Abstract
Objective
In rheumatoid arthritis, the enzyme 11 beta-hydroxysteroid dehydrogenase type 1 (11β-HSD1) is highly expressed at sites of inflammation, where it converts inactive glucocorticoids (GC) to their active counterparts. In conditions of GC excess it has been shown to be a critical regulator of muscle wasting and bone loss. Here we examine the contribution of 11β-HSD1 to the pathology of persistent chronic inflammatory disease.
Methods
To determine the contribution of 11β-HSD1 to joint inflammation, destruction and systemic bone loss associated with persistent inflammatory arthritis, we generated mice with global and mesenchymal specific 11β-HSD1 deletions in the TNF-transgenic (TNF-tg) model of chronic polyarthritis. Disease severity was determined by clinical scoring. Histology was assessed in formalin fixed sections and fluorescence-activated cell sorting (FACS) analysis of synovial tissue was performed. Local and systemic bone loss were measured by micro computed tomography (micro-CT). Measures of inflammation and bone metabolism were assessed in serum and in tibia mRNA.
Results
Global deletion of 11β-HSD1 drove an enhanced inflammatory phenotype, characterised by florid synovitis, joint destruction and systemic bone loss. This was associated with increased pannus invasion into subchondral bone, a marked polarisation towards pro-inflammatory M1 macrophages at sites of inflammation and increased osteoclast numbers. Targeted mesenchymal deletion of 11β-HSD1 failed to recapitulate this phenotype suggesting that 11β-HSD1 within leukocytes mediate its protective actions in vivo.
Conclusions
We demonstrate a fundamental role for 11β-HSD1 in the suppression of synovitis, joint destruction, and systemic bone loss. Whilst a role for 11β-HSD1 inhibitors has been proposed for metabolic complications in inflammatory diseases, our study suggests that this approach would greatly exacerbate disease severity.
Highlights
-
•
11β-HSD1 is critical in suppressing joint destruction and bone loss in chronic polyarthritis.
-
•
Global Deletion drives florid synovitis, joint destruction and systemic bone loss.
-
•
Characterised by a marked polarisation of macrophages towards an M1 phenotype.
-
•
Mesenchymal deletion does not reproduce the global KO phenotype.
-
•
Indicates that therapeutic inhibitors of 11β-HSD1 would exacerbate disease severity.
1. Introduction
The 11 beta-hydroxysteroid dehydrogenase (11β-HSD) type 1 enzyme determines tissue specific exposure to endogenous and therapeutic glucocorticoids (GCs). It is a bidirectional enzyme that converts inactive GCs to their active counterparts, conferring tissue-specific amplification and exposure to active endogenous and therapeutic GCs [1]. 11β-HSD1 was shown to be critical in mediating adverse metabolic complications of elevated GCs in vivo [2].
11β-HSD1 is highly expressed and active at sites of inflammation in diseases such as rheumatoid arthritis (RA), increasing local exposure to GCs [[3], [4], [5], [6]]. Resident mesenchymal derived populations such as fibroblast like synoviocytes (FLS) are important sites of 11β-HSD1 mediated GC activation in response to inflammation, which feeds back to suppress pro-inflammatory signalling in vitro [[3], [4], [5], [6], [7], [8], [9]]. 11β-HSD1 is also expressed in synovial leukocyte populations, including macrophages, lymphocytes and dendritic cells where it dampens pro-inflammatory signalling and promotes resolution [5,6,[10], [11], [12], [13], [14]].
The Tg197 (TNF-tg) mouse is a murine model of chronic polyarthritis with strong parallels with chronic inflammatory disease in humans [15] and is widely used to assess therapeutic interventions [[15], [16], [17]]. Consequently, this model has been invaluable in delineating the pathophysiology of RA, demonstrating the prominence of tumour necrosis factor alpha (TNFα) in the inflammatory cytokine cascade [18].To date, no study has examined the impact of global 11β-HSD1 deletion in models of chronic inflammatory arthritis. Therefore, we investigated the consequences of global and mesenchymal specific 11β-HSD1 deletion in the Tg197 (TNF-tg) murine model of chronic polyarthritis.
2. Materials and methods
2.1. Human TNFα transgenic mouse model and clinical scoring
Experiments were performed in compliance with guidelines governed by the UK Animal (Scientific Procedures) Act 1986 (project licence number 70/8582 or 70/8003) and approved by Birmingham Ethical Review Subcommittee. Tg197 mice (TNF-tg) that express stabilised human TNFα mRNA on a C57BL/6J strain background were obtained from Dr George Kollias (BSRC Fleming, Athens, Greece) [15]. Animals were scored for joint inflammation using a 16 point system 9,19: Clinical scores were calculated from measures of weight loss, behaviour, mobility, duration of joint swelling, mouse grimace and evidence of joint inflammation as previously reported [9,19]. At nine weeks, animals were culled and front paws, hind limbs and tibias collected.
2.2. Global and mesenchymal targeted deletion of 11β-HSD1 in the TNFα transgenic mouse
11β-HSD1 knock out (KO) animals with global 11β-HSD1 deletion were crossed with TNF-tg animals to generate TNF‐tg11βKO animals as previously described [9]. Breeding animals were maintained on anti-human TNFα monoclonal antibody (infliximab), as previously reported, to control inflammation and facilitate breeding [19]. Mesenchymal targeted 11β-HSD1 KO animals were created by crossing floxed HSD11B1 mice with Twist2-cre animals (where cre recombinase activity is reported to target mesenchymal derived cell populations such as osteoblasts, chondrocytes and FLS), to generate 11βHSD1flx/flx/Twist2cre animals [[20], [21], [22]]. These were crossed with TNF-tg animals to produce TNF-tg11βHSD1flx/flx/Twist2cre (TNF-tg11βflx/tw2cre) animals.
2.3. Analysis of mRNA abundance
Expression of mRNAs was determined using TaqMan® Gene Expression Assays (Thermo Fisher Scientific, Loughborough, UK). RNA was extracted from homogenised tibia following flushing of the bone marrow or from the bone marrow aspirate. Briefly, tibias were removed from the hind limbs and soft tissues removed. Tibias were powdered in liquid nitrogen. mRNA isolation was performed using an innuPREP RNA Mini Kit (Analytikjena, Cambridge). RNA was reverse transcribed using random hexamers (4311235, Multiscribe™, ThermoFisher Scientific) to generate cDNA. Gene expression was determined using species-specific probe sets for real time polymerase chain reaction (PCR) on an ABI7500 system (Applied Biosystems, Warrington, UK). mRNAs expression was normalised to that of 18S RNA. Data were obtained as cycle threshold (Ct) values to determine ΔCt values (Ct target – Ct 18S). Gene expression in arbitrary units (AU) was calculated from ΔCt values using the equation 1000x−2ΔCt.
2.4. Histological analysis of inflamed joints
Histochemistry was performed on paraffin-embedded 10 μm sections following decalcification in 0.5 M ethylenediaminetetraacetic acid (EDTA). Pannus size at the metatarsal-phalangeal joint interface was determined using Image J software as previously reported [19]. Sections were stained with safranin O and fast green to quantify cartilage thickness and proteoglycan loss across cartilage of the humerus/ulna interface using Image J software. Loss of safranin O staining across cartilage was quantified relative to total cartilage of the humerus/ulna interface. For all quantifications, the mean of data from three adjacent 10 μm sections cut from the centre of the joint were utilised to determine pannus size, cartilage thickness and proteoglycan loss.
2.5. Tissue digestion and flow cytometric analysis of synoviocytes
One Hind leg and one front paw per mouse was dissected and cleaned of tissue, separating the leg bones and keeping the surface of the knee joint intact. Joints were digested in 1.5 ml of RPMI containing 2% foetal calf serum (FCS), 2.5 mg/ml collagenase D (Roche) and 20 μg/ml DNase (Sigma‐Aldrich) for 45 min at 37 °C with agitation. Digests were filtered through a 100 μm strainer and cells kept on ice. Cells were centrifuged, red cells lysed for 5mins in ACK lysis buffer (Gibco) and cells counted. Cells were washed, filtered through 40 μm cell strainer, incubated with anti-CD16/CD32 blocking antibody (1:200; eBioscience) for 10 min at RT, followed by staining with antibody cocktail at 4 °C. Antibodies for membrane staining are outlined in Table 1. Data were acquired using a BD LSR Fortessa X20 and analysed using FlowJo software (FlowJo LLC). The following gating strategy was used for myeloid cells: Live cells were gated on CD45 +CD11b + cells. Neutrophils were identified as CD45 + CD11b + SiglecF- Ly6ghi, macrophages were CD45 + CD11b + SiglecF- Ly6g- CD64 + F4/80 + and M1 macrophages were CD45 + CD11b + SiglecF- Ly6g- CD64 + F4/80 + MHC Class II+. T cells were identified as live CD45 +CD3+. CD3+ cells were then stratified as CD4+ or CD8+ T cells. B cells were identified as CD45 + CD3-and CD19+.
Table 1.
Antibodies for FACS analysis of synovial leukocyte populations.
| Target | Label | Dilution | Manufacturer | Reference | Concentration |
|---|---|---|---|---|---|
| anti-CD45 | APC-CY7 | 1:400 | eBioscience | 47-0451-82 | 0.2 mg/ml |
| anti- CD11b | PerCP CY5.5 | 1:200 | BioLegend | 45-0112-82 | 0.2 mg/ml |
| anti-CD11c | FITC | 1:100 | eBioscience | 11-0114-82 | 0.5 mg/ml |
| anti-SiglecF | eFlour660 | 1:100 | eBioscience | 50-1702-80 | 0.2 mg/ml |
| anti-CD64 | PE-Cy7 | 1:100 | BioLegend | 139314 | 0.2 mg/ml |
| anti-Ly6g | PE Dazzle | 1:800 | BioLegend | 127648 | 0.2 mg/ml |
| anti-Ly6c | BV510 | 1:600 | BioLegend | 128033 | 0.2 mg/ml |
| anti-MHC Class II | BV711 | 1:800 | Biolegend | 107643 | 0.2 mg/ml |
| anti-F4/80 | PE labelled | 1:400 | BioLegend | 123110 | 0.2 mg/ml |
| anti-CD45 | APC-Cy7 | 1:400 | eBioscience | 47-0451-82 | 0.2 mg/ml |
| anti-CD3 | PE-Cy7 | 1:400 | eBioscience | 25-0031-82 | 0.2 mg/ml |
| anti-CD4 | Pacific blue | 1:600 | eBioscience | 116008 | 0.5 mg/ml |
| anti-CD19 | PE labelled | 1:400 | eBioscience | 12-0193-82 | 0.2 mg/ml |
| anti-CD8 | Texas red | 1:800 | eBioscience | MCD0817 | 0.2 mg/ml |
2.6. 11β-HSD1 activity
Steroid metabolism was determined by scanning thin layer chromatography following incubation with 3H–dehydrocorticosterone (Perkin Elmer, Beaconsfield, Buckinghamshire, UK for 6-h as previously described [5]. Thin-layer chromatography plates were analysed by Bioscan imager (Bioscan, Washington, DC, USA) to detect the formation of 3H –corticosterone. The fractional conversion of steroids was calculated as pmol product/per mg protein/hr.
2.7. Analysis of serum corticosterone, PINP and TRACP 5b by ELISA
Corticosterone levels in serum were measured using a commercially available sandwich enzyme linked immunoassay (ELISA) (R&D systems, Abingdon, UK) in accordance with manufacturer's instructions and data expressed as ng/ml. Serum PINP was determined using a commercially available sandwich ELISA (Immunodiagnostic Systems, Tyne & Wear, UK) in accordance with manufacturer's instructions and data expressed as ng/ml. Serum TRACP 5b was determined using a commercially available sandwich ELISA (Immunodiagnostic Systems, Tyne & Wear, UK) and data expressed as U/L.
2.8. Micro-CT (qualitative analysis)
Samples were imaged using a Skyscan 1172 micro-CT scanner (Bruker) using X-ray beam settings of 60 kV source voltage, 167 μA source current. Projections were taken every 0.45° at 600 ms exposure, with an image pixel size of 13.59 μm. Image volumes were reconstructed using the Feldkamp algorithm (NRecon 1.6.1.5, Bruker) applying beam hardening correction. A radiodensity range of −300 to 3000HU was chosen to isolate the bony structures from the imaging medium, CTAnalyser 1.12 software (SkyScan) was used to extract an isosurface mesh representation of reconstructed micro-CT slices. MeshLab 1.3.2 (open source software) was used to modify the raw meshes and samples were shaded in MeshLab using ambient occlusion. Meshes were scored as previously reported [19].
2.9. MicroCT morphometry analysis
Formalin-fixed tibias were scanned using a Skyscan 1172 x-ray microtomograph at 60 kV/167 μA with a 0.5 mm aluminium filter. Images were obtained at a 5 μm resolution with a rotation step of 0.45°. NRecon software was used to reconstruct the images. Trabecular bone parameters were analysed using CTAn Skyscan software. Regions of interest (ROI) were selected by highlighting trabecular bone regions for cross sectional slices of tibia (Fig. 2.1C) and bone architecture determined by quantifying trabecular bone parameters (bone volume to total volume (BV/TV), trabecular thickness (TT) and trabecular number (TN)) using CTAn software. 1.35 mm of trabecular bone (200 sections) was selected for trabecular bone analysis at the metaphyseal region near the growth plate. Extent was determined by the length of trabecular bone growth in each sample, which was calculated by multiplying slice number by pixel size of scanned image (13.5 μm). Meshlab software was used to process 3D meshes of tibiae.
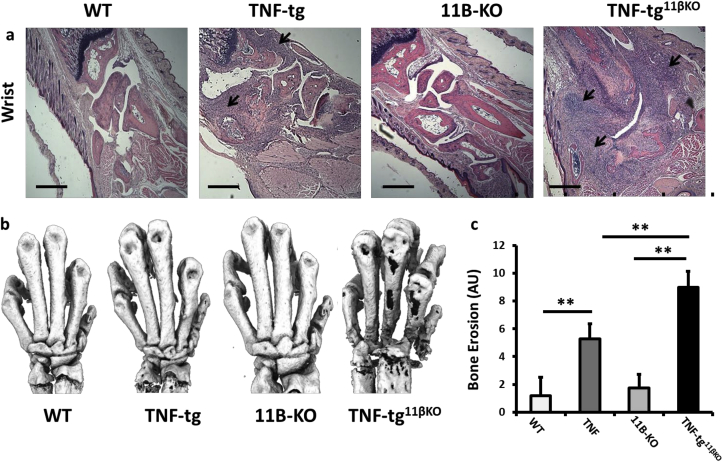
Fig. 2.
(a) Representative paraffin-embedded sections from the wrist below the ulna, stained with haematoxylin and eosin (scale bars 200 μm, arrows indicate regions of synovitis), (b) representative images of 3D reconstructions of hind paws using micro-CT, (c) quantification of cortical erosion in the bones of the ankle, metatarsals and phalanges in WT, TNF-tg, 11βKO, and TNF-tg11βKO mice. AU (Arbitrary units). Values are expressed as mean ± standard error of six animals per group. Statistical significance was determined using one way ANOVA with a Tukey post hoc analysis. *p < 0.05, **p < 0.005, ***p < 0.001.
2.10. Statistical analysis
Statistical significance was defined as P < 0.05 (*P < 0.05; **P < 0.005; ***P < 0.001) using either an unpaired Student's t-test or one way ANOVA with a Tukey post hoc analysis where a Gaussian distribution is identified, or a non-parametric Kruskal-Wallis test with a Dunn's Multiple Comparison absent.
3. Results
3.1. Global deletion of 11β-HSD1 greatly exacerbates polyarthritis
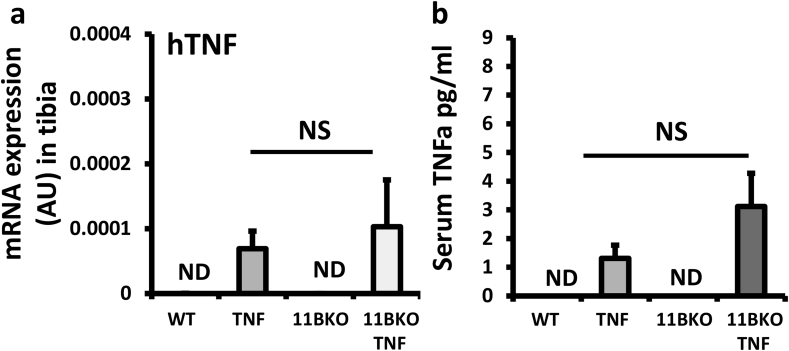
We crossed the TNF-tg mouse onto the 11β-HSD1 global KO mouse to generate TNF-tg animals with deletion of 11β-HSD1 (TNF‐tg11βKO). TNF‐tg11βKO animals developed marked increases in swelling and deformity of front paws relative to wild type, 11β-KO and TNF-tg counterparts (Fig. 1). GC activation was analysed in paw biopsies after dissection of skin, following dis-articulation of joints as previously reported [23]. Significant corticosterone generation was observed in wild type (WT) and TNF-tg animals but not 11β-KO and TNF‐tg11βKO mice (Fig. 1b). Serum measures of corticosterone remained consistent across WT, 11β-KO, TNF-tg and TNF‐tg11βKO mice (Fig. 1c). Human TNF transgene expression was assessed in tibia mRNA from all groups (Sup. 1a). TNF transgene was detected in both TNF-tg and TNF-tg11βKO mice and showed no difference between groups. This was consistent with serum levels of human TNFα in all groups (Sup. 1b).
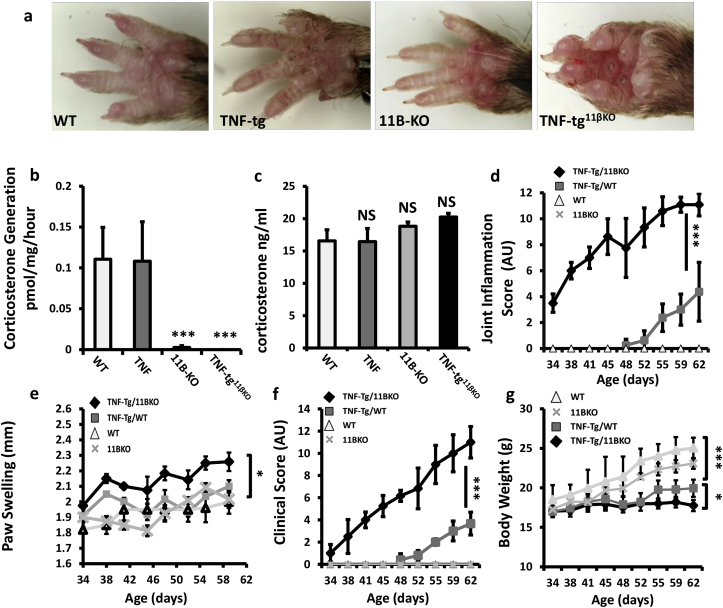
Fig. 1.
(a) Representative images of front paws from WT, TNF-tg, 11βKO, and TNF-tg11βKO mice at 9 weeks. (b) Corticosterone generation in front paw biopsies determined by scanning thin layer chromatography isolated from WT, TNF-tg, 11βKO, and TNF-tg11βKO mice at 9 weeks. (c) Serum corticosterone levels determined by ELISA; (d) scoring of joint inflammation, (e) front paw swelling (mm); (f) clinical scoring (weight, inflammation, grimace, behaviour, mobility, inflammation severity and duration) and (g) weight in WT, TNF-tg, 11βKO, and TNF-tg11βKO mice. AU (Arbitrary units). Values are expressed as mean ± standard error of six animals per group. Statistical significance was determined using one way ANOVA with a Tukey post hoc analysis. *p < 0.05, **p < 0.005, ***p < 0.001.
TNF‐tg11βKO mice developed inflammation from 34 days old compared to approximately 48 days in the TNF-tg animal with severity of inflammation significantly greater in the TNF-tg11βKO mouse at 62 days (Fig. 1d). This was supported by greater paw swelling in TNF-tg11βKO mice relative to TNF-tg mice (Fig. 1e). These data were reflected in clinical scores, where onset of disease activity was apparent from day 34 and significantly greater at day 62 (Fig. 1f). Whilst TNF-tg mice developed a significant reduction in body weight relative to wild type counterparts, TNF‐tg11βKO mice developed a more marked weight loss (Fig. 1g). Together these data confirm an important role for 11β-HSD1 in limiting the severity of paw swelling, local joint inflammation and measures of systemic inflammation in a model of chronic persistent arthritis.
3.2. Greater joint destruction in the TNF‐tg11βKO mouse
Histological analysis of tissue from TNF‐tg11βKO animals confirmed a marked increase in front paw joint synovitis relative to TNF-tg animals, with greater evidence of joint destruction and bone erosion determined by histology and micro-CT (Fig. 2a–c). Degradation of articular cartilage was also more marked in TNF‐tg11βKO (Sup. 2a-g). Together these data confirm an important role for 11β-HSD1 in protecting against local synovitis, joint destruction and juxta articular bone loss.
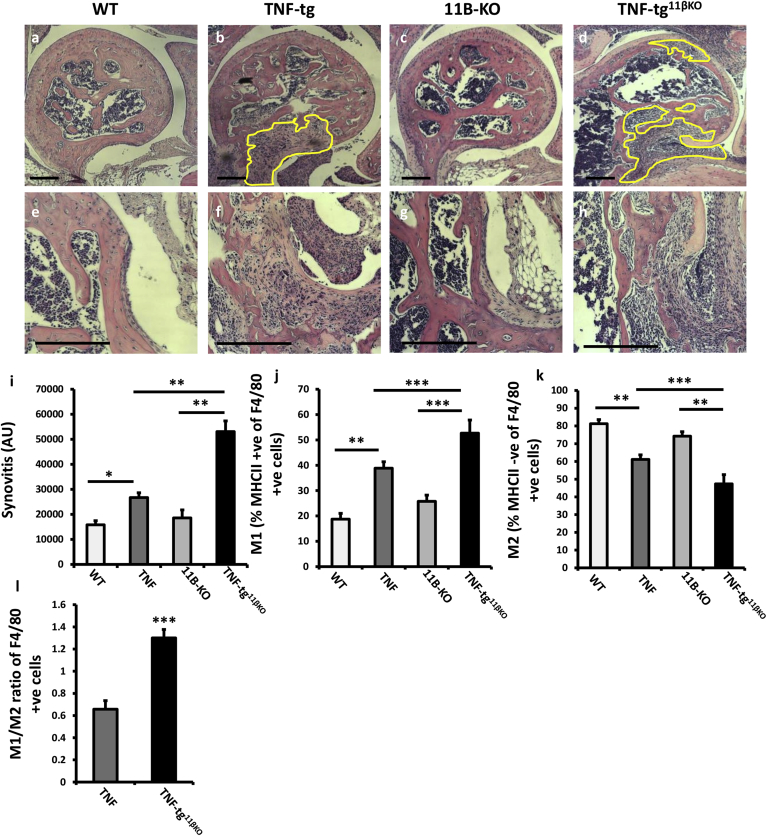
3.3. Synovitis is characterised by M1 polarisation in the TNF‐tg11βKO mouse
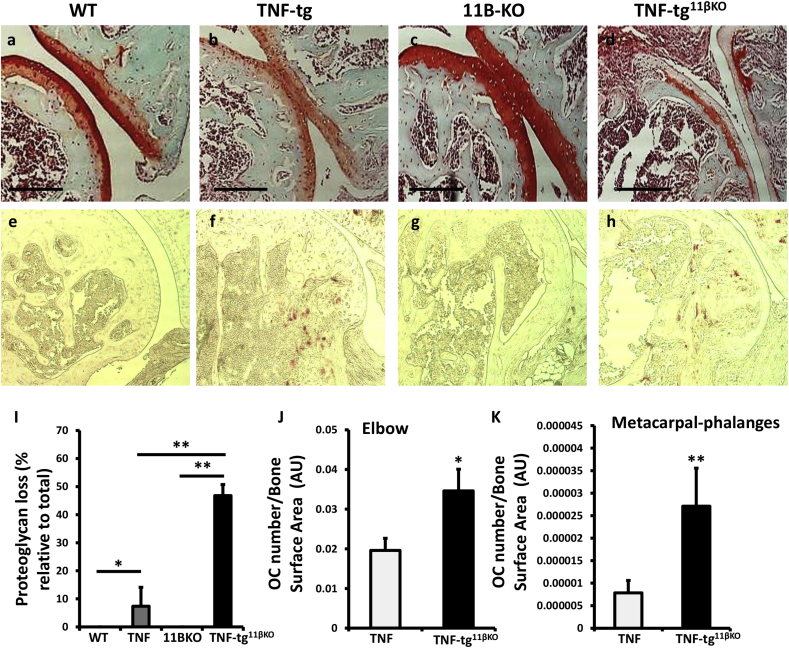
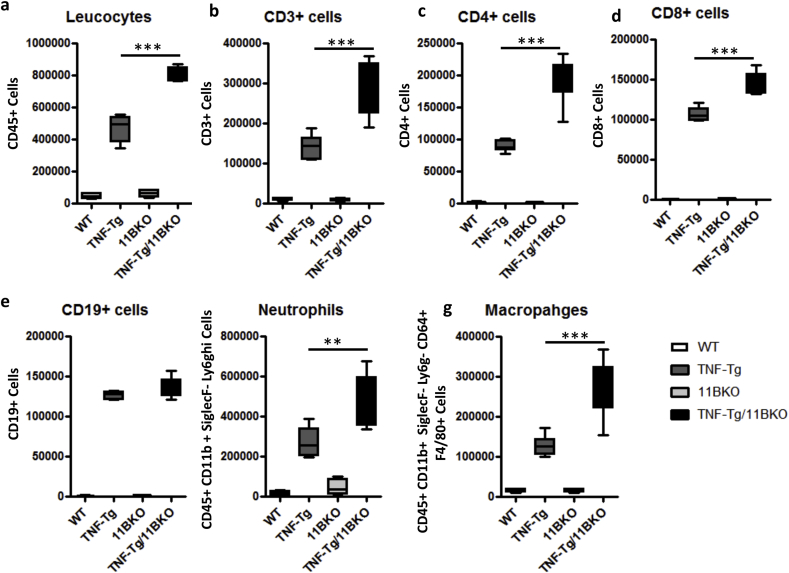
TNF‐tg11βKO animals showed greater pannus invasion into underlying bone compared with TNF-tg animals (Fig. 3a–h). FACS analysis of synovial leukocytes revealed a significant increase in CD3+, CD4+ and CD8+ T cells, macrophages and neutrophils in TNF‐tg11βKO mice relative to TNF-tg animals, whilst B cells numbers remained unchanged (Sup. 2a-e). To examine macrophage polarisation within the inflamed synovium, M1 and M2 were characterised within the F40/80 + population based on high and low MHCII expression as previously reported (Fig. 3j–l) [24,25]. This revealed a marked increase in M1 macrophages in the TNF‐tg11βKO mice relative to TNF-tg mice with a marked skewing of the M1/M2 ratio favouring M1 polarisation (Fig. 3j–l). These data confirm increased leukocyte infiltration and M1 polarisation in TNF‐tg11βKO mice.
Fig. 3.
(a–d) Representative images of paraffin-embedded ulna/humerus joint interface (20X) stained with haematoxylin and eosin (scale bars 200 μm, yellow region indicates synovial pannus formation), and (e–h) (40X) magnification of pannus invasion into subchondral bone (scale bars 200 μm), from WT, TNF-tg, 11βKO, and TNF-tg11βKO mice at 9 weeks. (i) Quantification of degree of synovitis in the ulna/humerus joint interface determined by image J and (j–l) FACS quantification of M1 and M2 polarisation in CD45 + CD11b + SiglecF- Ly6g- CD64 + F4/80 + murine macrophages determined by MHCIIhi and MHCIIlow respectively in WT, TNF-tg, 11βKO, and TNF-tg11βKO mice at 9 weeks. Values are expressed as mean ± standard error of six animals per group. AU (Arbitrary units). Statistical significance was determined using one way ANOVA with a Tukey post hoc analysis. *p < 0.05, **p < 0.005, ***p < 0.001. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.4. Systemic bone loss is exacerbated in the TNF‐tg11βKO mouse
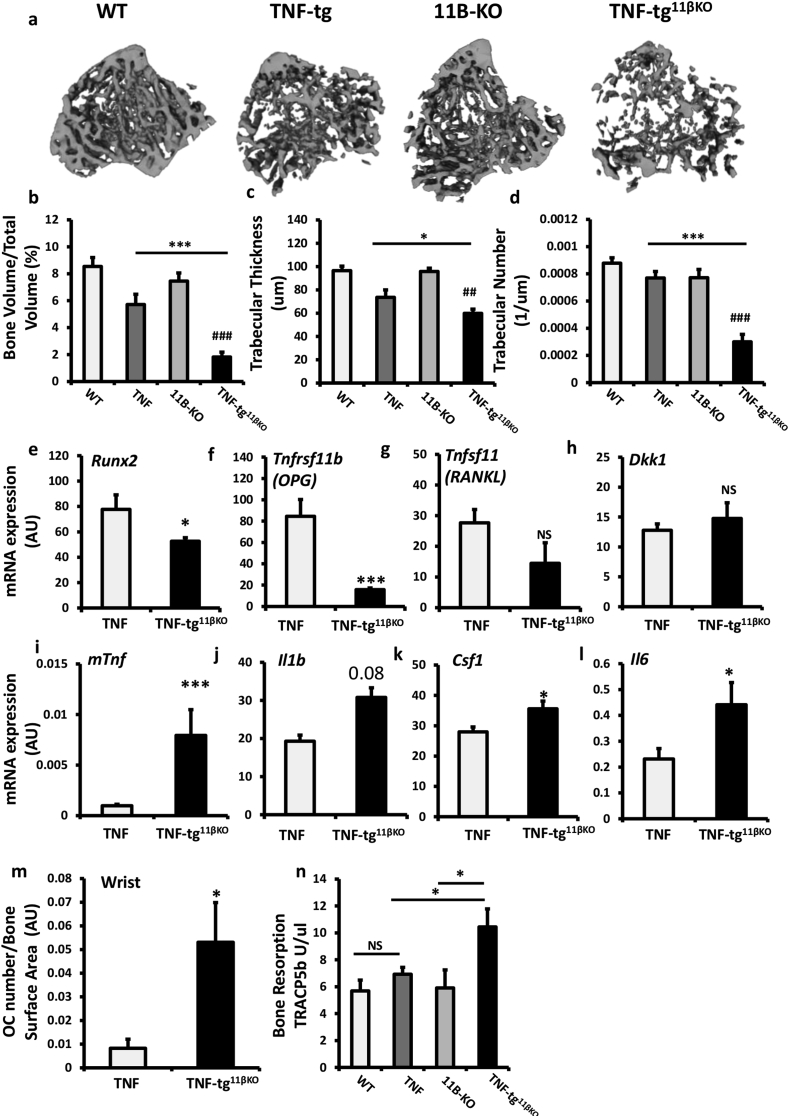
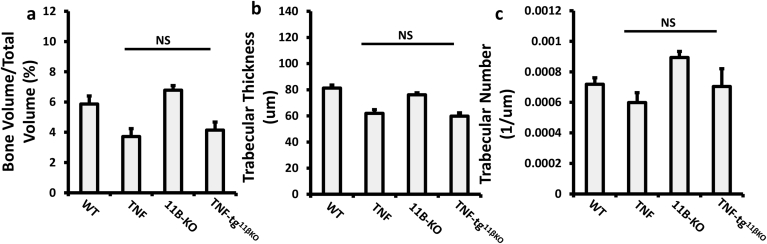
We examined the effects of global 11β-HSD1 deletion on systemic bone loss in the TNF-tg mouse by micro-CT analysis of trabecular bone. At five weeks, measures of bone density were reduced to a similar degree in both TNF-tg and TNF‐tg11βKO mice compared with WT (Sup. Fig. 4a–c). However, after five weeks, TNF‐tg11βKO animals developed marked reductions in all parameters relative to TNF-tg counterparts (Fig. 4a–d). mRNA analysis of tibia revealed a significant suppression of mature osteoblast markers runt-related transcription factor 2 (Runx2) and osteoprotegerin (Tnfrsf11b) in TNF‐tg11βKO animals (Fig. 4e,f). The pro-osteoclastogenic marker receptor activator of nuclear factor kappa-Β ligand (Tnfsf11) and the negative regulator of osteoblast differentiation dikkopf-1 (Dkk1) both remained stable (Fig. 4g,h). Analysis of serum procollagen type 1 N-terminal propeptide (P1NP) as a marker of systemic bone formation confirmed a significant reduction in osteoblast activity in the TNF‐tg11βKO mouse compared to TNF-tg animals (Fig. 4n).
Fig. 4.
(a) Representative images of 3D reconstructions of tibia trabecular bone using micro-CT from WT, TNF-tg, 11βKO, and TNF-tg11βKO mice at 9 weeks. (b) Bone volume to tissue volume (BV/TV), (c) trabecular thickness (TT) and (d) trabecular number (TN) determined by Meshlab software analysis of micro CT at 5 and 9 weeks in WT, TNF-tg, 11βKO, and TNF-tg11βKO mice. (e–l) Gene expression (AU) of Runx2, Tnfrsf11b, Tnfsf11, Dkk1, Tnf, Il1b, Csf1 and Il6 in homogenates of tibia determined by quantitative RT-PCR from TNF-tg and TNF-tg11βKO mice at 9 weeks. (m) Osteoclast numbers at the scaphoid/trapezoid interface normalised to bone surface area determined by image J analysis of TRAP stained paraffin-embedded sections of the wrist, (n) serum P1NP (ng/ml) and (o) serum TRAcP5B (U/μl) determined by ELISA in WT, TNF-tg, 11βKO, and TNF-tg11βKO mice at 9 weeks. AU (Arbitrary units). Values are expressed as mean ± standard error of six animals per group. Statistical significance was determined using one way ANOVA with a Tukey post hoc analysis. *p < 0.05, **p < 0.005, ***p < 0.001.
Analysis of the pro-inflammatory murine cytokine TNFα identified greater expression in the TNF‐tg11βKO mouse (8.7 fold increase, P < 0.001). In addition the pro-inflammatory, pro-osteoclastogenic cytokines interleukin-1 (Il1), macrophage colony-stimulating factor (Csf1) and interleukin-6 (Il6) all confirmed upregulation in TNF‐tg11βKO mice (Fig. 4i–l). Osteoclast numbers relative to bone surface area were significantly increased in TNF‐tg11βKO animals at the joint interface of the wrist, as well as in the elbow and metacarpophalangeal joints (Fig. 4m, Sup. 1e-k). Serum levels of the secreted, osteoclast-specific protein, tartrate-resistant acid phosphatase 5b (TRAcP5B), were significantly increased in TNF‐tg11βKO mice relative to TNF-tg and 11β-KO counterparts (Fig. 4o). Together, these data support a fundamental role for 11β-HSD1 in the protection of bone from inflammation-associated bone loss.
3.5. Inflammation, joint destruction and bone loss are not regulated by mesenchymal 11β-HSD1
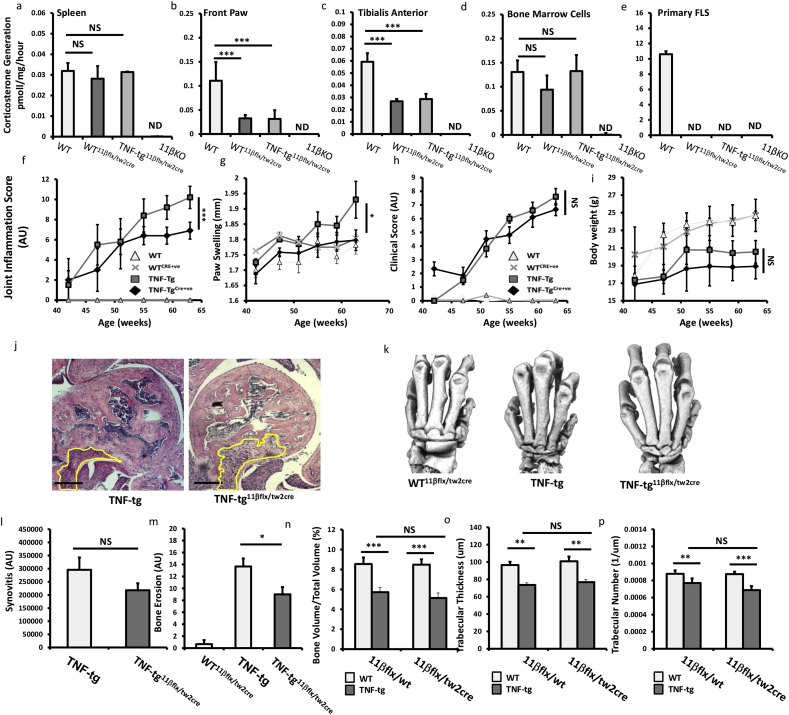
11β-HSD1 is potently upregulated in response to inflammation in mesenchymal derived cell populations, where it negatively regulates pro-inflammatory signalling in vitro [3,7,8]. We hypothesised that worsening inflammation in the TNF‐tg11βKO mouse was due to loss of GC production by FLS and other mesenchymal populations. Therefore, we generated a triple transgenic animal, crossing the TNF-tg mouse onto the HSD11B1 floxed/Twist2-cre to generate a TNF-tg11βflx/tw2cre mouse with mesenchymal cell-specific targeted deletion of 11β-HSD1. Ex vivo analysis of enzyme activity in tissues in WT11βflx/tw2cre and TNF-tg11βflx/tw2cre animals demonstrated that, whilst 11β-HSD1 activity was maintained within spleen and liver (Fig. 5a, Sup. Fig. 3a), activity was significantly suppressed within muscle, adipose and front paw (Fig. 5a–c, Sup Fig. 3b). Primary haematopoietic cells and leukocytes maintained normal 11β-HSD1 activity in WT11βflx/tw2cre and TNF-tg11βflx/tw2cre animals (Fig. 5d). However, 11β-HSD1 activity was totally abrogated in primary FLS (Fig. 5e), myocytes and osteoblasts (data not shown), indicating its deletion in mesenchymal derived cells from WT11βflx/tw2cre and TNF-tg11βflx/tw2cre animals. Clinical scores and body weights were similar in TNF-tg11βflx/tw2cre and TNF-tg mice, whilst a significant reduction in joint inflammation and paw swelling was identified in TNF-tg11βflx/tw2cre mice (Fig. 5f–i). Histological analysis of joint destruction in the elbows and phalanges of TNF-tg11βflx/tw2cre animals revealed similar patterns of bone erosion and synovitis compared with TNF-tg mice (Fig. 5j,l). Scoring of local bone erosions revealed significant reductions in scores in TNF-tg11βflx/tw2cre animals relative to TNF-tg (Fig. 5k,m). Analysis of BV/TV, TT and TN in trabeculae from tibia identified comparable systemic bone loss in both TNF-tg11βflx/tw2cre and TNF-tg animals (Fig. 5n–p). These data indicate that the exacerbated inflammatory phenotype reported in the TNF-tg11βKO mouse is not dependant on 11β-HSD1activity in mesenchymal derived cell populations such as FLS and osteoblasts.
Fig. 5.
(a–c) Corticosterone generation in ex vivo biopsies of spleen, front paw and tibialis anterior and in (d–e) primary cultures of bone marrow cells and fibroblast like synoviocytes (FLS) determined by scanning thin layer chromatography isolated from WT, TNF-tg, WTCre+ and TNF-tgCre+ at 9 weeks. (f) Scoring of joint inflammation, (g) front paw swelling (mm), (h) clinical scoring (weight, inflammation, grimace, behaviour, mobility, inflammation severity and duration), (i) body weights in WT, TNF-tg, WTCre+ and TNF-tgCre+ at 9 weeks. (j) Representative images of paraffin-embedded ulna/humerus joint interface (20X) stained with haematoxylin and eosin (scale bars 200 μm) (k) representative images of 3D reconstructions of hind paws using micro-CT, (l) quantification of degree of synovitis in the ulna/humerus joint interface determined by image J, (m) quantification of cortical erosion in the bones of the ankle, metatarsals and phalanges in WT, TNF-tg, WTCre+ and TNF-tgCre+ at 9 weeks. (n) Bone volume to tissue volume (BV/TV), (o) trabecular thickness (TT) and (p) trabecular number (TN) determined by Meshlab software analysis of micro CT at 5 and 9 weeks in WT, TNF-tg, WT11βflx/tw2cre and TNF-tg11βflx/tw2cre at 9 weeks. AU (Arbitrary units). Values are expressed as mean ± standard error of six animals per group. Statistical significance was determined using one way ANOVA with a Tukey post hoc analysis. *p < 0.05, **p < 0.005, ***p < 0.001.
4. Discussion
Studies examining the deletion of 11β-HSD1 in resolving models of inflammation have identified modest changes in peak inflammation with reduced leukocyte clearance, delayed resolution of inflammation and evidence of abnormal tissue repair [14,26]. We show that in chronic polyarthritis, 11β-HSD1 is a critical regulator of inflammation, with its absence greatly increasing synovitis, joint destruction and systemic bone loss.
During inflammatory arthritis, 11β-HSD1 is increased within synovial leukocytes and FLS [[4], [5], [6],27,28]. Its role in this context is complex, with some studies suggesting it possesses an important anti-inflammatory role [3,29]. Other groups have reported pro-inflammatory properties of 11β-HSD1 with selective inhibitors suppressing disease activity in models of LPS induced septic shock and collagen induced arthritis (CIA) [30,31].
Our study strongly supports a potent anti-inflammatory role for 11β-HSD1 in the context of chronic persistent arthritis, where its global deletion resulted in a marked increase in local inflammation, joint destruction and osteoporosis. This did not appear to be mediated by a direct increase in TNFα transgene expression in the TNF-tg11βKO mice, but rather, by an increase in downstream pro-inflammatory pathway activity with increased mRNA expression of cytokines such as IL-1β and IL-6. This would suggest that deletion of 11β-HSD1 has attenuated the well-established inhibitory role that GCs elicit on pro-inflammatory signalling pathways such as p38-MAPK and NF-κB, downstream of the TNFα receptor.
11β-HSD1 has been suggested to play an important role in regulating macrophage polarisation and function driving an anti-inflammatory, pro-resolution phenotype during macrophage maturation [26,[32], [33], [34]]. When we examined the synovial leukocyte infiltrate in TNF-tg11βKO mice we observed a marked increase in all leukocyte populations, with a shift in macrophages from M2 to M1 polarisation. These findings suggest that 11β-HSD1 supports M2 polarisation and thus phagocytic clearance of apoptotic and damaged cells in chronic inflammation. In future studies it will be of significant interest to examine the inflammatory phenotype in additional inflammatory models of polyarthritis, such as CIA and K/BxN serum induction model in conjunction with both global deletion of 11β-HSD1 as well as with targeted therapeutic inhibitors of this enzyme to further validate these findings.
Osteoporosis and increased fracture risk are prevalent in patients with chronic inflammatory arthritis [35,36]. The contribution of 11β-HSD1 to this process is of significant interest given the duality of therapeutic GC actions in vivo, where they suppress inflammation, but drive systemic osteoporosis. We observed a marked increase in systemic inflammatory bone loss in the TNF-tg11βKO mouse with reduced trabecular bone volume, trabecular number and thickness. Analysis of TNF-tg11βKO mice at five weeks confirmed that this was not a developmental phenotype. At nine weeks, bone loss in the TNF-tg11βKO mouse appeared to be driven by a decrease in bone formation and an increase in bone resorption. mRNA analysis of tibia confirmed a decrease in the osteoblast marker RUNX2 and Tnfrsf11b (osteoprotegerin) in conjunction with a marked increase in expression of the pro-osteoclastogenic cytokines TNFα, IL-1β, IL-6 and MCSF. Local GC activation by 11β-HSD1 thus plays a critical role in protecting against systemic bone loss in the context of persistent inflammation.
We have previously shown that mesenchymal derived cell populations including FLS, myocytes, osteoblasts and adipocytes all potently upregulate the expression of 11β-HSD1 in response to inflammation [7]. In this study we examined the contribution of this mesenchymal GC activation to synovitis, joint destruction and bone loss using a mesenchymal targeted 11β-HSD1 null mouse.
TNF-tg11βflx/tw2cre animals had a similar phenotype to TNF-tg mice, with comparable joint inflammation, joint destruction and systemic bone loss. Interestingly, these mice possessed a modest protection from local joint destruction that would support a catabolic role for 11β-HSD1 within the mesenchyme. Previous studies have reported that inhibition of GC signalling in mesenchymal derived osteoblasts and chondrocytes improves inflammation and joint destruction in acute models of polyarthritis, where GC signalling in osteoblasts and chondrocytes can possess pro-inflammatory actions, and act in an immune-stimulatory manner [[37], [38], [39]]. However, our data indicate that the marked increase in synovitis, joint destruction and bone loss reported in the global 11β-HSD1 null model of chronic polyarthritis are instead dependant on deletion of GC activation in non-mesenchymal cell populations such as macrophages and T cells.
5. Conclusion
We have demonstrated a fundamental role for GC activation via 11β-HSD1 in regulating joint inflammation, joint destruction and systemic bone loss in chronic inflammatory arthritis. Whilst a role for 11β-HSD1 inhibitors has been proposed in the treatment of metabolic complications of chronic inflammatory diseases this approach may severely exacerbate local and systemic features of disease.
Conflicts of interest disclosure
The authors reported no conflict of interests for the work described here.
Author contributions
RSH carried out experiments with support from CF, AJN, APC, GD and RB. RSH and KR supervised the project with support from MSC. RSH wrote the manuscript. All authors discussed the results and contributed to the final manuscript.
Acknowledgments
We would like to thank Professor George Kollias (Hellenic Pasteur Institute, Athens, Greece) for providing the hTNFtg mice. The Biomedical Services Unit (University of Birmingham) for supporting animal experiments and the Department of Musculoskeletal Pathology (Robert Aitken Institute, University of Birmingham) for embedding and cutting tissue for histology and Will Backhouse and Connar Westgate for support with analysis and quantification of histology. We would also like to acknowledge the Arthritis Research UK Centre of Excellence for the Pathogenesis of Rheumatoid Arthritis in supporting this work. This research was supported by the Arthritis Research UK grants (Reference: 19859 & 20843) and supported by the Arthritis Research UK Centre of Excellence for the Pathogenesis of Rheumatoid Arthritis and the MRC Arthritis Research UK Centre for Musculoskeletal Ageing Research.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.jaut.2018.05.010.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
Supplementary Fig. 1.
(a), Gene expression (AU) of hTNF in homogenates of tibia determined by quantitative RT-PCR from WT, TNF-tg, 11βKO, and TNF-tg11βKO mice at 9 weeks. (b) Serum levels of TNFα (human) (pg/ml) determined by ELISA in WT, TNF-tg, 11βKO, and TNF-tg11βKO mice at 9 weeks. Values are expressed as mean ± standard error of six animals per group. Statistical significance was determined using one way ANOVA with a Tukey's post hoc analysis.
Supplementary Fig. 2.
(a-d) Representative images of paraffin-embedded ulna/humerus joint interface (20X) stained with safranin O (scale bars 300 μm) from WT, TNF-tg, 11βKO, and TNF-tg11βKO mice at 9 weeks. (e-h) Representative images of paraffin-embedded ulna/humerus joint interface (20X) following TRAP staining to visualise osteoclasts. (i) Quantification of safranin O staining across cartilage of the ulna/humerus joint interface determined by image J. (j-k) Osteoclast numbers at the ulna/humerus and metacarpal/phalanges interface normalised to bone surface area determined by image J analysis of TRAP stained paraffin-embedded sections of the wrist, Values are expressed as mean ± standard error of six animals per group. AU (Arbitrary units). Statistical significance was determined using one way ANOVA with a Tukey post hoc analysis. *p < 0.05, **p < 0.005, ***p < 0.001.
Supplementary Fig. 3.
(a-g) FACS quantification of synoviocytes (CD45+leukocytes, CD45+CD3+ T Cells, CD45 + CD3+ CD4+ T cells, CD45 + CD3+ CD8+ T cells, CD45 + CD3− CD19 + B Cells, CD45 + CD11b + SiglecF- Ly6ghi neutrophils, and CD45 + CD11b + SiglecF- Ly6g- CD64 + F4/80 + macrophages) following tissue digestion of the joints from upper and lower hind leg bones from WT, TNF-tg, 11βKO, and TNF-tg11βKO mice at 9 weeks. Values are expressed as mean ± standard error of six animals per group. Statistical significance was determined using one way ANOVA with a Turkeys post hoc analysis. *p < 0.05, **p < 0.005, ***p < 0.001.
Supplementary Fig. 4.
(a), Bone volume to tissue volume (BV/TV), (b) trabecular thickness (TT) and (c) trabecular number (TN) determined by Meshlab software analysis of micro CT 5 weeks in WT, TNF-tg, 11βKO, and TNF-tg11βKO mice. Values are expressed as mean ± standard error of six animals per group. Statistical significance was determined using one way ANOVA with a Dunnett's post hoc analysis.
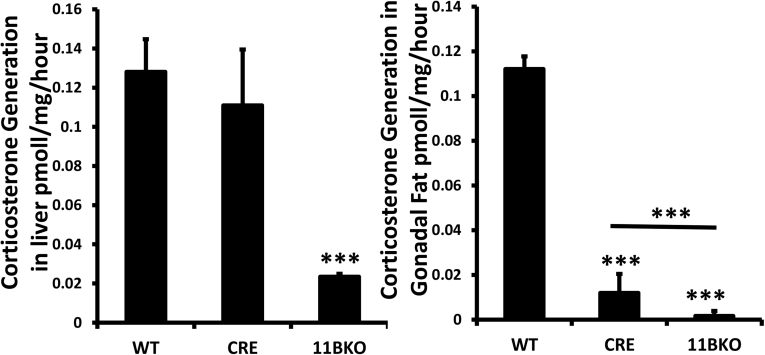
Supplementary Fig. 5.
(a-b) Corticosterone generation in ex vivo biopsies of liver and gonadal fat, determined by scanning thin layer chromatography isolated from WT, WTCre+ and 11βKO animals at 9 weeks. Values are expressed as mean ± standard error of three animals per group. Statistical significance was determined using one way ANOVA with a Tukey post hoc analysis. *p < 0.05, **p < 0.005, ***p < 0.001.
References
- 1.Stewart P.M., Krozowski Z.S. 11 beta-Hydroxysteroid dehydrogenase. Vitam. Horm. 1999;57:249–324. [PubMed] [Google Scholar]
- 2.Morgan S.A. 11beta-HSD1 is the major regulator of the tissue-specific effects of circulating glucocorticoid excess. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E2482–E2491. doi: 10.1073/pnas.1323681111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hardy R.S. Differential expression, function and response to inflammatory stimuli of 11beta-hydroxysteroid dehydrogenase type 1 in human fibroblasts: a mechanism for tissue-specific regulation of inflammation. Arth. Res. Ther. 2006;8:R108. doi: 10.1186/ar1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nanus D.E. TNFalpha regulates cortisol metabolism in vivo in patients with inflammatory arthritis. Ann. Rheum. Dis. 2015;74:464–469. doi: 10.1136/annrheumdis-2013-203926. [DOI] [PubMed] [Google Scholar]
- 5.Hardy R. Local and systemic glucocorticoid metabolism in inflammatory arthritis. Ann. Rheum. Dis. 2008;67:1204–1210. doi: 10.1136/ard.2008.090662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nanus D.E. Differential glucocorticoid metabolism in patients with persistent versus resolving inflammatory arthritis. Arthritis Res. Ther. 2015;17:121. doi: 10.1186/s13075-015-0633-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahasan M.M. Inflammatory regulation of glucocorticoid metabolism in mesenchymal stromal cells. Arthritis Rheum. 2012;64:2404–2413. doi: 10.1002/art.34414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaur K. Synergistic induction of local glucocorticoid generation by inflammatory cytokines and glucocorticoids: implications for inflammation associated bone loss. Ann. Rheum. Dis. 2010;69:1185–1190. doi: 10.1136/ard.2009.107466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hardy R.S. 11beta-hydroxysteroid dehydrogenase type 1 within muscle protects against the adverse effects of local inflammation. J. Pathol. 2016 doi: 10.1002/path.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thieringer R. 11 Beta-hydroxysteroid dehydrogenase type 1 is induced in human monocytes upon differentiation to macrophages. J. Immunol. 2001;167:30–35. doi: 10.4049/jimmunol.167.1.30. [DOI] [PubMed] [Google Scholar]
- 11.Freeman L. Expression of 11beta-hydroxysteroid dehydrogenase type 1 permits regulation of glucocorticoid bioavailability by human dendritic cells. Blood. 2005;106:2042–2049. doi: 10.1182/blood-2005-01-0186. [DOI] [PubMed] [Google Scholar]
- 12.Zhang T.Y., Ding X., Daynes R.A. The expression of 11 beta-hydroxysteroid dehydrogenase type I by lymphocytes provides a novel means for intracrine regulation of glucocorticoid activities. J. Immunol. 2005;174:879–889. doi: 10.4049/jimmunol.174.2.879. [DOI] [PubMed] [Google Scholar]
- 13.Ishii T. Augmentation of 11beta-hydroxysteroid dehydrogenase type 1 in LPS-activated J774.1 macrophages--role of 11beta-HSD1 in pro-inflammatory properties in macrophages. FEBS Lett. 2007;581:349–354. doi: 10.1016/j.febslet.2006.11.032. [DOI] [PubMed] [Google Scholar]
- 14.Gilmour J.S. Local amplification of glucocorticoids by 11 beta-hydroxysteroid dehydrogenase type 1 promotes macrophage phagocytosis of apoptotic leukocytes. J. Immunol. 2006;176:7605–7611. doi: 10.4049/jimmunol.176.12.7605. [DOI] [PubMed] [Google Scholar]
- 15.Keffer J. Transgenic mice expressing human tumour necrosis factor: a predictive genetic model of arthritis. EMBO J. 1991;10:4025–4031. doi: 10.1002/j.1460-2075.1991.tb04978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chu C.Q., Field M., Feldmann M., Maini R.N. Localization of tumor necrosis factor alpha in synovial tissues and at the cartilage-pannus junction in patients with rheumatoid arthritis. Arthritis Rheum. 1991;34:1125–1132. doi: 10.1002/art.1780340908. [DOI] [PubMed] [Google Scholar]
- 17.Elliott M.J. Randomised double-blind comparison of chimeric monoclonal antibody to tumour necrosis factor alpha (cA2) versus placebo in rheumatoid arthritis. Lancet. 1994;344:1105–1110. doi: 10.1016/s0140-6736(94)90628-9. [DOI] [PubMed] [Google Scholar]
- 18.Matsuno H. The role of TNF-alpha in the pathogenesis of inflammation and joint destruction in rheumatoid arthritis (RA): a study using a human RA/SCID mouse chimera. Rheumatology (Oxford) 2002;41:329–337. doi: 10.1093/rheumatology/41.3.329. [DOI] [PubMed] [Google Scholar]
- 19.Naylor A.J., Desanti G., Saghir A.N., Hardy R.S. TNFalpha depleting therapy improves fertility and animal welfare in TNFalpha-driven transgenic models of polyarthritis when administered in their routine breeding. Lab. Anim. 2017 doi: 10.1177/0023677217707985. 23677217707985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu K. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development. 2003;130:3063–3074. doi: 10.1242/dev.00491. [DOI] [PubMed] [Google Scholar]
- 21.Li A. Deletion of mesenchymal glucocorticoid receptor attenuates embryonic lung development and abdominal wall closure. PLoS One. 2013;8 doi: 10.1371/journal.pone.0063578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Semjonous N.M. Hexose-6-phosphate dehydrogenase contributes to skeletal muscle homeostasis independent of 11beta-hydroxysteroid dehydrogenase type 1. Endocrinology. 2011;152:93–102. doi: 10.1210/en.2010-0957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hardy R.S. Characterisation of fibroblast-like synoviocytes from a murine model of joint inflammation. Arthritis Res. Ther. 2013;15:R24. doi: 10.1186/ar4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Z. Th2 and eosinophil responses suppress inflammatory arthritis. Nat. Commun. 2016;7:11596. doi: 10.1038/ncomms11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Misharin A.V. Nonclassical Ly6C(-) monocytes drive the development of inflammatory arthritis in mice. Cell Rep. 2014;9:591–604. doi: 10.1016/j.celrep.2014.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coutinho A.E. 11beta-Hydroxysteroid dehydrogenase type 1, but not type 2, deficiency worsens acute inflammation and experimental arthritis in mice. Endocrinology. 2012;153:234–240. doi: 10.1210/en.2011-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy D., West H.F. Catabolism and interconversion of cortisol and cortisone in human synovial tissue in vitro. Ann. Rheum. Dis. 1969;28:637–643. doi: 10.1136/ard.28.6.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haas C.S. Identification of genes modulated in rheumatoid arthritis using complementary DNA microarray analysis of lymphoblastoid B cell lines from disease-discordant monozygotic twins. Arthritis Rheum. 2006;54:2047–2060. doi: 10.1002/art.21953. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt M. Reduced capacity for the reactivation of glucocorticoids in rheumatoid arthritis synovial cells: possible role of the sympathetic nervous system? Arthritis Rheum. 2005;52:1711–1720. doi: 10.1002/art.21091. [DOI] [PubMed] [Google Scholar]
- 30.Park S.B. Anti-inflammatory effect of a selective 11beta-hydroxysteroid dehydrogenase type 1 inhibitor via the stimulation of heme oxygenase-1 in LPS-activated mice and J774.1 murine macrophages. J. Pharmacol. Sci. 2016;131:241–250. doi: 10.1016/j.jphs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 31.Zhang L. 11beta-Hydroxysteroid dehydrogenase 1 inhibition attenuates collagen-induced arthritis. Int. Immunopharm. 2013;17:489–494. doi: 10.1016/j.intimp.2013.07.015. [DOI] [PubMed] [Google Scholar]
- 32.Ehrchen J. Glucocorticoids induce differentiation of a specifically activated, anti-inflammatory subtype of human monocytes. Blood. 2007;109:1265–1274. doi: 10.1182/blood-2006-02-001115. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y. Glucocorticoids promote nonphlogistic phagocytosis of apoptotic leukocytes. J. Immunol. 1999;162:3639–3646. [PubMed] [Google Scholar]
- 34.Varga G. Glucocorticoids induce an activated, anti-inflammatory monocyte subset in mice that resembles myeloid-derived suppressor cells. J. Leukoc. Biol. 2008;84:644–650. doi: 10.1189/jlb.1107768. [DOI] [PubMed] [Google Scholar]
- 35.Hooyman J.R., Melton L.J., 3rd, Nelson A.M., O'Fallon W.M., Riggs B.L. Fractures after rheumatoid arthritis. A population-based study. Arthritis Rheum. 1984;27:1353–1361. doi: 10.1002/art.1780271205. [DOI] [PubMed] [Google Scholar]
- 36.Laan R.F., van Riel P.L., van de Putte L.B. Bone mass in patients with rheumatoid arthritis. Ann. Rheum. Dis. 1992;51:826–832. doi: 10.1136/ard.51.6.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buttgereit F. Transgenic disruption of glucocorticoid signaling in mature osteoblasts and osteocytes attenuates K/BxN mouse serum-induced arthritis in vivo. Arthritis Rheum. 2009;60:1998–2007. doi: 10.1002/art.24619. [DOI] [PubMed] [Google Scholar]
- 38.Tu J. Endogenous glucocorticoid signaling in chondrocytes attenuates joint inflammation and damage. Faseb. J. 2018;32:478–487. doi: 10.1096/fj.201700659R. [DOI] [PubMed] [Google Scholar]
- 39.Tu J. Transgenic disruption of glucocorticoid signaling in osteoblasts attenuates joint inflammation in collagen antibody-induced arthritis. Am. J. Pathol. 2016;186:1293–1301. doi: 10.1016/j.ajpath.2015.12.025. [DOI] [PubMed] [Google Scholar]