Abstract
Emerging evidence indicated that many long non-coding (lnc)RNAs function in multiple biological processes and dysregulation of their expression can cause diseases. Most regulatory lncRNAs interact with biological macromolecules such as DNA, RNA, and protein. LncRNAs regulate gene expression through epigenetic modification, transcription, and posttranscription, through DNA methylation, histone modification, and chromatin remodeling. Interestingly, differential lncRNA expression profiles in human oocytes and cumulus cells was recently assessed, however, lncRNAs in human follicle development has not previously been described. In this study, transcriptome dynamics in human primordial, primary and small antral follicles were interrogated and revealed information of lncRNA genes. It is known that some lncRNAs form a complex with paraspeckle proteins and therefore, we extended our transcriptional analysis to include genes encoding paraspeckle proteins. Primordial, primary follicles and small antral follicles was isolated using laser capture micro-dissection from ovarian tissue donated by three women having ovarian tissue cryopreserved before chemotherapy. After RN sequencing, a bioinformatic class comparison was performed and primordial, primary and small antral follicles were found to express several lncRNA and genes encoding paraspeckle proteins. Of particular interest, we detected the lncRNAs XIST, NEAT1, NEAT2 (MALAT1), and GAS5. Moreover, we noted a high expression of FUS, TAF15, and EWS components of the paraspeckles, proteins that belong to the FET (previously TET) family of RNA-binding proteins and are implicated in central cellular processes such as regulation of gene expression, maintenance of genomic integrity, and mRNA/microRNA processing. We also interrogated the intra-ovarian localization of the FUS, TAF15, and EWS proteins using immunofluorescence. The presence and the dynamics of genes that encode lncRNA and paraspeckle proteins may suggest that these may mediate functions in the cyclic recruitment and differentiation of human follicles and could participate in biological processes known to be associated with lncRNAs and paraspeckle proteins, such as gene expression control, scaffold formation and epigenetic control through human follicle development. This comprehensive transcriptome analysis of lncRNAs and genes encoding paraspeckle proteins expressed in human follicles could potentially provide biomarkers of oocyte quality for the development of non-invasive tests to identify embryos with high developmental potential.
Keywords: human follicle, lncRNA, paraspeckle, fertility, treatment
Introduction
The nuclei of mammalian cells are highly organized and composed of distinct subnuclear structures termed nuclear bodies (Naganuma and Hirose, 2013; Yamazaki and Hirose, 2015). Paraspeckles are mammalian-specific sub-nuclear bodies built on long, non-protein-coding RNA (lncRNA), NEAT1 (nuclear-enriched abundant transcript 1), which assembles various protein components, including RNA-binding proteins of the DBHS (Drosophila behavior and human splicing) family. Paraspeckles have been proposed to control of several biological processes, such as stress responses, gene expression, and cellular differentiation. Human follicle development represents a continuous cyclic process throughout the reproductive lifespan of a woman and encompasses both cell growth and differentiation. Paraspeckles are among the most recently identified nuclear bodies and were first described in 2002 (Fox et al., 2002; Bond and Fox, 2009). The generation of paraspeckle sub-nuclear compartments has been extensively described (Naganuma and Hirose, 2013; Yamazaki and Hirose, 2015). Paraspeckles are sensitive to RNAse treatment, suggesting that their structures depend on RNAs for maintenance (Fox et al., 2002, 2005). Later the lncRNA NEAT1 was shown to be essential for paraspeckle formation, as a knockdown of the NEAT1 lncRNA function caused a disintegration of paraspeckles (Chen and Carmichael, 2009; Clemson et al., 2009; Sasaki et al., 2009; Sunwoo et al., 2009). Paraspeckle formation proceeds in conjunction with NEAT1 lncRNA biogenesis and involves the cooperation of multiple paraspeckle-localized RNA-binding proteins (Naganuma and Hirose, 2013; Yamazaki and Hirose, 2015). Currently about 40 proteins are known to assemble in paraspeckles (Naganuma et al., 2012). Paraspeckle proteins include DBHS (Drosophila melanogaster behavior, human splicing) proteins, PSPC1 (paraspeckle component 1), NONO (non-POU domain-containing octamer-binding), and SFPQ [splicing factor, proline- and glutamine-rich (also known as PSF (PTB-associated splicing factor)], RNA binding motif (RBM) 14, and CPSF6 (cleavage and polyadenylation specific factor 6) [Reviewed in (Yamazaki and Hirose, 2015)]. Many paraspeckle proteins are RNA binding proteins that contain an RNA recognition motif (RRM), a KH (hnRNP K homology) domain, a RGG (glycine-arginine-rich) box, or a zinc finger motif as the RNA-binding domain. The paraspeckle proteins NONO, SFPQ, RBM14, EWS, FUS, TAF15, and TDP-43 are RNA binding proteins that mediate transcription and RNA processing (Auboeuf et al., 2005).
The paraspeckle-localized FET family of RNA-binding proteins (Bertolotti et al., 1996) consists of FUS (TLS) (Crozat et al., 1993), EWS (Delattre et al., 1992), and TAF15 (TAFII68, TAF2N, RBP56) (Crozat et al., 1993). The proteins are structurally similar and contain a number of evolutionary conserved areas such as the RRM motif, the SYGQ-rich domain, a G rich domain, a RanBP2-type zinc finger motif, and the C-terminal RGG domain (Morohoshi et al., 1998; Guipaud et al., 2006; Nguyen et al., 2011; Chau et al., 2016). The FUS, EWS, and TAF15 proteins bind RNA as well as DNA and have both unique and overlapping functions. The human FET proteins are associated with transcription (Law et al., 2006), RNA splicing, microRNA (miRNA) processing, RNA transport, and the signaling and maintenance of genomic integrity (Schwartz et al., 2015).
Several paraspeckle proteins are disease-related. For instance, NONO, SFPQ, CPSF6, EWS, FUS, TAF15, DAZAP1, RBM3, SS18L1, WT1, BCL6, BCL11A, ZNF4444, and HNRNPH1 are implicated in various types of cancer (reviewed in Yamazaki and Hirose, 2015). Some paraspeckle proteins, such as TDP-13, FUS, EWS, TAF15, HNRNPA1, SS18L1, and SFPQ have been associated with neuro-degenerative diseases, such as amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) (Svetoni et al., 2016).
Paraspeckles have been described as nuclear sponges sequestering transcription factors and/or RNA-binding proteins such as lncRNAs. They are dynamic structures changing in size in response to ever changing cellular challenges/environment (Yamazaki and Hirose, 2015).
In addition to NEAT1, a number of lncRNAs localize to different subcellular compartments (Chen and Carmichael, 2010). MALAT1 (NEAT2 in human) is transcribed downstream of the NEAT1 gene and is found specifically associated with splicing speckles (Hutchinson et al., 2007). Moreover, lncRNA have also been implicated in stem cell pluripotency and in differentiation in mice (Dinger et al., 2008). Furthermore, roles for ncRNAs in cell fate decision have been explored (Ambasudhan et al., 2011; Yoo et al., 2011; Kurian et al., 2013).
Interestingly, lncRNAs have been shown to act as chromatin modifiers (Mercer et al., 2009) and potent regulators of histone methylation (Yamazaki and Hirose, 2015), including chromatin structure modeling and the integrity of subcellular compartments (Chen and Carmichael, 2010; Wang and Chang, 2011; Wang et al., 2011; Wapinski and Chang, 2011; Yan et al., 2012; Backofen and Vogel, 2014; Joh et al., 2014; Peschansky and Wahlestedt, 2014; Liu and Pan, 2015). A previous study showed that some human lncRNAs were bound to the polycomb repressive complex 2 (PRC2) and other chromatin-modifying complexes (Khalil et al., 2009).
Several lncRNA, including Xist, Tsix, and Xite contribute to X chromosome inactivation, the process of ensuring dosage regulation of X chromosome-expressed genes (Chow and Heard, 2009; Leeb et al., 2009) in a complex and highly controlled manner (Zhao et al., 2008). Furthermore, Xist transcription is required for maintenance of X-chromosome inactivation (Penny et al., 1996). Interestingly, another lncRNA, RepA, the reassembling part of the 5′UTR sequence of Xist, was found to associate indirectly with PRC2 (Zhao et al., 2008). The recruitment of PRC2 by RepA happens in competition with lncRNA Tsix, which acts as an antisense toward Xist, and the binding of RepA to PRC2 is inhibited by Tsix, and thus competes with RepA (Zhao et al., 2008).
In support of the developmental roles of lncRNA and paraspeckles, Neat1 knockout (KO) mice fail to become pregnant despite normal ovulation, which was found to be a caused by corpus luteum dysfunction and concomitant low progesterone (Nakagawa et al., 2014).
The developmental capacity of the matured oocyte for generating viable offspring is determined throughout follicle development in the ovary. The integrity of the oocytes is essential in maintaining the reproductive potential of the female. Pre-ovulatory oocyte maturation is a complex process resulting from multiple interactions between the oocyte and the surrounding follicular cells (Carabatsos et al., 2000; Adhikari and Liu, 2009; Binelli and Murphy, 2010; Reddy et al., 2010; Bonnet et al., 2013). The transition from primordial to primary follicle is a key first step event in follicle development, in which the primordial follicle is believed to have escaped the resting phase and has entered the follicular growth phase (Zuccotti et al., 2011). Subsequently, the cohort of follicles must remain activated in order to enter the secondary follicle stage, and a few continue to mature to the tertiary and antral follicle stages (McGee and Hsueh, 2000). Tertiary and antral follicles are characterized by the presence of a cavity known as the antrum, and have both granulosa and theca cells present. Tertiary follicles have an extensive network of gap junctions that permits the transfer of nutrients and regulatory signals between the oocyte and the granulosa cells (Espey, 1994). Only a small fraction of the ovarian follicles present in a fetal ovary will reach ovulation (Markström et al., 2002). Identifying the factors controlling follicle development may provide a basis for the fundamental mechanisms that regulate follicle activation and could potentially lead to new therapeutics in female reproduction as well as improvements in reproductive health and productivity in women of advanced maternal age (Baird et al., 2005). As paraspeckles and the regulatory molecules sequestered within them have been shown to be of importance in development, gene expression, and epigenetic control, these nuclear structures may prove essential in human fertility and infertility.
So far, only limited reports of the potential regulatory impact of short ncRNA in follicle development exist and our knowledge of the involvement of lncRNAs in human follicle development is almost non-existent (Wilhelmm and Bernard, 2016).
Therefore, in this study, the presence of lncRNAs were interrogated bioinformatically using RNA sequencing data representative of selected stages in human follicle development. We previously developed a method for isolating pure populations of oocytes from human primordial, intermediate and primary follicles using laser capture micro-dissection microscopy (Markholt et al., 2012). From these transcriptome data (Ernst et al., 2017, 2018), in silico extraction of data for lncRNAs. We identified the presence of the paraspeckle forming lncRNAs NEAT1 and NEAT2 as well as several other lncRNAs, such as XIST. As the discovery of NEAT1 and NEAT2 in early ovarian follicles suggested the presence of paraspeckle proteins, we further asked if genes encoding these proteins would also be present during human ovarian follicle development. We found the transcripts encoding the well-characterized FUS, EWS, and TAF15 highly expressed during early ovarian follicle development. We further employed immunohistochemistry in human ovary tissue to explore the presence and intraovarian localization of FUS, EWS, and TAF15 proteins to be present.
In summary, we identified the presence of several lncRNAs and genes encoding paraspeckle proteins not previously reported for human ovarian follicle development. This may hint that the functions of lncRNAs and paraspeckle proteins could indeed be relevant to oocyte physiology and development.
Materials and methods
Procurement of human ovarian cortex and isolation of oocytes and supportive somatic cells
We procured human ovarian cortex tissue from the Danish Cryopreservation Programme offering cryopreservation as means of fertility preservation prior to gonadotoxic chemotherapy (Rosendahl et al., 2011). Oocyte samples were obtained from ovarian cortical tissue procured from three patients who underwent unilateral oophorectomy prior to gonadotoxic treatment for a malignant disease (unrelated to any ovarian malignancies). Patients were normo-ovulatory, with normal reproductive hormones, and not received ovarian stimulation with exogenous gonadotropins. All methods were carried out in accordance with relevant guidelines and regulations, and The Central Denmark Region Committees on Biomedical Research Ethics and the Danish Data Protection Agency approved the study. Written informed consent was obtained from all participants before inclusion. Patients consented to the research conducted. In subjects undergoing oophorectomy, a small piece of the ovarian cortex is used for evaluating the ovarian reserve, and for research purposes (Danish Scientific Ethical Committee Approval Number: KF 299017 and J/KF/01/170/99) (Schmidt et al., 2003).
Laser capture micro-dissection (LCM)
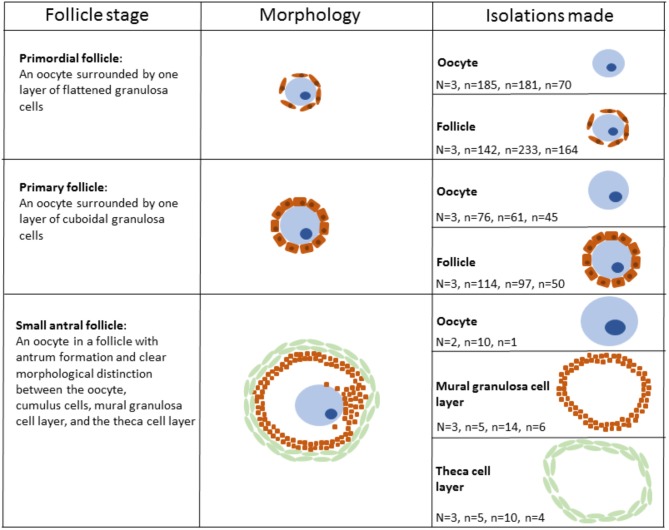
The LCM procedure to isolate staged oocytes and follicles was performed as previously described (Markholt et al., 2012; Ernst et al., 2017, 2018). Briefly, the ovarian cortical fragments, which had a size of 2 × 2 × 1 mm, were thawed and fixed by direct immersion into 4% paraformaldehyde (PFA) at 4°C for 4 h followed by dehydration and embedding in paraffin. Paraffin blocks were stored at −80°C until use. The blocks were cut into 15 μm thick sections on a microtome (Leica Microsystems, Wetzlar, Germany). Diethylpyrocarbonate (DEPC)-treated water was used in the microtome bath to avoid RNA degradation. The sections were mounted on RNase-free membrane glass slides (Molecular Devices, Sunnyvale, CA, USA) and immediately processed. Consecutively, the slides were de-paraffinized, stained, and dehydrated immediately before micro-dissection: Xylene (VWR—Bieog Berntsen, Herlev, Denmark) (5 min), 99.9% ethanol (Merck, Darmstadt, Germany) (5 min), 99.9% ethanol (5 min), 96% ethanol (5 min), 70% ethanol (5 min), DEPC-treated water (5 min), hematoxylin (Merck, Darmstadt, Germany) (5 min), DEPC-treated water (immersion), 70% ethanol (30 s), 96% ethanol (30 s), 99.9% ethanol (30 s), 99.9% ethanol (30 s), xylene (1 min), and xylene (5 min). All solutions were prepared with DEPC-treated water. LCM was performed using the Veritas™ Microdissection Instrument Model 704 (ArcturusXT™, Molecular Devices, Applied Biosystems®, Life Technologies, Foster City, CA, U.S.A.). The cells were isolated based on morphological appearance. Primordial oocytes were defined as an oocyte surrounded by 3–5 flattened pre-granulosa cells, and primary oocytes were defined as an oocyte surrounded by one layer of cuboidal granulosa cells. Antral follicles were defined as a follicle with an antral cavity. For the antral follicle to be eligible for isolation, we should be able to morphologically differentiate between the oocyte, the mural granulosa cells and the theca cell layer. In the antral stage the large size of the different compartments enabled us to isolate each compartment individually. An outline surrounding the cell(s) of interest was marked and subsequently cut using the ultraviolet laser. Following this the use of membrane glass slides (Arcturus® PEN Membrane Glass Slides, Applied Biosystems, Life Technologies, Foster City, CA, U.S.A.) enabled us to lift the isolate onto a sterile cap (Arcturus® CapSure® HS LCM Caps, Applied Biosystems, Life Technologies, Foster City, CA, U.S.A.) using infrared pulses. Isolated cells were inspected on the cap to ensure that no contamination from surrounding unwanted cells was present. From each of the three patients, several isolations were made (Table 1, Figure 1). RNA isolation, library preparation and sequencing, mapping and statistical analysis and bioinformatics were performed as described (Ernst et al., 2017, 2018).
Table 1.
Numbers of oocytes, follicles, and other somatic cells analyzed in RNA–seq. in three different patients.
| Cell type and Follicular Stages | No. of laser-collected cells from three patients respectively |
|---|---|
| Oocytes from primordial follicles | N = 3, n = 185, n = 181, n = 70 |
| Primordial follicles | N = 3, n = 142, n = 233, n = 164 |
| Oocytes from primary follicles | N = 3, n = 76, n = 61, n = 45 |
| Primary follicles | N = 3, n = 114, n = 97, n = 50 |
| Mural granulosa cell layers from small antral follicles | N = 3, n = 5, n = 14, n = 6 |
| Theca cell layers from small antral follicles | N = 3, n = 5, n = 10, n = 4 |
| Oocytes from small antral follicles*1 | N = 2, n = 10, n = 1 |
Based on duplicate samples.
Figure 1.
Schematic illustration of human follicular cells isolated using laser capture microdissection. Please note that the aspect ratio is arbitrary.
Library preparation and sequencing
RNA was extracted from the LCM-derived samples, converted to cDNA and subjected to linear amplification [Ovation® RNA-Seq System V2 kit (NuGen Inc., San Carlos, CA, U.S.A.)]. RNA-seq libraries (constructed from the output cDNA using Illumina TruSeq DNA Sample and Preparation kit (Illumina, San Diego, CA, USA, according to AROS Applied Biotechnology, now Eurofins (https://www.eurofinsgenomics.eu/). Integrity of libraries was verified on library yield via KAPA qPCR measurement, and Agilent Bioanalyzer 2100 peak size with a RNA 6000 Nano Lab Chip (Agilent Technologies, Santa Clare, CA, U.S.A.) during different library preparation stages. Sequencing was performed on an Illumina HiSeq2000 platform (Illumina Inc., San Diego, CA, U.S.A.) with 5 random samples per lane (AROS Applied Biotechnology).
Mapping and statistical analysis
Using Tophat (2.0.4), and Cufflinks (2.0.2) BAM files were generated to create a list of expressed transcripts in the samples. BWA (0.6.2) was subsequently used to map all readings to the human reference genome (hg19) using the transcript list as a filter so only readings mapping to RefSeq exons [incl. non-coding RNA, and mitochondrial RNA) overlapping with expressed transcripts were used. Expression of each gene in a given sample was normalized and transformed to a measurement of log2 (counts per million (CPM)]. On the basis of log2 (CPM), fragments per kilobase of exon per million fragments mapped (FPKM) values were calculated, and further filtered using custom analysis in R [R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org] (R Core Team, 2012).
Output from statistical analysis
The mean FPKM value for all ncRNA transcripts was calculated using a one-sample t-test on FPKM values for each identified transcript from patient triplicates (two for oocytes from small antral follicles) (Resource Data) (http://users-birc.au.dk/biopv/published_data/ernst_et_al_ncRNAs_2018/).
Cell Specific Consistently Expressed Genes (CSCEG) were defined as one-sample t-test p-value < 0.05 [(Resource Data), in gray]. In silico merging of transcriptomes from three patients was performed to account for biological variance. The transcripts in the CSCEG list were ranked based on p-value, with low p-value indicating a high degree of consistency in FPKM gene expression level between patients for the given isolate type. Full lists of ncRNAs detected in human follicle development is availiable (Supplementary Table 1).
Furthermore, we generated a list of all known paraspeckle proteins based on annotated protein-coding RNAs detected (Table 2) and non-coding RNAs (Table 3). Some of these data (oocytes and granulosa cells from primordial and primary follicles) has previously been published with a different focus in a global expression profile study (Ernst et al., 2017, 2018).
Table 2.
Expression of paraspeckle-protein encoding mRNAs in human follicle development.
| Ensemble | Symbol | Oocytes from primordial follicles, FPKM mean | t-test (p-value) | Primordial follicles, FPKM mean | t-test (p-value) |
|---|---|---|---|---|---|
| PRIMORDIAL FOLLICLES | |||||
| ENSG00000126705 | AHDC1 | ND | ND | 0.195 | 0.423 |
| ENSG00000011243 | AKAP8L | 2.094 | 0.366 | 0.347 | 0.423 |
| ENSG00000140488 | CELF6 | ND | ND | ND | ND |
| ENSG00000099622 | CIRBP | 1.826 | 0.053 | 2.486 | 0.059 |
| ENSG00000111605 | CPSF6 | 3.718 | 0.033 | 4.289 | 0.064 |
| ENSG00000149532 | CPSF7 | 2.089 | 0.351 | 2.739 | 0.003 |
| ENSG00000071626 | DAZAP1 | 1.817 | 0.408 | 1.368 | 0.192 |
| ENSG00000064195 | DLX3 | ND | ND | ND | ND |
| ENSG00000182944 | EWS/EWSR1*1 | 5.765 | 0.073 | 6.132 | 0.173 |
| ENSG00000119812 | FAM98A | 1.727 | 0.235 | 2.013 | 0.188 |
| ENSG00000182263 | FIGN | 0.284 | 0.187 | 2.842 | 0.184 |
| ENSG00000089280 | FUS*1 | 3.150 | 0.149 | 3.150 | 0.146 |
| ENSG00000139675 | HNRNPA1L2 | 0.029 | 0.423 | 0.060 | 0.423 |
| ENSG00000169813 | HNRNPF | 2.550 | 0.028 | 3.836 | 0.085 |
| ENSG00000169045 | HNRNPH1 | 3.410 | 0.017 | 6.310 | 0.002 |
| ENSG00000096746 | HNRNPH3 | 3.177 | 0.171 | 3.178 | 0.199 |
| ENSG00000165119 | HNRNPK | 5.365 | 0.068 | 5.511 | 0.060 |
| ENSG00000125944 | HNRNPR | 4.346 | 0.107 | 3.778 | 0.127 |
| ENSG00000105323 | HNRNPUL1 | 4.041 | 0.005 | 6.086 | 0.007 |
| ENSG00000176624 | MEX3C*1 | 1.365 | 0.204 | 3.570 | 0.160 |
| ENSG00000147140 | NONO*1 | 2.160 | 0.189 | 4.367 | 0.089 |
| ENSG00000167005 | NUDT21 | 4.365 | 0.155 | 5.401 | 0.065 |
| ENSG00000121390 | PSPC1 | 1.709 | 0.274 | 3.114 | 0.189 |
| ENSG00000102317 | RBM3 | 0.475 | 0.238 | 0.586 | 0.423 |
| ENSG00000268489 | RBM3 | ND | ND | ND | ND |
| ENSG00000173914 | RBM4B | 1.878 | 0.359 | 1.489 | 0.400 |
| ENSG00000076053 | RBM7 | 1.519 | 0.330 | 1.191 | 0.222 |
| ENSG00000244462 | RBM12 | 4.391 | 0.106 | 4.400 | 0.112 |
| ENSG00000239306 | RBM14 | 1.072 | 0.207 | 1.164 | 0.394 |
| ENSG00000147274 | RBMX | 4.218 | 0.117 | 3.981 | 0.122 |
| ENSG00000020633 | RUNX3 | ND | ND | 0.054 | 0.423 |
| ENSG00000116560 | SFPQ | 4.459 | 0.025 | 5.770 | 0.071 |
| ENSG00000188529 | SRSF10 | 1.465 | 0.373 | 3.245 | 0.109 |
| ENSG00000184402 | SS18L1 | 0.083 | 0.423 | 1.002 | 0.213 |
| ENSG00000172660 | TAF15*1 | 1.303 | 0.172 | 2.055 | 0.226 |
| ENSG00000143569 | UBAP2L | 7.856 | 0.006 | 7.315 | 0.006 |
| ENSG00000188177 | ZC3H6 | 6.564 | 0.002 | 2.015 | 0.365 |
| Oocytes from primary follicles | Primary follicles | ||||
| Ensemble | Symbol*1 | FPKM mean | t-test (p-value) | FPKM mean | t-test (p-value) |
| PRIMARY FOLLICLES | |||||
| ENSG00000198026 | ZNF335 | ND | ND | 1.216 | 0.423 |
| ENSG00000120948 | TARDBP | 5.408 | 0.131 | 4.119 | 0.112 |
| ENSG00000126705 | AHDC1 | 0.883 | 0.423 | ND | ND |
| ENSG00000011243 | AKAP8L | ND | ND | ND | ND |
| ENSG00000140488 | CELF6 | 1.933 | 0.186 | 4.296 | 0.196 |
| ENSG00000099622 | CIRBP | 2.339 | 0.359 | 1.864 | 0.423 |
| ENSG00000111605 | CPSF6 | 1.331 | 0.246 | 3.621 | 0.189 |
| ENSG00000149532 | CPSF7 | 0.208 | 0.423 | 1.386 | 0.399 |
| ENSG00000071626 | DAZAP1 | ND | ND | ND | ND |
| ENSG00000064195 | DLX3 | 4.564 | 0.087 | 5.184 | 0.185 |
| ENSG00000182944 | EWS/EWSR1*2,3 | 0.835 | 0.423 | 1.826 | 0.409 |
| ENSG00000119812 | FAM98A | 2.007 | 0.423 | 1.189 | 0.401 |
| ENSG00000182263 | FIGN | 2.666 | 0.066 | 2.451 | 0.268 |
| ENSG00000089280 | FUS*2,3 | 0.158 | 0.204 | 0.439 | 0.423 |
| ENSG00000139675 | HNRNPA1L2 | 3.494 | 0.280 | 3.308 | 0.147 |
| ENSG00000169813 | HNRNPF | 2.572 | 0.176 | 4.849 | 0.118 |
| ENSG00000169045 | HNRNPH1 | 6.586 | 0.022 | 2.636 | 0.287 |
| ENSG00000096746 | HNRNPH3 | 4.526 | 0.097 | 3.360 | 0.217 |
| ENSG00000165119 | HNRNPK | 0.554 | 0.244 | 3.590 | 0.195 |
| ENSG00000125944 | HNRNPR | 2.285 | 0.110 | 2.451 | 0.185 |
| ENSG00000105323 | HNRNPUL1 | 1.790 | 0.336 | 3.198 | 0.184 |
| ENSG00000176624 | MEX3C*1 | 3.151 | 0.046 | 3.493 | 0.167 |
| ENSG00000147140 | NONO*1 | 4.165 | 0.201 | 5.300 | 0.047 |
| ENSG00000167005 | NUDT21 | 1.994 | 0.312 | 3.174 | 0.199 |
| ENSG00000121390 | PSPC1 | 1.712 | 0.133 | 1.077 | 0.374 |
| ENSG00000102317 | RBM3 | ND | ND | ND | ND |
| ENSG00000268489 | RBM3 | 2.873 | 0.253 | 1.890 | 0.409 |
| ENSG00000173914 | RBM4B | 0.654 | 0.423 | 0.161 | 0.259 |
| ENSG00000076053 | RBM7 | 5.893 | 0.057 | 5.262 | 0.177 |
| ENSG00000244462 | RBM12 | 0.952 | 0.423 | 0.716 | 0.353 |
| ENSG00000239306 | RBM14 | 4.337 | 0.019 | 3.784 | 0.175 |
| ENSG00000147274 | RBMX | ND | ND | ND | ND |
| ENSG00000020633 | RUNX3 | 5.408 | 0.027 | 4.258 | 0.206 |
| ENSG00000116560 | SFPQ | 3.097 | 0.160 | 4.403 | 0.176 |
| ENSG00000188529 | SRSF10 | 0.564 | 0.423 | 0.5280 | 0.374 |
| ENSG00000184402 | SS18L1 | 0.411 | 0.423 | 2.030 | 0.306 |
| ENSG00000172660 | TAF15*1 | 7.527 | 0.013 | 7.630 | 0.002 |
| ENSG00000143569 | UBAP2L | 4.010 | 0.184 | 2.057 | 0.411 |
| ENSG00000188177 | ZC3H6 | ND | ND | 0.911 | 0.423 |
| ENSG00000198026 | ZNF335 | 1.576 | 0.378 | 2.515 | 0.393 |
| ENSG00000120948 | TARDBP | ND | ND | ND | ND |
| Mural garnulosa cell layer | Theca cell layer | ||||
| Ensemble | Symbol | FPKM mean | t-test (p-value) | FPKM mean | t-test (p-value) |
| SMALL ANTRAL FOLLICLES | |||||
| ENSG00000126705 | AHDC1 | 1.967 | 0.333 | 0.955 | 0.134 |
| ENSG00000011243 | AKAP8L | 1.100 | 0.132 | 2.477 | 0.103 |
| ENSG00000140488 | CELF6 | ND | ND | 0.181 | 0.423 |
| ENSG00000099622 | CIRBP | 3.468 | 0.017 | 4.523 | 0.021 |
| ENSG00000111605 | CPSF6 | 4.481 | 0.022 | 3.007 | 0.039 |
| ENSG00000149532 | CPSF7 | 4.775 | 0.001 | 1.925 | 0.075 |
| ENSG00000071626 | DAZAP1 | 1.899 | 0.063 | 1.419 | 0.045 |
| ENSG00000064195 | DLX3 | ND | ND | ND | ND |
| ENSG00000182944 | EWS/EWSR1 | 8.561 | 0.001 | 6.354 | 0.009 |
| ENSG00000119812 | FAM98A | 3.164 | 0.057 | 4.066 | 0.025 |
| ENSG00000182263 | FIGN | 3.326 | 0.018 | 3.210 | 0.023 |
| ENSG00000089280 | FUS | 4.624 | 0.003 | 3.323 | 0.077 |
| ENSG00000139675 | HNRNPA1L2 | 1.234 | 0.136 | 1.193 | 0.263 |
| ENSG00000169813 | HNRNPF | 3.712 | 0.004 | 4.046 | 0.001 |
| ENSG00000169045 | HNRNPH1 | 6.458 | 0.011 | 6.056 | 0.004 |
| ENSG00000096746 | HNRNPH3 | 4.316 | 0.033 | 3.635 | 0.014 |
| ENSG00000165119 | HNRNPK | 5.862 | 0.004 | 5.249 | 1.7237E-05 |
| ENSG00000125944 | HNRNPR | 5.096 | 0.011 | 4.504 | 0.014 |
| ENSG00000105323 | HNRNPUL1 | 6.769 | 0.002 | 5.058 | 0.049 |
| ENSG00000176624 | MEX3C | 5.705 | 0.006 | 2.639 | 0.036 |
| ENSG00000147140 | NONO | 6.395 | 0.002 | 5.664 | 0.027 |
| ENSG00000167005 | NUDT21 | 5.327 | 0.015 | 3.735 | 0.131 |
| ENSG00000121390 | PSPC1 | 2.929 | 0.086 | 4.015 | 0.003 |
| ENSG00000102317 | RBM3 | 3.057 | 0.104 | 4.013 | 0.033 |
| ENSG00000268489 | RBM3 | ND | ND | ND | ND |
| ENSG00000173914 | RBM4B | 2.125 | 0.140 | 1.097 | 0.078 |
| ENSG00000076053 | RBM7 | 1.148 | 0.190 | 0.728 | 0.200 |
| ENSG00000244462 | RBM12 | 5.530 | 0.028 | 4.575 | 0.017 |
| ENSG00000239306 | RBM14 | 0.882 | 0.028 | 1.247 | 0.192 |
| ENSG00000147274 | RBMX | 5.781 | 0.011 | 4.702 | 0.017 |
| ENSG00000020633 | RUNX3 | ND | ND | ND | ND |
| ENSG00000116560 | SFPQ | 6.759 | 0.009 | 5.362 | 0.014 |
| ENSG00000188529 | SRSF10 | 3.181 | 0.149 | 3.040 | 0.054 |
| ENSG00000184402 | SS18L1 | 0.916 | 0.317 | 0.856 | 0.212 |
| ENSG00000172660 | TAF15 | 3.400 | 0.060 | 3.245 | 0.001 |
| ENSG00000143569 | UBAP2L | 6.232 | 0.001 | 5.205 | 0.032 |
| ENSG00000188177 | ZC3H6 | 2.827 | 0.066 | 2.762 | 0.195 |
| ENSG00000198026 | ZNF335 | 0.811 | 0.110 | 0.143 | 0.245 |
| ENSG00000120948 | TARDBP | 4.411 | 0.028 | 3.083 | 0.097 |
| Oocytes from small antral follicles | |||||
| Ensemble | Symbol | FPKM mean | t-test (p-value) | ||
| ENSG00000126705 | AHDC1 | 0.416 | 0.5 | ||
| ENSG00000011243 | AKAP8L | ND | ND | ||
| ENSG00000140488 | CELF6 | ND | ND | ||
| ENSG00000099622 | CIRBP | 2.916 | 0.444 | ||
| ENSG00000111605 | CPSF6 | 0.871 | 0.272 | ||
| ENSG00000149532 | CPSF7 | 2.361 | 0.429 | ||
| ENSG00000071626 | DAZAP1 | 0.294 | 0.5 | ||
| ENSG00000064195 | DLX3 | ND | ND | ||
| ENSG00000182944 | EWS/EWSR1 | 8.638 | 0.149 | ||
| ENSG00000119812 | FAM98A | 0.307 | 0.320 | ||
| ENSG00000182263 | FIGN | 0.238 | 0.5 | ||
| ENSG00000089280 | FUS | 4.790 | 0.253 | ||
| ENSG00000139675 | HNRNPA1L2 | 0.190 | 0.5 | ||
| ENSG00000169813 | HNRNPF | 5.995 | 0.232 | ||
| ENSG00000169045 | HNRNPH1 | 4.243 | 0.050 | ||
| ENSG00000096746 | HNRNPH3 | 4.087 | 0.375 | ||
| ENSG00000165119 | HNRNPK | 3.696 | 0.359 | ||
| ENSG00000125944 | HNRNPR | 6.266 | 0.169 | ||
| ENSG00000105323 | HNRNPUL1 | 2.232 | 0.424 | ||
| ENSG00000176624 | MEX3C | 3.173 | 0.5 | ||
| ENSG00000147140 | NONO | 3.516 | 0.381 | ||
| ENSG00000167005 | NUDT21 | 3.383 | 0.452 | ||
| ENSG00000121390 | PSPC1 | 2.223 | 0.5 | ||
| ENSG00000102317 | RBM3 | 0.069 | 0.5 | ||
| ENSG00000268489 | RBM3 | ND | ND | ||
| ENSG00000173914 | RBM4B | 2.100 | 0.5 | ||
| ENSG00000076053 | RBM7 | ND | ND | ||
| ENSG00000244462 | RBM12 | 4.552 | 0.436 | ||
| ENSG00000239306 | RBM14 | 0.069 | 0.5 | ||
| ENSG00000147274 | RBMX | 3.804 | 0.258 | ||
| ENSG00000020633 | RUNX3 | ND | ND | ||
| ENSG00000116560 | SFPQ | 5.167 | 0.116 | ||
| ENSG00000188529 | SRSF10 | 3.538 | 0.5 | ||
| ENSG00000184402 | SS18L1 | ND | ND | ||
| ENSG00000172660 | TAF15 | 2.933 | 0.442 | ||
| ENSG00000143569 | UBAP2L | 3.791 | 0.457 | ||
| ENSG00000188177 | ZC3H6 | 4.146 | 0.189 | ||
| ENSG00000198026 | ZNF335 | 0.069 | 0.5 | ||
| ENSG00000120948 | TARDBP | 5.918 | 0.025 | ||
Table 3.
Long non-coding RNAs (p < 0.05) in human follicle development.
| Oocytes from primordial follicles | Primordial follicle | ||||
|---|---|---|---|---|---|
| Symbol*1 | FPKM mean | t-test (p-value) | Symbol | FPKM mean | t-test (p-value) |
| PRIMORDIAL FOLLICLES | |||||
| ADCY10P1 | 0.763 | 0.046 | BDNF-AS | 2.395 | 0.017 |
| LINC00485 | 2.7643 | 0.046 | FGD5-AS1 | 3.335 | 0.013 |
| LINC00924 | 0.171 | 0.015 | GLG1 | 2.640 | 0.007 |
| LINC01128 | 5.889 | 0.009 | LINC00221 | 3.800 | 0.012 |
| LINC01511 | 6.533 | 0.019 | LINC00485 | 2.770 | 0.007 |
| LOC100129434 | 6.859 | 0.0036 | LINC00707 | 4.801 | 0.041 |
| LOC100507557 | 3.783 | 0.005 | LINC01483 | 2.976 | 0.040 |
| LOC101927487 | 4.044 | 0.036 | LOC100129434 | 6.765 | 0.012 |
| LOC101928137 | 2.656 | 0.029 | LOC100506885 | 3.548 | 0.028 |
| LOC101929128 | 4.250 | 0.023 | LOC100507156 | 2.776 | 0.013 |
| LOC101929567 | 4.773 | 0.014 | LOC100507557 | 2.443 | 0.018 |
| LOC101929612 | 3.972 | 0.005 | LOC101926943 | 1.428 | 0.008 |
| LOC102467226 | 0.335 | 0.016 | LOC101927487 | 3.216 | 0.035 |
| LOC284798 | 2.494 | 0.002 | LOC101928137 | 1.999 | 0.012 |
| MALAT1 | 7.954 | 0.006 | LOC101929567 | 3.513 | 0.033 |
| MIR3609 | 5.490 | 0.012 | LOC101929612 | 3.197 | 0.005 |
| MIR99AHG | 0.262 | 0.016 | LOC440300 | 1.490 | 0.041 |
| NPY6R | 0.211 | 0.013 | LOC643201 | 4.491 | 0.034 |
| OIP5-AS1 | 4.190 | 0.029 | MALAT1 | 8.992 | 0.026 |
| RN7SK | 11.285 | 0.005 | MGC32805 | 0.219 | 0.039 |
| RN7SL2 | 8.794 | 0.012 | MIR4426 | 0.622 | 0.018 |
| RPS3A | 1.279 | 0.021 | OIP5-AS1 | 5.527 | 0.033 |
| UGDH-AS1 | 6.637 | 0.013 | RN7SK | 11.227 | 0.001 |
| XIST | 5.393 | 0.026 | RN7SL2 | 8.9514 | 0.001 |
| RPL13AP5 | 2.322 | 0.002 | |||
| RPL21P28 | 0.582 | 0.012 | |||
| RPS3A | 1.118 | 0.043 | |||
| SCARNA7 | 5.888 | 0.039 | |||
| SLC8A1-AS1 | 5.409 | 0.007 | |||
| SYN2 | 4.108 | 0.043 | |||
| TUNAR | 3.893 | 0.047 | |||
| UBXN8 | 2.491 | 0.050 | |||
| UGDH-AS1 | 6.382 | 0.006 | |||
| XIST | 8.236 | 0.003 | |||
| ZFAS1 | 4.296 | 0.014 | |||
| ZNF252P | 3.537 | 0.038 | |||
| Oocytes from primary follciles | Primary follicle | ||||
| Symbol | FPKM mean | t-test (p-value) | Symbol | FPKM mean | t-test (p-value) |
| PRIMARY FOLLICLES | |||||
| BCAR4 | 4.563 | 0.008 | CEACAM22P | 0.182 | 0.033 |
| LINC00485 | 1.902 | 0.002 | GLG1 | 1.270 | 0.046 |
| LINC00665 | 4.298 | 0.046 | KIZ | 4.874 | 0.014 |
| LINC01511 | 4.625 | 0.016 | LINC01511 | 5.425 | 0.028 |
| LOC100129434 | 6.444 | 0.011 | LOC100129434 | 5.792 | 0.047 |
| LOC100506885 | 2.187 | 0.039 | LOC100507557 | 1.854 | 0.041 |
| LOC101926943 | 1.548 | 0.044 | LOC101927487 | 3.822 | 0.007 |
| LOC101927337 | 2.226 | 0.014 | LOC101929567 | 4.053 | 0.049 |
| LOC101929491 | 0.330 | 0.004 | LOC101929612 | 3.456 | 0.010 |
| LOC101929567 | 4.287 | 0.008 | MALAT1 | 9.345 | 0.016 |
| LOC101929612 | 3.152 | 0.016 | NEXN-AS1 | 0.534 | 0.046 |
| LOC102546299 | 2.551 | 0.001 | OIP5-AS1 | 5.950 | 0.017 |
| MALAT1 | 9.614 | 0.001 | RN7SK | 11.308 | 0.004 |
| MEG3 | 0.220 | 0.024 | RN7SL2 | 8.743 | 0.001 |
| MIR3609 | 3.887 | 0.012 | RPL13AP5 | 2.877 | 0.003 |
| MIR4426 | 0.595 | 0.020 | RPL21P28 | 0.868 | 0.008 |
| RN7SK | 11.845 | 0.001 | RPL21P28 | 2.165 | 0.032 |
| RN7SL2 | 9.110 | 0.004 | SNORD89 | 1.513 | 0.034 |
| RPL13AP5 | 1.418 | 0.011 | UGDH-AS1 | 6.216 | 0.016 |
| RPL21P28 | 0.916 | 0.047 | XIST | 7.610 | 0.030 |
| RPS3A | 0.894 | 0.007 | ZFAS1 | 5.249 | 0.005 |
| SCARNA7 | 5.424 | 0.009 | |||
| UGDH-AS1 | 6.397 | 0.003 | |||
| XIST | 6.751 | 0.014 | |||
| ZNF518A | 2.983 | 0.018 | |||
| Mural granulosa cell | Theca cells | ||||
| Symbol | FPKM mean | t-test (p-value) | Symbol | FPKM mean | t-test (p-value) |
| SMALL ANTRAL | |||||
| ANP32AP1 | 0.683 | 0.009 | ANKRD36B | 2.277 | 0.020 |
| CASP8AP2 | 3.530 | 0.034 | BCYRN1 | 2.661 | 0.034 |
| CROCCP2 | 1.453 | 0.006 | BDNF-AS | 2.317 | 0.033 |
| EBLN3 | 3.812 | 0.002 | CD27-AS1 | 2.007 | 0.041 |
| FGD5-AS1 | 4.032 | 0.019 | CLEC2D | 1.283 | 0.027 |
| GOLGA6L5P | 1.750 | 0.015 | CSNK1A1 | 2.753 | 0.001 |
| H3F3AP4 | 2.121 | 0.027 | CTBP1-AS2 | 2.457 | 0.028 |
| LINC00657 | 5.467 | 0.009 | FGD5-AS1 | 3.506 | 0.045 |
| LINC01128 | 4.413 | 0.018 | FLJ42627 | 0.239 | 0.000 |
| LINC01420 | 2.152 | 0.012 | H3F3AP4 | 2.185 | 0.014 |
| LOC100129434 | 3.831 | 0.025 | HCG18 | 3.976 | 0.035 |
| LOC100131564 | 2.425 | 0.024 | HERC2P3 | 0.111 | 0.017 |
| LOC100507557 | 1.904 | 0.028 | LINC00485 | 1.467 | 0.049 |
| LOC101927027 | 1.875 | 0.003 | LINC00657 | 4.740 | 0.009 |
| LOC101929124 | 3.0195 | 0.010 | LINC01128 | 3.643 | 0.020 |
| LOC101929612 | 3.418 | 0.007 | LINC01133 | 0.111 | 0.017 |
| LOC102477328 | 0.135 | 0.020 | LOC100129434 | 4.893 | 0.008 |
| LOC150776 | 3.000 | 0.002 | LOC101929612 | 3.455 | 0.001 |
| LOC643201 | 3.740 | 0.013 | LOC102724699 | 0.420 | 0.021 |
| LOC646762 | 2.431 | 0.008 | MAGI2-AS3 | 2.791 | 0.020 |
| LOC728554 | 1.619 | 0.023 | MALAT1 | 10.73 | 0.000 |
| MALAT1 | 9.495 | 0.006 | MEG3 | 7.602 | 0.001 |
| MIR3609 | 5.033 | 0.001 | MIR3609 | 4.974 | 0.009 |
| MIR99AHG | 3.823 | 0.014 | MIR4426 | 0.504 | 0.043 |
| OIP5-AS1 | 5.926 | 0.003 | MIR99AHG | 5.083 | 0.008 |
| PCBP1-AS1 | 2.541 | 0.029 | NCBP2-AS2 | 0.824 | 0.001 |
| PIGBOS1 | 1.804 | 0.011 | NEXN-AS1 | 0.735 | 0.025 |
| PKI55 | 4.600 | 0.000 | NKAPP1 | 3.545 | 0.032 |
| RN7SK | 10.395 | 0.002 | OIP5-AS1 | 4.636 | 0.039 |
| RN7SL2 | 8.088 | 0.004 | PCBP1-AS1 | 4.558 | 0.009 |
| RPL13AP5 | 2.509 | 0.006 | PDIA3P1 | 0.450 | 0.016 |
| RPL21P28 | 2.293 | 0.012 | PGM5P2 | 1.302 | 0.028 |
| RPL21P28 | 0.589 | 0.026 | PKI55 | 4.791 | 0.024 |
| RPL34P6 | 0.405 | 0.013 | PTOV1-AS1 | 2.060 | 0.001 |
| RPS3A | 2.326 | 0.001 | RN7SK | 10.57 | 0.005 |
| SCAND2P | 1.312 | 0.014 | RN7SL2 | 8.740 | 0.006 |
| SCARNA7 | 5.104 | 0.016 | RNU4-2 | 4.075 | 0.007 |
| SNHG17 | 3.099 | 0.018 | RPL13AP5 | 2.816 | 0.011 |
| TMEM120A | 3.042 | 0.002 | RPL21P28 | 2.840 | 0.006 |
| TUG1 | 6.0234 | 0.003 | RPL21P28 | 1.081 | 0.017 |
| UGDH-AS1 | 3.874 | 0.031 | RPL34P6 | 0.844 | 0.019 |
| XIST | 8.056 | 0.004 | RPS3A | 1.846 | 0.010 |
| ZFAS1 | 5.124 | 0.005 | SCARNA7 | 5.588 | 0.026 |
| ZNF761 | 4.206 | 0.017 | SDHAP2 | 2.223 | 0.001 |
| ZNF826P | 0.990 | 0.018 | SH3BP5-AS1 | 2.069 | 0.008 |
| SNORA23 | 0.659 | 0.007 | |||
| SNORA79 | 1.713 | 0.039 | |||
| SNORD89 | 2.354 | 0.017 | |||
| SPON1 | 3.979 | 0.030 | |||
| THUMPD3-AS1 | 0.829 | 0.049 | |||
| TMEM120A | 1.996 | 0.042 | |||
| TUG1 | 6.154 | 0.008 | |||
| UBXN8 | 4.509 | 0.006 | |||
| UGDH-AS1 | 4.596 | 0.000 | |||
| XIST | 8.474 | 0.001 | |||
| ZFAS1 | 4.881 | 0.013 | |||
| ZNF252P | 1.822 | 0.012 | |||
| ZNF518A | 2.901 | 0.020 | |||
| ZSCAN26 | 3.800 | 0.007 | |||
| Oocyte from small antral follicle | |||||
| Symbol | FPKM mean | t-test (p-value) | |||
| LOC100653061 | 5.033 | 0.033 | |||
| MALAT1 | 7.219 | 0.027 | |||
| OIP5-AS1 | 0.842 | 0.007 | |||
| PMS2CL | 0.481 | 0.008 | |||
| RN7SK | 10.140 | 0.021 | |||
| ROR1-AS1 | 0.481 | 0.008 | |||
Genes alphabetically sorted.
*2Presented in heat map (Figure 2).
Immunofluorescence microscopy
Human ovarian cortical tissue was cut in 5 μm sections and mounted on glass slides. Dehydration and antigen retrieval was performed as described elsewhere (Stubbs et al., 2005) followed by serum block (30 min), then the primary antibody; (1/200) anti-TAFII68 Rabbit pAb (Bethyl Laboratories, #IHC-00094), (1/500) anti-FUS Rabbit pAb (Bethyl Laboratories, #A300-302A), or (1/200) anti-EWS Rabbit pAb (Bethyl Laboratories, #IHC-00086) overnight at 4°C. The sections were then incubated in a 1:700 dilution of secondary antibody (Donkey-anti-Rabbit) conjugated with Alexa Fluor 488 Dye (Life Technologies). Finally, sections were incubated in 1/7500 Hoechst (Life Technologies) followed by mounting with Dako Fluorescent Mounting Medium (Agilent Technologies, Santa Clara, CA, U.S.A.) and analyzed using a LSM510 laser-scanning confocal microscope using a 63x C-Apochromat water immersion objective NA 1.2 (Carl Zeiss, Göttingen, Germany) and ZEN 2011 software (Carl Zeiss, Göttingen, Germany).
Results
Laser-isolation of oocytes and somatic cells during human follicle development
Specific isolates of oocytes and follicles (oocytes with surrounding somatic granulosa cells) from the primordial and primary stage, respectively, as well as oocytes, mural granulosa cells, and theca cells from small antral follicles were collected via Laser Capture Microdissection (LCM). Each stage was isolated on the basis of stringent morphological criteria (Gougeon, 1996) (Figure 1). Primordial follicles were defined as an oocyte surrounded by one layer of flattened granulosa cells (Figure 1) and primary follicles were defined as an oocyte surrounded by a single layer of cubic granulosa cells (Figure 1). Small antral follicles were defined based on the presence of a follicular antrum with a clear distinction between the oocyte, the mural granulosa cells and the theca cell layer (Figure 1) The samples (1,473 isolates) of cells from primordial, primary, and small antral follicles were pooled into 20 samples (Table 1, Figure 1). These 20 samples were then subjected to RNA sequencing using the IlluminaHiSeq2000 sequencing platform (Illumina Inc., San Diego, CA, U.S.A.) at an external sequencing facility (AROS Applied Biotechnology, Aarhus, Denmark). We previously validated the expression pattern for various RNAs in the present RNA seq. dataset using RT-qPCR (Ernst et al., 2017, 2018). The RNA sequencing yielded on average 35.3 million reads per sample (range: 31.8–39.6 million reads) and was mapped to the human genome (hg19) (average number of reads mapped: 31.7 million, range: 29.4–34.0). Gene expression was calculated as FPKM by a custom R script (Ernst et al., 2017, 2018).
Transcriptional profiles of genes encoding paraspeckle proteins across different follicle stages
The expression of 39 genes encoding paraspeckle proteins (Naganuma et al., 2012) was interrogated during human follicle development (Table 2). The highest expression of paraspeckle genes in the primordial follicle stage, based on FPKM values, were EWS, HNRNPK, ZC3H6, UBAP2L, and TARDBP (Table 2). Several other genes encoding paraspeckle proteins were present (e.g., MEX3C, FUS, TAF15, CPSF6, NUDT21, RBM12, RBMX, DLX3).
Interestingly, the expression of ZC3H6 appears to be downregulated from primordial to primary follicles, indicating a specific function associated with the primordial follicle. The EXSR1 gene expression remains high and upregulated in small antral follicles. NONO was noted to be upregulated as follicle development advances, with the highest expression detected in the somatic cells in the small antral follicle (Table 2).
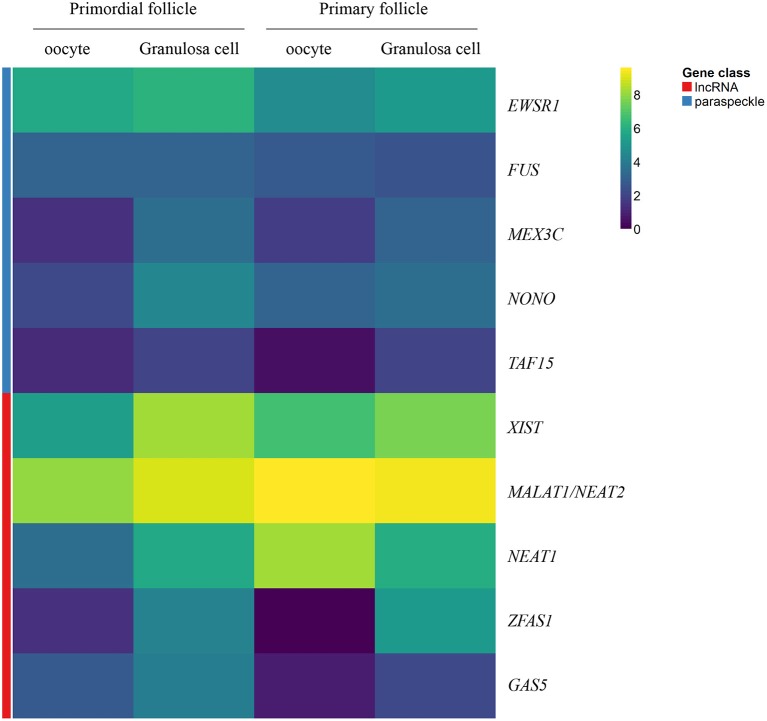
A heatmap of FPKM data for selected genes encoding paraspeckle genes was generated to show the expression for the two different cell-stages in isolates - and the correlation between cell-specific isolates (Figure 2).
Figure 2.
Heatmap representation of selected genes encoding paraspeckle proteins and lncRNA genes. Heatmap of gene expression in oocytes and granulosa cells during the primordial-to-primary follicle transition. The heatmap includes the paraspeckle-encoding genes (blue bar) MEX3C, NONO, FUS, EWS, TAF15 transcripts, and the lncRNAs (red bar) NEAT1, MALAT1 (NEAT2), XIST, ZFAS1, GAS5. Color code reflects average FPKM values.
Intra-ovarian distribution of paraspeckle proteins TAF15, EWS, and FUS
The gene products of TAF15, EWS, and FUS were selected for immunofluorescent staining (IMF) (bold in Table 2) to reveal their localization in human ovarian sections.
The TAF15 translational product was expressed in both oocytes and follicles from primordial, primary, and small antral stages, with a particular high expression in oocytes from primordial follicles, as well as in primordial follicles (Table 2). We interrogated the TAF15 protein using a specific antibody toward TAF15. This showed detection of the TAF15 protein in both oocyte and granulosa cells of primordial (Figure 3A), primary (Figure 3B), secondary (Figure 3C), as well as small pre-antral/early antral follicles (Figures 3D–F). As the TAF15 protein appears detectable in both oocytes and the surrounding somatic cells are in line with the RNA sequencing data, gene expression and its translational product appears coupled.
Figure 3.
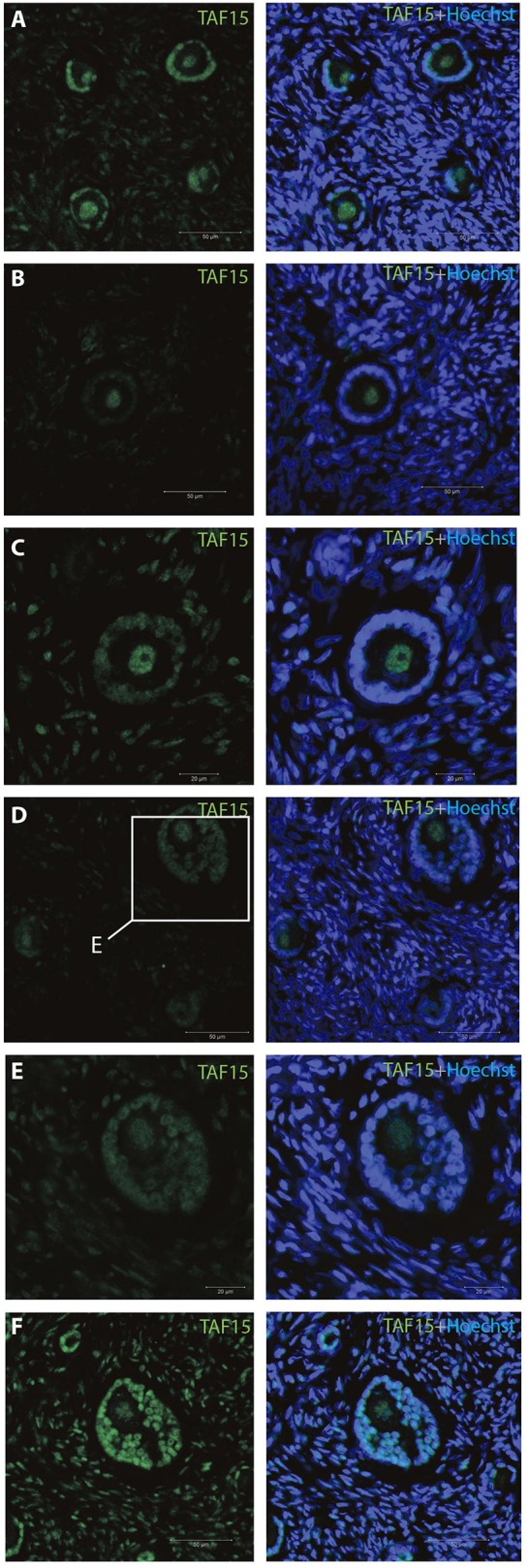
Intra-ovarian distribution of TAF15 in human primordial and primary follicles. This showed detection of the TAF15 protein in both oocyte and granulosa cells of (A) primordial, (B) primary, and (C) secondary, as well as (D–F) small pre-antral/early antral follicles. Hoechst staining identifies the nucleus of cells. Scale bars; 30 μm.
The IMF of EWS showed that EWS is present in both oocytes and the surrounding granulosa cells, and in primordial and primary follicles (Figures 4A–C). The EWS transcript was found highly expressed in these early stages of follicle development, and thus the RNA expression appears coupled to its translational protein product.
Figure 4.
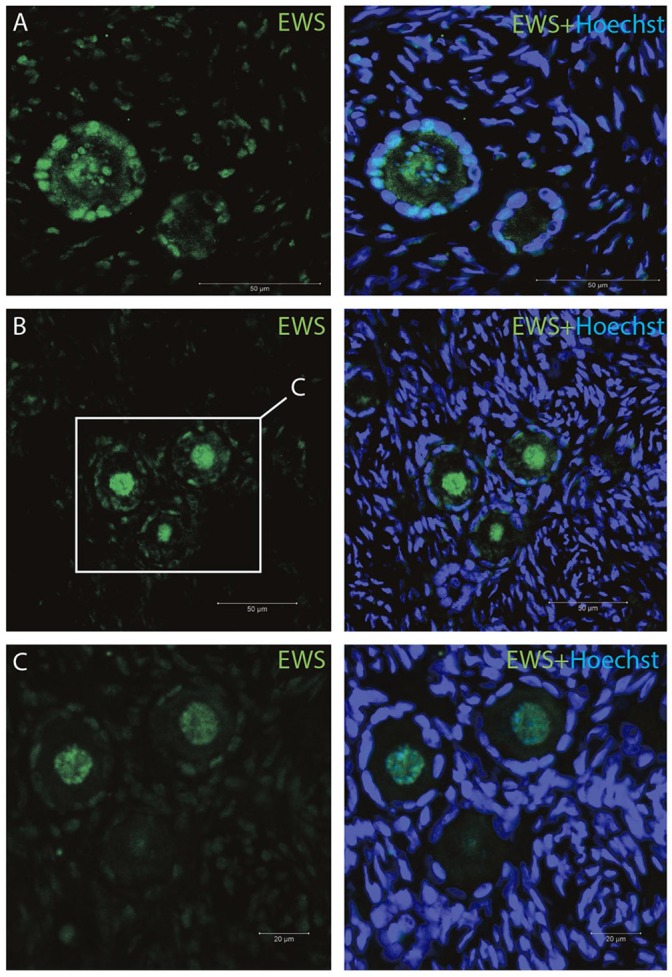
Intra-ovarian distribution of EWS in human primordial and primary follicles (A-C). EWS is present in both oocytes and the surrounding granulosa cells in primordial and primary follicles. Hoechst staining identifies the nucleus of cells. Scale bars; 30 μm.
The FUS transcript was also highly expressed during early follicle development (Table 2), and as we interrogated its protein using IMF, found that the FUS protein was detectable in primordial follicles, (Figure 5A), primary follicles (Figures 5B, C), as well as in late pre-antral/early antral follicles (Figure 5C).
Figure 5.
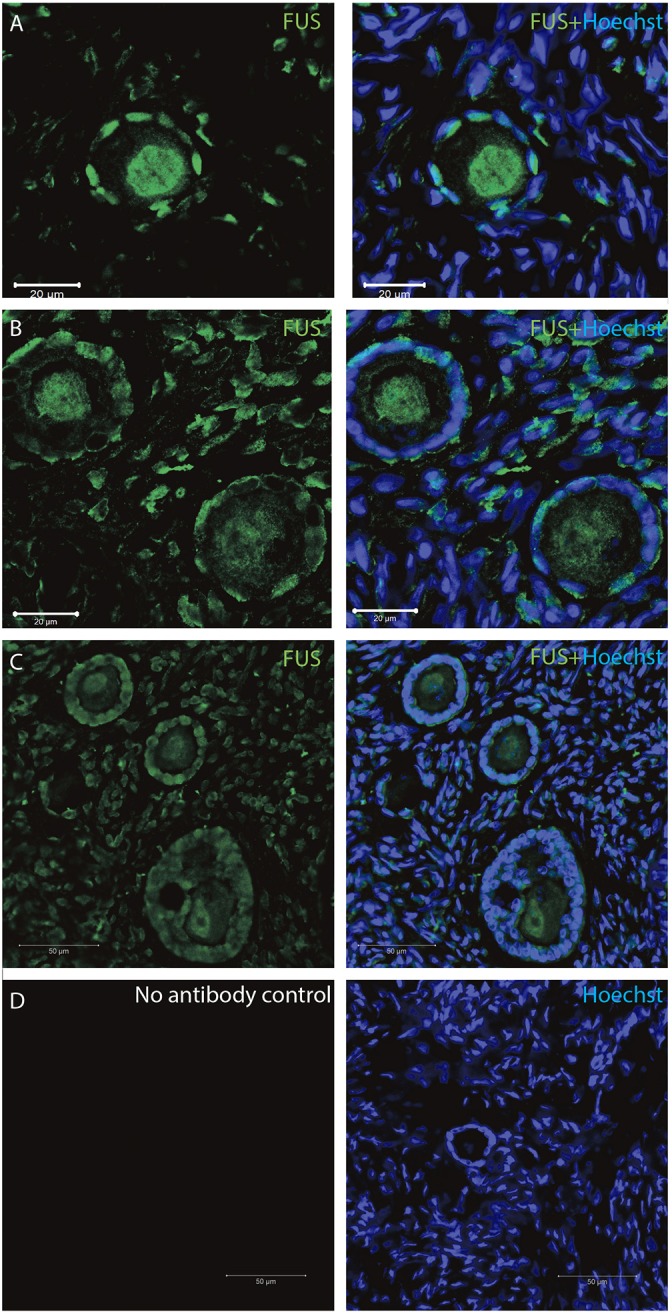
Intra-ovarian distribution of FUS in human primordial, and primary follicles. The FUS protein was detectable in (A) primordial follicles, (B,C) primary follicles (C) late pre-antral/early antral follicles. All samples were compared to a (D) no-antibody control. Hoechst staining identifies the nucleus of cells. Scale bars; 30 μm.
All samples were compared to a no-antibody control, which did not detect any signal (Figure 5D).
We detected nuclear localization of TAF15, EWS and FUS, with evidence of speckle-like structures in an infrequent manner distributed throughout the cells.
Non-coding RNAs
ncRNA genes produce functional RNA molecules rather than encoding proteins (Eddy, 2001). The groups of ncRNA are diverse and include, for instance, short and long ncRNAs as well as micro (mi)RNA, snoRNA, scaRNA, SRP RNA, and antisense RNA. The presence of ncRNAs during human follicle development was analyzed in transcriptomes representative for oocytes and follicles from primordial and primary follicles as well as oocytes, mural granulosa cell layers, and theca cell layers of small antral follicles [Figure 1, Table 1, (Resource Data)]. Genes encoding ncRNAs was identified using the Ensembl gene annotation version GRCh37.p13.
Long ncRNAs
Most ncRNAs longer than 200 nucleotides are referred to as ‘long non-coding RNAs’ (lncRNAs). Although the estimated number of different types of human lncRNAs has ranged from 5,400 to 53,000 (Palazzo and Lee, 2015), these ncRNAs appear to comprise functions for the control of various levels of gene expression in physiology and development, including chromatin architecture/epigenetic memory, transcription, RNA splicing, editing, translation, and turnover (Mattick and Makunin, 2006). In this study the presence of lncRNAs was interrogated (Table 3).
Interestingly, we detected the lncRNA XIST during human follicle development in both oocytes and follicle samples (Table 3). It should be noted, however, that XIST in the oocytes from small antral follicles did not display a cell-specific consistently expressed expression pattern but was noted used a less stringent p-value (Resource Data), which was likewise observed for TSIX (Table 3). The fact that a less stringent p-value was needed to detect this transcript in the oocytes from small antral follicles was expected, as these oocytes were rarely found in the ovarian biopsies, and thus these oocyte samples are less represented (Table 1, Figure 1).
MALAT1 (NEAT2) was detected throughout all the included stages (Resource Data), indicating the need for this paraspeckle-forming protein during human follicle development. Interestingly, several lncRNAs with no biological functions annotated were noted (Resource Data). While the expression of several lncRNA genes (OIP5-AS1, RN7SK, RN7SL2) was present in all samples tested, others appeared to be cell- and stage-specifically (GLG1, KIZ, BCAR4, EBLN3) expressed (Resource Data).
The lncRNA ZFAS1 appears to be restricted to somatic cells, e.g., the mural granuloma cell layer and the theca cell layer in the small antral follicle (Resource Data). Interestingly, the ROR1-AS1 seems to be specific to the oocyte from small antral follicles (Resource Data). We found lncRNA Growth Arrest Specific 5 (GAS5) expressed in oocytes from primordial follicles, as well as a high expression in primordial follicles, somewhat lower in primary follicles, and in turn high in the mural granulosa cell layer and the theca cell layer from small antral follicles (Resource Data).
A heatmap of FPKM data for selected lncRNA genes was generated to show the expression for the two different cell-stages (primordial versus primary) in isolates - and the correlation between cell-specific (oocyte versus granulosa cell) isolates (Figure 2).
Discussion
Extensive efforts to gain deeper understanding of RNA biology have yielded evidence of the diverse structural and regulatory roles in protecting chromosome integrity, maintaining genomic architecture, X chromosome inactivation, imprinting, transcription, translation and epigenetic regulation (Khorkova et al., 2015). Bioinformatics analysis of chromatin marks in intergenic DNA regions and of expressed sequence tags (ESTs) predicts the existence of more than 5,000 long noncoding RNA (lncRNA) genes in the human genome (Gomez et al., 2013). Some studies have found the number of lncRNAs to exceed that of protein-coding genes (Bouckenheimer et al., 2016; Hon et al., 2017). In our transcriptome study of lncRNA, we applied a strict filter to only consider transcript that were consistently expressed in our samples. This was applied as a major limiting factor is the number of patients included in the study. In oocytes and granulosa cells from primordial and primary follicles, 20, 33, and 20 and 19 lncRNAs were noted expressed (using a cut of value of 1 FPKM). Interestingly comparing this to the number of protein coding transcripts in the same stages (oocytes and granulosa cells from primordial and primary follicles showed 1099, 1695, and 1046 and 815, SSCEG, respectively (Ernst et al., 2017, 2018), it is noteworthy that few lncRNAs compared to the protein coding transcript are present during these early stages in human follicle development.
Strict filters in the bioinformatic management was applied to this study to ensure the most precise outcome from the global transcriptome analysis. Previous studies validated selected candidates by qPCR analysis (Ernst et al., 2017, 2018). Moreover, the analysis contains several DEG-lists based on both SSCEGs and non-SSCEGs and caution in the analysis of fold of change for DEG transcripts is recommended. Importantly, this study analyzed the presence of transcripts, and whether a gene is translated or its protein product present, is unknown. Using single cell techniques, we confirmed the presence of selected paraspeckle proteins using immunohistochemistry.
In a few well-studied cases, such as AIR, XIST, and HOTAIR, these lncRNAs have been shown to operate at the transcriptional level by binding to proteins in histone-modifying complexes and targeting them to particular genes (Nagano et al., 2008; Chu et al., 2011; Jeon and Lee, 2011; Wang and Chang, 2011). A role for lncRNAs in human follicle development has not previously been described (Wilhelmm and Bernard, 2016) although their potential involvement has been suggested (Zhao and Rajkovic, 2008; Bouckenheimer et al., 2016). Differential lncRNA expression profiles in human oocytes and cumulus cells was recently analyzed (Bouckenheimer et al., 2018), which determined the lncRNA expression profiles of human MII oocytes (BCAR4, C3orf56, TUNAR, OOEP-AS1, CASC18, and LINC01118) and cumulus cells (NEAT1, MALAT1, ANXA2P2, MEG3, IL6STP1, and VIM-AS1).
The presence of the paraspeckle-forming NEAT1 and MALAT1 (NEAT2) indicates that paraspeckles are actively formed and present during human follicle development. Paraspeckle formation is initiated by transcription of the NEAT1 chromosomal locus and proceeds in conjunction with NEAT1 lncRNA biogenesis and a subsequent assembly step involving >39 paraspeckle proteins (PSPs). Interestingly, a study has shown that subunits of SWItch/Sucrose NonFermentable (SWI/SNF) chromatin-remodeling complexes were identified as paraspeckle components that interact with PSPs and NEAT1 lncRNA (Kawaguchi et al., 2015). In particular, it was shown by electron microscopy that SWI/SNF complexes were enriched in paraspeckle subdomains depleted of chromatin. Interestingly, and consistent with this, it was found that the arginine methyltransferase CARM1 (coactivator-associated arginine methyltransferase 1) promotes the nuclear export of mRNAs that contain inverted Alu elements in their 3' untranslated region by methylating the paraspeckle component p54(nrb), which reduces the binding of p54(nrb) to the inverted Alu elements. It also down-regulated the synthesis of NEAT1. This in turn inhibited paraspeckle formation (Elbarbary and Maquat, 2015).
The lncRNA XIST was present at high levels throughout the stages tested during human follicle development. To ensure X-linked gene dosage compensation between females (XX) and males (XY), one X chromosome randomly undergoes X chromosome inactivation (XCI) in female cells (Lyon, 1961). The human XIST (Brown et al., 1991a,b) and mouse Xist (Borsani et al., 1991; Brockdorff et al., 1991) lncRNAs accumulate over the X chromosome. X chromosomal inactivation is tightly regulated throughout development with XIST as a key regulator involved in the establishment of several layers of repressive epigenetic modifications. These reported functions of XIST are consistent with our observation that this gene is highly transcribed during human follicle development and reveals that XIST lncRNA is present already from the dormant primordial stage of human follicle development. The functional role of XIST during early follicle development remains to be elucidated, and this may include early marks of maternal imprinting and dosage regulation.
The lncRNA ZFAS1 appears specific to somatic cells during human follicle development. ZFAS1 has been described as being upregulated in different cancer types (Askarian-Amiri et al., 2011; Li et al., 2015; Nie et al., 2016; Thorenoor et al., 2016) and is involved in cell apoptosis and cell cycle control. It was recently shown that the action of ZFAS1 occurred through interaction with EZH2 and LSD1/CoREST in order to repress the underlying targets KLF2 and NKD2 transcription (Nie et al., 2016). The epigenetic dysregulation of central granulosa cell factors such as FOXL2 are involved in the development of granulosa cell tumors (Xu et al., 2016), which is possible through EZH2 interaction. Furthermore, prominent roles for FOXL2 include control of primordial follicle activation (Schmidt et al., 2004).
It remains to be tested if lncRNA ZFAS1 functions to regulate transcriptional control in follicle development, and this may have an effect on granulosa cell proliferation and cell cycle control in the human follicle.
We identified the potential tumor suppressor lncRNA growth arrest specific 5 (GAS5), expressed particularly in the primordial stage, as well as in primary follicles and in mural granulosa cell layers and the theca cell layers. Part of the GAS5 RNA structure mimics the glucocorticoid response element, enabling it to bind the DNA binding domain of the glucocorticoid receptor, thus inhibiting glucocorticoid induced transcription. GAS5 is further thought to regulate transcriptional activity of the androgen receptor. In line with this, GAS5 lncRNA has been found to repress the AR/androgen complex from binding to target through sequestering, thus repressing transcription (Wang and Lee, 2009). Of further interest, GAS5 lncRNA has been found to supress the AKT/mTOR signaling pathway in prostate cancer cells (Yacqub-Usman et al., 2015). As previous studies have shown that activated AKT/mTOR signaling increases primordial follicle activation (Makker et al., 2014), we suggest that GAS5 expression in the primordial follicle may be involved in primordial follicle dormancy and survival. Recently, the GAS5 was found to promote proliferation and survival of female germline stem cells in vitro (Wang et al., 2018). The functional involvement of GAS5 in normal and aberrant human follicle development remains to be determined.
Increasing evidence supports a central role for ncRNA in numerous aspects of chromatin function (Názer and Lei, 2014). Interestingly, it has long been appreciated that ncRNAs are central components of the dosage compensation machinery, and recent work has elucidated how various ncRNAs contribute to Polycomb Group (PcG) and chromatin insulator activities (reviewed in Názer and Lei, 2014). The PcG proteins are required for the adequate development of multicellular organisms, functioning to preserve pluripotency and/or cellular identity. Their main function however is to repress the expression of genes that would otherwise promote differentiation into other cell types (reviewed in Simon and Kingston, 2013).
The precise role of lncRNAs in chromatin modifications during human follicle development remains to be elucidated. However, our data suggests that several distinct lncRNAs are present and that they probably have separate functions in order to secure follicle integrity and development.
We found the protein-coding MEX3C gene present during human follicle development. However, the role of the gene product, MEX3C, is unknown. The MEX3BM isoform and the E3 ubiquitin ligase DZIP3 are bought together with their substrates (Ataxin-1 and Snurportin-1) by the lncRNA HOTAIR, accelerating their degradation (Khorkova et al., 2015), thus lncRNA-mediated regulation also affects protein stability (reviewed in Khorkova et al., 2015). Furthermore, proteasomal inhibition causes upregulation of paraspeckle-associated lncRNA NEAT1, which in turn protects fibroblasts from cell death triggered by proteasome inhibition (Khorkova et al., 2015). Interestingly, it was found that a MEX3 homolog is required for differentiation during planarian stem cell lineage development (Zhu et al., 2015). In this study it was shown that MEX3-1 was required for generating differentiated cells of multiple lineages, while restricting the size of the stem cell compartment. This indicates that MEX3-1 functions as a cell fate regulator (Zhu et al., 2015). The presence of the MEX3C transcript during human follicle development has not been functionally addressed, and future studies should reveal whether MEX3C has a pivotal role in cell commitment and/or differentiation in the selection of the dominant follicle.
The upregulated expression profile of NONO during follicle development suggests that this protein is under tight control. NONO deficiency led to upregulation of PSPC1, which replaces NONO in a stable complex with SFPQ (Li et al., 2014). The knockdown of PSPC1 in a Nono-deficient background led to severe radio-sensitivity and delayed resolution of double stranded break (DSB) repair foci. From this it can be concluded that NONO or related proteins are critical for DSB repair (Li et al., 2014). The complex of NONO with SFPQ and PSPC1 served a multipurpose scaffold, including frequently identified engagement at almost every step of gene regulation, and including, but not limited to, transcriptional regulation, RNA processing and transport, and DNA repair (Knott et al., 2016). Interestingly, a report has investigated the inner cell mass marker OCT4 and its gene expression patterns, as well as CpG sites methylation profiles during embryonic stem (ES) cell differentiation into neurons (Park et al., 2013). It was found that NONO binds to the CpG island of the Oct4 promoter and positively regulates Oct4 gene expression in ES cells (Park et al., 2013), thus indicating a role in cell lineage during early development. The future role of NONO during human follicle development and how this might participate in regulating gene expression and/or DNA repair will be important steps toward the understanding of the capacity of the human ovarian follicles.
Several lines of evidence suggest paraspeckle proteins to be essential in cell fate determination, which is highly relevant for early developmental processes (Yamazaki and Hirose, 2015).
FUS, EWS, and TAF15 are structurally similar multifunctional proteins that were initially discovered in the process of characterization of fusion oncogenes in human sarcomas and leukemias. As they are implicated in numerous central cellular processes such as gene regulation, genomic integrity maintenance and mRNA/microRNA processing, it is therefore not surprising to find them in many cellular contexts and in different cell types and tissues. The expression profile of the FET proteins were characterized in both the human (Andersson et al., 2008) and porcine (Blechingberg et al., 2012b) developing brain. The FET proteins are expressed in most human tissues and are localized mainly in the cell nucleus (Andersson et al., 2008), but are also found in the cytoplasm (Zinszner et al., 1997; Belyanskaya et al., 2001; Jobert et al., 2009). This is supported by the fact that the functions of hnRNPs include nucleocytoplasmic shuttling (Bedford and Clarke, 2009; Yu, 2011). Interestingly, FUS, EWS, and TAF15 has previously revealed a cell-specific expression pattern (Andersson et al., 2008), and processes such a heat shock and/or oxidative stress induce the re-localization of these proteins to stress granules (Andersson et al., 2008; Blechingberg et al., 2012a). The fact that we observed infrequent staining in nuclear and cytoplasmic localizations supports the activity of these FET proteins.
The FET proteins also frequently exhibit gene translocation in human cancers (Paronetto, 2013; Campos-Melo et al., 2014). Emerging evidence demonstrates their physical interactions with DNA Damage Response proteins (Kai, 2016) and thus suggests their involvement in the maintenance of genome stability. Interestingly, it was recently proposed that FET proteins are involved in the maintenance of lifespan, cellular stress resistance, and neuronal integrity (Therrien et al., 2016).
It has been shown that FUS interact directly with NEAT1 lncRNA, reducing the expression of FUS, and subsequently causing cell apoptosis. In combination with miR-548ar-3p, this regulates breast cancer cell apoptosis (Ke et al., 2016).
Conclusion
We identified the presence of lncRNAs as well as the genes encoding the paraspeckle proteins, offering insights into how their transcripts are expressed during human follicle development. The study is descriptive in nature. As a proof of concept, we probed for the intracellular presence and localization of three selected paraspeckle proteins. It remains to be determined for several other proteins encoding by genes noted, as well as lncRNAs. In particular, our study indicates that they may be involved in cellular processes such as cell differentiation and cell integrity. This could be accomplished by their ability to control gene expression, epigenetics and mRNA turnover during follicle development.
Author contributions
EE and KL-H conceived the study. PV performed bioinformatics analysis. EE, JN, and MI performed IMF. EE and KL-H analyzed RNA sequencing data and wrote the manuscript. All authors approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank the clinical, paramedical and laboratory team of the Fertility Clinic, Aarhus University Hospital. We thank Lykke-Hartmann laboratory (AU) for scientific discussions. Anders Heuck is also thanked for excellent laboratory work.
Footnotes
Funding. EE was supported by a Ph.D. scholarship from Health Faculty, Aarhus University. This work was further supported by grants from Kong Christian Den Tiendes Fond (to EE and KL-H), Th. Maigaards Eft. Fru Lily Benthine Lunds Fond and Fonden til Lægevidenskabens Fremme (to KL-H). KL-H was further supported by a grant from the Novo Nordisk Foundation (NNF16OC0022480 to KL-H).
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2018.00078/full#supplementary-material
Full lists of ncRNAs detected in human follicle development [oocytes from primordial follicles (tab 1), primordial follicles (tab 2), oocytes from primary follicles (tab 3), mural granulosa cell layers from small antral follicles (tab 4), theca cell layers from small antral follicles (tab 5), oocytes from small antral follicles (tab 6)]. Cell specific consistently expressed genes (CSCEG) (one-sample t-test p-value < 0.05 in gray).
References
- Adhikari D., Liu K. (2009). Molecular mechanisms underlying the activation of mammalian primordial follicles. Endocr. Rev. 30, 438–464. 10.1210/er.2008-0048 [DOI] [PubMed] [Google Scholar]
- Ambasudhan R., Talantova M., Coleman R., Yuan X., Zhu S., Lipton S. A., et al. (2011). Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell 9, 113–118. 10.1016/j.stem.2011.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson M. K., Ståhlberg A., Arvidsson Y., Olofsson A., Semb H., Stenman G., et al. (2008). The multifunctional FUS, EWS and TAF15 proto-oncoproteins show cell type-specific expression patterns and involvement in cell spreading and stress response. BMC Cell Biol. 9:37. 10.1186/1471-2121-9-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askarian-Amiri M. E., Crawford J., French J. D., Smart C. E., Smith M. A., Clark M. B., et al. (2011). SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA 17, 878–891. 10.1261/rna.2528811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auboeuf D., Dowhan D. H., Dutertre M., Martin N., Berget S. M., O'Malley B. W. (2005). A subset of nuclear receptor coregulators act as coupling proteins during synthesis and maturation of RNA transcripts. Mol. Cell. Biol. 25, 5307–5316. 10.1128/MCB.25.13.5307-5316.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backofen R., Vogel T. (2014). Biological and bioinformatical approaches to study crosstalk of long-non-coding RNAs and chromatin-modifying proteins. Cell Tissue Res. 356, 507–526. 10.1007/s00441-014-1885-x [DOI] [PubMed] [Google Scholar]
- Baird D. T., Collins J., Egozcue J., Evers L. H., Gianaroli L., Leridon H., et al. (2005). Fertility and ageing. Hum. Reprod. Update 11, 261–276. 10.1093/humupd/dmi006 [DOI] [PubMed] [Google Scholar]
- Bedford M. T., Clarke S. G. (2009). Protein arginine methylation in mammals: who, what, and why. Mol. Cell. 33, 1–13. 10.1016/j.molcel.2008.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyanskaya L. L., Gehrig P. M., Gehring H. (2001). Exposure on cell surface and extensive arginine methylation of ewing sarcoma (EWS) protein. J. Biol. Chem. 276, 18681–18687. 10.1074/jbc.M011446200 [DOI] [PubMed] [Google Scholar]
- Bertolotti A., Lutz Y., Heard D. J., Chambon P., Tora L. (1996). hTAF(II)68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. EMBO J. 15, 5022–5031. [PMC free article] [PubMed] [Google Scholar]
- Binelli M., Murphy B. D. (2010). Coordinated regulation of follicle development by germ and somatic cells. Reprod. Fertil. Dev. 22, 1–12. 10.1071/RD09218 [DOI] [PubMed] [Google Scholar]
- Blechingberg J., Luo Y., Bolund L., Damgaard C. K., Nielsen A. L. (2012a). Gene expression responses to FUS, EWS, and TAF15 reduction and stress granule sequestration analyses identifies FET-protein non-redundant functions. PLoS ONE 7:e46251. 10.1371/journal.pone.0046251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blechingberg J., Holm I. E., Nielsen A. L. (2012b). Characterization and expression analysis in the developing embryonic brain of the porcine FET family: FUS, EWS, and TAF15. Gene 493, 27–35. 10.1016/j.gene.2011.11.038 [DOI] [PubMed] [Google Scholar]
- Bond C. S., Fox A. H. (2009). Paraspeckles: nuclear bodies built on long noncoding RNA. J. Cell Biol. 186, 637–644. 10.1083/jcb.200906113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet A., Cabau C., Bouchez O., Sarry J., Marsaud N., Foissac S., et al. (2013). An overview of gene expression dynamics during early ovarian folliculogenesis: specificity of follicular compartments and bi-directional dialog. BMC Genomics 14:904. 10.1186/1471-2164-14-904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani G., Tonlorenzi R., Simmler M. C., Dandolo L., Arnaud D., Capra V., et al. (1991). Characterization of a murine gene expressed from the inactive X chromosome. Nature 351, 325–329. 10.1038/351325a0 [DOI] [PubMed] [Google Scholar]
- Bouckenheimer J., Assou S., Riquier S., Hou C., Philippe N., Sansac C., et al. (2016). Long non-coding RNAs in human early embryonic development and their potential in ART. Hum. Reprod. Update 23, 19–40. 10.1093/humupd/dmw035 [DOI] [PubMed] [Google Scholar]
- Bouckenheimer J., Fauque P., Lecellier C. H., Bruno C., Commes T., Lemaître J. M., et al. (2018). Differential long non-coding RNA expression profiles in human oocytes and cumulus cells. Sci. Rep. 8:2202. 10.1038/s41598-018-20727-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N., Ashworth A., Kay G. F., Cooper P., Smith S., McCabe V. M., et al. (1991). Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature 351, 329–331. 10.1038/351329a0 [DOI] [PubMed] [Google Scholar]
- Brown C. J., Ballabio A., Rupert J. L., Lafreniere R. G., Grompe M., Tonlorenzi R., et al. (1991a). A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349, 38–44. 10.1038/349038a0 [DOI] [PubMed] [Google Scholar]
- Brown C. J., Lafreniere R. G., Powers V. E., Sebastio G., Ballabio A., Pettigrew A. L., et al. (1991b). Localization of the X inactivation centre on the human X chromosome in Xq13. Nature 349, 82–84. 10.1038/349082a0 [DOI] [PubMed] [Google Scholar]
- Campos-Melo D., Droppelmann C. A., Volkening K., Strong M. J. (2014). RNA-binding proteins as molecular links between cancer and neurodegeneration. Biogerontology 15, 587–610. 10.1007/s10522-014-9531-2 [DOI] [PubMed] [Google Scholar]
- Carabatsos M. J., Sellitto C., Goodenough D. A., Albertini D. F. (2000). Oocyte-granulosa cell heterologous gap junctions are required for the coordination of nuclear and cytoplasmic meiotic competence. Dev. Biol. 226, 167–179. 10.1006/dbio.2000.9863 [DOI] [PubMed] [Google Scholar]
- Chau B. L., Ng K. P., Li K. K., Lee K. A. (2016). RGG boxes within the TET/FET family of RNA-binding proteins are functionally distinct. Transcription 7, 141–151. 10.1080/21541264.2016.1183071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. L., Carmichael G. G. (2009). Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol. Cell 35, 467–478. 10.1016/j.molcel.2009.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. L., Carmichael G. G. (2010). Decoding the function of nuclear long non-coding RNAs. Curr. Opin. Cell Biol. 22, 357–364. 10.1016/j.ceb.2010.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow J., Heard E. (2009). X inactivation and the complexities of silencing a sex chromosome. Curr. Opin. Cell Biol. 21, 359–366. 10.1016/j.ceb.2009.04.012 [DOI] [PubMed] [Google Scholar]
- Chu C., Qu K., Zhong F. L., Artandi S. E., Chang H. Y. (2011). Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 44, 667–678. 10.1016/j.molcel.2011.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson C. M., Hutchinson J. N., Sara S. A., Ensminger A. W., Fox A. H., Chess A., et al. (2009). An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 33, 717–726. 10.1016/j.molcel.2009.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozat A., Aman P., Mandahl N., Ron D. (1993). Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature 363, 640–644. 10.1038/363640a0 [DOI] [PubMed] [Google Scholar]
- Delattre O., Zucman J., Plougastel B., Desmaze C., Melot T., Peter M., et al. (1992). Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature 359, 162–165. 10.1038/359162a0 [DOI] [PubMed] [Google Scholar]
- Dinger M. E., Amaral P. P., Mercer T. R., Pang K. C., Bruce S. J., Gardiner B. B., et al. (2008). Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 18, 1433–1445. 10.1101/gr.078378.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S. R. (2001). Non-coding RNA genes and the modern RNA world. Nat. Rev. Genet. 2, 919–929. 10.1038/35103511 [DOI] [PubMed] [Google Scholar]
- Elbarbary R. A., Maquat L. E. (2015). CARMing down the SINEs of anarchy: two paths to freedom from paraspeckle detention. Genes Dev. 29, 687–689. 10.1101/gad.261438.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst E. H., Grøndahl M. L., Grund S., Hardy K., Heuck A., Sunde L., et al. (2017). Dormancy and activation of human oocytes from primordial and primary follicles: molecular clues to oocyte regulation. Hum. Reprod. 32, 1684–1700. 10.1093/humrep/dex238 [DOI] [PubMed] [Google Scholar]
- Ernst E. H., Franks S., Hardy K., Villesen P., Lykke-Hartmann K. (2018). Granulosa cells from human primordial and primary follicles show differential global gene expression profiles. Hum Reprod. 33, 666–679. 10.1093/humrep/dey011 [DOI] [PubMed] [Google Scholar]
- Espey L. L. (1994). Current status of the hypothesis that mammalian ovulation is comparable to an inflammatory reaction. Biol. Reprod. 50, 233–238. 10.1095/biolreprod50.2.233 [DOI] [PubMed] [Google Scholar]
- Fox A. H., Lam Y. W., Leung A. K., Lyon C. E., Andersen J., Mann M., et al. (2002). Paraspeckles: a novel nuclear domain. Curr. Biol. 12, 13–25. 10.1016/S0960-9822(01)00632-7 [DOI] [PubMed] [Google Scholar]
- Fox A. H., Bond C. S., Lamond A. I. (2005). P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol. Biol. Cell. 16, 5304–5315. 10.1091/mbc.e05-06-0587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez J. A., Wapinski O. L., Yang Y. W., Bureau J. F., Gopinath S., Monack D. M., et al. (2013). The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell 152, 743–754. 10.1016/j.cell.2013.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gougeon A. (1996). Regulation of ovarian follicular development in primates: facts and hypotheses. Endocr. Rev. 17, 121–155. 10.1210/edrv-17-2-121 [DOI] [PubMed] [Google Scholar]
- Guipaud O., Guillonneau F., Labas V., Praseuth D., Rossier J., Lopez B., et al. (2006). An in vitro enzymatic assay coupled to proteomics analysis reveals a new DNA processing activity for Ewing sarcoma and TAF(II)68 proteins. Proteomics 6, 5962–5972. 10.1002/pmic.200600259 [DOI] [PubMed] [Google Scholar]
- Hon C. C., Ramilowski J. A., Harshbarger J., Bertin N., Rackham O. J., Gough J., et al. (2017). An atlas of human long non-coding RNAs with accurate 5' ends. Nature 543, 199–204. 10.1038/nature21374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson J. N., Ensminger A. W., Clemson C. M., Lynch C. R., Lawrence J. B., Chess A. (2007). A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 8:39. 10.1186/1471-2164-8-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y., Lee J. T. (2011). YY1 tethers Xist RNA to the inactive X nucleation center. Cell 146, 119–133. 10.1016/j.cell.2011.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobert L., Argentini M., Tora L. (2009). PRMT1 mediated methylation of TAF15 is required for its positive gene regulatory function. Exp. Cell Res. 315, 1273–1286. 10.1016/j.yexcr.2008.12.008 [DOI] [PubMed] [Google Scholar]
- Joh R. I., Palmieri C. M., Hill I. T., Motamedi M. (2014). Regulation of histone methylation by noncoding RNAs. Biochim. Biophys. Acta 1839, 1385–1394. 10.1016/j.bbagrm.2014.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai M. (2016). Roles of RNA-Binding Proteins in DNA Damage Response. Int. J. Mol. Sci. 17:310. 10.3390/ijms17030310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi T., Tanigawa A., Naganuma T., Ohkawa Y., Souquere S., Pierron G., et al. (2015). SWI/SNF chromatin-remodeling complexes function in noncoding RNA-dependent assembly of nuclear bodies. Proc. Natl. Acad. Sci. U.S.A. 112, 4304–4309. 10.1073/pnas.1423819112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke H., Zhao L., Feng X., Xu H., Zou L., Yang Q., et al. (2016). NEAT1 is required for survival of breast cancer cells through FUS, and miR-548. Gene Regul Syst Bio. 10(Suppl. 1), 11–7. 10.4137/GRSB.S29414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil A. M., Guttman M., Huarte M., Garber M., Raj A., Rivea Morales D., et al. (2009). Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U.S.A. 106, 11667–11672. 10.1073/pnas.0904715106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khorkova O., Hsiao J., Wahlestedt C. (2015). Basic biology and therapeutic implications of lncRNA. Adv. Drug Deliv. Rev. 87, 15–24. 10.1016/j.addr.2015.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott G. J., Bond C. S., Fox A. H. (2016). The DBHS proteins SFPQ, NONO and PSPC1: a multipurpose molecular scaffold. Nucleic Acids Res. 44, 3989–4004. 10.1093/nar/gkw271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurian L., Sancho-Martinez I., Nivet E., Aguirre A., Moon K., Pendaries C., et al. (2013). Conversion of human fibroblasts to angioblast-like progenitor cells. Nat. Methods 10, 77–83. 10.1038/nmeth.2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law W. J., Cann K. L., Hicks G. G. (2006). TLS, EWS and TAF15: a model for transcriptional integration of gene expression. Brief. Funct. Genomic. Proteomic 5, 8–14. 10.1093/bfgp/ell015 [DOI] [PubMed] [Google Scholar]
- Leeb M., Steffen P. A., Wutz A. (2009). X chromosome inactivation sparked by non-coding RNAs. RNA Biol. 6, 94–99. 10.4161/rna.6.2.7716 [DOI] [PubMed] [Google Scholar]
- Li S., Li Z., Shu F. J., Xiong H., Phillips A. C., Dynan W. S. (2014). Double-strand break repair deficiency in NONO knockout murine embryonic fibroblasts and compensation by spontaneous upregulation of the PSPC1 paralog. Nucleic Acids Res. 42, 9771–9780. 10.1093/nar/gku650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Xie J., Shen C., Cheng D., Shi Y., Wu Z., et al. (2015). Amplification of Long Noncoding RNA ZFAS1 Promotes Metastasis in Hepatocellular Carcinoma. Cancer Res. 75, 3181–3191. 10.1158/0008-5472.CAN-14-3721 [DOI] [PubMed] [Google Scholar]
- Liu N., Pan T. (2015). RNA epigenetics. Transl. Res. 165, 28–35. 10.1016/j.trsl.2014.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon M. F. (1961). Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190, 372–373. 10.1038/190372a0 [DOI] [PubMed] [Google Scholar]
- Makker A., Goel M. M., Mahdi A. A. (2014). PI3K/PTEN/Akt and TSC/mTOR signaling pathways, ovarian dysfunction, and infertility: an update. J. Mol. Endocrinol. 53, R103–R118. 10.1530/JME-14-0220 [DOI] [PubMed] [Google Scholar]
- Markholt S., Grøndahl M. L., Ernst E. H., Andersen C. Y., Ernst E., Lykke-Hartmann K. (2012). Global gene analysis of oocytes from early stages in human folliculogenesis shows high expression of novel genes in reproduction. Mol. Hum. Reprod. 18, 96–110. 10.1093/molehr/gar083 [DOI] [PubMed] [Google Scholar]
- Markström E., Svensson E., Shao R., Svanberg B., Billig H. (2002). Survival factors regulating ovarian apoptosis – dependence on follicle differentiation. Reproduction 123, 23–30. 10.1530/rep.0.1230023 [DOI] [PubMed] [Google Scholar]
- Mattick J. S., Makunin I. V. (2006). Non-coding RNA. Hum Mol Genet. 15, R17–R29. 10.1093/hmg/ddl046 [DOI] [PubMed] [Google Scholar]
- McGee E. A., Hsueh A. J. (2000). Initial and cyclic recruitment of ovarian follicles. Endocr. Rev. 21, 200–214. 10.1210/er.21.2.200 [DOI] [PubMed] [Google Scholar]
- Mercer T. R., Dinger M. E., Mattick J. S. (2009). Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 10, 155–159. 10.1038/nrg2521 [DOI] [PubMed] [Google Scholar]
- Morohoshi F., Ootsuka Y., Arai K., Ichikawa H., Mitani S., Munakata N., et al. (1998). Genomic structure of the human RBP56/hTAFII68 and FUS/TLS genes. Gene 221, 191–198. 10.1016/S0378-1119(98)00463-6 [DOI] [PubMed] [Google Scholar]
- Nagano T., Mitchell J. A., Sanz L. A., Pauler F. M., Ferguson-Smith A. C., Feil R., et al. (2008). The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322, 1717–1720. 10.1126/science.1163802 [DOI] [PubMed] [Google Scholar]
- Naganuma T., Hirose T. (2013). Paraspeckle formation during the biogenesis of long non-coding RNAs. RNA Biol. 10, 456–461. 10.4161/rna.23547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naganuma T., Nakagawa S., Tanigawa A., Sasaki Y. F., Goshima N., Hirose T. (2012). Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 31, 4020–4034. 10.1038/emboj.2012.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S., Shimada M., Yanaka K., Mito M., Arai T., Takahashi E., et al. (2014). The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development 141, 4618–4627. 10.1242/dev.110544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Názer E., Lei E. P. (2014). Modulation of chromatin modifying complexes by noncoding RNAs in trans. Curr. Opin. Genet. Dev. 25, 68–73. 10.1016/j.gde.2013.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen C. D., Mansfield R. E., Leung W., Vaz P. M., Loughlin F. E., Grant R. P., et al. (2011). Characterization of a family of RanBP2-type zinc fingers that can recognize single-stranded RNA. J. Mol. Biol. 407, 273–283. 10.1016/j.jmb.2010.12.041 [DOI] [PubMed] [Google Scholar]
- Nie F., Yu X., Huang M., Wang Y., Xie M., Ma H., et al. (2016). Long noncoding RNA ZFAS1 promotes gastric cancer cells proliferation by epigenetically repressing KLF2 and NKD2 expression. Oncotarget 8, 38227–38238. 10.18632/oncotarget.9611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo A. F., Lee E. S. (2015). Non-coding RNA: what is functional and what is junk? Front. Genet. 6:2. 10.3389/fgene.2015.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y., Lee J. M., Hwang M. Y., Son G. H., Geum D. (2013). NonO binds to the CpG island of oct4 promoter and functions as a transcriptional activator of oct4 gene expression. Mol. Cells 35, 61–69. 10.1007/s10059-013-2273-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paronetto M. P. (2013). Ewing sarcoma protein: a key player in human cancer. Int. J. Cell Biol. 2013:642853. 10.1155/2013/642853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny G. D., Kay G. F., Sheardown S. A., Rastan S., Brockdorff N. (1996). Requirement for Xist in X chromosome inactivation. Nature 379, 131–137. 10.1038/379131a0 [DOI] [PubMed] [Google Scholar]
- Peschansky V. J., Wahlestedt C. (2014). Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics 9, 3–12. 10.4161/epi.27473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2012). R: A Language and Environment for STATISTICAL COMPUTING. R Vienna: Foundation for Statistical Computing; Available online at: http://www.R-project.org/. [Google Scholar]
- Reddy P., Zheng W., Liu K. (2010). Mechanisms maintaining the dormancy and survival of mammalian primordial follicles. Trends Endocrinol. Metab. 21, 96–103. 10.1016/j.tem.2009.10.001 [DOI] [PubMed] [Google Scholar]
- Rosendahl M., Schmidt K. T., Ernst E., Rasmussen P. E., Loft A., Byskov A. G., et al. (2011). Cryopreservation of ovarian tissue for a decade in Denmark: a view of the technique. Reprod. Biomed. Online 22, 162–171. 10.1016/j.rbmo.2010.10.015 [DOI] [PubMed] [Google Scholar]
- Sasaki Y. T., Ideue T., Sano M., Mituyama T., Hirose T. (2009). MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl. Acad. Sci. U.S.A. 106, 2525–2530. 10.1073/pnas.0807899106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt K. L., Byskov A. G., Nyboe Andersen A., Müller J., Yding Andersen C. (2003). Density and distribution of primordial follicles in single pieces of cortex from 21 patients and in individual pieces of cortex from three entire human ovaries. Hum. Reprod. 18, 1158–1164. 10.1093/humrep/deg246 [DOI] [PubMed] [Google Scholar]
- Schmidt D., Ovitt C. E., Anlag K., Fehsenfeld S., Gredsted L., Treier A. C., et al. (2004). The murine winged-helix transcription factor Foxl2 is required for granulosa cell differentiation and ovary maintenance. Development 131, 933–942. 10.1242/dev.00969 [DOI] [PubMed] [Google Scholar]
- Schwartz J. C., Cech T. R., Parker R. R. (2015). Biochemical properties and biological functions of FET proteins. Annu. Rev. Biochem. 84, 355–379. 10.1146/annurev-biochem-060614-034325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J. A., Kingston R. E. (2013). Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell. 49, 808–824. 10.1016/j.molcel.2013.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs S. A., Hardy K., Da Silva-Buttkus P., Stark J., Webber L. J., Flanagan A. M., et al. (2005). Anti-mullerian hormone protein expression is reduced during the initial stages of follicle development in human polycystic ovaries. J. Clin. Endocrinol. Metab. 90, 5536–5543. 10.1210/jc.2005-0907 [DOI] [PubMed] [Google Scholar]
- Sunwoo H., Dinger M. E., Wilusz J. E., Amaral P. P., Mattick J. S., Spector D. L. (2009). MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 19, 347–359. 10.1101/gr.087775.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svetoni F., Frisone P., Paronetto M. P. (2016). Role of FET proteins in neurodegenerative disorders. RNA Biol. 13, 1089–1102. 10.1080/15476286.2016.1211225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therrien M., Rouleau G. A., Dion P. A., Parker J. A. (2016). FET proteins regulate lifespan and neuronal integrity. Sci. Rep. 6:25159. 10.1038/srep25159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorenoor N., Faltejskova-Vychytilova P., Hombach S., Mlcochova J., Kretz M., Svoboda M., et al. (2016). Long non-coding RNA ZFAS1 interacts with CDK1 and is involved in p53-dependent cell cycle control and apoptosis in colorectal cancer. Oncotarget 7, 622–637. 10.18632/oncotarget.5807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. C., Chang H. Y. (2011). Molecular mechanisms of long noncoding RNAs. Mol. Cell. 43, 904–914. 10.1016/j.molcel.2011.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Lee C. G. (2009). MicroRNA and cancer–focus on apoptosis. J. Cell. Mol. Med. 13, 12–23. 10.1111/j.1582-4934.2008.00510.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Song X., Glass C. K., Rosenfeld M. G. (2011). The long arm of long noncoding RNAs: roles as sensors regulating gene transcriptional programs. Cold Spring Harb. Perspect. Biol. 3:a003756. 10.1101/cshperspect.a003756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Gong X., Tian G. G., Hou C., Zhu X., Pei X., et al. (2018). Long noncoding RNA growth arrest-specific 5 promotes proliferation and survival of female germline stem cells in vitro. Gene 653, 14–21. 10.1016/j.gene.2018.02.021 [DOI] [PubMed] [Google Scholar]
- Wapinski O., Chang H. Y. (2011). Long noncoding RNAs and human disease. Trends Cell Biol. 21, 354–361. 10.1016/j.tcb.2011.04.001 [DOI] [PubMed] [Google Scholar]
- Wilhelmm D., Bernard P. (2016). Non-coding RNAs and the Reproductive System. Adv Exp Med Biol. 886, V–Vi. 10.1007/978-94-017-7417-8 [DOI] [PubMed] [Google Scholar]
- Xu Y., Li X., Wang H., Xie P., Yan X., Bai Y., et al. (2016). Hypermethylation of CDH13, DKK3 and FOXL2 promoters and the expression of EZH2 in ovary granulosa cell tumors. Mol. Med. Rep. 14, 2739–2745. 10.3892/mmr.2016.5521 [DOI] [PubMed] [Google Scholar]
- Yacqub-Usman K., Pickard M. R., Williams G. T. (2015). Reciprocal regulation of GAS5 lncRNA levels and mTOR inhibitor action in prostate cancer cells. Prostate 75, 693–705. 10.1002/pros.22952 [DOI] [PubMed] [Google Scholar]
- Yamazaki T., Hirose T. (2015). The building process of the functional paraspeckle with long non-coding RNAs. Front. Biosci. 7, 1–41. 10.2741/e715 [DOI] [PubMed] [Google Scholar]
- Yan B., Wang Z. H., Guo J. T. (2012). The research strategies for probing the function of long noncoding RNAs. Genomics 99, 76–80. 10.1016/j.ygeno.2011.12.002 [DOI] [PubMed] [Google Scholar]
- Yoo A. S., Sun A. X., Li L., Shcheglovitov A., Portmann T., Li Y., et al. (2011). MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476, 228–231. 10.1038/nature10323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M. C. (2011). The role of protein arginine methylation in mRNP dynamics. Mol. Biol. Int. 2011:163827. 10.4061/2011/163827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Rajkovic A. (2008). MicroRNAs and mammalian ovarian development. Semin. Reprod. Med. 26, 461–468. 10.1055/s-0028-1096126 [DOI] [PubMed] [Google Scholar]
- Zhao J., Sun B. K., Erwin J. A., Song J. J., Lee J. T. (2008). Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322, 750–756. 10.1126/science.1163045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S. J., Hallows S. E., Currie K. W., Xu C., Pearson B. J. (2015). A mex3 homolog is required for differentiation during planarian stem cell lineage development. eLife 4:e07025. 10.7554/eLife.07025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinszner H., Sok J., Immanuel D., Yin Y., Ron D. (1997). TLS (FUS) binds RNA in vivo and engages in nucleo-cytoplasmic shuttling. J Cell Sci. 110 (Pt 15), 1741–1750. [DOI] [PubMed] [Google Scholar]
- Zuccotti M., Merico V., Cecconi S., Redi C. A., Garagna S. (2011). What does it take to make a developmentally competent mammalian egg? Hum. Reprod. Update 17, 525–540. 10.1093/humupd/dmr009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full lists of ncRNAs detected in human follicle development [oocytes from primordial follicles (tab 1), primordial follicles (tab 2), oocytes from primary follicles (tab 3), mural granulosa cell layers from small antral follicles (tab 4), theca cell layers from small antral follicles (tab 5), oocytes from small antral follicles (tab 6)]. Cell specific consistently expressed genes (CSCEG) (one-sample t-test p-value < 0.05 in gray).