Abstract
Background
Shigellosis remains a serious public health problem and an important cause of morbidity and mortality worldwide. The aim of this study was to characterize fliC and the genetic relatedness of Shigella spp. isolated during a one-year period from children in a suspected outbreak in Tehran, Iran.
Methods and results
Fifty Shigella spp. were isolated from 3779 stool samples of children with diarrhea (prevalence rate: 1.32%). Among the isolates, 92% were characterized as Shigella sonnei, while 6% and 2% were identified as S. flexneri and S. boydii, respectively. S. dysenteriae was not recovered from the patients. All isolates were negative for fliC except for Shigella standard strains. The enterobacterial repetitive intergenic consensus-polymerase chain reaction (ERIC-PCR) profiles allowed differentiating the 50 isolates into 5 ERIC types, which were grouped into five clusters (ET1–ET5). Computer-assisted clustering of the strains showed a high degree of similarity among the isolates.
Conclusion
In conclusion, given the clonal correlation of the Shigella strains isolated in this study and the lack of fliC among them, we propose that probably a single or limited fliC-defected Shigella clone spread and caused the outbreak.
Keywords: Shigella spp, ERIC-PCR, fliC, Outbreak
Introduction
Shigellosis is an infectious disease and a health problem in both developed and developing countries. It is also a major cause of diarrheal diseases among children in Iran.1 Shigella spp. are highly specific human pathogens with variable epidemiological and pathological features.2 Based on their biochemical characteristics and antigenic properties, Shigella spp. are classified to four species or serogroups: Shigella dysenteriae (group A), S. flexneri (group B), S. boydii (group C), and S. sonnei (group D).
Although the genomic information of Shigella has described this species as non-flagellated organisms, controversial published data have shown cryptic flagellin genes for S. flexneri and S. sonnei strains.3, 4 In addition, a study using electron microscopy proved that prototypic strains of all four Shigella species possess flagella.5
The organism has a well-documented ability to cause sustained outbreaks from a wide variety of sources and high rates of infection in populations, making accurate typing of patient isolates mandatory for understanding the transmission dynamics and control of further spread.6
A wide variety of typing systems has been developed to examine and compare isolates of Shigella spp. for outbreak inspection and taxonomic characterization. Bacterial genomes are generally considered to be continually reorganizing with numerous families of short interspersed repetitive sequences. Enterobacterial repetitive intergenic consensus (ERIC) sequences are described as intergenic repetitive units, different from most other bacterial repeats in being distributed or present among a wider range of species. ERIC sequences are imperfect palindromes of 127 bp7 and were first described in Escherichia coli, Salmonella enterica serovar Typhimurium, other Enterobacteriaceae members, and Vibrio cholerae.
The success of ERIC-PCR as a simplified typing method for a wide and progressively expanding number of organisms makes this method suitable for hospital-based epidemiology or localized epidemiology, where a highly recreated typing method that requires minimal disease-specific reagents is an expedient characteristic.7
The aim of this study was to characterize fliC and genetic relatedness of Shigella spp. isolated during a 1-year period from children in a suspected outbreak in Tehran, Iran.
Materials and methods
Ethical approval was obtained from the Faculty of Medical Sciences, Tarbiat Modares University.
Bacterial strains
A total of 3779 stool specimens were collected from outpatient children, younger than 5 years, with diarrhea characterized by ≥3 episodes of loose or watery stools with or without blood, mucus, and stomach cramps referred to two major hospital laboratories from November 2012 to October 2013. For the preliminary isolation of Shigella spp., the specimens were cultured on selective agar plates including Salmonella-Shigella agar and XLD agar (Merck, Hamburg, Germany).8 Putative Shigella organisms were selected and preliminarily characterized by biochemical tests including TSI, SIM, ODC, LIA, Simmons Citrate and Urea agar and confirmed by an API-20E strip kit according to manufacturer instructions (API-bioMérieux, Inc., La Balme les Grottes, France).9 The phenotypic biochemical profiles of the isolates were compared with the World Health Organization (WHO) criteria for biochemical identification of Shigella spp.10 S. sonnei ATCC 9290, S. flexneri ATCC 12022, S. boydii, and S. dysenteriae control strains were kindly provided by the WHO and used as controls in each assay.
DNA preparation, molecular confirmation of Shigella spp., and serotyping
The isolates were identified by biochemical tests and confirmed by PCR for ipaH with a specific set of primers (Table 1). Additional primer pairs that amplify wbgZ and rfc specifically for S. sonnei and S. flexneri, respectively, were used for identifying the two most commonly found Shigella species. All PCR reactions were performed with DNA preparations obtained by the boiling method.
Table 1.
Oligonucleotide primers used in this study.
| Primer name (target) | Sequence (5′ to 3′) | Product size (bp) | Annealing temperature | Reference |
|---|---|---|---|---|
| Shigella (ipaH) | GTTCCTTGACCGCCTTTCCGATACCGTC GCCGGTCAGCCACCCTCTGAGAGTAC |
619 | 55 °C | 24 |
| Shigella sonnei (wbgZ) | TCTGAATATGCCCTCTACGCT GACAGAGCCCGAAGAACCG |
430 | 56 °C | 25 |
| Shigella flexneri (rfc) | TTTATG GCTTCTTTGTCG GC CTG CGTGATCCGACCATG |
537 | 56 °C | 25 |
| ERIC | ATGTAAGCTCCTGGGGATTCAC AAGTAAGTGACTGGGGTGAGCG |
– | 54 °C | 12 |
| fliC-truncate | AACTCCCTCATTGACACCTTT ATCCATTACACCTTCAGCG |
1500 | 58 °C | This study |
| fliC | GGGTGGAAACCCAATACG GCGCATCAGGCAATTTGG | 1714 | 58 °C | 26 |
Freshly cultured plates of Shigella isolates confirmed with molecular identification of ipaH were prepared for agglutination on glass slides by commercially available polyclonal antisera (Baharafshan Institute of Research & Development, Tehran, Iran) using the standard slide agglutination method.11
Investigation of fliC
Two sets of primer pairs were used for amplifying fliC with regard to the complete and truncated flagellin sequences in GenBank. Primer names, their sequences, and amplification conditions are presented in Table 1.
CLC software (http://www.clcbio.com) was used for designing specific primer pairs for truncated fliC according to the genome sequence of S. sonnei ATCC 9290 (GenBank accession number HE616528).
ERIC PCR performance and analysis
ERIC-PCR was performed on Shigella strains according to Versalovic et al. (1991). Each PCR reaction (25 μL) contained 10× buffer 2.5 μL, dNTPs (2.5 mmol/L) 2.5 μL, Taq DNA polymerase (5 U/μL) 0.5 μL, MgCl2 (25 mmol/L) 1.5 μL, primers (100 mol/L) each 1 μL, template DNA 1 μL, and addition of distilled water to 25 μL. All PCR reactions were performed in the following program: initial denaturation of 94 °C for 5 min and 35 cycles of 94 °C for 1 min, 54 °C for 1 min, and 72 °C for 5 min followed by a final extension of 72 °C for 10 min. Primer sequences are presented in Table 1.
The amplicon was electrophoresed on 2% agarose gel with DNA safe stain along with a 1-kb DNA ladder. An image was captured for further analysis with Gel compare II software version 4.0 (Applied Maths, Sint-Martens-Latem, Belgium). The clustering of the isolates was performed on the basis of Unweighted Pair Group Method with Arithmetic Mean (UPGMA) analysis and Dice similarity coefficient.
Results
Molecular confirmation of Shigella spp. isolates and detection of flagellin genes
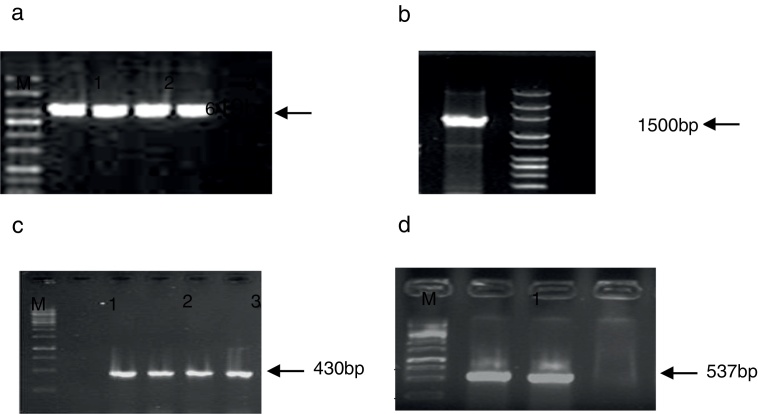
Based on the biochemical and molecular analyses, 50 Shigella spp. were isolated from 3779 stool samples of children with diarrhea (prevalence rate: 1.32%). All suspected Shigella spp. (by biochemical tests) were positive for ipaH and showed a fragment of 619 bp, which was further confirmed by sequencing (Fig. 1). Among the isolates, 46 (92%) were characterized as S. sonnei and 3 (6%) as S. flexneri by serogrouping, which was confirmed by amplifying 430-bp and 537-bp bands related to wbgz and rfc, respectively. Among the isolates, S. boydii accounted for 2%, while S. dysenteriae was not recovered from the patients. All isolates were negative for fliC except for standard Shigella strains (Fig. 1).
Fig. 1.
PCR amplification of ipaH gene. Lane 1: positive control; Lane 2–4: positive isolates; Lane 5: negative control; M: mid-range DNA ladder (100 bp to 3 kb). PCR amplification of fliC gene. Lane 1: Shigella flexneri ATCC1202 and M: mid range DNA ladder (100 bp to 3 kb). PCR amplification of wbgZ gene in Shigella spp. L. Lane 1: negative control; Lane 2: positive control; Lane 3–5: positive isolates; M: 1 kb DNA ladder. PCR amplification of rfc gene in Shigella spp. Lane 1: positive control; Lane 2: positive isolate; Lane 3: negative control; M: 1 kb DNA ladder.
ERIC PCR analysis
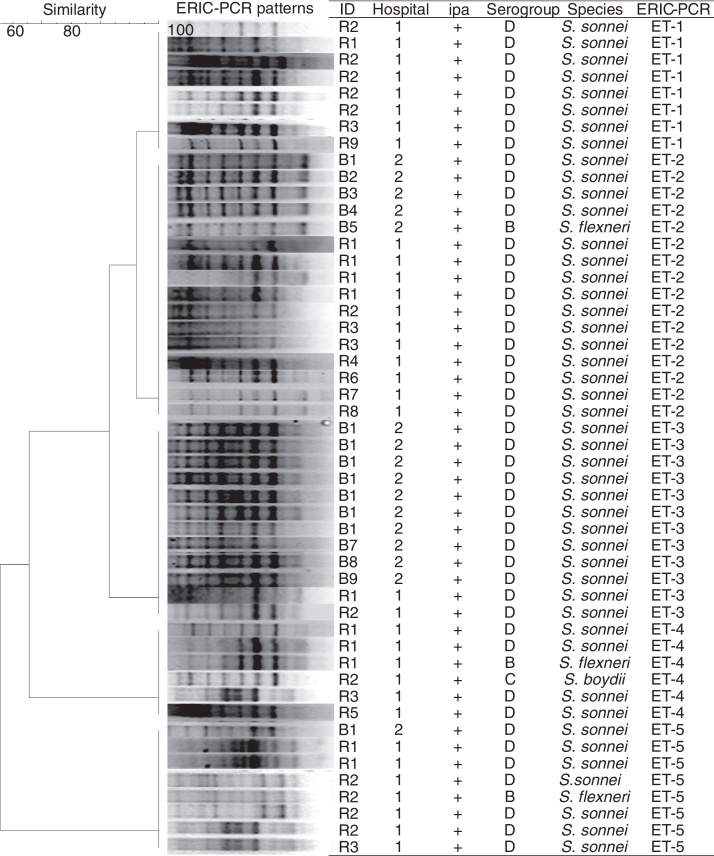
The genotyping profiles of 50 Shigella strains according to ERIC-PCR fingerprinting are shown in Fig. 2. The ERIC-PCR profiles allowed differentiating 50 isolates into 5 ERIC types, which were grouped into 5 clusters (ET1–ET5) (Fig. 2). All 50 Shigella spp. under analysis produced 7–9 amplicons ranging from 500 to 3200 bp. Computer-assisted clustering of the strains showed a high degree of similarity among the isolates, with similarity indices ranging from 92.4% for the most diverse isolates to 100% for identical isolates.
Fig. 2.
Dendrogram generated from enterobacterial repetitive intergenic consensus-polymerase chain reaction (ERIC-PCR) banding pattern of 50 Shigella spp. isolated from children with diarrhea. The similarity analysis was performed with Dice coefficient and UPGMA method.
Discussion
The number of intestinal infections caused by Shigella spp. is increasing continuously and is turning into a prominent public health concern worldwide. The prevalence rate of Shigella spp. in this study was 1.32% with S. sonnei constituting the most prevalent species (92% of all isolates). The data from other studies have shown S. flexneri as the most frequent Shigella spp. in developing countries and in Iran.13, 14, 15 In the last decades, most of incidences of shigellosis in Iran were caused by S. flexneri; however, recent studies have revealed a growing tendency of shigellosis caused by S. sonnei.16, 17 The present results upon comparison with those from the studies in developed countries where S. sonnei is the most prevalent causative agent of shigellosis reflect the industrialization of the capital city, Tehran, and the improvement of hygiene levels in Iran.17
Although the standard Shigella standard strains carried fliC gene, in this study, fliC was not detected in the clinical isolates despite several attempts using two sets of primers. The absence of fliC, even in the truncated form,18 among all isolates under study may be due to mutational or recombinational events within the coding sequence of this gene. These changes may have caused nucleotide changes in the primer-annealing sequence or deletion of the entire gene from the Shigella genome. In a study by Coimbra et al. (2001), all Shigella serotypes amplified the fliC region except for the reference strain S. boydii serotype 12 NCDC266-59, which failed to give a PCR product.18 The data from the present study along with limited observations on the existence and expression of fliC in Shigella spp. make further studies necessary to track the changes in fliC in the genome of Shigella spp.
Several methods exist for investigating the molecular diversity of bacteria in the community and hospitals. ERIC-PCR has a proven discriminatory power and is a quick and relatively easy technique, making it useful for routine epidemiological investigations. Using the ERIC-PCR technique, genetic relatedness among Shigella strains isolated from pediatric patients in Iran was investigated and the isolates were differentiated into five clusters (ET1–ET5) in this study. The isolates showed less than 6-band differences in their profile, which could be considered to be a confirmation for the incidence of a Shigella outbreak. Ranjbar et al. (2013) showed the presence of limited ERIC-PCR profiles among S. sonnei isolates recovered during 2008–2010, with most isolates showing an identical banding pattern, confirming the epidemic nature of the S. sonnei strain leading to several outbreaks annually in Iran.19 ERIC-PCR has been also used for typing the epidemic isolates of Shigella spp. and other enteric bacteria for outbreak detection in many parts of the world, including Iran.20, 21, 22, 23
Interestingly, S. flexneri isolates in this study showed an identical banding pattern with the pattern presented by S. sonnei isolates, which may suggest that inter-species differences do not necessarily affect the ERIC-PCR banding profiles of Shigella strains.
Conclusion
In conclusion, given the clonal correlation of the Shigella strains isolated in this study and the lack of fliC among them, we propose that probably a single or limited fliC-defected Shigella clone spread and caused the outbreak.
Authors’ contribution
BB designed the study and drafted the manuscript. NA participated in the molecular tests. FF helped in designing the study and participated in statistical analysis.
Conflicts of interest
Authors declare no conflicts of interest.
Acknowledgments
The authors would like to thank the Pediatric Infection Research Center, Mofid Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran.
Associate Editor: Roxane Maria Fontes Piazza
References
- 1.Eftekhari N., Bakhshi B., Pourshafie M.R. Genetic diversity of Shigella spp. and their integron content. Foodborne Pathog Dis. 2013;10(3):237–242. doi: 10.1089/fpd.2012.1250. [DOI] [PubMed] [Google Scholar]
- 2.Yang F., Yang J., Zhang X. Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic Acids Res. 2005;33(19):6445–6458. doi: 10.1093/nar/gki954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki T., Nunez G. A role for Nod-like receptors in autophagy induced by Shigella infection. Autophagy. 2008;4(1):73–75. doi: 10.4161/auto.5101. [DOI] [PubMed] [Google Scholar]
- 4.Tominaga A., Mahmoud M.A.H., Mukaihara T., Enomoto M. Molecular characterization of intact, but cryptic, flagellin genes in the genus Shigella. Mol Microbiol. 1994;12(2):277–285. doi: 10.1111/j.1365-2958.1994.tb01016.x. [DOI] [PubMed] [Google Scholar]
- 5.Girón J.A. Expression of flagella and motility by Shigella. Mol Microbiol. 1995;18(1):63–75. doi: 10.1111/j.1365-2958.1995.mmi_18010063.x. [DOI] [PubMed] [Google Scholar]
- 6.Liu P.Y., Lau Y.J., Hu B.S. Analysis of clonal relationships among isolates of Shigella sonnei by different molecular typing methods. J Clin Microbiol. 1995;33(7):1779–1783. doi: 10.1128/jcm.33.7.1779-1783.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson L.A., Sharp P.M. Enterobacterial repetitive intergenic consensus (ERIC) sequences in Escherichia coli: evolution and implications for ERIC-PCR. Mol Biol Evolut. 2006;23(6):1156–1168. doi: 10.1093/molbev/msj125. [DOI] [PubMed] [Google Scholar]
- 8.Ewing W.H. Elsevier Science Publishing Co. Inc.; 1986. Edwards and Ewing's Identification of Enterobacteriaceae. [Google Scholar]
- 9.Aldridge K.E., Gardner B.B., Clark S.J., Matsen J.M. Comparison of Micro-ID, API 20E, and conventional media systems in identification of Enterobacteriaceae. J Clin Microbiol. 1978;7(6):507–513. doi: 10.1128/jcm.7.6.507-513.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koneman E.W., Allen S.D., Janda W., Schreckenberger P., Winn W. JC Lippincott; 1988. Diagnostic Microbiology. [Google Scholar]
- 11.Guardabassi L., Dijkshoorn L., Collard J.-M., Olsen J., Dalsgaard A. Distribution and in-vitro transfer of tetracycline resistance determinants in clinical and aquatic Acinetobacter strains. J Med Microbiol. 2000;49(10):929–936. doi: 10.1099/0022-1317-49-10-929. [DOI] [PubMed] [Google Scholar]
- 12.Versalovic J., Koeuth T., Lupski R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial enomes. Nucleic Acids Res. 1991;19(24):6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MoezArdalan K., Zali M.R., Dallal M.M., Hemami M.R., Salmanzadeh-Ahrabi S. Prevalence and pattern of antimicrobial resistance of Shigella species among patients with acute diarrhoea in Karaj, Tehran, Iran. J Health Popul Nutr. 2003;21(2):96–102. [PubMed] [Google Scholar]
- 14.Nikkah J., Mehr-Movahead A. Antibiotic resistance among Shigella species isolated in Tehran, Iran. Ann Trop Med Parasitol. 1988;82(5):481–483. doi: 10.1080/00034983.1988.11812280. [DOI] [PubMed] [Google Scholar]
- 15.A foodborne outbreak of Shigella sonnei infection in Europe. Commun Dis Rep CDR Wkl. 1994;4(25):115. [PubMed] [Google Scholar]
- 16.Farshad S., Sheikhi R., Japoni A., Basiri E., Alborzi A. Characterization of Shigella strains in Iran by plasmid profile analysis and PCR amplification of ipa genes. J Clin Microbiol. 2006;44(8):2879–2883. doi: 10.1128/JCM.00310-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ranjbar R., Dallal M.M.S., Talebi M., Pourshafie M.R. Increased isolation and characterization of Shigella sonnei obtained from hospitalized children in Tehran, Iran. J Health Popul Nutr. 2008;26(4):426. doi: 10.3329/jhpn.v26i4.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coimbra R.S., Lefevre M., Grimont F., Grimont P.A. Clonal relationships among Shigella serotypes suggested by cryptic flagellin gene polymorphism. J Clin Microbiol. 2001;39(2):670–674. doi: 10.1128/JCM.39.2.670-674.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ranjbar R., Mirsaeed Ghazi F. Antibiotic sensitivity patterns and molecular typing of Shigella sonnei strains using ERIC-PCR. Iran J Public Health. 2013;42(10):1151–1157. [PMC free article] [PubMed] [Google Scholar]
- 20.Najibi S., Bakhshi B., Fallahzad S. Distribution of class 1 integrons among enteropathogenic Escherichia coli. Can J Microbiol. 2012;58(5):637–643. doi: 10.1139/w2012-035. [DOI] [PubMed] [Google Scholar]
- 21.Aliabad N.H., Bakhshi B., Pourshafie M.R., Sharifnia A., Ghorbani M. Molecular diversity of CTX prophage in Vibrio cholerae. Lett Appl Microbiol. 2012;55(1):27–32. doi: 10.1111/j.1472-765X.2012.03253.x. [DOI] [PubMed] [Google Scholar]
- 22.Saxena M.K., Singh V.P., Lakhcharua B.D., Taj G., Sharma B. Strain differentiation of Indian isolates of Salmonella by ERIC-PCR. Res Vet Sci. 2002;73(3):313–314. doi: 10.1016/s0034-5288(02)00030-9. [DOI] [PubMed] [Google Scholar]
- 23.Waturangi D.E., Joanito I., Yogi Y., Thomas S. Use of REP- and ERIC-PCR to reveal genetic heterogeneity of Vibrio cholerae from edible ice in Jakarta, Indonesia. Gut Pathog. 2012;4(1):2. doi: 10.1186/1757-4749-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sethabutr O., Venkatesan M., Murphy G.S., Eampokalap B., Hoge C.W., Echeverria P. Detection of Shigellae and enteroinvasive Escherichia coli by amplification of the invasion plasmid antigen H DNA sequence in patients with dysentery. J Infect Dis. 1993;167(2):458–461. doi: 10.1093/infdis/167.2.458. [DOI] [PubMed] [Google Scholar]
- 25.Ojha S.C., Yean Yean C., Ismail A., Banga Singh K.-K. A pentaplex PCR assay for the detection and differentiation of Shigella species. BioMed Res Int. 2013;2013 doi: 10.1155/2013/412370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L., Rothemund D., Curd H., Reeves P.R. Sequence diversity of the Escherichia coli H7 fliC genes: implication for a DNA-based typing scheme for E. coli O157:H7. J Clin Microbiol. 2000;38(5):1786–1790. doi: 10.1128/jcm.38.5.1786-1790.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]