Abstract
Protein-protein interactions (PPI) were once considered “undruggable”, but clinical successes, driven by advanced methods in drug discovery, have challenged that notion. Here, we review the last three years of literature on PPI inhibitors to understand what is working and why. From the 66 recently reported PPI inhibitors, we found that the average molecular weight was significantly greater than 500 Da, but that this trend was driven, in large part, by the contribution of peptide-based compounds. Despite differences in average molecular weight, we found that compounds based on small molecules or peptides were almost equally likely to be potent inhibitors (KD < 1 μM). Finally, we found PPIs with buried surface area (BSA) less than 2000 Å2 were more likely to be inhibited by small molecules, while PPIs with larger BSA values were typically inhibited by peptides. PPIs with BSA values over 4000 Å2 seemed to create a particular challenge, especially for orthosteric small molecules. Thus, it seems important to choose the inhibitor scaffold based on the properties of the target interactions. Moreover, this survey suggests a (more nuanced) conclusion to the question of whether PPIs are good drug targets; namely, that some PPIs are readily “druggable” given the right choice of scaffold, while others still seem to deserve the “undruggable” moniker.
Introduction
Protein-protein interactions (PPIs) are potential drug targets for a broad range of therapeutic areas, such as oncology [1, 2*, 3, 4*, 5], immune-checkpoints for cancer immunotherapy [6], tropical infectious diseases [7*], neurological disorders [8], heart failure [9] and inflammation and oxidative stress [10]. Interest in these targets is further heightened by the fact that modern proteomics studies have shown that there are an estimated 650,000 PPIs, compared to only ~20,000 protein coding genes [11]. At the same time, gene-editing methods are making it possible to create point mutations within the genomes of mammalian cells [12*], allowing validation of individual PPIs as putative drug targets with unprecedented precision. Finally, because the interfaces are often less conserved than active sites, PPI inhibitors are also commonly thought to have a greater opportunity for being selective [13].
This current enthusiasm is in contrast with the attitude twenty years ago, when PPIs were commonly regarded to as “undruggable”. This conclusion was based, in part, on a growing number of crystal structures that showed protein interfaces with daunting large (1,000–2,000 Å2) and flat interfacial areas, when compared to traditional targets, such as enzyme active sites (~300–500 Å2) [4]. However, even at that time, “undruggable” was an admittedly broad designation, because peptide hormones that act at PPIs had long been approved as clinical drugs [14]. The 1990s also saw the approval of small molecule PPI modulators (sometimes stabilizers instead of inhibitors), such as Tirofiban and various taxanes [15–17]. What were the common features of these early compounds? How were they able to escape the dogma? On commonality amongst the early PPI inhibitors is that they were based on natural products. While this feature likely helped the molecules overcome the challenges of inhibiting PPIs, it also gave them poor oral bioavailability and low cell-permeability. Thus, these successes did not clarify whether PPIs inhibitors could play an important role across the landscape of drug discovery. Fortunately, the number of PPI inhibitors that are either approved or under late-stage clinical investigation has expanded over the years. One milestone was the 2016 approval of the Bcl-2 inhibitor, Venclexta (ABT-199). This compound was derived from a fragment-based screen [18*] (rather than a natural product) and it is orally active for the treatment of chronic lymphocytic leukemia [19*]. Now, the possibilities of targeting a range of PPIs with compounds from different sources seems increasingly likely.
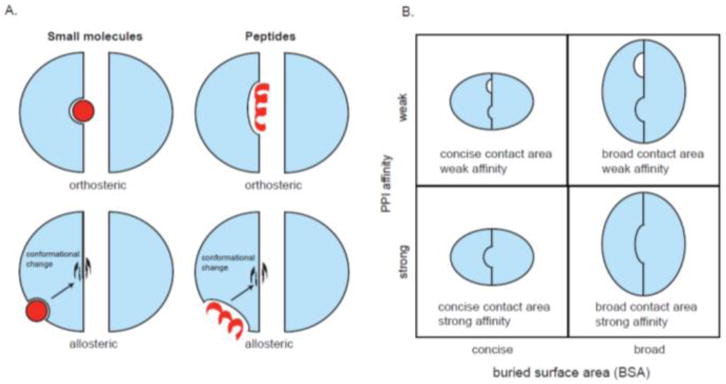
What drove this evolution from “undruggable” to increasingly common? One significant development was greater basic knowledge of PPI structure and energetics. Early observations by Arkin, Wells and others suggested that the interaction energy (ΔGbind) of PPIs was often not evenly distributed across the entire buried surface area (BSA). Rather, mutagenesis showed that there are “hot spots” that confer a disproportionate amount of the ΔGbind [17,20]. By placing molecules at these sites, orthosteric (i.e. competitive) inhibitors with relatively low molecular mass values could be created, even if the PPI itself had a comparatively large BSA (Fig 1A). Another major advance was the realization of the plasticity of many protein interaction surfaces [4]. Indeed, molecules that take advantage of conformational changes are sometimes able to inhibit seemingly intractable PPIs [21,22]. In some cases, these molecules bind at the interface itself and remodel the topology of the contact surface. However, other compounds bind to distal sites to influence the PPI. For example, JG-98 binds with a sub-micromolar affinity to an allosteric site on Hsp70, disrupting an interaction with BAG3 that is >20 Å away. This interaction has a BSA of 4,473 Å2 and it involves two different subdomains [23*], so it is difficult to imagine how an orthosteric inhibitor might be capable of doing that job. Other technological advances have focused on creating inhibitors from non-traditional scaffolds, often inspired by natural products. For example, new methods for creating cell-permeable peptides, such as helical regions that are covalently “stapled” along the backbone to improve their cLogP and proteolytic stability [11,24*,25] or polycationic “tags” to improve passive permeability [26], have seen significant progress in the last decade. Other advances have included semi-synthetic [27] and synthetic methods [28,29] for creating a greater range of biologically active macrocycles [30] and advanced methods for creating peptide-inspired foldamers [31]. Now, an entire zoo of scaffolds - small molecules, peptides and others - are available for targeting PPIs. These approaches are further supplemented by the emergence of fundamentally new techniques, such as proteolysis targeting chimera (PROTAC) molecules, in which the target protein is degraded instead of trying to physically block its interactions [32*]. Given the rise of so many new approaches for inhibiting PPIs, it seems worth performing a retrospective analysis to find out what is working.
Figure 1.
Schematic properties of PPI inhibitors. (A) Small molecules or peptides can inhibit PPIs by binding to the interface (orthosteric) or a distal site (allosteric). Peptides are often derived from natural protein partners and can be either structured (e.g. helical) or disordered (e.g. random coil). (B) Categorization of PPIs based on buried surface area (BSA) and the affinity of the PPI creates four types of interactions. In the last 20+ years of PPI inhibitor discovery, the majority of reported inhibitors have been directed at the concise/strong qudrant (bottom left). These compounds also tend to have the best potency values.See the text for references.
Here, we present a survey of inhibitors targeting PPIs, with a special focus on those described since 2015. Our hypothesis is that the field of PPI inhibitors may be mature enough that “rules” could emerge from an overview of the types of strategies, targets and molecules being deployed. Indeed, in retrospective studies performed in 2012 and 2015 [13,33,34], putative PPI targets were divided into four categories based on their affinity (KD) and BSA values (Fig 1B). When known inhibitors were placed into these quadrants, it became clear that some PPIs were easier to inhibit than others. For example, the majority (~80%) of reported inhibitors (at that time) were found to target PPIs that have a small, concise surface area (<2000 Å2) and a strong interaction ΔGbind (KD < 200 nM). These compounds were also the most potent, suggesting that there was simply more binding energy available. In contrast, the other categories of PPIs seemed to be less amenable to inhibitors (~20% of the total), including those with: (a) weak affinity and small, concise BSA values or (b) those with a large BSA and tight affinity. At the extreme end, there were no reported inhibitors of PPIs within the category characterized as having weak interactions over a large surface area. Together, this analysis seems to roughly match biophysical intuition. For example, PPIs with a concise surface area and tight affinity would be likely to have closely spaced “hot spots” and be most amenable to orthosteric inhibition by low molecular mass, high ligand efficiency (LE) compounds. Conversely, large surface areas or weak affinities both create theoretical challenges in the pursuit of inhibitors, requiring either larger molecules (> 500 Da) or possibly allosteric mechanisms (see Fig 1A).
In this review, we were particularly interested in learning whether PPI inhibitors derived from small molecules were being used to target different types of PPIs than those derived from peptides or other scaffolds. In other words, with the rise in new technologies for making and discovering PPI inhibitors, we wondered whether any of them had become more/less dominant in recent years. We were also interested in whether recent examples might continue the early trends about non-equivalent “druggability” of PPIs or whether the difficult categories were becoming more amenable to inhibition as technology develops. More broadly, it seemed important to periodically re-evaluate these questions (since 2015) in an effort to understand what is working and what gaps remain. Before continuing with this task, we first provide brief vignettes of some recent case studies, sequestered into sections based on whether the inhibitor was derived from a small molecule, peptide or alternative scaffold.
Recent Examples of Small-Molecules as Inhibitors of Protein-Protein Interactions
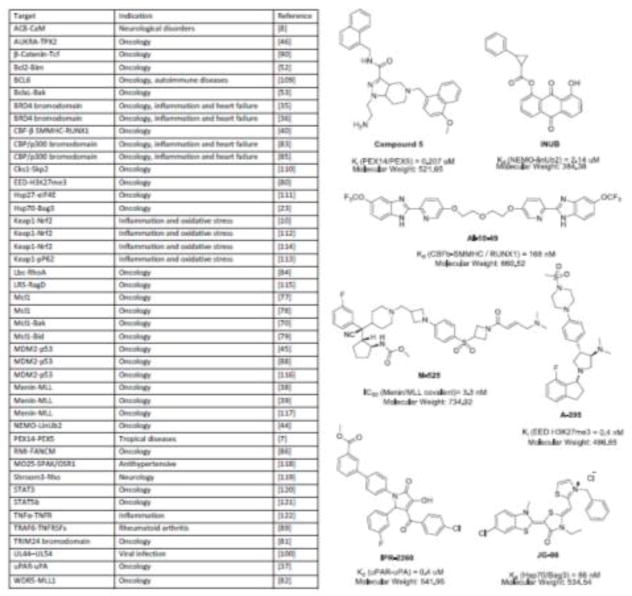
Small-molecules clearly have a number of advantages compared to other potential therapeutic modalities, including metabolic stability, bio-distribution, shelf life, manufacturing cost, pharmacokinetics and permeability [35,36]. In addition, there are well-established tools available to support chemical discovery and optimization, including computer-assisted drug design (CADD) and high-throughput screening infrastructure. However, it seems possible that small molecules might have some disadvantages when PPIs are the target, because of their relatively limited surface area. Here, we briefly summarize small molecule-based inhibitors that have been described in the last few years. We don’t intend this discussion to be inclusive of the targets or the approaches, but; rather, to give a sense of the scope of the discovery efforts (Fig 2) and illustrate some themes.
Figure 2.
Targets, disease indications and chemical structures of select small molecule PPI inhibitors
The concept of hotspots at orthosteric sites is continuing to influence modern PPI inhibitor design. For example, Liu et al. developed a series of pyrrolinones to target a hotspot in the uPAR-uPA interaction [37]. Interestingly, they found that large jumps in potency were gained when the series made an unexpected cation-pi interaction with an arginine in uPAR, which was outside the anticipated hotspot region but helped favorably position the scaffold. Structure-based approaches to target hotspots have also been used to create low nanomolar inhibitors of menin-MLL interactions [38,39*], which were potent in mouse models of leukemias caused by MLL translocations. In another compelling study in an entirely distinct biological system, Dawidowski et al. targeted the PPI between PEX14 and PEX5 that is required for peroxisome maturation in Trypanosoma species [7*]. In this study, they performed two, sequential screening campaigns: firstly, an in silico 3D pharmacophore-based search followed by docking; secondly, an NMR-based fragment screening from their in-house 1500 compound library to further exclude off-target effects and reduce toxicity of the initial hit. This effort produced sub-micromolar inhibitors (~ 0.2 to 0.5 μM) of the PPI that mimicked the natural hotspot in the contact region and validated PEX14-PEX5 as a new target for the treatment of parasitic infections by Trypanosomes. It also highlights the interdisciplinary nature of modern PPI inhibitor programs, incorporating biophysical methods, disease models and CBDD. For some types of PPIs, especially those with concise BSA values, this combination has created a well-worn pathway to small molecule-based inhibitors.
Transcription factor PPIs were also popular, and very challenging, targets in the last few years. For example, Illendula et. al. developed the first small-molecule targeting the transcription factor fusion CBFβ-SMMHC [40*]. They used a fluorescence resonance energy transfer (FRET) assay to screen the National Cancer Institute’s Diversity Set; revealing the most potent molecule, AI-10-49, as a bivalent small-molecule that can restore transcriptional activity of RUNX1 in acute myeloid leukemia (AML), and selectively induce cancer cell death in vivo. In some ways, the PPIs of transcription factors, such as CBFβ-SMMHC, are prime examples of the most challenging targets. They are dynamic, polar and often lack detailed structural information [41]. In the CBFβ-SMMHC example, the active compound acts at an allosteric pocket, perhaps creating a template for future studies. Covalent inhibitors of other transcription factors have also been reported [42,43], providing a potentially complementary strategy.
Another theme in the recent literature is a continued development of chemical screening libraries that are enriched for PPI inhibitors. Essentially, these collections tend to be more “natural product-like”, having higher average molecular mass and more stereocenters. For example, Vincendeau et al. screened a natural product library to identify an anthraquinone derivative that inhibits binding of NEMO-Ubiquitin, a large interface area of 4520 Å2 [44]. Similarly, inhibitors inspired by natural products also led to the discovery of the first non-azaphilone containing chlorofusin as an inhibitor of MDM2/p53 [45]. Another approach is to assemble subsets of compounds that are cherry-picked from traditional screening decks. Venkitaraman et al. took this strategy to build 17000 rationally-selected compounds, yielding an inhibitor of the PPI between AURKA and TPX2 [46]. These molecules were found to act at an allosteric site instead of directly binding to the PPI interface, a growing trend in kinase inhibitor programs that is often used to generate more selectivity [47].
Recent Examples of Peptides as Inhibitors of Protein-Protein Interactions
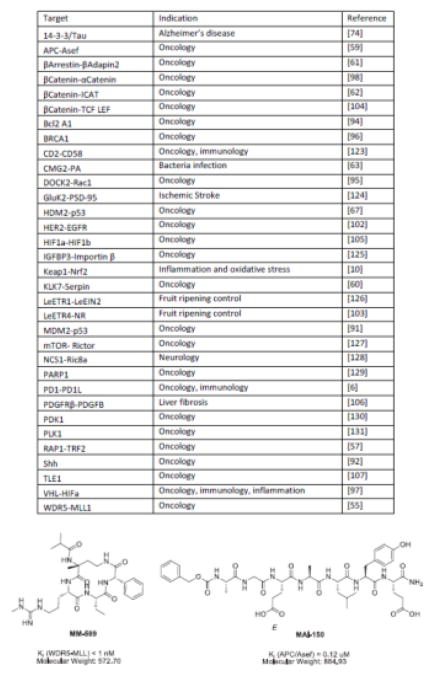
It is logical to consider peptides as potential inhibitors of PPIs, as they can be mimics of the natural interaction. Indeed, many groups have designed peptide-based inhibitors (Fig 3) using information gleaned from co-crystal structures of the protein targets [48,49]. However, poor membrane permeability and rapid metabolic instability are often major limitations to their clinical application. Accordingly, advances that tackle these pharmacological problems, such as macrocycles [50], short peptide mimetics [51], introduction of non-natural amino acids [52,53], conformational restricted cyclized peptides [24,54,55], and non-peptide mimetics of α-helical peptides [56] are important milestones.
Figure 3.
Targets, disease indications and chemical structures of select peptide-based PPI inhibitors
Recent years have seen peptide-based inhibitors used in many different indications and often as first-in-class inhibitors. For example, Milroy et al. were inspired by co-crystal structure of the PPI stabilizer fusicoccin A to develop an inhibitor of tau binding to 14-3-3 for the potential treatment of Alzheimer’s disease [56]. Ran et al. employed triazole-stapling strategy to make a 300-times more potent TRF2-based peptide, which blocked a previously under-explored RAP1-TRF2 interaction in the shelterin complex [57]. Ran et al. rationally designed and screened a peptide library to identify a molecule that inhibits the transcription factor: heat shock factor 1 (HSF1) [58]. Although HSF1 is a large, topologically complex protein, the authors found that optimized peptides derived from the natural inhibitory region, HR-C, limit DNA binding. Zhang et al. rationally designed peptidomimetics that potently bind to APC at its interface with Asef. They further optimized several peptides by attaching a transcription trans-activating (TAT) sequence to increase cell permeability and used a cellular thermal-shift assay (CETSA) to show that they disrupt APC-Asef interactions in colorectal cancer cells [59]. Jendrny and Beck-Sickinger took a different approach to modify their peptide-based inhibitor of serpin protease PPIs, by grafting it into a loop of sunflower trypsin inhibitor (SFTI-1) [60]. This chimeric SFTI-1 stabilized the active conformer of the inhibitor and increased its stability. In contrast, an interesting discovery from Giralt et al. indicated that a stable secondary structure might not be required in all cases, as they found that flexible structures had greater potency compared to those in helical conformations [61].
Finally, it is worth noting that peptide-based discovery programs have the advantage of established screening technologies, such as phage display and SICLOPPS, that continue to evolve. Phage display was used by Bertoldo et al. to identify macrocyclic, rather than linear, peptides that bound to a previous difficult region of beta-catenin [62]. Male et al. screened a 3.2 million member SICLOPPS library to identify a peptide that binds to the CMG2 receptor required for uptake of bacterial-derived toxins [63]. Interestingly, the top hits from that screen were linear peptides, rather than the macrocycles that are typically expected from SICLOPPS screens [64*]. New methods for chemically modifying peptides on phage are also emerging [65], promising to diversify the functionality of these materials.
PPI Inhibitors from Other Miscellaneous Classes
One of the other recent trends in PPI inhibitor discovery is the use of molecules that don’t readily fit into the designation of small molecules or peptides. For example, organometallic complexes [66–69], foldamer helix mimetics [70,71], peptoids [72] and monobodies [73] have provided supplementary approaches to target PPIs. Several bifunctional molecules of high molecular mass were also reported [74,75], and we have placed them into this category because of their non-tradition size. Whether any of these molecules can be further optimized into drug candidates is still in question, given the limited in vivo studies thus far; however, such strategies may be particularly effective as research tools. One counter-intuitive example is worth pointing out: In the process of studying the mechanism of a false positive screening hit, Lumb et. al. solved the X-ray crystal structure of an ordered conglomerate of an aggregated small-molecule, bound to TNF-α [76*]. Such a compound would normally be termed a pan-assay interference (PAINS) concern, so one wonders how many other false positives have this surprisingly specific interface with their protein target. Although the molecule is likely not a lead candidate, its contacts with TNF- might help inform future studies.
Analysis of Recent PPI Inhibitors: Molecular Mass, Potency and BSA
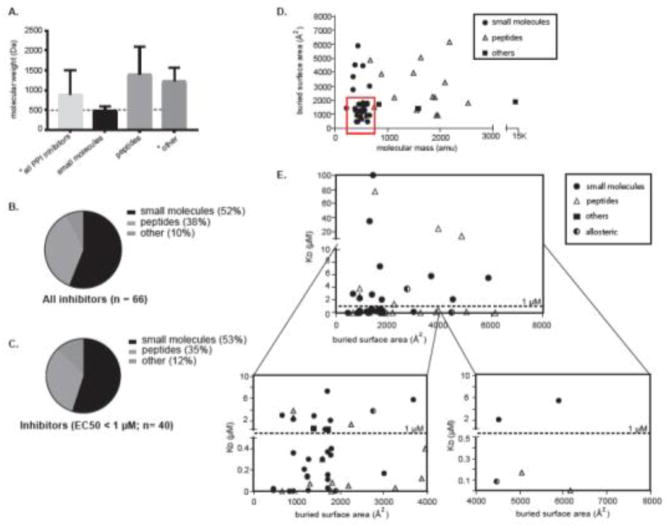
From these case studies, it seems that different types of scaffolds (i.e. small molecules, peptides, other) are all being generated and that they are being directed against a wide range of PPI targets and indications. To more quantitatively address this question, we performed a PubMed search for PPI inhibitors in the period from Jan 1st 2015 to March 1st 2018. From this list of ~140 examples, we excluded molecules that only had reported cellular activity (e.g. EC50) and only used those clear biophysical KD or Ki values. This search criteria yielded 66 compounds [6,7,23,35–38,40,44–46,55,57,59,60,62,63,70–75,77–131]. Each of these inhibitors was then manually categorized as being either a small molecule, peptide or belonging to neither group. Overall, the average molecular mass of these compounds was significantly larger (~800 Da) than the typical Lipinski range (Fig 4A). However, the small molecule subset largely conformed to the 500 Da cutoff, while the peptides and other categories were almost entirely above this value (see Fig 4A). Next, we prepared a pie chart to analyze which types of scaffolds were being explored (Fig 4B). From the pool of 66 inhibitors, it was clear that small-molecules are the dominant strategy, with over 50% of total cases. Peptides were also quite common, with just under 40%, while only ~10% were in the miscellaneous category. Then, the compounds with the most promising potency values (KD < 1 μM) were extracted, giving a pool of the 40 best inhibitors. Interestingly, these compounds had approximately the same distribution as the total pool (Fig 4C), suggesting that sub-micromolar inhibitors can be identified using any of the approaches.
Figure 4.
Relative properties and composition of the technologies used to target PPIs (reported 1/15 to 3/18). (A)The average molecular mass of the three categories of PPI inhibitors. Error bars represent the standard error of the mean (SEM). The dotted line is set to 500 Da. * monobody excluded (B) The relative contriibution of the three inhibitor categories, calculated from the 66 literature examples with reported Kd/Ki value. (B) An analysis of the subset of 40 cases that have reported Kd/Ki values < 1 μM. (D) Distribution of PPI inhibitors based on their molecular weight and BSA of the target interface. The red box signifies the most drug-like inhibitors (e.g. those with the lowest mass and best potency).
(E) Distribution of recent PPI inhibitors (1/15 to 3/18). Each compound was manually designated as either a small molecule, peptide or miscellaneous. In addition, the mechanism-of-action was designated as orthosteric (solid color) or allosteric (split color). For each compound, its potency (Kd or Ki) was plotted against buried surface area (BSA) of the target PPI. The bottom two quadrants (the most potent molecules) are shown as close-ups for clarity.
We were also interested in whether the type of scaffold (e.g. small molecule, peptide) impacted the type of PPI that was targeted. To approach this question, we calculated the BSA values for 50 of the 66 PPI interfaces that had sufficient structural information, using the reported PDB structures and UCSF Chimera [132]. When we plotted the molecular weight of the inhibitor against the calculated BSA values, we found that more than 80% of the small molecule were inhibiting PPIs with interfacial area below 2000 Å2 (Fig 5). Conversely, ~50% of the peptides were directed against PPIs with BSA > 2000 Å2, suggesting that they tend to be better for inhibiting large interactions.
Finally, we wanted to understand the relationships between inhibitor potency and the BSA of the target PPI. A plot shows that the majority of potent inhibitors (KD < 1 μM) seemed to be clustered against targets with low BSA (Fig 6), consistent with pre-2015 studies [13]. Thus, concise PPIs seemed to be the most amenable to the discovery of inhibitors. However, it is interesting to note that sub-micromolar inhibitors of large interfaces were also identified. For targets in the range between 2000 Å2 and 4000 Å2, there were 5 potent, reported inhibitors and there were even three inhibitors of targets in the BSA range >4000 Å2. Interestingly, inhibitors of these large targets tended to be peptide-based. For example, two of the inhibitors of large (>4000 Å2 BSA) interfaces were cyclic peptides targeting DOCK2-Rac1 (6160 Å2) and Shh-HHIP (5048 Å2), respectively [92,95]. Overall, 6/8 inhibitors of large PPIs were peptides. In addition, one of the small molecules is known to have an allosteric mechanism (Hsp70-BAG3; 4473 Å2) [23]). Together, these collective findings suggest that orthosteric small molecules might be more challenging to develop against these types of PPIs.
Summary and Prospectus
The last few years have continued to remove the undeserved stigma of PPIs being “undruggable”. At the same time, it is becoming increasingly clear that some PPIs are more difficult to tackle than others and that the original concerns may sometimes be justified. Thus, it seems that a more nuanced and sophisticated answer to the question of whether PPIs can be inhibited is that all targets are not equal. For each individual system, its own idiosyncratic features (i.e. are high resolution crystal structures available? do conformational changes accompany binding? is there a natural ligand to start with? what is the BSA?) will likely dictate whether a small molecule, peptide or other approach might be best and whether the search will ultimately be successful in producing a sub-micromolar inhibitor. For example, from our analysis here, we would argue that peptide-based compounds might be preferred for PPIs with large BSA values (>2000 Å2). We would also conclude that small molecules are still the go-to method for amenable PPIs, such as those with low BSA.
What is next for PPI inhibitor discovery? How do we improve discovery, especially for the PPIs with large BSA values? One exciting speculation is that cryo-EM methods may allow atomistic resolution of previously inaccessible protein complexes. For example, Merk et al. have been able to identify the binding site of ligands in isocitrate dehydrogenase, lactate dehydrogenase and glutamate dehydrogenase at < 4 Å resolution [133*], potentially opening the way to CBDD or other strategies for structure-guided design. Recent advances in the computational identification of cryptic binding sites may also create opportunities for the rational development of allosteric inhibitors [134,135]. Finally, it seems that PROTACS molecules or covalent inhibitors [136] might be fundamentally new ways target the currently “undruggable” systems. More broadly, retrospective analyses, such as this one, might reveal the gaps: the problems that lack a reliable, current solution. These are likely the topics that need the most innovation.
Acknowledgments
The authors apologize to colleagues whose work we were not able to adequately highlight. The authors thank Dr. Chao-Yie Yang (University Michigan) for help in extracting BSA values from the PDB. Our work on PPI inhibitors is supported by NIH grant R01NS059690.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nature Reviews Drug Discovery. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- *2.Nero TL, Morton CJ, Holien JK, Wielens J, Parker MW. Oncogenic protein interfaces: small molecules, big challenges. Nat Rev Cancer. 2014;14:248–262. doi: 10.1038/nrc3690. A comprehensive review on small-molecule drugs targeting PPIs with oncological significance. [DOI] [PubMed] [Google Scholar]
- 3.Arkin MR, Whitty A. The road less traveled: modulating signal transduction enzymes by inhibiting their protein-protein interactions. Curr Opin Chem Biol. 2009;13:284–290. doi: 10.1016/j.cbpa.2009.05.125. [DOI] [PubMed] [Google Scholar]
- *4.Arkin MR, Tang Y, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing toward the reality. Chem Biol. 2014;21:1102–1114. doi: 10.1016/j.chembiol.2014.09.001. A seminal review of PPI inhibitors, which includes categorization based on the conformation of the binding moieties. A great place to start for those new to the field. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Z, Ivanov AA, Su R, Gonzalez-Pecchi V, Qi Q, Liu S, Webber P, McMillan E, Rusnak L, Pham C, et al. The OncoPPi network of cancer-focused protein-protein interactions to inform biological insights and therapeutic strategies. Nat Commun. 2017;8:14356. doi: 10.1038/ncomms14356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Q, Quan LN, Lyu JK, He ZH, Wang X, Meng JJ, Zhao ZJ, Zhu LL, Liu XF, Li HL. Discovery of peptide inhibitors targeting human programmed death 1 (PD-1) receptor. Oncotarget. 2016;7:64967–64976. doi: 10.18632/oncotarget.11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *7.Dawidowski M, Emmanouilidis L, Kalel VC, Tripsianes K, Schorpp K, Hadian K, Kaiser M, Maser P, Kolonko M, Tanghe S, et al. Inhibitors of PEX14 disrupt protein import into glycosomes and kill Trypanosoma parasites. Science. 2017;355:1416. doi: 10.1126/science.aal1807. A tour-de-force in high throughput experimental and computational methods applied to an important and difficult PPI. [DOI] [PubMed] [Google Scholar]
- 8.Hayes MP, Soto-Velasquez M, Fowler CA, Watts VJ, Roman DL. Identification of FDA-Approved Small Molecules Capable of Disrupting the Calmodulin-Adenylyl Cyclase 8 Interaction through Direct Binding to Calmodulin. ACS Chem Neurosci. 2018;9:346–357. doi: 10.1021/acschemneuro.7b00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anand P, Brown JD, Lin CY, Qi J, Zhang R, Artero PC, Alaiti MA, Bullard J, Alazem K, Margulies KB, et al. BET bromodomains mediate transcriptional pause release in heart failure. Cell. 2013;154:569–582. doi: 10.1016/j.cell.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu MC, Tan SJ, Ji JA, Chen ZY, Yuan ZW, You QD, Jiang ZY. Polar Recognition Group Study of Keap1-Nrf2 Protein-Protein Interaction Inhibitors. ACS Med Chem Lett. 2016;7:835–840. doi: 10.1021/acsmedchemlett.5b00407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Azzarito V, Long K, Murphy NS, Wilson AJ. Inhibition of alpha-helix-mediated protein-protein interactions using designed molecules. Nature Chemistry. 2013;5:161–173. doi: 10.1038/nchem.1568. [DOI] [PubMed] [Google Scholar]
- *12.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. Genome editting methods are promisign to transform the way that we validate targets, includign PPI targets, in mammalian cells. This manuscript describes one powerful way to create point mutations and test ideas of molecular interactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cesa LC, Mapp AK, Gestwicki JE. Direct and Propagated Effects of Small Molecules on Protein-Protein Interaction Networks. Front Bioeng Biotechnol. 2015;3:119. doi: 10.3389/fbioe.2015.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pollak MN, Schernhammer ES, Hankinson SE. Insulin-like growth factors and neoplasia. Nature Reviews Cancer. 2004;4:505–518. doi: 10.1038/nrc1387. [DOI] [PubMed] [Google Scholar]
- 15.Hartman GD, Egbertson MS, Halczenko W, Laswell WL, Duggan ME, Smith RL, Naylor AM, Manno PD, Lynch RJ, Zhang GX, et al. Nonpeptide Fibrinogen Receptor Antagonists .1. Discovery and Design of Exosite Inhibitors. Journal of Medicinal Chemistry. 1992;35:4640–4642. doi: 10.1021/jm00102a020. [DOI] [PubMed] [Google Scholar]
- 16.Two i.v antiplatelet agents marketed for coronary disease. American Journal of Health-System Pharmacy. 1998;55:1440. doi: 10.1093/ajhp/55.14.1440. [DOI] [PubMed] [Google Scholar]
- 17.Schiff PB, Horwitz SB. Taxol Stabilizes Microtubules in Mouse Fibroblast Cells. Proceedings of the National Academy of Sciences of the United States of America-Biological Sciences. 1980;77:1561–1565. doi: 10.1073/pnas.77.3.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *18.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. Describes the use of fragment-based screening to build what would become the first truly synthetic, FDA-approved PPI inhibitor (see below). The methods described in this work have become standard in the field. [DOI] [PubMed] [Google Scholar]
- *19.Venetoclax (Venclexta) for Chronic Lymphocytic Leukemia. Medical Letter on Drugs and Therapeutics. 2016;58:101–102. After reading reference 18, it is worth reading this description of the first FDA-approved PPI inhibitor: Venetoclax and its clinical applications. As a series, these manuscripts show how clever discovery strategies and perseverance can yield first-in-class drugs. [PubMed] [Google Scholar]
- 20.Cukuroglu E, Engin HB, Gursoy A, Keskin O. Hot spots in protein-protein interfaces: Towards drug discovery. Progress in Biophysics & Molecular Biology. 2014;116:165–173. doi: 10.1016/j.pbiomolbio.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 21.Higueruelo AP, Jubb H, Blundell TL. Protein-protein interactions as druggable targets: recent technological advances. Current Opinion in Pharmacology. 2013;13:791–796. doi: 10.1016/j.coph.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 22.Wilson CG, Arkin MR. Small-molecule inhibitors of IL-2/IL-2R: lessons learned and applied. Curr Top Microbiol Immunol. 2011;348:25–59. doi: 10.1007/82_2010_93. [DOI] [PubMed] [Google Scholar]
- *23.Li X, Colvin T, Rauch JN, Acosta-Alvear D, Kampmann M, Dunyak B, Hann B, Aftab BT, Murnane M, Cho M, et al. Validation of the Hsp70-Bag3 protein-protein interaction as a potential therapeutic target in cancer. Mol Cancer Ther. 2015;14:642–648. doi: 10.1158/1535-7163.MCT-14-0650. It is often difficult to find small molecule inhibitors of PPIs with large buried surface area. In this example, an allosteric molecule was shown to inhibit a PPI with a BSA of >4,400 Å2. The binding site is small and compact and disrupts the PPI at a linear distance of over 20 Å. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *24.Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, Wright RD, Wagner G, Verdine GL, Korsmeyer SJ. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science. 2004;305:1466–1470. doi: 10.1126/science.1099191. An important landmark, as this work describes the first cell-permeable and in vivo active stapled peptide. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qian Z, Rhodes CA, McCroskey LC, Wen J, Appiah-Kubi G, Wang DJ, Guttridge DC, Pei D. Enhancing the Cell Permeability and Metabolic Stability of Peptidyl Drugs by Reversible Bicyclization. Angew Chem Int Ed Engl. 2017;56:1525–1529. doi: 10.1002/anie.201610888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang NJ, Hinner MJ. Getting across the cell membrane: an overview for small molecules, peptides, and proteins. Methods Mol Biol. 2015;1266:29–53. doi: 10.1007/978-1-4939-2272-7_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lowell AN, DeMars MD, 2nd, Slocum ST, Yu F, Anand K, Chemler JA, Korakavi N, Priessnitz JK, Park SR, Koch AA, et al. Chemoenzymatic Total Synthesis and Structural Diversification of Tylactone-Based Macrolide Antibiotics through Late-Stage Polyketide Assembly, Tailoring, and C-H Functionalization. J Am Chem Soc. 2017;139:7913–7920. doi: 10.1021/jacs.7b02875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen X, Nguyen TT, Koh MJ, Xu D, Speed AW, Schrock RR, Hoveyda AH. Kinetically E-selective macrocyclic ring-closing metathesis. Nature. 2017;541:380–385. doi: 10.1038/nature20800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bockus AT, Schwochert JA, Pye CR, Townsend CE, Sok V, Bednarek MA, Lokey RS. Going Out on a Limb: Delineating The Effects of beta-Branching, N-Methylation, and Side Chain Size on the Passive Permeability, Solubility, and Flexibility of Sanguinamide A Analogues. J Med Chem. 2015;58:7409–7418. doi: 10.1021/acs.jmedchem.5b00919. [DOI] [PubMed] [Google Scholar]
- 30.Villar EA, Beglov D, Chennamadhavuni S, Porco JA, Jr, Kozakov D, Vajda S, Whitty A. How proteins bind macrocycles. Nat Chem Biol. 2014;10:723–731. doi: 10.1038/nchembio.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Checco JW, Gellman SH. Targeting recognition surfaces on natural proteins with peptidic foldamers. Curr Opin Struct Biol. 2016;39:96–105. doi: 10.1016/j.sbi.2016.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *32.Churcher I. Protac-Induced Protein Degradation in Drug Discovery: Breaking the Rules or Just Making New Ones? Journal of Medicinal Chemistry. 2018;61:444–452. doi: 10.1021/acs.jmedchem.7b01272. A perspective on proteolysis targeting chimera (PROTAC), a potential approach to PPI inhibitors that is not discussed in detail here, but is worth a closer look. [DOI] [PubMed] [Google Scholar]
- 33.Smith MC, Gestwicki JE. Features of protein-protein interactions that translate into potent inhibitors: topology, surface area and affinity. Expert Rev Mol Med. 2012;14:e16. doi: 10.1017/erm.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson AD, Dugan A, Gestwicki JE, Mapp AK. Fine-tuning multiprotein complexes using small molecules. ACS Chem Biol. 2012;7:1311–1320. doi: 10.1021/cb300255p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ran X, Zhao Y, Liu L, Bai L, Yang CY, Zhou B, Meagher JL, Chinnaswamy K, Stuckey JA, Wang S. Structure-Based Design of gamma-Carboline Analogues as Potent and Specific BET Bromodomain Inhibitors. J Med Chem. 2015;58:4927–4939. doi: 10.1021/acs.jmedchem.5b00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Y, Bai L, Liu L, McEachern D, Stuckey JA, Meagher JL, Yang CY, Ran X, Zhou B, Hu Y, et al. Structure-Based Discovery of 4-(6-Methoxy-2-methyl-4-(quinolin-4-yl)-9H-pyrimido[4,5-b]indol-7-yl)-3,5-dimethy lisoxazole (CD161) as a Potent and Orally Bioavailable BET Bromodomain Inhibitor. J Med Chem. 2017;60:3887–3901. doi: 10.1021/acs.jmedchem.7b00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu D, Xu D, Liu M, Knabe WE, Yuan C, Zhou D, Huang M, Meroueh SO. Small Molecules Engage Hot Spots through Cooperative Binding To Inhibit a Tight Protein-Protein Interaction. Biochemistry. 2017;56:1768–1784. doi: 10.1021/acs.biochem.6b01039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borkin D, He S, Miao H, Kempinska K, Pollock J, Chase J, Purohit T, Malik B, Zhao T, Wang J, et al. Pharmacologic inhibition of the Menin-MLL interaction blocks progression of MLL leukemia in vivo. Cancer Cell. 2015;27:589–602. doi: 10.1016/j.ccell.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *39.Xu S, Aguilar A, Xu T, Zheng K, Huang L, Stuckey J, Chinnaswamy K, Bernard D, Fernandez-Salas E, Liu L, et al. Design of the First-in-Class, Highly Potent Irreversible Inhibitor Targeting the Menin-MLL Protein-Protein Interaction. Angew Chem Int Ed Engl. 2018;57:1601–1605. doi: 10.1002/anie.201711828. Covalent molecules are one of the important, new strategies for PPI inhibitor design. This work is a good place to start in learning how one approaches this possibility. [DOI] [PubMed] [Google Scholar]
- *40.Illendula A, Pulikkan JA, Zong HL, Grembecka J, Xue LT, Sen S, Zhou YP, Boulton A, Kuntimaddi A, Gao Y, et al. A small-molecule inhibitor of the aberrant transcription factor CBF beta-SMMHC delays leukemia in mice. Science. 2015;347:779–784. doi: 10.1126/science.aaa0314. The pursuit of PPI inhibitors often requires full integration of medicinal chemistry, structural biology, a suite of cell-based assays and careful animal work. This manuscript is a good place to find a complete story of how a small molecule inhibitor targeting transcription factor PPIs was developed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pricer R, Gestwicki JE, Mapp AK. From Fuzzy to Function: The New Frontier of Protein Protein Interactions. Accounts of Chemical Research. 2017;50:584–589. doi: 10.1021/acs.accounts.6b00565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang NK, Lodge JM, Fierke CA, Mapp AK. Dissecting allosteric effects of activator-coactivator complexes using a covalent small molecule ligand. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:12061–12066. doi: 10.1073/pnas.1406033111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dugan A, Majmudar CY, Pricer R, Niessen S, Lancia JK, Fung HYH, Cravatt BF, Mapp AK. Discovery of Enzymatic Targets of Transcriptional Activators via in Vivo Covalent Chemical Capture. Journal of the American Chemical Society. 2016;138:12629–12635. doi: 10.1021/jacs.6b07680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vincendeau M, Hadian K, Messias AC, Brenke JK, Halander J, Griesbach R, Greczmiel U, Bertossi A, Stehle R, Nagel D, et al. Inhibition of Canonical NF-kappaB Signaling by a Small Molecule Targeting NEMO-Ubiquitin Interaction. Sci Rep. 2016;6:18934. doi: 10.1038/srep18934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cominetti MMD, Goffin SA, Raffel E, Turner KD, Ramoutar JC, O’Connell MA, Howell LA, Searcey M. Identification of a new p53/MDM2 inhibitor motif inspired by studies of chlorofusin. Bioorganic & Medicinal Chemistry Letters. 2015;25:4878–4880. doi: 10.1016/j.bmcl.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 46.Janecek M, Rossmann M, Sharma P, Emery A, Huggins DJ, Stockwell SR, Stokes JE, Tan YS, Almeida EG, Hardwick B, et al. Allosteric modulation of AURKA kinase activity by a small-molecule inhibitor of its protein-protein interaction with TPX2. Sci Rep. 2016;6:28528. doi: 10.1038/srep28528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu P, Clausen MH, Nielsen TE. Allosteric small-molecule kinase inhibitors. Pharmacology & Therapeutics. 2015;156:59–68. doi: 10.1016/j.pharmthera.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 48.Bullock BN, Jochim AL, Arora PS. Assessing helical protein interfaces for inhibitor design. J Am Chem Soc. 2011;133:14220–14223. doi: 10.1021/ja206074j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siegert TR, Bird MJ, Makwana KM, Kritzer JA. Analysis of Loops that Mediate Protein-Protein Interactions and Translation into Submicromolar Inhibitors. J Am Chem Soc. 2016;138:12876–12884. doi: 10.1021/jacs.6b05656. [DOI] [PubMed] [Google Scholar]
- 50.Dougherty PG, Qian Z, Pei D. Macrocycles as protein-protein interaction inhibitors. Biochem J. 2017;474:1109–1125. doi: 10.1042/BCJ20160619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cai Q, Sun HY, Peng YF, Lu JF, Nikolovska-Coleska Z, McEachern D, Liu L, Qiu S, Yang CY, Miller R, et al. A Potent and Orally Active Antagonist (SM-406/AT-406) of Multiple Inhibitor of Apoptosis Proteins (IAPs) in Clinical Development for Cancer Treatment. Journal of Medicinal Chemistry. 2011;54:2714–2726. doi: 10.1021/jm101505d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boersma MD, Haase HS, Peterson-Kaufman KJ, Lee EF, Clarke OB, Colman PM, Smith BJ, Horne WS, Fairlie WD, Gellman SH. Evaluation of Diverse alpha/beta-Backbone Patterns for Functional alpha-Helix Mimicry: Analogues of the Bim BH3 Domain. Journal of the American Chemical Society. 2012;134:315–323. doi: 10.1021/ja207148m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yap JL, Cao X, Vanommeslaeghe K, Jung KY, Peddaboina C, Wilder PT, Nan A, MacKerell AD, Jr, Smythe WR, Fletcher S. Relaxation of the rigid backbone of an oligoamide-foldamer-based alpha-helix mimetic: identification of potent Bcl-xL inhibitors. Org Biomol Chem. 2012;10:2928–2933. doi: 10.1039/c2ob07125h. [DOI] [PubMed] [Google Scholar]
- 54.Kawamoto SA, Coleska A, Ran X, Yi H, Yang CY, Wang S. Design of triazole-stapled BCL9 alpha-helical peptides to target the beta-catenin/B-cell CLL/lymphoma 9 (BCL9) protein-protein interaction. J Med Chem. 2012;55:1137–1146. doi: 10.1021/jm201125d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karatas H, Li Y, Liu L, Ji J, Lee S, Chen Y, Yang J, Huang L, Bernard D, Xu J, et al. Discovery of a Highly Potent, Cell-Permeable Macrocyclic Peptidomimetic (MM-589) Targeting the WD Repeat Domain 5 Protein (WDR5)-Mixed Lineage Leukemia (MLL) Protein-Protein Interaction. J Med Chem. 2017;60:4818–4839. doi: 10.1021/acs.jmedchem.6b01796. [DOI] [PubMed] [Google Scholar]
- 56.Lee JH, Zhang Q, Jo S, Chai SC, Oh M, Im W, Lu H, Lim HS. Novel pyrrolopyrimidine-based alpha-helix mimetics: cell-permeable inhibitors of protein-protein interactions. J Am Chem Soc. 2011;133:676–679. doi: 10.1021/ja108230s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ran X, Liu L, Yang CY, Lu J, Chen Y, Lei M, Wang S. Design of High-Affinity Stapled Peptides To Target the Repressor Activator Protein 1 (RAP1)/Telomeric Repeat-Binding Factor 2 (TRF2) Protein-Protein Interaction in the Shelterin Complex. J Med Chem. 2016;59:328–334. doi: 10.1021/acs.jmedchem.5b01465. [DOI] [PubMed] [Google Scholar]
- 58.Ran X, Burchfiel ET, Dong B, Rettko NJ, Dunyak BM, Shao H, Thiele DJ, Gestwicki JE. Rational design and screening of peptide-based inhibitors of heat shock factor 1 (HSF1) Bioorg Med Chem. 2018 doi: 10.1016/j.bmc.2018.04.018. [DOI] [PMC free article] [PubMed]
- 59.Jiang HM, Deng R, Yang XY, Shang JL, Lu SY, Zhao YL, Song K, Liu XY, Zhang QF, Chen Y, et al. Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration. Nature Chemical Biology. 2017;13:994. doi: 10.1038/nchembio.2442. [DOI] [PubMed] [Google Scholar]
- 60.Jendrny C, Beck-Sickinger AG. Inhibition of Kallikrein-Related Peptidases 7 and 5 by Grafting Serpin Reactive-Center Loop Sequences onto Sunflower Trypsin Inhibitor-1 (SFTI-1) Chembiochem. 2016;17:719–726. doi: 10.1002/cbic.201500539. [DOI] [PubMed] [Google Scholar]
- 61.Martin-Quiros A, Nevola L, Eckelt K, Madurga S, Gorostiza P, Giralt E. Absence of a stable secondary structure is not a limitation for photoswitchable inhibitors of beta-arrestin/beta-Adaptin 2 protein-protein interaction. Chem Biol. 2015;22:31–37. doi: 10.1016/j.chembiol.2014.10.022. [DOI] [PubMed] [Google Scholar]
- 62.Bertoldo D, Khan MM, Dessen P, Held W, Huelsken J, Heinis C. Phage Selection of Peptide Macrocycles against beta-Catenin To Interfere with Wnt Signaling. ChemMedChem. 2016;11:834–839. doi: 10.1002/cmdc.201500557. [DOI] [PubMed] [Google Scholar]
- 63.Male AL, Forafonov F, Cuda F, Zhang G, Zheng S, Oyston PCF, Chen PR, Williamson ED, Tavassoli A. Targeting Bacillus anthracis toxicity with a genetically selected inhibitor of the PA/CMG2 protein-protein interaction. Sci Rep. 2017;7:3104. doi: 10.1038/s41598-017-03253-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *64.Tavassoli A. SICLOPPS cyclic peptide libraries in drug discovery. Curr Opin Chem Biol. 2017;38:30–35. doi: 10.1016/j.cbpa.2017.02.016. Due to space constraints, we were not able to provide much technical detail into how SICLOPPS screens are designed or variations on the technology. This recent review provides a good entry point. [DOI] [PubMed] [Google Scholar]
- 65.He B, Tjhung KF, Bennett NJ, Chou Y, Rau A, Huang J, Derda R. Compositional Bias in Naive and Chemically-modified Phage-Displayed Libraries uncovered by Paired-end Deep Sequencing. Sci Rep. 2018;8:1214. doi: 10.1038/s41598-018-19439-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhong HJ, Lu LH, Leung KH, Wong CCL, Peng C, Yan SC, Ma DL, Cai ZW, Wang HMD, Leung CH. An iridium(III)-based irreversible protein-protein interaction inhibitor of BRD4 as a potent anticancer agent. Chemical Science. 2015;6:5400–5408. doi: 10.1039/c5sc02321a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu LJ, He BY, Miles JA, Wang WH, Mao ZF, Che WI, Lu JJ, Chen XP, Wilson AJ, Ma DL, et al. Inhibition of the p53/hDM2 protein-protein interaction by cyclometallated iridium(III) compounds. Oncotarget. 2016;7:13965–13975. doi: 10.18632/oncotarget.7369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang C, Wang WH, Li GD, Zhong HJ, Dong ZZ, Wong CY, Kwong DWJ, Ma DL, Leung CH. Anticancer osmium complex inhibitors of the HIF-1 alpha and p300 protein-protein interaction. Scientific Reports. 2017:7. doi: 10.1038/srep42860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kang TS, Mao ZF, Ng CT, Wang MD, Wang WH, Wang CM, Lee SMY, Wang YT, Leung CH, Ma DL. Identification of an Iridium(III)-Based Inhibitor of Tumor Necrosis Factor-alpha. Journal of Medicinal Chemistry. 2016;59:4026–4031. doi: 10.1021/acs.jmedchem.6b00112. [DOI] [PubMed] [Google Scholar]
- 70.Drennen B, Scheenstra JA, Yap JL, Chen L, Lanning ME, Roth BM, Wilder PT, Fletcher S. Structural Re-engineering of the alpha-Helix Mimetic JY-1-106 into Small Molecules: Disruption of the Mcl-1-Bak-BH3 Protein-Protein Interaction with 2,6-Di-Substituted Nicotinates. ChemMedChem. 2016;11:827–833. doi: 10.1002/cmdc.201500461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee JH, Oh M, Kim HS, Lee H, Im W, Lim HS. Converting One-Face alpha-Helix Mimetics into Amphiphilic alpha-Helix Mimetics as Potent Inhibitors of Protein-Protein Interactions. Acs Combinatorial Science. 2016;18:36–42. doi: 10.1021/acscombsci.5b00080. [DOI] [PubMed] [Google Scholar]
- 72.Oh M, Lee JH, Moon H, Hyun YJ, Lim HS. A Chemical Inhibitor of the Skp2/p300 Interaction that Promotes p53-Mediated Apoptosis. Angewandte Chemie-International Edition. 2016;55:602–606. doi: 10.1002/anie.201508716. [DOI] [PubMed] [Google Scholar]
- 73.Wojcik J, Lamontanara AJ, Grabe G, Koide A, Akin L, Gerig B, Hantschel O, Koide S. Allosteric Inhibition of Bcr-Abl Kinase by High Affinity Monobody Inhibitors Directed to the Src Homology 2 (SH2)-Kinase Interface. Journal of Biological Chemistry. 2016;291:8836–8847. doi: 10.1074/jbc.M115.707901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Parvatkar P, Kato N, Uesugi M, Sato S, Ohkanda J. Intracellular Generation of a Diterpene-Peptide Conjugate that Inhibits 14-3-3-Mediated Interactions. Journal of the American Chemical Society. 2015;137:15624–15627. doi: 10.1021/jacs.5b09817. [DOI] [PubMed] [Google Scholar]
- 75.Tallorin L, Finzel K, Nguyen QG, Beld J, La Clair JJ, Burkart MD. Trapping of the Enoyl-Acyl Carrier Protein Reductase-Acyl Carrier Protein Interaction. J Am Chem Soc. 2016;138:3962–3965. doi: 10.1021/jacs.5b13456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *76.Blevitt JM, Hack MD, Herman KL, Jackson PF, Krawczuk PJ, Lebsack AD, Liu AX, Mirzadegan T, Nelen MI, Patrick AN, et al. Structural Basis of Small-Molecule Aggregate Induced Inhibition of a Protein-Protein Interaction. J Med Chem. 2017;60:3511–3517. doi: 10.1021/acs.jmedchem.6b01836. A fascinating case study on how an "artifact" PPI inhibitor can have a remarkably complex mechanism-of-action. [DOI] [PubMed] [Google Scholar]
- 77.Whiting E, Raje MR, Chauhan J, Wilder PT, Van Eker D, Hughes SJ, Bowen NG, Vickers GEA, Fenimore IC, Fletcher S. Discovery of Mcl-1 inhibitors based on a thiazolidine-2,4-dione scaffold. Bioorg Med Chem Lett. 2017 doi: 10.1016/j.bmcl.2017.11.023. [DOI] [PubMed] [Google Scholar]
- 78.Liu J, Tian Z, Zhou N, Liu X, Liao C, Lei B, Li J, Zhang S, Chen H. Targeting the apoptotic Mcl-1-PUMA interface with a dual-acting compound. Oncotarget. 2017;8:54236–54242. doi: 10.18632/oncotarget.17294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lanning ME, Yu W, Yap JL, Chauhan J, Chen L, Whiting E, Pidugu LS, Atkinson T, Bailey H, Li W, et al. Structure-based design of N-substituted 1-hydroxy-4-sulfamoyl-2-naphthoates as selective inhibitors of the Mcl-1 oncoprotein. Eur J Med Chem. 2016;113:273–292. doi: 10.1016/j.ejmech.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Curtin ML, Pliushchev MA, Li HQ, Torrent M, Dietrich JD, Jakob CG, Zhu H, Zhao H, Wang Y, Ji Z, et al. SAR of amino pyrrolidines as potent and novel protein-protein interaction inhibitors of the PRC2 complex through EED binding. Bioorg Med Chem Lett. 2017;27:1576–1583. doi: 10.1016/j.bmcl.2017.02.030. [DOI] [PubMed] [Google Scholar]
- 81.Liu J, Li F, Bao H, Jiang Y, Zhang S, Ma R, Gao J, Wu J, Ruan K. The polar warhead of a TRIM24 bromodomain inhibitor rearranges a water-mediated interaction network. FEBS J. 2017;284:1082–1095. doi: 10.1111/febs.14041. [DOI] [PubMed] [Google Scholar]
- 82.Li DD, Wang ZH, Chen WL, Xie YY, You QD, Guo XK. Structure-based design of ester compounds to inhibit MLL complex catalytic activity by targeting mixed lineage leukemia 1 (MLL1)-WDR5 interaction. Bioorg Med Chem. 2016;24:6109–6118. doi: 10.1016/j.bmc.2016.09.073. [DOI] [PubMed] [Google Scholar]
- 83.Popp TA, Tallant C, Rogers C, Fedorov O, Brennan PE, Muller S, Knapp S, Bracher F. Development of Selective CBP/P300 Benzoxazepine Bromodomain Inhibitors. J Med Chem. 2016;59:8889–8912. doi: 10.1021/acs.jmedchem.6b00774. [DOI] [PubMed] [Google Scholar]
- 84.Diviani D, Raimondi F, Del Vescovo CD, Dreyer E, Reggi E, Osman H, Ruggieri L, Gonano C, Cavin S, Box CL, et al. Small-Molecule Protein-Protein Interaction Inhibitor of Oncogenic Rho Signaling. Cell Chem Biol. 2016;23:1135–1146. doi: 10.1016/j.chembiol.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 85.Picaud S, Fedorov O, Thanasopoulou A, Leonards K, Jones K, Meier J, Olzscha H, Monteiro O, Martin S, Philpott M, et al. Generation of a Selective Small Molecule Inhibitor of the CBP/p300 Bromodomain for Leukemia Therapy. Cancer Res. 2015;75:5106–5119. doi: 10.1158/0008-5472.CAN-15-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Voter AF, Manthei KA, Keck JL. A High-Throughput Screening Strategy to Identify Protein-Protein Interaction Inhibitors That Block the Fanconi Anemia DNA Repair Pathway. J Biomol Screen. 2016;21:626–633. doi: 10.1177/1087057116635503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gal M, Bloch I, Shechter N, Romanenko O, Shir OM. Efficient Isothermal Titration Calorimetry Technique Identifies Direct Interaction of Small Molecule Inhibitors with the Target Protein. Comb Chem High Throughput Screen. 2016;19:4–13. doi: 10.2174/1386207319666151203001529. [DOI] [PubMed] [Google Scholar]
- 88.Holzer P, Masuya K, Furet P, Kallen J, Valat-Stachyra T, Ferretti S, Berghausen J, Bouisset-Leonard M, Buschmann N, Pissot-Soldermann C, et al. Discovery of a Dihydroisoquinolinone Derivative (NVP-CGM097): A Highly Potent and Selective MDM2 Inhibitor Undergoing Phase 1 Clinical Trials in p53wt Tumors. J Med Chem. 2015;58:6348–6358. doi: 10.1021/acs.jmedchem.5b00810. [DOI] [PubMed] [Google Scholar]
- 89.Moriya J, Takeuchi K, Tai K, Arai K, Kobayashi N, Yoneda N, Fukunishi Y, Inoue A, Kihara M, Murakami T, et al. Structure-Based Development of a Protein-Protein Interaction Inhibitor Targeting Tumor Necrosis Factor Receptor-Associated Factor 6. J Med Chem. 2015;58:5674–5683. doi: 10.1021/acs.jmedchem.5b00778. [DOI] [PubMed] [Google Scholar]
- 90.Catrow JL, Zhang Y, Zhang M, Ji H. Discovery of Selective Small-Molecule Inhibitors for the beta-Catenin/T-Cell Factor Protein-Protein Interaction through the Optimization of the Acyl Hydrazone Moiety. J Med Chem. 2015;58:4678–4692. doi: 10.1021/acs.jmedchem.5b00223. [DOI] [PubMed] [Google Scholar]
- 91.Morimoto J, Hosono Y, Sando S. Isolation of a peptide containing d-amino acid residues that inhibits the alpha-helix-mediated p53-MDM2 interaction from a one-bead one-compound library. Bioorg Med Chem Lett. 2018 doi: 10.1016/j.bmcl.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 92.Owens AE, de Paola I, Hansen WA, Liu YW, Khare SD, Fasan R. Design and Evolution of a Macrocyclic Peptide Inhibitor of the Sonic Hedgehog/Patched Interaction. Journal of the American Chemical Society. 2017;139:12559–12568. doi: 10.1021/jacs.7b06087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sogabe S, Sakamoto K, Kamada Y, Kadotani A, Fukuda Y, Sakamoto JI. Discovery of a Kelch-like ECH-associated protein 1-inhibitory tetrapeptide and its structural characterization. Biochem Biophys Res Commun. 2017;486:620–625. doi: 10.1016/j.bbrc.2017.03.038. [DOI] [PubMed] [Google Scholar]
- 94.de Araujo AD, Lim J, Good AC, Skerlj RT, Fairlie DP. Electrophilic Helical Peptides That Bond Covalently, Irreversibly, and Selectively in a Protein-Protein Interaction Site. ACS Med Chem Lett. 2017;8:22–26. doi: 10.1021/acsmedchemlett.6b00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sakamoto K, Adachi Y, Komoike Y, Kamada Y, Koyama R, Fukuda Y, Kadotani A, Asami T, Sakamoto J. Novel DOCK2-selective inhibitory peptide that suppresses B-cell line migration. Biochemical and Biophysical Research Communications. 2017;483:183–190. doi: 10.1016/j.bbrc.2016.12.170. [DOI] [PubMed] [Google Scholar]
- 96.Na Z, Pan S, Uttamchandani M, Yao SQ. Protein-Protein Interaction Inhibitors of BRCA1 Discovered Using Small Molecule Microarrays. Methods Mol Biol. 2017;1518:139–156. doi: 10.1007/978-1-4939-6584-7_10. [DOI] [PubMed] [Google Scholar]
- 97.Frost J, Galdeano C, Soares P, Gadd MS, Grzes KM, Ellis L, Epemolu O, Shimamura S, Bantscheff M, Grandi P, et al. Potent and selective chemical probe of hypoxic signalling downstream of HIF-alpha hydroxylation via VHL inhibition. Nat Commun. 2016;7:13312. doi: 10.1038/ncomms13312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kawakami T, Ogawa K, Hatta T, Goshima N, Natsume T. Directed Evolution of a Cyclized Peptoid-Peptide Chimera against a Cell-Free Expressed Protein and Proteomic Profiling of the Interacting Proteins to Create a Protein-Protein Interaction Inhibitor. ACS Chem Biol. 2016;11:1569–1577. doi: 10.1021/acschembio.5b01014. [DOI] [PubMed] [Google Scholar]
- 99.Nishihara T, Kitada H, Fujiwara D, Fujii I. Macrocyclization and labeling of helix-loop-helix peptide with intramolecular bis-thioether linkage. Biopolymers. 2016;106:415–421. doi: 10.1002/bip.22826. [DOI] [PubMed] [Google Scholar]
- 100.Chen H, Coseno M, Ficarro SB, Mansueto MS, Komazin-Meredith G, Boissel S, Filman DJ, Marto JA, Hogle JM, Coen DM. A Small Covalent Allosteric Inhibitor of Human Cytomegalovirus DNA Polymerase Subunit Interactions. ACS Infect Dis. 2017;3:112–118. doi: 10.1021/acsinfecdis.6b00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.He Y, Selvaraju S, Curtin ML, Jakob CG, Zhu H, Comess KM, Shaw B, The J, Lima-Fernandes E, Szewczyk MM, et al. The EED protein-protein interaction inhibitor A-395 inactivates the PRC2 complex. Nat Chem Biol. 2017;13:389–395. doi: 10.1038/nchembio.2306. [DOI] [PubMed] [Google Scholar]
- 102.Kanthala SP, Liu YY, Singh S, Sable R, Pallerla S, Jois SD. A peptidomimetic with a chiral switch is an inhibitor of epidermal growth factor receptor heterodimerization. Oncotarget. 2017;8:74244–74262. doi: 10.18632/oncotarget.19013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kessenbrock M, Klein SM, Muller L, Hunsche M, Noga G, Groth G. Novel Protein-Protein Inhibitor Based Approach to Control Plant Ethylene Responses: Synthetic Peptides for Ripening Control. Front Plant Sci. 2017;8:1528. doi: 10.3389/fpls.2017.01528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dietrich L, Rathmer B, Ewan K, Bange T, Heinrichs S, Dale TC, Schade D, Grossmann TN. Cell Permeable Stapled Peptide Inhibitor of Wnt Signaling that Targets beta-Catenin Protein-Protein Interactions. Cell Chem Biol. 2017;24:958–968 e955. doi: 10.1016/j.chembiol.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 105.Mistry IN, Tavassoli A. Reprogramming the Transcriptional Response to Hypoxia with a Chromosomally Encoded Cyclic Peptide HIF-1 Inhibitor. ACS Synth Biol. 2017;6:518–527. doi: 10.1021/acssynbio.6b00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang X, Wu X, Zhang A, Wang S, Hu C, Chen W, Shen Y, Tan R, Sun Y, Xu Q. Targeting the PDGF-B/PDGFR-beta Interface with Destruxin A5 to Selectively Block PDGF-BB/PDGFR-betabeta Signaling and Attenuate Liver Fibrosis. EBioMedicine. 2016;7:146–156. doi: 10.1016/j.ebiom.2016.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.McGrath S, Tortorici M, Drouin L, Solanki S, Vidler L, Westwood I, Gimeson P, Van Montfort R, Hoelder S. Structure-Enabled Discovery of a Stapled Peptide Inhibitor to Target the Oncogenic Transcriptional Repressor TLE1. Chemistry. 2017;23:9577–9584. doi: 10.1002/chem.201700747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Scharow A, Knappe D, Reindl W, Hoffmann R, Berg T. Development of Bifunctional Inhibitors of Polo-Like Kinase 1 with Low-Nanomolar Activities Against the Polo-Box Domain. Chembiochem. 2016;17:759–767. doi: 10.1002/cbic.201500535. [DOI] [PubMed] [Google Scholar]
- 109.Leeman-Neill RJ, Bhagat G. BCL6 as a therapeutic target for lymphoma. Expert Opin Ther Targets. 2018;22:143–152. doi: 10.1080/14728222.2018.1420782. [DOI] [PubMed] [Google Scholar]
- 110.Singh R, Sran A, Carroll DC, Huang J, Tsvetkov L, Zhou X, Sheung J, McLaughlin J, Issakani SD, Payan DG, et al. Developing structure-activity relationships from an HTS hit for inhibition of the Cks1-Skp2 protein-protein interaction. Bioorg Med Chem Lett. 2015;25:5199–5202. doi: 10.1016/j.bmcl.2015.09.067. [DOI] [PubMed] [Google Scholar]
- 111.Hajer Z, Claudia A, Erik L, Sara K, Maurizio F, Ridha O, David T, Michel C, Olivier S, Sabrina P, et al. Targeting Hsp27/eIF4E interaction with phenazine compound: a promising alternative for castration-resistant prostate cancer treatment. Oncotarget. 2017;8:77317–77329. doi: 10.18632/oncotarget.20469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yasuda D, Yuasa A, Obata R, Nakajima M, Takahashi K, Ohe T, Ichimura Y, Komatsu M, Yamamoto M, Imamura R, et al. Discovery of benzo[g]indoles as a novel class of non-covalent Keap1-Nrf2 protein-protein interaction inhibitor. Bioorganic & Medicinal Chemistry Letters. 2017;27:5006–5009. doi: 10.1016/j.bmcl.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 113.Yasuda D, Nakajima M, Yuasa A, Obata R, Takahashi K, Ohe T, Ichimura Y, Komatsu M, Yamamoto M, Imamura R, et al. Synthesis of Keap1-phosphorylated p62 and Keap1-Nrf2 protein-protein interaction inhibitors and their inhibitory activity. Bioorg Med Chem Lett. 2016;26:5956–5959. doi: 10.1016/j.bmcl.2016.10.083. [DOI] [PubMed] [Google Scholar]
- 114.Jiang ZY, Xu LL, Lu MC, Chen ZY, Yuan ZW, Xu XL, Guo XK, Zhang XJ, Sun HP, You QD. Structure-Activity and Structure-Property Relationship and Exploratory in Vivo Evaluation of the Nanomolar Keap1-Nrf2 Protein-Protein Interaction Inhibitor. Journal of Medicinal Chemistry. 2015;58:6410–6421. doi: 10.1021/acs.jmedchem.5b00185. [DOI] [PubMed] [Google Scholar]
- 115.Kim J, Jung J, Koo J, Cho W, Lee WS, Kim C, Park W, Park SB. Diversity-oriented synthetic strategy for developing a chemical modulator of protein-protein interaction. Nat Commun. 2016;7:13196. doi: 10.1038/ncomms13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Grigoreva TA, Novikova DS, Petukhov AV, Gureev MA, Garabadzhiu AV, Melino G, Barlev NA, Tribulovich VG. Proapoptotic modification of substituted isoindolinones as MDM2-p53 inhibitors. Bioorganic & Medicinal Chemistry Letters. 2017;27:5197–5202. doi: 10.1016/j.bmcl.2017.10.049. [DOI] [PubMed] [Google Scholar]
- 117.Ren J, Xu W, Tang L, Su MB, Chen DQ, Chen YL, Zang Y, Li J, Shen JK, Zhou YB, et al. Design and synthesis of benzylpiperidine inhibitors targeting the menin-MLL1 interface. Bioorganic & Medicinal Chemistry Letters. 2016;26:4472–4476. doi: 10.1016/j.bmcl.2016.07.074. [DOI] [PubMed] [Google Scholar]
- 118.Kadri H, Alamri MA, Navratilova IH, Alderwick LJ, Simpkins NS, Mehellou Y. Towards the Development of Small-Molecule MO25 Binders as Potential Indirect SPAK/OSR1 Kinase Inhibitors. Chembiochem. 2017;18:460–465. doi: 10.1002/cbic.201600620. [DOI] [PubMed] [Google Scholar]
- 119.Dickson HM, Wilbur A, Reinke AA, Young MA, Vojtek AB. Targeted inhibition of the Shroom3-Rho kinase protein-protein interaction circumvents Nogo66 to promote axon outgrowth. Bmc Neuroscience. 2015:16. doi: 10.1186/s12868-015-0171-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Leung KH, Liu LJ, Lin S, Lu LH, Zhong HJ, Susanti D, Rao WD, Wang MD, Che WI, Chan DSH, et al. Discovery of a small-molecule inhibitor of STAT3 by ligand-based pharmacophore screening. Methods. 2015;71:38–43. doi: 10.1016/j.ymeth.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 121.Elumalai N, Berg A, Natarajan K, Scharow A, Berg T. Nanomolar Inhibitors of the Transcription Factor STAT5b with High Selectivity over STAT5a. Angewandte Chemie-International Edition. 2015;54:4758–4763. doi: 10.1002/anie.201410672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mouhsine H, Guillemain H, Moreau G, Fourati N, Zerrouki C, Baron B, Desallais L, Gizzi P, Ben Nasr N, Perrier J, et al. Identification of an in vivo orally active dual-binding protein-protein interaction inhibitor targeting TNF alpha through combined in silico/in vitro/in vivo screening. Scientific Reports. 2017:7. doi: 10.1038/s41598-017-03427-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sable R, Durek T, Taneja V, Craik DJ, Pallerla S, Gauthier T, Jois S. Constrained Cyclic Peptides as Immunomodulatory Inhibitors of the CD2:CD58 Protein-Protein Interaction. ACS Chem Biol. 2016;11:2366–2374. doi: 10.1021/acschembio.6b00486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yin XH, Yan JZ, Yang G, Chen L, Xu XF, Hong XP, Wu SL, Hou XY, Zhang GY. PDZ1 inhibitor peptide protects neurons against ischemia via inhibiting GluK2-PSD-95-module-mediated Fas signaling pathway. Brain Research. 2016;1637:64–70. doi: 10.1016/j.brainres.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 125.Njomen E, Evans HG, Gedara SH, Heyl DL. Humanin Peptide Binds to Insulin-Like Growth Factor-Binding Protein 3 (IGFBP3) and Regulates Its Interaction with Importin-beta. Protein Pept Lett. 2015;22:869–876. doi: 10.2174/0929866522666150728114955. [DOI] [PubMed] [Google Scholar]
- 126.Bisson MM, Kessenbrock M, Muller L, Hofmann A, Schmitz F, Cristescu SM, Groth G. Peptides interfering with protein-protein interactions in the ethylene signaling pathway delay tomato fruit ripening. Sci Rep. 2016;6:30634. doi: 10.1038/srep30634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Benavides-Serrato A, Lee J, Holmes B, Landon KA, Bashir T, Jung ME, Lichtenstein A, Gera J. Specific blockade of Rictor-mTOR association inhibits mTORC2 activity and is cytotoxic in glioblastoma. Plos One. 2017:12. doi: 10.1371/journal.pone.0176599. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 128.Mansilla A, Chaves-Sanjuan A, Campillo NE, Semelidou O, Martinez-Gonzalez L, Infantes L, Gonzalez-Rubio JM, Gil C, Conde S, Skoulakis EMC, et al. Interference of the complex between NCS-1 and Ric8a with phenothiazines regulates synaptic function and is an approach for fragile X syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:E999–E1008. doi: 10.1073/pnas.1611089114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Na ZK, Peng B, Ng SK, Pan SJ, Lee JS, Shen HM, Yao SQ. A Small-Molecule Protein-Protein Interaction Inhibitor of PARP1 That Targets Its BRCT Domain. Angewandte Chemie-International Edition. 2015;54:2515–2519. doi: 10.1002/anie.201410678. [DOI] [PubMed] [Google Scholar]
- 130.Kroon E, Schulze JO, Suss E, Camacho CJ, Biondi RM, Domling A. Discovery of a Potent Allosteric Kinase Modulator by Combining Computational and Synthetic Methods. Angewandte Chemie-International Edition. 2015;54:13933–13936. doi: 10.1002/anie.201506310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Narvaez AJ, Ber S, Crooks A, Emery A, Hardwick B, Almeida EG, Huggins DJ, Perera D, Roberts-Thomson M, Azzarelli R, et al. Modulating Protein-Protein Interactions of the Mitotic Polo-like Kinases to Target Mutant KRAS. Cell Chemical Biology. 2017;24:1017. doi: 10.1016/j.chembiol.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- *133.Merk A, Bartesaghi A, Banerjee S, Falconieri V, Rao P, Davis MI, Pragani R, Boxer MB, Earl LA, Milne JLS, et al. Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery. Cell. 2016;165:1698–1707. doi: 10.1016/j.cell.2016.05.040. A tour-de-force that opens the possibility of structure-based design on targets that are inaccessible by crystallography or NMR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ghanakota P, Carlson HA. Moving Beyond Active-Site Detection: MixMD Applied to Allosteric Systems. J Phys Chem B. 2016;120:8685–8695. doi: 10.1021/acs.jpcb.6b03515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cimermancic P, Weinkam P, Rettenmaier TJ, Bichmann L, Keedy DA, Woldeyes RA, Schneidman-Duhovny D, Demerdash ON, Mitchell JC, Wells JA, et al. CryptoSite: Expanding the Druggable Proteome by Characterization and Prediction of Cryptic Binding Sites. J Mol Biol. 2016;428:709–719. doi: 10.1016/j.jmb.2016.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Singh J, Petter RC, Baillie TA, Whitty A. The resurgence of covalent drugs. Nat Rev Drug Discov. 2011;10:307–317. doi: 10.1038/nrd3410. [DOI] [PubMed] [Google Scholar]