Abstract
Self-propagating gene drive technologies have a number of desirable characteristics that warrant their development for the control of insect pest and vector populations, such as the malaria-transmitting mosquitoes. Theoretically easy to deploy and self-sustaining, these tools may be used to generate cost-effective interventions that benefit society without obvious bias related to wealth, age or education. Their species-specific design offers the potential to reduce environmental risks and aim to be compatible and complementary with other control strategies, potentially expediting the elimination and eradication of malaria. A number of strategies have been proposed for gene-drive based control of the malaria mosquito and recent demonstrations have shown proof-of-principle in the laboratory. Though several technical, ethical and regulatory challenges remain, none appear insurmountable if research continues in a step-wise and open manner.
Keywords: Gene drive, malaria, mosquito, genetic vector control, CRISPR, genome editing
Genetic control of mosquitoes
Mosquitoes are considered to be the most dangerous animals on earth. Through the transmission of deadly pathogens, they are believed to cause more than 700,000 deaths each year and are considered a major underlying cause of poverty in developing countries. Although more than 3000 mosquito species have been described, only a handful are able to transmit human disease. Species within the genera Aedes, Anopheles and Culex are responsible for the most deadly and debilitating mosquito-borne diseases including dengue fever, Zika, yellow fever, encephalitis and malaria. Malaria, exclusively transmitted by a limited group of Anopheles mosquitoes, is the most devastating of these diseases and is thought to cause 445,000 deaths each year, killing a child under the age of 5 every two minutes. Control measures based upon antiparasitic drugs, vaccines, bed-nets, insecticides and the destruction of vector habitats have contributed to a progressive decrease in vector-borne diseases to the extent that regional disease elimination has been achieved in several countries [1]. Control based upon these strategies alone is unlikely to achieve global eradication because the scale of the problem far exceeds the capabilities of existing or planned infrastructure, even with substantially increased funding. Moreover, the spread of resistance to insecticides and drugs threatens to reduce the efficacy of front-line interventions such that even maintaining the current levels of disease transmission is expected to cost significantly more in the coming years [2–4]. As a result, interest has increased for genetic control that aims to reduce the harmful impact of pest and vector populations through the dissemination of desirable traits by mating or inheritance. As applied to vector control, these traits can be designed to reduce the capacity of a vector to spread human disease or to reduce its capacity to reproduce, known respectively as population replacement and population suppression. Another important consideration is the persistence of the modification in the target population to distinguish between self-limiting, where the modification tends to disappear, and self-sustaining genetic strategies in which the modification is designed to persist and even spread within the target population. Though self-limiting strategies such as the sterile insect technique (SIT) and release of insects carrying a dominant sterile (RIDL) have been used successfully for local control of insect pest and vector species, they require a continuous mass release of insects that is simply not feasible across large geographic regions or in areas where vector and parasite populations are particularly high [5–9].
Genetic control of malaria
Over 15 years ago, a team at Imperial College London lead by Professor Andrea Crisanti was able to modify the genome of the Anopheles mosquito for the first time and two years later its full genome sequence was made publicly available ushering in a new era in the fight against malaria [10,11]. Together, these tools facilitated the identification of key genetic factors such as those involved in mosquito fertility, viability or vectorial capacity. As the molecular genetics underlying these traits were gradually uncovered, novel strategies to disseminate them throughout wild insect populations were also developed. The problem, which has long been considered, stems from the realization that without the means to spread a genetic modification, it may be unfeasible to rear and release enough modified insects to alter a relevant fraction of the wild population. Nevertheless, engineered traits are not easily disseminated throughout natural populations because of the tendency for natural selection to remove them over time, and because many of these traits are specifically designed to incur a fitness cost that will prevent their spread.
Gene drive
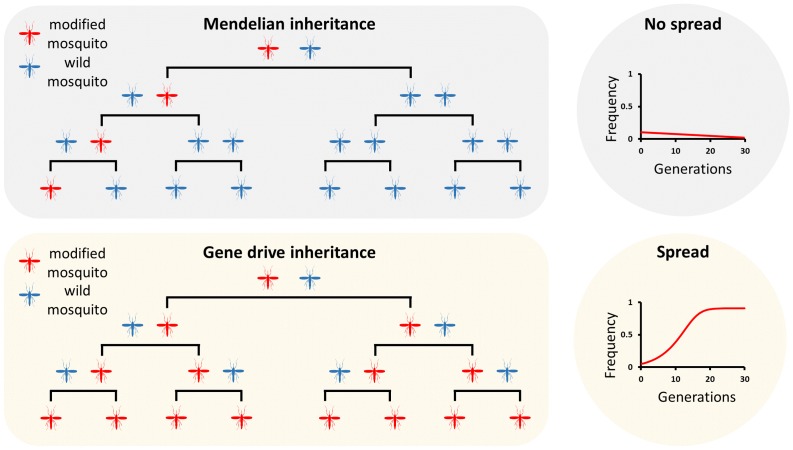
In the 1940s and 1960s, three researchers postulated the first strategies to control insect pest and vector populations using a self-sustaining mechanism to spread a desirable modification. Alexander Sergeevich Serebrovsky at Moscow University and Chris Curtis at the University of Bristol realized that certain chromosomal translocations could spread through and eliminate a population if they cause heterozygous inferiority – a strategy that later became known as genetic underdominance [12,13]. Around the same time at Imperial College London, Bill Hamilton postulated that a genetically encoded sex-ratio distorter could extinguish a population if, for example, a generator of extreme male bias were linked to the Y-chromosome [14]. These strategies, and the many others that have since been proposed, are now collectively known as gene drive, a phenomenon where one or more genetic elements bias their inheritance above and beyond what is predicted by Mendelian genetics therefore increasing their frequency within a population over generations. The modification that is spread through the population is therefore self-sustaining and can be designed to reduce the harmful impact of insect pest and vector populations (Figure 1).
Figure 1.
Mendelian inheritance vs. gene drive inheritance. In mosquitoes, as for any other sexually reproducing organism, genetic elements in heterozygosis have a 50 percent chance of being inherited by the progeny and therefore its frequency remains constant in the population, or more likely, it is gradually lost if the transgene carries a cost (upper panel). A gene drive results in most or all progeny of heterozygotes receiving the driving genetic element, this allows the modification to spread rapidly throughout the population over a few generations (lower panel).
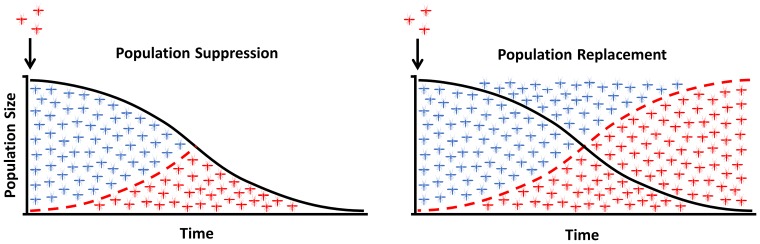
Gene drives are ideally suited for vector control as the short generation time of insects allows them to spread to fixation very quickly even if released from a low initial frequency [6]. In theory, a gene drive could be engineered to reduce either the potential of an insect vector population to transmit disease (replacement) or its potential to reproduce (suppression/elimination). Vector-borne diseases may be completely eliminated if a sufficiently high proportion of the natural vector population is immunized, or if the number of insects is brought below the threshold required to sustain disease transmission (Figure 2).
Figure 2.
Population suppression versus population replacement. In both cases the modified insects are released at a low initial frequency and spread in the population over generations. The modification can be designed to interfere with the mosquito reproduction or viability, aiming to the eliminate or suppress the population to levels that do not support disease transmission (left panel). Alternatively, the modification can be engineered to replace the vector population with insects unable to effectively transmit disease (right panel).
Gene drive systems
Ever since, the potential applications to vector control were first highlighted, researchers have looked to naturally occurring selfish genetic elements from which to draw inspiration. Among these: transposable elements, heritable microorganisms, genetic underdominance, Maternal Effect Dominant Embryonic Arrest (MEDEA), meiotic drives and the natural ‘genetic scissors’ homing endonucleases are discussed briefly here.
Transposable elements
The transposable element P was the first natural gene drive to be extensively studied. Discovered first as the cause of hybrid dysgenesis between certain laboratory and naturally isolated strains of Drosophila melanogaster, it was found to spread throughout natural populations of the fruit fly in the late twentieth Century. It has a very simple structure consisting of a transposase gene within short inverted repeat sequences that, together, allow it to duplicate itself throughout the genome. In doing so, the P element ensures that it is inherited by a disproportionately high frequency of the progeny. The selfish nature of P led some researchers to propose the use of transposases to engineer vector populations as a form of genetic control [12]. Since then, several genetic elements have been identified with characteristics similar to the P [15].
Heritable microorganisms
There are a class of naturally occurring microbiota that are heritable and some of these, including the most notably bacteria of the Wolbachia genus, show a natural ability to spread through natural populations. The Wolbachia are transmitted from one individual to the other in such a way that the entire progeny of an infected female will be infected. Eggs deposited by infected females always produce viable progeny whether they are fertilized by sperm from infected or non-infected males. Conversely, non-infected females do not produce viable progeny when fertilized by sperm from infected males and therefore, non-infected progeny are progressively eliminated from the population [16]. Depending on the specific release program, Wolbachia can be used to suppress a population, or to reduce its capacity to transmit human disease as a result of its fortuitous ability to reduce arbovirus transmission. Wolbachia based control of Aedes mosquitoes have successfully entered field testing in Australia, where the infection has maintained a moderate spread at two release sites [17–19]. In theory, transposons and Wolbachia could be used to spread the genes of mosquitoes that alter their reproductive capacity or make them unable to transmit the malaria parasite, however neither system has found practical applications for malaria control thus far. Nevertheless, two recent reports have demonstrated that natural and modified bacteria with antiparasitic properties can spread throughout laboratory populations of the mosquito [20,21].
Genetic underdominance
Underdominant genetic traits cause infertility or reduced fitness when they are in heterozygosity compared to their homozygotes counterparts, such that a population will naturally select for homozygosity of whichever allele is initially more frequent [12,13]. Therefore, an underdominant element can invade a population if seeded above the critical threshold required for spread. Though genetic underdominance is found naturally in mutually incompatible translocations, it is also possible to engineer underdominance using reciprocal killer-rescue, where the ‘killer’ gene codes for toxicity and the ‘rescue’ codes for protection from the toxic effect, or by inducing chromosomal rearrangements [22–25]. To date, proof-of-principle of underdominant population transformation systems have been shown in the fruit fly Drosophila melanogaster, suggesting that translocation-based synthetic systems may be engineered for the local population replacement of other vector species [25,26].
Maternal effect dominant embryonic arrest (MEDEA)
Selfish genetic elements called Maternal Effect Dominant Embryonic Arrest (MEDEA) are able to bias their own inheritance by selectively killing non-drive offspring using a toxin-antidote rescue system. They were first discovered because of their spread throughout natural populations of the flour beetle Tribolium castaneum and the toxin-antidote system was eventually used to create the first synthetic drive system in the fruit fly [27–29]. Since then, a number of similar strategies have been proposed that vary in the arrangement and mechanism of killer-rescue components, as well as the release threshold predicted for these to drive population replacement or population suppression [30–33]. Most recently, the strategy has been developed for use in the crop pest Drosophila suzukii using highly regulated maternal expression of a toxic miRNA that targets the zygotically-required myd88 gene. The toxicity is rescued by a drive-encoded copy of the myd88 gene that is expressed in the zygote [34]. Mathematical models indicate that Medea elements carrying transgenes that cause female-specific lethality in response to specific environmental signals could be used to generate population suppression [30].
Meiotic drive and sex distortion
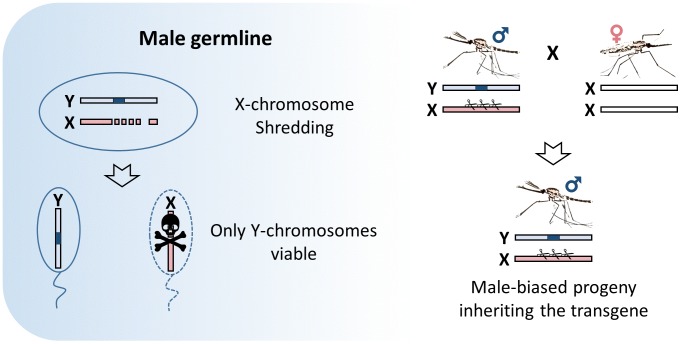
Meiotic drive elements act by impairing the transmission of specific gametes during meiosis by, for example, selectively removing the gametes that do not carry the drive element. The generation of extreme male sex ratio distortion has been proposed as a powerful tool to suppress or eliminate pest populations, and the presence of naturally occurring sex distortion in Aedes and Culex has generated increasing interest in the approach [14,35–37]. In 2003, Burt proposed a strategy where an engineered Y-chromosome, the determinant of maleness in Anopheles mosquitoes, could favor its own inheritance such that it can propagate an extreme male bias throughout a natural mosquito population to cause its ultimate suppression through the progressive reduction of females [6]. In this system, the Y-chromosome is engineered to carry an endonuclease that selectively cuts multiple sites exclusively within the X-chromosome during male meiosis such that only Y-bearing sperm are able to fertilize eggs. In 2007, the naturally occurring endonuclease I-PpoI was used to selectively target Anopheles X-inked ribosomal DNA (rDNA) repeats, and a few years later it was possible to generate extreme male bias by restricting its enzymatic activity to male meiosis [38–40]. The strategy has also been proven to work using a CRISPR-based nuclease that can be widely applicable to other insect species [41]. However, it has not yet been possible to link the sex distorter to the Y-chromosome as would be required to spread [14,42–44] (Figure 3).
Figure 3.
Meiotic Y-drive. An endonuclease (blue block), placed onto the mosquito Y-chromosome, is expressed during male meiosis to cut a multicopy target sequence on the X-chromosome. The shredding of the X-chromosome favors the unaffected Y-carrying sperm and results in the production of a male-biased progeny.
Homing gene drives
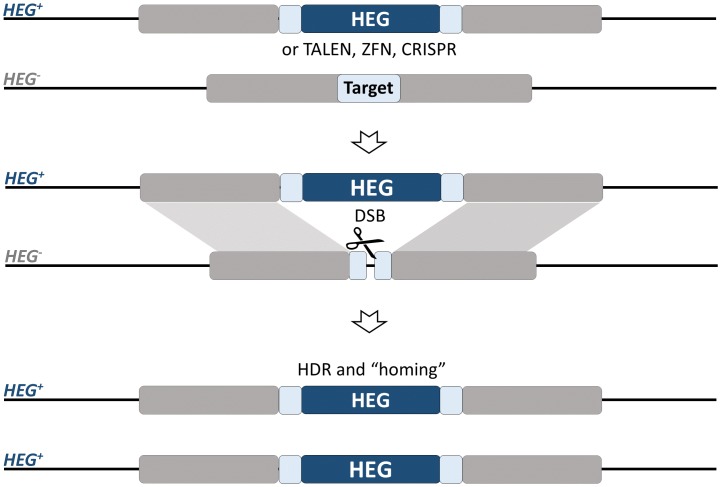
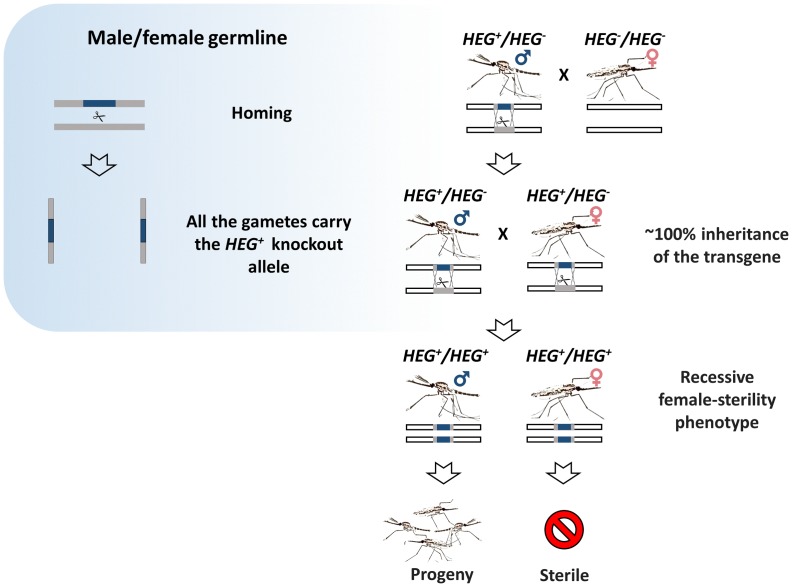
Homing endonuclease genes (HEGs) are selfish genes that are found throughout microbial life. They increase their allele frequency by cleaving rare or unique DNA targets within the host cell and, taking advantage of the cellular DNA repair machinery, use the HEG+ allele as a template for the homology-directed repair (HDR). During this process, known as ‘homing’, the HEG- allele is converted to HEG+ allowing the homing endonucleases to be transmitted at super-Mendelian rates. The ‘homing’ is unidirectional as the recognition sequence, once modified to contain the HEG, can no longer be recognized or cleaved by the nuclease. As such, HEGs cause an irreversible conversion from heterozygosity to HEG homozygosity, and this results in their biased inheritance. Because the endonuclease has high specificity for a 20–30 bp sequence, they do not tend to cleave anywhere other than their genomic target [45–49]. This has inspired another ground-breaking approach for the genetic control of natural populations of pest vectors. Drawing upon the natural system used by HEGs, Burt suggested that if homing may be allowed to proceed in the germline of a heterozygote then a disproportionately high fraction of its progeny will contain the element. If, for example, the homing reaction causes full conversion in the germline, then a heterozygote would produce 100% modified progeny instead of the 50% predicted by Mendelian inheritance (Figure 4). Because of this bias, a driving endonuclease gene is self-sustaining in a population as heterozygotes spread the element at the detriment of unmodified wild-type chromosomes. Genetic loads sufficient to eradicate a pest population can be imposed if the HEG is designed to target a gene that is essential or required for female fertility (Figure 5). Moreover, a driving endonuclease could be linked to an antiparasitic effector that would spread with the driving endonuclease gene [6].
Figure 4.
Homing gene-drive. The HEG (or TALEN, ZFN, CRISPR) is inserted within the target gene and expressed under a germline-specific promoter. The endonucleases can be designed to disrupt essential mosquito genes, genes required for reproduction or those involved with parasite development within the mosquito (grey block). Alternatively, the HEG can be linked to an antiparasitic effector. Homology-directed repair (HDR) of the double-strand break generated by the endonuclease leads to copying and ‘homing’ of the HEG+ allele.
Figure 5.
Gene drive system targeting female reproduction. The confinement of homing to the germline leads to super-Mendelian inheritance of the homing construct (in blue) that reduce the number of fertile females by targeting a haplosufficient, somatic female-fertility genes (in gray).
In 2011, research in the laboratories of Crisanti, Russell and Burt led to the first demonstration of homing-based gene drives by placing the naturally occurring I-SceI homing endonuclease gene within an artificially inserted target site in the genome of mosquitoes and flies [50,51]. Moderate levels of homing allowed transmission to be biased by up to 60% in these studies, surpassing the minimum requirements predicted for use in vector control [43]. Several groups then embarked upon a concerted effort to re-engineer naturally occurring HEGs to target mosquito genes [52–56].
In theory, any endonuclease with a sufficiently long recognition sequence could be repurposed to function as a gene drive [6]; programmable zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) had been developed as easy-to-engineer alternatives [6,57,58]. Though the modular nature of ZFNs and TALENs simplifies the engineering of endonucleases to target mosquito genes, it also makes them poorly suited to build gene drives because of their tendency to recombine during homing [59].
By fortunate coincidence, the engineering of TALENs was quickly followed by the development of bacterial adaptive immune system Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR associated proteins (Cas) (CRISPR/Cas or CRISPR) that has since revolutionized the process of genome engineering [60]. The CRISPR system, as it is most commonly used today, is based upon an endonuclease, called Cas9, directed to its genomic target by a short guide RNA that can be easily modified to target almost any site in the genome. The potential to adapt CRISPR for use in gene drives was highlighted almost immediately, and several groups were quick to demonstrate CRISPR-based homing in yeast and fruit flies before gene drives built for population replacement and population suppression were demonstrated in Anopheles stephensi and Anopheles gambiae respectively [61–65]. In contrast to previous gene drives built using HEGs, TALENs or ZFNs, CRISPR-based gene drives showed almost complete transmission bias in all demonstrations of the technology.
Resistance to driving endonuclease genes
As with any strategy aimed at reducing the vector or parasite population, gene drives are at risk of resistance developing that can lessen or reverse the beneficial impact. Both molecular and behavioral traits can account for resistance and these include epigenetic silencing, RNA interference, assortative mating, co-evolution of the vector and parasite and, perhaps most importantly, target site mutations that are resistant to cleavage [66–68]. For drives designed to spread an effector, there is the additional risk that the effector is lost leaving the empty drive to continue spreading at the exclusion of the intact drive.
In the best-case scenario, the few generations required by some drives to spread from low frequency to near fixation may simply outpace the creation and selection of many resistant traits, such as those requiring several adaptations. Nevertheless, significant push-back should be anticipated in the form of nuclease-resistant targets that block homing at little or no cost to the host [6,33,42,43,61,69,70]. Nuclease-resistant alleles may already be common in the wild, single nucleotide polymorphisms (SNPs) are estimated to occur every 2.2 bases on average in the genome of Anopheles gambiae, and most variants would be expected to block the homing reaction [71]. Even at sites with minimal natural variation, resistant alleles could be generated by the nuclease itself as a by-product of error-prone end-joining. Several studies have found that repair by non-homologous end joining (NHEJ) and microhomology-mediated end-joining (MMEJ) generate high levels of resistance in the early embryo, where error-prone repair seems to dominate over homologous recombination [63–65,72–74]. Nevertheless, if the target is required for viability or fertility then cleavage-resistant mutants will only stop the drive from spreading if they also restore some or all of the original function. In a caged release of gene drive, these restorative resistant alleles were quickly selected from the wider pool of diverse mutants generated by the nuclease [75]. Mathematical modelling predicts that none of the drive systems developed thus far would sustain an invasion into natural populations without further measures to mitigate resistance [65,76].
Strategies to limit target site resistance
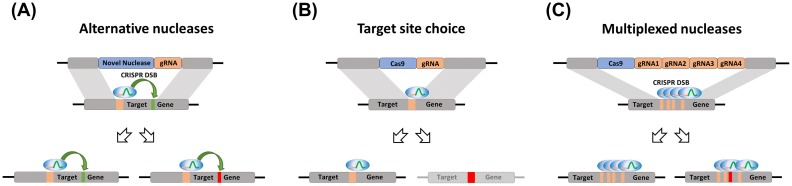
Since drives based upon homing endonucleases were first postulated, Burt also suggested a number of strategies to mitigate against target site resistance [6]. These range from restricting the nuclease expression to defined germline tissues, to the careful analysis of target sites that could reduce the pool of potentially resistance mutants and the targeting of multiple sequences to increase tolerance to individual resistant target sequences (Figure 6 and Box 1).
Figure 6.
Strategies to limit target site resistance. Schematic representation of some of the alternatives proposed to reduce the chance of resistance to homing-gene drives. (a) Alternative nucleases such as FokI-dCas9 fusion proteins retain the ability to cleave a sequence of DNA proximal to the target sites. Cleavage-induced mutations or pre-existing polymorphisms at the cut site, and outside the target, do not prevent further cleavage and homing [78]. (b) Target sites can be selected based on nucleotide sequence conservation or the likelihood that micro-homology mediated end-joining will generate null alleles. Mutations that may prevent cleavage are mostly null and get removed from population. (c) Multiple gRNA-expression cassettes or nuclease able to process its own CRISPR-RNA (crRNA), such as Cpf1, can be used to target multiple sites within the target gene. Mutations that prevent cleavage must simultaneously occur at all target sites to inhibit homing. Mutation occurring in only some of the targets can be removed during the homing-process stimulated by the cleavage of the intact sites.
Box 1
-
(a)
Combined release. Several gene drives can be programmed to target independent loci in the genome and, akin to combination drug therapy, a combined release would reduce the impact of resistance to any one of these drives [6]
-
(b)
Multiplexing. Individual gene drives can be multiplexed to target several sites in the genome so that there is redundancy in the sequence-specific recognition of target genes [61,77].
-
(c)
Novel endonucleases. Novel or engineered CRISPR endonucleases, such as dCas9-FokI and Cpf1, may show more tolerance of sequence variation at the cut site so that nuclease-induced mutations are less likely to prevent secondary cleavage [78,79].
-
(d)
Rational target site selection. Target sites within essential genes can be selected on the basis that non-resistant null alleles are more frequently generated after repair by micro-homology mediated end-joining [74].
-
(e)
Regulation of nuclease expression. Tightly regulated germline-specific promoters, modifications that reduce mRNA or protein stability, or synthetic regulatory circuits can be used to limit nuclease deposition into the embryo that may cause the majority of resistant mutations [65,73].
-
(f)
New strategies. Several new strategies have been proposed that should slow the rise of resistance whilst altering wild populations including daisy-chain, where a number of genetic elements are combined to drive each other while losing their linkage until they are unable to spread, and daisyfield drives, where multiple copies of a ‘booster’ element are introduced in repetitive regions of the genome. Other approaches include combining homing-based drives with a killer-rescue system that can remove non-drive progeny and help preserve cargo [80,81].
By choosing targets that are highly conserved in nature, resistance should mainly arise from the target site misrepair and, presumably, the absence of natural variation at these sites would be indicative of strong sequence constraints. The degenerate nature of the genetic code means that, for most protein-coding genes, there will be an array of sequence variants that do not alter the final encoded protein but could easily escape recognition by the nuclease. There are, however, genes that depend upon the nucleic acid sequence for their functionality – for example non-coding RNA or the targets of transcription factors. By targeting these sequences, it may be possible to substantially reduce the fraction of mutations that are also restorative. Furthermore, the range of end-joining mutants generated by CRISPR-based drives seems to be biased towards deletions expected to occur as a result of microhomology-mediated end-joining (MMEJ). The predictability of MMEJ mutations may allow target sites to be selected on the basis that, in the absence of homing, MMEJ is expected to generate out-of-frame or otherwise non-functional mutants [75].
Another potentially remedy is the use of several gene drives that could be targeted to different sequences such that the likelihood of resistance occurring to all of them would be reduced. This would be especially effective if these sites are dispersed throughout a single gene because the close proximity of targets would slow the process of recombination necessary to bring resistant sequences together on a single chromosome. Each drive could be further improved if there is redundancy in its target site recognition – however there must be a balance between increasing the tolerance of a nuclease to mutant targets and limiting promiscuous cleavage at genomic off-targets that resemble the target elsewhere in the genome. Natural homing endonuclease genes have evolved this type of tolerance in the 3rd base ‘wobble’ positions that could otherwise confer resistance, however this nuanced recognition is not possible using CRISPR. In 2014, Esvelt suggested that CRISPR-based drives could achieve similar redundancy through the use of multiple gRNAs in a single drive, each targeted to different sites within the same gene [61]. This has the added benefit that partially resistant alleles are actively removed from the population because, as long as one or more targets remain cleavable, homing can convert the entire locus to a drive allele. Multiplexed strategies may suffer from additional toxicity due to increased on- or off-target DNA damage; this may as well be exacerbated by an increased dose of nuclease that could result from using several nuclease-expression cassettes. Moreover, these elements may be more prone to breakdown in the process in homing by recombining along the repeated elements such as the gRNA scaffold or promoter. Along similar rationale, alternative nucleases could also reduce resistant mutations if the site of nuclease recognition were separated from the site of cleavage. In this scenario, mutations at the cleavage site would be re-cleavable if they do not extend to the nuclease recognition site, and several nucleases may already be suitable for this including Cpf1 nucleases, dCas9-FokI fusions and TALENs [58,78,79].
The above strategies focus upon reducing the range of mutant sequences that would confer resistance, however there are several strategies to limit their generation in the first instance. Several studies have shown that nuclease deposition into the embryo results in high levels of end-joining as opposed to homing that tends to be preferred in the post-embryonic germline, and tight temporal and spatial regulation of the nuclease could limit this activity [73,75].More complex drive systems such as daisy chain and daisy field drives may also limit the accumulation of resistant mutations because the system comprises several layers of nucleases that each target independent sites [80]. By coupling any of these drives with a poison-antidote system, individuals are killed if they inherit a resistant allele instead of the antidote-containing drive [7].
Off-target mutagenesis
Homing-based gene drives have the potential to generate unintended and heritable off-target mutations that may accumulate and result in fitness costs that impairs their potential to spread. Moreover, these mutations could be stably propagated by nuclease-induced gene conversion. Currently, we know very little about how frequently off-target mutations are generated by a gene drive and what the potential impact could be. The likelihood of generating an off-target mutation will mostly depend upon the activity, specificity, dose and spatio-temporal regulation of the nuclease, the sequence of its target and flanking region, the presence and frequency of genomic sequences resembling the target, and the accessibility of these sites to the nuclease and DNA repair machinery. Fortunately there are a number of systems that could be used to investigate off-target mutagenesis across the entire genome, most notably the in vivo assays Di-Genome Seq and GUIDE-SEQ, and the in vitro assay CIRLCE-SEQ [82–84]. Future designs for gene drive will undoubtedly include alternatives to Streptococcus pyogenes Cas9 that show improved on-target specificity such as natural or engineered high fidelity variants, Cpf1 nucleases shown to generate fewer off-target mutations or nickase/dimerising nucleases that require the cooperative binding at two independent sites and may be able to initiate the homing reaction [78,85–89].
Confinement, containment and scaling-up
Gene drive experiments must be safeguarded to minimize the risk of accidental release and spread into wild populations using strategies for confinement and containment that provide biological and physical barriers respectively [90].
To some extent, gene drives provide their own level of confinement. Gene drives are naturally confined to a particular species or subset of closely related species because most species are ipso facto reproductively isolated and, with the exception of heritable organisms, gene drives necessarily spread by genetic inheritance. The extreme specificity with which synthetic endonucleases can recognize and cleave a target sequence may allow homing-based drives to be designed to target sequences that are specific to a species, subspecies or possibly even local populations. Nevertheless, the specificity afforded by gRNAs is mutable and could, in theory, adapt to generate gene drives that can cleave closely related sequences in non-target populations within the target species.
To ensure gene drive research is carried out responsibly, laboratory experiments should be designed to combine different elements of containment and confinement, many of which have been described in detail elsewhere [90,91]. First and foremost, all gene drive research should be carried out by trained professionals using robust physical barriers such as triple-nested containers within secure and restricted access rooms [92]. Additional layers of security should be included where possible such as molecular containment that ensures that only laboratory-contained populations are susceptible to the drive by, for example, separating driver components or targeting synthetic sequences that are not found in natural populations [50,93]. Where possible, researchers should strive for reproductive and ecological confinement by using organisms that cannot breed with natural populations or by restricting experiments to geographical locations that are outside of the habitable range respectively.
Beyond addressing the technical limitations to improve efficacy, a careful monitoring in physically, environmentally and biologically confined large enclosures will be essential before moving to the field. Large cage testing allows to: I) reproduce releases and performances of the gene drives in environmental conditions more similar to the natural, such as temperature, humidity and light cycle; II) stimulate natural mosquito behaviors, otherwise not stimulated in small cages studies, like assortative mating and male swarming; III) evaluate fitness and competitiveness of the genetically engineered strains in consecutive, defined or overlapping generations of a larger number of mosquitoes; IV) accurate validation of modelling predictions of gene drive dynamics; V) increase the statistical power to detect rare events such as resistance or off-target effects. Laboratory experiments should be carried out alongside field studies that interrogate the genetic composition of mosquito species at potential release sites to help understand genetic heterogeneity, distribution and gene flow prior to release [94,95]. This will serve to inform release strategies and post-release monitoring that must follow any release into the wild.
Reversal and recall of driving endonucleases
The risk of unforeseen consequences to the ecosystem or spread outside of the intended area following intentional or unintentional release has pushed many to consider strategies to slow, reverse or completely recall gene drives. For suppressive drives that cause sterility or inviability in a population, there will be strong selection for target-site resistance that cause the drive to be lost [6,68,76]. Current designs for gene drive aim to prevent this type of resistance from occurring naturally, therefore, insects could be engineered to carry synthetic resistance and released as a countermeasure. If the drive population is geographically isolated, the release of resistant insects could be planned to create a buffer zone that protects against further spread as the drive population is eventually extinguished [6,42]. A resistant buffer zone could also prevent further spread of a replacement drive. However, because replacement drives do not confer major fitness costs, resistance would not replace the drive where it already exists.
Strategies have been proposed to speed up the process and include several designs for ‘reversal drives’. ‘Immunizing reversal drives’ can spread on their own and inactivate the first drive in the process, however these will also pose their own risk [61,93,96]. Potentially safer alternatives have been proposed that do not spread or spread only in the presence of the original drive and include split-drive type elements such as CATCHA (Cas9-triggered chain ablation), ERACR (elements for reversing the autocatalytic chain reaction), CHACR (constructs hitchhiking on the autocatalytic chain reaction) and e-CHACR [97,98]. Depending upon the specific parameters of the drive and countermeasure, modelling predicts that successful drive removal will depend upon the timing and number of released insects and can sometimes result in stable equilibria or oscillatory dynamics in which the drive is never completely removed [96].
Regulation, ethical challenges and public acceptance
Gene drives pose a unique challenge for regulators because they have great potential to alter natural populations in the absence of continued human intervention and without a comparator technology from which precedent can be drawn. As it stands, regulation for the technology is not in place and so research has undergone a degree of self-governance led by the scientific community, research institutes and funders – who have advocated strict containment and confinement [61,65,90,92,99]. Following several important proof-of-principle experiments, the scientific community has called for clarity as to how gene drive research should be regulated [99]. In response, the National Academy of Sciences has published a report containing clear guidelines for safe and ethical research that encourages self-governance alongside government regulation that will evaluate gene drives on a case-by-case basis [91]. In this way, prudent regulatory decisions can be made on the basis of efficacy, safety and ethical concerns related to an individual gene drive product rather than forcing a precipitate judgement on the use of gene drives that may prematurely stifle research.
Gene drives are capable of disseminating a genetic modification throughout an entire population or species within a time-frame that is unprecedented in history. As such, their development and potential use raises ethical considerations. Once a drive is released, there is currently no practical strategy to limit its geographical spread or to recall it and they may intentionally or unintentionally alter the natural ecosystem. In some cases, the successful use of gene drive may reduce the harmful impact that extensive vector and pest control campaigns may have on the ecosystem. For example, gene drives being developed for malaria control have the potential to target the few species that transmit malaria without affecting more than 3000 mosquito species that do not transmit human diseases. The expectation being that the species-specificity afforded by gene drive-based vector control should reduce dependency on mass spraying of insecticides or other broad-spectrum interventions. The decision to use gene drive technologies must therefore seek broad consensus from the nations that may be affected, and these decisions must be based upon rigorous evidence. Moreover, informed consent must extend beyond governments and regulators to include local communities who will potentially benefit from its use. A successful deployment program requires an extensive and scrupulous public engagement to communicate the key concepts and potential risks, and this must take place well-before any planned release of a drive.
Concluding remarks
Gene drives-based strategies have a number of advantages that make them particularly suited for malaria control, most notably their self-sustaining nature. For instance, these approaches can offer a solution for malarial zones that are difficult to reach with conventional interventions as they do not require continuous or mass releases. This would also negate the requirement for substantial infrastructure that may be unfeasible in the vast rural areas of Africa where the burden of malaria is most severe. Because gene drives spread by mating and rely upon highly specific target site recognition, the technology can potentially be restricted to single species or perhaps even local populations. Together, these features depict this technology as the most targeted and cost-effective intervention currently under development with the potential to benefit entire communities irrespective of their socio-economic conditions. Nevertheless, gene drives should not be seen as a silver bullet but rather, a complementary intervention that can be used alongside other malaria control strategies such as bed-nets, antimalarial drugs and vaccines. Target site resistance and complex population dynamics may mean that gene drives do not reach fixation, that their spread is spatially limited, or that they fail to become established at all. Whilst ongoing research is geared towards overcoming these barriers, recent studies have aimed to outline the best practices for laboratory testing and eventual release to ensure the technology is safe and efficacious [92,100].
To conclude, gene drives may represent a powerful tool to reduce infectious diseases and their related economic and ecological burden worldwide. Enormous progress has been made over the last decades and more will be required to overcome the challenges still lying ahead. Eventual decisions on their use will require evaluation on a case-by-case basis that balances the ethical and ecological implications with the potential benefits in agreement with the communities and the countries that are affected by the disease.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This work was supported by Foundation for the National Institutes of Health through the Vector-Based Control of Transmission: Discovery Research (VCTR) program of the Grand Challenges in Global Health initiative of the Bill & Melinda Gates Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- [1].WHO | Eliminating malaria [Internet]. WHO . [cited 2018 Feb 2]. Available from: http://www.who.int/malaria/publications/atoz/eliminating-malaria/en/. [Google Scholar]
- [2].WHO | Global vector control response [Internet]. WHO. [cited 2017 Sep 29]. Available from: http://www.who.int/malaria/areas/vector_control/global-vector-control-response/en/.
- [3].WHO | World malaria report 2017 [Internet]. WHO [cited 2017 Dec 3]. Available from: http://www.who.int/malaria/publications/world-malaria-report-2017/report/en/.
- [4].WHO | A global brief on vector-borne diseases [Internet]. WHO. [cited 2017 Sep 29]. Available from: http://www.who.int/campaigns/world-health-day/2014/global-brief/en/.
- [5].Knipling EF. Sterile-male method of population control: successful with some insects, the method may also be effective when applied to other noxious animals. Science. 1959;130:902–904. [DOI] [PubMed] [Google Scholar]
- [6].Burt A. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc R Soc B Biol Sci. 2003;270:921–928. 10.1098/rspb.2002.2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gould F, Schliekelman P. Population genetics of autocidal control and strain replacement. Annu Rev Entomol. 2003;49:193–217. [DOI] [PubMed] [Google Scholar]
- [8].Alphey L. Genetic control of mosquitoes. Annu Rev Entomol. 2014;59:205–224. 10.1146/annurev-ento-011613-162002 [DOI] [PubMed] [Google Scholar]
- [9].Gabrieli P, Smidler A, Catteruccia F. Engineering the control of mosquito-borne infectious diseases. Genome Biol. 2014;15:535. 10.1186/s13059-014-0535-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Catteruccia F, Nolan T, Loukeris TG, et al. Stable germline transformation of the malaria mosquito Anopheles stephensi. Nature. 2000;405:35016096. [DOI] [PubMed] [Google Scholar]
- [11].Holt RA, Subramanian GM, Halpern A, et al. The genome sequence of the malaria mosquito Anopheles gambiae .Science .2002;298:129–149. 10.1126/science.1076181 [DOI] [PubMed] [Google Scholar]
- [12].Curtis CF. Possible use of translocations to fix desirable genes in insect pest populations. Nature. 1968;218:368. 10.1038/218368a0 [DOI] [PubMed] [Google Scholar]
- [13].Serebrovsky A. On the possibility of a new method for the control of insect pests. Sterile-Male Tech. Erad. Control Harmful Insects Proc Panel Appl Sterile-Male Tech Erad Control Harmful Species Insects Organised Jt. FAOIAEA Div At Energy Food Agric. Vienna: 27–31 May 1968. 1969;123–237. [Google Scholar]
- [14].Hamilton WD. Extraordinary sex ratios. Science. 1967;156:477–488. 10.1126/science.156.3774.477 [DOI] [PubMed] [Google Scholar]
- [15].Donald C. Rio, Sharmistha Majumdar*.. P transposable elements in Drosophila and other eukaryotic organisms. Microbiol Spectr. 2015;3(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. 2008;6:741. 10.1038/nrmicro1969 [DOI] [PubMed] [Google Scholar]
- [17].Hoffmann AA, Iturbe-Ormaetxe I, Callahan AG, et al. Stability of the wMel Wolbachia Infection following Invasion into Aedes aegypti Populations. In: Rasgon JL, editor. PLoS Negl Trop Dis. 2014;8:e3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Schmidt TL, Barton NH, Rašić G, et al. Local introduction and heterogeneous spatial spread of dengue-suppressing Wolbachia through an urban population of Aedes aegypti. PLOS Biol. 2017;15:e2001894. 10.1371/journal.pbio.2001894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Jiggins FM. The spread of Wolbachia through mosquito populations. PLOS Biol. 2017;15:e2002780. 10.1371/journal.pbio.2002780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Pike A, Dong Y, Dizaji NB, et al. Changes in the microbiota cause genetically modified Anopheles to spread in a population. Science. 2017;357:1396–1399. 10.1126/science.aak9691 [DOI] [PubMed] [Google Scholar]
- [21].Wang S, Dos-Santos ALA, Huang W, et al. Driving mosquito refractoriness to Plasmodium falciparum with engineered symbiotic bacteria. Science. 2017;357:1399–1402. 10.1126/science.aan5478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Davis S, Bax N, Grewe P. Engineered underdominance allows efficient and economical introgression of traits into pest populations. J Theor Biol. 2001;212:83–98. [DOI] [PubMed] [Google Scholar]
- [23].Akbari OS, Matzen KD, Marshall JM, et al. A synthetic gene drive system for local, reversible modification and suppression of insect populations. Curr Biol. 2013;23:671–677. 10.1016/j.cub.2013.02.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Maddalo D, Manchado E, Concepcion CP, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516:423. 10.1038/nature13902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Buchman AB, Ivy T, Marshall JM, et al. Engineered reciprocal chromosome translocations drive high threshold, reversible population replacement in Drosophila. bioRxiv. 2016;088393. [DOI] [PubMed] [Google Scholar]
- [26].Reeves RG, Bryk J, Altrock PM, et al. First Steps towards Underdominant Genetic Transformation of Insect Populations. In: Franz AWE, editor. PLoS ONE. 2014;9:e97557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wade MJ, Beeman RW. The population dynamics of maternal-effect selfish genes. Genetics. 1994;138:1309–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Beeman RW, Friesen KS, Denell RE. Maternal-effect selfish genes in flour beetles. Science. 1992;256:89–92. 10.1126/science.1566060 [DOI] [PubMed] [Google Scholar]
- [29].Chen C-H, Huang H, Ward CM, et al. A synthetic maternal-effect selfish genetic element drives population replacement in drosophila. Science. 2007;316:597–600. 10.1126/science.1138595 [DOI] [PubMed] [Google Scholar]
- [30].Akbari OS, Chen C-H, Marshall JM, et al. Novel synthetic medea selfish genetic elements drive population replacement in drosophila ; a theoretical exploration of medea -dependent population suppression. ACS Synth Biol. 2014;3:915–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Marshall JM, Hay BA. General principles of single-construct chromosomal gene drive. Evol Int J Org Evol. 2012;66:2150–2166. 10.1111/evo.2012.66.issue-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Marshall JM, Hay BA. Inverse Medea as a novel gene drive system for local population replacement: a theoretical analysis. J Hered. 2011;102:336–341. 10.1093/jhered/esr019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Champer J, Buchman A, Akbari OS. Cheating evolution: engineering gene drives to manipulate the fate of wild populations. Nat Rev Genet. 2016;17:146–159. 10.1038/nrg.2015.34 [DOI] [PubMed] [Google Scholar]
- [34].Buchman A, Marshall J, Ostrovski D, et al. Synthetically engineered medea gene drive system in the Worldwide Crop Pest, D. suzukii. bioRxiv. 2017;162255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Schliekelman P, Ellner S, Gould F. Pest control by genetic manipulation of sex ratio. J Econ Entomol. 2005;98:18–34. 10.1093/jee/98.1.18 [DOI] [PubMed] [Google Scholar]
- [36].Hickey WA, Craig GB. Genetic distortion of sex ratio in a mosquito. AEDES AEGYPTI. Genetics. 1966;53:1177–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sweeny TL, Barr AR. Sex ratio distortion caused by meiotic drive in a mosquito.Culex pipiens L. Genetics. 1978;88:427–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Windbichler N, Papathanos PA, Catteruccia F, et al. Homing endonuclease mediated gene targeting in Anopheles gambiae cells and embryos. Nucleic Acids Res. 2007;35:5922–5933. 10.1093/nar/gkm632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Windbichler N, Papathanos PA, Crisanti A. Targeting the X chromosome during spermatogenesis induces Y chromosome transmission ratio distortion and early dominant embryo lethality in Anopheles gambiae. In: Stern DL, editor. PLoS Genet. 2008;4:e1000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Roberto Galizi, Lindsey A. Doyle, Miriam Menichelli, et al. A synthetic sex ratio distortion system for the control of the human malaria mosquito. Nat Commun. 2014;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Roberto Galizi, Andrew Hammond, Kyros Kyrou, et al.. A CRISPR-Cas9 sex-ratio distortion system for genetic control. Sci Rep. 2016;6(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Deredec A, Burt A, Godfray HCJ. The population genetics of using homing endonuclease genes in vector and pest management. Genetics. 2008;179:2013–2026. 10.1534/genetics.108.089037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Deredec A, Godfray HCJ, Burt A. Requirements for effective malaria control with homing endonuclease genes. Proc Natl Acad Sci. 2011;108:E874–E880. 10.1073/pnas.1110717108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Beaghton A, Beaghton PJ, Burt A. Gene drive through a landscape: reaction–diffusion models of population suppression and elimination by a sex ratio distorter. Theor Popul Biol. 2016;108:51–69. 10.1016/j.tpb.2015.11.005 [DOI] [PubMed] [Google Scholar]
- [45].Chevalier BS, Stoddard BL. Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. 2001;29:3757–3774. 10.1093/nar/29.18.3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Goddard MR, Greig D, Burt A. Outcrossed sex allows a selfish gene to invade yeast populations. Proc R Soc Lond B Biol Sci. 2001;268:2537–2542. 10.1098/rspb.2001.1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Belfort M, Perlman PS. Mechanisms of intron mobility. J Biol Chem. 1995;270:30237–30240. 10.1074/jbc.270.51.30237 [DOI] [PubMed] [Google Scholar]
- [48].Belfort M, Roberts RJ. Homing endonucleases: keeping the house in order. Nucleic Acids Res. 1997;25:3379–3388. 10.1093/nar/25.17.3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Stoddard BL. Homing endonuclease structure and function. Q Rev Biophys. 2005;38:49–95. [DOI] [PubMed] [Google Scholar]
- [50].Windbichler N, Menichelli M, Papathanos PA, et al. A synthetic homing endonuclease-based gene drive system in the human malaria mosquito. Nature. 2011;473:212–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Chan Y-S, Naujoks DA, Huen DS, et al. Insect population control by homing endonuclease-based gene drive: an evaluation in drosophila melanogaster. Genetics. 2011;188:33–44. 10.1534/genetics.111.127506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Chan Y-S, Huen DS, Glauert R, et al. Optimising homing endonuclease gene drive performance in a semi-refractory species: the drosophila melanogaster experience. PLoS ONE. 2013;8:e54130. 10.1371/journal.pone.0054130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Thyme SB, Boissel SJS, Arshiya Quadri S, et al. Reprogramming homing endonuclease specificity through computational design and directed evolution. Nucleic Acids Res. 2014;42:2564–2576. 10.1093/nar/gkt1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Takeuchi R, Lambert AR, Mak AN-S, et al. Tapping natural reservoirs of homing endonucleases for targeted gene modification. Proc Natl Acad Sci. 2011;108:13077–13082. 10.1073/pnas.1107719108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].BL Homing endonucleases from mobile group I introns: discovery to genome engineering. Mob. DNA. 2014;5:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Werther R, Hallinan JP, Lambert AR, et al. Crystallographic analyses illustrate significant plasticity and efficient recoding of meganuclease target specificity. Nucleic Acids Res. 2017;45:8621–8634. 10.1093/nar/gkx544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Kim HJ, Lee HJ, Kim H, et al. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009;19:1279–1288. 10.1101/gr.089417.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Miller JC, Tan S, Qiao G, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. 10.1038/nbt.1755 [DOI] [PubMed] [Google Scholar]
- [59].Simoni A, Siniscalchi C, Chan Y-S, et al. Development of synthetic selfish elements based on modular nucleases in Drosophila melanogaster. Nucleic Acids Res. 2014;42:7461–7472. 10.1093/nar/gku387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Esvelt KM, Smidler AL, Catteruccia F, et al. Emerging Technology: Concerning RNA-guided gene drives for the alteration of wild populations. eLife. 2014;3:e03401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].DiCarlo JE, Norville JE, Mali P, et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41:4336–4343. 10.1093/nar/gkt135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Gantz VM, Jasinskiene N, Tatarenkova O, et al. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc Natl Acad Sci. 2015;112:E6736–E6743. 10.1073/pnas.1521077112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Gantz VM, Bier E. The mutagenic chain reaction: a method for converting heterozygous to homozygous mutations. Science. 2015;348:442–444. 10.1126/science.aaa5945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Hammond A, Galizi R, Kyrou K, et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat Biotechnol. 2016;34:78–83. 10.1038/nbt.3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Sinkins SP, Gould F. Gene drive systems for insect disease vectors. Nat Rev Genet. 2006;7:427–435. 10.1038/nrg1870 [DOI] [PubMed] [Google Scholar]
- [67].Burt A. Heritable strategies for controlling insect vectors of disease. Phil Trans R Soc B. 2014;369:20130432. 10.1098/rstb.2013.0432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Bull JJ. Lethal gene drive selects inbreeding. Evol Med Public Health. 2016;2017:1–16. 10.1093/emph/eow030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Leftwich PT, Bolton M, Chapman T. Evolutionary biology and genetic techniques for insect control. Evol Appl. 2016;9:212–230. 10.1111/eva.12280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Lindholm AK, Dyer KA, Firman RC, et al. The ecology and evolutionary dynamics of meiotic drive. Trends Ecol Evol. 2016;31:315–326. 10.1016/j.tree.2016.02.001 [DOI] [PubMed] [Google Scholar]
- [71].Miles A, Harding NJ, Bottà G, et al. Genetic diversity of the African malaria vector Anopheles gambiae. Nature [Internet]. 2017. [cited 2017 Dec 4]; Available from: http://www.nature.com/doifinder/10.1038/nature24995. [DOI] [PMC free article] [PubMed]
- [72].Truong LN, Li Y, Shi LZ, et al. Microhomology-mediated End Joining and Homologous Recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc Natl Acad Sci USA. 2013;110:7720–7725. 10.1073/pnas.1213431110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Champer J, Reeves R, Oh SY, et al. Novel CRISPR/Cas9 gene drive constructs reveal insights into mechanisms of resistance allele formation and drive efficiency in genetically diverse populations. PLoS Genet. 2017;13:e1006796. 10.1371/journal.pgen.1006796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Hailong Wang, Xingzhi Xu. . Microhomology-mediated end joining: new players join the team. Cell Biosci. 2017;7(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Hammond AM, Kyrou K, Bruttini M, et al. The creation and selection of mutations resistant to a gene drive over multiple generations in the malaria mosquito. PLOS Genet. 2017;13:e1007039. 10.1371/journal.pgen.1007039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Unckless RL, Clark AG, Messer PW. Evolution of resistance against CRISPR/Cas9 gene drive. Genetics. 2017;205:827–841. 10.1534/genetics.116.197285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Marshall JM, Buchman A, Sánchez CHM, et al.. Overcoming evolved resistance to population-suppressing homing-based gene drives. Sci Rep. [Internet]. 2017. [cited 2017 Dec 4]; 7. Available from: http://www.nature.com/articles/s41598-017-02744-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Tsai SQ, Wyvekens N, Khayter C, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32:569. 10.1038/nbt.2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Zetsche B, Gootenberg JS, Abudayyeh OO, et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015;163:759–771. 10.1016/j.cell.2015.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Noble C, Min J, Olejarz J, et al. Daisy-chain gene drives for the alteration of local populations. bioRxiv. 2016;057307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Marshall J, Akbari O. Can CRISPR-based gene drive be confined in the wild? A question for molecular and population biology. bioRxiv. 2017;173914. [DOI] [PubMed] [Google Scholar]
- [82].Kim D, Bae S, Park J, et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. 2015;12:237. 10.1038/nmeth.3284 [DOI] [PubMed] [Google Scholar]
- [83].Tsai SQ, Zheng Z, Nguyen NT, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187. 10.1038/nbt.3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Tsai SQ, Nguyen NT, Malagon-Lopez J, et al. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR–Cas9 nuclease off-targets. Nat Methods. 2017;14:607. 10.1038/nmeth.4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Kleinstiver BP, Pattanayak V, Prew MS, et al. High-fidelity CRISPR-Cas9 variants with undetectable genome-wide off-targets. Nature. 2016;529:490–495. 10.1038/nature16526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Kleinstiver BP, Tsai SQ, Prew MS, et al. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat Biotechnol. 2016;34:869. 10.1038/nbt.3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Slaymaker IM, Gao L, Zetsche B, et al. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. 10.1126/science.aad5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Shen B, Zhang W, Zhang J, et al. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods. 2014;11:399. 10.1038/nmeth.2857 [DOI] [PubMed] [Google Scholar]
- [89].Kim D, Kim J, Hur JK, et al. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat Biotechnol. 2016;34:863. 10.1038/nbt.3609 [DOI] [PubMed] [Google Scholar]
- [90].Akbari OS, Bellen HJ, Bier E, et al. Safeguarding gene drive experiments in the laboratory. Science. 2015;349:927–929. 10.1126/science.aac7932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Committee on Gene Drive Research in Non-Human Organisms: Recommendations for Responsible Conduct, Board on Life Sciences, Division on Earth and Life Studies, et al Gene Drives on the Horizon: Advancing Science, Navigating Uncertainty, and Aligning Research with Public Values [Internet]. Washington, DC: National Academies Press; 2016. [cited 2017 Dec 4]. Available from: http://www.nap.edu/catalog/23405. [PubMed] [Google Scholar]
- [92].Adelman Z, Akbari O, Bauer J, et al. Rules of the road for insect gene drive research and testing [Internet]. Nat Biotechnol. 2017. [cited 2017 Dec 4]. Available from: https://www.nature.com/articles/nbt.3926. [DOI] [PMC free article] [PubMed]
- [93].DiCarlo JE, Chavez A, Dietz SL, et al. Safeguarding CRISPR-Cas9 gene drives in yeast. Nat Biotechnol. 2015;33:1250. 10.1038/nbt.3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Klein TA, Windbichler N, Deredec A, et al. Infertility resulting from transgenic I-PpoI male Anopheles gambiae in large cage trials. Pathog Glob Health. 2012;106:20–31. 10.1179/2047773212Y.0000000003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Luca Facchinelli, Laura Valerio, Rosemary S Lees, et al. . Stimulating Anopheles gambiae swarms in the laboratory: application for behavioural and fitness studies. Malar J. 2015;14(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Vella MR, Gunning CE, Lloyd AL, et al. Evaluating strategies for reversing CRISPR-Cas9 gene drives. Sci Rep. 2017;7:11038. 10.1038/s41598-017-10633-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Wu B, Luo L, Gao XJ. Cas9-triggered chain ablation of cas9 as a gene drive brake [Internet]. Nat Biotechnol. 2016. [cited 2017 Dec 4]. Available from: https://www.nature.com/articles/nbt.3444. [DOI] [PMC free article] [PubMed]
- [98].Gantz VM, Bier E. The dawn of active genetics. BioEssays. 2016;38:50–63. 10.1002/bies.201500102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Oye KA, Esvelt K, Appleton E, et al. Regulating gene drives. Science. 2014;345:626–628. 10.1126/science.1254287 [DOI] [PubMed] [Google Scholar]
- [100].Benedict MQ, Burt A, Capurro ML, et al. Recommendations for laboratory containment and management of gene drive systems in arthropods. Vector-Borne Zoonotic Dis. [Internet]. 2017. [cited 2017 Dec 4]; Available from: http://online.liebertpub.com/doi/10.1089/vbz.2017.2121. [DOI] [PMC free article] [PubMed]