Abstract
ARHGEF9, also known as Collybistin, a guanine nucleotide exchange factor for Rho family GTPases, is thought to play an essential role in the mammalian brain. In this study, we prepared a specific polyclonal antibody against ARHGEF9, anti-ARHGEF9, and carried out expression analyses with mouse tissues especially brain. Western blotting analyses demonstrated tissue-dependent expression profiles of ARHGEF9 in the young adult mouse, and strongly suggested a role during brain development. Immunohistochemical analyses revealed developmental stage-dependent expression profiles of ARHGEF9 in cerebral cortex, hippocampus and cerebellum. ARHGEF9 exhibited partial localization at dendritic spines in cultured hippocampal neurons. From the obtained results, anti-ARHGEF9 was found to be a useful tool for biochemical and cell biological analyses of ARHGEF9.
Keywords: ARHGEF9, guanine nucleotide exchange factor (GEF), antibody, cerebral cortex, neuron
I. Introduction
ARHGEF9, also known as Collybistin, is a Dbl (diffuse B-cell lymphoma)-family guanine nucleotide exchange factor (GEF) for Rho small GTPases [9]. ARHGEF9 has been reported to regulate the recruitment of gephyrin, the main scaffolding protein essential for clustering glycine and GABAA receptors at the cell surface [5, 9, 11, 14, 15, 17]. The critical role of ARHGEF9 in brain development is supposed by mutations associated with neurodevelopmental disorders such as epilepsy and mental retardation [1, 5, 8, 10, 16, 20, 24]. Considering the involvement in the etiology of neurodevelopmental disorders, ARHGEF9 is thought to play a crucial role during brain development. However, precise expression analyses of ARHGEF9 have not so far been conducted.
In this report, we produced a rabbit polyclonal antibody for ARHGEF9 and performed expression analyses of the protein. In western blotting analyses, ARHGEF9 was detected in a wide variety of mouse tissues including the central nervous system (CNS), and this molecule was expressed in a developmental stage-dependent manner in the brain. We then performed immunohistochemical analyses with mouse brain slices including cerebral cortex, hippocampus and cerebellum at different developmental stages. Subcellular distribution of ARHGEF9 was also examined with primary cultured hippocampal neurons with immunofluorescence analyses. The obtained results suggest function(s) of ARHGEF9 during development of the CNS.
II. Materials and Methods
Ethics statement
For the animal experiments, we followed the Fundamental Guidelines for Proper Conduct of Animal Experiments and Related Activity in Academic Research Institution under the jurisdiction of the Ministry of Education, Culture, Sports, Science and Technology, Japan. All protocols for animal handling and treatment were reviewed and approved by the Animal Care and Use Committee of Institute for Developmental Research, Aichi Human Service Center (approval number; M-10).
Plasmids
Mouse (m)ARHGEF9-v1 cDNA was obtained from Kazusa DNA Institute and constructed into pCAG-Myc vector (Addgene Inc., Cambridge, MA, USA). For RNAi experiments, pSuper-RNAi-puro vector (OligoEngine, Seattle, WA, USA) was designed to target 2 distinct coding sequences in mARHGEF9-v1 cDNA (pSuper-mARHGEF9#1: 5'-GCTTTCAAGCTTCATAACA-3', 1171–1189; pSuper-mARHGEF9#2: 5'-GGTACAAGAAGATGAAAAA-3', 1279–1297). Numbers indicate the positions from translational start sites. All constructs were verified by DNA sequencing.
Antibodies
Maltose-binding protein (MBP)-fused mARHGEF9-v1 C-terminal fragment (aa 331–516) was expressed in E. coli, affinity-purified using amylose resin (New England BioLabs Inc., Ipswich, MA, USA) and used as an antigen. After preimmune bleeding, 2 rabbits were subcutaneously immunized with the antigen (200 μg) in complete Freund’s adjuvant. We then boosted twice every 4 weeks with 200 μg of antigen with incomplete Freund’s adjuvant. Anti-ARHGEF9#1 and #2 sera were obtained 1 week after the final boost, and affinity-purified on a column to which the antigen had been conjugated. Since purified anti-ARHGEF9#1 and #2 showed similar quality in western blotting, we used anti-ARHGEF9#1 in the present study.
The Monoclonal mouse anti-glial fibrillary acidic protein (GFAP), anti-Tau-1 and anti-MAP2 were from Santa Cruz Biotech (Santa Cruz Biotechnology, Santa Cruz, CA, USA), Chemicon International Inc. (Temecula, CA, USA) and Sigma-Aldrich (St Louis, MO, USA), respectively. Polyclonal rabbit antibodies against Sept11 (a cytoskeleton-related protein) and Myc-tag were prepared as described [4, 13].
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting
Whole tissue or cell extracts were solubilized in SDS-sample loading buffer (50 mM Tris/pH 6.8, 2.5% SDS, 4% glycerol, 1.5% mercaptoethanol, 0.03% bromophenol blue), fractionated by SDS-PAGE (10% gel) and transferred to a nitrocellulose membrane using a transfer apparatus. After blocking with 0.5% nonfat milk in TBST (20 mM Tris/pH 7.6, 140 mM NaCl, 0.1% Tween 20) for 60 min, the membrane was washed once with TBST and incubated with antibodies against ARHGEF9 (1:1000), Sept11 (1:1000) or Myc-tag (1:10000) at room temperature for overnight. Membranes were washed 3 times for 10 min and incubated with horseradish peroxidase-conjugated anti-mouse or anti-rabbit antibodies (1:10000 dilution) (Amersham Biosciences Corp., Little Chalfont, UK) for 1 hr. Blots were washed with TBST 3 times and developed with the ECL system (Amersham Biosciences Corp.) according to the manufacturer’s protocol [2, 21].
Preparation of extracts from mouse tissues
ICR mice were purchased from Japan SLC (Shizuoka, Japan) and raised in our laboratory. Cytosol and membrane extracts of various tissues were prepared from postnatal day 40 (P40) mice of either sex as described previously [7]. Briefly, tissues were homogenized with 10 vol. of 50 mM Tris/pH 7.5 containing 0.1 M NaF, 5 mM EDTA, 1 mM Na3VO4, 10 μg/ml aprotinin and 10 μg/ml leupeptin (buffer S). Each suspension was sonicated at 0°C for 1 min and centrifuged at 125,000 g for 20 min at 4°C. The supernatants were used as cytosolic extracts and the pellets were washed once by sonication and centrifugation with buffer S and then solubilized with buffer S containing 2% SDS. In some experiments, brain tissues were directly homogenized with buffer S containing 2% SDS to obtain the whole tissue extracts [6]. Protein concentration was estimated with a micro bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific, Waltham, MA, USA) with bovine serum albumin (BSA) as a standard. Western blotting was done as described earlier.
COS7 cell culture and transfection
COS7 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum. Cells were transfected with Lipofectamine 2000 (Thermo Fisher Scientific, Carlsbad, CA, USA). After 48 h, transfected cells were treated with 10% trichloroacetic acid to inactivate protein degradation enzymes for 5 min on ice. After rinsing the plate with PBS three times, cells were harvested with SDS-sample loading buffer and subjected SDS-PAGE followed by western blotting [6, 22].
Immunohistochemical analyses
Young adult mice (P40) were perfused transcardially with saline and then with phosphate-buffered 10% formalin. Brains were removed and processed for paraffin embedding. Paraffinized 3 μm-sections were made and mounted on slide glasses. Endogenous peroxidase activity in sections was blocked by incubation in 3% hydrogen peroxide for 5 min. Antigen retrieval was performed by the Pascal heat-induced target retrieval system (DAKO Tokyo, Japan) with 10 mM citrate buffer (pH 6.0). After treatment with 10 mM phosphate-buffered saline (PBS/pH 7.4) containing 2% BSA for 60 min, the slides were incubated with anti-ARHGEF9 (dilution at 1:100 in 2% BSA/PBS) overnight at 4°C. ARHGEF9 was then detected with immunoperoxidase polymer reagents Histofine Simple Stain MAX-PO (R) (424142, Nichirei Bioscience, Tokyo, Japan) for 30 min. The peroxidase binding sites were detected by staining with 3,3'-diaminobenzidine in 50 mM Tris (pH 7.6). Counterstaining was performed using Mayer’s hematoxylin. Images were captured with BZ-9000 microscope (Keyence, Osaka, Japan).
Primary hippocampal neuron culture and immunofluorescence
Mouse primary hippocampal neurons were isolated as follows. Briefly, hippocampal tissue was isolated from embryonic day 16 (E16) mouse pups. Then, gentle dissociation of neuronal cells was performed with trypsin (0.25%) and mechanical disruption was done to separate cells from connective tissue while providing minimum damage to individual neuronal cells [19]. The isolated neurons were cultured essentially as described [3, 6]. Transient transfection was carried out using Lipofectamine 2000 (Life Technologies). Immunofluorescence analysis was done as described [6]. Briefly, cells grown on 13-mm coverslips were fixed in 3.7% formaldehyde in PBS for 15 min, then treated with 0.2% Triton-X 100 for 5 min. 4',6-diamidino-2-phenylindole (DAPI; Nichirei Bioscience, Tokyo, Japan) and anti-ARHGEF9 were used for staining of DNA and ARHGEF9, respectively. Alexa Fluor 488-labeled IgG (Life Technologies) was used as a secondary antibody. Images were captured using FV-1000 confocal laser microscope (Olympus, Tokyo, Japan).
III. Results
Production and characterization of anti-ARHGEF9 antibody
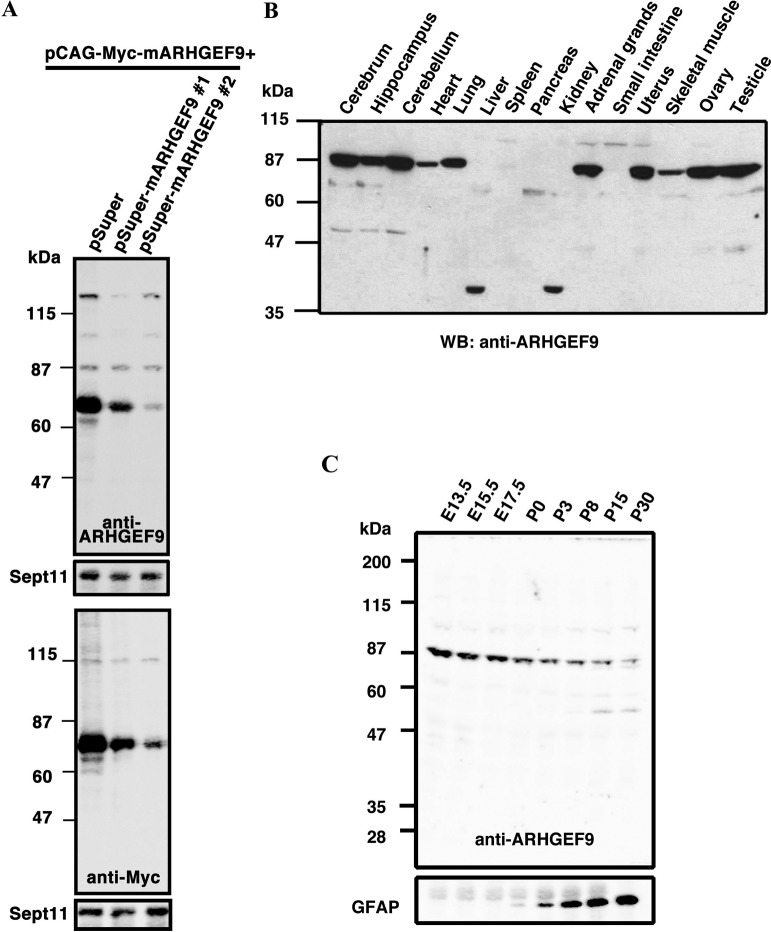
We prepared a rabbit polyclonal antibody, anti-ARHGEF9, against a C-terminal fragment of ARHGEF9, and affinity-purified it. While anti-ARHGEF9 detected Myc-tagged ARHGEF9 expressed in COS7 cells, the immunoreactivity was significantly reduced when Myc-ARHGEF9 expression was silenced by two pSuper-mARHGEF9 RNAi vectors targeting distinct ARHGEF9-coding regions (Fig. 1A, upper large panel). The blot with anti-Myc antibody was also demonstrated (Fig. 1A, lower large panel). Sept11 was visualized as a loading control in each experiment. From these results, anti-ARHGEF9 was considered to recognize ARHGEF9 specifically.
Fig. 1.
Production of an affinity-purified antibody for ARHGEF9, anti-ARHGEF9. (A) COS7 cells were transfected with pCAG-Myc-ARHGEF9 (0.2 μg) together with pSuper control vector or pSuper-mARHGEF9#1 or #2 (1.2 μg each). After 48 h, cells were harvested and whole cell lysates (20 μg of protein) were subjected to western blotting with anti-ARHGEF9 (upper large panel) or anti-Myc (lower large panel). The blots were reprobed with anti-Sept11 (cropped panels). Molecular size markers were shown at left. (B) Cytosolic (10 μg of protein per lane) fractions from young adult mouse (P40) organs were separated by SDS-PAGE (10% gel) and then subjected to western blotting with anti-ARHGEF9. Molecular size markers are shown at left. (C) Developmental changes of ARHGEF9 protein level during corticogenesis. Extracts (20 μg of proteins) at various developmental stages were subjected to western blotting with anti-ARHGEF9 (upper panel). The blot was reprobed with anti-GFAP to confirm the tissue differentiation (lower panel).
Expression profile of ARHGEF9 in mouse tissues
We then performed western blotting analyses to observe the tissue distribution of ARHGEF9. As shown in Fig. 1B, anti-ARHGEF9 detected protein bands in the cytosol fraction of a variety of mouse tissues tested. A protein with a molecular mass of ~87 kDa was detected in all tissues except liver, spleen, pancreas, kidney and small intestine. Given that molecular weight of ARHGEF9-v1 (516 aa) is calculated to be 60,929, the higher molecular mass may be a posttranslationally modified form or yet unidentified isoform. While a ~38 kDa band was detected in liver and kidney, ARHGEF9 was little expressed in spleen, pancreas and small intestine. The ~38 kDa protein is possible to be a degradation product during sample preparation. Further genetic analyses of mouse ARHGEF9 gene as well as analyses with isoform-specific antibodies should be required to address this issue. As for the insoluble membrane fraction, no obvious protein bands were detected in all tissues tested (data not shown).
To gain some insight into the involvement of ARHGEF9 in neuronal development, we conducted expression analyses with the whole tissue extracts of mouse brains prepared from various developmental stages. ARHGEF9 with ~87 kDa was strongly detected at embryonic day (E)13.5 and then gradually decreased throughout the developmental process to postnatal day (P)30 (Fig. 1C, upper panel). A ~50 kD protein was faintly detected from P15. It remains to be clarified whether this is a splicing isoform expressed in a developmental stage dependent manner or a degradation product during sample preparation. GFAP was visualized as a differentiation marker (Fig. 1C, lower panel).
Immunohistochemical analyses of ARHGEF9 during mouse brain development
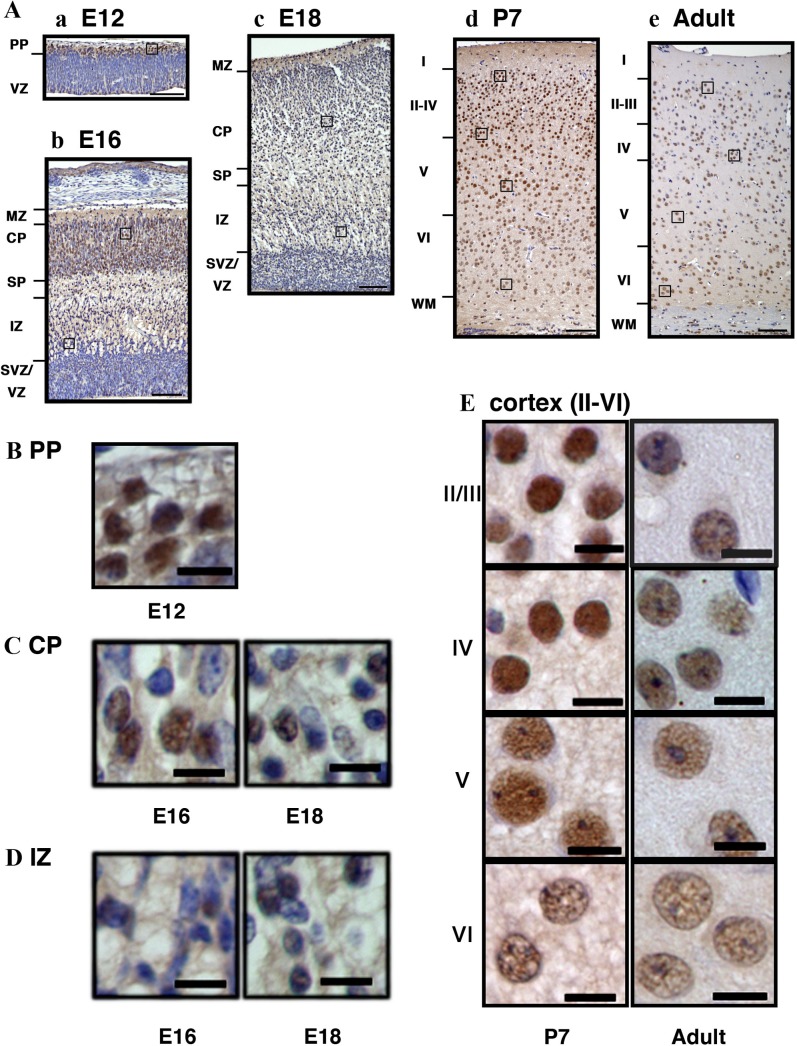
To determine the localization of ARHGEF9 during brain development, we carried out immunohistochemical analyses with mouse brain sections at E12, E16, E18, P7 and P40. ARHGEF9 was detected relatively strongly in the preplate (PP) and cortical plate (CP) at E12 and E16, respectively (Fig. 2Aa and Ab). Around E18, abovementioned ARHGEF9 expression disappeared and the protein came to be distributed evenly throughout the cortex (Fig. 2Ac). At P7, ARHGEF9 was detected across the cortical layers but relatively strong expression at the nucleus was observed. In the young adult stage (P40), the distribution pattern was similar to that at P7 although the intensity became overall lower. When the intracellular localization was analyzed with magnified images, ARHGEF9 appeared to be enriched in the nucleus at all time points tested (Fig. 2B–E). It is notable that many ARHGEF9-negative cells were observed especially in the CP and IZ during the embryonic stage (Fig. 2C and D). In the meantime, dense nuclear staining of ARHGEF9 was visualized in layer II–IV at P7, when compared to layer V–VI neurons with larger nucleus (Fig. 2E). Further analyses are required for identification of ARHGEF9-negative cells as well as positive ones observed during the developmental process. From these immunohistochemical data, ARHGEF9 was found to be expressed spatiotemporally during corticogenesis.
Fig. 2.
Immunohistochemical analyses of ARHGEF9 in mouse cerebral cortex. (A) Cerebral cortex sections of the somatosensory area at E12 (a), E16 (b), E18 (c), P7 (d) and P40 (e) were stained with anti-ARHGEF9. PP, preplate; VZ, ventricular zone; MZ, marginal zone; CP, cortical plate; IZ, intermediate zone; SVZ, subventricular zone; WM, white matter. (B) Cells at the PP (boxed area in Aa) were magnified. (C) Cells at the CP (boxed areas in Ab and Ac) were magnified. (D) Cells at the IZ (boxed areas in Ab and Ac) were magnified. (E) Cells at different layers of cerebral cortex (boxed areas in Ad and Ae) were magnified. Bars = 100 μm (A) and 10 μm (B–E).
Expression of ARHGEF9 in hyppocampus
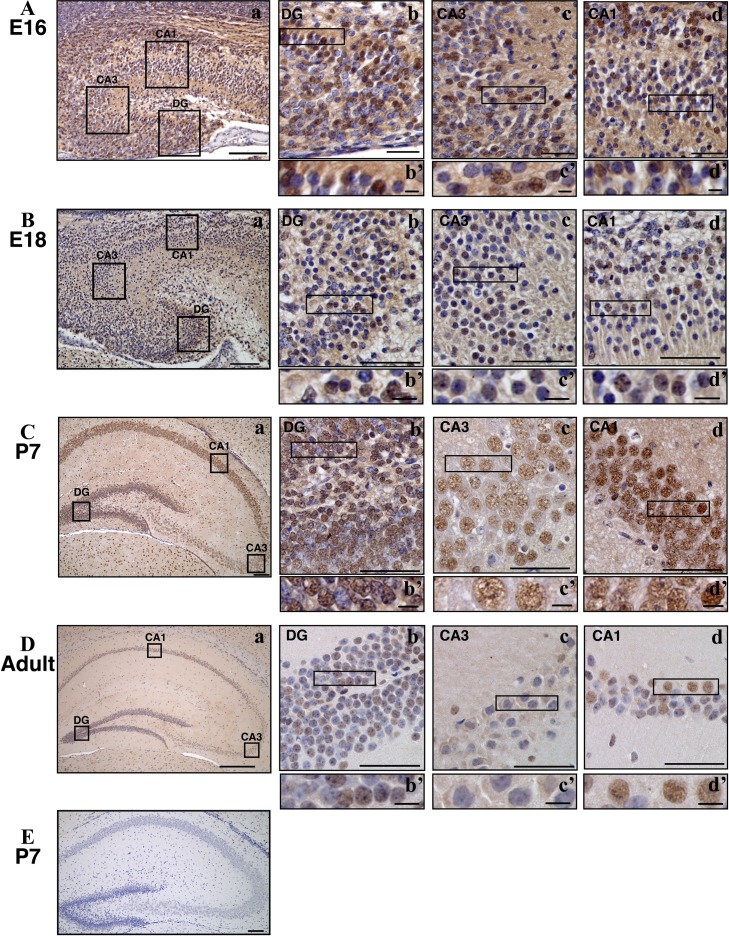
We then examined the distribution of ARHGEF9 in hippocampus at various developmental stages. At E16 and E18, strong nuclear staining was observed in CA1, CA3 and dentate gyrus, although ARHGEF9-negative cells were also observed (Fig. 3A and B). Identities of these cells remain to be clarified. At P7, ARHGEF9 was expressed in most neurons in CA1, CA3 and dentate gyrus (Fig. 3C). While the staining signals relatively strong at dentate gyrus and moderate in CA1, neurons in CA3 showed relatively weak signal. This tendency was eminent at P40 while ARHGEF9 expression became overall weaker in hippocampus at this stage (Fig. 3D). In addition, ARHGEF9-negative cells were prominent especially in CA3 region at P40 (Fig. 3D). Expression profile of ARHGEF9 in hippocampus seems to recapitulate that observed in the western blotting analyses.
Fig. 3.
Immunohistochemical analyses of ARHGEF9 in mouse hippocampus. Distribution of ARHGEF9 in CA1, CA3 and dentate gyrus (DG) were shown at E16 (A), E18 (B), P7 (C) and P40 (D). Double-staining with anti-ARHGEF9 and horseradish peroxidase/diaminobenzidine (HRP/DAB) was carried out with sagittal (A, B) and coronal sections (C, D). Boxed areas in (a) were magnified (b), (c) and (d). Boxed areas in (b), (c) and (d) were magnified (b’), (c’) and (d’), respectively. Bars = 100 μm (A–Ca, and E), 300 μm (Da), 50 μm (b, c, d) and 20 μm (b’, c’, d’). (E) A hippocampal section (P7) was stained without anti-ARHGEF9 for a negative control.
Expression of ARHGEF9 in cerebellum
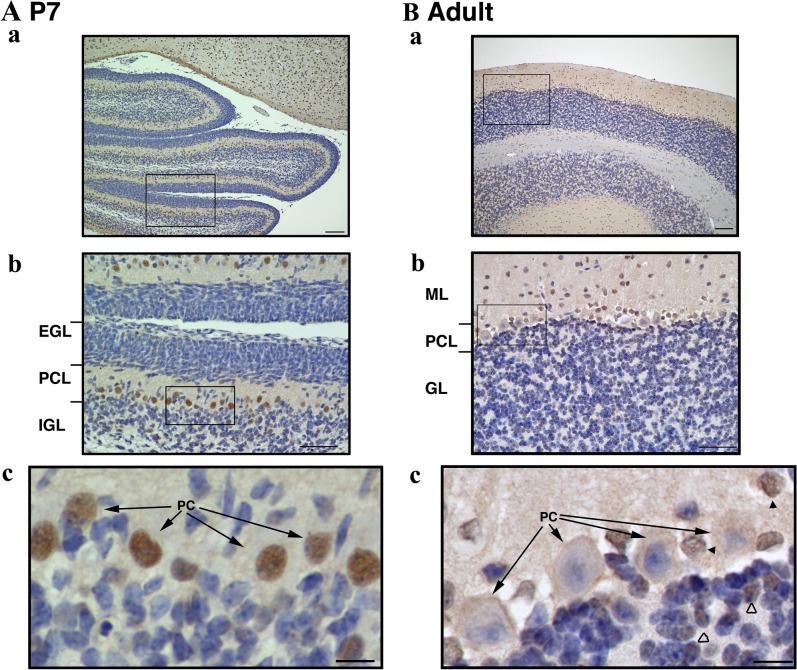
We next examined the distribution of ARHGEF9 in mouse cerebellum. When ARHGEF9 was stained with a developing cerebellar slice at P7, it was detected in the nucleus, but not in the cytoplasm, of Purkinje cells (Fig. 4A). Staining signal was detected in very limited number of cells in the outer and inner granular layers at P7. In the young adult stage (P40), the cortex consists of three layers: molecular layer, purkinje cells and granular layer, respectively from outermost to innermost. Notably, the strong signal at the nucleus of purkinje cells was disappeared at P40 (Fig. 4B, arrows). Instead, ARHGEF9 was detected in the nucleus of cells in the molecular and granular layers as indicated by black and white arrowheads, respectively (Fig. 4Bc). These cells might be basket cells in the molecular layer and a portion of granule cells, although further analyses are required to identify their cell types.
Fig. 4.
Immunohistochemical analyses of ARHGEF9 in mouse cerebellum. Double-staining with anti-ARHGEF9 and HRP/DAB was carried out with coronal sections at P7 (A) and P40 (B). EGL, external granular layer; PCL, Purkinje cell layer; IGL, internal granular layer; ML, molecular layer; GL, granular layer; PC, Purkinje cell. Boxed areas in (a) and (b) were magnified in (b) and (c), respectively. Bars = 100 μm (a), 50 μm (b) and 10 μm (c).
Subcellular localization of ARHGEF9 in primary cultured hippocampal neurons
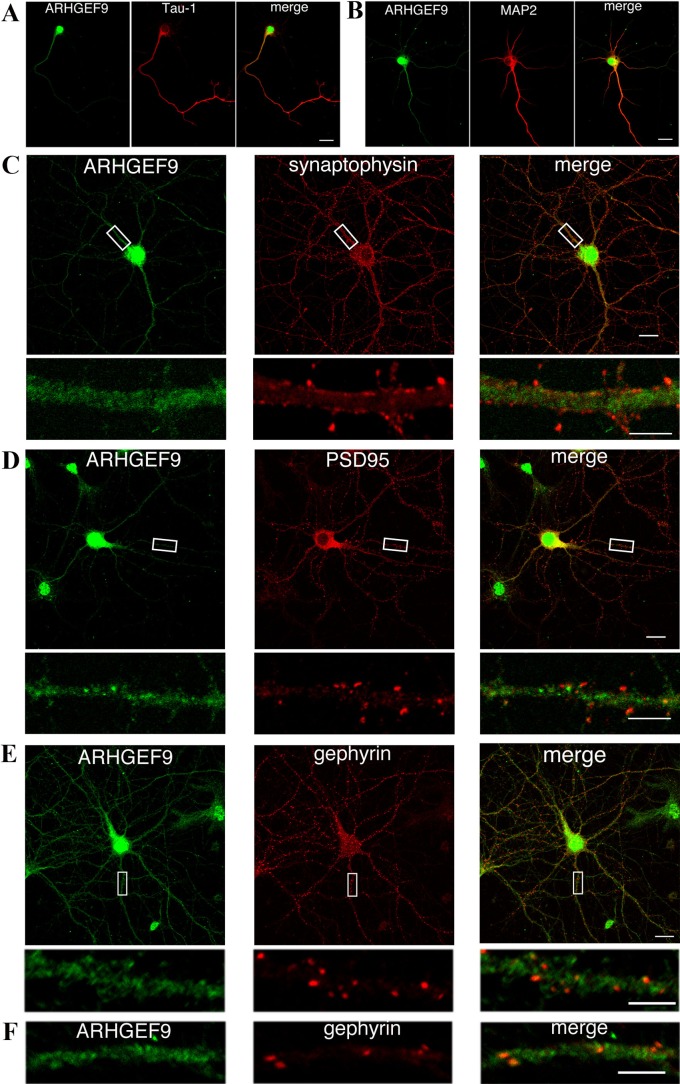
To obtain some insight into the function of ARHGEF9, we looked into the localization of ARHGEF9 in mouse primary cultured hippocampal neurons. In immature neurons of 3 days in vitro (div), ARHGEF9 was distributed dominantly in the nucleus while moderate expression was observed in the perinuclear region and the proximal region of Tau-1-positive axon (Fig. 5A). In 6 div neurons, ARHGEF9 came to be observed in MAP2-positive dendrites in addition to the nuclear and cytoplasmic localization (Fig. 5B). In matured 13 div neurons, while ARHGEF9 was still located in the nucleus and cytoplasm, it was diffusely distributed in dendrites with only partial colocalization with a presynaptic marker synaptophysin and an excitatory postsynaptic marker PSD95 (Fig. 5C and D). When the neurons were stained with anti-ARHGEF9 together with anti-Gephyrin, a maker for inhibitory postsynapses, their partial colocalization was observed in dendrites, suggesting that ARHGEF9 interacts with Gephyrin at inhibitory synapses (Fig. 5E). The colocalization was also observed in cortical neurons (Fig. 5F).
Fig. 5.
Localization of ARHGEF9 in primary cultured mouse hippocampal and cortical neurons. (A and B) Neurons cultured for 3 (A) or 6 days (B) in vitro were double-stained for ARHGEF9 with Tau-1 (A) or MAP2 (B). Merged images were also shown. Bar = 20 μm. (C–E) Neurons cultured for 13 days were double-stained for ARHGEF9 with synaptophysin (C), PSD95 (D) or Gephyrin (E). Boxed areas in the upper panels were magnified in the lower panels. Bars = 20 μm (upper panels) and 5 μm (lower panels). (F) Cortical neurons cultured for 13–14 days in vitro were double-stained for ARHGEF9 with Gephyrin and a dendrite was magnified. Merged images were also shown. Bar = 5 μm.
IV. Discussion
In the present study, we generated a specific polyclonal antibody for ARHGEF9, and performed some biochemical and morphological characterization of the molecule during mouse brain development. Although the molecular mass of ARHGEF9 is predicted to ~60 kDa from the reported cDNA sequence data [9, 14, 23], anti-ARHGEF9 detected a protein band with ~87 kDa in western blotting analyses. We assume that posttranslational modification(s) may change apparent molecular mass of ARHGEF9. Alternatively, the 87 kDa protein may be yet unidentified ARHGEF9 isoform expressed in a tissue- and cell type-specific manner. Further genetic analyses of ARHGEF9 are required to identify its possible isoforms. Consistent with the results here with immunohistochemical analyses, ARHGEF9 mRNAs have been reported to be highly expressed in the CP rather than the SVZ/VZ at E14 and E18, and cortical layer in young adult mice, suggesting that ARHGEF9 is induced in postmitotic neurons [12].
Partial colocalization of ARHGEF9 with PSD95 might indicate a role of ARHGEF9 in synaptic functions such as protein transport [18]. Since ARHGEF9-negative cells dotted cerebral cortex and hippocampus during corticogenesis, further analyses are essential to identify these cell types. In addition, it remains to determine the entity of synaptophysin/PSD95-negative punctate staining of ARHGEF9 in dendrites of cultured hippocampal neurons (Fig. 5D).
Although ARHGEF9 was detected in axon, dendrite, soma and nucleus in the primary cultured hippocampal neurons (Fig. 5), this localization profile is different from that in immunohistochemical analyses (Fig. 2). Possible explanation of the discrepancy was the difference of staining conditions and methods.
We consider that anti-ARHGEF9 will contribute to future histological, cell biological and biochemical analyses of ARHGEF9. This antibody also may be useful for pathophysiological analyses of ARHGEF9 in neurodevelopmental disorders such as epilepsy and ID [1, 5, 8, 10, 16, 20, 24].
V. Abbreviations
CP, cortical plate; DG, dentate gyrus; div, days in vitro; EGL, external granular layer; GL, granular layer; GST, glutathione S-transferase; IGL, internal granular layer; IZ, intermediate zone; ML, molecular layer; MZ, marginal zone; PC, Purkinje cell; PCL, Purkinje cell layer; PP, preplate; SDS, sodium dodecyl sulfate; SP, subplate; SVZ, subventricular zone; VZ, ventricular zone; WM, white matter.
VI. Conflicts of Interest
The authors declare no conflicts of interest.
VII. Acknowledgments
This work was supported in part by JSPS KAKENHI Grant (grant no. 16J06511, 23590124, 16K07211 and 17K16294), a grant-in-aid of the Practical Research Project for Rare/Intractable Diseases from Japan Agency for Medical Research and Development (AMED) (17bm0804009h0201), and Takeda Science Foundation.
VIII. References
- 1.Alber M., Kalscheuer V. M., Marco E., Sherr E., Lesca G., Till M., Gradek G., Wiesener A., Korenke C., Mercier S., Becker F., Yamamoto T., Scherer S. W., Marshall C. R., Walker S., Dutta U. R., Dalal A. B., Suckow V., Jamali P., Kahrizi K., Najmabadi H. and Minassian B. A. (2017) ARHGEF9 disease: Phenotype clarification and genotype-phenotype correlation. Neurol. Genet. 3; e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamada N., Ito H., Nishijo T., Iwamoto I., Morishita R., Tabata H., Momiyama T. and Nagata K-I. (2016) Essential role of the nuclear isoform of RBFOX1, a candidate gene for autism spectrum disorders, in the brain development. Sci. Rep. 6; 30805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamada N., Iwamoto I., Tabata H. and Nagata K-I. (2017) MUNC18-1 gene abnormalities are involved in neurodevelopmental disorders through defective cortical architecture during brain development. Acta Neuropathol. Commun. 5; 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanai N., Nagata K-I., Kawajiri A., Shiromizu T., Saitoh N., Hasegawa Y., Murakami S. and Inagaki M. (2004) Biochemical and cell biological characterization of a mammalian septin, Sept11. FEBS Lett. 568; 83–88. [DOI] [PubMed] [Google Scholar]
- 5.Harvey K., Duguid I. C., Alldred M. J., Beatty S. E., Ward H., Keep N. H., Lingenfelter S. E., Pearce B. R., Lundgren J., Owen M. J., Smart T. G., Lüscher B., Rees M. I. and Harvey R. J. (2004) The GDP-GTP exchange factor collybistin: an essential determinant of neuronal gephyrin clustering. J. Neurosci. 24; 5816–5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inaguma Y., Ito H., Iwamoto I., Matsumoto A., Yamagata T., Tabata H. and Nagata K-I. (2016) Morphological characterization of Class III phosphoinositide 3-kinase during mouse brain development. Med. Mol. Morphol. 49; 28–33. [DOI] [PubMed] [Google Scholar]
- 7.Ito H., Mizuno M., Noguchi K., Morishita R., Iwamoto I., Hara A. and Nagata K-I. (2018) Expression analysis of Phactr1 (phosphatase and actin regulator 1) during mouse brain development. Neurosci. Res. 128; 50–57. [DOI] [PubMed] [Google Scholar]
- 8.Kalscheuer V. M., Musante L., Fang C., Hoffmann K., Fuchs C., Carta E., Deas E., Venkateswarlu K., Menzel C., Ullmann R., Tommerup N., Dalprà L., Tzschach A., Selicorni A., Lüscher B., Ropers H. H., Harvey K. and Harvey R. J. (2009) A balanced chromosomal translocation disrupting ARHGEF9 is associated with epilepsy, anxiety, aggression, and mental retardation. Hum. Mutat. 30; 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kins S., Betz H. and Kirsch J. (2000) Collybistin, a newly identified brain-specific GEF, induces submembrane clustering of gephrin. Nat. Neurosci. 3; 22–29. [DOI] [PubMed] [Google Scholar]
- 10.Klein K. M., Pendziwiat M., Eilam A., Gilad R., Blatt I., Rosenow F., Kanaan M., Heibig I., Afawi Z. and Israeli-Palestinian Epilepsy Family Consortium. (2017) The phenotypic spectrum of ARHGEF9 includes intellectual disability, focal epilepsy and febrile seizures. J. Neurol. 264; 1421–1425. [DOI] [PubMed] [Google Scholar]
- 11.Kneussel M. and Betz H. (2000) Receptors, gephyrin and gephyrin-associated proteins: novel insights into the assembly of inhibitory postsynaptic membrane specializations. J. Physiol. 525; 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kneussel M., Engelkamp D. and Betz H. (2001) Distribution of transcripts for the brain-specific GDP/GTP exchange factor collybistin in the developing mouse brain. Eur. J. Neurosci. 13; 487–492. [DOI] [PubMed] [Google Scholar]
- 13.Mizutani Y., Ito H., Iwamoto I., Morishita R., Kanoh H., Seishima M. and Nagata K-I. (2013) Possible role of a septin, SEPT1, in spreading in squamous cell carcinoma DJM-1 cells. Biol. Chem. 394; 281–290. [DOI] [PubMed] [Google Scholar]
- 14.Papadopoulos T., Korte M., Eulenburg V., Kubota H., Retiounskaia M., Harvey R. J., Harvey K., O’Sullivan G. A., Laube B., Hülsmann S., Geiger J. R. P. and Betz H. (2007) Impaired GABAergic transmission and alterd hippocampal synaptic plasticity in collybistin-deficient mice. EMBO J. 26; 3888–3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papadopoulos T., Eulenburg V., Reddy-Alla S., Mansuy I. M., Li Y. and Betz H. (2008) Collybistin is required for both the formation and maintenance of GABAergic postsynapses in the hippocampus. Mol. Cell. Neurosci. 39; 161–169. [DOI] [PubMed] [Google Scholar]
- 16.Papadopoulos T., Schemm R., Grubmüller H. and Brose N. (2015) Lipid binding defects and perturbed synaptogenic activity of a Collybistin R290H mutant that causes epilepsy and intellectual disability. J. Biol. Chem. 290; 8256–8270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papadopoulos T., Rhee H. J., Subramanian D., Paraskevopoulou F., Mueller R., Schultz C., Brose N., Rhee J-S. and Betz H. (2017) Endosomal phosphatidylinositol 3-phosphate promotes gephyrin clustering and GABAergic neurotransmission at inhibitory postsynapses. J. Biol. Chem. 292; 1160–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patrizi A., Viltono L., Frola E., Harvey K., Harvey R. J. and Sassoe-Pognetto M. (2012) Selective localization of collybistin at a subset of inhibitory synapses in brain circuits. J. Comp. Neurol. 520; 130–141. [DOI] [PubMed] [Google Scholar]
- 19.Seibenhener M. L. and Wooten M. W. (2012) Isolation and culture of hippocampal neurons from prenatal mice. J. Vis. Exp. 26; 3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimoijma K., Sugawara M., Shichiji M., Mukaida S., Takayama R., Imai K. and Yamamoto T. (2011) Loss-of-function mutation of collybistin is responsible for X-linked mental retardation associated with epilepsy. J. Hum. Genet. 56; 561–565. [DOI] [PubMed] [Google Scholar]
- 21.Shiotsu N., Kawamoto T., Kawai M., Ikegame M., Torii Y., Sasaki H. and Yamamoto T. (2017) Morphological and functional analysis of the tight junction in the palatal epithelium of mouse. Acta Histochem. Cytochem. 50; 119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shirafuji T., Ueyama T., Tanaka S., Hide I., Saito N. and Sakai N. (2017) Validation of anti-CSPα, SNAP25, tyrosine hydroxylase, ubiquitin, cleaved caspase3, and pSer PKC motif antibodies for utilization in western blotting. Acta Histochem. Cytochem. 50; 177–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soykan T., Schneeberger D., Tria G., Buechner C., Bader N., Svergun D., Tessmer I., Poulopoulos A., Papadopoulos T., Varoqueaux F., Schindelin H. and Brose N. (2014) A conformational switch in collybistin determines the differentiation of inhibitory postsynapses. EMBO J. 33; 2113–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J. Y., Zhou P., Wang J., Tang B., Su T., Liu X. R. Li, B. M., Meng H., Shi Y. W., Yi Y. H., He N. and Liao W. P. (2018) ARHGEF9 mutations in epileptic encephalopathy/intellectual disability: toward understanding the mechanism underlying phenotypic variation. Neurogenetics 19; 9–16. [DOI] [PubMed] [Google Scholar]