Abstract
Use of proteomic strategies to identify a risk classifier that estimates probability of distant recurrence in early-stage hormone receptor (HR)-positive breast cancer is relevant to physiological cellular function and therefore to intrinsic tumor biology. We used a 298-sample retrospective training set to develop an immunohistochemistry-based novel risk classifier called CanAssist-Breast (CAB) which combines 5 prognostically relevant biomarkers and 3 clinico-pathological parameters to arrive at probability of distant recurrence within 5 years from diagnosis. Five selected biomarkers, namely, CD44, ABCC4, ABCC11, N-cadherin, and pan-cadherin, were chosen based on their role in tumor metastasis. The chosen biomarkers represent the hallmarks of cancer and are distinct from other proliferation and gene expression–based prognostic signatures. The 3 clinico-pathological parameters integrated into the machine learning–based CAB algorithm are tumor size, tumor grade, and node status. These features are used to calculate a “CAB risk score” that classifies patients into low- or high-risk groups and predicts probability of distant recurrence in 5 years. Independent clinical validation of CAB in a retrospective study comprising 196 patients indicated that distant metastasis-free survival (DMFS) was significantly different in the 2 risk groups. The difference in DMFS between the low- and high-risk categories was 19% in the validation cohort (P = .0002). In multivariate analysis, CAB risk score was the most significant independent predictor of distant recurrence with a hazard ratio of 4.3 (P = .0003). CanAssist-Breast is a precise and unique machine learning–based proteomic risk-classifier that can assist in risk stratification of patients with early-stage HR+ breast cancer.
Keywords: early breast cancer, prognosis, recurrence risk classification, immunohistochemistry, machine learning
Introduction
The molecular analysis of breast cancer has demonstrated the existence of several different subtypes such as estrogen receptor (ER)-positive or negative, progesterone receptor (PR)-positive or negative human epidermal growth factor receptor 2 (HER2)-positive or negative or triple negative (ER−, PR−, and HER2−) that present with varied clinical behavior and response to therapy.1 Several prognostic assays that are based on gene expression profiling of tumor tissue complement these molecular subtypes and address certain unmet clinical needs, such as the identification of patient subgroups with low risk of developing distant metastasis who can be spared adjuvant chemotherapy.2-5 These tests focus primarily on proliferation and do not consider the role of the tumor microenvironment and cross-talk between various signaling pathways in progression of disease.6 Furthermore, transcriptional abundance of a gene does not necessarily correlate with its protein expression,7 and posttranslational modifications of proteins are not captured by gene expression analysis. Qualitative and quantitative examination of protein expression in a cell allows the study of specific cellular responses and functions, including the visual examination of proteins in various cellular locations by immunohistochemistry (IHC). All of this is critical to identification of novel drug targets, the ultimate goal of personalized breast cancer therapeutics.
With this goal in mind, we identified and validated a novel proteomic risk-classifier of metastasis in early-stage ER+ breast cancer. We used the hallmarks of cancer8,9 as a guideline in our marker selection strategy to shortlist markers with prognostic relevance in breast cancer progression that regulated 1 of the 6 critical hallmarks. We selected several such markers and studied the association between their expression and clinical outcome with respect to distant metastasis in early-stage hormone receptor–positive (HR+) breast cancer. We shortlisted 5 markers and combined them with 3 clinico-pathological parameters—tumor size, tumor grade, and node status in a binary classifier that predicts risk of distant recurrence within 5 years. We used both biomarkers and clinico-pathological parameters as studies have shown that combining the 2 enables more accurate prediction of prognosis,10-12 and therefore better clinical decision making. Here, we describe the development, pilot clinical validation, and independent prognostic ability of a novel biomarker based-risk classifier reflective of the metastatic potential of the tumor in early-stage HR+ breast cancer. This classifier, called CanAssist-Breast (CAB), predicts risk of distant metastasis within 5 years.
Methods
Ethics statement
This multicentric study was performed in accordance with the Declaration of Helsinki and approved by the Institutional Review Board (IRB) and/or Institutional Ethics Committee (IEC) of participating hospitals. The study was performed with the approval of Bangalore Ethics Committee (registration number: ECR/87/Indt/KA/2013), an independent ethics committee registered with Central Drugs Standard Control Organization, Government of India. Patient information was anonymized and de-identified prior to analysis.
Patients and tumor samples
Inclusion criteria for the study were as follows:
Hormone receptor–positive tumors;
Stage I, II, III disease;
Information available about tumor type and treatment taken. This included information on age and calendar year of diagnosis, type of surgery, tumor (size, tumor grade, histopathologic type), node status, details of radiation therapy, details of hormone therapy taken including drug prescribed, and duration of therapy. Details of chemotherapy including type of regimen, number of cycles, dosage and dates of treatment.
Minimum 5 years of follow-up since diagnosis. Clinical follow-up included dates and methods of annual follow-up, or distant recurrence—detection and treatment, and date of last visit, if death—cause and date of death.
At least 30% or more invasive tumor content in the formalin-fixed paraffin-embedded (FFPE) block for adequate assessment of IHC staining on several sections;
Less than 50% necrosis and hemorrhagic content in the FFPE block.
Exclusion criteria for the study were as follows:
Hormone receptor–negative tumors;
Less than 5 years clinical follow-up since diagnosis unless a distant recurrence occurred within 5 years;
Incomplete information about type of tumor and treatment taken;
Evidence of local recurrence including chest wall, ipsilateral, or contralateral tumors;
Patients treated with neoadjuvant therapy;
Samples with <30% invasive tumor content;
Improperly fixed tumors;
Samples with >50% necrotic tissue content;
Patients with metastatic disease at diagnosis (M1).
The FFPE blocks of 298 patients with Stage I, II, and III breast carcinoma, ER+/PR+, HER2+/−, with a minimum of 5-year follow-up and containing at least 30% tumor were used for development of CAB, whereas an additional 196 samples were used for clinical validation.
Study end points
The event of interest in the study was distant recurrence. If a recurrence occurred within 5 years, the date of recurrence was considered the end of the study for that patient. If no recurrence occurred within 5 years, the patient was considered disease free at the end point. Any distant recurrences after 5 years were censored for the purpose of the study. Success criteria for the study were defined as statistically significant separation of patients into low-risk or high-risk for recurrence groups in Kaplan-Meier survival analysis.
Sample size estimation
Sample size needed to validate the proteomic risk-classifier was estimated based on relative risk using data from the training set after accounting for overfitting. To achieve a power of 80%, we estimated a sample size of at least 138 patients with 24 events.
IHC staining
The IHC analysis is semiautomated and performed as follows. The FFPE tissues are sectioned into 3- to 5-µ slices using a Leica microtome (#RM2125RTS). Poly-l-Lysine–coated slides (PathnSitu, India) were used for taking sections. The sections are fixed on glass slides by placing them in a hot-air oven (Apollo Scientific, India) at 55°C for 1 hour. The slides are then deparaffinized with xylene (Fisher Scientific, USA) solution twice for 15 minutes each. Slides are rehydrated by washing twice with 100% alcohol for 5 minutes followed by 2 washes with 70% alcohol for 5 minutes and finally with demineralized water (Nice Cat # D1505) for 5 minutes. Antigen retrieval is performed for each antibody as per the manufacturer’s instructions. Following antigen retrieval, slides are cooled completely to room temperature in the same buffer. On attaining room temperature, the slides are washed in demineralized water for 5 minutes. After wiping extra moisture on the slide with a tissue, the tumor section is marked with a PAP pen. The rest of the steps are performed using the Novolink Polymer Secondary Kit (Leica, Biosystems, Germany; RE-7280K). Peroxidase block is added to each slide and incubated for 5 minutes. Slides are washed with wash buffer (10 mM TBS-Tween 20, pH 7.4) twice, for 5 minutes each. After washing, the protein block is added and slides are incubated for 5 minutes. Slides are washed with wash buffer twice, for 5 minutes each. Dilution of primary antibody is performed as per the manufacturer’s instructions for all antibodies. All antibodies are obtained from commercial vendors (details in Supplementary Methods). Slides are incubated for 1 hour in a humidifying chamber with antibody. After primary antibody incubation, slides are washed with wash buffer twice, for 5 minutes each. Postprimary solution is added to the slides, and incubated for 30 minutes, followed by 2 washes with wash buffer as described previously. Following this, slides are incubated with polymer for 30 minutes and then washed twice with wash buffer. Peroxidase activity is developed using 3,3′-diaminobenzidine (DAB) working solution for 5 minutes, following which the slides are rinsed with demineralized water for 2 minutes. Sections are counterstained with hematoxylin (Fisher Scientific, USA) for 8 minutes and rinsed in demineralized water for 8 minutes. The slides are subsequently dehydrated with 70%, 95%, and 100% alcohol, each for 5 minutes. They are dried at room temperature and then incubated in xylene for 5 minutes. Slides are dried and mounted with D.P.X. Mountant (Nice Chemicals, India; Product # D30475).
IHC grading
The IHC grading was performed as follows:
All markers were assessed for % and intensity of the staining on membrane or cytoplasm or nucleus.
Percentage of cells stained can range from 0% to 100%. Percentage staining is derived by assessing number of tumor cells stained in roughly about 100 cells per field. The percentage staining may vary from field to field in a given slide and hence an average of all the scores after scanning the entire slide is given as the final % value.
Intensity of staining can vary from 1 to 3: 1 being weak (light brown), 2 being intermediate (medium brown), and 3 being strong intensity (dark brown). The intensity of staining observed in most of the tumor tissue section was recorded (eg, if 15% of the tissue was found to be stained with intensity 1, and 85% with intensity 2, the recorded intensity was 2).
Risk score generation
Each biomarker that is part of the CAB classifier is graded quantitatively on a scale of 0 to 100 for % of staining on membrane (CD44, ABCC4, and ABCC11) or cytoplasm (N-cadherin and pan-cadherin) along with intensity of staining (scale of 0-3) by trained pathologists. The % staining or intensity of the 5 biomarkers is then used by the machine learning–based classifier along with the values of 3 clinical parameters to compute the risk score. Clinical parameters are used by the risk classifier as categorical variables—tumor size T1, T2, or T3; tumor grade 1, 2, or 3; and node status N0, N1, N2, or N3. The CAB risk classifier generates risk scores on a scale of 0 to 100. Using a prespecified threshold of 15.5, patients are classified into low-risk or high risk for recurrence.
Patient demographics
Patients were categorized into various subgroups based on factors such as age (<50, >50), tumor size (T1, T2, T3+T4), tumor grade (1—well differentiated, 2—moderately differentiated, 3—poorly differentiated), and node status (no nodes positive N0, 1-3 nodes positive N1, >4 nodes positive N2+N3) to study patient demographics.
Results
Selection of breast cancer prognostic markers
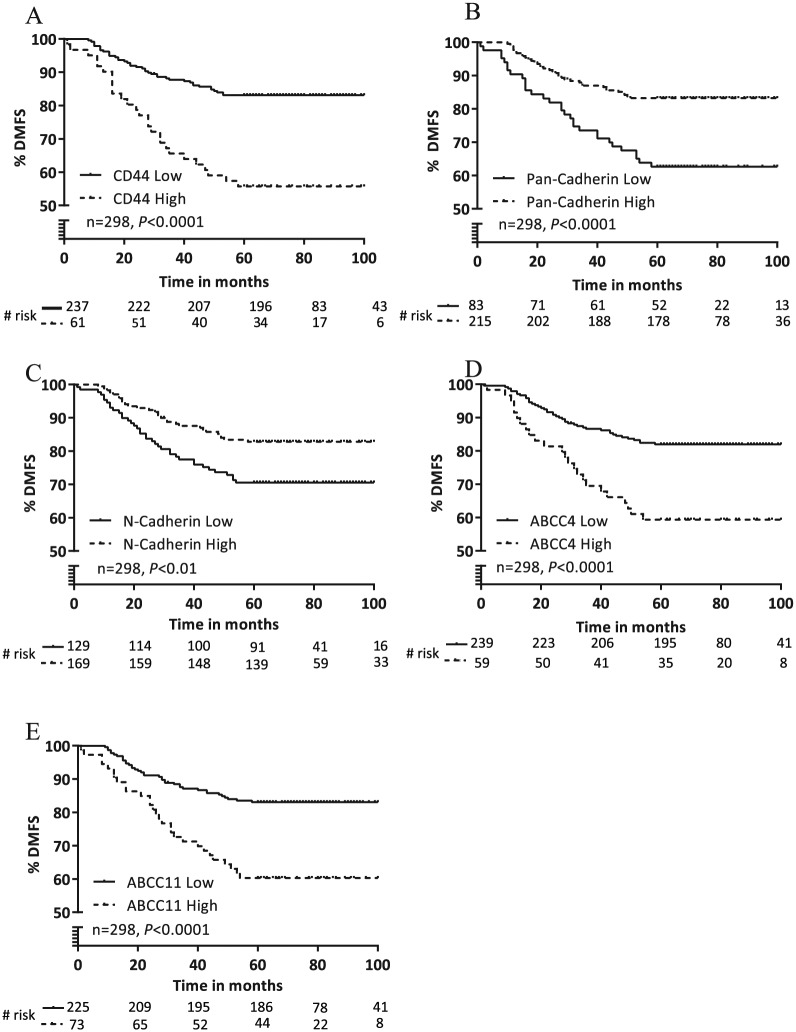
Cancer metastasis involves multiple steps, and the critical steps with some of the key biomarkers involved in each step are listed in Table 1. We chose to examine some of the biomarkers (Supplementary Methods) listed in Table 1 as representatives of the steps involved in metastasis. We selected biomarkers which were not part of other risk stratifying tests and had robust proteomic tools available to examine the expression. We analyzed these markers by performing IHC in our 298 sample training set (Table 3) that comprised samples from patients who were recurrence free at 5 years (n = 230) or recurred at a distant site within 5 years (n = 68). Each biomarker was assessed for % and intensity of staining in the membrane or cytoplasm or nucleus depending on the biomarker. We prioritized the biomarkers by univariate ranking based on absolute value of Pearson correlation coefficient between IHC expression of the marker and outcome (recurrence or no recurrence) and chose the top 5 markers (Table 2). In order of the rank assigned by Pearson correlation, we tested the ability of these markers to prognosticate patients into distinct distant metastasis-free survival (DMFS) groups by Kaplan-Meier survival analysis. To this end, we divided the training set into 2 groups based on low or high IHC expression for each biomarker and correlated the differential expression of each marker with DMFS (Figure 1). Low or high expression of the top 5 markers CD44, ABCC4, ABCC11, N-cadherin, and pan-cadherin showed significant correlation with DMFS (Figure 1A to E). We found that high membrane expression of the cancer stem cell marker CD44 correlated with poor metastasis-free survival (Figure 1A). Patients with high membrane expression of CD44 (DMFS: 55%) were significantly separated from patients with low membrane expression of CD44 (DMFS: 83%). The average recurrence rate in patients with low CD44 expression was 16%, compared with 44% in patients with high CD44 expression—a 2.75-fold increase in recurrence rate. Similarly, we found that high membrane expression of the 2 adenosine triphosphate (ATP) drug transporters, ABCC4 and ABCC11, also correlated with worse metastasis-free survival (Figure 1D and E). Patients with high membrane expression of ABCC4 (DMFS: 59%) or ABCC11 (DMFS: 60%) had 2.2-fold higher rate of distant recurrence compared with patients with low membrane expression of these proteins. We also observed that low expression of the cadherin markers, N-cadherin and pan-cadherin, were associated with poor prognosis in the training set (Figure 1B and C). Patients with low expression of N-cadherin (DMFS: 70%) and pan-cadherin (DMFS: 62%) had ~1.7-fold higher rate of distant metastasis compared with patients with high expression of N-cadherin (DMFS: 82%) and pan-cadherin (DMFS: 83%). Representative images of low and high expression of each of the 5 biomarkers are provided in Supplementary Material (Figure S1).
Table 1.
The critical steps and associated biomarkers involved in cancer progression.
| Hallmark of cancer | Biomarker |
|---|---|
| Self-sufficiency in growth signals | Ki67, FOXA1, IFITM1, GATA3, c-Myc, IGFBP3, FOXP1, FOXP3 |
| Insensitivity to antigrowth signals | ABCG2, ABCC4, ABCC11, Nrf2, PI3K, Akt |
| Evading apoptosis | MAGE-A9, MAGE-A11, BAG1, Apaf1, BCL2 |
| Limitless replicative potential | CD44, CD24, SOX2, Oct3, NANOG, NESTIN, KLF4, ALDH1A1, CD133, CD90 (THY-1), CD15, CD61, hTERT |
| Sustained angiogenesis | HIF1α, HIF2α, XBP1, TIE2, FGF, ANG1, VEGFR1, VEGFR2, CXCR1, MMP8 |
| Tissue invasion and metastasis | P-cadherin, N-cadherin, E-cadherin, β-catenin, APC, EpCAM, FOXA1, KLK6, CxCR4, CD147, HSP70, Integrinb-6, EGFR |
Table 3.
The demographics and patient characteristics of the training and validation cohorts.
| Training cohort (n = 298) | Validation cohort (n = 196) | |||
|---|---|---|---|---|
| No. of samples | % of samples | No. of samples | % of samples | |
| Age | ||||
| <50 | 142 | 47.6 | 95 | 48.5 |
| >50 | 156 | 52.3 | 101 | 51.5 |
| Tumor size | ||||
| T1 | 41 | 13.7 | 23 | 11.7 |
| T2 | 230 | 77.1 | 160 | 81.6 |
| T3+T4 | 27 | 9.1 | 13 | 6.6 |
| Tumor grade | ||||
| Well differentiated | 22 | 7.3 | 18 | 9.2 |
| Moderately differentiated | 150 | 50.3 | 115 | 58.6 |
| Poorly differentiated | 126 | 42.2 | 63 | 32.1 |
| Node status | ||||
| N0 | 122 | 40.9 | 95 | 48.5 |
| N1 | 107 | 35.9 | 63 | 32.1 |
| N2+N3 | 69 | 23.1 | 38 | 19.4 |
Table 2.
The ranking of the top 5 biomarkers by Pearson correlation coefficient.
| Marker name | Pearson correlation coefficient |
|---|---|
| CD44 | 0.26 |
| Pan-cadherin | 0.24 |
| N-cadherin | 0.21 |
| ABCC11 | 0.20 |
| ABCC4 | 0.19 |
Figure 1.
Correlation of the expression of the top 5 ranked biomarkers with DMFS in the training set. (A) Kaplan-Meier survival analysis of distant recurrence in the training cohort (n = 298) analyzed by low versus high staining of CD44 in the cell membrane. (B) Kaplan-Meier survival analysis of distant recurrence in the training cohort (n = 298) analyzed by low versus high staining of ABCC4 in the cell membrane. (C) Kaplan-Meier survival analysis of distant recurrence in the training cohort (n = 298) analyzed by low versus high staining of ABCC11 in the cell membrane. (D) Kaplan-Meier survival analysis of distant recurrence in the training cohort (n = 298) analyzed by low versus high staining of N-cadherin in the cell cytoplasm. (E) Kaplan-Meier survival analysis of distant recurrence in the training cohort (n = 298) analyzed by low versus high staining of pan-cadherin in the cell cytoplasm.
Classifier development
The top 5 biomarkers identified were integrated with 3 well-studied clinico-pathological parameters—tumor size, tumor grade, and node status into a classifier that produces a risk score (on a scale of 0-100) to classify patients into low- or high-risk groups for distant recurrence. The classifier was developed using the 298-sample training set, and the clinical characteristics of the patients in this cohort are defined in Table 3. The training set is representative of the demographics of breast cancer in the Indian population and comprises 47% patients under the age of 50 and 77% patients with T2 disease.13
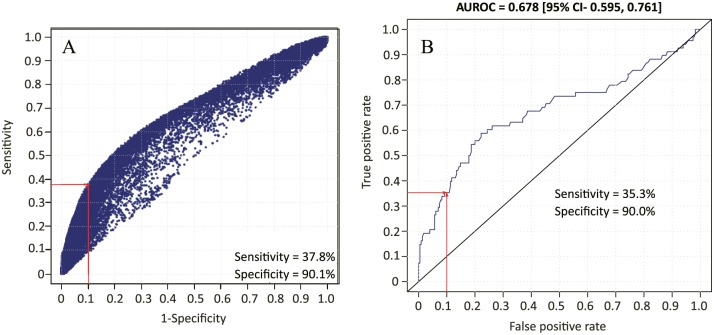
To choose the best method to develop the classifier, we evaluated multiple machine learning techniques including (1) Support Vector Machine (SVM) with linear and Radial Basis Function (RBF) kernel, (2) Random Forest (RF), (3) Elastic Net (ESL), (4) multilayer perceptron (MLP), and (5) normal mixture modeling. Classifier selection was facilitated using ROG (receiver operating graph) whereby each classifier was represented by its cross-validation sensitivity versus 1-specificity. The selection criterion was maximum achievable sensitivity at specificity >90%. The RBF-SVM (Figure 2A) proved superior to the other classifiers (Supplementary Figure S2) according to the chosen criteria of 90% specificity and maximum sensitivity and was therefore selected for classifier assessment. The Supplementary Figure S2 shows ROG for ESL, RF, MLP, and linear SVM. Normal mixture model performed poorly (data not shown). The chosen RBF-SVM classifier was assessed further by application of cross-validation criteria by performing repeated 10-fold nested cross-validation (NCV). The NCV performance was analyzed by means of a receiver operating characteristic plot which determined that the classifier performance was acceptable and all parameters were within the predefined limits, including the achievement of specificity >90% and area under the curve of 0.67 (Figure 2B). The threshold for discrimination between low and high risks was set at the risk score of 15.5 that corresponded to a 9% probability of distant recurrence (Figure S3).
Figure 2.
Classifier development. (A) Receiver operating graph analysis of various RBF-SVM classifiers that were tested. For each potential classifier, we plotted cross-validation sensitivity versus 1-specificity. The intersection of the red lines indicates the chosen classifier. (B) Receiver operating characteristic analysis of 10-fold nested cross-validation for distant metastasis-free survival by the chosen RBF-SVM classifier. RBF-SVM indicates Radial Basis Function-Support Vector Machine.
Prognostic performance of the CAB classifier
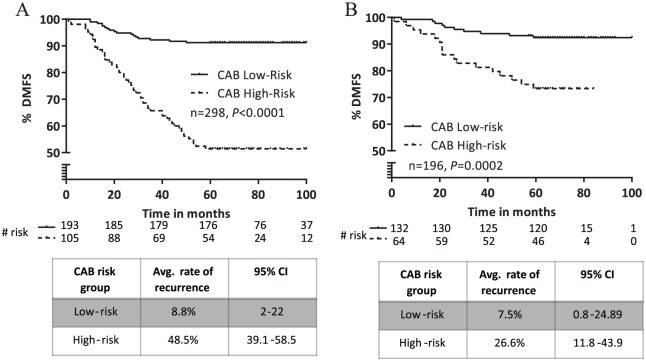
The CAB risk score in training set ranged from 0 to 100. Patients in the training set were classified into low- or high-risk groups using the CAB risk score. Accordingly, 193 patients (64%) were called low risk and 105 patients (36%) were called high risk (Figure 3A). The Kaplan-Meier survival curve showed a statistically significant difference in DMFS between the low- and high-risk groups (P < .0001). The 5-year probability estimates of DMFS in the low- and high-risk groups were 91% and 51%, respectively. The average rate of distant recurrence was ~5.5 times higher in the high-risk group (48.5%) versus the low-risk group (8.8%). This result demonstrates that the CAB risk score can significantly differentiate patients at low or high risk of distant metastasis. We tested the association between the CAB risk score and distant metastasis using the Cox proportional hazards model–based multivariate analysis (Table 4). Multivariate analysis comparing the performance of the CAB risk score with respect to other prognostic clinical variables found that the CAB risk score correlated significantly with distant metastasis (hazard ratio: 6.82, 95% confidence interval [CI]: 3.9-11.8, P < .0001), whereas the other tested clinical variables (age, ER+, PR+, and disease stage) did not show significant correlation (Table 4).
Figure 3.
Risk classification by CAB. (A) Kaplan-Meier plot of distant recurrence in the training set (n = 298) stratified by CAB into low- or high-risk groups. (B) Kaplan-Meier survival analysis of distant recurrence in the validation set (n = 196) stratified by CAB into low- or high-risk groups. CAB indicates CanAssist-Breast.
Table 4.
Multivariate analysis of the CAB risk score and clinico-pathological parameters for distant metastasis-free survival in the training set.
| Covariate | Hazard ratio | P value | 95% CI |
|---|---|---|---|
| Age | 0.69 | .15 | 0.42–1.12 |
| ER+ | 1.49 | .19 | 0.81–2.72 |
| PR+ | 1.45 | .13 | 0.89–2.36 |
| Stage | 1.45 | .42 | 0.46–4.30 |
| CAB risk score | 6.82 | <.0001 | 3.92–11.84 |
Abbreviations: CAB, CanAssist-Breast; CI, confidence interval; ER, estrogen receptor; PR, progesterone receptor.
Independent validation of the CAB classifier
The CAB classifier was independently validated in a cohort of 196 patients. Patient demographics and clinical characteristics of the validation set are provided in Table 3. Similar to the training set, the validation cohort also had ~48% patients under 50 years of age. Approximately 81% patients had T2 disease and 48% patients were node negative at diagnosis. The CAB risk score ranged from 2.1 to 100 in the validation set. Kaplan-Meier survival analysis of CAB risk score discrimination in the validation set demonstrated statistically significant difference in DMFS between the low- and high-risk groups (P = .0002; Figure 3B). About 132 patients (68%) were called low risk and 64 patients (32%) were called high risk by CAB in the validation set. The 5-year probability estimates of DMFS in the low- and high-risk groups were 92% and 73%, respectively. The average rate of distant recurrence was ~3.5 times higher in the high-risk group (26.6%) versus the low-risk group (7.5%). Multivariate analysis of the validation set demonstrated that the CAB risk score correlated significantly with distant metastasis (hazard ratio: 4.37, 95% CI 1.99-9.61, P = .0003), whereas the other tested clinical parameters including age, % ER staining and disease stage did not show significant correlation (Table 5). % PR staining also showed significant correlation with distant metastasis; however, the CAB risk score exhibited stronger association with distant metastasis and was shown to be the best independent predictor of prognosis in the validation set (Table 5). Taken together, these results indicate that the CAB risk classifier can accurately place patients into low- or high-risk groups based on their probability of distant recurrence within 5 years.
Table 5.
Multivariate analysis of the CAB risk score and clinico-pathological parameters for distant metastasis-free survival in the validation set.
| Covariate | Hazard ratio | P value | 95% CI |
|---|---|---|---|
| Age | 1.25 | .56 | 0.58–2.69 |
| ER+ | 0.59 | .36 | 0.19–1.81 |
| PR+ | 2.50 | .02 | 1.11–5.6 |
| Stage | 1.06 | .92 | 0.31–3.57 |
| CAB risk score | 4.37 | .0003 | 1.99–9.61 |
Abbreviations: CAB, CanAssist-Breast; CI, confidence interval; ER, estrogen receptor; PR, progesterone receptor.
Discussion
In this article, we describe the development and clinical validation of CAB—a risk-classifier in patients with early-stage HR+ breast cancer that measures the protein expression of 5 biomarkers and uses clinical information from 3 clinico-pathological parameters, tumor size, tumor grade, and node status, to arrive at a risk estimate for distant recurrence within 5 years. Studies have shown that random gene expression signatures can be associated with breast cancer outcome owing to the confounding effect of proliferation genes which comprise more than 50% of the breast cancer transcriptome.14 It is therefore critical to select biomarkers from pathways other than proliferation and delineate their role in cancer progression.14 We took a hypothesis driven approach to solve this problem by choosing biomarkers that characterize several important biological processes in cancer. CAB uses the IHC-based expression of 5 biomarkers, including CD44, ABCC4, ABCC11, N-cadherin, pan-cadherin, that regulate important steps in metastasis of cancer. The CD44-high–expressing cells within a breast tumor are breast cancer stem cells that fingerprint aggressive disease15 by effecting cancer stem cell self-renewal and loss of cell adhesion.16,17 Altered expression of the 2 ATP drug transporters, ABCC4 and ABCC11, can lead to chemotherapy drug resistance, and therefore poorer overall survival18 by causing insensitivity to antigrowth signals.18,19 Altered expression of the cadherin proteins are known to be associated with the epithelial-mesenchymal transition of cancer cells that facilitates invasion of the primary tumor into distant secondary sites.20,21
Studies have also shown that using the nominal P value from a Cox proportional hazards analysis to develop a metastatic signature is potentially misleading14; we therefore used machine learning technology22 to build CAB—a binary risk classifier that differentiates patients into distinct low- and high-risk groups for distant recurrence with >90% specificity. Analysis of the performance of CAB in the training set showed ~40% risk difference between the low- and high-risk groups. Prognostic classifiers display better performance in the training set owing to model overfitting23; therefore, we performed an independent validation study to confirm the prognostic value of CAB. Our validation study showed that an absolute difference of 19% in DMFS between the CAB low- and high-risk groups, with a 3.5-fold higher rate of distant recurrences in patients called for recurrence. CanAssist-Breast was also the best independent predictor of distant metastasis in a multivariate analysis that compared it with other prognostic clinical parameters (hazard ratio: 4.3, P = .0003).
It is established that the messenger RNA (mRNA) and protein levels of a marker may not correlate,7 complicating the prognostic relevance of gene expression–based signatures. Studies examining HER2 expression by IHC and the 21-gene assay have shown that relying on mRNA expression alone could lead to false-negative results in up to 12% of patients with breast cancer.24 Various biomarkers including CD44 and ABCC11 have been shown to exhibit differences in protein and mRNA level.25,26 Our approach of measuring protein expression using the gold standard IHC technique overcomes these limitations. Moreover, as opposed to gene expression, measuring protein expression also leads to identification of potentially druggable targets. CD44, a component of CAB, is currently in clinical trials for various cancers such as lung and colorectal cancer.10 Its role as a potential therapeutic target in breast cancer could be the subject of further investigation.
This study has some limitations. First, ours was a study performed on patients treated with chemotherapy. Chemotherapy benefit rates in early-stage breast cancer are known to be in the range of 3% to 5%27,28 and may confound the analysis of DMFS. A future study to validate the performance of CAB in a cohort patients treated with hormone therapy alone is ongoing and would address this limitation adequately. Second, the size of validation cohort should ideally be much larger than the size of the training cohort, which is not the case in our pilot validation study. Further larger studies required to confirm these results are currently ongoing and will yield a much larger sample size in the near future. Finally, to provide level I evidence of utility as a prognostic classifier, CAB needs to be validated in samples from a suitable randomized controlled trial29 such as the validation of the 21-gene assay in samples from the NSABP-B20 study.30
The clinical utility of a risk classifier comes from its predictive proficiency which is demonstrated by the ability of the test to predict benefit of chemotherapy.31 We are currently conducting a study to test the ability of CAB to predict benefit of chemotherapy.
Based on the pilot validation study described in this article, we believe CAB adds significant value to predicting prognosis in patients with early-stage HR+ breast cancer. In conclusion, here we present CAB—a prognostic risk classifier that (1) uses IHC to interrogate protein expression of various hallmarks of cancer, (2) was developed using machine learning, and (3) and is the only risk classifier validated in a cohort of Indian patients.
Supplemental Material
Supplemental material, uomf_june_25_2018_supplementary_material for Development of a Novel Proteomic Risk-Classifier for Prognostication of Patients With Early-Stage Hormone Receptor–Positive Breast Cancer by Charusheila Ramkumar, Ljubomir Buturovic, Sukriti Malpani, Arun Attuluri Kumar, Chetana Basavaraj, Chandra Prakash, Lekshmi Madhav, Dinesh Chandra Doval, Anurag Mehta and Manjiri M Bakre in Biomarker Insights
Acknowledgments
The authors thank Ms S. Kanaldekar, Dr N. Krishnamoorthy, Dr N. Naidu, Ms Prathima R and Mr. Dinesh Babu for help with experimentation and pathological assessment. They also thank Dr P. Patil of Manipal Hospital, Bangalore and Dr S. Gupta of Tata Memorial Hospital, Mumbai for insightful discussions, Dr J. Jain of Sapien Biosciences for help with sample acquisition of Apollo Hospitals. They acknowledge the contribution of all clinicians, study co-ordinators and hospitals who participated in the development and validation study including; Dr P. Patil and Dr S. Mishra, Manipal Hospital; Dr R. Ananthkrishnan and Dr Rajkumar, GKNM Hospital, Coimbatore; Dr Karmarkar and Dr A. Chitale, Jaslok Hospital, Mumbai, Dr D G Vijay, HCG Ahmedabad; R. Kumar and Dr M Ghosh, HCG, Bangalore; Dr S. Mathews, Bangalore Baptist Hospital; Dr A. Brooks, Drexel Medical Centre, Philadelphia; Dr A. Pais and Dr Poovamma, Mazumdar Shaw Cancer Hospital, Bangalore; Dr H. Chaturvedi and Dr U. Mukherjee, Max Super speciality Hospital, Delhi; Dr A. Bapat, Virtua Hospital, Voorhees.
Footnotes
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The company received private funding for the research, authorship, and/or publication of this article.
Declaration of conflicting interests:The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: All authors except D.C.D. and A.M. are current or former employees/consultants at OncoStem Diagnostics Private Limited which has developed the CanAssist-Breast risk classifier. M.M.B. and C.R. are co-inventors on a patent application related to this article. All other authors have no other competing interests to declare.
Author Contributions: MMB conceived and designed the study. CR, SM and MMB analyzed, interpreted the data and drafted the manuscript. CR and LB made all the figures for the manuscript. LB performed all the statistical analysis. AKA, CP, and LM were involved in data acquisition and analysis. C.B. performed all the histopathological analysis. DCD and AM participated in clinical discussions and helped with the clinical study.
References
- 1. Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. [DOI] [PubMed] [Google Scholar]
- 2. Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. http://www.ncbi.nlm.nih.gov/pubmed/15591335. [DOI] [PubMed] [Google Scholar]
- 3. Buyse M, Loi S, van’t Veer L, et al. Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst. 2006;98:1183–1192. [DOI] [PubMed] [Google Scholar]
- 4. Bernard PS, Parker JS, Mullins M, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dubsky P, Brase JC, Jakesz R, et al. The EndoPredict score provides prognostic information on late distant metastases in ER+/HER2− breast cancer patients. Br J Cancer. 2013;109:2959–2964. http://www.nature.com/doifinder/10.1038/bjc.2013.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Karagiannis GS, Goswami S, Jones JG, Oktay MH, Condeelis JS. Signatures of breast cancer metastasis at a glance. J Cell Sci. 2016;129:1751–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Crameri R, Schulz-Knappe PZH. The future of post-genomic biology at the proteomic level: an outlook. Comb Chem High Throughput Screen. 2005;8:807–810. [DOI] [PubMed] [Google Scholar]
- 8. Hanahan D, Weinberg R. The hallmarks of cancer. Cell. 2000;100:57–70. [DOI] [PubMed] [Google Scholar]
- 9. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. [DOI] [PubMed] [Google Scholar]
- 10. Sahin IH, Klostergaard J. CD44 as a drug delivery target in human cancers: where are we now? Expert Opin Ther Targets. 2015;19:1587–1591. [DOI] [PubMed] [Google Scholar]
- 11. Zhao X, Rødland EA, Sørlie T, et al. Systematic assessment of prognostic gene signatures for breast cancer shows distinct influence of time and ER status. BMC Cancer. 2014;14:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sestak I, Buus R, Cuzick J, et al. Comparison of the performance of 6 prognostic signatures for estrogen receptor-positive breast cancer: a secondary analysis of a randomized clinical trial. JAMA Oncol. 2018;4:545–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ambroise M, Ghosh M, Mallikarjuna VS, Kurian A. Immunohistochemical profile of breast cancer patients at a tertiary care hospital in South India. Asian Pac J Cancer Prev. 2011;12:625–629. [PubMed] [Google Scholar]
- 14. Venet D, Dumont JE, Detours V. Most random gene expression signatures are significantly associated with breast cancer outcome. PLoS Comput Biol. 2011;7:e1002240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu R, Wang X, Chen GY, et al. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med. 2007;356:217–226. [DOI] [PubMed] [Google Scholar]
- 16. Thapa R, Wilson GD. The importance of CD44 as a stem cell biomarker and therapeutic target in cancer. Stem Cells Int. 2016;2016:2087204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kittaneh M, Montero AJ. Molecular profiling for breast cancer: a comprehensive review. Biomark Cancer. 2013;5:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fletcher JI, Williams RT, Henderson MJ, Norris MD, Haber M. ABC transporters as mediators of drug resistance and contributors to cancer cell biology. Drug Resist Updat. 2016;26:1–9. [DOI] [PubMed] [Google Scholar]
- 19. Chen Z, Shi T, Zhang L, et al. Mammalian drug efflux transporters of the ATP binding cassette (ABC) family in multidrug resistance: a review of the past decade. Cancer Lett. 2016;370:153–164. [DOI] [PubMed] [Google Scholar]
- 20. Cowin P, Rowlands TM, Hatsell SJ. Cadherins and catenins in breast cancer. Curr Opin Cell Biol. 2005;17:499–508. [DOI] [PubMed] [Google Scholar]
- 21. Hazan RB, Qiao R, Keren R, Badano ISK. Cadherin switch in tumor progression. Ann N Y Acad Sci. 2004;1014:155–163. [DOI] [PubMed] [Google Scholar]
- 22. Statnikov A, Wang L, Aliferis CF. A comprehensive comparison of random forests and support vector machines for microarray-based cancer classification. BMC Bioinformatics. 2008;9:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cawley GC, Talbot NL. On over-fitting in model selection and subsequent selection bias in performance evaluation. J Mach Learn Res. 2010;20:79–107. [Google Scholar]
- 24. Dabbs DJ, Klein ME, Mohsin SK, Tubbs RR, Shuai Y, Bhargava R. High false-negative rate of HER2 quantitative reverse transcription polymerase chain reaction of the oncotype DX test: an independent quality assurance study. J Clin Oncol. 2011;29:4279–4285. [DOI] [PubMed] [Google Scholar]
- 25. Woodman AC, Sugiyama M, Yoshida K, et al. Analysis of anomalous CD44 gene expression in human breast, bladder, and colon cancer and correlation of observed mRNA and protein isoforms. Am J Pathol. 1996;149:1519–1530. [PMC free article] [PubMed] [Google Scholar]
- 26. Sosonkina N, Nakashima M, Ohta T, Niikawa N, Starenki D. Down-regulation of ABCC11 protein (MRP8) in human breast cancer. Exp Oncol. 2011;33:42–46. [PubMed] [Google Scholar]
- 27. Early Breast Cancer Trialists’ Collaborative Group (EBCTCG). Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet (London, England). 2005;365:1687–717. http://www.ncbi.nlm.nih.gov/pubmed/15894097. [DOI] [PubMed] [Google Scholar]
- 28. Early Breast Cancer Trialists’ Collaborative Group (EBCTCG). Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet. 2011;378:771–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Simon RM, Paik S, Hayes DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst. 2009;101:1446–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Paik S, Tang G, Shak S, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol. 2006;24:3726–3734. [DOI] [PubMed] [Google Scholar]
- 31. Hudis CA. Biology before anatomy in early breast cancer—precisely the point. N Engl J Med. 2015;373:2079–2080. http://www.nejm.org/doi/full/10.1056/NEJMe1512092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, uomf_june_25_2018_supplementary_material for Development of a Novel Proteomic Risk-Classifier for Prognostication of Patients With Early-Stage Hormone Receptor–Positive Breast Cancer by Charusheila Ramkumar, Ljubomir Buturovic, Sukriti Malpani, Arun Attuluri Kumar, Chetana Basavaraj, Chandra Prakash, Lekshmi Madhav, Dinesh Chandra Doval, Anurag Mehta and Manjiri M Bakre in Biomarker Insights