Although members of the Slfn family have been implicated in the regulation of type I interferon (IFN) responses, the mechanisms by which they mediate their effects remain unknown. In the present study, we provide evidence that targeted disruption of the Slfn2 gene leads to increased transcription of IFN-stimulated genes (ISGs) and enhanced type I IFN-mediated antiviral responses.
KEYWORDS: NF-κB, Slfn2, antiviral responses, interferon, signal transduction
ABSTRACT
Although members of the Slfn family have been implicated in the regulation of type I interferon (IFN) responses, the mechanisms by which they mediate their effects remain unknown. In the present study, we provide evidence that targeted disruption of the Slfn2 gene leads to increased transcription of IFN-stimulated genes (ISGs) and enhanced type I IFN-mediated antiviral responses. We demonstrate that Slfn2 interacts with protein phosphatase 6 regulatory subunit 1 (PPP6R1), leading to reduced type I IFN-induced activation of nuclear factor kappa B (NF-κB) signaling, resulting in reduced expression of ISGs. Altogether, these data suggest a novel mechanism by which Slfn2 controls ISG expression and provide evidence for a critical role for Slfn2 in the regulation of IFN-mediated biological responses.
INTRODUCTION
The type I interferons (IFNs), IFN-α and IFN-β, are potent cytokines that inhibit both viral infections and cell growth (1, 2). Binding of type I IFNs to their cognate cell surface receptor, IFNAR, induces activation of the canonical Janus-activated kinase (JAK)/signal transducer and activator of transcription (STAT) signaling pathway, leading to STAT1 and STAT2 phosphorylation and their dimerization and association with interferon regulatory factor 9 (IRF9), forming the IFN-stimulated gene factor 3 (ISGF3) complex. Once active, this complex translocates into the nucleus and binds interferon-sensitive response elements (ISREs) present in the promoter region of IFN-stimulated genes (ISGs), leading to their expression (3–5). Additionally, IFN engagement of IFNAR activates noncanonical signaling pathways that regulate transcription of ISGs through activation of other transcription factors, including nuclear factor kappa B (NF-κB), IRF1, IRF8, IRF3, ATF-2, retinoic acid/retinoic acid receptor (RA/RAR) complex, RA/retinoic X receptor complex, and STAT5/CRK-like proto-oncogene (Crkl) complex (3). IFN-α/β-induced activation of the phosphatidylinositol 3-kinase (PI3K)/AKT signaling pathway activates NF-κB, which in turn regulates transcription of ISGs (6–9). Under physiological conditions, NF-κB is located in the cytoplasm bound to its specific inhibitor IκB complex (10, 11). Signal-induced activation of IκB kinase (IKK) leads to phosphorylation of the IκB regulatory domain and, subsequently, ubiquitination and degradation of the IκB complex by the proteasome (10, 11). Under these conditions, NF-κB becomes active and translocates into the nucleus, where it promotes gene transcription (10, 11).
The Schlafen (Slfn) proteins were first identified in mice and found to be differently expressed during thymocyte maturation and T cell activation (12). There are 10 members of the mouse Slfn family, divided into three groups: group I comprises Slfn1, Slfn1L, and Slfn2; group II comprises Slfn3 and Slfn4; and group III comprises Slfn5, Slfn8, Slfn9, Slfn10, and Slfn14 (13). All murine and human SLFN proteins contain a highly conserved domain, the SLFN box, located next to the ATP/GTP binding AAA domain, whose function is still not well understood (13, 14). Groups II and III contain a SWADL domain, defined by five specific amino acids (Ser-Trp-Ala-Asp-Leu), and only Slfn proteins within group III exhibit a C-terminal extension with a motif homologous to that of superfamily I of DNA/RNA helicases (13, 14). Previously, we demonstrated that some Slfn proteins are induced by type I IFNs and that Slfn2 plays a key role in the generation of IFN-α-induced antiproliferative responses in NIH 3T3 cells (15). In the present study, we sought to define the mechanism of action of Slfn2 in type I IFN signaling. Our results demonstrate that Slfn2 is a negative regulator of ISG expression through regulation of the NF-κB signaling pathway.
RESULTS
Absence of Slfn2 expression enhances IFN-β-inducible transcription of select ISGs and IFN-dependent antiviral responses.
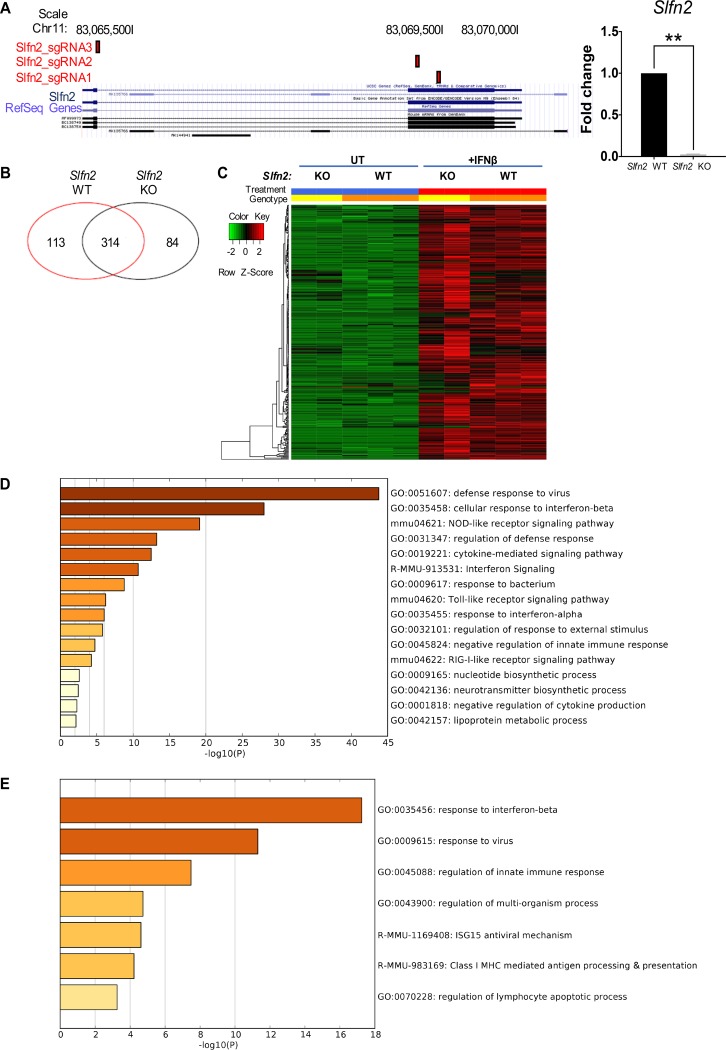
To elucidate the role of Slfn2 in the generation of type I IFN-mediated responses, we generated Slfn2 knockout (KO) mouse embryonic fibroblasts (MEFs) using CRISPR (clustered regularly interspaced short palindromic repeat)/Cas9 technology. We cotransfected MEFs with a pool of three plasmids, each carrying target-specific 20-nucleotide guide RNAs (gRNAs) specific for the Slfn2 gene (Fig. 1A, left) and with a Cas9 nuclease homology-directed repair (HDR) plasmid, which incorporates red fluorescent protein (RFP) and a puromycin resistance gene, for selection of cells where Cas9-induced DNA cleavage has occurred. We validated the generation of Slfn2 KO MEFs, comparing the expression of Slfn2 between wild-type (WT) and Slfn2 KO MEFs by quantitative reverse transcription (qRT)-PCR analysis (Fig. 1A, right).
FIG 1.
Loss of Slfn2 enhances transcription of select type I IFN-stimulated genes. (A) Slfn2 KO MEFs were generated using CRISPR/Cas9 technology. (Left) Schematic representation of the three single guide RNA (sgRNA) target sites located in the Slfn2 locus (red rectangles). (Right) Slfn2 mRNA expression in Slfn2 WT and Slfn2 KO MEFs was determined by qRT-PCR using Slfn2-specific primers, and Gapdh was used for normalization and as an internal control. The graph shows means and standard errors (SE) of the results of three independent experiments. Statistical analyses were performed using a ratio-paired one-tailed t test (**, P < 0.01). (B and C) Slfn2 WT and KO MEFs were left untreated (UT) or were treated with mouse IFN-β for 6 h. Gene expression profiles of Slfn2 WT and KO MEFs, untreated and treated with IFN-β, were compared in three independent experiments for Slfn2 WT MEFs and in two independent experiments for Slfn2 KO MEFs, using MouseWG-6 v2.0 Expression BeadChips (Illumina) and Illumina iScan. (B) Venn diagram showing the differentially expressed genes in Slfn2 WT and Slfn2 KO MEFs after IFN-β treatment. (C) Heat map depicting the differential expression of ISGs in Slfn2 WT and KO MEFs. (D) Ontology analysis of the ISGs induced at least 2-fold more by IFN-β in Slfn2 KO MEFs than in Slfn2 WT MEFs. (E) Ontology analysis of the ISGs induced at least 2-fold more by IFN-β in Slfn2 WT MEFs than in Slfn2 KO MEFs.
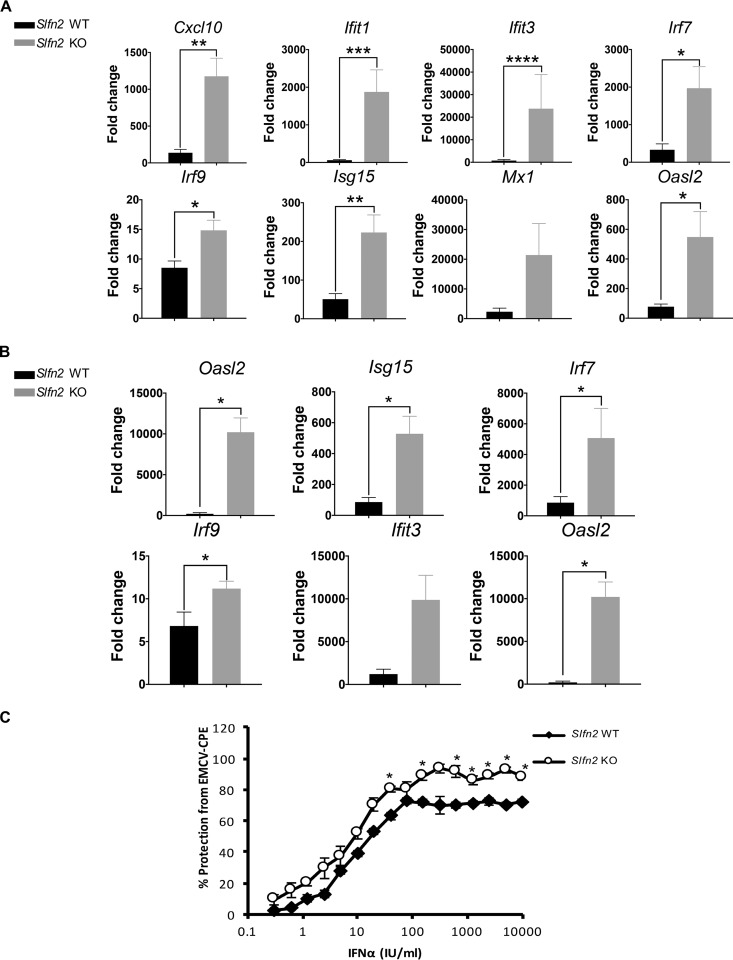
Next, we examined the effects of Slfn2 on the regulation of ISGs by specifically analyzing the differential expression of type I IFN-inducible genes in untreated versus IFN-β-treated Slfn2 WT and KO MEFs, using RNA-sequencing (RNA-seq) analysis. Comparison of the transcriptomic profile between untreated and IFN-β-treated cells revealed differential expression of type I IFN-inducible genes in the Slfn2 WT and KO MEFs: IFN-inducible expression of 113 unique genes in Slfn2 WT MEFs and 84 unique genes in Slfn2 KO MEFs (Fig. 1B; see Tables S1 and S2 in the supplemental material). Additionally, 314 genes were found to be induced by IFN-β treatment in both Slfn2 WT and KO MEFs (Fig. 1B and C; see Table S3 in the supplemental material). Importantly, among them, 80 ISGs were found to be induced at levels at least 2-fold higher in IFN-β-treated Slfn2 KO MEFs than in Slfn2 WT MEFs, and 52 ISGs were found to be induced at levels at least 2-fold higher in IFN-β-treated Slfn2 WT MEFs than in Slfn2 KO MEFs (see Table S3 in the supplemental material; highlighted in red and blue, respectively). Ontology analysis of these two groups of genes revealed that 39 genes are involved in the defense response to virus in Slfn2 KO MEFs (Fig. 1D; see Data Set S1 in the supplemental material), whereas only 17 genes were found to be involved in response to virus in Slfn2 WT MEFs (Fig. 1E; see Data Set S2 in the supplemental material). Ontology analysis of the genes differentially induced by IFN-β treatment only in Slfn2 WT or in Slfn2 KO MEFs (see Tables S1 and S2 in the supplemental material) revealed that they are involved in different biological functions and molecular mechanisms, such as regulation of cell matrix adhesion in Slfn2 KO MEFs and protein methylation in Slfn2 WT MEFs (see Data Sets S3 and S4 in the supplemental material). Next, we validated the RNA-sequencing results by qRT-PCR analysis of untreated versus IFN-β-treated Slfn2 WT and Slfn2 KO MEFs (pooled and single clones) (Fig. 2A and B). We show that the Cxcl10, Ifit1, Ifit3, Irf7, Irf9, Isg15, Mx1, and Oasl2 genes were significantly induced to higher levels in the cells with targeted Slfn2 disruption. Viewed together, our results suggest that Slfn2 may have a key role in the transcriptional regulation of ISGs. To determine whether increased type I IFN-induced expression of select antiviral ISGs in Slfn2 KO MEFs correlates with more effective type I IFN-induced antiviral responses, we examined IFN-α-inducible antiviral responses against encephalomyocarditis virus (EMCV) in Slfn2 WT versus Slfn2 KO MEFs. We provide evidence that Slfn2 KO MEFs are more sensitive to the antiviral effects of IFN-α than Slfn2 WT MEFs (Fig. 2C). Of note, in our previous study, we showed that Slfn2 knockdown/depletion did not affect IFN-α-induced antiviral responses (13). In contrast, in the present study, complete knockout of Slfn2 by the CRISPR/Cas9 methodology led to complete depletion of the Sfln2 gene; thus, the absence of Slfn2 is associated with enhanced expression of immune-related ISGs and enhanced IFN-α-induced antiviral responses.
FIG 2.
Loss of Slfn2 enhances transcription of antiviral ISGs and increases IFN-inducible antiviral responses. (A) Slfn2 WT and KO MEFs were either left untreated or treated with mouse IFN-β for 6 h. qRT-PCR analyses of the relative mRNA expression of the indicated genes after IFN-β stimulation are shown. The expression levels of the indicated genes were determined using Gapdh for normalization and as an internal control. The data are expressed as fold change over untreated samples, and the graphs represent means and SE of the results of three independent experiments (Mx1 and Oasl2) and four independent experiments (Cxcl10, Ifit1, Ifit3, Irf7, Irf9, and Isg15). Statistical analyses were performed using a one-tailed ratio-paired t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). (B) Slfn2 WT MEFs and Slfn2 KO MEFs derived from selected single clones were left untreated or were treated with mouse IFN-β for 6 h. qRT-PCR analyses of the relative mRNA expression of ISGs after IFN-β stimulation are shown. The expression levels of the indicated genes were determined using Gapdh for normalization and as an internal control. The data are expressed as fold change over untreated samples, and the graphs represent means and SE of three independent experiments (Ifit3 and Oasl2) and five independent experiments (Cxcl10, Isg15, Irf7, and Irf9). Statistical analysis was performed using a one-tailed ratio-paired t test (*, P < 0.05). (C) Slfn2 WT and Slfn2 KO MEFS were treated with mouse IFN-α for 16 h, as indicated, and then challenged with EMCV. EMCV-induced CPE was determined 24 h later. The values shown represent means and SE of the results of four independent experiments. Statistical analyses were performed using a two-tailed unpaired t test (*, P < 0.05).
Increased type I IFN-induced transcription of select ISGs in Slfn2 KO MEFs is not dependent on activation of the JAK/STAT signaling pathway.
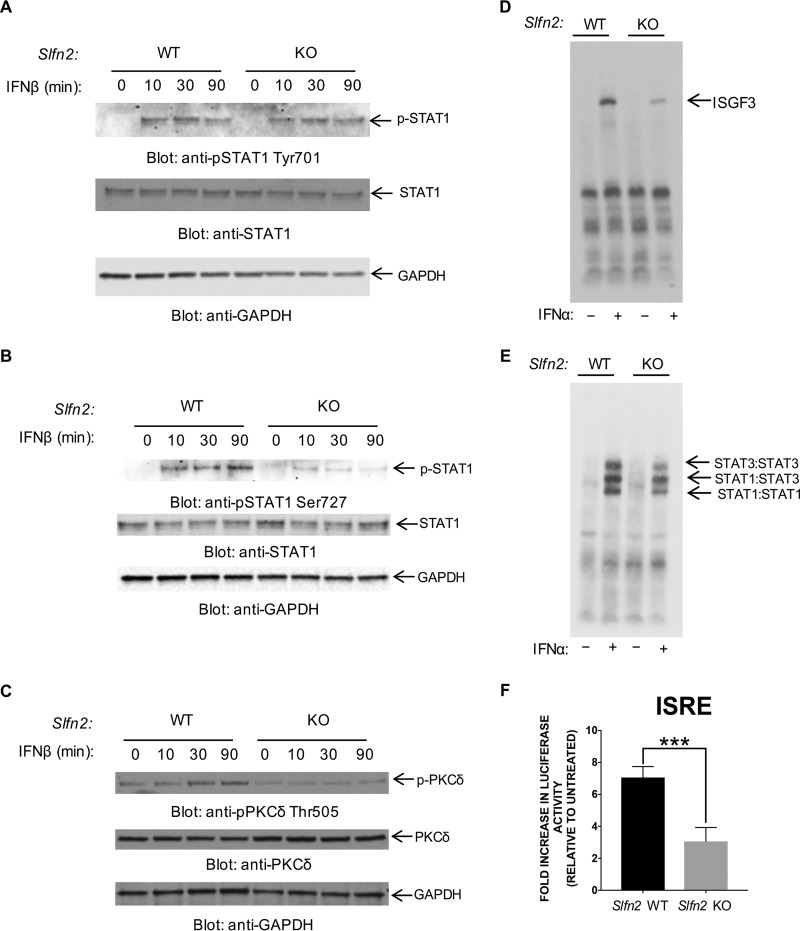
To investigate whether the increase in select IFN-β-inducible ISG expression in Slfn2 KO MEFs is due to increased STAT1 activity in the cells, we treated Slfn2 WT and Slfn2 KO MEFs with IFN-β in a time course study and analyzed the phosphorylation levels of STAT1 by immunoblotting. Our results showed that IFN-β-mediated phosphorylation of STAT1 on tyrosine 701 was similar in Slfn2 WT and Slfn2 KO MEFs (Fig. 3A). Surprisingly, IFN-β-induced phosphorylation of STAT1 on serine 727 was defective in Slfn2 KO MEFs compared to Slfn2 WT MEFs (Fig. 3B). Previously, we and others have shown that protein kinase C delta (PKCδ) is required for type II IFN-induced phosphorylation of STAT1 on serine 727 (16, 17). Accordingly, we examined IFN-β-inducible PKCδ phosphorylation and observed less PKCδ phosphorylation in Slfn2 KO MEFs than in Slfn2 WT MEFs (Fig. 3C). We next conducted a series of electrophoretic mobility shift assays (EMSAs) employing both ISRE and sis-inducible element (SIE) DNA elements and lysates from IFN-α treated MEFs. Lysates from IFN-α-treated Slfn2 KO MEFs resulted in reduced ISGF3-ISRE complex formation in the cells than lysates from IFN-α-treated Slfn2 WT MEFs (Fig. 3D). Moreover, we also observed reduced binding of STAT1 homodimers, STAT3 homodimers, and STAT1-STAT3 heterodimers to SIE in Slfn2 KO MEFs compared to Slfn2 WT MEFs following IFN-α treatment (Fig. 3E). To determine whether reduced ISGF3-ISRE complex formation in Slfn2 KO MEFs correlated with reduced IFN-β-dependent ISRE enhancer activity in the promoters of ISGs, we stably transduced both Slfn2 WT and Slfn2 KO MEFs with a luciferase reporter lentiviral vector containing an ISRE that drives transcription of the luciferase gene and measured luciferase activity in untreated and IFN-β-treated cells. Our results demonstrate that IFN-β-mediated ISRE-luciferase activity is reduced in Slfn2 KO MEFs compared to Slfn2 WT MEFs (Fig. 3F). Viewed together, our data demonstrate that the enhanced IFN-β-induced expression of select ISGs we observed in Slfn2 KO MEFs is neither dependent on nor associated with activation of the classical type I IFN-induced JAK/STAT signaling pathway.
FIG 3.
Slfn2 activity is required for IFN-β-mediated activation of JAK/STAT signaling. (A to C) Slfn2 WT and Slfn2 KO MEFs were serum starved overnight and then treated with mouse IFN-β, as indicated. Equal amounts of total cell lysates were prepared and resolved by SDS-PAGE and immunoblotted with the indicated antibodies. (D and E) Slfn2 WT and Slfn2 KO MEFs were either left untreated or treated for 15 min with mouse IFN-α, as indicated. Nuclear extracts were prepared and reacted with radiolabeled ISRE (D) and SIE (E) probes. Protein-probe complexes were resolved by native gel electrophoresis and visualized by autoradiography. (F) Slfn2 WT and Slfn2 KO MEFs were stably transduced with an ISRE-luciferase reporter construct. The cells were either left untreated or treated with mouse IFN-β for 6 h, followed by measurement of luciferase activity. The data are expressed as fold increase of luciferase activity in response to IFN-β treatment over control, untreated samples for each condition. The graph shows means and standard errors of the mean (SEM) of the results of four independent experiments. Statistical analyses were performed using a two-tailed unpaired t test (***, P < 0.001).
Slfn2 controls type I IFN-induced activation of NF-κB.
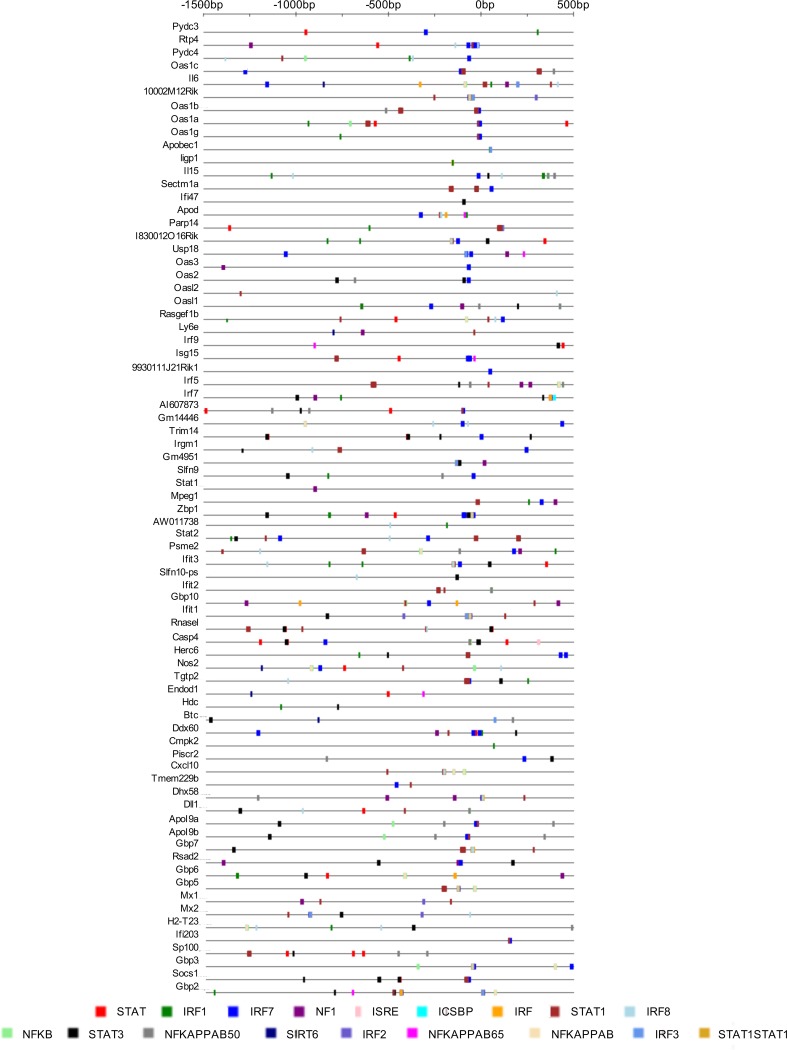
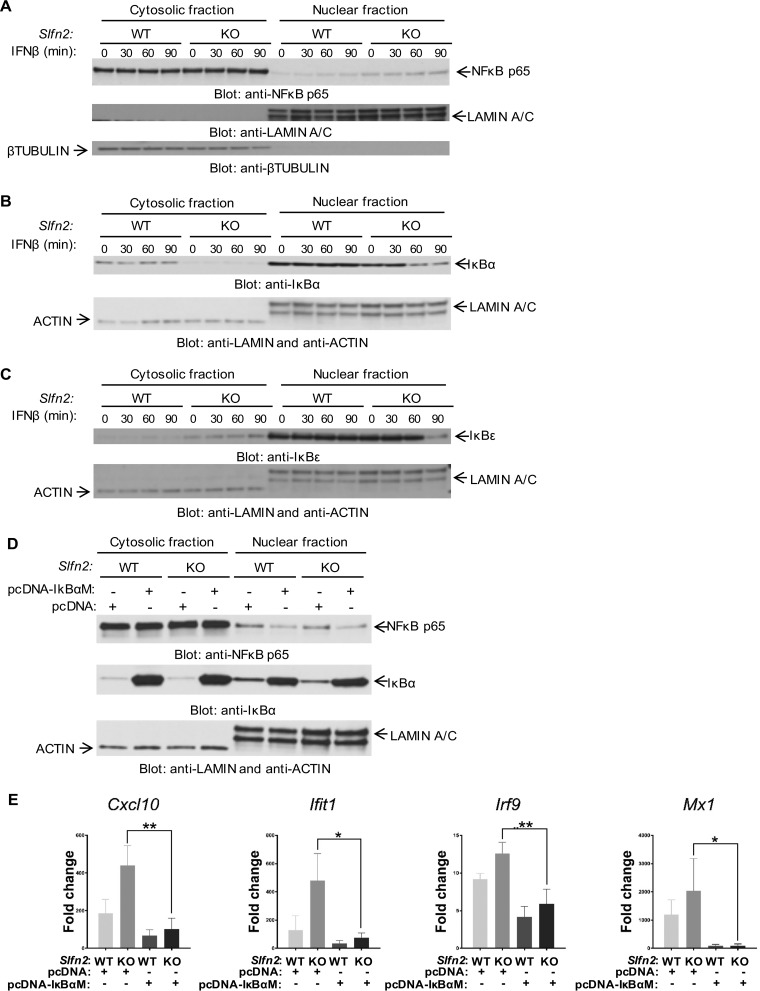
To investigate which transcription factors (TFs) might be responsible for the enhanced IFN-inducible ISG expression we observed in Slfn2 KO MEFs, we analyzed the predicted TF binding sites present in the promoter regions of the genes that were found to be induced greater by IFN-β in Slfn2 KO MEFs (see Table S3 in the supplemental material; highlighted in red) than in Slfn2 WT MEFs, using the Interferome database (18). We identified different binding sites for the NF-κB family of TFs in the promoter regions of the genes (Fig. 4). Interestingly, many of these genes present NF-κB binding sites but not ISRE binding sites, suggesting that loss of Slfn2 may alter regulation by IFN of ISRE-independent NF-κB target genes (Fig. 4). Studies have shown that IFN-β induces activation of NF-κB through both canonical and noncanonical pathways and that the promoter regions of ISGs have binding sites for NF-κB (9, 19). Accordingly, we next investigated the role of Slfn2 in type I IFN-induced activation of the NF-κB signaling pathway. We provide evidence that Slfn2 KO MEFs have increased nuclear levels of NF-κB p65 compared to Slfn2 WT MEFs (Fig. 5A). Additionally, the levels of the NF-κB inhibitor IκBα are reduced in the cytoplasmic fractions of Slfn2 KO MEFs compared to Slfn2 WT MEFs (Fig. 5B), and the levels of the NF-κB inhibitor IκBε are increased in the cytoplasmic fractions of Slfn2 KO MEFs compared to Slfn2 WT MEFs (Fig. 5C). Next, to determine whether the higher levels of expression of IFN-β-induced ISGs in Slfn2 KO MEFs are due to increased IFN-β-mediated NF-κB p65/p50 activation in these cells, we overexpressed the dominant-negative mutant IκBαM in Slfn2 WT and Slfn2 KO MEFs (Fig. 5D). The IκBαM mutant protein is not susceptible to phosphorylation at the N-terminal Ser32 and Ser36, which have been replaced by alanines, thereby inhibiting the translocation of NF-κB p65/p50 to the nucleus (Fig. 5D). Analyses of mRNA expression of Cxcl10, Ifit1, Irf9, and Mx1 after IFN-β treatment in Slfn2 WT and Slfn2 KO MEFs transfected with an empty-vector plasmid (pcDNA) or an IκBαM mutant plasmid showed that the increased IFN-β-induced expression of these genes in Slfn2 KO MEFs is lost when NF-κB p65/p50 activation is inhibited (Fig. 5E). Together, these results suggest that Slfn2 inhibits IFN-β-mediated transcription of ISGs by regulating the activation of NF-κB signaling.
FIG 4.
Transcription factor binding sites present in the promoter regions of genes that were more highly induced in IFN-β-treated Slfn2 KO MEFs than in Slfn2 WT MEFs. Shown is analysis of the predicted transcription factor binding sites present in the promoter regions of the genes that were induced by IFN-β at least 2-fold more in Slfn2 KO MEFs than in Slfn2 WT MEFs in our RNA-seq analysis using the Interferome database (http://www.interferome.org). All the predicted binding sites for the different transcription factors are indicated.
FIG 5.
Higher expression of IFN-β-induced ISGs in Slfn2 KO MEFs is NF-κB dependent. (A to C) Slfn2 WT and Slfn2 KO MEFs were serum starved overnight and then treated with mouse IFN-β, as indicated. Cellular lysates were prepared and fractionated into cytosol and nuclear fractions, and then equal amounts of lysates were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. (D) Slfn2 WT and Slfn2 KO MEFs were transfected with pcDNA-IκBαM vector or pcDNA empty-vector plasmids. Twenty-four hours later, cellular lysates were prepared and fractionated into cytosol and nuclear fractions, and then equal amounts of lysates were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. (E) Slfn2 WT and Slfn2 KO MEFs were transfected with pcDNA-IκBαM vector or pcDNA empty-vector plasmids. Twenty-four hours after transfection, the cells were treated with mouse IFN-β for 6 h. qRT-PCR analyses of the relative mRNA expression of the indicated genes after IFN-β stimulation are shown. The expression levels of the indicated genes were determined using Gapdh for normalization and as an internal control. The data are expressed as fold change over untreated samples, and the graphs represent means and SE of the results of three independent experiments. Statistical analyses were performed using ordinary one-way ANOVA, followed by Tukey's multiple-comparison test. P values between IFN-β-treated pcDNA- and IκBαM-transfected Slfn2 KO MEFs are shown (*, P < 0.05; **, P < 0.01).
Slfn2 binds PPP6R1, a regulator of NF-κB signaling, controlling IFN-β-induced expression of ISGs.
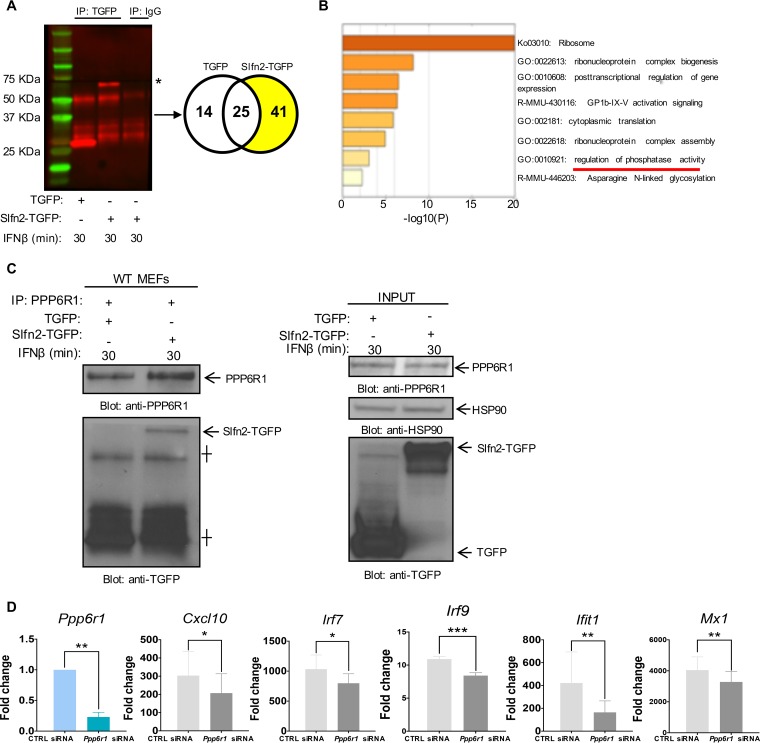
To identify the mechanism by which loss of Slfn2 expression enhances IFN-β-dependent NF-κB activation, we investigated which proteins interact with Slfn2 in the presence of IFN-β by performing mass spectrometry (MS). For this, we overexpressed turbo-green fluorescent protein (TGFP)-tagged Slfn2 protein (Slfn2-TGFP) in NIH 3T3 cells, immunoprecipitated the complexes formed with Slfn2 using anti-TGFP antibody, and then analyzed them by mass spectrometry (Fig. 6A). Our proteomic studies revealed 41 putative Slfn2 binding partners in the presence of IFN-β stimulation (Fig. 6A and B and Table 1). Of these, serine/threonine protein phosphatase 6 regulatory subunit 1 (PPP6R1) has previously been identified as involved in the PP6-mediated dephosphorylation of IκBε, inhibiting its degradation in response to treatment with tumor necrosis factor alpha (TNF-α) (20). Ontology analysis of the 41 putative Slfn2 binding partners revealed that PPP6R1 is involved in regulation of phosphatase activity (Fig. 6B). To confirm our mass spectrometry results, we immunoprecipitated PPP6R1 from IFN-β-treated Slfn2 WT MEFs transfected with Slfn2-TGFP- or TGFP-expressing plasmids and confirmed that PPP6R1 interacts with Slfn2, but not with TGFP, by immunoblotting analysis (Fig. 6C). Previous studies have shown that PPP6R1 binds IκBε and that knocking down of PPP6R1 enhances IκBε degradation after TNF-α treatment (20, 21). Phosphorylation and degradation of the IκBs is the critical step that leads to activation of the NF-κB family of TFs (10). To examine whether the reduced interaction between Slfn2 and PPP6R1 could be responsible for the higher IFN-β-induced activation of NF-κB, leading to enhanced expression of ISGs in Slfn2 KO MEFs, we knocked down Ppp6r1 in Slfn2 KO MEFs, treated the cells with IFN-β for 6 h, and then evaluated gene expression for Cxcl10, Irf7, Irf9, Ifit1, and Mx1 by qRT-PCR analysis (Fig. 6D). Our results revealed that knockdown of Ppp6r1 in Slfn2 KO MEFs decreases IFN-β-induced expression of these ISGs (Fig. 6D), suggesting that the interaction between Slfn2 and PPP6R1 affects the activation of IFN-inducible NF-κB signaling and subsequent transcription of ISGs.
FIG 6.
Slfn2 interacts with PPP6R1, regulating IFN-β-mediated ISG expression. (A) NIH 3T3 cells were transfected with Slfn2-TGFP- or TGFP-overexpressing plasmids and then treated with mouse IFN-β for 30 min. Cell lysates were prepared and then immunoprecipitated (IP) with either TGFP-specific antibody or control nonimmune rabbit IgG, as indicated. (Left) Fifty micrograms of the immunoprecipitated proteins was resolved by SDS-PAGE and immunoblotted with anti-TGFP antibody. The asterisk indicates expression of Slfn2-TGFP. (Right) Three milligrams of the immunoprecipitated proteins was subjected to mass spectrometry analysis. The Venn diagram shows the number of proteins identified by mass spectrometry as putative interactors of Slfn2, highlighted in yellow. (B) Ontology analysis of the 41 proteins bound to Slfn2 as detected by mass spectrometry. Shown is a heat map of enriched terms across input protein lists. Colors reflect P values. (C) Slfn2 WT MEFs were transfected with Slfn2-TGFP- or TGFP-overexpressing vector plasmids and treated with mouse IFN-β for 30 min. Cell lysates were prepared, and then equal amounts of proteins were immunoprecipitated with anti-PPP6R1-specific antibody. (Left) Immunoprecipitated proteins were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. The crosses indicate heavy and light chains. (Right) Total cell lysates (INPUT) derived from the same experiment were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. (D) Slfn2 KO MEFs were transfected with either Ppp6r1 siRNA or control (CTRL) siRNA. Twenty-four hours after transfection, the cells were treated with mouse IFN-β for 6 h. qRT-PCR analyses of the relative mRNA expression of the indicated genes after IFN-β stimulation are shown. The expression levels of the indicated genes were determined using Gapdh for normalization and as an internal control. The data are expressed as fold change over untreated samples, and the graphs represent means and SE of the results of four independent experiments. Statistical analyses were performed using a one-tailed ratio-paired t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
TABLE 1.
List of putative protein interactors of Slfn2 identified by mass spectrometrya
| No. | Identified protein | Accession no. | Molecular mass (kDa) |
|---|---|---|---|
| 1 | Alpha-enolase OS = M. musculus GN = Eno1 PE = 1 SV = 3 | ENOA_MOUSE | 47 |
| 2 | CAD protein | PYR1_MOUSE | 243 |
| 3 | Caprin-1 | CAPR1_MOUSE | 78 |
| 4 | Collagen alpha-1(I) chain | CO1A1_MOUSE | 138 |
| 5 | Collagen alpha-2(l) chain | CO1A2_MOUSE | 130 |
| 6 | Cytoplasmic dynein 1 heavy chain 1 | DYHC1_MOUSE | 532 |
| 7 | Endoplasmin | ENPL_MOUSE | 92 |
| 8 | Enhancer of mRNA-decapping protein 4 | EDC4_MOUSE | 152 |
| 9 | Filamin-A | FLNA_MOUSE | 281 |
| 10 | Gamma-adducin | ADDG_MOUSE | 79 |
| 11 | Heterogeneous nuclear ribonucleoprotein A0 | ROA0_MOUSE | 31 |
| 12 | Importin subunit beta-1 | IMB1_MOUSE | 97 |
| 13 | Interferon-induced protein with tetratricopeptide repeats 1 | IFIT1_MOUSE | 54 |
| 14 | La-related protein 1 | LARP1_MOUSE | 121 |
| 15 | Lupus La protein homolog | LA_MOUSE | 48 |
| 16 | Myosin light polypeptide 6 | MYL6_MOUSE | 17 |
| 17 | N-Acetylglucosamine-6-phosphate deacetylase | NAGA_MOUSE | 44 |
| 18 | Protein argonaute 2 | AGO2_MOUSE | 97 |
| 19 | Protein C21orf2 homolog | CU002_MOUSE | 28 |
| 20 | Putative helicase MOV-10 | MOV10_MOUSE | 114 |
| 21 | RNA-binding motif, single-stranded-interacting protein 1 | RBMS1_MOUSE | 44 |
| 22 | Serine/threonine-protein phosphatase 6 regulatory subunit 1 | PP6R1_MOUSE | 95 |
| 23 | T-complex protein 1 subunit eta | TCPH_MOUSE | 60 |
| 24 | Transcriptional activator protein Pur-alpha | PURA_MOUSE | 35 |
| 25 | Transitional endoplasmic reticulum ATPase | TERA_MOUSE | 89 |
| 26 | tRNA selenocysteine 1-associated protein 1 | TSAP1_MOUSE | 32 |
| 27 | UDP-glucose:glycoprotein glucosyltransferase 1 | UGGG1_MOUSE | 176 |
| 28 | 40S ribosomal protein S24 | RS24_MOUSE | 15 |
| 29 | 40S ribosomal protein S29 | RS29_MOUSE | 7 |
| 30 | 40S ribosomal protein S30 | RS30_MOUSE | 7 |
| 31 | 60S acidic ribosomal protein P2 | RLA2_MOUSE | 12 |
| 32 | 60S ribosomal protein L10a | RL10A_MOUSE | 25 |
| 33 | 60S ribosomal protein L13a | RL13A_MOUSE | 23 |
| 34 | 60S ribosomal protein L18a | RL18A_MOUSE | 21 |
| 35 | 60S ribosomal protein L28 | RL28_MOUSE | 16 |
| 36 | 60S ribosomal protein L29 | RL29_MOUSE | 18 |
| 37 | 60S ribosomal protein L35 | RL35_MOUSE | 15 |
| 38 | 60S ribosomal protein L37a | RL37A_MOUSE | 10 |
| 39 | 60S ribosomal protein L38 | RL38_MOUSE | 8 |
| 40 | 60S ribosomal protein L5 | RL5_MOUSE | 34 |
| 41 | 60S ribosomal protein L7 | RL7_MOUSE | 31 |
PPP6R1 information is in boldface.
DISCUSSION
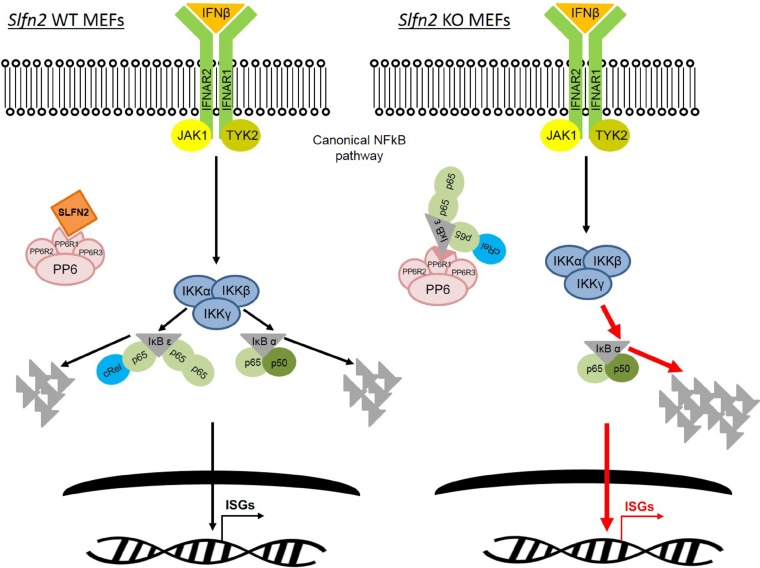
Understanding the roles of Slfn2 in different physiological/pathological contexts is important, since Slfn2 has been found to regulate distinct signaling pathways and to affect molecular mechanisms in a cell-type-specific/tissue-dependent manner (22). For example, in naive T cells, Slfn2 was shown to regulate quiescence by preventing endoplasmic reticulum stress that in turn regulates cholesterol levels (23). This ensures naive T cells remain quiescent yet poised for activation by specific stimuli, such as a microbial invasion (23, 24). In primary mouse neurons, type I IFN-induced antiviral responses against neurotropic viruses were defective and heterogeneous, due to a reduced induction of 15 ISGs, including Slfn2, in the cells compared to primary fibroblasts derived from the same mice (25). In osteoclast precursors, receptor activator of NF-κB ligand (RANKL) selectively induces Slfn2 expression, which is required for osteoclastogenesis, by regulating c-Jun activation and expression of NFATc1 and c-Jun (26). In previous studies, we provided evidence for type I IFN-inducible expression of Slfn1, Slfn2, Slfn3, Slfn4, Slfn5, Slfn8, and Slfn9 in murine renal cell carcinoma cells, melanoma cells, and fibroblasts (15, 27). These mouse Slfn proteins are classified as ISGs, but the mechanism(s) by which they regulate type I IFN responses is poorly understood (27). In the present study, we investigated the role of Slfn2 in type I IFN signaling. IFN-α/β activation of IFNAR leads to activation of JAK/STAT signaling, which results in the expression of STAT-dependent ISGs (2, 5, 28, 29). Studies have shown that type I IFN treatment induces degradation of IκBα, increasing NF-κB p65/p50-mediated transcription of antiviral ISGs, such as Mx1 and Nmi (8). Moreover, different groups have provided evidence that type I IFNs and TNF-α work together to protect the host from lethal virus infection by enhancing transcription of antiviral ISGs through activation of both JAK/STAT and NF-κB signaling pathways (19, 30). Interestingly, our data suggest that Slfn2 plays a key role in the regulation/balance of type I IFN-mediated activation of STAT1 and NF-κB. High circulating levels of IFN and/or ISGs have been linked to autoimmune diseases and type I interferonopathies (31, 32). Additionally, type I IFN signaling is induced by a DNA damage response and may contribute to the accumulation of DNA damage and promote cellular senescence and premature aging (33). Characterization of the specific role of Slfn2 in the tight regulation of type I IFN responses, beyond an antiviral response, and in preventing deleterious effects of type I IFNs on specific cell types or tissues is warranted. In our studies, IFN-treated Slfn2 WT MEFs exhibited increased activation of JAK/STAT signaling but decreased activation of NF-κB signaling compared to IFN-treated Slfn2 KO MEFs. Our results suggest that Slfn2 interacts with PPP6R1, possibly preventing its binding to IκBε. Following IFN engagement of IFNAR, the IKK complex, composed of IKKα, IKKβ, and IKKγ, phosphorylates both NF-κB inhibitors IκBα and IκBε. Once phosphorylated, IκB inhibitor molecules are ubiquitinated, leading to their degradation by the proteasome, enabling NF-κB homodimers (p65/p65) and heterodimers (cRel/p65 and p65/p50) to translocate into the nucleus and activate gene transcription, including that of ISGs (Fig. 7) (10, 34, 35). In contrast, in the absence of Sfln2 expression, PPP6R1 binds to IκBε, preventing its phosphorylation by the IKK complex during IFN stimulation. Under these conditions, the IKK complex predominantly phosphorylates IκBα, which is then degraded by the proteasome, leading to increased levels of free NF-κB p65/p50 complex, which translocates into the nucleus, resulting in higher transcription of select ISGs (Fig. 7) (9, 34, 35).
FIG 7.
Role of Slfn2 in type I IFN signaling. Shown is a schematic representation of a proposed mechanism by which Slfn2 controls expression of ISGs.
In a recent study, we identified human SLFN5 as a transcriptional corepressor of STAT1-induced ISG transcription (36). Viewed together, our studies provide evidence that SLFN proteins are involved in the regulation of both canonical and noncanonical type I IFN-inducible signaling pathways. IFN engagement of IFNAR activates STAT3, leading to the induction of the PI3K/AKT signaling pathway, which in turn induces activation of NF-κB (7). In Daudi cells, with defective STAT3 expression, type I IFN treatment fails to activate NF-κB, resulting in defective type I IFN-mediated antiviral responses (7, 37, 38). Our results show that Slfn2 KO MEFs are more sensitive to type I IFN-mediated antiviral responses, possibly because these cells exhibit higher levels of activated NF-κB p65/p50 heterodimers in their nuclei than Slfn2 WT MEFs, thereby leading to enhanced transcription of antiviral ISGs.
NF-κB signaling has been implicated in the pathogenesis of many human diseases, including inflammatory diseases, cancers, autoimmune diseases, and virus infections (11, 39). The clinical use of type I IFNs for the treatment of cancers and virus infections has not yielded the kinds of outcomes that the promise reflected in in vitro and/or in vivo studies suggested (4, 40, 41). Targeting SLFN proteins could be a mechanism to enhance type I IFN responses, mediated by modulation of NF-κB signaling.
MATERIALS AND METHODS
Cell culture.
MEFs were isolated from C57/BL6 mice and cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and antibiotics. NIH 3T3 cells were purchased from the ATCC (CRL-1658) and were cultured in DMEM supplemented with 10% bovine serum (BS). The cells were cultured at 37°C and 5% CO2 and tested for mycoplasma contamination.
Reagents.
Recombinant mouse IFN-β was provided by Biogen. The antibodies against phospho-PKCδ (p-PKCδ) (Thr 505; number 9374S), PKCδ (number 9616S), p-STAT1 (Ser 727; number 8826S), p-STAT1 (Tyr 701; number 7649S), pSTAT3 (Tyr 705; number 9131S), lamin A/C (number 2032S), NF-κB–p65 (number 8242S), and NF-κB–p50 (number 13586S) were obtained from Cell Signaling. The antibodies against actin (number sc-8432), STAT1 (number sc-346), STAT3 (number sc-482), IκBα (number sc-203), IκBε (number sc-7275), and HSP90 (number sc-7941) were obtained from Santa Cruz Biotechnology. The antibody against TGFP (number PA5-22688) was obtained from Thermo Fisher, anti-GAPDH (anti-glyceraldehyde-3-phosphate dehydrogenase) (number MAB374) was obtained from Millipore. Control small interfering RNA A (siRNA-A) (number sc-37007) and mouse PPP6R1 siRNA (number sc-106531) were from Santa Cruz Biotechnology. pcDNA3.0-IκBαM was kindly provided by Francesca Zazzeroni (University of L'Aquila, L'Aquila, Italy) and was previously described (42).
Generation of Slfn2 KO MEFs using CRISPR/Cas9 technology.
Slfn2 KO MEFs were generated as described in our previous study (36). Briefly, 2 μg of the Slfn2 CRISPR/Cas9 KO plasmid (m2; number sc423017) and 2 μg of the Slfn2 HDR plasmid (number sc423017-HDR; Santa Cruz Biotechnology) were cotransfected into MEFs using the Lipofectamine 3000 transfection reagent (Thermo Fisher), according to the manufacturer's instructions. Forty-eight hours after transfection, the MEFs were cultured in the presence of puromycin (1 μg/ml), and 2 weeks later, the puromycin-resistant cells were expanded and RFP-expressing MEFs were sorted by flow cytometry. We obtained single clones using flow cytometry cell-sorting technology. Deletion of the Slfn2 gene was confirmed by qRT-PCR using Slfn2-specific primers and by RNA-sequencing analysis.
Protein mass spectrometry analysis.
NIH 3T3 cells (6 × 106) were seeded in 15-cm plates. Next, the cells were transfected with either pCMV6-TGFP-Slfn2 or pCMV6-TGFP using the Lipofectamine 3000 reagent (Thermo Fisher) according to the manufacturer's protocol. Twenty-four hours later, the cells were treated with mouse IFN-β at 104 IU/ml for the indicated time, followed by coimmunoprecipitation: 3 mg of total cell lysates was incubated overnight at 4°C with anti-TGFP antibody as indicated, followed by incubation at 4°C for 1 h with protein G-Sepharose 4 Fast Flow beads (GE Healthcare). The same procedure was followed using anti-rabbit IgG antibody (BD Pharmigen; number 550875) as a control. The beads were then washed three times with Nonidet P-40 buffer. Protein complexes were eluted from the beads and submitted for mass spectrometry analysis. Samples were run on an SDS-PAGE gel, and a gel lane was divided into 10 gel fractions based on molecular weight before performing in-gel digestion. The gel bands were washed in 100 mM ammonium bicarbonate (AmBic)-acetonitrile (ACN) and reduced with 10 mM dithiothreitol at 50°C for 30 min. Cysteines were alkylated with 100 mM iodoacetamide in the dark for 30 min at room temperature. The gel bands were washed in 100 mM AmBic-ACN prior to adding 600 ng trypsin for overnight incubation at 37°C. Supernatant that contained peptides was saved in a new tube. The gel bands were washed with 50% ACN-5% formic acid (FA) at room temperature for 10 min with gentle shaking, and the eluates were collected. The wash step was repeated using 80% ACN-5% FA and 100% ACN, and all the eluates were then subjected to SpeedVac drying. After lyophilization, peptides were reconstituted with 5% ACN-0.1% FA in water and injected onto a trap column (150-μm inside diameter [i.d.] by 3 cm packed in house with a ReproSil C18aq high-pressure liquid chromatography [HPLC] column [3 μm]) coupled with a nanobore analytical column (a 75-μm i.d. by 10.5-cm PicoChip column packed with ReproSil C18aq [1.9 μm]; New Objectives). Samples were separated in a linear gradient of solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid in ACN) over 45 min, using an Easy nano-liquid chromatography (LC) instrument (ThermoFisher Scientific). MS data were obtained on an LTQ Velos Orbitrap mass spectrometer (ThermoFisher Scientific). The data collected were analyzed using Mascot (Matrix Science) 2.5 against the UniProt database, and the results were reported at a 1% false-discovery rate (FDR) in Scaffold 4.5 (Proteome Software). We considered Slfn2 putative interactor proteins to be those found to bind only Slfn2, excluding proteins that were bound to the IgG and TGFP negative controls.
RNA-sequencing analysis. (i) Library construction and sequencing.
Library construction and sequencing were performed at the Genomics Core facility at the University of Chicago. RNA quality and quantity were first determined with the Agilent Bioanalyzer 2100, accepting RNA integrity numbers (RIN) of >7 and quantities of 100 ng or more per sample. The total-RNA samples were treated with DNase I to degrade any possible DNA contamination, followed by rRNA removal using RiboZero (Epicentre, Madison, WI). Directional libraries were prepared using an Illumina TruSeq Stranded Total RNA HT sample preparation kit (Illumina, San Diego, CA) according to the manufacturer's instructions. The barcoded libraries were amplified by PCR, and the resulting cDNA libraries were quantified using quantitative PCR (qPCR). Equimolar amounts of each cDNA library were pooled and sequenced on an Illumina HiSeq2500, with four libraries multiplexed per lane.
(ii) Transcriptome analysis.
The quality of DNA reads, in fastq format, was evaluated using FastQC. Adapters were trimmed, and reads of poor quality or aligning to rRNA sequences were filtered. The cleaned reads were aligned to the Mus musculus genome (mm10) using STAR (43). Read counts for each gene were calculated using htseq-count (44) in conjunction with a gene annotation file for mm10 obtained from the University of California—Santa Cruz (UCSC) (http://genome.ucsc.edu). Differential expression was determined using DESeq2 (45). The cutoff for determining significantly differentially expressed genes was an FDR-adjusted P value of less than 0.05.
Enriched ontology cluster analyses.
Gene lists (GenBank accession numbers) from corresponding RNA-seq results and protein lists identified from corresponding mass spectrometry were converted to gene lists and submitted to the Metascape database (http://metascape.org) (46, 47). Metascape first identified all statistically enriched terms, and accumulative hypergeometric P values and enrichment factors were calculated and used for filtering. The remaining significant terms were then hierarchically clustered into a tree based on kappa-statistical similarities among their gene memberships, and a kappa score of 0.3 was applied as the threshold to cast the tree into term clusters.
Antiviral assays.
The antiviral effects of mouse IFN-α on Slfn2 WT and KO MEFs were determined by challenging cells with EMCV, and the extent of virus-induced cytopathic effect (CPE) was determined, as previously described (48).
Luciferase assays.
Slfn2 WT and KO MEFs carrying the pGF1-ISRE-Luc reporter vector were generated as previously described (36). Slfn2 WT and KO MEFs stably transfected with the ISRE luciferase reporter were seeded (500 cells per well) in triplicate in a 96-well plate. The next day, cells were either left untreated or treated with 5 × 103 IU/ml of mouse IFN-β for 6 h and then lysed using 100 μl of cell culture lysis reagent (Promega). Luciferase activity was then measured using a Cytation 3 plate reader (BioTek Instruments Inc.) after addition of 100 μl of luciferase assay substrate (Promega), according to the manufacturer's protocol.
Overexpression system and siRNA-mediated knockdown.
The Slfn2 (NM_011408) cDNA clone was purchased from Origene. The Slfn2 open reading frame (ORF) was cloned directly into an SgfI-MluI-digested pCMV6-AC-TGFP plasmid (Origene). pCMV6-AC-TGFP was used as a control vector. NIH 3T3 cells or MEFs (6 × 106 cells) were seeded in 15-cm plates. Next, the cells were transfected with either pCMV6-TGFP-Slfn2 or pCMV6-TGFP, using Lipofectamine 3000 reagent (Thermo Fisher), according to the manufacturer's protocol. Twenty-four hours later, the cells were treated with mouse IFN-β at 104 IU/ml for the indicated time, followed by coimmunoprecipitation and immunoblot analyses. Slfn2 WT and KO MEFs (6 × 106) were seeded in duplicate in 15-cm plates. The next day, the MEFs were transfected with either pcDNA3.0-IκBαM or pcDNA3.0 as a control, using Turbofect (Thermo Fisher) according to the manufacturer's protocol. Twenty-four hours later, the cells were treated with 5 × 103 IU/ml of mouse IFN-β for the indicated time, followed by RNA isolation and qRT-PCR analysis.
Immunoprecipitations and immunoblotting.
MEFs or NIH 3T3 cells were starved overnight in DMEM containing 0.25% FBS and then treated with 104 IU/ml of mouse IFN-β in DMEM containing 0.25% FBS for the indicated times. After treatment, the cells were lysed in Nonidet P-40 buffer (40 mM HEPES, pH 7.5, containing 0.1% Nonidet P-40, 120 mM sodium chloride, 1 mM EDTA, 10 mM sodium pyrophosphate, 50 mM NaF, and 10 mM B-glycerophosphate) to which protease and phosphatase inhibitors were added (Millipore). For immunoblot analyses, lysates were resolved by SDS-PAGE (Bio-Rad), transferred to Immobilon-P polyvinylidene difluoride (PVDF) membranes (Millipore), and then probed with primary and secondary horseradish peroxidase (HRP)-conjugated antibodies. After reaction with Amersham ECL detection reagent (GE Healthcare), the blots were visualized for the indicated proteins using a ChemiDoc MP imager (Bio-Rad) or autoradiography. For coimmunoprecipitation analyses, total cell lysates were incubated overnight at 4°C with anti-TGFP antibody or anti-PPP6R1, as indicated, followed by incubation at 4°C for 1 h with protein G-Sepharose 4 Fast Flow beads (GE Healthcare). The same procedure was followed using anti-rabbit IgG antibody (BD Pharmingen; number 550875) as a control. The beads were then washed three times with Nonidet P-40 buffer. Protein complexes were eluted from the beads, resolved by SDS-PAGE, and processed for immunoblot analysis.
Real-time qRT-PCR.
Slfn2 WT and KO MEFs (1.5 × 106) were seeded in 10-cm plates. One day later, MEFs were either left untreated or treated with 5 × 103 IU/ml of mouse IFN-β for 6 h. Total RNA was isolated using an RNeasy minikit (Qiagen) and retrotranscribed using an Omniscript reverse transcription kit (Qiagen). TaqMan qRT-PCR was performed using commercially available primers (Thermo Fisher): Gapdh, Mm99999915_g1; Slfn2, Mm 00488307_m1; Cxcl10, Mm00445235_m1; Ifit1, Mm00515153_m1; Ifit3, Mm01704846_s1; Irf7, Mm00516793_g1; Irf9, Mm00492679_m1; Oasl2, Mm00496187_m1; Isg15, Mm01705338_s1; Mx1, Mm01218004_m1; and Ppp6r1, Mm00556520_m1.
Mobility shift assays.
Slfn2 WT and KO MEFs were either left untreated or treated with 104 IU/ml of murine IFN-α for 15 min. The cells were washed twice with ice-cold phosphate-buffered saline and resuspended in hypotonic buffer (12 mM HEPES, pH 7.9, 4 mM Tris, pH 7.9, 0.6 mM EDTA, 10 mM KCl, 5 mM MgCl2, 1 mM Na3VO4, 1 mM Na4P2O7, 1 mM NaF, 0.6 mM dithiothreitol [DTT], 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 10 mg/ml aprotinin, 2 mg/ml leupeptin, 2 mg/ml pepstatin A). Following a 10-min incubation, the cells were supplemented with 0.2% Triton X-100 and disrupted by repeated passage through a 25-gauge needle; then, the suspensions were centrifuged at 12,000 × g for 40 s. The pellets were incubated in high-salt buffer for 30 min and clarified by centrifugation at 12,000 × g for 20 min. Hypotonic buffer contained 12 mM HEPES, pH 7.9, 4 mM Tris, pH 7.9, 0.6 mM EDTA, 300 mM KCl, 5 mM MgCl2, 1 mM Na3VO4, 1 mM Na4P2O7, 1 mM NaF, 0.6 mM DTT, 0.5 mM PMSF, 10 mg/ml aprotinin, 2 mg/ml leupeptin, 2 mg/ml pepstatin A, and 20% glycerol. The protein concentration was determined using the Bio-Rad protein assay, and 10 μg of extracts was analyzed by EMSA. The extracts were incubated with 32P-end-labeled double-stranded oligonucleotides representing the SIE of the human c-fos promoter (5′-AGCTTCATTTCCCGTAAATCCCT-3′) and the ISRE of the human 2-5A synthetase promoter (5′-AGCTTCCCTTCTGAGGAAACGAAACCA-3′) in the presence of 2 μg of herring sperm DNA in EMSA buffer for 20 min. Protein-probe complexes were resolved on 4.5% polyacrylamide gels, dried, and exposed to X-ray film. The EMSA buffer contained 13 mM HEPES, pH 7.9, 65 mM NaCl, 0.15 mM EDTA, 0.06 mM EGTA, 1 mM DTT, and 15% Ficoll.
Statistical analysis.
Data were analyzed using Prism v6.0 (GraphPad Software, Inc.). Student's t test or a ratio-paired t test was used for comparison of one observation between two groups, and one-way analysis of variance (ANOVA) was used to compare more than two groups, followed by Tukey's multiple-comparison test, as indicated in the figure legends. Differences were considered statistically significant when P values were less than 0.05.
Accession number(s).
The raw RNA-seq data have been uploaded to the NIH GEO Web portal under accession number GSE109991.
Supplementary Material
ACKNOWLEDGMENTS
We thank Francesca Zazzeroni (Department of Biotechnological and Applied Clinical Sciences, University of L'Aquila, L'Aquila, Italy) for the pcDNA3.0-IκBαM plasmid. RNA-sequencing analyses were performed by the NUSeq Core of Northwestern University.
This work was supported in part by NIH grants CA161196, CA77816, and CA155566 and by grant I01CX000916 from the Department of Veterans Affairs. A.D.A. was supported in part by NIH/NCI grant T32 CA070085, and D.S. was supported in part by NIH/NCI grant T32 CA080621. E.N.F. is a Tier 1 Canada Research Chair in Women's Health and Immunology. Proteomics services were performed by the Northwestern Proteomics Core Facility, generously supported by NCI CCSG P30 CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center, and the National Resource for Translational and Developmental Proteomics, supported by P41 GM108569. Flow cytometry cell sorting was performed on a BD FACS Aria SORP system, purchased through the support of NIH 1S10OD011996-01, and this work was supported by the Northwestern University Flow Cytometry Core Facility, supported by a Cancer Center Support Grant (NCI CA060553).
We declare that we have no competing financial interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00053-18.
REFERENCES
- 1.Gonzalez-Navajas JM, Lee J, David M, Raz E. 2012. Immunomodulatory functions of type I interferons. Nat Rev Immunol 12:125–135. doi: 10.1038/nri3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Platanias LC. 2005. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol 5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 3.Wang W, Xu L, Su J, Peppelenbosch MP, Pan Q. 2017. Transcriptional regulation of antiviral interferon-stimulated genes. Trends Microbiol 25:573–584. doi: 10.1016/j.tim.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fish EN, Platanias LC. 2014. Interferon receptor signaling in malignancy: a network of cellular pathways defining biological outcomes. Mol Cancer Res 12:1691–1703. doi: 10.1158/1541-7786.MCR-14-0450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stark GR, Cheon H, Wang Y. 2018. Responses to cytokines and interferons that depend upon JAKs and STATs. Cold Spring Harb Perspect Biol 10:a028555. doi: 10.1101/cshperspect.a028555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang CH, Murti A, Pfeffer SR, Basu L, Kim JG, Pfeffer LM. 2000. IFNalpha/beta promotes cell survival by activating NF-kappa B. Proc Natl Acad Sci U S A 97:13631–13636. doi: 10.1073/pnas.250477397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang CH, Murti A, Pfeffer SR, Kim JG, Donner DB, Pfeffer LM. 2001. Interferon alpha /beta promotes cell survival by activating nuclear factor kappa B through phosphatidylinositol 3-kinase and Akt. J Biol Chem 276:13756–13761. doi: 10.1074/jbc.M011006200. [DOI] [PubMed] [Google Scholar]
- 8.Pfeffer LM, Kim JG, Pfeffer SR, Carrigan DJ, Baker DP, Wei L, Homayouni R. 2004. Role of nuclear factor-kappaB in the antiviral action of interferon and interferon-regulated gene expression. J Biol Chem 279:31304–31311. doi: 10.1074/jbc.M308975200. [DOI] [PubMed] [Google Scholar]
- 9.Pfeffer LM. 2011. The role of nuclear factor kappaB in the interferon response. J Interferon Cytokine Res 31:553–559. doi: 10.1089/jir.2011.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayden MS, Ghosh S. 2012. NF-kappaB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev 26:203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiDonato JA, Mercurio F, Karin M. 2012. NF-kappaB and the link between inflammation and cancer. Immunol Rev 246:379–400. doi: 10.1111/j.1600-065X.2012.01099.x. [DOI] [PubMed] [Google Scholar]
- 12.Schwarz DA, Katayama CD, Hedrick SM. 1998. Schlafen, a new family of growth regulatory genes that affect thymocyte development. Immunity 9:657–668. doi: 10.1016/S1074-7613(00)80663-9. [DOI] [PubMed] [Google Scholar]
- 13.Mavrommatis E, Fish EN, Platanias LC. 2013. The Schlafen family of proteins and their regulation by interferons. J Interferon Cytokine Res 33:206–210. doi: 10.1089/jir.2012.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel VB, Yu Y, Das JK, Patel BB, Majumdar AP. 2009. Schlafen-3: a novel regulator of intestinal differentiation. Biochem Biophys Res Commun 388:752–756. doi: 10.1016/j.bbrc.2009.08.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katsoulidis E, Carayol N, Woodard J, Konieczna I, Majchrzak-Kita B, Jordan A, Sassano A, Eklund EA, Fish EN, Platanias LC. 2009. Role of Schlafen 2 (SLFN2) in the generation of interferon alpha-induced growth inhibitory responses. J Biol Chem 284:25051–25064. doi: 10.1074/jbc.M109.030445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwon MJ, Yao Y, Walter MJ, Holtzman MJ, Chang CH. 2007. Role of PKCdelta in IFN-gamma-inducible CIITA gene expression. Mol Immunol 44:2841–2849. doi: 10.1016/j.molimm.2007.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deb DK, Sassano A, Lekmine F, Majchrzak B, Verma A, Kambhampati S, Uddin S, Rahman A, Fish EN, Platanias LC. 2003. Activation of protein kinase C delta by IFN-gamma. J Immunol 171:267–273. doi: 10.4049/jimmunol.171.1.267. [DOI] [PubMed] [Google Scholar]
- 18.Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H, Chapman R, Hertzog PJ. 2013. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res 41:D1040–D1046. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang W, Xu L, Brandsma JH, Wang Y, Hakim MS, Zhou X, Yin Y, Fuhler GM, van der Laan LJ, van der Woude CJ, Sprengers D, Metselaar HJ, Smits R, Poot RA, Peppelenbosch MP, Pan Q. 2016. Convergent transcription of interferon-stimulated genes by TNF-alpha and IFN-alpha augments antiviral activity against HCV and HEV. Sci Rep 6:25482. doi: 10.1038/srep25482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stefansson B, Brautigan DL. 2006. Protein phosphatase 6 subunit with conserved Sit4-associated protein domain targets IkappaBepsilon. J Biol Chem 281:22624–22634. doi: 10.1074/jbc.M601772200. [DOI] [PubMed] [Google Scholar]
- 21.Stefansson B, Ohama T, Daugherty AE, Brautigan DL. 2008. Protein phosphatase 6 regulatory subunits composed of ankyrin repeat domains. Biochemistry 47:1442–1451. doi: 10.1021/bi7022877. [DOI] [PubMed] [Google Scholar]
- 22.Liu F, Zhou P, Wang Q, Zhang M, Li D. 2018. The Schlafen family: complex roles in different cell types and virus replication. Cell Biol Int 42:2–8. doi: 10.1002/cbin.10778. [DOI] [PubMed] [Google Scholar]
- 23.Omar I, Rom O, Aviram M, Cohen-Daniel L, Gebre AK, Parks JS, Berger M. 2017. Slfn2 mutation-induced loss of T-cell quiescence leads to elevated de novo sterol synthesis. Immunology 152:484–493. doi: 10.1111/imm.12785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berger M, Krebs P, Crozat K, Li X, Croker BA, Siggs OM, Popkin D, Du X, Lawson BR, Theofilopoulos AN, Xia Y, Khovananth K, Moresco EM, Satoh T, Takeuchi O, Akira S, Beutler B. 2010. An Slfn2 mutation causes lymphoid and myeloid immunodeficiency due to loss of immune cell quiescence. Nat Immunol 11:335–343. doi: 10.1038/ni.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kreit M, Paul S, Knoops L, De Cock A, Sorgeloos F, Michiels T. 2014. Inefficient type I interferon-mediated antiviral protection of primary mouse neurons is associated with the lack of apolipoprotein l9 expression. J Virol 88:3874–3884. doi: 10.1128/JVI.03018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee NK, Choi HK, Yoo HJ, Shin J, Lee SY. 2008. RANKL-induced Schlafen2 is a positive regulator of osteoclastogenesis. Cell Signal 20:2302–2308. doi: 10.1016/j.cellsig.2008.08.019. [DOI] [PubMed] [Google Scholar]
- 27.Mavrommatis E, Arslan AD, Sassano A, Hua Y, Kroczynska B, Platanias LC. 2013. Expression and regulatory effects of murine Schlafen (Slfn) genes in malignant melanoma and renal cell carcinoma. J Biol Chem 288:33006–33015. doi: 10.1074/jbc.M113.460741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darnell JE Jr, Kerr IM, Stark GR. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 29.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. 1998. How cells respond to interferons. Annu Rev Biochem 67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 30.Rubio D, Xu RH, Remakus S, Krouse TE, Truckenmiller ME, Thapa RJ, Balachandran S, Alcami A, Norbury CC, Sigal LJ. 2013. Crosstalk between the type 1 interferon and nuclear factor kappa B pathways confers resistance to a lethal virus infection. Cell Host Microbe 13:701–710. doi: 10.1016/j.chom.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trinchieri G. 2010. Type I interferon: friend or foe? J Exp Med 207:2053–2063. doi: 10.1084/jem.20101664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodero MP, Crow YJ. 2016. Type I interferon-mediated monogenic autoinflammation: the type I interferonopathies, a conceptual overview. J Exp Med 213:2527–2538. doi: 10.1084/jem.20161596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu Q, Katlinskaya YV, Carbone CJ, Zhao B, Katlinski KV, Zheng H, Guha M, Li N, Chen Q, Yang T, Lengner CJ, Greenberg RA, Johnson FB, Fuchs SY. 2015. DNA-damage-induced type I interferon promotes senescence and inhibits stem cell function. Cell Rep 11:785–797. doi: 10.1016/j.celrep.2015.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simeonidis S, Liang S, Chen G, Thanos D. 1997. Cloning and functional characterization of mouse IkappaBepsilon. Proc Natl Acad Sci U S A 94:14372–14377. doi: 10.1073/pnas.94.26.14372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whiteside ST, Israel A. 1997. I kappa B proteins: structure, function and regulation. Semin Cancer Biol 8:75–82. doi: 10.1006/scbi.1997.0058. [DOI] [PubMed] [Google Scholar]
- 36.Arslan AD, Sassano A, Saleiro D, Lisowski P, Kosciuczuk EM, Fischietti M, Eckerdt F, Fish EN, Platanias LC. 2017. Human SLFN5 is a transcriptional co-repressor of STAT1-mediated interferon responses and promotes the malignant phenotype in glioblastoma. Oncogene 36:6006–6019. doi: 10.1038/onc.2017.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfeffer LM, Mullersman JE, Pfeffer SR, Murti A, Shi W, Yang CH. 1997. STAT3 as an adapter to couple phosphatidylinositol 3-kinase to the IFNAR1 chain of the type I interferon receptor. Science 276:1418–1420. doi: 10.1126/science.276.5317.1418. [DOI] [PubMed] [Google Scholar]
- 38.Yang CH, Murti A, Pfeffer LM. 1998. STAT3 complements defects in an interferon-resistant cell line: evidence for an essential role for STAT3 in interferon signaling and biological activities. Proc Natl Acad Sci U S A 95:5568–5572. doi: 10.1073/pnas.95.10.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarkar FH, Li Y, Wang Z, Kong D. 2008. NF-kappaB signaling pathway and its therapeutic implications in human diseases. Int Rev Immunol 27:293–319. doi: 10.1080/08830180802276179. [DOI] [PubMed] [Google Scholar]
- 40.Saleiro D, Kosciuczuk EM, Platanias LC. 2016. Beyond autophagy: new roles for ULK1 in immune signaling and interferon responses. Cytokine Growth Factor Rev 29:17–22. doi: 10.1016/j.cytogfr.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Platanias LC, Fish EN. 1999. Signaling pathways activated by interferons. Exp Hematol 27:1583–1592. doi: 10.1016/S0301-472X(99)00109-5. [DOI] [PubMed] [Google Scholar]
- 42.De Smaele E, Zazzeroni F, Papa S, Nguyen DU, Jin R, Jones J, Cong R, Franzoso G. 2001. Induction of gadd45beta by NF-kappaB downregulates pro-apoptotic JNK signalling. Nature 414:308–313. doi: 10.1038/35104560. [DOI] [PubMed] [Google Scholar]
- 43.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soonthornvacharin S, Rodriguez-Frandsen A, Zhou Y, Galvez F, Huffmaster NJ, Tripathi S, Balasubramaniam VR, Inoue A, de Castro E, Moulton H, Stein DA, Sanchez-Aparicio MT, De Jesus PD, Nguyen Q, Konig R, Krogan NJ, Garcia-Sastre A, Yoh SM, Chanda SK. 2017. Systems-based analysis of RIG-I-dependent signalling identifies KHSRP as an inhibitor of RIG-I receptor activation. Nat Microbiol 2:17022. doi: 10.1038/nmicrobiol.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tripathi S, Pohl MO, Zhou Y, Rodriguez-Frandsen A, Wang G, Stein DA, Moulton HM, DeJesus P, Che J, Mulder LC, Yanguez E, Andenmatten D, Pache L, Manicassamy B, Albrecht RA, Gonzalez MG, Nguyen Q, Brass A, Elledge S, White M, Shapira S, Hacohen N, Karlas A, Meyer TF, Shales M, Gatorano A, Johnson JR, Jang G, Johnson T, Verschueren E, Sanders D, Krogan N, Shaw M, Konig R, Stertz S, Garcia-Sastre A, Chanda SK. 2015. Meta- and orthogonal integration of influenza “OMICs” data defines a role for UBR4 in virus budding. Cell Host Microbe 18:723–735. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saleiro D, Mehrotra S, Kroczynska B, Beauchamp EM, Lisowski P, Majchrzak-Kita B, Bhagat TD, Stein BL, McMahon B, Altman JK, Kosciuczuk EM, Baker DP, Jie C, Jafari N, Thompson CB, Levine RL, Fish EN, Verma AK, Platanias LC. 2015. Central role of ULK1 in type I interferon signaling. Cell Rep 11:605–617. doi: 10.1016/j.celrep.2015.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.