Abstract
Ebolaviruses, marburgviruses, and henipaviruses are zoonotic pathogens belonging to the Filoviridae and Paramyxoviridae families. They exemplify viruses that continue to spill over into the human population, causing outbreaks characterized by high mortality and significant clinical sequelae in survivors of infection. There are currently no approved small molecule therapeutics for use in humans against these viruses. In this study, we evaluated the antiviral activity of the nucleoside analog 4′-azidocytidine (4′N3-C, R1479) and its 2′-monofluoro- and 2′-difluoro-modified analogs (2′F-4′N3-C and 2′diF-4′N3-C) against representative paramyxoviruses (Nipah virus, Hendra virus, measles virus, and human parainfluenza virus 3) and filoviruses (Ebola virus, Sudan virus, and Ravn virus). We observed enhanced antiviral activity against paramyxoviruses with both 2′diF-4′N3-C and 2′F-4′N3-C compared to R1479. On the other hand, while R1479 and 2′diF-4′N3-C inhibited filoviruses similarly to paramyxoviruses, we observed 10-fold lower filovirus inhibition by 2′F-4′N3-C. To our knowledge, this is the first study to compare the susceptibility of paramyxoviruses and filoviruses to R1479 and its 2′-fluoro-modified analogs. The activity of these compounds against negative-strand RNA viruses endorses the development of 4′-modified nucleoside analogs as broad-spectrum therapeutics against zoonotic viruses of public health importance.
Keywords: Nucleoside analog, Filovirus, Ebola virus, Paramyxovirus, Henipavirus, Nipah virus, R1479, Antiviral
Highlights
-
•
2′-modified analogs of R1479 (2′F-4′N3-C and 2′diF-4′N3-C) are more potent than R1479 against paramyxoviruses.
-
•
R1479 and 2′diF-4′N3-C inhibited filoviruses similarly to paramyxoviruses, but 2′F-4′N3-C was 10-fold less potent.
-
•
R1479, 2′F-4′N3-C, and 2′diF-4′N3-C reduced henipavirus and filovirus infectious virus yields by 101.5–105.5-fold.
1. Introduction
In the decades prior to 2014, human outbreaks of emerging zoonotic viruses causing fatal hemorrhagic fever, such as Ebola virus (EBOV), or encephalitis, such as Nipah virus (NiV), have been relatively sporadic and small in terms of case numbers, ranging from isolated single cases to several hundred cases (CDC, 2017, Luby and Gurley, 2012). The historically largest EBOV outbreak of 2013–2016 highlighted the relative paucity of efficacious therapeutics and vaccines available to deploy against such pathogens, despite extensive pre-clinical studies demonstrating efficacy of therapeutic antibodies and vaccines for these biosafety level-4 pathogens (Broder et al., 2016, Geisbert et al., 2014, Mendoza et al., 2017, Mire et al., 2016, Qiu et al., 2014). To identify therapeutics that could be repurposed against EBOV, in vitro screens of libraries of FDA-approved compounds have been conducted, screening, among others, antiviral nucleoside and nucleotide analogs (Madrid et al., 2015, Veljkovic et al., 2015, Welch et al., 2016). Over the last 30 years, the development of antiviral nucleoside and nucleotide analogs was primarily directed towards combating viruses responsible for chronic infections such as human immunodeficiency virus, herpes viruses, and hepatitis viruses (Ray and Hitchcock, 2009). In 2006, Klumpp and colleagues first demonstrated the in vitro activity of 4′-azidocytidine (4′N3-C, R1479) against hepatitis C virus (HCV), a single-stranded, positive-sense RNA virus (Klumpp et al., 2006). The development of R1479 and its prodrug balapiravir was halted following findings of toxicity and low efficacy of these compounds in clinical trials for treating HCV and Dengue virus (Nelson et al., 2012, Nguyen et al., 2013). Despite this, the in vitro activity of R1479 against diverse flaviviruses of public health importance, such as Dengue virus and tick-borne encephalitis virus, suggested that it may be a template for developing modified analogs with antiviral activity (Chen et al., 2014, Eyer et al., 2016). Interestingly, R1479 and other 4′-modified analogs have recently been shown to also inhibit respiratory syncytial virus (RSV), a single-stranded, negative-sense RNA virus (Clarke et al., 2015, Deval et al., 2015, Wang et al., 2015). Following those studies, we described potent antiviral activity of R1479 against representative members of the Paramyxoviridae family, including the henipaviruses, NiV and Hendra virus (HeV) (Hotard et al., 2017). Given the in vitro antiviral properties of R1479 and its 2′-mono and 2′-difluoro analogs (2′F-4′N3-C and 2′diF-4′N3-C, respectively) against RSV (Deval et al., 2015), and the highly conserved nucleotide binding domains shared across Pneumoviridae, Paramyxoviridae, and Filoviridae families (Lo et al., 2017), we evaluated and compared the antiviral potencies of these cytidine analogs against representative paramyxoviruses and filoviruses including the 2014 Makona variant of EBOV (Albarino et al., 2015). Our study documents the susceptibility of paramyxoviruses and filoviruses to R1479 and its 2′-fluoro-modified analogs, and reinforces the prospect of developing 4′-modified nucleoside analogs as potential broad-spectrum therapeutics against RNA viruses of public health importance.
2. Materials and methods
2.1. Biosafety
All work with infectious virus was performed in Class 2 Biosafety cabinets, and all work utilizing live Nipah virus (NiV), Hendra virus (HeV), Ebola virus (EBOV), Sudan virus (SUDV), Ravn virus (RAVV), Marburg virus (MARV), and Rift Valley Fever virus (RVFV) was conducted in a BSL-4 laboratory at the Centers for Disease Control and Prevention (CDC; Atlanta, GA).
2.2. Cells, viruses, and compounds
HeLa, SK-N-MC, and NCI-H358 cells were purchased from the American Type Tissue Culture Collection (ATCC, Manassas, VA, USA). HeLa and SK-N-MC cells were propagated in Dulbecco's modified Eagle medium (DMEM; Life Technologies, Carlsbad, CA, USA) supplemented with 10% (vol/vol) fetal calf serum (FCS; Hyclone; Thermo Scientific, Waltham, MA, USA) and penicillin-streptomycin (Life Technologies). NCI-H358 cells were propagated in Roswell Park Memorial Institute medium (RPMI 1640), supplemented with 10% FCS. Huh7 cells were obtained from Apath, LLC (Brooklyn, NY, USA), and propagated in DMEM supplemented with 10% FCS and 1× nonessential amino acids (Life Technologies). Normal human small airway epithelial cells (SAECs) were purchased from ATCC and propagated in Airway Epithelial Cell Basal medium supplemented with the Bronchial Epithelial Cell Growth Kit (ATCC).
NiV (Malaysian genotype), recombinant NiV Malaysian genotype expressing ZsGreen1 fluorescent protein (NiV-GFP2AM) (Lo et al., 2014), HeV, recombinant Measles virus (MV) (Edmonston-Zagreb strain) expressing enhanced green fluorescent protein) (rMVEZEGFP(3)) (Rennick et al., 2015), EBOV (Makona variant), recombinant EBOV (Mayinga variant) (representative of Ebolavirus genus), SUDV (Gulu variant) (Sanchez and Rollin, 2005), RAVV (Ravn variant) (Johnson et al., 1996), and recombinant RVFV expressing enhanced green fluorescent protein (RVFV-GFP, ZH501) (Bird et al., 2007) were propagated in either Vero E6 (ATCC CRL-1586) or Vero (ATCC CCL-81) cells, and were quantitated by 50% tissue culture infections dose (TCID50) assay using the Reed and Muench method (Reed and Muench, 1938). Recombinant EBOV Makona variant expressing ZsGreen1 (EBOV-ZsG) (Albarino et al., 2015) and recombinant MARV (Bat371 variant) expressing ZsGreen1 (MARV-ZsG) was propagated and quantitated as above using Huh7 cells. Recombinant respiratory syncytial virus (RSV) expressing enhanced green fluorescent protein (rgRSV224) (Hallak et al., 2000) was propagated and quantitated as above using HeLa cells. Recombinant human parainfluenza virus 3 expressing enhanced green fluorescent protein (hPIV3-GFP) (JS strain) (Zhang et al., 2005) was obtained from ViraTree and was propagated and quantitated as above using Vero cells.
R1479, 2′F-4′N3-C, 2′diF-4′N3-C (Fig. 1 ) and their corresponding triphosphates were synthesized at Alios BioPharma.
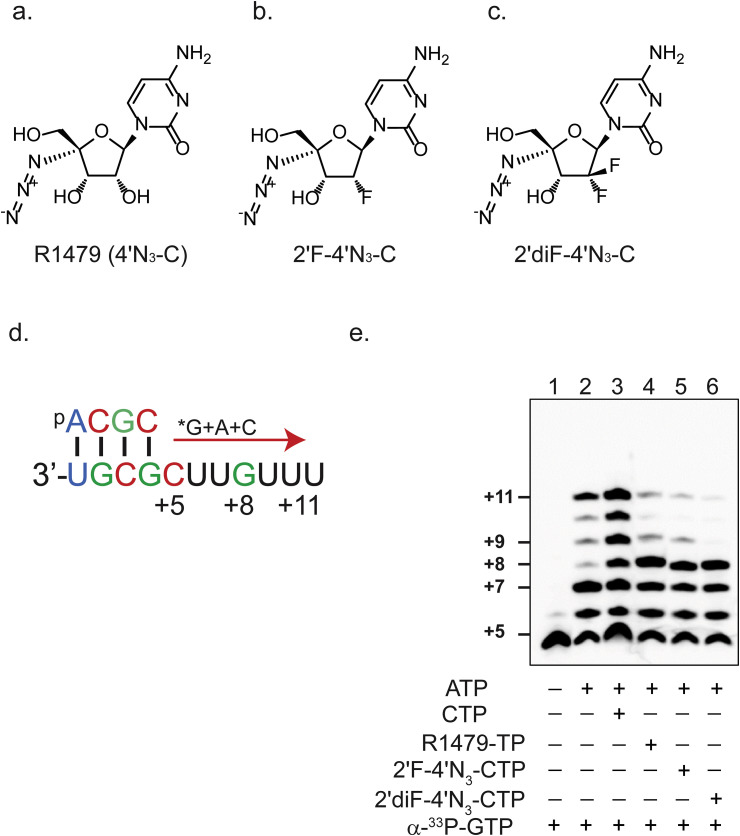
Fig. 1.
Inhibition of RSV polymerase activity by 4′-modified cytidine analogs. Chemical structures of (a) 4′-azidocytidine (R1479), (b) 2′-monofluoro-4′-azidocytidine (2′F-N3-C), and (c) 2′-difluoro-4′-azidocytidine (2′diF-4′N3-C). (d) Schematic of template-directed primer (5′-ACGC) extension assay. *G above the red arrow indicates radioactively labeled guanosine triphosphate (GTP). Similarly, A and C above the red arrow indicate triphosphorylated forms of adenosine (ATP) and cytidine (CTP), respectively. Numbers specify nucleotide positions relative to the first nucleotide of the final primer extension product. (e). Primer extension assay performed with the presence of 33P-radiolabeled GTP alone (lane 1), with ATP (lane 2), and with varying combinations of CTP (lane 3), R1479-TP (lane 4), 2′F-4′N3-CTP (lane 5), or 2′diF-4′N3-CTP (lane 6), as indicated by + and - signs.
2.3. Single nucleotide incorporation by recombinant RSV L-P
Recombinant RSV L-P was produced through the co-expression of RSV L and P proteins in a baculovirus expression system, according to previously described procedures (Noton et al., 2012). The reaction mixture was composed of 0.2 μM modified template sequence (UGCGCUUGUUU), 0.2 μM recombinant RSV L-P polymerase, 200 μM 5′-pACGC primer, buffer (20 mM Tris pH 7.5, 10 mM KCl, 6 mM MgCl2, 2 mM DTT, 0.01% Triton, 10% DMSO), and (α-33P)-GTP with a final volume of 10 μL. The reaction was started through the addition of NTPs, incubated at 30 °C for 30 min, and quenched through the addition of gel loading buffer (Ambion). Samples were run for 1.5 h at 80 W in a 22.5% polyacrylamide urea sequencing gel. The gel was then exposed to a phosphor-screen, and scanned.
2.4. Measurement of nucleoside triphosphate (NTP) formation
R1479 triphosphate (TP) formation was measured in HeLa, Huh7, SK-N-MC, and NCI-H358 cells. 2′F-4′N3-CTP, and 2′diF-4′N3-CTP formation were measured in HeLa and NCI-H358 only. The cells were maintained and cultured at Alios BioPharma. The cells were seeded in six-well plates at 1.5 × 106 cells/well with their corresponding media and incubated overnight in a cell culture incubator at 37 °C and 5% CO2 before use. In the experiment, 50 μM of R1479, 2′F-4′N3-C, or 2′diF-4′N3-C was added to each well, incubated for 24 h at 37 °C and 5% CO2. At the end of the incubation, the medium was removed and cells were washed twice with cold 0.9% sodium chloride in water. The cells were lysed with methanol/water (70%/30%, v/v), and the extracted supernatant was dried and reconstituted in 1 mM ammonium phosphate before LC/MS/MS analysis to determine the corresponding R1479-TP, 2′F-4′N3-CTP, and 2′diF-4′N3-CTP levels (Chen et al., 2009). The concentrations of the R1479-TP, 2′F-4′N3-CTP, and 2′diF-4′N3-CTP were normalized by the number of cells and reported as picomoles per million cells.
2.5. Recombinant reporter virus assays
All viruses expressing fluorescent (GFP, ZsGreen) proteins were assayed for fluorescence by using an H1 Synergy plate reader (Biotek). NCI-H358 cells or SAECs were seeded at 2 × 104 cells per well in black opaque 96-well plates (Corning 3619, Corning, NY) or Perkin-Elmer CellCarrier Ultra plates (Waltham, MA) and compounds were added to the assay plates for 1 h. Assay plates were transferred to the BSL-4 suite (where appropriate), and infected with 0.25–0.5 TCID50 per cell of the respective virus, and were read between 48 and 168 h post-infection (hpi) depending on the virus used. Fluorescence signal from DMSO-treated infected cells were set as 100% GFP. 50% effective concentrations (EC50) were calculated using four-parameter variable slope non-linear regression fitting of mean values of assays performed in quadruplicate (Graphpad Prism 6, La Jolla, CA).
2.6. Cytopathic effect (CPE) inhibition and cell viability assays
NCI-H358 cells or SAECs were seeded at 2 × 104 cells per well in white opaque 96-well plates, and compounds were added to the assay plates. Assay plates were transferred to the BSL-4 suite and infected with 0.25–0.5 TCID50 per cell, and were analyzed with CellTiter-Glo 2.0 (Promega, Madison, WI) between 72 and 96 h post-infection (pi) in a HD1 Synergy plate reader. Values were normalized to uninfected cell controls according to % viability as follows: % viability = [(specific value-reference value)/(DMSO control value – reference value)] × 100. Reference values were derived from control wells without cells. Uninfected cell control values (after subtraction of reference values) were set at 100% inhibition of CPE. EC50 values were calculated using four-parameter variable slope non-linear regression fitting of values. The CellTiter-Glo 2.0 assay was also used to determine viability of uninfected NCI-H358 cells treated with 3-fold serial dilutions of the compounds for 72 h (3 days) or 168 h (7 days). Values were normalized to DMSO controls according to % viability as follows: % viability = [(specific value-reference value)/(DMSO control value – reference value)] × 100. Reference values were derived from control wells without cells. DMSO control values (after subtraction of reference values) were set at 100% viability. 50% viability/cytotoxicity (CC50) values were calculated using four-parameter variable slope non-linear regression fitting of mean values derived from quadruplicate samples.
2.7. Quantitative focus-forming unit (FFU) assay
To measure compound inhibition of filovirus infection and spread, 2 × 104 NCI-H358 cells or SAECs treated with compound were infected with 0.5 TCID50 of EBOV, SUDV, RAVV, or MARV-ZsG. At 7 days pi, cells were fixed in 10% formalin supplemented with 0.2% Triton-X detergent, and stained with primary rabbit anti-EBOV polyclonal serum (1:2000), and after several washes the corresponding anti-rabbit Dylight 488 conjugated secondary antibody (Bethyl Laboratories, Bethesda, MD) was added (1:1000) for 1 h. After 3 washes, filovirus infection-induced focus forming units (FFU) were measured in each well of the 96-well plate (CellCarrier-96, Perkin Elmer, Waltham, MA) using a Cytation5 cell imaging multi-mode reader paired with Gen5 software (Biotek, Winooski, VT). For MARV-ZsG, GFP+ cells were counted directly at 3 days pi without need for fixation and antibody staining. A 2.74× objective lens was used to take 12 overlapping images encompassing each entire well, which were assembled and then analyzed for FFUs ranging in size from 15-200 μm, and which had a relative fluorescence signal that was above 2000. The camera gain was set at 15.6, with an integration time of 306 msec, using an LED intensity setting of 10. The average number of background cell counts were subtracted from each well to give normalized cells counts, and any negative values were adjusted to “0”. For analysis of each experimental replicate, the highest number of positive counts was regarded as 100%, while 0 counts was used for 0% positivity. Following this normalization, data were fitted to a 4-parameter variable slope non-linear regression fitting of mean values derived from quadruplicate samples.
2.8. Infectious virus yield reduction assays
To measure compound inhibition of infectious filovirus yield, 2 × 104 NCI-H358 cells were infected with 0.5 TCID50 of SUDV or RAVV per cell for 1 h. Virus inoculum was then removed, cells were washed once with phosphate buffered saline, and replaced with culture medium containing respective compound in a 10-point 3-fold dilution series. At 7 days pi, supernatants were harvested, serially diluted (10-fold) and mixed with 104 Vero cells per well in 96 well plates. At day 5 pi, plates were fixed with 10% formalin supplemented with 0.2% Triton-X detergent, and stained with primary rabbit anti-EBOV serum, and after several washes the corresponding anti-rabbit Dylight 488 conjugated secondary antibody was added. Plates were visually assayed using a microscope for fluorescent cells, and quantitated by 50% tissue TCID50 assay using the Reed and Muench method (Reed and Muench, 1938). EC50 values were calculated using four-parameter variable slope non-linear regression fitting of mean values derived from quadruplicate samples. Similarly, to measure compound inhibition of infectious henipavirus yield, 2 × 104 NCI-H358 cells were infected with 0.25 TCID50 of NiV or HeV per cell for 1 h. Virus inoculum was then removed, cells were washed once with phosphate buffered saline, and replaced with culture medium containing compound in a 10-point 3-fold dilution series. At 48 h pi supernatants were harvested, serially diluted, and mixed with 104 Vero cells per well in 96-well plates. At day 5 pi, plates were visually assayed for CPE, and then virus titers were quantitated by 50% tissue TCID50 assay using the Reed and Muench method (Reed and Muench, 1938). EC50 values were calculated using four-parameter variable slope non-linear regression fitting of mean values derived from quadruplicate samples.
3. Results
3.1. Triphosphate forms of R1479 and its 2′-mono- and 2′-difluoro-modified analogs inhibit RSV polymerase activity by acting as RNA chain terminators
To confirm the antiviral mechanism of action of R1479 and its 2′-mono and 2′-difluoro-modified analogs (depicted in Fig. 1a, b, and c respectively), we first performed cell-free in vitro RSV polymerase assays. The RSV polymerase forms a dimer of the L and P protein (RSV L-P) that accepts RNA templates with a primer to generate short RNA synthesis products (Fig. 1d) (Noton et al., 2012). RSV L-P extends the primer by 1 base with the addition of (α-33P)-GTP (Fig. 1e, lane 1) and fully it extends by 7 bases with the addition of ATP and CTP (Fig. 1e, lane 3). We individually substituted the 3 cytidine analogs in place of natural CTP to understand their potential effects as non-obligatory chain terminators in an RNA synthesis assay (Fig. 1e, lanes 4–6). R1479-TP, 2′F-4′N3-CTP, and 2′diF-4′N3-CTP terminated RNA synthesis by RSV L-P at position +8, and were recognized almost equally by the enzyme. This is consistent with observations of other 2′ and 4′-modified cytidine analogs that inhibit RSV polymerase in similar assays (Deval et al., 2015).
3.2. Intracellular phosphorylation levels of R1479, 2′F-4′N3-C, and 2′diF-4′N3-C are highly dependent on cell type
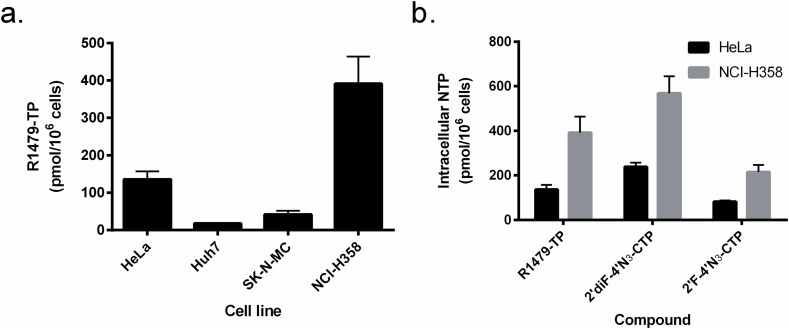
In a previous study, we observed different levels of R1479 antiviral activity against henipaviruses depending on the cell line used, with consistently lower R1479 antiviral activity in HeLa cells than in a human epithelial lung carcinoma cell line (NCI-H358) (Hotard et al., 2017). To investigate whether this difference was due to cellular phosphorylation levels of R1479, we measured intracellular phosphorylation of R1479 in 4 human cell lines: HeLa, a cervical adenocarcinoma line; Huh7, a hepatoma line; SK-N-MC, a neuroepithelioma line; and NCI-H358. Each cell line was treated with R1479 for 24 h, and intracellular triphosphate levels were then measured using tandem liquid chromatography/mass spectrometry (LC/MS/MS) and quantitated using synthetic standards. Levels of the corresponding triphosphate (R1479-TP) were measured in each of the 4 cell lines, with highest R1479-TP levels seen in NCI-H358 cells (391 ± 72 pmol/million cells; Fig. 2 a). Phosphorylation of R1479 was lowest in Huh7 cells and SK-N-MC cells (18 ± 0.6 and 42 ± 10 pmol/million cells, respectively), and intermediate in HeLa cells (136 ± 21 pmol/million cells). In addition to R1479, which contains a 4′-azido substitution, we examined the effects of 2′-monofluoro or 2′-difluoro substitutions on NTP levels using the 2 cell lines with the highest NTP levels. HeLa and NCI-H358 cells were treated with 2′F-4′N3-C and 2′diF-4′N3-C for 24 h at 50 μM, and NTP formation was measured and quantified as previously described (Fig. 2b). In both cell lines, 2′diF-4′N3-C levels were highest, followed by R1479 and then 2′F-4′N3-C, but higher overall NTP levels of all 3 nucleosides were seen in NCI-H358 cells. In light of these results, we decided to use NCI-H358 cells for our cell-based viral infection assays.
Fig. 2.
Quantification and comparison of cellular nucleoside triphosphate (TP) levels of R1479, 2′F-4′N3-C, and 2′diF-4′N3-C. (a). R1479-TP levels were measured in 4 different cell types (HeLa, Huh7, SK-N-MC, and NCI-H358) after 24 h of treatment at 50 μM. (b) Levels of triphosphorylated R1479, 2′F-4′N3-C, and 2′diF-4′N3-C in HeLa cells (black bars) and NCI-H358 cells (gray bars) after 24 h of treatment at 50 μM. Graphs and error bars indicate mean values and standard deviations for each compound in each cell line performed at minimum in biological triplicates.
3.3. R1479, 2′F-4′N3-C, and 2′diF-4′N3-C show minimal cytotoxicity in NCI-H358 cells
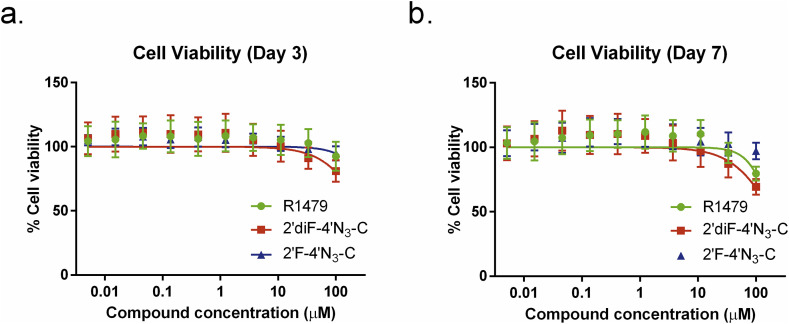
To ensure that the antiviral effects we observed in this study were not due to any cytotoxic effects attributed to the compounds themselves, we measured the viability of NCI-H358 cells continually incubated with varying dilutions of each compound for 3 and 7 days (Fig. 3 ). All 3 compounds produced low cytotoxicity 3 days post treatment, and R1479 and 2′diF-4′N3-C resulted only in mild cytotoxicity at the highest concentration used (100 μM) after 7 days of treatment (Fig. 4 b). Even 7 days post treatment, 50% cell cytotoxicity (CC50) levels were not reached.
Fig. 3.
R1479, 2′F-4′N3-C, and 2′diF-4′N3-C do not cause significant cell cytotoxicity. Representative dose response curves measuring cell viability of uninfected cells treated with R1479 (green circles), 2′F-4′N3-C (blue triangles), and 2′diF-4′N3-C (red squares) measured at (a) 72 h post treatment or (b) 168 h post treatment. Cell viability was measured using CellTiter-Glo 2.0 assay reagent. Luminescence levels (indicative of cellular ATP levels as a surrogate marker of cell viability) assayed in DMSO-treated, uninfected cells were set as 100% cell viability. Dose response curves were fitted to the mean value of experiments performed in quadruplicate for each concentration in the 10-point 3-fold dilution series using a 4-parameter non-linear logistic regression curve with variable slope. Viability assay was repeated once for the 72 h time point.
Fig. 4.
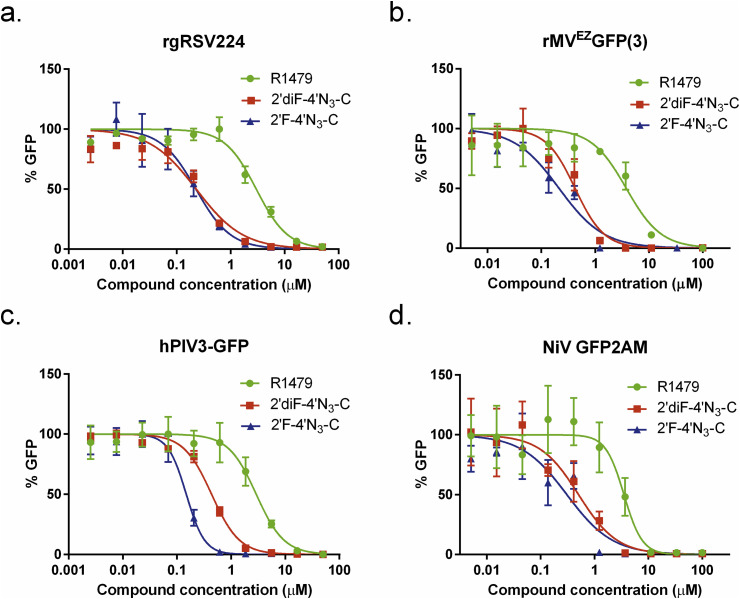
Antiviral activity of R1479, 2'′F-4′N3-C, and 2′diF-4′N3-C against reporter pneumo- and paramyxoviruses. Representative dose response curves for R1479 (green circles), 2′F-4′N3-C (blue triangles), and 2′diF-4′N3-C (red squares) against recombinant reporter (a) respiratory syncytial virus (rgRSV224), (b) measles virus (rMVEZ(3)GFP), (c) human parainfluenza virus 3 (hPIV3-GFP), and (d) Nipah virus (NiV-GFP2AM). Data points and error bars indicate the mean value and standard deviation of 4 biological replicates, and are representative of at least 2 independent experiments for each compound.
3.4. 2′F-4′N3-C and 2′diF-4′N3-C show superior antiviral activity compared to R1479 against recombinant reporter NiV, RSV, measles virus, and human parainfluenza virus 3
We initially evaluated the antiviral activity of R1479, 2′F-4′N3-C, and 2′diF-4′N3-C against recombinant reporter RSV (rgRSV224; (Hallak et al., 2000), measles virus (rMVEZEGFP(3); (Rennick et al., 2015), human parainfluenza virus-3 (hPIV3-GFP; (Zhang et al., 2005), and NiV (NiV-GFP2AM (Lo et al., 2014), expressing green fluorescent reporter proteins (Fig. 4). Whereas EC50 values for R1479 against all 4 viruses were consistently in the single-digit micromolar range, both 2′F-4′N3-C and 2′diF-4′N3-C inhibited each virus 3- to 20-fold more potently than R1479 (Table 1 ). 2′F-4′N3-C and 2′diF-4′N3-C inhibited rgRSV224 and rMVEZEGFP(3) at similar concentrations (Table 1), but 2′F-4′N3-C was measurably more potent than 2′diF-4′N3-C against hPIV3-GFP (Fig. 4c, Table 1). 2′F-4′N3-C also inhibited NiV-GFP2AM moderately more strongly than 2′diF-4′N3-C as measured by both a GFP reporter assay (Fig. 4d) and CPE inhibition assay (data not shown; Table 1).
Table 1.
Mean antiviral activity and selective indices of R1479, 2′F-4′N3-C, and 2′diF-4′N3-C in NCI-H358 cells.
| Family | Virus | Species/Variant | Assay | EC50 (μM) [SI] |
||
|---|---|---|---|---|---|---|
| R1479 | 2′diF-4′N3-C | 2′F-4′N3-C | ||||
| Paramyoxviridae | NiV | Rec. M-GFP2AM | REP | 3.1 ± 0.36 [>32] | 0.37 ± 0.11 [>270] | 0.18 ± 0.02 [>555] |
| CPE | 2.4 ± 0.22 [>41] | 0.37 ± 0.09 [>270] | 0.23 ± 0.02 [>434] | |||
| M-1999 | CPE | 2.9 ± 0.95 [>34] | 0.48 ± 0.19 [>208] | 0.29 ± 0.10 [>344] | ||
| VTR | 1.5 [>66]* | 0.36 [>277] | 0.14 [>714] | |||
| HeV | 1996 | CPE | 2.1 ± 0.57 [>47] | 0.57 ± 0.11 [>175] | 0.37 ± 0.09 [>270] | |
| VTR | 2.4 [>41]* | 0.15 [>666] | 0.15 [>666] | |||
| hPIV3 | Rec. JS-GFP | REP | 3.2 ± 0.50 [>32] | 0.44 ± 0.03 [>227] | 0.16 ± 0.02 [>625] | |
| MV | Rec. rMVEZGFP(3) | REP | 1.9 ± 0.12 [>52] | 0.34 ± 0.07 [>294] | 0.27 ± 0.04 [>370] | |
| Pneumoviridae | RSV | Rec. rgRSV224 (A2) | REP | 3.3 ± 0.39 [>30] | 0.21 ± 0.04 [>476] | 0.28 ± 0.05 [>357] |
| Filoviridae | EBOV | Rec. Mayinga-GFP | REP | 2.6 ± 0.85 [>38] | 0.30 ± 0.02 [>333] | 7.79 ± 4.2 [>12] |
| Rec. Makona-ZsG | REP | 2.1 ± 0.07 [>47] | 0.33 ± 0.03 [>303] | 3.85 ± 0.09 [>25] | ||
| Makona | FFU | 2.1 ± 0.02 [>47] | 0.60 ± 0.08 [>166] | ND [ND] | ||
| SUDV | Gulu | FFU | 2.5 [40] | 0.70 ± 0.08 [>142] | 9.57 [>10] | |
| VTR | 0.92 [>108] | 0.16 [>625] | 0.30 [>333] | |||
| RAVV | Ravn | FFU | 7.4 ± 1.6 [>13] | 0.73 ± 0.16 [>136] | 10.8 ± 4.9 [>9] | |
| VTR | 3.0 [>33] | 0.16 [>625] | 6.07 [>16] | |||
| Phenuiviridae | RVFV | Rec. ZH501-GFP | REP | >50 [ND] | 44.6 [>2] | >50 [ND] |
EC50, 50% effective inhibition concentration; SI, selective index = EC50/CC50; REP, reporter; CPE, cytopathic effect; VTR, virus titer reduction; FFU, focus-forming unit; ND, not determined; Rec, recombinant; *, data adapted from (Hotard et al., 2017). Mean values with ±standard deviation values were derived from a minimum of 2 independent experiments performed in biological quadruplicate.
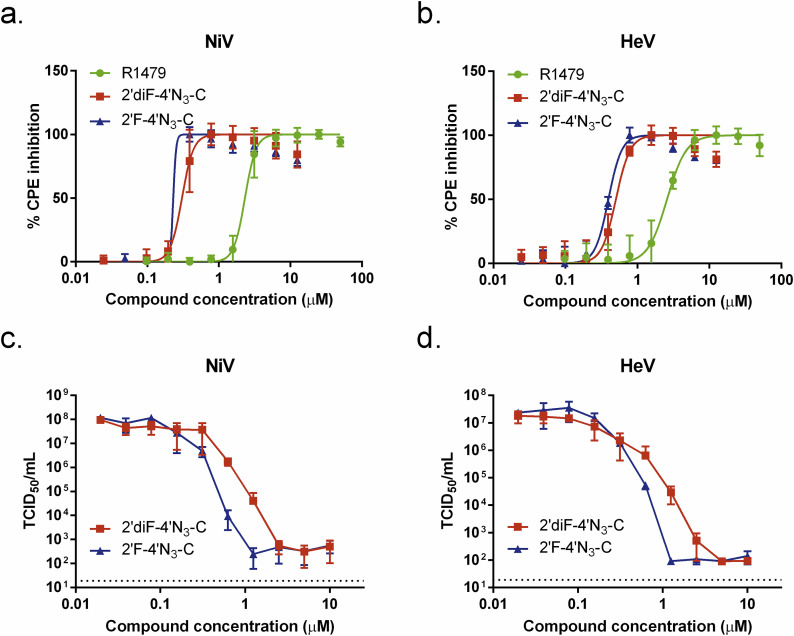
3.5. 2′F-4′N3-C and 2′diF-4′N3-C reduce both henipavirus-induced CPE and infectious virus yield more strongly than does R1479
We further evaluated the antiviral activity of 2′F-4′N3-C and 2′diF-4′N3-C against wild-type NiV and HeV by measuring the inhibition of henipavirus-induced CPE and infectious virus yield (Fig. 5 ). Similar to what was observed for NiV-GFP2AM, 2′F-4′N3-C and 2′diF-4′N3-C inhibited NiV- and HeV-induced CPE with significantly greater potency than R1479, with 2′F-4′N3-C being marginally more potent than 2′diF-4′N3-C (Fig. 5a and b). Accordingly, ∼1.5 μM 2′F-4′N3-C and ∼3 μM 2′diF-4′N3-C reduced infectious henipavirus yields by 4–5 orders of magnitude (Fig. 5c and d), whereas 6–10 μM R1479 was required for similar virus yield reduction (Hotard et al., 2017). Although in this assay, EC50 values for 2′F-4′N3-C and 2′diF-4′N3-C were identical against HeV and were within 3-fold against NiV (Table 1), 2′F-4′N3-C caused a sharper decline in both NiV and HeV titers than did 2′diF-4′N3-C (Fig. 5c and d).
Fig. 5.
R1479, 2′F-4′N3-C, and 2′diF-4′N3-C reduce henipavirus cytopathic effect (CPE) and infectious virus yield. Representative dose response curves for R1479 (green circles), 2′F-4′N3-C (blue triangles), and 2′diF-4′N3-C (red squares) against wild-type (a) NiV and (b) Hendra virus (HeV) as measured by CPE determination. CPE assays were independently repeated at least 3 times for each compound for both NiV and HeV. Reduction of infectious NiV (c) and HeV (d) yield by 2′F-4′N3-C and 2′diF-4′N3-C. TCID50/mL values (in log scale) for each data point represent the mean of triplicate (2′diF-4′N3-C) or quadruplicate (2′F-4′N3-C) infection experiments for each concentration in the 10-point, 3-fold dilution series of each compound performed once. Dotted line indicates limit of detection. Error bars indicate standard deviation of the mean values.
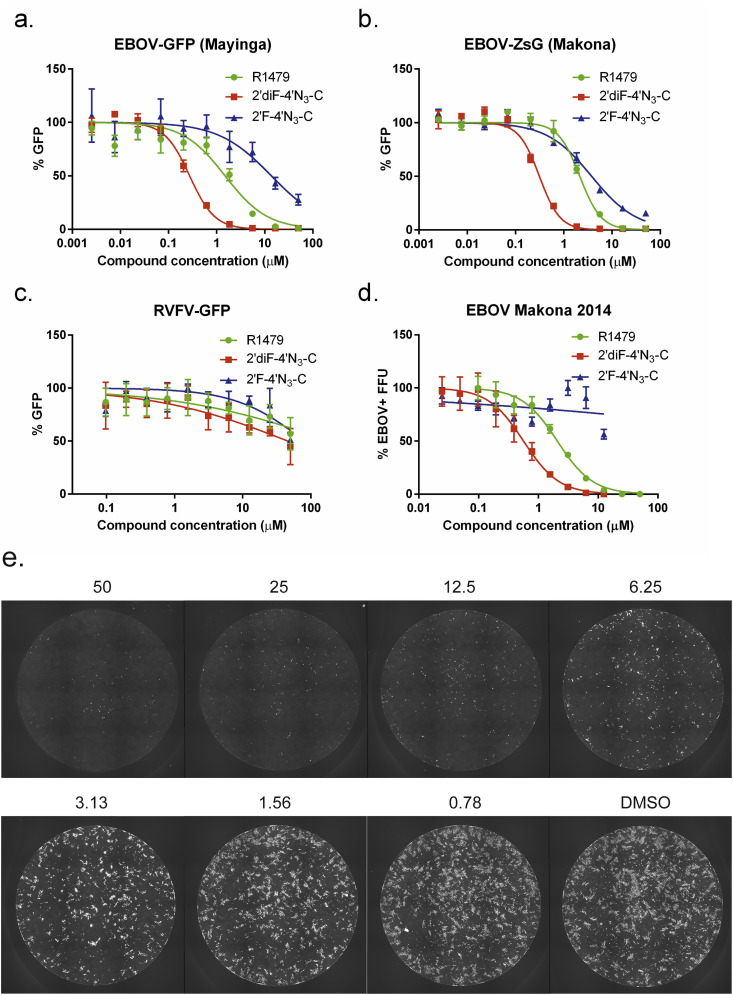
3.6. EBOV variants are susceptible to inhibition by R1479 and 2′diF-4′N3-C, but are less susceptible to 2′F-4′N3-C inhibition
Recent work describing the broad-spectrum activity of adenosine nucleotide analog GS-5734 across Paramyxoviridae, Pneumoviridae, and Filoviridae families documented conserved amino acids in the nucleotide-binding domains of polymerases of these viruses, which may explain their susceptibility to similar compounds (Lo et al., 2017). In light of this work, alongside other studies documenting the antiviral activity of R1479, 2′F-4′N3-C, and 2′diF-4′N3-C against both HCV and RSV (Deval et al., 2015, Klumpp et al., 2006, Smith et al., 2007, Smith et al., 2009, Wang et al., 2015), we tested the antiviral activities of these compounds against recombinant reporter Mayinga and Makona EBOV variants (genus Ebolavirus, species Zaire ebolavirus) expressing green fluorescent proteins (Albarino et al., 2015, Towner et al., 2005) (Fig. 6 a and b). R1479 and 2′diF-4′N3-C inhibited both EBOV variants similarly to henipaviruses, with approximate EC50 values of 2 μM and 0.3 μM, respectively (Table 1). In contrast to what was observed for henipaviruses, however, 2′F-4′N3-C was 10 times less potent against EBOV, with EC50 values ranging from 3–8 μM (Table 1). All 3 compounds only negligibly inhibited a reporter RVFV, which belongs to the family Phenuiviridae, outside of the order Mononegavirales (Fig. 6c).
Fig. 6.
R1479, 2′F-4′N3-C, and 2′diF-4′N3-C inhibit Ebola virus (EBOV) variants but not Rift Valley fever virus (RVFV). Representative dose-response curves for R1479 (green circles), 2′F-4′N3-C (blue triangles), and 2′diF-4′N3-C (red squares) against recombinant reporter (a) EBOV-Mayinga variant, (b) EBOV-Makona variant, and (c) RVFV expressing green fluorescent proteins. (d) Representative dose-response curves of the same compounds against wild-type EBOV (2014 Makona variant) using a focus forming unit (FFU) assay. FFU assays against EBOV were repeated once for R1479, twice for 2′diF-4′N3-C, and was performed once for 2′F-4′N3-C. (e) Representative composite micrographs of FFU assays in individual wells of a 96-well plate of R1479-treated, EBOV-infected, formalin-inactivated NCI-H358 cells stained with anti-EBOV polyclonal sera and Dylight-488 conjugated secondary antibody. Concentrations of R1479 (in μM) are indicated above each pictured well.
To confirm our observations, we tested the compounds against wild-type EBOV (Makona variant) and measured their inhibition of EBOV infection and spread. By using an image-based plate reader, we quantified EBOV infection by the percentage of EBOV antigen-positive focus-forming units (FFUs) detected (Fig. 6d–e). We observed dose-dependent inhibition of FFU formation with R1479 and 2′diF-4′N3-C treatment, but did not detect significant inhibition by 2′F-4′N3-C (Fig. 6d, Table 1).
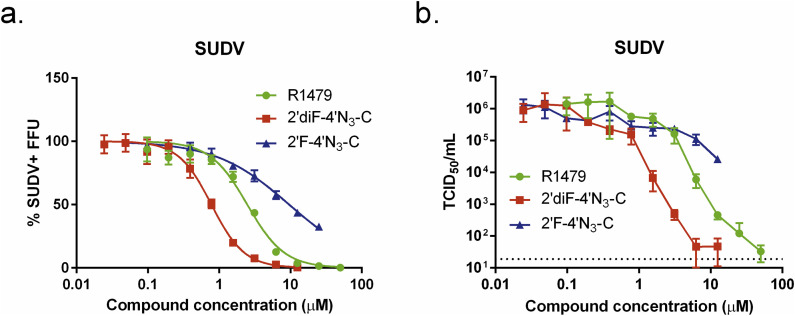
3.7. R1479 and 2′diF-4′N3-C potently inhibit SUDV
We then tested the 3 compounds against the Gulu variant of SUDV (genus Ebolavirus, species Sudan ebolavirus), a phylogenetically divergent relative of EBOV which prior to 2014 was responsible for the largest filovirus outbreak in humans (Lamunu et al., 2004, Sanchez and Rollin, 2005). R1479 and 2′diF-4′N3-C inhibited SUDV with efficacy similar to that observed with EBOVs by the FFU assay (Fig. 7 a), with EC50 values of 2.5 μM (R1479) and 0.7 μM (2′diF-4′N3-C). Accordingly, 2′F-4′N3-C inhibited SUDV less potently than the other compounds, with an EC50 value of approximately 10 μM (Table 1). We further evaluated the antiviral activity of these 3 cytidine analogs against SUDV by measuring infectious virus yield (Fig. 7b). Both R1479 and 2′diF-4′N3-C reduced infectious SUDV titers by > 4 orders of magnitude at concentrations of 25 μM and 6.25 μM, respectively, with respective EC50 values of 0.92 μM and 0.16 μM. On the other hand, 2′F-4′N3-C only reduced infectious virus yield by approximately 1.5 orders of magnitude at the highest concentration used in the assay (12.5 μM) (Fig. 7b).
Fig. 7.
R1479, 2′F-4′N3-C, and 2′diF-4′N3-C inhibit Sudan virus (SUDV). (a) Representative dose-response curves for R1479 (green circles), 2′F-4′N3-C (blue triangles), and 2′diF-4′N3-C (red squares) against SUDV (Gulu variant) using a focus forming unit (FFU) assay. FFU assays for R1479 and 2′F-4′N3-C were performed once, and was repeated once for 2′diF-4′N3-C. (b) Reduction of infectious SUDV yield (in log scale) by R1479, 2′diF-4′N3-C, and 2′F-4′N3-C. TCID50/mL values (in log scale) for each data point represent the mean of quadruplicate infections for each concentration in the 10-point, 3-fold dilution series for each compound performed once.
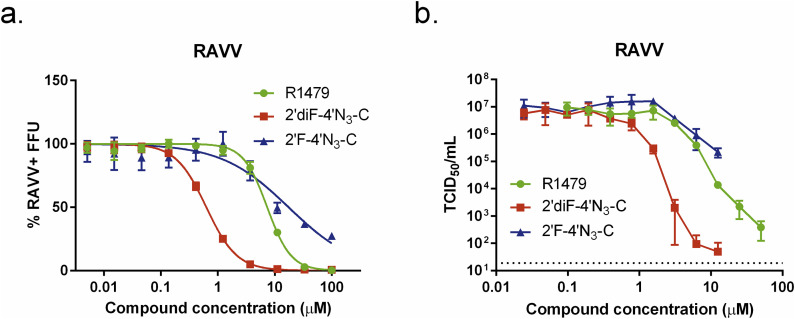
3.8. R1479 and 2′diF-4′N3-C potently inhibit RAVV
Having determined the antiviral activity of these compounds against phylogenetically divergent members of the genus Ebolavirus, we proceeded to test them against another filovirus, Ravn virus (RAVV) (genus Marburgvirus, species Marburg marburgvirus), which, when compared with EBOV and SUDV, was determined to be the most phylogenetically divergent member of the genus Marburgvirus (∼67–68% nucleotide divergence) (Towner et al., 2006). Using our FFU assay, we observed that R1479 inhibited RAVV with 3-fold lower potency than EBOV or SUDV, with an EC50 value of 7.4 μM (Fig. 8 a, Table 1). 2′diF-4′N3-C and 2′F-4′N3-C, however, inhibited RAVV similarly to EBOV and SUDV, with EC50 values of 0.73 μM and 10.8 μM, respectively. We then measured infectious RAVV production in the presence of the 3 compounds. Similar to what we observed for SUDV, both R1479 and 2′diF-4′N3-C reduced infectious RAVV titers by over 4 orders of magnitude at concentrations of 50 μM and 6.25 μM, respectively, with respective EC50 values of 3 μM (R1479) and 0.16 μM (2′diF-4′N3-C; Fig. 8b, Table 1). 2′F-4′N3-C reduced RAVV titers by roughly 1.5 orders of magnitude at the highest concentration used (25 μM), with an EC50 value of 6 μM (Fig. 8b, Table 1).
Fig. 8.
R1479, 2′F-4′N3-C, and 2′diF-4′N3-C inhibit Marburgvirus Ravn virus (RAVV). (a) Representative dose-response curves of R1479 (green circles), 2′F-4′N3-C (blue triangles), and 2′diF-4′N3-C (red squares) against RAVV using an FFU assay. FFU assays were independently repeated four times for R1479, twice for 2′F-4′N3-C, and three times for 2′diF-4′N3-C. Dose response curves were fitted to the mean value of experiments performed in quadruplicate for each concentration in the 10-point, 3-fold dilution series using a 4-parameter non-linear logistic regression curve with variable slope. (b) Reduction of infectious RAVV yield (in log scale) by R1479, 2′diF-4′N3-C, and 2′F-4′N3-C. TCID50/mL values for each data point represent the mean of quadruplicate infections for each concentration in the 10-point, 3-fold dilution series of each compound performed once.
3.9. 2′F-4′N3-C potently inhibits NiV in human primary small airway epithelial cells
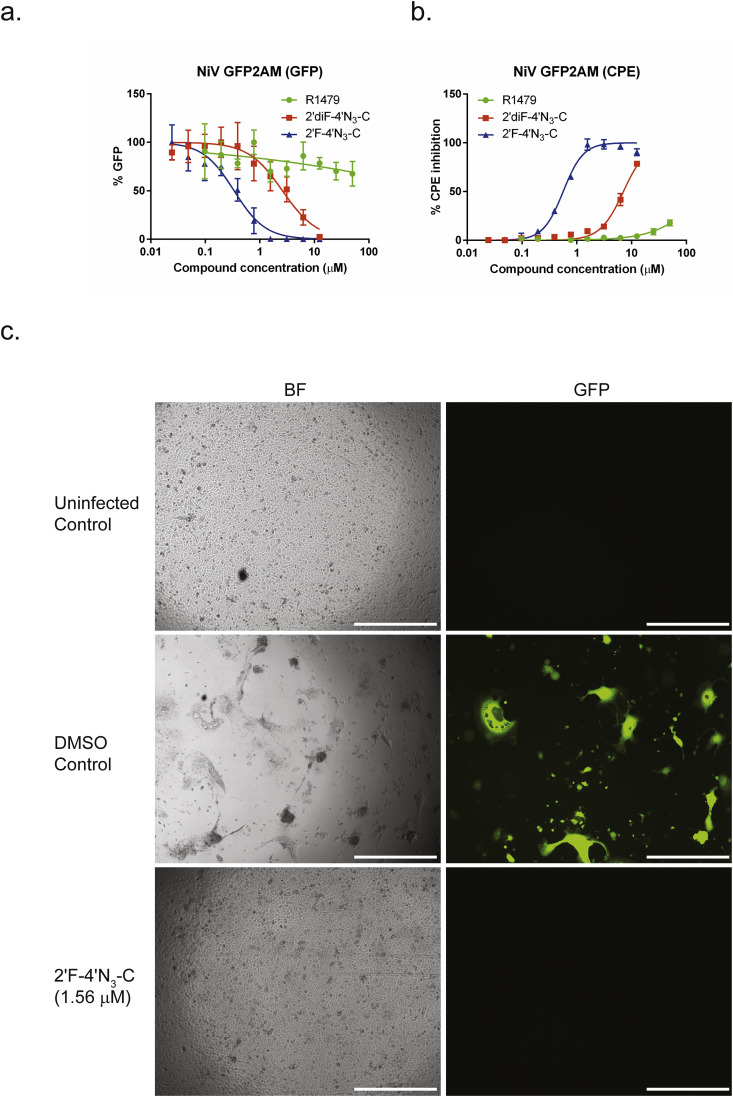
To verify the antiviral activity observed in the NCI-H358 cell line, we performed reporter and CPE assays for NiV using human primary small airway epithelial cells (SAECs), which have been shown to support robust NiV replication (Escaffre et al., 2013). For both the reporter and CPE assays which utilized the recombinant reporter NiV-GFP2AM virus, we observed potent inhibition by 2′F-4′N3-C, with respective EC50 values of 0.34 and 0.56 μM (Fig. 9 a–b, Table 2 ). The inhibition of both GFP and CPE could be visually observed, as treatment with 2′F-4′N3-C at 1.56 μM completely abolished GFP expression as well as CPE in the SAECs (Fig. 9c). On the other hand, we observed markedly lower antiviral activity for R1479 and 2′diF-4′N3-C, which indicates that their respective intracellular NTP levels were likely lower than their levels observed in NCI-H358 cells.
Fig. 9.
2′F-4′N3-C potently inhibits reporter NiV gene expression (GFP) and cytopathic effect (CPE) in human primary small airway epithelial cells. Representative dose response curves for R1479 (green circles), 2′F-4′N3-C (blue triangles), and 2′diF-4′N3-C (red squares) against recombinant reporter NiV (NiVGFP2AM) by measuring (a) GFP expression at 48 h post-infection, and by measuring (b) CPE inhibition at 72 h post-infection. Data points and error bars indicate the mean value and standard deviation of 4 biological replicates. (c) Bright field (BF) and fluorescence (GFP) microscopy of uninfected control, DMSO treated NiV-GFP2AM infected control, and 2′F-4′N3-C treated NiV-GFP2AM infected SAECs. Concentration of 2′F-4′N3-C is indicated in μM. White bar indicates distance of 500 μm.
Table 2.
Mean antiviral activity of R1479, 2′F-4′N3-C, and 2′diF-4′N3-C in primary human small airway epithelial cells.
| Family | Virus | Species/Variant | Assay | EC50 (μM) [SI] |
||
|---|---|---|---|---|---|---|
| R1479 | 2′diF-4′N3-C | 2′F-4′N3-C | ||||
| Paramyxoviridae | NiV | Rec. M-GFP2AM | REP | >50 [ND] | 2.61 [>19] | 0.34 [>147] |
| CPE | >50 [ND] | 7.09 [>7] | 0.56 [>89] | |||
| Filoviridae | MARV | Rec. Bat371-ZsG | FFU | >50 [ND] | 3.59 ± 0.32 [>13] | >50 [ND] |
EC50, 50% effective inhibition concentration; SI, selective index = EC50/CC50; REP, reporter; CPE, cytopathic effect; FFU, focus-forming unit; ND, not determined; Rec, recombinant. Mean values with ±standard deviation values were derived from a minimum of 2 independent experiments performed in biological quadruplicate.
3.10. 2′diF-4′N3-C inhibits Marburg virus infection in human primary small airway epithelial cells
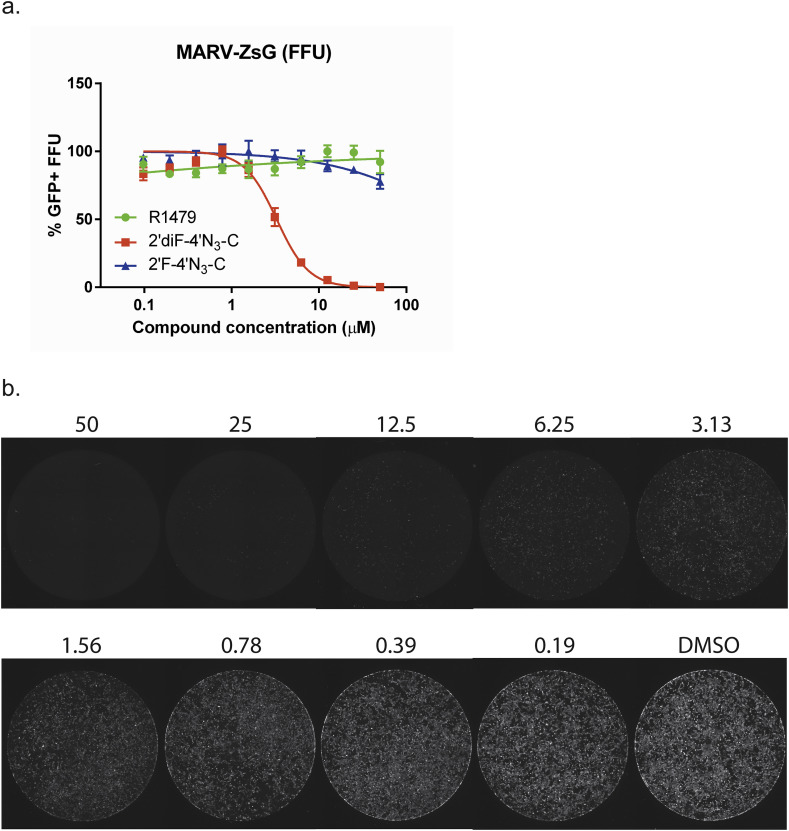
Since filovirus-pseudotyped lentiviruses can efficiently transduce airway epithelial cells in vivo (Kobinger et al., 2001), we proceeded to evaluate the antiviral activities of these nucleosides in SAECs against a reporter Marburg virus (MARV-ZsG) expressing a green fluorescent protein. In cells treated with serial dilutions of 2′diF-4′N3-C, we observed a dose-dependent inhibition of MARV infection by FFU assay; whereas R1479 and 2′F-4′N3-C did not show any significant inhibition (Fig. 10 a–b, Table 2). The comparatively higher EC50 value for 2′diF-4′N3-C against MARV-ZsG in SAECs (3.59 μM) than what was observed for RAVV in NCI-H358 cells (0.73 μM) is likely due to lower intracellular NTP levels.
Fig. 10.
2′diF-4′N3-C inhibits reporter Marburg virus (MARV) infection in human primary small airway epithelial cells. (a) Representative dose response curves for R1479 (green circles), 2′F-4′N3-C (blue triangles), and 2′diF-4′N3-C (red squares) against recombinant reporter MARV expressing a green fluorescent protein (ZsGreen1) using a focus forming unit (FFU) assay. FFU assays for R1479 and 2′F-4′N3-C were performed once, and was repeated once for 2′diF-4′N3-C. Data points and error bars indicate the mean value and standard deviation of 4 biological replicates. (b) Representative composite micrographs of MARV GFP+ FFUs in individual wells of a 96-well plate of 2′diF-4′N3-C -treated, MARV-infected SAECs. Concentrations of 2′diF-4′N3-C (in μM) are indicated above each pictured well.
4. Discussion
In this study, we characterized the antiviral properties of R1479 and its analogs 2′F-4′N3-C and 2′diF-4′N3-C using both cell-free and cell-based assays. Despite the presence of 3′-OH group in each analog, all 3 compounds behaved like conventional non-obligatory RNA synthesis chain terminators in cell-free RSV polymerase assays (Deval et al., 2015). In order for nucleoside analogs to retain their antiviral function inside a cell, multiple cellular kinases are required to convert them to the active triphosphate form (Ray and Hitchcock, 2009). To better understand the cell type-dependent variability we previously observed with R1479 antiviral activity (Hotard et al., 2017), we measured cellular levels of R1479-TP in 4 different cell lines, and observed 2 to 3-fold higher levels of R1479-TP, 2′F-4′N3-CTP, and 2′diF-4′N3-CTP in NCI-H358 cells than in HeLa cells. The comparatively elevated NTP levels observed in NCI-H358 cells may be attributed to higher expression levels of uridine-cytidine kinases, which catalyze the presumed initial rate-limiting step of phosphorylating uridine and cytidine nucleosides (Van Rompay et al., 2001). Characterizing the NCI-H358 cell line as the optimal line to use in this study highlights the importance of confirming cellular triphosphate levels when conducting antiviral screens of nucleoside analog libraries. We have observed significantly decreased antiviral activity for R1479, 2′F-4′N3-C, and 2′diF-4′N3-C in Huh7 cells against reporter EBOVs (data not shown), which is likely explained by overall lower phosphorylation levels of these cytidine analogs in this cell line (Fig. 2a). The higher potency of 2′F-4′N3-C, and 2′diF-4′N3-C over R1479 against paramyxoviruses, however, cannot solely be attributed to NTP levels, since R1479-TP levels were higher than 2′F-4′N3-CTP levels in these cells (Fig. 2b). Moreover, the change in rank order of anti-filoviral potency in the cell-based assays suggests that the differences in antiviral activity are due to differential recognition, or discrimination, of these NTP analogs by viral polymerases. Our gel-based biochemical assay using RSV polymerase provides qualitative results and informs mainly on the mechanism of action of each compound, demonstrating substrate recognition and chain termination (Fig. 1). It does not provide enough information to rank order potencies and/or discrimination between the three nucleotides (Deval et al., 2015). Evaluation of these compounds in primary human SAECs confirmed the potent anti-NiV activity of 2′F-4′N3-C (Fig. 9) and the anti-MARV activity of 2′diF-4′N3-C (Fig. 10). Our study did not include two other related nucleosides 2′-deoxy-2′-β-fluoro-4′-azidocytidine (RO-0622) and 2′-deoxy-2′-β-hydroxy-4′-azidocytidine (RO-9187) (Klumpp et al., 2008) because they had little to no antiviral activity against both NiV and EBOV, respectively (data not shown). Our results warrant further studies comparing the antiviral potency of these NTP analogs against recombinant paramyxovirus and filovirus polymerases, although technical challenges have precluded such work only until very recently (Jordan et al., 2018). The ability of these cytidine analogs to reduce henipavirus and filovirus infectious yields by over 4 log TCID50/mL mirrors viral inhibition by GS-441524 (a 1′-cyano-modified adenosine analog) and its phosphoramidate nucleotide prodrug GS-5734 (Lo et al., 2017). GS-5734 inhibited filoviruses and coronaviruses in non-human primate and mouse models, respectively, and is currently in phase 2 clinical trials for treatment of EBOV disease (EVD) (Sheahan et al., 2017, Warren et al., 2016). Favipiravir, also known at T-705, is a broad-spectrum small molecule nucleoside precursor originally developed to treat influenza A virus; it also decreased EBOV infectious yield by 4 logTCID50/mL, albeit at high concentrations (1000 μM) by a yet-undefined mechanism (Baranovich et al., 2013, Oestereich et al., 2014). BCX4430 is an adenosine nucleoside analog with variable in vitro broad-spectrum activity across 8 virus families including Filoviridae, and is currently in phase 1 clinical trials for treating EVD (Taylor et al., 2016, Warren et al., 2014). Although BCX4430 inhibits HCV polymerase similarly to conventional chain terminators and also reduces infectious filovirus yield, its mechanism of action against negative-strand viruses is unclear. While neither R1479 nor its 2′-fluoro-modified analogs are viable candidates for antiviral therapeutics due to variable incorporation by host mitochondrial RNA and DNA polymerases (Arnold et al., 2012, Clarke et al., 2015), they provide an initial comparison of filo- and paramyxovirus susceptibility to 4′-modified nucleosides. Our study provides evidence supporting the development of 4′-modified nucleosides with near-equivalent potency not only against 3 Mononegavirales order families, but also against the positive-sense Flaviviridae family. Developing a small-molecule, broad-spectrum therapeutic to treat phylogenetically divergent viral infections that share significant epidemiological and clinical overlap (e.g., EBOV and yellow fever virus; NiV and Japanese encephalitis virus) would benefit clinicians with no access to rapid diagnostics amidst an outbreak of these serious viral infections.
Acknowledgments
We thank the nucleoside chemistry group at Alios for synthesizing the compounds and their NTPs, and Julian Symons and Leo Beigelman from Alios for supporting this work. We thank Mike Flint and Tatyana Klimova for helpful comments in reviewing the manuscript. We also thank Cesar A. Albarino for providing the MARV-ZsG virus. The findings and conclusions in this report are those of the authors and do not necessarily represent those of the Centers for Disease Control and Prevention. This work was financially supported by CDC core funding.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.antiviral.2018.03.009.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Albarino C.G., Wiggleton Guerrero L., Lo M.K., Nichol S.T., Towner J.S. Development of a reverse genetics system to generate a recombinant Ebola virus Makona expressing a green fluorescent protein. Virology. 2015;484:259–264. doi: 10.1016/j.virol.2015.06.013. [DOI] [PubMed] [Google Scholar]
- Arnold J.J., Sharma S.D., Feng J.Y., Ray A.S., Smidansky E.D., Kireeva M.L., Cho A., Perry J., Vela J.E., Park Y., Xu Y., Tian Y., Babusis D., Barauskus O., Peterson B.R., Gnatt A., Kashlev M., Zhong W., Cameron C.E. Sensitivity of mitochondrial transcription and resistance of RNA polymerase II dependent nuclear transcription to antiviral ribonucleosides. PLoS Pathog. 2012;8:e1003030. doi: 10.1371/journal.ppat.1003030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranovich T., Wong S.S., Armstrong J., Marjuki H., Webby R.J., Webster R.G., Govorkova E.A. T-705 (favipiravir) induces lethal mutagenesis in influenza A H1N1 viruses in vitro. J. Virol. 2013;87:3741–3751. doi: 10.1128/JVI.02346-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird B.H., Albarino C.G., Nichol S.T. Rift Valley fever virus lacking NSm proteins retains high virulence in vivo and may provide a model of human delayed onset neurologic disease. Virology. 2007;362:10–15. doi: 10.1016/j.virol.2007.01.046. [DOI] [PubMed] [Google Scholar]
- Broder C.C., Weir D.L., Reid P.A. Hendra virus and Nipah virus animal vaccines. Vaccine. 2016;34:3525–3534. doi: 10.1016/j.vaccine.2016.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC . 2017. Outbreaks Chronology: Ebola Virus Disease. [Google Scholar]
- Chen P., Liu Z., Liu S., Xie Z., Aimiuwu J., Pang J., Klisovic R., Blum W., Grever M.R., Marcucci G., Chan K.K. A LC-MS/MS method for the analysis of intracellular nucleoside triphosphate levels. Pharm. Res. 2009;26:1504–1515. doi: 10.1007/s11095-009-9863-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.L., Abdul Ghafar N., Karuna R., Fu Y., Lim S.P., Schul W., Gu F., Herve M., Yokohama F., Wang G., Cerny D., Fink K., Blasco F., Shi P.Y. Activation of peripheral blood mononuclear cells by dengue virus infection depotentiates balapiravir. J. Virol. 2014;88:1740–1747. doi: 10.1128/JVI.02841-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke M.O., Mackman R., Byun D., Hui H., Barauskas O., Birkus G., Chun B.K., Doerffler E., Feng J., Karki K., Lee G., Perron M., Siegel D., Swaminathan S., Lee W. Discovery of beta-D-2'-deoxy-2'-alpha-fluoro-4'-alpha-cyano-5-aza-7,9-dideaza adenosine as a potent nucleoside inhibitor of respiratory syncytial virus with excellent selectivity over mitochondrial RNA and DNA polymerases. Bioorg Med. Chem. Lett. 2015;25:2484–2487. doi: 10.1016/j.bmcl.2015.04.073. [DOI] [PubMed] [Google Scholar]
- Deval J., Hong J., Wang G., Taylor J., Smith L.K., Fung A., Stevens S.K., Liu H., Jin Z., Dyatkina N., Prhavc M., Stoycheva A.D., Serebryany V., Liu J., Smith D.B., Tam Y., Zhang Q., Moore M.L., Fearns R., Chanda S.M., Blatt L.M., Symons J.A., Beigelman L. Molecular basis for the selective inhibition of respiratory syncytial virus RNA polymerase by 2'-fluoro-4'-chloromethyl-cytidine triphosphate. PLoS Pathog. 2015;11:e1004995. doi: 10.1371/journal.ppat.1004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escaffre O., Borisevich V., Carmical J.R., Prusak D., Prescott J., Feldmann H., Rockx B. Henipavirus pathogenesis in human respiratory epithelial cells. J. Virol. 2013;87:3284–3294. doi: 10.1128/JVI.02576-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyer L., Smidkova M., Nencka R., Neca J., Kastl T., Palus M., De Clercq E., Ruzek D. Structure-activity relationships of nucleoside analogues for inhibition of tick-borne encephalitis virus. Antivir. Res. 2016;133:119–129. doi: 10.1016/j.antiviral.2016.07.018. [DOI] [PubMed] [Google Scholar]
- Geisbert T.W., Mire C.E., Geisbert J.B., Chan Y.P., Agans K.N., Feldmann F., Fenton K.A., Zhu Z., Dimitrov D.S., Scott D.P., Bossart K.N., Feldmann H., Broder C.C. Therapeutic treatment of Nipah virus infection in nonhuman primates with a neutralizing human monoclonal antibody. Sci. Transl. Med. 2014;6 doi: 10.1126/scitranslmed.3008929. 242ra282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallak L.K., Spillmann D., Collins P.L., Peeples M.E. Glycosaminoglycan sulfation requirements for respiratory syncytial virus infection. J. Virol. 2000;74:10508–10513. doi: 10.1128/jvi.74.22.10508-10513.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotard A.L., He B., Nichol S.T., Spiropoulou C.F., Lo M.K. 4'-Azidocytidine (R1479) inhibits henipaviruses and other paramyxoviruses with high potency. Antivir. Res. 2017;144:147–152. doi: 10.1016/j.antiviral.2017.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E.D., Johnson B.K., Silverstein D., Tukei P., Geisbert T.W., Sanchez A.N., Jahrling P.B. Characterization of a new Marburg virus isolated from a 1987 fatal case in Kenya. Arch. Virol. Suppl. 1996;11:101–114. doi: 10.1007/978-3-7091-7482-1_10. [DOI] [PubMed] [Google Scholar]
- Jordan P.C., Liu C., Raynaud P., Lo M.K., Spiropoulou C.F., Symons J.A., Beigelman L., Deval J. Initiation, extension, and termination of RNA synthesis by a paramyxovirus polymerase. PLOS Pathog. 2018;14:e1006889. doi: 10.1371/journal.ppat.1006889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp K., Kalayanov G., Ma H., Le Pogam S., Leveque V., Jiang W.R., Inocencio N., De Witte A., Rajyaguru S., Tai E., Chanda S., Irwin M.R., Sund C., Winqist A., Maltseva T., Eriksson S., Usova E., Smith M., Alker A., Najera I., Cammack N., Martin J.A., Johansson N.G., Smith D.B. 2'-deoxy-4'-azido nucleoside analogs are highly potent inhibitors of hepatitis C virus replication despite the lack of 2'-alpha-hydroxyl groups. J. Biol. Chem. 2008;283:2167–2175. doi: 10.1074/jbc.M708929200. [DOI] [PubMed] [Google Scholar]
- Klumpp K., Leveque V., Le Pogam S., Ma H., Jiang W.R., Kang H., Granycome C., Singer M., Laxton C., Hang J.Q., Sarma K., Smith D.B., Heindl D., Hobbs C.J., Merrett J.H., Symons J., Cammack N., Martin J.A., Devos R., Najera I. The novel nucleoside analog R1479 (4'-azidocytidine) is a potent inhibitor of NS5B-dependent RNA synthesis and hepatitis C virus replication in cell culture. J. Biol. Chem. 2006;281:3793–3799. doi: 10.1074/jbc.M510195200. [DOI] [PubMed] [Google Scholar]
- Kobinger G.P., Weiner D.J., Yu Q.C., Wilson J.M. Filovirus-pseudotyped lentiviral vector can efficiently and stably transduce airway epithelia in vivo. Nat. Biotechnol. 2001;19:225–230. doi: 10.1038/85664. [DOI] [PubMed] [Google Scholar]
- Lamunu M., Lutwama J.J., Kamugisha J., Opio A., Nambooze J., Ndayimirije N., Okware S. Containing a haemorrhagic fever epidemic: the Ebola experience in Uganda (October 2000-January 2001) Int. J. Infect. Dis. 2004;8:27–37. doi: 10.1016/j.ijid.2003.04.001. [DOI] [PubMed] [Google Scholar]
- Lo M.K., Jordan R., Arvey A., Sudhamsu J., Shrivastava-Ranjan P., Hotard A.L., Flint M., McMullan L.K., Siegel D., Clarke M.O., Mackman R.L., Hui H.C., Perron M., Ray A.S., Cihlar T., Nichol S.T., Spiropoulou C.F. GS-5734 and its parent nucleoside analog inhibit Filo-, Pneumo-, and Paramyxoviruses. Sci. Rep. 2017;7:43395. doi: 10.1038/srep43395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo M.K., Nichol S.T., Spiropoulou C.F. Evaluation of luciferase and GFP-expressing Nipah viruses for rapid quantitative antiviral screening. Antivir. Res. 2014;106:53–60. doi: 10.1016/j.antiviral.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby S.P., Gurley E.S. Epidemiology of henipavirus disease in humans. Curr. Top. Microbiol. Immunol. 2012;359:25–40. doi: 10.1007/82_2012_207. [DOI] [PubMed] [Google Scholar]
- Madrid P.B., Panchal R.G., Warren T.K., Shurtleff A.C., Endsley A.N., Green C.E., Kolokoltsov A., Davey R., Manger I.D., Gilfillan L., Bavari S., Tanga M.J. Evaluation of Ebola virus inhibitors for drug repurposing. ACS Infect. Dis. 2015;1:317–326. doi: 10.1021/acsinfecdis.5b00030. [DOI] [PubMed] [Google Scholar]
- Mendoza E.J., Racine T., Kobinger G.P. The ongoing evolution of antibody-based treatments for Ebola virus infection. Immunotherapy. 2017;9:435–450. doi: 10.2217/imt-2017-0010. [DOI] [PubMed] [Google Scholar]
- Mire C.E., Geisbert T.W., Feldmann H., Marzi A. Ebola virus vaccines - reality or fiction? Expert Rev. Vaccines. 2016;15:1421–1430. doi: 10.1080/14760584.2016.1178068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D.R., Zeuzem S., Andreone P., Ferenci P., Herring R., Jensen D.M., Marcellin P., Pockros P.J., Rodriguez-Torres M., Rossaro L., Rustgi V.K., Sepe T., Sulkowski M., Thomason I.R., Yoshida E.M., Chan A., Hill G. Balapiravir plus peginterferon alfa-2a (40KD)/ribavirin in a randomized trial of hepatitis C genotype 1 patients. Ann. Hepatol. 2012;11:15–31. [PMC free article] [PubMed] [Google Scholar]
- Nguyen N.M., Tran C.N., Phung L.K., Duong K.T., Huynh Hle A., Farrar J., Nguyen Q.T., Tran H.T., Nguyen C.V., Merson L., Hoang L.T., Hibberd M.L., Aw P.P., Wilm A., Nagarajan N., Nguyen D.T., Pham M.P., Nguyen T.T., Javanbakht H., Klumpp K., Hammond J., Petric R., Wolbers M., Nguyen C.T., Simmons C.P. A randomized, double-blind placebo controlled trial of balapiravir, a polymerase inhibitor, in adult dengue patients. J. Infect. Dis. 2013;207:1442–1450. doi: 10.1093/infdis/jis470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noton S.L., Deflube L.R., Tremaglio C.Z., Fearns R. The respiratory syncytial virus polymerase has multiple RNA synthesis activities at the promoter. PLoS Pathog. 2012;8:e1002980. doi: 10.1371/journal.ppat.1002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestereich L., Ludtke A., Wurr S., Rieger T., Munoz-Fontela C., Gunther S. Successful treatment of advanced Ebola virus infection with T-705 (favipiravir) in a small animal model. Antivir. Res. 2014;105:17–21. doi: 10.1016/j.antiviral.2014.02.014. [DOI] [PubMed] [Google Scholar]
- Qiu X., Wong G., Audet J., Bello A., Fernando L., Alimonti J.B., Fausther-Bovendo H., Wei H., Aviles J., Hiatt E., Johnson A., Morton J., Swope K., Bohorov O., Bohorova N., Goodman C., Kim D., Pauly M.H., Velasco J., Pettitt J., Olinger G.G., Whaley K., Xu B., Strong J.E., Zeitlin L., Kobinger G.P. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature. 2014;514:47–53. doi: 10.1038/nature13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A.S., Hitchcock M.J.M. Metabolism of antiviral nucleosides and nucleotides. In: Lafemina R.L., editor. Antiviral Research: Strategies in Antiviral Drug Discovery. ASM Press; Washington, DC: 2009. [Google Scholar]
- Reed L.J., Muench H. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 1938;27:493–497. [Google Scholar]
- Rennick L.J., de Vries R.D., Carsillo T.J., Lemon K., van Amerongen G., Ludlow M., Nguyen D.T., Yuksel S., Verburgh R.J., Haddock P., McQuaid S., Duprex W.P., de Swart R.L. Live-attenuated measles virus vaccine targets dendritic cells and macrophages in muscle of nonhuman primates. J. Virol. 2015;89:2192–2200. doi: 10.1128/JVI.02924-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A., Rollin P.E. Complete genome sequence of an Ebola virus (Sudan species) responsible for a 2000 outbreak of human disease in Uganda. Virus Res. 2005;113:16–25. doi: 10.1016/j.virusres.2005.03.028. [DOI] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Graham R.L., Menachery V.D., Gralinski L.E., Case J.B., Leist S.R., Pyrc K., Feng J.Y., Trantcheva I., Bannister R., Park Y., Babusis D., Clarke M.O., Mackman R.L., Spahn J.E., Palmiotti C.A., Siegel D., Ray A.S., Cihlar T., Jordan R., Denison M.R., Baric R.S. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.B., Kalayanov G., Sund C., Winqvist A., Maltseva T., Leveque V.J., Rajyaguru S., Le Pogam S., Najera I., Benkestock K., Zhou X.X., Kaiser A.C., Maag H., Cammack N., Martin J.A., Swallow S., Johansson N.G., Klumpp K., Smith M. The design, synthesis, and antiviral activity of monofluoro and difluoro analogues of 4'-azidocytidine against hepatitis C virus replication: the discovery of 4'-azido-2'-deoxy-2'-fluorocytidine and 4'-azido-2'-dideoxy-2',2'-difluorocytidine. J. Med. Chem. 2009;52:2971–2978. doi: 10.1021/jm801595c. [DOI] [PubMed] [Google Scholar]
- Smith D.B., Martin J.A., Klumpp K., Baker S.J., Blomgren P.A., Devos R., Granycome C., Hang J., Hobbs C.J., Jiang W.R., Laxton C., Le Pogam S., Leveque V., Ma H., Maile G., Merrett J.H., Pichota A., Sarma K., Smith M., Swallow S., Symons J., Vesey D., Najera I., Cammack N. Design, synthesis, and antiviral properties of 4'-substituted ribonucleosides as inhibitors of hepatitis C virus replication: the discovery of R1479. Bioorg Med. Chem. Lett. 2007;17:2570–2576. doi: 10.1016/j.bmcl.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Taylor R., Kotian P., Warren T., Panchal R., Bavari S., Julander J., Dobo S., Rose A., El-Kattan Y., Taubenheim B., Babu Y., Sheridan W.P. BCX4430-A broad-spectrum antiviral adenosine nucleoside analog under development for the treatment of Ebola virus disease. J. Infect. Public Health. 2016;9:220–226. doi: 10.1016/j.jiph.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner J.S., Khristova M.L., Sealy T.K., Vincent M.J., Erickson B.R., Bawiec D.A., Hartman A.L., Comer J.A., Zaki S.R., Stroher U., Gomes da Silva F., del Castillo F., Rollin P.E., Ksiazek T.G., Nichol S.T. Marburgvirus genomics and association with a large hemorrhagic fever outbreak in Angola. J. Virol. 2006;80:6497–6516. doi: 10.1128/JVI.00069-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner J.S., Paragas J., Dover J.E., Gupta M., Goldsmith C.S., Huggins J.W., Nichol S.T. Generation of eGFP expressing recombinant Zaire ebolavirus for analysis of early pathogenesis events and high-throughput antiviral drug screening. Virology. 2005;332:20–27. doi: 10.1016/j.virol.2004.10.048. [DOI] [PubMed] [Google Scholar]
- Van Rompay A.R., Norda A., Linden K., Johansson M., Karlsson A. Phosphorylation of uridine and cytidine nucleoside analogs by two human uridine-cytidine kinases. Mol. Pharmacol. 2001;59:1181–1186. doi: 10.1124/mol.59.5.1181. [DOI] [PubMed] [Google Scholar]
- Veljkovic V., Loiseau P.M., Figadere B., Glisic S., Veljkovic N., Perovic V.R., Cavanaugh D.P., Branch D.R. Virtual screen for repurposing approved and experimental drugs for candidate inhibitors of EBOLA virus infection. F1000Res. 2015;4:34. doi: 10.12688/f1000research.6110.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Deval J., Hong J., Dyatkina N., Prhavc M., Taylor J., Fung A., Jin Z., Stevens S.K., Serebryany V., Liu J., Zhang Q., Tam Y., Chanda S.M., Smith D.B., Symons J.A., Blatt L.M., Beigelman L. Discovery of 4'-chloromethyl-2'-deoxy-3',5'-di-O-isobutyryl-2'-fluorocytidine (ALS-8176), a first-in-class RSV polymerase inhibitor for treatment of human respiratory syncytial virus infection. J. Med. Chem. 2015;58:1862–1878. doi: 10.1021/jm5017279. [DOI] [PubMed] [Google Scholar]
- Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H.C., Larson N., Strickley R., Wells J., Stuthman K.S., Van Tongeren S.A., Garza N.L., Donnelly G., Shurtleff A.C., Retterer C.J., Gharaibeh D., Zamani R., Kenny T., Eaton B.P., Grimes E., Welch L.S., Gomba L., Wilhelmsen C.L., Nichols D.K., Nuss J.E., Nagle E.R., Kugelman J.R., Palacios G., Doerffler E., Neville S., Carra E., Clarke M.O., Zhang L., Lew W., Ross B., Wang Q., Chun K., Wolfe L., Babusis D., Park Y., Stray K.M., Trancheva I., Feng J.Y., Barauskas O., Xu Y., Wong P., Braun M.R., Flint M., McMullan L.K., Chen S.S., Fearns R., Swaminathan S., Mayers D.L., Spiropoulou C.F., Lee W.A., Nichol S.T., Cihlar T., Bavari S. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren T.K., Wells J., Panchal R.G., Stuthman K.S., Garza N.L., Van Tongeren S.A., Dong L., Retterer C.J., Eaton B.P., Pegoraro G., Honnold S., Bantia S., Kotian P., Chen X., Taubenheim B.R., Welch L.S., Minning D.M., Babu Y.S., Sheridan W.P., Bavari S. Protection against filovirus diseases by a novel broad-spectrum nucleoside analogue BCX4430. Nature. 2014;508:402–405. doi: 10.1038/nature13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch S.R., Guerrero L.W., Chakrabarti A.K., McMullan L.K., Flint M., Bluemling G.R., Painter G.R., Nichol S.T., Spiropoulou C.F., Albarino C.G. Lassa and Ebola virus inhibitors identified using minigenome and recombinant virus reporter systems. Antivir. Res. 2016;136:9–18. doi: 10.1016/j.antiviral.2016.10.007. [DOI] [PubMed] [Google Scholar]
- Zhang L., Bukreyev A., Thompson C.I., Watson B., Peeples M.E., Collins P.L., Pickles R.J. Infection of ciliated cells by human parainfluenza virus type 3 in an in vitro model of human airway epithelium. J. Virol. 2005;79:1113–1124. doi: 10.1128/JVI.79.2.1113-1124.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.