-bottomed plate. Cells were then centrifuged at 350g for 3 min at 4 °C and resuspended in 100 µl of blocking buffer (1× HBSS, 5% BSA, 20 mM HEPES, pH 7.4). After 30 min incubation on ice, cells were incubated with mouse monoclonal 9E10 antibody (prepared in-house) targeted to the c-myc epitope tag at 5 µg/ml in assay buffer (1× HBSS, 0.1% BSA, 20 mM HEPES, pH 7.4) for 90 min on ice. Cells were then washed twice with assay buffer and incubated with a secondary goat anti-mouse IgG antibody conjugated to Alexa Fluor 647 (1 µg/ml, Molecular Probes, Invitrogen) for 60 min on ice. Following two washes, cells were resuspended in assay buffer containing 1 µM Sytox blue (Thermo Fisher Scientific). The fluorescence signal was quantified using a FACSCanto II flow cytometer (BD Biosciences).
2.6. Data analysis
All data were analysed using GraphPad Prism 7 (San Diego, CA). Inhibition binding data between ACh and the radioligand antagonist, [3H]NMS, were analysed according to a one-site binding model [33]. Binding interaction studies between orthosteric agonists and allosteric modulators were fitted to the following allosteric ternary complex model [23] (Eq. (1)):
| (1) |
where Bmax is the total number of receptors, [D], [B] and [I] denote the concentrations of radioligand, allosteric ligand, and orthosteric ligand, respectively, and KD, KB and KI represent their respective equilibrium dissociation constants. α′ and α are the cooperativity factors between the allosteric ligand and radioligand or orthosteric ligand, respectively. Values of α or α′ > 1 denote positive cooperativity, values between 0 and 1 denote negative cooperativity, and a value of 1 indicates neutral cooperativity.
To estimate intrinsic efficacy of ACh in IP1 accumulation assays at the WT or mutant M1 mAChRs, the following operational model of agonism [34] (Eq. (2)) was used:
| (2) |
where Em is the maximal possible system response and Basal is the response in the absence of agonist. [A] denotes the concentration of ligand, and KA represents its equilibrium dissociation constants. τ denotes the intrinsic efficacy of the ligand, which incorporates the total receptor density and the efficiency of stimulus-response coupling.
Functional interaction studies between orthosteric agonists and allosteric modulators in IP1 assays were analysed using the following operational model of allosterism and agonism [35] (Eq. (3)):
| (3) |
where Em is the maximal possible system response, and Basal is the response in the absence of agonist. KB is the equilibrium dissociation constant of allosteric ligand, and EC50 is the concentration of orthosteric agonist required to achieve half maximal response. [A] and [B] denote concentrations of orthosteric and allosteric ligands, respectively. α and β denote allosteric effects on orthosteric ligand binding affinity and efficacy, respectively, and τB denotes the efficacy of allosteric ligand. This model assumes that ACh is a full agonist at the receptor in both the absence/presence of modulator and/or there is no efficacy modulation (i.e., β = 1). As shown in the Results, both of these assumptions were met depending on the experimental protocol, and thus the β parameter was constrained to 1.
All affinity, efficacy and cooperativity values were estimated as logarithms [36], and where appropriate, were compared using unpaired Student’s t-test or one-way analysis of variance (ANOVA) with Dunnett’s multiple comparison test. A p value < 0.05 was considered statistically significant.
3. Results
3.1. Novel PAMs behave according to a two-state model of allostery at the M1 mAChR
Equilibrium [3H]NMS binding interaction studies between ACh and increasing concentrations of BQCA, PF-06767832, VU6004256 or MIPS1780 were performed at the M1 mAChR to determine the affinity (KB) of each PAM and their binding cooperativity (α) with the endogenous agonist ACh or the radiolabelled antagonist [3H]NMS, using an allosteric ternary complex model (Eq. (1)). As shown in Fig. 2, all PAMs displayed high negative cooperativity with the radioligand, but strong positive cooperativity with ACh (Table 1). Consistent with the results of previous studies [21], [22], [26], PF-06767832, VU6004256 and MIPS1780 have higher affinities at the M1 mAChR compared to BQCA (Table 1). However, the affinity of BQCA in our study is higher than previously reported values [18], [27], [31], [37] for this compound. This could be due to different experimental conditions including radiolabelled antagonist used, membrane vs. whole cell binding-based assays, incubation time and temperature.
Fig. 2.
BQCA, PF-06767832, VU6004256 or MIPS1780 display high positive cooperativity with the endogenous agonist ACh and negative cooperativity with the antagonist [3H]NMS at the M1 mAChR. Inhibition of [3H]NMS (0.3 nM) binding by ACh in the presence of increasing concentrations of BQCA (A), PF-06767832 (B), VU6004256 (C) or MIPS1780 (D). Data points represent the mean + S.E.M. of four experiments performed in duplicate. The curves were generated by fitting the data to an allosteric ternary complex model (Eq. (1)). Binding parameter estimates from these experiments are listed in Table 1.
Table 1.
Binding parameters for the interaction between ACh and M1 PAMs, BQCA, PF-06767832, VU6004256 or MIPS1780, at the WT and mutant M1 mAChRs. Values represent the mean ± S.E.M. of four experiments performed in duplicate, and were estimated from binding interaction studies using an allosteric ternary complex model (Eq. (1)). The pKA of [3H]NMS was constrained to the values listed in Table 3. ND, not determined.
| PAMs | pKBa | Logα (α)NMSb | Logα (α)AChc | |
|---|---|---|---|---|
| BQCA | WT | 5.78 ± 0.05 | −1.07 ± 0.10 (0.06) |
1.32 ± 0.11 (13.5) |
| F772.56I | 5.82 ± 0.06 | −0.90 ± 0.07 (0.08) |
1.31 ± 0.12 (13.5) |
|
| Y822.61A | 5.76 ± 0.06 | −0.84 ± 0.06 (0.11) |
1.58 ± 0.11 (30.2) |
|
| Y179ECL2A | ND | ND | ND | |
| W4007.35A | ND | ND | ND | |
| PF-06767832 | WT | 6.51 ± 0.03 | −1.05 ± 0.06 (0.06) |
1.40 ± 0.08 (16.2) |
| F772.56I | 6.38 ± 0.07 | −1.02 ± 0.12 (0.06) |
1.83 ± 0.14* (44.7) |
|
| Y822.61A | 5.61 ± 0.12* | −0.16 ± 0.02 (0.62) |
1.53 ± 0.11 (30.9) |
|
| Y179ECL2A | ND | ND | ND | |
| W4007.35A | ND | ND | ND | |
| VU6004256 | WT | 6.32 ± 0.03 | = −2 | 1.40 ± 0.09 (15.5) |
| F772.56I | 6.38 ± 0.03 | = −2 | 1.45 ± 0.10 (14.1) |
|
| Y822.61A | 5.65 ± 0.19* | −0.09 ± 0.03 (0.76) |
1.08 ± 0.13 (11.2) |
|
| Y179ECL2A | ND | ND | ND | |
| W4007.35A | ND | ND | ND | |
| MIPS1780 | WT | 6.18 ± 0.03 | = −2 | 1.60 ± 0.08 (25.7) |
| F772.56I | 6.18 ± 0.03 | = −2 | 1.60 ± 0.09 (25.7) |
|
| Y822.61A | 5.00 ± 0.24* | −0.16 ± 0.06 (0.69) |
1.66 ± 0.19 (45.7) |
|
| Y179ECL2A | ND | ND | ND | |
| W4007.35A | ND | ND | ND | |
Negative logarithm of the allosteric modulator equilibrium dissociation constant.
Logarithm of binding cooperativity between [3H]NMS and each modulator. Where determined as the preferred model by F-test, logαNMS was constrained to −2, consistent with high negative cooperativity between the two ligands.
Logarithm of binding cooperativity between ACh and each modulator.
Significantly different compared to WT, p < 0.05, one-way ANOVA with Dunnett’s post-hoc test.
3.2. Novel PAMs modulate ACh affinity at the M1 mAChR
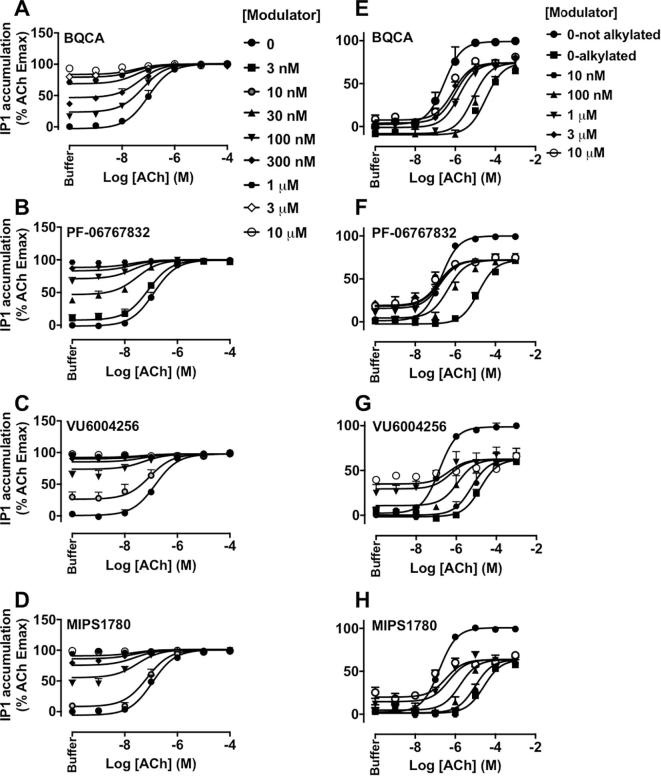
The effects of each modulator on ACh-stimulated IP1 accumulation were then investigated. BQCA, PF-06767832, VU6004256 or MIPS1780 robustly enhanced the response to ACh, as indicated by a leftward shift in the ACh concentration response curve, while also stimulating IP1 accumulation in their own right (Fig. 3A–D), indicating that they each act as M1 mAChR PAM-agonists in our cell line. The functional cooperativity values (αβ) for PF-06767832, VU6004256 or MIPS1780 with ACh were higher than that of BQCA (Table 2). Because ACh was a full agonist in the absence or presence of each PAM, it was unclear whether the allosteric enhancement of ACh potency was due to effects on orthosteric agonist affinity, intrinsic efficacy, or both. Thus, to differentiate these possibilities, IP1 interaction studies between ACh and each modulator were also performed under alkylation conditions with PBZ to reduce levels of receptor reserve (Fig. 3E–H). Concentration-response curves to ACh were first generated under pre-treatment of CHO-hM1 cells with vehicle or increasing concentrations of PBZ for 30 min (followed by extensive washout) to determine the concentration of PBZ required to reduce receptor availability by approx. 50% for ACh (data not shown). Pre-treatment with 10 µM PBZ reduced the potency and maximal response to ACh in IP1 accumulation assays to the level of a partial agonist. This provided a substantial window to unmask any additional effects of the PAMs on ACh maximal response. However, as noted (Fig. 3E–H), BQCA, PF-06767832, VU6004256 or MIPS1780 increased the potency of ACh with no effect on the maximal agonist response, and with αβ values similar to those obtained without PBZ pre-treatment (Table 2). Collectively, these findings indicate that the PAMs exert their allosteric modulatory effects on agonist affinity rather than efficacy (i.e., β = 1).
Fig. 3.
BQCA, PF-06767832, VU6004256 or MIPS1780, modulate the affinity but not efficacy of ACh at the M1 mAChR. Modulation of ACh-stimulated IP1 accumulation by BQCA (A, E), PF-06767832 (B, F), VU6004256 (C, G) or MIPS1780 (D, H) without PBZ (left panels) or with PBZ (10 µM) pre-treatment (right panels) in CHO-hM1 cells. Data points represent the mean + S.E.M. of at least four experiments performed in duplicate. The vehicle-treated curves for ACh under non-alkylation conditions in panels E – H are from a global fit to a three-parameter logistic equation, whereas all other curves in the same panels (under alkylated conditions), as well as the curves in panels A–D, were globally fitted to an operational model of allosterism (Eq. (2)). Functional cooperativity (logαβ) estimates from these experiments are listed in Table 2.
Table 2.
Functional cooperativity (logαβ) estimates for the interactions between ACh and M1 PAMs in IP1 accumulation assays with or without pre-treatment with PBZ in CHO-hM1 cells. The values were estimated by fitting the data to an operational model of allosterism (Eq. (3)), and represent the mean ± S.E.M. of at least four experiments performed in duplicate. The pKB of each modulator was constrained to the values listed in Table 1. The β parameter was constrained to 1 to indicate lack of efficacy modulation. Logα β values were not significantly different between non-alkylated and alkylated experiments (p > 0.05, unpaired Student’s t-test, degree of freedom 6).
| PAM | Logαβ (αβ) |
|
|---|---|---|
| Non-alkylated | Alkylated with PBZ | |
| BQCA | 1.34 ± 0.10 (22) | 1.79 ± 0.11 (62) |
| PF-06767832 | 1.92 ± 0.11 (83) | 2.16 ± 0.11 (144) |
| VU6004256 | 1.78 ± 0.18 (60) | 1.96 ± 0.22 (91) |
| MIPS1780 | 1.77 ± 0.14 (59) | 2.07 ± 0.13 (117) |
3.3. Effects of amino acid substitutions on receptor expression and affinity of orthosteric ligands at the M1 mAChR
The similar mechanism of action observed in aforementioned experiments for the M1 PAMs suggest that PF-06767832, VU6004256 and MIPS1780 may bind to a similar binding site as that occupied by BQCA. To further investigate this, and to identify key amino acid residues that govern the binding affinity, efficacy and cooperativity of the novel M1 PAMs with ACh, we adopted a structure–function approach. Five key amino acid residues from distinct locations at the M1 mAChR, F772.56, Y822.61 in TM2, Y179 in ECL2, Y3816.51 in TM6 and W4007.35 in TM7 (residues are numbered using the Ballesteros-Weinstein numbering system [38]), previously reported to be involved in the binding of orthosteric, allosteric or bitopic ligands at the M1 mAChR, were selected [17], [27], [32], [39], [40].
Whole cell [3H]NMS saturation binding assays were first performed to determine the equilibrium dissociation constant of the radiolabelled antagonist (KD) and the cell surface receptor expression (Bmax) of the mutant M1 mAChRs relative to WT (302549 ± 32197 sites/cell). No [3H]NMS binding was observed at Y3816.51A (data not shown), therefore, together with the lack of detectable ACh response in functional assays, this mutation was excluded from subsequent experiments. The affinity of [3H]NMS was unchanged at F772.56I, Y822.61A, Y179ECL2A or W4007.35A, however, all the mutations caused a significant reduction in the levels of receptor expression (Table 3). Flow cytometry of antibody binding to the c-myc epitope was also performed to detect immunolabeled cell surface-expressed receptors. The estimated normalized values from saturation binding assays agreed well with the normalized values obtained from flow cytometry experiments (Table 3). The affinity of ACh (KI) was estimated from [3H]NMS inhibition binding assays using a one-site binding model, and was unaltered at F772.56I or Y179ECL2A, mutant receptors, whereas it was significantly reduced at Y822.61A or W4007.35A mutant receptors compared with the WT value (Table 3), consistent with our previous observations [31], [32].
Table 3.
Levels of expression of the mutant M1 mAChRs relative to WT in whole cell saturation binding assays and flow cytometric analysis, and equilibrium inhibition binding parameters for [3H]NMS and ACh at the WT and mutant M1 mAChRs. Values represent the mean ± S.E.M. of four experiments performed in duplicate.
| Receptor expressiona |
pKAb | pKIc | ||
|---|---|---|---|---|
| [3H]NMS binding | Flow cytometry | |||
| WT | 9.63 ± 0.14 | 5.38 ± 0.03 | ||
| F772.56I | 69.2 ± 7.9* | 62.0 ± 5.2* | 9.65 ± 0.17 | 5.41 ± 0.02 |
| Y822.61A | 40.0 ± 7.6* | 38.9 ± 2.6* | 9.28 ± 0.08 | 5.03 ± 0.05* |
| Y179ECL2A | 57.8 ± 11* | 57.4 ± 6.0* | 9.57 ± 0.18 | 5.23 ± 0.04 |
| W4007.35A | 22.1 ± 5.6* | 27.0 ± 1.9* | 9.80 ± 0.17 | 4.85 ± 0.05* |
Values represent the percentage of expression relative to the WT receptor expression. Normalised data were not significantly different between estimates of cell surface expression from saturation binding versus flow cytometry of fluorescent antibody labelling of epitope tagged receptors. p > 0.05, unpaired Student’s t-test, degree of freedom 6.
Negative logarithm of the [3H]NMS equilibrium dissociation constant.
Negative logarithm of ACh equilibrium dissociation constant.
Significantly different compared to WT, p < 0.05, one-way ANOVA with Dunnett’s post-hoc test.
3.4. Effects of amino acid substitutions on the binding of PAMs at the M1 mAChR
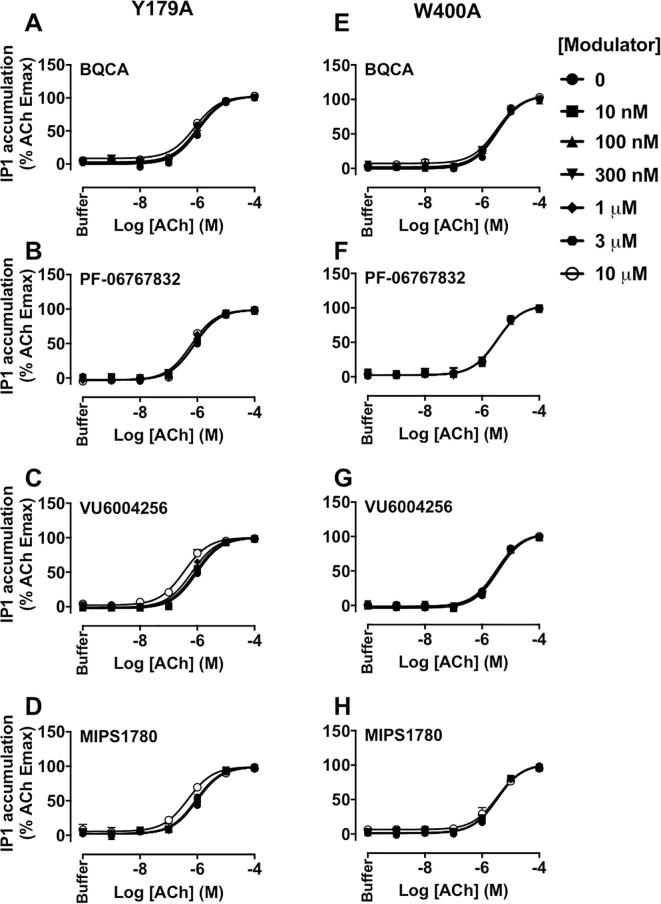
Equilibrium binding interaction studies were then performed to determine the effects of mutations on the affinity and binding cooperativity of M1 PAMs with ACh. At the Y179ECL2A or W4007.35A mutants, the modulatory effects of BQCA, PF-06767832, VU6004256 or MIPS1780 on ACh inhibition of [3H]NMS binding were completely abolished, and hence, their affinity and their binding cooperativity with ACh could not be estimated (Table 1 and Fig. 4), indicating a vital role of these two residues that is common to the actions of all PAM chemotypes tested.
Fig. 4.
The binding of M1 PAMs, BQCA, PF-06767832, VU6004256 or MIPS1780 is abolished at the Y179ECL2A or W4007.35A M1 mAChRs. Inhibition of [3H]NMS (0.3 nM) binding by ACh in the presence of increasing concentrations of BQCA (A, E), PF-06767832 (B, F), VU6004256 (C, G) or MIPS1780 (D, H) at the Y179ECL2A (left panels) or W4007.35A (right panels) mutations. Data points represent the mean + S.E.M. of four experiments performed in duplicate. The curves were generated by fitting the data to an allosteric ternary complex model (Eq. (1)). Binding parameter estimates from these experiments are listed in Table 1.
F772.56 was previously reported to be important for the binding and agonist activity of bitopic ligands i.e., extended hybrid molecules that concomitantly engage both orthosteric and allosteric sites [32], [39], [40]. As shown in Table 1 and Fig. 5A–D, the F772.56I mutation did not alter the affinity of the PAMs when compared with WT. The binding cooperativity between ACh and the PAMs was also unchanged, with the exception of PF-06767832, which displayed a significant, albeit small, increase in cooperativity with ACh at this mutation (Table 1). Collectively, these data suggest that a bitopic mechanism of action is unlikely for these compounds although the residue at position 2.56 may play some role in the transmission of cooperativity depending on the nature of the interacting ligands. Interestingly, at Y822.61A, the affinities of PF-06767832, VU6004256 or MIPS1780, but not BQCA, were significantly reduced (Fig. 5E–H), suggesting that the PAMs may be adopting slightly different binding poses within a shared pocket. Furthermore, while the binding cooperativity with [3H]NMS of PF-06767832, VU6004256 or MIPS1780, but not BQCA, was abolished at Y822.61A, the binding cooperativity values between ACh and the PAMs were not different from the WT values (Table 1).
Fig. 5.
Effects of F772.56I and Y822.61A mutations on the binding affinity and cooperativity of M1 PAMs, BQCA, PF-06767832, VU6004256 or MIPS1780, with ACh at the M1 mAChR. Inhibition of [3H]NMS binding by ACh in the presence of increasing concentrations of BQCA (A, E), PF-06767832 (B, F), VU6004256 (C, G) or MIPS1780 (D, H) at the F772.56I (left panels) or Y822.61A (right panels) mutations. Assays were performed using KA concentrations of [3H]NMS (0.3 nM and 0.6 nM for F772.56I and Y822.61A, respectively). Data points represent the mean + S.E.M. of four experiments performed in duplicate. The curves were generated by fitting the data to an allosteric ternary complex model (Eq. (1)). Binding parameter estimates from these experiments are listed in Table 1.
3.5. Effects of amino acid substitutions on signaling properties of PAMs at the M1 mAChR
We next determined the effects of selected mutations on the potency (pEC50) and efficacy (τA) of ACh at each mutant receptor in parallel to the WT M1 mAChR in IP1 accumulation assays. To account for the effect of varying receptor expression levels of the different constructs on the efficacy of ACh, the estimated τA values were corrected for receptor expression relative to the WT M1 mAChR Bmax value. As shown in Table 4, while all mutations reduced ACh potency, when corrected for expression relative to WT, the estimated operational efficacy of the cognate agonist was significantly increased at the Y822.61A and the W4007.35A mutants, indicating an important role of these residues in the transmission of signaling efficacy. In contrast, efficacy was unchanged at the F772.56I or Y179ECL2A mutant receptors.
Table 4.
Potency (pEC50) and efficacy (τA) of ACh in IP1 accumulation assays at the WT and mutant M1 mAChRs. Values represent the mean ± S.E.M. of at least four experiments performed in duplicate. Potency values were estimated using a three-parameter logistic equation, and efficacy values were quantified according to an operational model of agonism (Eq. (2)). The pKA of ACh at each mutant M1 mAChR was constrained to the corresponding binding affinity (pKI), listed in Table 3.
| pEC50a | LogτA (τA)b | |
|---|---|---|
| WT | 6.82 ± 0.03 | 1.47 ± 0.02 (30) |
| F772.56I | 6.52 ± 0.06* | 1.67 ± 0.03 (47) |
| Y822.61A | 5.72 ± 0.04* | 2.39 ± 0.08* (245) |
| Y179ECL2A | 5.90 ± 0.03* | 1.42 ± 0.06 (26) |
| W4007.35A | 5.43 ± 0.04* | 3.78 ± 0.17* (6025) |
Negative logarithm of the agonist concentration required to produce half the maximal response, estimated using a three-parameter logistic equation.
Logarithm of the functional efficacy of ACh, estimated via fitting to an operational model of agonism (Eq. (1)) and then corrected for receptor expression levels relative to WT.
Significantly different compared to WT, p < 0.05, one-way ANOVA with Dunnett’s post-hoc test.
Finally, the effects of mutations on the efficacy (τB) of the PAMs and their functional cooperativity with ACh were investigated in IP1 interaction experiments. The estimated τB values were corrected for receptor expression of mutant receptors relative to the WT M1 mAChR Bmax value. At the WT M1 mAChR, BQCA, PF-06767832, VU6004256 or MIPS1780 behaved as PAM-agonists (Table 5 and Fig. 3A–D). The Y179ECL2A or W4007.35A mutations resulted in a loss of ACh potentiation by all PAMs tested (Fig. 6), in agreement with the binding experiments, again highlighting a common and critical role for these residues irrespective of the PAM scaffold. The functional efficacy of BQCA, PF-06767832 or MIPS1780 was increased, whereas the efficacy of VU6004256 was unchanged at the F772.56I mutation. However, their cooperativities with ACh were not different at this construct compared to the WT M1 mAChR (Table 5 and Fig. 7A–D). As with the interpretation of the binding interaction studies (Table 1), the modest effects of this mutation argue against a bitopic mechanism of action for the novel chemotypes. At the Y822.61A construct, the functional cooperativity values of PAMs with ACh were similar to the WT values. Interestingly, this mutation had differential effects on the efficacy of the PAMs, causing a significant decrease in VU6004256 efficacy while increasing the efficacy of BQCA or MIPS1780, and not altering PF-06767832 efficacy relative to WT (Table 5 and Fig. 7E–H).
Table 5.
Operational model parameters for IP1 interaction assays between ACh and M1 PAMs, BQCA, PF-06767832, VU6004256 or MIPS1780, at the WT and mutant M1 mAChRs. Values represent the mean ± S.E.M. of at least four experiments performed in duplicate, and were estimated using an operational model of allosterism (Eq. (2)). The pKB of each modulator was constrained to the corresponding binding affinity, listed in Table 1. The β parameter was constrained to 1 to indicate lack of efficacy modulation. ND, not determined (no ACh modulation).
| PAMs | Logαβ (αβ)a | LogτB (τB)b | |
|---|---|---|---|
| BQCA | WT | 1.34 ± 0.10 (22) | 0.78 ± 0.03 (6) |
| F772.56I | 1.40 ± 0.15 (25) | 1.03 ± 0.06* (11) | |
| Y822.61A | 1.66 ± 0.11 (46) | 1.30 ± 0.07* (20) | |
| Y179ECL2A | ND | ND | |
| W4007.35A | ND | ND | |
| PF-06767832 | WT | 1.92 ± 0.11 (83) | 1.01 ± 0.03 (10) |
| F772.56I | 2.05 ± 0.13 (112) | 1.51 ± 0.07* (32) | |
| Y822.61A | 1.66 ± 0.12 (46) | 0.95 ± 0.12 (9) | |
| Y179ECL2A | ND | ND | |
| W4007.35A | ND | ND | |
| VU6004256 | WT | 1.78 ± 0.18 (60) | 1.24 ± 0.05 (17) |
| F772.56I | 1.71 ± 0.15 (51) | 1.43 ± 0.06 (27) | |
| Y822.61A | 1.29 ± 0.10 (19) | 0.50 ± 0.10* (3) | |
| Y179ECL2A | ND | ND | |
| W4007.35A | ND | ND | |
| MIPS1780 | WT | 1.77 ± 0.14 (59) | 1.00 ± 0.05 (10) |
| F772.56I | 1.80 ± 0.15 (63) | 1.52 ± 0.06* (33) | |
| Y822.61A | 1.96 ± 0.14 (91) | 1.85 ± 0.12* (71) | |
| Y179ECL2A | ND | ND | |
| W4007.35A | ND | ND | |
Logarithm of functional cooperativity between ACh and each modulator.
Logarithm of functional efficacy of the modulator corrected for receptor expression levels relative to WT.
Significantly different compared to WT, p < 0.05, one-way ANOVA with Dunnett’s post-hoc test.
Fig. 6.
The modulation of ACh-stimulated IP1 accumulation by M1 PAMs, BQCA, PF-06767832, VU6004256 or MIPS1780 is abolished at the Y179ECL2A or W4007.35A M1 mAChRs. IP1 interaction assays between ACh and increasing concentrations of BQCA (A, E), PF-06767832 (B, F), VU6004256 (C, G) or MIPS1780 (D, H) at the Y179ECL2A (left panels) or W4007.35A (right panels) mutations. Data points represent the mean + S.E.M. of four experiments performed in duplicate. The curves were generated by fitting the data to an operational model of allosterism (Eq. (3)). Operational parameter estimates from these experiments are listed in Table 5.
Fig. 7.
Effects of F772.56I and Y822.61A mutations on the efficacy and functional cooperativity of M1 PAMs, BQCA, PF-06767832, VU6004256 or MIPS1780, with ACh at the M1 mAChR. IP1 interaction assays between ACh and increasing concentrations of BQCA (A, E), PF-06767832 (B, F), VU6004256 (C, G) or MIPS1780 (D, H) at the F772.56I (left panels) or Y822.61A (right panels) mutations. Data points represent the mean + S.E.M. of four experiments performed in duplicate. The curves were generated by fitting the data to an operational model of allosterism (Eq. (3)). Operational parameter estimates from these experiments are listed in Table 5.
4. Discussion
Subtype selective allosteric modulation of M1 mAChRs has been increasingly explored as a potential approach for the treatment of AD and other cognitive deficit disorders [7], [41]. The discovery of BQCA, a highly selective M1 mAChR PAM [17] led to development of newer classes of M1 PAMs with different pharmacological and pharmacokinetic properties, including PF-06767832 [21], VU6004256 [24] and MIPS1780 [26]. Despite the distinct chemical scaffolds of these PAMs, the current study provides strong support for a common mode of binding to that seen with BQCA, and a similar mode of action whereby cooperativity arises predominantly from modulation of ACh affinity. This has implications for the discovery of novel M1 mAChR-targeting PAMs with improved drug-like properties to facilitate translational studies.
There is a substantial body of pharmacology data that supports the existence of at least two allosteric sites on the mAChRs. By far the best studied has been the so-called “common” allosteric binding site, located in an extracellular vestibule above the orthosteric pocket and predominantly comprised of residues in ECL2, TM2 and TM7 [42]. This region is recognized by prototypical and well-studied negative allosteric modulators (NAMs), such as gallamine and C7/3-phth [43], but also by PAMs such as BQCA (M1) [27] and LY2033298 (M2 and M4; [23], [44], [45]). The existence of this vestibular site has been more recently validated directly through structural and computational studies of the M1–M4 mAChRs [42], [46], [47], [48], [49]. In contrast, the location of the “second” allosteric site that binds indolocarbazole and benzimidazole modulators is currently unknown, although one study has suggested a potential intracellular pocket [28], [50], [51].
To elaborate our understanding of novel M1 mAChR PAMs, we adopted two complementary approaches to characterize the nature of the allosteric action and likely binding mode of these compounds. The first approach characterised affinity, relative efficacy and cooperativity with orthosteric ligands using a combination of cell-based assays of M1 mAChR-mediated IP1 accumulation and radioligand binding. Moreover, we extended this analysis to account for the contribution of receptor reserve in the observed pharmacology. We have previously shown that the irreversible receptor alkylating agent, PBZ, precludes the binding of ACh directly, or indirectly prevents the binding of PAMs to the allosteric site via high negative cooperativity [52]. In either instance, the net effect is a reduction in receptor reserve such that the functional affinity and relative efficacy of each agent as a direct activator of the M1 mAChR can be determined. Moreover, interaction studies between ACh and each PAM under these same conditions, where ACh would be acting as essentially a partial agonist, provided a powerful functional approach to directly determine whether the PAMs exert effects on agonist affinity, intrinsic efficacy, or both as part of their allosteric mechanism of action. Accordingly, three key outcomes were obtained from the functional and binding experiments. First, each of the PAMs has the potential to display intrinsic allosteric agonism (i.e., as PAM-agonists), second, allosteric effects were mediated primarily through the enhancement of agonist binding affinity and, third, all modulators exhibited high positive cooperativity with ACh but high negative cooperativity with the antagonist, [3H]NMS. These observations mirror those previously noted with BQCA and the related BQZ-12 (3-((1S,2S)-2-hydroxycyclohexyl)-6-((6-(1-methyl-1H-pyrazol-4-yl)pyridin-3-yl)methyl)benzo[h]quinazolin-4(3H)-one)), [18], [31], [52], [53]. Collectively, our findings suggest that PAMs acting via the “common” allosteric pocket are likely to mimic the two-state mode of allosteric effect described for BQCA [19], [54]. The work also supports the importance of understanding relative intrinsic agonism of modulators, as the observed degree of PAM-agonism can be highly system dependent [54], and this property could contribute to undesirable over-activation of a target GPCR by novel classes of PAMs.
In the second series of studies, we probed for binding site location via mutational analysis of key residues previously implicated in the actions of prototypical “common” site modulators or bitopic ligands. Our previous mutagenesis and molecular modelling predicted important roles for Y179ECL2 and W4007.35 in the stability of BQCA binding via hydrophobic π-π interactions [27]. We had also confirmed the importance of these residues for BQZ-12 binding, indicating a shared binding site for both ligands [31]. In the current study, we also found that Y179ECL2 and W4007.35 are absolutely critical for the binding and activity of PF-06767832, VU6004256 and MIPS1780, consistent with the binding of these PAMs to a similar site as that of BQCA or BQZ-12. Further support for these residues, as well as for Y822.61, was noted in a prior homology model of the M1 mAChR bound to a structurally related compound to PF-06767832 (compound 11 in [21]). We had also previously found that Y822.61 was involved in transmission of positive cooperativity and/or modulator binding affinity in studies focusing on BQCA and BQZ-12 [27], [31]. Our current findings with PF-06767832, VU6004256 and MIPS1780 are also in agreement with a broad role for this residue either in modulator binding affinity and/or transmission of cooperativity. The reduced binding cooperativity of these modulators with [3H]NMS but not with ACh at Y822.61A indicates the key role of this residue in the transmission of cooperativity specifically with the antagonist and not the agonist. The differences in which pharmacological behaviours are modified (i.e., affinity, cooperativity or efficacy), likely relate to differences in the poses each PAM adopts within the common allosteric pocket. Our data suggest that Y822.61 may form key π-π interactions with the thiazole (in PF-06767832) or pyrazole (in VU6004256 and MIPS1780) pendent group, whereas Y179ECL2 and W4007.35 interact with the core of these compounds as well as BQCA, which lacks an extended pendent group. However, this needs to be confirmed in future studies by additional structure-activity relationships and molecular modelling. In contrast to the aforementioned key residues, only modest effects of the F772.56I mutation on the M1 PAMs were noted. Previous studies had highlighted a vital role for this TM2 residue in the action of extended, bitopic, ligands [32], [39], [40]. The lack of observed effect in the current study argues against such a mode of interaction for the novel chemotypes investigated herein. Ideally, an additional test of our hypothesis for a common binding site for different PAM chemotypes at the M1 mAChR would be a direct competition assay between each PAM and a neutral, or near neutral, allosteric ligand binding to the same site with high affinity. However, to our knowledge, no such allosteric ligand has thus far been identified for the M1 mAChR.
Collectively, our findings support a model whereby PF-06767832, VU6004256 and MIPS1780 share a similar binding site with BQCA, located within the “common” extracellular vestibule region. However, additional studies are required to better understand the structural basis of selectivity of these PAM scaffolds. For instance, W7.35 is conserved across the five mAChR subtypes, Y179ECL2 is present in both M1 and M2 mAChRs, and Y2.61 is conserved across all but the M3 subtype indicating complexity beyond sequence conservation in the subtype selective actions of the modulators. One intriguing possibility is that the exquisite degrees of selectivity achieved by allosteric modulators of GPCRs arise from a combination of both allosteric pocket residues and energetically preferred dynamic networks that underlie transmission of cooperativity between orthosteric and allosteric sites [20], [46], [55], [56]. In addition, despite the commonalities in mechanism proposed for the different classes of M1 PAMs described in our study, it should be noted that different modes of GPCR allosteric modulation occur, including differential effects on efficacy in addition to affinity, as well as the potential for pathway-biased modulation [23], [45], [52], [57], highlighting additional mechanistic questions for the field. Our current study was limited to analysis of the relative effects on a single signaling endpoint and additional work is required to understand the extent to which biased modulation may or may not occur. Nonetheless, and as evidenced by very recent advances in structure-based approaches to discovering new allosteric modulators [42], [58], [59], novel insights into the structural basis of M1 allosteric modulator binding and activity can facilitate the rational design of new PAMs as drug-like candidates.
5. Conflict of interest
The authors declare no conflict of interest.
Acknowledgments
This work was supported by National Health and Medical Research Council (NHMRC) Program Grant (APP1055134) and Wellcome Trust Collaborative Research Award (201529/Z/16/Z). AC is a Senior Principal, and PMS a Principal, Research Fellow of the National Health and Medical Research Council of Australia. CV is supported by a Future Fellowship from the Australian Research Council.
References
- 1.Fredriksson R., Lagerström M.C., Lundin L.-G., Schiöth H.B. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 2003;63(6):1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 2.Bymaster F.P., McKinzie D.L., Felder C.C., Wess J. Use of M1–M5 muscarinic receptor knockout mice as novel tools to delineate the physiological roles of the muscarinic cholinergic system. Neurochem. Res. 2003;28(3):437–442. doi: 10.1023/a:1022844517200. [DOI] [PubMed] [Google Scholar]
- 3.Lanzafame A., Christopoulos A., Mitchelson F. Cellular signaling mechanisms for muscarinic acetylcholine receptors. Recept. Channels. 2003;9(4):241–260. [PubMed] [Google Scholar]
- 4.Levey A.I. Immunological localization of m1–m5 muscarinic acetylcholine receptors in peripheral tissues and brain. Life Sci. 1993;52(5–6):441–448. doi: 10.1016/0024-3205(93)90300-r. [DOI] [PubMed] [Google Scholar]
- 5.Wess J. Muscarinic acetylcholine receptor knockout mice: novel phenotypes and clinical implications. Annu. Rev. Pharmacol. Toxicol. 2004;44:423–450. doi: 10.1146/annurev.pharmtox.44.101802.121622. [DOI] [PubMed] [Google Scholar]
- 6.Foster D.J., Choi D.L., Conn P.J., Rook J.M. Activation of M1 and M4 muscarinic receptors as potential treatments for Alzheimer’s disease and schizophrenia. Neuropsychiatr. Dis. Treat. 2014;10:183–191. doi: 10.2147/NDT.S55104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melancon B.J., Tarr J.C., Panarese J.D., Wood M.R., Lindsley C.W. Allosteric modulation of the M 1 muscarinic acetylcholine receptor: improving cognition and a potential treatment for schizophrenia and Alzheimer's disease. Drug Discov. Today. 2013;18(23):1185–1199. doi: 10.1016/j.drudis.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kruse A.C., Kobilka B.K., Gautam D., Sexton P.M., Christopoulos A., Wess J. Muscarinic acetylcholine receptors: novel opportunities for drug development. Nat. Rev. Drug Discov. 2014;13(7):549–560. doi: 10.1038/nrd4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartus R.T., Dean R.R., Beer B., Lippa A.S. The cholinergic hypothesis of geriatric memory dysfunction. Science. 1982;217(4558):408–414. doi: 10.1126/science.7046051. [DOI] [PubMed] [Google Scholar]
- 10.Crews L., Masliah E. Molecular mechanisms of neurodegeneration in Alzheimer’s disease. Hum. Mol. Genet. 2010;19(R1):R12–R20. doi: 10.1093/hmg/ddq160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolfe M.S. Therapeutic strategies for Alzheimer’s disease. Nat. Rev. Drug Discov. 2002;1(11):859. doi: 10.1038/nrd938. [DOI] [PubMed] [Google Scholar]
- 12.Giacobini E. Cholinesterase inhibitors: new roles and therapeutic alternatives. Pharmacol. Res. 2004;50(4):433–440. doi: 10.1016/j.phrs.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 13.Bodick N.C., Offen W.W., Levey A.I., Cutler N.R., Gauthier S.G., Satlin A. Effects of xanomeline, a selective muscarinic receptor agonist, on cognitive function and behavioral symptoms in Alzheimer disease. Arch. Neurol. 1997;54(4):465–473. doi: 10.1001/archneur.1997.00550160091022. [DOI] [PubMed] [Google Scholar]
- 14.Mirza N.R., Peters D., Sparks R.G. Xanomeline and the antipsychotic potential of muscarinic receptor subtype selective agonists. CNS Drug Rev. 2003;9(2):159–186. doi: 10.1111/j.1527-3458.2003.tb00247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conn P.J., Christopoulos A., Lindsley C.W. Allosteric modulators of GPCRs: a novel approach for the treatment of CNS disorders. Nat. Rev. Drug Discov. 2009;8(1):41–54. doi: 10.1038/nrd2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christopoulos A. Allosteric binding sites on cell-surface receptors: novel targets for drug discovery. Nat. Rev. Drug Discov. 2002;1(3):198–210. doi: 10.1038/nrd746. [DOI] [PubMed] [Google Scholar]
- 17.Ma L., Seager M.A., Wittmann M., Jacobson M., Bickel D., Burno M. Selective activation of the M1 muscarinic acetylcholine receptor achieved by allosteric potentiation. Proc. Natl. Acad. Sci. U.S.A. 2009;106(37):15950–15955. doi: 10.1073/pnas.0900903106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Canals M., Lane J.R., Wen A., Scammells P.J., Sexton P.M., Christopoulos A. A Monod-Wyman-Changeux mechanism can explain G protein-coupled receptor (GPCR) allosteric modulation. J. Biol. Chem. 2012;287(1):650–659. doi: 10.1074/jbc.M111.314278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuduk S.D., Chang R.K., Di Marco C.N., Pitts D.R., Greshock T.J., Ma L. Discovery of a selective allosteric M1 receptor modulator with suitable development properties based on a quinolizidinone carboxylic acid scaffold. J. Med. Chem. 2011;54(13):4773–4780. doi: 10.1021/jm200400m. [DOI] [PubMed] [Google Scholar]
- 20.Christopoulos A. Advances in G protein-coupled receptor allostery: from function to structure. Mol. Pharmacol. 2014;86(5):463–478. doi: 10.1124/mol.114.094342. [DOI] [PubMed] [Google Scholar]
- 21.Davoren J.E., Lee C.-W., Garnsey M., Brodney M.A., Cordes J., Dlugolenski K. Discovery of the potent and selective M1 PAM-agonist N-[(3 R, 4 S)-3-hydroxytetrahydro-2 H-pyran-4-yl]-5-methyl-4-[4-(1, 3-thiazol-4-yl) benzyl] pyridine-2-carboxamide (PF-06767832): Evaluation of efficacy and cholinergic side effects. J. Med. Chem. 2016;59(13):6313–6328. doi: 10.1021/acs.jmedchem.6b00544. [DOI] [PubMed] [Google Scholar]
- 22.Davoren J.E., Garnsey M., Pettersen B., Brodney M.A., Edgerton J.R., Fortin J.P. Design and synthesis of γ-and δ-lactam M1 positive allosteric modulators (PAMs): convulsion and cholinergic toxicity of an M1-selective PAM with weak agonist activity. J. Med. Chem. 2017;60(15):6649–6663. doi: 10.1021/acs.jmedchem.7b00597. [DOI] [PubMed] [Google Scholar]
- 23.Leach K., Loiacono R.E., Felder C.C., McKinzie D.L., Mogg A., Shaw D.B. Molecular mechanisms of action and in vivo validation of an M4 muscarinic acetylcholine receptor allosteric modulator with potential antipsychotic properties. Neuropsychopharmacology. 2010;35(4):855–869. doi: 10.1038/npp.2009.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grannan M.D., Mielnik C.A., Moran S.P., Gould R.W., Ball J., Lu Z. Prefrontal cortex-mediated impairments in a genetic model of NMDA receptor hypofunction are reversed by the novel M1 PAM VU6004256. ACS Chem. Neurosci. 2016;7(12):1706–1716. doi: 10.1021/acschemneuro.6b00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rook J.M., Abe M., Cho H.P., Nance K.D., Luscombe V.B., Adams J.J. Diverse effects on M1 signaling and adverse effect liability within a series of M1 Ago-PAMs. ACS Chem. Neurosci. 2017;8(4):866–883. doi: 10.1021/acschemneuro.6b00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mistry S.N., Jörg M., Lim H., Vinh N.B., Sexton P.M., Capuano B. 4-Phenylpyridin-2-one derivatives: a novel class of positive allosteric modulator of the M1 muscarinic acetylcholine receptor. J. Med. Chem. 2015;59(1):388–409. doi: 10.1021/acs.jmedchem.5b01562. [DOI] [PubMed] [Google Scholar]
- 27.Abdul-Ridha A., López L., Keov P., Thal D.M., Mistry S.N., Sexton P.M. Molecular determinants of allosteric modulation at the M1 muscarinic acetylcholine receptor. J. Biol. Chem. 2014;289(9):6067–6079. doi: 10.1074/jbc.M113.539080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lazareno S., Popham A., Birdsall N.J. Allosteric interactions of staurosporine and other indolocarbazoles with N-[methyl-3H] scopolamine and acetylcholine at muscarinic receptor subtypes: identification of a second allosteric site. Mol. Pharmacol. 2000;58(1):194–207. doi: 10.1124/mol.58.1.194. [DOI] [PubMed] [Google Scholar]
- 29.Birdsall N.J., Lazareno S., Popham A., Saldanha J. Multiple allosteric sites on muscarinic receptors. Life Sci. 2001;68(22):2517–2524. doi: 10.1016/s0024-3205(01)01047-5. [DOI] [PubMed] [Google Scholar]
- 30.Espinoza-Fonseca L.M., Trujillo-Ferrara J.G. The existence of a second allosteric site on the M 1 muscarinic acetylcholine receptor and its implications for drug design. Bioorg. Med. Chem. Lett. 2006;16(5):1217–1220. doi: 10.1016/j.bmcl.2005.11.097. [DOI] [PubMed] [Google Scholar]
- 31.Abdul-Ridha A., Lane J.R., Mistry S.N., López L., Sexton P.M., Scammells P.J. Mechanistic insights into allosteric structure-function relationships at the M1 muscarinic acetylcholine receptor. J. Biol. Chem. 2014;289(48):33701–33711. doi: 10.1074/jbc.M114.604967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keov P., López L., Devine S.M., Valant C., Lane J.R., Scammells P.J. Molecular mechanisms of bitopic ligand engagement with the M1 muscarinic acetylcholine receptor. J. Biol. Chem. 2014;289(34):23817–23837. doi: 10.1074/jbc.M114.582874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Motulsky H., Christopoulos A. Oxford University Press; 2004. Fitting Models to Biological Data Using Linear and Nonlinear Regression: A Practical Guide to Curve Fitting. [Google Scholar]
- 34.Black J., Leff P. Operational models of pharmacological agonism. Proc. R. Soc. Lond. B. Biol. Sci. 1983:141–162. doi: 10.1098/rspb.1983.0093. [DOI] [PubMed] [Google Scholar]
- 35.Aurelio L., Valant C., Flynn B.L., Sexton P.M., Christopoulos A., Scammells P.J. Allosteric modulators of the adenosine A1 receptor: synthesis and pharmacological evaluation of 4-substituted 2-amino-3-benzoylthiophenes. J. Med. Chem. 2009;52(14):4543–4547. doi: 10.1021/jm9002582. [DOI] [PubMed] [Google Scholar]
- 36.Christopoulos A. Assessing the distribution of parameters in models of ligand–receptor interaction: to log or not to log. Trends Pharmacol. Sci. 1998;19(9):351–357. doi: 10.1016/s0165-6147(98)01240-1. [DOI] [PubMed] [Google Scholar]
- 37.Mistry S.N., Lim H., Jörg M., Capuano B., Christopoulos A., Lane J.R. Novel fused arylpyrimidinone based allosteric modulators of the M1 muscarinic acetylcholine receptor. ACS Chem. Neurosci. 2016;7(5):647–661. doi: 10.1021/acschemneuro.6b00018. [DOI] [PubMed] [Google Scholar]
- 38.Ballesteros J.A., Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 1995;25:366–428. [Google Scholar]
- 39.Jacobson M.A., Kreatsoulas C., Pascarella D.M., O'Brien J.A., Sur C. The M1 muscarinic receptor allosteric agonists AC-42 and 1-[1′-(2-methylbenzyl)-1, 4′-bipiperidin-4-yl]-1, 3-dihydro-2H-benzimidazol-2-one bind to a unique site distinct from the acetylcholine orthosteric site. Mol. Pharmacol. 2010;78(4):648–657. doi: 10.1124/mol.110.065771. [DOI] [PubMed] [Google Scholar]
- 40.Avlani V.A., Langmead C.J., Guida E., Wood M.D., Tehan B.G., Herdon H.J. Orthosteric and allosteric modes of interaction of novel selective agonists of the M1 muscarinic acetylcholine receptor. Mol. Pharmacol. 2010;78(1):94–104. doi: 10.1124/mol.110.064345. [DOI] [PubMed] [Google Scholar]
- 41.Conn P.J., Jones C.K., Lindsley C.W. Subtype-selective allosteric modulators of muscarinic receptors for the treatment of CNS disorders. Trends Pharmacol. Sci. 2009;30(3):148–155. doi: 10.1016/j.tips.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dror R.O., Green H.F., Valant C., Borhani D.W., Valcourt J.R., Pan A.C. Structural basis for modulation of a G-protein-coupled receptor by allosteric drugs. Nature. 2013;503(7475):295–299. doi: 10.1038/nature12595. [DOI] [PubMed] [Google Scholar]
- 43.Lanzafame A., Christopoulos A., Mitchelson F. Three allosteric modulators act at a common site, distinct from that of competitive antagonists, at muscarinic acetylcholine M2 receptors. J. Pharmacol. Exp. Ther. 1997;282(1):278–285. [PubMed] [Google Scholar]
- 44.Nawaratne V., Leach K., Felder C.C., Sexton P.M., Christopoulos A. Structural determinants of allosteric agonism and modulation at the M4 muscarinic acetylcholine receptor: Identification of ligand-specific and global activation mechanisms. J. Biol. Chem. 2010;285(25):19012–19021. doi: 10.1074/jbc.M110.125096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valant C., Felder C.C., Sexton P.M., Christopoulos A. Probe dependence in the allosteric modulation of a G protein-coupled receptor: Implications for detection and validation of allosteric ligand effects. Mol. Pharmacol. 2012;81(1):41–52. doi: 10.1124/mol.111.074872. [DOI] [PubMed] [Google Scholar]
- 46.Thal D.M., Sun B., Feng D., Nawaratne V., Leach K., Felder C.C. Crystal structures of the M1 and M4 muscarinic acetylcholine receptors. Nature. 2016;531(7594):335–340. doi: 10.1038/nature17188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kruse A.C., Hu J., Kobilka B.K., Wess J. Muscarinic acetylcholine receptor X-ray structures: potential implications for drug development. Curr. Opin. Pharmacol. 2014;16:24–30. doi: 10.1016/j.coph.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haga K., Kruse A.C., Asada H., Yurugi-Kobayashi T., Shiroishi M., Zhang C. Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature. 2012;482(7386):547–551. doi: 10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kruse A.C., Hu J., Pan A.C., Arlow D.H., Rosenbaum D.M., Rosemond E. Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature. 2012;482(7386):552–556. doi: 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lanzafame A.A., Sexton P.M., Christopoulos A. Interaction studies of multiple binding sites on m4 muscarinic acetylcholine receptors. Mol. Pharmacol. 2006;70(2):736–746. doi: 10.1124/mol.106.024711. [DOI] [PubMed] [Google Scholar]
- 51.Lazareno S., Popham A., Birdsall N. Analogs of WIN 62,577 define a second allosteric site on muscarinic receptors. Mol. Pharmacol. 2002;62(6):1492–1505. doi: 10.1124/mol.62.6.1492. [DOI] [PubMed] [Google Scholar]
- 52.Berizzi A.E., Gentry P.R., Rueda P., Den Hoedt S., Sexton P.M., Langmead C.J. Molecular mechanisms of action of M5 muscarinic acetylcholine receptor allosteric modulators. Mol. Pharmacol. 2016;90(4):427–436. doi: 10.1124/mol.116.104182. [DOI] [PubMed] [Google Scholar]
- 53.Kuduk S.D., Beshore D.C. Novel M1 allosteric ligands: a patent review. Expert Opin. Ther. Pat. 2012;22(12):1385–1398. doi: 10.1517/13543776.2012.731395. [DOI] [PubMed] [Google Scholar]
- 54.Keov P., Sexton P.M., Christopoulos A. Allosteric modulation of G protein-coupled receptors: a pharmacological perspective. Neuropharmacology. 2011;60(1):24–35. doi: 10.1016/j.neuropharm.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 55.Lazareno S., Doležal V., Popham A., Birdsall N. Thiochrome enhances acetylcholine affinity at muscarinic M4 receptors: receptor subtype selectivity via cooperativity rather than affinity. Mol. Pharmacol. 2004;65(1):257–266. doi: 10.1124/mol.65.1.257. [DOI] [PubMed] [Google Scholar]
- 56.Suratman S., Leach K., Sexton P., Felder C., Loiacono R., Christopoulos A. Impact of species variability and ‘probe-dependence’on the detection and in vivo validation of allosteric modulation at the M4 muscarinic acetylcholine receptor. Br. J. Pharmacol. 2011;162(7):1659–1670. doi: 10.1111/j.1476-5381.2010.01184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marlo J.E., Niswender C.M., Days E.L., Bridges T.M., Xiang Y., Rodriguez A.L. Discovery and characterization of novel allosteric potentiators of M1 muscarinic receptors reveals multiple modes of activity. Mol. Pharmacol. 2009;75(3):577–588. doi: 10.1124/mol.108.052886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Korczynska M., Clark M.J., Valant C., Xu J., Von Moo E., Albold S. Structure-based discovery of selective positive allosteric modulators of antagonists for the M2 muscarinic acetylcholine receptor. Proc. Natl. Acad. Sci. U.S.A. 2018;201718037 doi: 10.1073/pnas.1718037115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miao Y., Goldfeld D.A., Von Moo E., Sexton P.M., Christopoulos A., McCammon J.A. Accelerated structure-based design of chemically diverse allosteric modulators of a muscarinic G protein-coupled receptor. Proc. Natl. Acad. Sci. U.S.A. 2016;113(38):E5675–E5684. doi: 10.1073/pnas.1612353113. [DOI] [PMC free article] [PubMed] [Google Scholar]