Abstract
Several studies have suggested that long non-coding RNA (lncRNA) gene polymorphisms are associated with cancer risk. In the present study, we conducted a meta-analysis related to studies on the association between lncRNA single-nucleotide polymorphisms (SNPs) and the overall risk of cancer. A total of 12 SNPs in five common lncRNA genes were finally included in the meta-analysis. In the lncRNA antisense non-coding RNA (ncRNA) in the INK4 locus (ANRIL), the rs1333048 A/C, rs4977574 A/G, and rs10757278 A/G polymorphisms, but not rs1333045 C/T, were correlated with overall cancer risk. Our study also demonstrated that other SNPs were correlated with overall cancer risk, namely, metastasis-associated lung adenocarcinoma transcript 1 (MALAT1, rs619586 A/G), HOXA distal transcript antisense RNA (HOTTIP, rs1859168 A/C), and highly up-regulated in liver cancer (HULC, rs7763881 A/C). Moreover, four prostate cancer-associated ncRNA 1 (PRNCR1, rs16901946 G/A, rs13252298 G/A, rs1016343 T/C, and rs1456315 G/A) SNPs were in association with cancer risk. No association was found between the PRNCR1 (rs7007694 C/T) SNP and the risk of cancer. In conclusion, our results suggest that several studied lncRNA SNPs are associated with overall cancer risk. Therefore, they might be potential predictive biomarkers for the risk of cancer. More studies based on larger sample sizes and more lncRNA SNPs are warranted to confirm these findings.
Keywords: Cancer, LncRNA, Polymorphisms
Introduction
As a new class of functional non-coding RNAs (ncRNAs), long ncRNAs (lncRNAs) are made up of over 200 nts and lack the ability of protein coding [1]. Recently, the association between lncRNA and human diseases, especially cancer, has been widely investigated. Compared with other ncRNAs, lncRNAs play an important role in numerous vital activities of cell, including the regulation of epigenetic modifications, cell cycle, cell differentiation, and stress response [2]. The most important function of lncRNA is involvement in the tumorigenesis as proto-oncogene [3] or anti-oncogene [4]. Moreover, the differential expression of lncRNA may facilitate tumor cell proliferation, invasion, and metastasis [5].
Currently, single nucleotide polymorphisms (SNPs) are the most common genetic variants of concern and universally present in lncRNA genes. It is predicted that the expression and function of lncRNAs are affected by SNPs [6]. Studies have also suggested that polymorphism in lncRNA may influence the process of splicing and stability of mRNA conformation, leading to the modification of their interacting partners [7]. To date, several studies have assessed the associations amongst more than 20 lncRNA polymorphisms and susceptibility of cancers, but the results are inconsistent.
In the present study, we conducted a meta-analysis of epidemiological studies to explore the associations between five lncRNA SNPs and overall cancer risk. Furthermore, our study may shed some light on the biomarkers for predicting cancer risk.
Materials and methods
Publication search
A computerized literature search was performed in the Medline, PubMed, Web of Science, and Embase database up to 6 Februrary 2018. The search strategy included the terms (‘lncRNA’ or ‘long non-coding RNA’) and (‘polymorphisms’ or ‘variants’ or ‘variation’ or ‘SNP’) and (‘cancer’ or ‘carcinoma’ or ‘tumor’ or ‘neoplasm’). To be eligible for inclusion in the meta-analysis, a study must meet the following criteria: (i) case–control study or cohort study; (ii) assessing the association between lncRNA SNPs and cancer risk; (iii) having an available genotype or allele frequency for estimating an odds ratio (OR) with 95% confidence interval (95% CI) or hazard ratio (HR) with 95% CI; and (iv) genotype frequencies in controls being consistent with those expected from Hardy–Weinberg equilibrium (HWE) (P>0.05). The exclusion criteria were: (i) duplicate studies; (ii) not relevant to cancer or lncRNA SNPs; or (iii) no available data and the authors could not be contacted.
Data extraction and quality assessment
Two investigators (X.H. and W.Z.) evaluated the eligibility of all retrieved studies and extracted the relevant data independently. Extracted databases were then cross-checked between the two authors to rule out any discrepancy. Disagreement was resolved by consulting with the third investigator (Z.S.). The study quality was assessed in accordance with the Newcastle–Ottawa Scale (NOS) (Supplementary Table S1). Eight items were extracted, and each item scored 1. The total scores ranged from 0 to 8. If the scores were ≥7, then the study was considered to be of high quality.
Statistical analysis
The statistical analysis was performed using STATA 14. Estimates were summarized as ORs with 95% CIs for each study (P<0.05 was considered statistically significant). The genotype frequencies of the lncRNA polymorphisms for the HWE were calculated for the controls using the chi-square test, and P<0.05 was considered as significant disequilibrium. The between-study heterogeneity was evaluated by using the chi-square test and the I2 statistic. An I2 value of >50% of the I2 statistic was considered to indicate significant heterogeneity [8]. When a significant heterogeneity existed across the included studies, a random-effects model was used for the analysis. Otherwise, the fixed-effects model was used. Subgroup analyses were performed to detect the source of heterogeneity. As to genotype comparison, the risks of the heterozygote and variant homozygote compared with the wild-type homozygote were estimated respectively. Then we evaluated the dominant and recessive effects of the variant allele (heterozygote + variant homozygote compared with wild-type homozygote and variant homozygote compared with heterozygote + wild-type homozygote), respectively. Begg’s rank correlation and Egger’s linear regression method were used to assess the publication bias statistically. A two-tailed P-value <0.05 implies a statistically significant publication bias [9,10]. We further conducted sensitivity analyses to substantiate the stability of results and detect the potential source of heterogeneity.
Results
Characteristics of the eligible studies
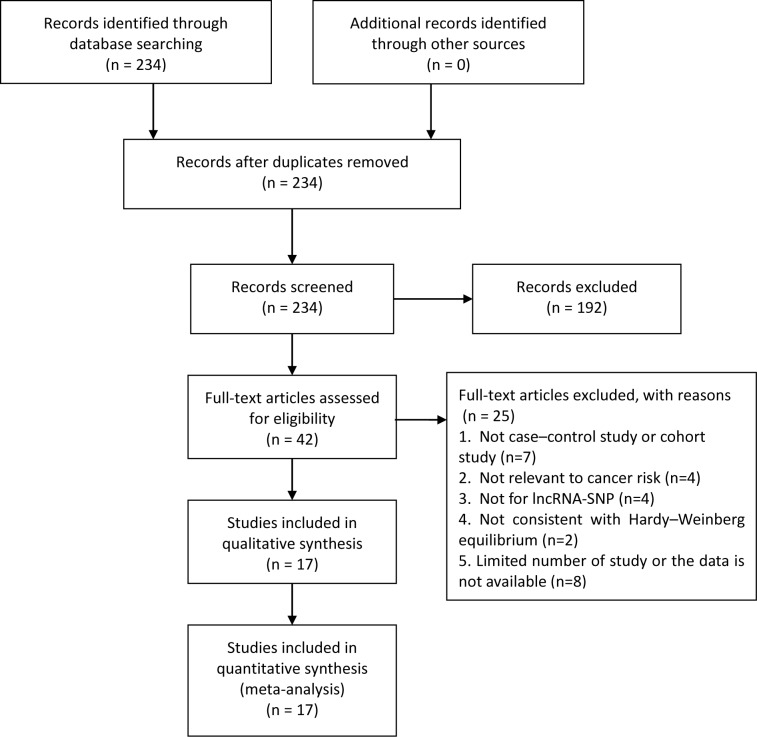
Finally, a total of 234 articles were included in the meta-analysis, 42 case–control studies that met our inclusion criteria were included in quantitative synthesis, and 17 of them involving 9548 cases and 9828 controls were included in our meta-analysis (Figure 1). Table 1 lists the characteristics of the eligible studies. Amongst the 17 case–control studies, the control groups of 9 were hospital-based and 8 were population-based. Genotyping methods included tetra-primer amplification refractory mutation system (T-ARMS)-PCR (2), MALDI-TOF MS (1), PCR-restriction fragment length polymorphism (RFLP) (5), created restriction site (CRS)-RFLP (1), TaqMan (3), MassARRAY (4), multiplex PCR-based Invader assay (1), and SNPlex Genotyping System (1) (Table 1). Table 2 presents the genotype frequency distributions of a total 19 SNPs in five lncRNA genes (antisense ncRNA in the INK4 locus (ANRIL), metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), HOXA distal transcript antisense RNA (HOTTIP), highly up-regulated in liver cancer (HULC), and prostate cancer-associated ncRNA 1 (PRNCR1)) involved in the 17 eligible studies. After removal of those records for which PHWE<0.05, seven SNPs were found to be only based on one single eligible study. They were ANRIL rs2151280, MALAT1 rs3200401, MALAT1 rs7927113, MALAT1 rs1194338, HOTTIP rs5883064, PRNCR1 rs7841060, and PRNCR1 rs7463708. Therefore, the remaining 12 lncRNA SNPs were included in our final calculation (Table 2).
Figure 1. The studies identified in this meta-analysis based on the inclusion and exclusion criteria.
Table 1. Characteristics of eligible studies.
| Number | First author | Year | Country | Ethnicity | Sample size | Source of control groups | Genotyping method | Adjusted factors | Citation | |
|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | |||||||||
| 1 | Khorshidi et al. | 2017 | Iran | Asian | 122 | 200 | PB | T-ARMS-PCR | Age | [11] |
| 2 | Kang et al. | 2015 | China | Asian | 380 | 380 | HB | MALDI-TOF MS | Age, sex, and drinking status | [12] |
| 3 | Taheri et al. | 2017 | Iran | Asian | 125 | 220 | PB | T-ARMS-PCR | Age, BMI, and smoking history | [13] |
| 4 | Peng et al. | 2017 | China | Asian | 487 | 489 | PB | PCR-RFLP, CRS-RFLP | Age | [14] |
| 5 | Liu et al. | 2012 | China | Asian | 1300 | 1344 | PB | TaqMan Assay-PCR | Age, sex, smoking rate, and HBV chronic infection | [15] |
| 6 | Li et al. | 2017 | China | Asian | 821 | 857 | HB | MassARRAY | Age, sex, BMI, smoking, and alcohol drinking | [16] |
| 7 | Gong et al. | 2016 | China | Asian | 498 | 213 | HB | MassARRAY | Age and sex | [17] |
| 8 | Hu et al. | 2017 | China | Asian | 921 | 921 | PB | TaqMan Assay-PCR | Age, sex, and area of residence | [18] |
| 9 | Shaker et al. | 2017 | Egypt | Caucasian | 120 | 96 | PB | TaqMan Assay-PCR | Age and sex | [19] |
| 10 | He et al. | 2017 | China | Asian | 494 | 494 | HB | MassARRAY | Helicobacter pylori infection rate, age, sex, and smoking and drinking status | [20] |
| 11 | Duan et al. | 2017 | China | Asian | 470 | 470 | HB | PCR-RFLP | Age, sex, and drinking | [6] |
| 12 | Li et al. | 2016 | China | Asian | 219 | 394 | HB | PCR-RFLP | Age and sex | [21] |
| 13 | Sattarifard et al. | 2017 | Iran | Asian | 178 | 180 | HB | PCR-RFLP | Age | [22] |
| 14 | Li et al. | 2013 | China | Asian | 313 | 595 | HB | PCR-RFLP | Age and sex | [23] |
| 15 | Chung et al. | 2010 | Japan | Asian | 1504 | 1554 | HB | Multiplex PCR-based Invader | NM | [24] |
| 16 | Salinas et al. | 2008 | U.S.A. | Caucasian | 1308 | 1266 | PB | SNPlex genotyping system | Age | [25] |
| 17 | Zheng et al. | 2010 | China | Asian | 288 | 155 | PB | MassARRAY | Age, sex, and BMI | [26] |
Abbreviations: BMI, body mass index; HB, hospital based; NM, not mentioned; PB, population based.
Table 2. Genotype frequency distributions of lncRNA SNPs studied in included studies.
| First author | Year | lncRNA | SNPs | Type of cancer | Sample size | Case | Control | P for HWE | Quality score | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Homozygote wild | Heterozygote | Homozygote variant | Homozygote wild | Heterozygote | Homozygote variant | |||||||
| Khorshidi et al. | 2017 | ANRIL | rs1333045 (C/T) | Breast cancer | 122 | 200 | 31 | 52 | 39 | 57 | 100 | 43 | 0.944 | 7 |
| ANRIL | rs1333048 (A/C) | Breast cancer | 122 | 200 | 39 | 51 | 32 | 51 | 97 | 52 | 0.672 | |||
| ANRIL | rs4977574 (A/G) | Breast cancer | 122 | 200 | 61 | 44 | 17 | 81 | 93 | 26 | 0.931 | |||
| ANRIL | rs10757278 (A/G) | Breast cancer | 122 | 200 | 38 | 62 | 22 | 74 | 100 | 26 | 0.387 | |||
| Kang et al. | 2015 | ANRIL | rs2151280 (C/T)1 | ESCC | 380 | 380 | 57 | 153 | 161 | 43 | 173 | 154 | 0.595 | 8 |
| Taheri et al. | 2017 | ANRIL | rs1333045 (C/T) | Prostate cancer | 125 | 220 | 41 | 61 | 23 | 75 | 102 | 43 | 0.435 | 7 |
| ANRIL | rs1333048 (A/C) | Prostate cancer | 148 | 220 | 25 | 65 | 58 | 101 | 88 | 31 | 0.103 | |||
| ANRIL | rs4977574 (A/G) | Prostate cancer | 114 | 220 | 55 | 46 | 13 | 79 | 109 | 32 | 0.570 | |||
| ANRIL | rs10757278 (A/G) | Prostate cancer | 132 | 220 | 14 | 65 | 53 | 95 | 93 | 32 | 0.241 | |||
| Peng et al. | 2017 | MALAT1 | rs3200401 (T/C)1 | Breast cancer | 487 | 489 | 357 | 120 | 10 | 338 | 145 | 6 | 0.057 | 8 |
| MALAT1 | rs619586 (A/G) | Breast cancer | 487 | 489 | 415 | 65 | 7 | 386 | 93 | 10 | 0.124 | |||
| MALAT1 | rs7927113 (A/G)1 | Breast cancer | 487 | 489 | 476 | 10 | 1 | 469 | 19 | 1 | 0.096 | |||
| Liu et al. | 2012 | MALAT1 | rs619586 (A/G) | HCC | 1300 | 1344 | 1094 | 169 | 5 | 1115 | 205 | 10 | 0.864 | 8 |
| Li et al. | 2017 | MALAT1 | rs1194338 (A/C)1 | CRC | 821 | 857 | 389 | 357 | 72 | 381 | 377 | 95 | 0.905 | 8 |
| Gong et al. | 2016 | HOTTIP | rs5883064 (C/T)1 | Lung cancer | 491 | 206 | 161 | 252 | 78 | 89 | 87 | 30 | 0.252 | 8 |
| HOTTIP | rs1859168 (A/C) | Lung cancer | 491 | 210 | 151 | 254 | 86 | 85 | 94 | 31 | 0.549 | |||
| Hu et al. | 2017 | HOTTIP | rs1859168 (A/C) | Pancreatic cancer | 921 | 921 | 239 | 497 | 185 | 364 | 421 | 136 | 0.428 | 8 |
| Duan et al. | 2017 | HOTTIP | rs1859168 (A/C) | Gastric cancer | 455 | 451 | 141 | 117 | 191 | 102 | 210 | 139 | 0.185 | 8 |
| Kang et al. | 2015 | HULC | rs7763881 (A/C) | ESCC | 380 | 380 | 122 | 168 | 84 | 99 | 195 | 81 | 0.412 | 8 |
| Shaker et al. | 2017 | HULC | rs7763881 (A/C) | CRC | 120 | 96 | 32 | 88 | 0 | 12 | 84 | 0 | 0.0002 | 6 |
| Liu et al. | 2012 | HULC | rs7763881 (A/C) | HCC | 1300 | 1344 | 377 | 617 | 283 | 333 | 695 | 288 | 0.057 | 8 |
| He et al. | 2017 | PRNCR1 | rs16901946 (A/G) | Gastric cancer | 494 | 494 | 261 | 203 | 30 | 301 | 176 | 17 | 0.153 | 8 |
| PRNCR1 | rs13252298 (A/G) | Gastric cancer | 494 | 494 | 236 | 215 | 43 | 209 | 235 | 50 | 0.173 | |||
| PRNCR1 | rs7463708 (G/T)1 | Gastric cancer | 494 | 494 | 241 | 209 | 44 | 228 | 209 | 57 | 0.390 | |||
| PRNCR1 | rs7007694 (C/T) | Gastric cancer | 494 | 494 | 264 | 199 | 31 | 272 | 198 | 24 | 0.111 | |||
| Li et al. | 2016 | PRNCR1 | rs16901946 (A/G) | Gastric cancer | 219 | 394 | 125 | 92 | 2 | 230 | 135 | 29 | 0.144 | 8 |
| PRNCR1 | rs13252298 (A/G) | Gastric cancer | 219 | 394 | 88 | 107 | 24 | 198 | 161 | 35 | 0.781 | |||
| PRNCR1 | rs7007694 (C/T) | Gastric cancer | 219 | 394 | 142 | 72 | 5 | 214 | 159 | 21 | 0.219 | |||
| PRNCR1 | rs1016343 (C/T) | Gastric cancer | 219 | 394 | 78 | 109 | 32 | 140 | 176 | 78 | 0.096 | |||
| PRNCR1 | rs1456315 (A/G) | Gastric cancer | 219 | 394 | 109 | 103 | 7 | 179 | 177 | 38 | 0.546 | |||
| Sattarifard et al. | 2017 | PRNCR1 | rs13252298 (A/G) | Prostate cancer | 178 | 179 | 33 | 107 | 38 | 25 | 141 | 13 | 0.0002 | 7 |
| PRNCR1 | rs1456315 (A/G) | Prostate cancer | 178 | 180 | 30 | 148 | 0 | 92 | 88 | 0 | 0.0002 | |||
| PRNCR1 | rs7007694 (C/T) | Prostate cancer | 178 | 180 | 150 | 28 | 0 | 139 | 41 | 0 | 0.085 | |||
| PRNCR1 | rs7841060 (G/T)1 | Prostate cancer | 178 | 180 | 29 | 149 | 0 | 96 | 84 | 0 | 0.0002 | |||
| Li et al. | 2013 | PRNCR1 | rs1016343 (C/T) | CRC | 313 | 595 | 117 | 156 | 40 | 227 | 276 | 92 | 0.593 | 8 |
| PRNCR1 | rs13252298 (A/G) | CRC | 313 | 595 | 166 | 121 | 26 | 264 | 270 | 61 | 0.508 | |||
| PRNCR1 | rs16901946 (A/G) | CRC | 313 | 595 | 175 | 117 | 138 | 338 | 232 | 257 | 0.0002 | |||
| PRNCR1 | rs1456315 (A/G) | CRC | 313 | 595 | 167 | 119 | 27 | 294 | 262 | 39 | 0.055 | |||
| PRNCR1 | rs7007694 (C/T) | CRC | 313 | 595 | 184 | 107 | 22 | 362 | 208 | 25 | 0.474 | |||
| Chung et al. | 2010 | PRNCR1 | rs1016343 (C/T) | Prostate cancer | 1504 | 1554 | 650 | 667 | 185 | 841 | 608 | 103 | 0.624 | 7 |
| PRNCR1 | rs13252298 (A/G) | Prostate cancer | 1504 | 1554 | 808 | 556 | 137 | 609 | 737 | 204 | 0.416 | |||
| PRNCR1 | rs16901946 (A/G) | Prostate cancer | 1504 | 1554 | 690 | 637 | 177 | 783 | 645 | 126 | 0.671 | |||
| PRNCR1 | rs1456315 (A/G) | Prostate cancer | 1504 | 1554 | 905 | 495 | 104 | 663 | 703 | 187 | 0.975 | |||
| PRNCR1 | rs7007694 (C/T) | Prostate cancer | 1504 | 1554 | 656 | 650 | 191 | 700 | 684 | 170 | 0.880 | |||
| Salinas et al. | 2008 | PRNCR1 | rs1456315 (A/G) | Prostate cancer | 1308 | 1266 | 464 | 598 | 192 | 401 | 605 | 227 | 0.964 | 7 |
| PRNCR1 | rs1016343 (C/T) | Prostate cancer | 1253 | 1233 | 711 | 454 | 88 | 796 | 385 | 52 | 0.529 | |||
| Zheng et al. | 2010 | PRNCR1 | rs1016343 (C/T) | Prostate cancer | 284 | 147 | 76 | 159 | 49 | 66 | 65 | 16 | 0.999 | 7 |
Abbreviations: CRC, colorectal cancer; EOC, epithelial ovarian cancer; ESCC, esophageal squamous cell carcinoma; HCC, hepatocellular carcinoma.
Not included due to the limited number of studies for this lncRNA locus.
Not included because the P of the HWE was <0.05.
Quantitative data synthesis of 12 SNPs in five highly studied lncRNA genes
Four SNPs in ANRIL
First, we calculated the pooled ORs of all eligible studies to estimate the association between the four SNPs in ANRIL and overall cancer risk. The rs1333045 C/T polymorphism was not associated with cancer; and the rs1333048 A/C, rs4977574 A/G, and rs10757278 A/G polymorphisms were associated with overall cancer risk. The rs1333048 A/C polymorphism was associated with increased overall risk of cancer in all genetic models (C compared with A: P=0.000, OR = 2.06, 95% CI = 1.64–2.57; CC compared with AA: P=0.000, OR = 4.26, 95% CI = 2.67–6.78; AC compared with AA: P=0.049, OR = 1.45, 95% CI = 1.00–2.10; dominant model: P=0.001, OR = 1.80, 95% CI = 1.28–2.51; recessive model: P=0.000, OR = 2.01, 95% CI = 1.42–2.84). For the rs4977574 A/G polymorphism, both the heterozygote type AG and the dominant model were associated with decreased overall risk of cancer compared with the wild-type AA (AG compared with AA: P=0.006, OR = 0.62, 95% CI = 0.44–0.87; dominant model: P=0.007, OR = 0.64, 95% CI = 0.46–0.88). However, both the mutation type GG and the allelic model were associated with increased overall risk of cancer (GG compared with AA: P=0.000, OR = 2.40, 95% CI = 1.60–3.59; G compared with A: P=0.000, OR = 1.68, 95% CI = 1.35–2.08). For the rs10757278 A/G polymorphism, the heterozygote type AG, the dominant model, and the recessive model were associated with increased overall risk of cancer (AG compared with AA: P=0.000, OR = 2.13, 95% CI = 1.45–3.12; dominant model: P=0.000, OR = 2.58, 95% CI = 1.80–3.69; recessive model: P=0.000, OR = 2.64, 95% CI = 1.79–3.88). Nevertheless, the allelic model was associated with decreased overall risk of cancer (G compared with A: P=0.030, OR = 0.77, 95% CI = 0.60–0.97, Table 3).
Table 3. Meta-analysis of the association between common SNPs and cancer risk.
| Stratification | n | Allelic model | Mutation homozygote compared with wild-type | Heterozygote compared with wild-type | Dominant model | Recessive model | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | I2 (%) | OR (95% CI) | P | I2 (%) | OR (95% CI) | P | I2 (%) | OR (95% CI) | P | I2 (%) | OR (95% CI) | P | I2 (%) | ||
| ANRIL | ||||||||||||||||
| rs1333048 (A/C) | 2 | 2.06 (1.64– 2.57) | 0.0001 | 94.3 | 4.26 (2.67– 6.78) | 0.0001 | 93.1 | 1.45 (1.00– 2.10) | 0.0491 | 93.0 | 1.80 (1.28– 2.51) | 0.0011 | 95.7 | 2.01 (1.42– 2.84) | 0.000a | 92.7 |
| rs4977574 (A/G) | 2 | 1.68 (1.35– 2.08) | 0.0001 | 96.7 | 2.40 (1.60– 3.59) | 0.0001 | 96.1 | 0.62 (0.44– 0.87) | 0.006 | 0.0 | 0.64 (0.46– 0.88) | 0.007 | 0.0 | 0.91 (0.57– 1.46) | 0.693 | 0.0 |
| rs10757278 (A/G) | 2 | 0.77 (0.60– 0.97) | 0.030 | 0.0 | 0.72 (0.43– 1.18) | 0.192 | 0.0 | 2.13 (1.45– 3.12) | 0.0001 | 90.7 | 2.58 (1.80– 3.69) | 0.0001 | 93.9 | 2.64 (1.79– 3.88) | 0.0001 | 82.7 |
| rs1333045 (C/T) | 2 | 1.15 (0.92– 1.43) | 0.236 | 27.7 | 1.29 (0.83– 1.99) | 0.260 | 28.5 | 1.03 (0.71– 1.48) | 0.874 | 0.0 | 1.11 (0.79– 1.56) | 0.556 | 0.0 | 1.30 (0.89– 1.88) | 0.1751 | 60.4 |
| MALAT1 | ||||||||||||||||
| rs619586 (A/G) | 2 | 0.77 (0.65– 0.92) | 0.003 | 9.7 | 0.58 (0.28– 1.20) | 0.141 | 0.0 | 0.78 (0.65– 0.94) | 0.009 | 33.5 | 0.77 (0.64– 0.92) | 0.0041 | 27.9 | 0.61 (0.30– 1.26) | 0.180 | 0.0 |
| HOTTIP | ||||||||||||||||
| rs1859168 (A/C) | 3 | 1.32 (1.19– 1.45) | 0.0001 | 75.2 | 1.54 (1.27– 1.87) | 0.0001 | 81.8 | 1.24 (1.06– 1.45) | 0.0061 | 96.4 | 1.37 (1.19– 1.59) | 0.0001 | 94.3 | 1.49 (1.26– 1.76) | 0.000 | 0.0 |
| HULC | ||||||||||||||||
| rs7763881 (A/C) | 3 | 0.91 (0.83– 0.99) | 0.040 | 0.0 | 0.86 (0.71– 1.05) | 0.132 | 0.0 | 0.74 (0.63– 0.86) | 0.000 | 41.3 | 0.77 (0.66– 0.89) | 0.000 | 45.2 | 1.02 (0.87– 1.21) | 0.776 | 0.0 |
| PRNCR1 | ||||||||||||||||
| rs16901946 (G/A) | 3 | 1.15 (1.06– 1.25) | 0.0011 | 66.4 | 1.26 (1.06–1.50) | 0.0081 | 82.6 | 1.15 (1.03– 1.28) | 0.017 | 0.0 | 1.17 (1.06– 1.30) | 0.003 | 21.6 | 1.21 (1.03–1.43) | 0.0191 | 81.7 |
| Type of cancer | ||||||||||||||||
| Gastric cancer | 2 | 1.15 (0.97– 1.35) | 0.1041 | 83.8 | 0.96 (0.59– 1.56) | 0.8711 | 92.4 | 1.30 (1.06– 1.60) | 0.013 | 0.0 | 1.26 (1.03– 1.54) | 0.025 | 40.7 | 0.86 (0.53– 1.39) | 0.5331 | 92.4 |
| rs13252298 (G/A) | 4 | 0.78 (0.72– 0.85) | 0.0001 | 89.2 | 0.68 (0.56– 0.81) | 0.0001 | 81.6 | 0.69 (0.62– 0.77) | 0.0001 | 85.1 | 0.81 (0.73– 0.90) | 0.0001 | 73.7 | 0.85 (0.72– 1.01) | 0.0651 | 82.7 |
| Type of cancer | ||||||||||||||||
| Gastric cancer | 2 | 1.00 (0.86– 1.16) | 0.9941 | 86.6 | 0.99 (0.69– 1.41) | 0.9451 | 72.1 | 1.01 (0.82– 1.25) | 0.9231 | 86.7 | 1.01 (0.83– 1.23) | 0.9451 | 88.5 | 0.98 (0.70– 1.38) | 0.921 | 21.1 |
| rs7007694 (C/T) | 5 | 1.03 (0.95– 1.12) | 0.5221 | 69.0 | 1.19 (0.98– 1.45) | 0.0861 | 58.4 | 0.96 (0.86– 1.07) | 0.443 | 42.5 | 0.99 (0.89– 1.10) | 0.8481 | 61.0 | 1.19 (0.98– 1.44) | 0.070 | 49.9 |
| Type of cancer | ||||||||||||||||
| Gastric cancer | 2 | 0.92 (0.78– 1.09) | 0.3321 | 85.8 | 0.92 (0.58– 1.47) | 0.7301 | 80.4 | 0.89 (0.73– 1.10) | 0.2801 | 71.6 | 0.89 (0.73– 1.09) | 0.2691 | 81.7 | 0.96 (0.60– 1.52) | 0.8531 | 75.0 |
| Prostate cancer | 2 | 1.05 (0.95– 1.16) | 0.3711 | 69.6 | 1.20 (0.95– 1.51) | 0.126 | 0.98 (0.85– 1.13) | 0.7691 | 64.0 | 1.02 (0.88– 1.16) | 0.8321 | 69.2 | 1.19 (0.96– 1.48) | 0.120 | ||
| rs1016343 (T/C) | 5 | 1.31 (1.22– 1.41) | 0.0001 | 85.2 | 1.67 (1.41– 1.97) | 0.0001 | 86.0 | 1.35 (1.22– 1.49) | 0.000 | 47.2 | 1.41 (1.28– 1.55) | 0.0001 | 73.1 | 1.42 (1.21– 1.66) | 0.0001 | 84.5 |
| Ethnicity | ||||||||||||||||
| Asian | 4 | 1.30 (1.19– 1.41) | 0.0001 | 88.7 | 1.60 (1.33– 1.94) | 0.0001 | 89.3 | 1.37 (1.21– 1.54) | 0.0001 | 59.8 | 1.42 (1.26– 1.59) | 0.0001 | 79.8 | 1.35 (1.13– 1.61) | 0.0011 | 87.7 |
| Type of cancer | ||||||||||||||||
| Prostate cancer | 3 | 1.45 (1.34– 1.57) | 0.000 | 1.9 | 2.21 (1.81– 2.70) | 0.000 | 0.0 | 1.41 (1.27– 1.57) | 0.000 | 49.3 | 1.51 (1.37– 1.68) | 0.0001 | 55.6 | 1.86 (1.54– 2.26) | 0.000 | 0.0 |
| rs1456315 (G/A) | 4 | 0.77 (0.72– 0.83) | 0.0001 | 94.6 | 0.59 (0.49– 0.69) | 0.0001 | 85.5 | 0.76 (0.68– 0.83) | 0.0001 | 95.4 | 0.72 (0.66– 0.79) | 0.0001 | 95.7 | 0.69 (0.59– 0.81) | 0.0001 | 80.8 |
| Ethnicity | ||||||||||||||||
| Asian | 3 | 0.72 (0.66– 0.79) | 0.0001 | 95.7 | 0.48 (0.39– 0.60) | 0.0001 | 86.4 | 0.71 (0.63– 0.80) | 0.0001 | 96.4 | 0.68 (0.61– 0.76) | 0.0001 | 96.6 | 0.60 (0.49– 0.75) | 0.0001 | 84.2 |
| Type of cancer | ||||||||||||||||
| Prostate cancer | 2 | 0.75 (0.70– 0.81) | 0.0001 | 97.2 | 0.56 (0.47– 0.67) | 0.0001 | 90.7 | 0.73 (0.66– 0.82) | 0.0001 | 97.6 | 0.69 (0.63– 0.77) | 0.0001 | 97.8 | 0.68 (0.58– 0.80) | 0.0001 | 81.6 |
The results are in bold if P<0.05.
P was calculated by random model.
One SNP in MALAT1
The meta-analysis showed that MALAT1 rs619586 A/G polymorphism was associated with overall cancer risk. For the rs619586 A/G polymorphism, the allelic model, the heterozygote type AG and the dominant model were associated with decreased overall risk of cancer compared with the wild-type AA (G compared with A: P=0.003, OR = 0.77, 95% CI = 0.65–0.92; AG compared with AA: P=0.009, OR = 0.78, 95% CI = 0.65–0.94; dominant model: P=0.004, OR = 0.77, 95% CI = 0.64–0.92, Table 3).
One SNP in HOTTIP
Our results suggested that the HOTTIP rs1859168 A/C polymorphism was associated with increased overall risk of cancer in all genetic models (C compared with A: P=0.000, OR = 1.32, 95% CI = 1.19–1.45; CC compared with AA: P=0.000, OR = 1.54, 95% CI = 1.27–1.87; AC compared with AA: P=0.006, OR = 1.24, 95% CI = 1.06–1.45; dominant model: P= 0.000, OR = 1.37, 95% CI = 1.19–1.59; recessive model: P=0.000, OR = 1.49, 95% CI = 1.26–1.76, Table 3).
One SNP in HULC
In the present study, the allelic model, the heterozygote type AC, and the dominant model of HULC rs7763881 A/C polymorphism were associated with decreased overall risk of cancer compared with the wild-type AA (C compared with A: P=0.040, OR = 0.91, 95% CI = 0.83–0.99; AC compared with AA: P=0.000, OR = 0.74, 95% CI = 0.63–0.86; dominant model: P=0.000, OR = 0.77, 95% CI = 0.66–0.89, Table 3).
Five SNPs in PRNCR1
The pooled OR and stratified analyses showed that amongst the five PRNCR1 SNPs included in the meta-analysis, only rs16901946 G/A, rs13252298 G/A, rs1016343 T/C, and rs1456315 G/A were associated with cancer risk, while the association of the rs7007694 C/T was not statistically significant (P>0.05).
The rs16901946 G/A polymorphism was associated with increased overall risk of cancer in all genetic models (A compared with G: P=0.001, OR = 1.15, 95% CI = 1.06–1.25; AA compared with GG: P=0.008, OR = 1.26, 95% CI = 1.06–1.50; AG compared with GG: P=0.017, OR = 1.15, 95% CI = 1.03–1.28; dominant model: P=0.003, OR = 1.17, 95% CI = 1.06–1.30; recessive model: P=0.019, OR = 1.21, 95% CI = 1.03–1.43).
For the rs13252298 G/A polymorphism, the allelic model, the mutation type AA, the heterozygote type AG, and the dominant model were associated with decreased overall risk of cancer compared with the wild-type GG (A compared with G: P=0.000, OR = 0.78, 95% CI = 0.72–0.85; AA compared with GG: P=0.000, OR = 0.68, 95% CI = 0.56–0.81; AG compared with GG: P=0.000, OR = 0.69, 95% CI = 0.62–0.77; dominant model: P=0.000, OR = 0.81, 95% CI = 0.73–0.90).
Additionally, the rs1016343 T/C polymorphism was associated with increased overall risk of cancer in all genetic models (C compared with T: P=0.000, OR = 1.31, 95% CI = 1.22–1.41; CC compared with TT: P=0.000, OR = 1.67, 95% CI = 1.41–1.97; CT compared with TT: P=0.000, OR = 1.35, 95% CI = 1.22–1.49; dominant model: P=0.000, OR = 1.41, 95% CI = 1.28–1.55; recessive model: P=0.000, OR = 1.42, 95% CI = 1.21–1.66).
The rs1456315 G/A polymorphism was associated with decreased overall risk of cancer in all genetic models (A compared with G: P=0.000, OR = 0.77, 95% CI = 0.72–0.83; AA compared with GG: P=0.000, OR = 0.59, 95% CI = 0.49–0.69; AG compared with GG: P=0.000, OR = 0.76, 95% CI = 0.68–0.83; dominant model: P=0.000, OR = 0.72, 95% CI = 0.66–0.79; recessive model: P=0.000, OR = 0.69, 95% CI = 0.59–0.81, Table 3).
Due to heterogeneity, we performed stratified analyses based on ethnicity and cancer type. Stratified analyses based on cancer type showed a significant association between the rs16901946 G/A polymorphism and increased risk of gastric cancer in the heterozygote type AG and the dominant model. In the Asian subgroup, the rs1016343 T/C polymorphism was associated with increased cancer risk in all genetic models. When stratified with cancer type, a significant association between the rs1456315 G/A polymorphism and decreased risk of prostate cancer was observed in our study (Table 3).
Heterogeneity
There was interstudy heterogeneity (slight, moderate, or severe) in the overall comparison and the subgroup analyses (Table 3). We subsequently performed sensitivity analyses to explore the influence of an individual study on the pooled results by estimating the sensitivity before and after the removal of the study from the analysis. Some ORs and 95% CIs ranged from insignificantly to statistically significant after individual studies were removed (Supplementary Table S2).
Publication bias
We used Begg’s test and Egger’s test to evaluate potential publication bias of the included studies. No statistically significant publication bias was indicated in any of the genetic models for all lncRNA SNPs (Table 4).
Table 4. The results of Begg’s and Egger’s test for the publication bias.
| Comparison type | Begg’s test | Egger’s test | ||
|---|---|---|---|---|
| Z-value | P-value | Z-value | P-value | |
| ANRIL rs1333048 (A/C) | ||||
| Allelic model | 0.00 | 1.000 | NA | NA |
| Mutation homozygote compared with wild-type | 0.00 | 1.000 | NA | NA |
| Heterozygote compared with wild-type | 0.00 | 1.000 | NA | NA |
| Dominant model | 0.00 | 1.000 | NA | NA |
| Recessive model | 0.00 | 1.000 | NA | NA |
| ANRIL rs4977574 (A/G) | ||||
| Allelic model | 0.00 | 1.000 | NA | NA |
| Mutation homozygote compared with wild-type | 0.00 | 1.000 | NA | NA |
| Heterozygote compared with wild-type | 0.00 | 1.000 | NA | NA |
| Dominant model | 0.00 | 1.000 | NA | NA |
| Recessive model | 0.00 | 1.000 | NA | NA |
| ANRIL rs10757278 (A/G) | ||||
| Allelic model | 0.00 | 1.000 | NA | NA |
| Mutation homozygote compared with wild-type | 0.00 | 1.000 | NA | NA |
| Heterozygote compared with wild-type | 0.00 | 1.000 | NA | NA |
| Dominant model | 0.00 | 1.000 | NA | NA |
| Recessive model | 0.00 | 1.000 | NA | NA |
| ANRIL rs1333045 (C/T) | ||||
| Allelic model | 0.00 | 1.000 | NA | NA |
| Mutation homozygote compared with wild-type | 0.00 | 1.000 | NA | NA |
| Heterozygote compared with wild-type | 0.00 | 1.000 | NA | NA |
| Dominant model | 0.00 | 1.000 | NA | NA |
| Recessive model | 0.00 | 1.000 | NA | NA |
| MALAT1 rs619586 (A/G) | ||||
| Allelic model | 0.00 | 1.000 | NA | NA |
| Mutation homozygote compared with wild-type | 0.00 | 1.000 | NA | NA |
| Heterozygote compared with wild-type | 0.00 | 1.000 | NA | NA |
| Dominant model | 0.00 | 1.000 | NA | NA |
| Recessive model | 0.00 | 1.000 | NA | NA |
| HOTTIP rs1859168 (A/C) | ||||
| Allelic model | 0.00 | 1.000 | −0.86 | 0.548 |
| Mutation homozygote compared with wild-type | 0.00 | 1.000 | −0.46 | 0.725 |
| Heterozygote compared with wild-type | 0.00 | 1.000 | −1.02 | 0.494 |
| Dominant model | 0.00 | 1.000 | −0.91 | 0.531 |
| Recessive model | 0.00 | 1.000 | −0.75 | 0.590 |
| HULC rs7763881 (A/C) | ||||
| Allelic model | 1.04 | 0.296 | −3.13 | 0.197 |
| Mutation homozygote compared with wild-type | 0.00 | 1.000 | NA | NA |
| Heterozygote compared with wild-type | 1.04 | 0.296 | −9.06 | 0.070 |
| Dominant model | 1.04 | 0.296 | −5.60 | 0.113 |
| Recessive model | 0.00 | 1.000 | NA | NA |
| PRNCR1 rs16901946 (G/A) | ||||
| Allelic model | 0.34 | 0.734 | −0.71 | 0.553 |
| Mutation homozygote compared with wild-type | 0.34 | 0.734 | −0.71 | 0.553 |
| Heterozygote compared with wild-type | −0.34 | 1.000 | 0.38 | 0.742 |
| Dominant model | −0.34 | 1.000 | −0.27 | 0.810 |
| Recessive model | 0.34 | 0.734 | −0.19 | 0.867 |
| PRNCR1 rs13252298 (G/A) | ||||
| Allelic model | 1.22 | 0.221 | 3.30 | 0.046 |
| Mutation homozygote compared with wild-type | 1.71 | 0.086 | 3.34 | 0.044 |
| Heterozygote compared with wild-type | 0.24 | 0.806 | 1.07 | 0.363 |
| Dominant model | 0.73 | 0.462 | 0.70 | 0.535 |
| Recessive model | 1.71 | 0.086 | 1.82 | 0.166 |
| PRNCR1 rs7007694 (C/T) | ||||
| Allelic model | 0.73 | 0.462 | −1.42 | 0.251 |
| Mutation homozygote compared with wild-type | −0.34 | 1.000 | −0.10 | 0.933 |
| Heterozygote compared with wild-type | 1.71 | 0.086 | −1.96 | 0.145 |
| Dominant model | 1.22 | 0.221 | −1.70 | 0.188 |
| Recessive model | −0.34 | 1.000 | −0.04 | 0.974 |
| PRNCR1 rs1016343 (T/C) | ||||
| Allelic model | 0.24 | 0.806 | −0.87 | 0.450 |
| Mutation homozygote compared with wild-type | 0.24 | 0.806 | −0.83 | 0.467 |
| Heterozygote compared with wild-type | −0.24 | 1.000 | 0.25 | 0.820 |
| Dominant model | −0.24 | 1.000 | −0.15 | 0.888 |
| Recessive model | 0.73 | 0.462 | −1.29 | 0.288 |
| PRNCR1 rs1456315 (G/A) | ||||
| Allelic model | 1.22 | 0.221 | 1.74 | 0.181 |
| Mutation homozygote compared with wild-type | −0.24 | 1.000 | 0.27 | 0.810 |
| Heterozygote compared with wild-type | 1.71 | 0.086 | 2.07 | 0.130 |
| Dominant model | 1.71 | 0.086 | 2.10 | 0.127 |
| Recessive model | −0.24 | 1.000 | 0.20 | 0.862 |
Abbreviation: NA, not available.
Discussion
It is known to all that over 20 lncRNA polymorphisms are associated with susceptibility of cancer. In recent studies, most of meta-analyses were conducted to focus on the association between lncRNA HOTAIR [27,28] or lncRNA ZNRD1-AS1 [28] or lncRNA POLR2E [29] or lncRNA H19 [28,30] polymorphisms and cancer risk. For example, the study of Lv et al. [28] included only four common lncRNA genes such as H19, HOTAIR, ZNRD1-AS1, and PRNCR1. However, more lncRNA polymorphisms with larger sample sizes are warranted. Therefore, a total of 12 SNPs in five common lncRNA genes were finally included in our study. In addition, our study was the first meta-analysis to show the significant association between the lncRNA ANRIL, MALAT1, HOTTIP, and HULC polymorphisms and cancer risk. Compared with the studies of Lv et al. [28] and Chu et al. [29], we decided to include more eligible studies related to lncRNA PRNCR1 genes according to the inclusion and exclusion criteria. Therefore, we included a larger size of cancer patients with more SNPs of lncRNA PRNCR1 into our study to confirm the results. More importantly, discussions about underlying mechanisms of each gene and the related polymorphisms were included in our study. It might help readers better understand the function of different lncRNA genes in cancer. Our study provides theoretical bases and research clues for future studies.
The ANRIL SNPs
Chromosome region 9p21 is a hotspot for disease-associated polymorphisms and encodes three tumor suppressors, namely p16INK4a, p14ARF, and p15INK4b, and the lncRNA ANRIL [31]. ANRIL is 3.8-kb long and expressed on the reverse strand. It has been shown to bind to and recruit polycomb repression complex 2 (PRC2) to repress the expression of p15INK4B [32]. Further study showed that SNPs can disrupt ANRIL splicing and result in a circular transcript that is resistant to RNase digestion [7]. The circularized transcripts affect the normal function of ANRIL and INK4/ARF expression. For example, rs1333048 has been shown to be associated with the level of highly sensitive C-reactive protein (hsCRP), which is a biomarker for systemic inflammation [33] and breast cancer susceptibility [34]. And previous results have revealed that rs4977574 is significantly associated with the risk of coronary artery disease [35]. Moreover, rs10757278 has been reported to increase the ANRIL variant EU741058 expression which contains exons 1–5 of the long transcript [36]. In addition, this SNP might modulate the ANRIL binding site for the transcription factor STAT1, which in turn regulates ANRIL expression [37]. In conclusion, three SNPs in ANRIL (rs1333048 A/C, rs4977574 A/G, and rs10757278 A/G) can be used to determine cancer risk.
The MALAT1 SNPs
MALAT1 is located in chromosome 11q13, which is over 8000 nts long. It is enriched in nuclear speckles in interphase cells and concentrates in mitotic interchromatin granule clusters. And it is co-localized with pre-mRNA-splicing factor SF2/ASF and CC3 antigen in the nuclear speckles [38]. It is reported that lncRNA MALAT1 could regulate the expression through modulating transcription and the processing of post-transcriptional pre-mRNA in various genes [39]. Zhuo et al. [40] suggested that rs619586 SNP could bind with miR-214 directly and suppress the expression of MALAT1. Several studies revealed that MALAT1 has an elevated expression and was associated with a higher risk and poorer survival in many kinds of cancers [41]. Our study showed that MALAT1 rs619586 A/G polymorphism was potential predictive biomarker of overall cancer risk.
The HOTTIP SNPs
HOTTIP is an antisense non-coding transcript located at the 5′-end of the HOXA gene cluster. The previous study showed that rs1859168 might change the expression level of HOTTIP by affecting transcription factor binding sites [17]. Furthermore, RNAfold web server also revealed that rs1859168 could alter the centroid secondary structure and minimum free energy. It might also influence the folding of HOTTIP and its function [17]. Further studies are warranted to explore the specific mechanisms. Our results suggested that the HOTTIP rs1859168 A/C polymorphism was associated with increased overall risk of cancer. Although the detailed mechanisms underlying the association of SNP in HOTTIP with cancer susceptibility are unclear, these findings could provide a new insight into understanding the genetic factors of cancer susceptibility and carcinogenesis.
The HULC SNPs
The lncRNA HULC is approximately 1.6 k nucleotide long and contains two exons but not translated [42]. Some studies have reported that HULC is highly up-regulated in hepatocellular carcinoma (HCC) and colorectal cancer (CRC) that metastasized to livers [42,43]. Rs7763881 SNP changing from A to C in HULC gene was located in the 6p24.3 region. Based on the Hapmap database, all the SNPs in HULC are in high linkage disequilibrium (LD). For example, rs7763881 was in complete LD with rs1328867 (r2 = 1), which is located in the promoter region of HULC. Additionally, the wild-type allele T of rs1328867 is predicted to bind with some transcription factors including C-Myc [15]. It has been identified that C-Myc is critical in the regulation of the growth, differentiation, and apoptosis of both normal and neoplastic liver cells [44]. In conclusion, HULC rs7763881 A/C polymorphism was associated with decreased overall risk of cancer.
The PRNCR1 SNPs
The lncRNA PRNCR1, also referred to as PCAT8 and CARLo3, is transcribed from the ‘gene desert’ region of chromosome 8q24 (128.14–128.28 Mb) [24]. It has been stated that PRNCR1 is involved in the development of prostate cancer by activating androgen receptor (AR) [45]. Moreover, lncRNA PRNCR1 SNPs were observed to be risk of diverse cancers [21–23]. It might affect the predicted secondary structure of PRNCR1 mRNA, altering the stability of PRNCR1 or the mRNA conformation, and giving rise to the modification of its interacting partners [24]. All the PRNCR1 polymorphisms in the exon region might result in the mechanism [28]. More specific mechanisms are warranted to be explored in further studies. Amongst the five PRNCR1 SNPs included in our study, rs16901946 G/A, rs13252298 G/A, rs1016343 T/C, and rs1456315 G/A could be predictive biomarkers of cancer risk.
Limitations
Although this meta-analysis revealed the significant association between lncRNA polymorphisms and cancer risk, however, some limitations still should be acknowledged. First, the number of subjects in the included studies is relatively small, which might result in a lack of statistical power and prevent a meaningful analysis of the results. Second, in stratified analyses based on ethnicity and cancer type, we failed to perform further subgroup analysis because of limited relevant reports. Third, only English articles were included in our study and it may result in publication bias. Finally, study of the association between lncRNA polymorphisms and cancer risk remains an emerging field, we concluded only representative SNPs in our study. Therefore, additional prospective studies with larger sample sizes including other polymorphisms are warranted.
Summary and future directions
We systematically reviewed studies on the association between lncRNA SNPs and overall cancer risk, and used the available data to perform a meta-analysis of 19 SNPs in five common lncRNA genes. The results suggest that the association between lncRNA SNPs and cancer risk can be categorized into four types: (i) complete association, where polymorphisms are significantly associated with risk of overall cancer in all genetic models, including ANRIL rs1333048, HOTTIP rs1859168, PRNCR1 rs16901946, PRNCR1 rs1016343, and PRNCR1 rs1456315; (ii) ANRIL rs4977574, ANRIL rs10757278, MALAT1 rs619586, HULC rs7763881, and PRNCR1 rs13252298 polymorphisms are only associated with cancer risk in some genetic models; (iii) no association, where the association of polymorphisms with cancer risk are not statistically significant, including ANRIL rs1333045 and PRNCR1 rs7007694; (iv) failed to be quantitatively synthesized due to limited studies. Therefore, the lncRNA SNPs provide more alternatives for biomarkers that can predict cancer risk.
More attention should be paid to several research directions in the future studies. First, more lncRNA polymorphisms and other aspects of cancer including chemotherapeutic susceptibility, metastasis, and relapse should be explored. Second, functional studies are needed to clarify the underlying mechanisms of lncRNA polymorphism in the tumorigenesis. Finally, the extensive clinical application of lncRNA polymorphisms requires further study.
Supporting information
Supplementary Table S1. Quality assessment of eligible studies (Newcastle-Ottawa Scale).
Supplementary Table S2. The results of ORs and 95% CI of sensitivity analysis.
Abbreviations
- ANRIL
antisense non-coding RNA in the INK4 locus
- CRS
created restriction site
- HOTTIP
HOXA distal transcript antisense RNA
- HULC
highly up-regulated in liver cancer
- HWE
Hardy–Weinberg equilibrium
- LD
linkage disequilibrium
- lncRNA
long non-coding RNA
- MALAT1
metastasis-associated lung adenocarcinoma transcript 1
- ncRNA
non-coding RNA
- NOS
Newcastle–Ottawa scale
- OR
odds ratio
- PRNCR1
prostate cancer-associated non-coding RNA 1
- RFLP
restriction fragment length polymorphism
- SNP
single-nucleotide polymorphism
- T-ARMS
tetra-primer amplification refractory mutation system
- 95% CI
95% confidence interval
Appendix A: supplementary data
Supplementary data associated with this article can be found, in the online version.
Supplementary Table S1. Quality assessment of eligible studies NOS.
Supplementary Table S2. The results of ORs and 95% CI of sensitivity analysis.
Funding
This work was supported by the National Key Research and Development Program of China [grant number 2016YFC1100100]; the Major Research Plan of National Natural Science Foundation of China [grant number 91649204]; and the National Natural Science Foundation of China [grant number 81572203].
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
Author contribution
Z.S. and X.H. conceived and designed the study. X.H. and W.Z. performed data collection and management. X.H. performed data analysis. Z.S. and X.H. wrote the paper. All the authors reviewed the manuscript.
References
- 1.Ponting C.P., Oliver P.L. and Reik W. (2009) Evolution and functions of long noncoding RNAs. Cell 136, 629–641 10.1016/j.cell.2009.02.006 [DOI] [PubMed] [Google Scholar]
- 2.Brosnan C.A. and Voinnet O. (2009) The long and the short of noncoding RNAs. Curr. Opin. Cell Biol. 21, 416–425 10.1016/j.ceb.2009.04.001 [DOI] [PubMed] [Google Scholar]
- 3.Li L., Feng T.T., Lian Y.Y., Zhang G.F., Garen A. and Song X. (2009) Role of human noncoding RNAs in the control of tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 106, 12956–12961 10.1073/pnas.0906005106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang X., Rice K., Wang Y.Y., Chen W.D., Zhong Y., Nakayama Y. et al. (2010) Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: isoform structure, expression, and functions. Endocrinology 151, 939–947 10.1210/en.2009-0657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsai M.C., Spitale R.C. and Chang H.Y. (2011) Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 71, 3–7 10.1158/0008-5472.CAN-10-2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duan F., Jiang J., Song C., Wang P., Ye H., Dai L. et al. (2018) Functional long non-coding RNAs associated with gastric cancer susceptibility and evaluation of the epidemiological efficacy in a central Chinese population. Gene 646, 227–233 10.1016/j.gene.2017.12.063 [DOI] [PubMed] [Google Scholar]
- 7.Burd C.E., Jeck W.R., Liu Y., Sanoff H.K., Wang Z. and Sharpless N.E. (2010) Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 6, e1001233 10.1371/journal.pgen.1001233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egger M., Davey Smith G., Schneider M. and Minder C. (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315, 629–634 10.1136/bmj.315.7109.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ioannidis J.P.A. (2013) Clarifications on the application and interpretation of the test for excess significance and its extensions. J. Math. Psychol. 57, 184–187 10.1016/j.jmp.2013.03.002 [DOI] [Google Scholar]
- 10.Ioannidis J.P.A. and Trikalinos T.A. (2007) An exploratory test for an excess of significant findings. Clin. Trials 4, 245–253 10.1177/1740774507079441 [DOI] [PubMed] [Google Scholar]
- 11.Khorshidi H.R., Taheri M., Noroozi R., Sarrafzadeh S., Sayad A. and Ghafouri-Fard S. (2017) ANRIL genetic variants in Iranian breast cancer patients. Cell J. 19 (Suppl. 1), 72–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang M.Q., Sang Y.H., Gu H.Y., Zheng L., Wang L.M., Liu C. et al. (2015) Long noncoding RNAs POLR2E rs3787016 C/T and HULC rs7763881 A/C polymorphisms are associated with decreased risk of esophageal cancer. Tumour Biol. 36, 6401–6408 10.1007/s13277-015-3328-z [DOI] [PubMed] [Google Scholar]
- 13.Taheri M., Pouresmaeili F., Omrani M.D., Habibi M., Sarrafzadeh S., Noroozi R. et al. (2017) Association of ANRIL gene polymorphisms with prostate cancer and benign prostatic hyperplasia in an Iranian population. Biomark. Med. 11, 413–422 10.2217/bmm-2016-0378 [DOI] [PubMed] [Google Scholar]
- 14.Peng R., Luo C., Guo Q., Cao J., Yang Q., Dong K. et al. (2018) Association analyses of genetic variants in long non-coding RNA MALAT1 with breast cancer susceptibility and mRNA expression of MALAT1 in Chinese Han population. Gene 642, 241–248 10.1016/j.gene.2017.11.013 [DOI] [PubMed] [Google Scholar]
- 15.Liu Y., Pan S.D., Liu L., Zhai X.J., Liu J.B., Wen J. et al. (2012) A genetic variant in long non-coding RNA HULC contributes to risk of HBV-related hepatocellular carcinoma in a Chinese population. PLoS ONE 7, e35145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y.J., Bao C.Z., Gu S.M., Ye D., Jing F.Y., Fan C.H. et al. (2017) Associations between novel genetic variants in the promoter region of MALAT1 and risk of colorectal cancer. Oncotarget 8, 92604–92614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gong W.J., Yin J.Y., Li X.P., Fang C., Xiao D., Zhang W. et al. (2016) Association of well-characterized lung cancer lncRNA polymorphisms with lung cancer susceptibility and platinum-based chemotherapy response. Tumour Biol. 37, 8349–8358 10.1007/s13277-015-4497-5 [DOI] [PubMed] [Google Scholar]
- 18.Hu P.H., Qiao O., Wang J., Li J., Jin H., Li Z.L. et al. (2017) rs1859168 A >C polymorphism regulates HOTTIP expression and reduces risk of pancreatic cancer in a Chinese population. World J. Surg. Oncol. 15, 10.1186/s12957-017-1218-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaker O.G., Senousy M.A. and Elbaz E.M. (2017) Association of rs6983267 at 8q24, HULC rs7763881 polymorphisms and serum lncRNAs CCAT2 and HULC with colorectal cancer in Egyptian patients. Sci. Rep. 7, 10.1038/s41598-017-16500-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He B.S., Sun H.L., Xu T., Pan Y.Q., Lin K., Gao T.Y. et al. (2017) Association of genetic polymorphisms in the LncRNAs with gastric cancer risk in a Chinese population. J. Cancer 8, 531–536 10.7150/jca.17519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li L.J., Jia F., Bai P., Liang Y.D., Sun R.F., Yuan F. et al. (2016) Association between polymorphisms in long non-coding RNA PRNCR1 in 8q24 and risk of gastric cancer. Tumour Biol. 37, 299–303 10.1007/s13277-015-3750-2 [DOI] [PubMed] [Google Scholar]
- 22.Sattarifard H., Hashemi M., Hassanzarei S., Narouie B. and Bahari G. (2017) Association between genetic polymorphisms of long non-coding RNA PRNCR1 and prostate cancer risk in a sample of the Iranian population. Mol. Clin. Oncol. 7, 1152–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L.J., Sun R.F., Liang Y.D., Pan X.M., Li Z.H. and Bai P. (2013) Association between polymorphisms in long non-coding RNA PRNCR1 in 8q24 and risk of colorectal cancer. J. Exp. Clin. Cancer Res. 32, 104 10.1186/1756-9966-32-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chung S.Y., Nakagawa H., Uemura M., Piao L., Ashikawa K., Hosono N. et al. (2011) Association of a novel long non-coding RNA in 8q24 with prostate cancer susceptibility. Cancer Sci. 102, 245–252 10.1111/j.1349-7006.2010.01737.x [DOI] [PubMed] [Google Scholar]
- 25.Salinas C.A., Kwon E., Carlson C.S., Koopmeiners J.S., Feng Z., Karyadi D.M. et al. (2008) Multiple independent genetic variants in the 8q24 region are associated with prostate cancer risk. Cancer Epidemiol. Biomarkers Prev. 17, 1203–1213 10.1158/1055-9965.EPI-07-2811 [DOI] [PubMed] [Google Scholar]
- 26.Zheng S.L., Hsing A.W., Sun J., Chu L.W., Yu K., Li G. et al. (2010) Association of 17 prostate cancer susceptibility loci with prostate cancer risk in Chinese men. Prostate 70, 425–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu X., Duan Q. and Zhang J. (2017) Quantitative assessment of lncRNA HOTAIR polymorphisms and cancer risk in Chinese population: a meta-analysis based on 26,810 subjects. Oncotarget 8, 59698–59708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lv Z., Xu Q. and Yuan Y. (2017) A systematic review and meta-analysis of the association between long non-coding RNA polymorphisms and cancer risk. Mutat. Res. 771, 1–14 10.1016/j.mrrev.2016.10.002 [DOI] [PubMed] [Google Scholar]
- 29.Chu H., Chen Y., Yuan Q., Hua Q., Zhang X., Wang M. et al. (2017) The HOTAIR, PRNCR1 and POLR2E polymorphisms are associated with cancer risk: a meta-analysis. Oncotarget 8, 43271–43283 10.18632/oncotarget.14920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X.F., Yin X.H., Cai J.W., Wang M.J., Zeng Y.Q., Li M. et al. (2017) Significant association between lncRNA H19 polymorphisms and cancer susceptibility: a meta-analysis. Oncotarget 8, 45143–45153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Royds J.A., Pilbrow A.P., Ahn A., Morrin H.R., Frampton C., Russell I.A. et al. (2015) The rs11515 polymorphism is more frequent and associated with aggressive breast tumors with increased ANRIL and decreased p16 (INK4a) expression. Front. Oncol. 5, 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kotake Y., Nakagawa T., Kitagawa K., Suzuki S., Liu N., Kitagawa M. et al. (2011) Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15 (INK4B) tumor suppressor gene. Oncogene 30, 1956–1962 10.1038/onc.2010.568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teeuw W.J., Laine M.L., Bizzarro S. and Loos B.G. (2015) A lead ANRIL polymorphism is associated with elevated CRP levels in periodontitis: a pilot case-control study. PLoS ONE 10, e0137335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frydenberg H., Thune I., Lofterod T., Mortensen E.S., Eggen A.E., Risberg T. et al. (2016) Pre-diagnostic high-sensitive C-reactive protein and breast cancer risk, recurrence, and survival. Breast Cancer Res. Treat. 155, 345–354 10.1007/s10549-015-3671-1 [DOI] [PubMed] [Google Scholar]
- 35.Huang Y., Ye H., Hong Q., Xu X., Jiang D., Xu L. et al. (2014) Association of CDKN2BAS polymorphism rs4977574 with coronary heart disease: a case-control study and a meta-analysis. Int. J. Mol. Sci. 15, 17478–17492 10.3390/ijms151017478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holdt L.M., Beutner F., Scholz M., Gielen S., Gabel G., Bergert H. et al. (2010) ANRIL expression is associated with atherosclerosis risk at chromosome 9p21. Arterioscler. Thromb. Vasc. Biol. 30, 620–627 10.1161/ATVBAHA.109.196832 [DOI] [PubMed] [Google Scholar]
- 37.Harismendy O., Notani D., Song X., Rahim N.G., Tanasa B., Heintzman N. et al. (2011) 9p21 DNA variants associated with coronary artery disease impair interferon-gamma signalling response. Nature 470, 264–268 10.1038/nature09753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bernard D., Prasanth K.V., Tripathi V., Colasse S., Nakamura T., Xuan Z.Y. et al. (2010) A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 29, 3082–3093 10.1038/emboj.2010.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tripathi V., Ellis J.D., Shen Z., Song D.Y., Pan Q., Watt AT. et al. (2010) The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39, 925–938 10.1016/j.molcel.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhuo Y.F., Zeng Q.C., Zhang P., Li G.Y., Xie Q. and Cheng Y. (2017) Functional polymorphism of lncRNA MALAT1 contributes to pulmonary arterial hypertension susceptibility in Chinese people. Clin. Chem. Lab. Med. 55, 38–46 10.1515/cclm-2016-0056 [DOI] [PubMed] [Google Scholar]
- 41.Tian X.L. and Xu G.X. (2015) Clinical value of lncRNA MALAT1 as a prognostic marker in human cancer: systematic review and meta-analysis. BMJ Open 5, 10.1136/bmjopen-2015-008653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Panzitt K., Tschernatsch M.M.O., Guelly C., Moustafa T., Stradner M., Strohmaier H.M. et al. (2007) Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology 132, 330–342 10.1053/j.gastro.2006.08.026 [DOI] [PubMed] [Google Scholar]
- 43.Matouk I.J., Abbasi I., Hochberg A., Galun E., Dweik H. and Akkawi M. (2009) Highly upregulated in liver cancer noncoding RNA is overexpressed in hepatic colorectal metastasis. Eur. J. Gastroenterol. Hepatol. 21, 688–692 10.1097/MEG.0b013e328306a3a2 [DOI] [PubMed] [Google Scholar]
- 44.Lin C.P., Liu C.R., Lee C.N., Chan T.S. and Liu H.E. (2010) Targeting c-Myc as a novel approach for hepatocellular carcinoma. World J. Hepatol. 2, 16–20 10.4254/wjh.v2.i1.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang L.Q., Lin C.R., Jin C.Y., Yang J.C., Tanasa B., Li W.B. et al. (2013) lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature 500, 598–602 10.1038/nature12451 [DOI] [PMC free article] [PubMed] [Google Scholar]