Summary
Embryonic stem cells (ESCs) display heterogeneous expression of pluripotency factors such as Nanog when cultured with serum and leukemia inhibitory factor (LIF). In contrast, dual inhibition of the signaling kinases GSK3 and MEK (2i) converts ESC cultures into a state with more uniform and high Nanog expression. However, it is so far unclear whether 2i acts through an inductive or selective mechanism. Here, we use continuous time-lapse imaging to quantify the dynamics of death, proliferation, and Nanog expression in mouse ESCs after 2i addition. We show that 2i has a dual effect: it both leads to increased cell death of Nanog low ESCs (selective effect) and induces and maintains high Nanog levels (inductive effect) in single ESCs. Genetic manipulation further showed that presence of NANOG protein is important for cell viability in 2i medium. This demonstrates complex Nanog-dependent effects of 2i treatment on ESC cultures.
Keywords: 2i, Gsk3, Mek, embryonic stem cells, pluripotency, Nanog, cell heterogeneity, continuous time-lapse imaging, single-cell tracking, cell fate
Graphical Abstract

Highlights
-
•
Continuous long-term single-cell quantification of 2i effects on murine ESCs
-
•
2i enriches for a Nanog high population through a selective cell death effect
-
•
2i also upregulates Nanog expression and prevents its downregulation
-
•
The viability of Nanog−/− cells is compromised in 2i
Schroeder and colleagues show that GSK3 and MEK inhibition (2i) has several simultaneous effects on murine ESC cultures. They applied long-term time-lapse imaging to quantify cell death, proliferation, and Nanog expression dynamics in single ESCs. This demonstrated that 2i treatment of ESCs leads to more uniform and high Nanog expression due to both selective and inductive effects.
Introduction
Under the proper culture conditions, embryonic stem cells (ESCs) can self-renew indefinitely or differentiate into cell types of all germ layers (pluripotency). The maintenance of the pluripotent state is controlled by a network of transcription factors (TFs; Orkin and Hochedlinger, 2011). The expression levels of several of those TFs are heterogeneous in individual pluripotent ESCs in traditionally used serum plus leukemia inhibitory factor (LIF) culture conditions (SerumLIF; Chambers et al., 2007, Torres-Padilla and Chambers, 2014, Toyooka et al., 2008). Nanog, an extensively studied key pluripotency regulator (Chambers et al., 2003, Mitsui et al., 2003, Torres-Padilla and Chambers, 2014), has been shown to fluctuate between low and high expression in pluripotent ESCs (Abranches et al., 2014, Chambers et al., 2007, Filipczyk et al., 2015, Kalmar et al., 2009, Singer et al., 2014).
Dual inhibition of the signaling kinases GSK3 and MEK (2i) together with LIF promotes self-renewal of mouse ESCs in a state more similar to the pluripotent preimplantation epiblast (Boroviak et al., 2014, Ying et al., 2008). In 2i conditions, ESC cultures are more homogeneous with a more uniform and high Nanog expression (Abranches et al., 2014, Filipczyk et al., 2013, Singer et al., 2014, Wray et al., 2010). It is usually assumed that this shift in the Nanog distribution occurs because 2i upregulates Nanog expression in individual ESCs. Some publications suggest that GSK3 and MEK inhibitors increase Nanog transcription, protein synthesis, or stability (Hansen and van Oudenaarden, 2013, Kim et al., 2014, Miyanari and Torres-Padilla, 2012, Ochiai et al., 2014, Sanchez-Ripoll et al., 2013, Singer et al., 2014), and induce Nanog expression during the reprogramming to naive pluripotency (Silva et al., 2008, Theunissen et al., 2011).
However, these conclusions are based on Nanog quantifications of population averages and/or long after the start of 2i treatment. A reduction in Nanog decreases after 2i treatment was shown in single cells, but only in a qualitative way, and Nanog-dependent cell deaths were not quantified (Cannon et al., 2015). A selective effect of 2i cannot, therefore, be ruled out (Abranches et al., 2014, Boroviak et al., 2014, Etzrodt et al., 2014, Rieger et al., 2009). In this situation, 2i treatment would lead to the death or reduced proliferation specifically of Nanog low-expressing ESCs, thus enriching cultures for a uniform Nanog high-expressing population. 2i treatment would thus not have an effect on Nanog expression at the single-cell level but would differentially influence survival and/or proliferation speed in ESC subsets. In line with a potential selective effect of 2i, epiblast stem cells, which are in an alternative pluripotent state, as well as somatic cells at early stages of reprogramming, die or differentiate in 2i conditions (Guo et al., 2009, Silva et al., 2008). A potential selective 2i effect has also been assumed to assess the exit from the ESC state in vitro (Betschinger et al., 2013).
Given the widespread use of 2i treatment for research of the molecular control of pluripotency, it is therefore important to quantitatively clarify whether 2i effects on ESCs are in fact inductive or selective, or whether both effects simultaneously contribute to the observed homogenization of Nanog expression in ESC populations. Here, we performed continuous time-lapse imaging of Nanog reporter mouse ESC lines and quantified the dynamics of 2i-induced cell death events, cell proliferation rates, and Nanog expression (Etzrodt and Schroeder, 2017, Skylaki et al., 2016).
Results
Inductive and Selective 2i Effects Can Be Distinguished by Continuous Single-Cell Quantification
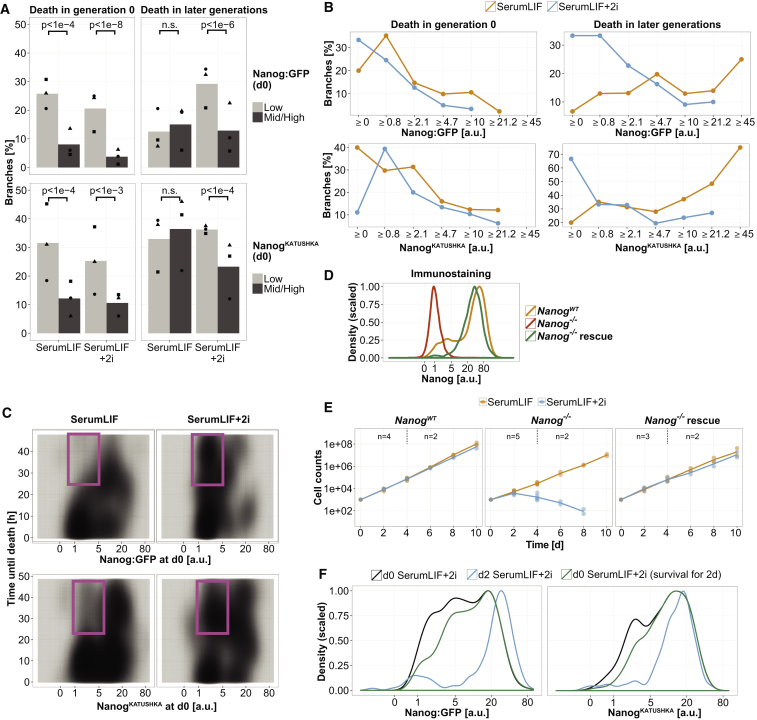
We confirmed that dual GSK3/MEK inhibition reduces the number of Nanog low-expressing cells using two different reporter ESC lines: a Nanog:GFP cell line (NG4) reporting transcription from a transgenic Nanog promoter (Schaniel et al., 2009), and a NanogKATUSHKA knockin cell line reporting endogenous NANOG protein levels from one Nanog allele (Filipczyk et al., 2013; Figure 1A). We aimed to distinguish whether 2i either induces or maintains high Nanog levels, or rather selects for Nanog high cells (Figure 1B). Hence, we applied continuous time-lapse imaging to track individual cells and quantified their Nanog expression after plating in SerumLIF or SerumLIF+2i. We confirmed that our experimental conditions, like the use of E-cadherin for plate coating, were largely neutral to the cells (Figures S1A–S1C). To obtain a representative dataset of many different clonal colonies, we tracked one random sister cell after each cell division, resulting in one branch per tree (a total of 1,383 independent branches; see Figure S1D for counts of individual experiments). To check for a potential selective effect of 2i, we measured the Nanog level in each tracked cell at the start of the movie (d0) and analyzed whether it or its progeny survived for 2 days (d2) when about 50% of the cells were in generation 3. To test for an inductive effect, we calculated the fold change of Nanog expression at d2 over d0 in each surviving branch. To distinguish between cell deaths induced by 2i and not by cell splitting, we quantified early deaths before the first cell division (in generation 0) and later deaths separately (Figure 1C; e.g., cells 12 and 13 in Video S1).
Figure 1.
Analysis of 2i Effects on Nanog Expression by Continuous Single-Cell Quantification
(A) Effects of 2i treatment on Nanog expression in ESC populations. Flow-cytometry analysis at the experiment start (d0) and after 2 days (d2) in SerumLIF with or without 2i. Wild-type ESCs were used as control for cell autofluorescence. One of three representative experiments is shown.
(B) Question of the study: at the single-cell level, the Nanog distribution change after 2i treatment can be explained either by induction and maintenance of high Nanog expression or by a negative selection against low Nanog-expressing cells.
(C) Experimental setup. Four representative trees are shown for the three groups we distinguish. Tracked cells are indicated by black circles (one random branch per tree, 1,383 branches). Pink crosses indicate cell death. Green bars indicate each of five time points when Nanog expression fold changes were calculated (d2 over d0) and cell survival was determined (d2). Question marks indicate cells that were not tracked. The first cell of a tree is defined as generation 0.
(D) Quantification of Nanog expression with image analysis in d0 cells and surviving d2 cells reproduced the Nanog distribution change in 2i between d2 and d0 (Nanog:GFP line: 343 branches in SerumLIF and 361 in SerumLIF+2i; NanogKATUSHKA line: 331 branches in SerumLIF and 348 in SerumLIF+2i; data were pooled from three independent experiments). The selection of cells at d0 was intentionally biased toward low-expressing cells to obtain higher numbers of Nanog low ESCs for robust statistical analysis. See also Figure S1F. Low/mid/high compartments, indicated by dashed lines, were defined by arbitrary thresholds and used the same way in Figures 3B, S3A, and 4A.
In (A) and (D), the smooth density estimate of each distribution was scaled to a maximum value of 1 (same for Figures S1B, S1C, S1F, 2D, 2F, and S2A). See also Figure S1.
The reduction of Nanog low cells after 2i addition within 2 days was also observed by our imaging approach in both serum-containing (Figure 1D) and serum-free (Figure S1F) basal medium. Of note, this occurred independently of potential selective proliferation effects: since we only analyzed one branch per tree, each cell at d0 contributed exactly one cell to the measured d2 distribution when the branch was surviving. In addition, we compared proliferation (determined by the cell generation of a branch at d2) with Nanog expression at d0 for each surviving branch, and found that they were weakly positively correlated. However, this was independent of 2i treatment (Spearman correlation for Nanog:GFP = 0.36 ± 0.13/0.45 ± 0.13, and for NanogKATUSHKA = 0.27 ± 0.06/0.29 ± 0.07, without/with 2i, respectively; mean ± SD, n = 3 independent experiments). Thus, 2i treatment does not influence the proliferation differences between Nanog low and high cells and hence, a selective proliferation effect cannot explain the observed changes in Nanog expression distributions after 2i treatment.
2i Selects for Nanog High-Expressing ESCs
We next analyzed whether 2i selectively induces cell death in Nanog low-expressing cells. The cell population dying immediately before the first cell division was strongly enriched for Nanog low cells (Figure S2A), independently of 2i treatment. These Nanog low-specific cell deaths are constantly introduced, likely by cell splitting of ESC cultures, irrespective of 2i. In contrast, ESCs that would die in later generations were enriched for Nanog low cells in SerumLIF+2i compared with SerumLIF (Figure S2A). For statistical analysis, we divided the Nanog distribution into low and mid/high cells based on this differential enrichment. Nanog low cells had significantly higher death rates than mid/high cells in generation 0, regardless of 2i (Figure 2A). This difference was also observed with conventional gelatin plate coating or in serum-free 2i + LIF medium (Figures S2B and S2C). In later generations, cell death rates of Nanog low cells were significantly higher than those of mid/high cells only after 2i addition (Figure 2A). This 2i-specific selective effect also occurred in serum-free 2i + LIF medium (Figure S2C). Furthermore, it did not depend on the threshold we had chosen. It was also seen when dividing Nanog levels into seven equally spaced bins. Indeed, Nanog levels and cell death rates are negatively correlated in later generations in SerumLIF+2i, with no or a slightly positive correlation in SerumLIF (Figure 2B). When analyzing the time until death, we found a late-dying population generated from Nanog low ESCs (between ∼25 and 48 hr), which was only present in SerumLIF+2i (Figure 2C). This further validates a 2i-specific selective effect against Nanog low ESCs. Of note, cell death rates were higher for NanogKATUSHKA than for Nanog:GFP ESCs, indicating additional unknown factors influencing cell survival. Importantly, however, 2i led to increased survival of high cells compared with low cells in both reporter lines, and thus, Nanog high cells were overall enriched by a selective effect of 2i.
Figure 2.
Addition of 2i Selects for Nanog High-Expressing Cells
(A) Nanog low cells have higher death rates than mid/high cells before the first cell division irrespective of 2i treatment. In addition, only 2i induces death of Nanog low cells in later generations (Cochran-Mantel-Haenszel chi-squared test; means [n = 3 experiments] are shown as bars). n.s., not significant. See also Figures S2A–S2C and Supplemental Experimental Procedures.
(B) Death rates in generation 0 (2i independent) and 2i-specific death rates in later generations gradually decrease with increasing Nanog levels. Cells at d0 were binned for Nanog levels and fractions of dying branches were determined for each bin. No data points for bins without branches are shown. See also Figure S2D for absolute branch counts.
(C) 2D density plots of dying cells reveal a subpopulation of Nanog low cells dying after ∼25–48 hr only present in SerumLIF+2i (magenta boxes).
(D) Quantitative immunostaining shows that the Nanog−/− rescue cell line, where Nanog is expressed from a constitutive promoter in a Nanog−/− background, expresses NANOG protein at endogenous levels. One representative experiment is shown (n > 7,900 cells per condition, pooled from two technical replicates).
(E) Proliferation of the Nanog−/− cell population collapses 2 days after 2i addition but is rescued by constitutive Nanog expression. Means are shown as lines and individual experiments as points. NanogWT denotes Nanog:GFP reporter cells that have both endogenous wild-type Nanog alleles. Nanog−/− cells were only cultured for 8 days in 2i due to scarcity of cells. See also Figure S2E.
(F) The selective effect of 2i cannot fully explain the measured Nanog distribution change in SerumLIF+2i. The Nanog distribution at d0 of branches surviving for 2 days in SerumLIF+2i (green; from Figure S2A) is compared with the measured SerumLIF+2i distributions at d0 (black) and d2 (blue; from Figure 1D). See text for further explanations and Figure S2A for d0 Nanog distributions of the corresponding dying populations.
In (B), (C), and (F), data were pooled from three independent experiments. See also Figure S2.
To distinguish whether Nanog expression is just a predictor for cell death probability or whether NANOG itself is important for cell viability in 2i conditions, we genetically controlled Nanog levels. We compared Nanog:GFP cells that contain both wild-type Nanog alleles (here referred to as NanogWT) with Nanog−/− cells, where both Nanog alleles are knocked out (Chambers et al., 2007), and Nanog−/− rescue cells, where a transgene constitutively expressing Nanog is stably integrated into Nanog−/− cells. We confirmed by quantitative immunostaining that NANOG was absent in Nanog−/− cells and that the Nanog−/− rescue cell line expressed NANOG at wild-type levels (Figure 2D). We then measured the proliferation of the cell populations with or without 2i every 2 days. While NanogWT cell proliferation was normal independent of 2i, we observed a drastic reduction of Nanog−/− cell counts after the first cell splitting in SerumLIF+2i, but not in SerumLIF. Nanog−/− cell counts continuously decreased in 2i, but Nanog−/− rescue cells proliferated similarly to wild-type cells in both conditions. This demonstrates that NANOG protein is important for cell viability in 2i (Figure 2E). We also observed a strong reduction of Nanog−/− cell counts in serum-free 2i + LIF medium, but the population did not fully collapse and morphologically normal colonies were predominant after 10 days, indicating that NANOG protein is important but can be dispensable under certain conditions even in the presence of 2i (Figure S2E).
We then addressed the question of whether the observed cell death-based selective effect is sufficient to explain the Nanog distribution change after 2i addition in our time-lapse data. We therefore determined the d0 Nanog distribution of surviving branches in SerumLIF+2i. This distribution is equivalent to a theoretical d2 Nanog distribution generated exclusively by selective cell death effects and without changes in Nanog expression of individual cells (Figure 2F). We then compared the SerumLIF+2i d0 distribution of surviving branches with the measured SerumLIF+2i d2 distribution (which includes Nanog expression dynamics in single cells) and found that Nanog low and mid cells were much more decreased in the latter (Figure 2F). Therefore, a selective cell death effect of 2i against Nanog low-expressing ESCs exists, but is not sufficient to explain the enrichment of Nanog high ESCs in 2i-treated populations.
2i Also Induces and Maintains High Nanog Expression in Single ESCs
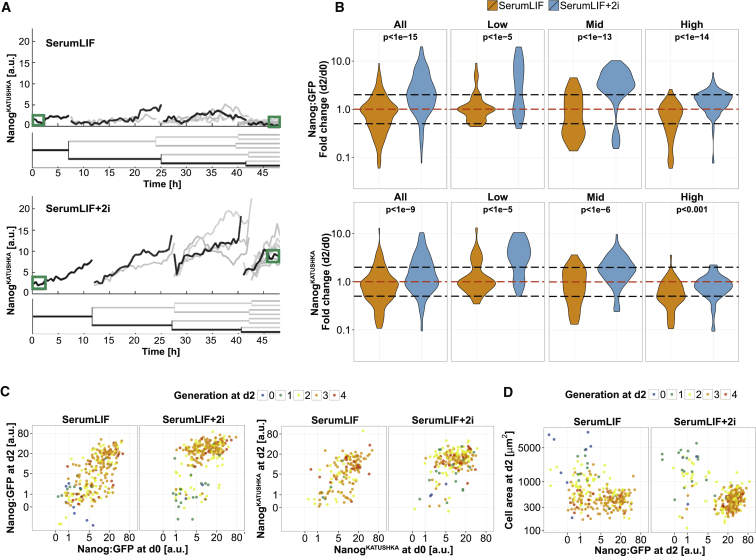
Thus, we tested how dual GSK3/MEK inhibition changes the Nanog expression dynamics in individual cells by continuous single-cell quantification (Figure 3A). We found many trees with strongly increased Nanog expression within 2 days in 2i (Figure 3A; Videos S2 and S3). To better detect differences of Nanog dynamics within the mid/high compartment, we binned ESCs into Nanog low, mid, and high expression at d0 (Figures 1D, S1F, 3B, and 4A). Nanog expression was significantly increased after 2i addition in single-cell branches, regardless of whether we pooled all cells or analyzed the low, mid, and high compartments individually (Figure 3B). Nanog was also significantly upregulated in low and mid cells after switching to serum-free 2i + LIF medium (Figure S3A). Importantly, many observed Nanog changes were higher than 2-fold and therefore could not be explained by cell-cycle effects alone. Addition of 2i to SerumLIF induced high Nanog expression in low and mid cells, and maintained this state in already high Nanog-expressing cells. Thus, most surviving cells were Nanog high after 2 days, except a minor fraction that did not upregulate, or even downregulate Nanog in 2i (Figure 3C). Those cells also showed a reduced cell proliferation speed and large sizes at d2 (Figure 3D). In contrast, the major Nanog low cell population present in SerumLIF, with cell areas (<1,000 μm2) and proliferation rates (at least in generation 2 at d2) similar to those in Nanog high cells, was strongly reduced after 2i addition (65%/21% of Nanog low cells or 22%/3% of all cells at d2, without/with 2i; pooled from three independent Nanog:GFP experiments), indicating that the remaining low cells in SerumLIF+2i are distinct from most of the Nanog low cells in SerumLIF. Additionally, we found that Nanog:GFP low cells with a large cell area often expressed only very low levels of the pluripotency markers POU5F1 (previously known as OCT4), SOX2, and KLF4, and endogenous NANOG protein. In contrast, morphologically normal Nanog low ESCs in SerumLIF conditions, which also had low NANOG protein, typically expressed high POU5F1 and SOX2 levels (Figure S3B). We conclude that 2i generally increases Nanog expression in single cells but some, especially very low Nanog-expressing cells, do not respond in the same way.
Figure 3.
2i Treatment Induces Nanog Upregulation and Prevents Its Downregulation in Single Mouse ESCs
(A) Representative example trees of low NanogKATUSHKA-expressing cells in SerumLIF or SerumLIF+2i. Single branches are highlighted in black. Green boxes indicate the time points at d0 and d2 that were used for Nanog quantification in (B) to (D). See also Figure S4A.
(B) Log10 fold changes of Nanog expression in single-cell branches show that 2i upregulates Nanog expression and prevents its downregulation. Surviving cells were either all pooled or binned by Nanog levels at d0. The red dashed line indicates Nanog maintenance and black dashed lines indicate 2-fold changes. p values were calculated using the Wilcoxon rank-sum test. See also Figure S3A.
(C) Induction and maintenance of a high Nanog state after 2i addition in most cell branches. A minor fraction characterized by strongly reduced proliferation does not enter a Nanog high state after 2i addition.
(D) After 2 days in SerumLIF+2i two cell populations exist, which are distinct in Nanog levels, cell area, and cell proliferation, but a Nanog low population with small cell area and high cell proliferation present in SerumLIF is nearly absent.
In (B) to (D), data were pooled from three independent experiments. See also Figure S3.
Figure 4.
Nanog Upregulation Dynamics in SerumLIF+2i Depends on Nanog Levels Rather than Cell Survival
(A) Cells with distinct Nanog levels show different Nanog upregulation kinetics in SerumLIF+2i. Original data points and smoothed curves of mean Nanog levels of low, mid, and high compartments (grouped at d0) are shown over time.
(B) Strategy to analyze Nanog upregulation kinetics before cell death. An example branch is shown for visualization. The maximum Nanog level (max), its fold change compared with d0, and the associated time point as well as the first time point where a fold change was bigger than 2 (>2-fold) were determined (yellow boxes). Those extracted parameters were used for (C) and (D) as well as Figures S4B and S4C. See also Supplemental Experimental Procedures.
(C) A large fraction of Nanog low cells upregulates Nanog more than 2-fold (red dashed line) before cell death in SerumLIF+2i (Nanog:GFP [61%, n = 28 branches] and NanogKATUSHKA [51%, n = 37 branches]). Cells that died in generation 0 were excluded from analysis.
(D) Nanog is upregulated on average in low cells in SerumLIF+2i independent of survival. The dataset was filtered on dying branches that upregulated Nanog >2-fold until death and surviving branches that upregulated Nanog >2-fold from d0 to d2. Cells that died in generation 0 were excluded from analysis. Time points containing less than 9 branches are not shown.
(E) Summary of cell fates of low (l), mid (m), and high (h) Nanog-expressing cells in SerumLIF −/+ 2i. “Downregulation” and “Upregulation” include greater than 2-fold Nanog decreases or increases, respectively. All dying cells are contained in the two “Death” groups and all surviving cells in the other groups. Means ± SEM are shown (n = 3).
In (A), (C), and (D), data were pooled from three independent experiments. See also Figure S4.
Kinetics and Concurrence of 2i Effects
We next checked the temporal order of selective and inductive 2i effects. We analyzed the mean Nanog expression dynamics within different Nanog expression compartments. We found that the Nanog:GFP reporter, which measures transcriptional activity, showed a consistent and rapid GFP upregulation after 2i addition compared with the control. This was apparent immediately in Nanog low cells and after around 3–6 hr in the other compartments. In contrast, the dynamics of the fusion-protein reporter NanogKATUSHKA varied strongly between compartments (Figure 4A). Here, Nanog levels started to increase in low cells also immediately after 2i addition, but only after about 1 day in mid cells. In Nanog high cells, levels even dropped initially in a 2i-independent fashion and started to increase again after about 1.5 days compared with SerumLIF.
Of note, this subpopulation analysis is able to disentangle the typical dynamics of larger groups well, but individual cells can still deviate substantially from the averages shown. For example, we found very variable Nanog onset times, maximum Nanog levels, and occurrences of fluctuations within single Nanog low branches in SerumLIF+2i (Figure S4A). Fluctuations were more prominent for NanogKATUSHKA than Nanog:GFP, which might be a result of transcriptional bursting (Ochiai et al., 2014) or post-transcriptional Nanog regulation.
We then checked whether the inductive 2i effect also occurs in branches that eventually die. Our focus was on Nanog low cells that died in later generations after the first cell division in SerumLIF+2i. We first established a method that allowed us to detect when and how much Nanog was upregulated in those branches before cell death (Figure 4B). We found that in over 50% of those branches Nanog had increased more than 2-fold any time before death (Figure 4C). This threshold was reached for Nanog:GFP after around 10 hr, similarly to a comparable subset of surviving branches (Figure S4B). Also, the average Nanog:GFP upregulation dynamics were the same as in the surviving branches (Figure 4D). NanogKATUSHKA upregulation appeared slightly later and weaker in the dying branches, but qualitatively comparable with the surviving ones. We also observed that dying branches that survived longer upregulated Nanog more and that upregulation reached its maximum shortly before cell death (Figure S4C).
We have described above that the selective 2i effect predominantly occurs after the first cell division (Figure 2A), starting after around 25 hr (Figure 2C), and thus later than the inductive effect, which already becomes detectable after a few hours (Figures 4A and S4B). Together, those preliminary data show that the inductive 2i effect also occurs in about half of the dying Nanog low branches. Nevertheless it remains uncertain whether those are exactly the branches where 2i was the reason for death. This complexity is further illustrated by two examples, where Nanog induction, cell death, or ceased proliferation co-occur either in the same cells or separately in different cells within one colony (Videos S1 and S4). We provide a quantitative overview of the main findings from our time-lapse data in Figure 4E.
Discussion
We quantitatively assessed both the selective and inductive effects of 2i treatment on changing Nanog expression in ESC populations at the single-cell level. Both selective and inductive effects exist in heterogeneous ESC cultures. The high resolution of our continuous and quantitative single-cell data allowed us to distinguish a 2i-specific selective effect against Nanog low cells from the generally reduced survival and proliferation potential of Nanog low cells (Abranches et al., 2014, Chambers et al., 2007, Kalmar et al., 2009). A previous time-lapse-based study failed to detect this selective effect because cell death events were neither observed over several cell generations nor correlated with Nanog expression. In addition, Nanog induction in Nanog low cells was not analyzed possibly because the reporter used did not faithfully report Nanog protein expression in low cells (Cannon et al., 2015).
It has been previously mentioned that Nanog−/− ESCs expand more efficiently in 2i + LIF than in SerumLIF medium, but without supporting data (Silva et al., 2009). In contrast, we observed that morphologically normal Nanog−/− colonies persisted in serum-free 2i + LIF medium for at least 10 days, but a switch to 2i caused severe viability defects, and in combination with serum even a continued collapse of the culture. We demonstrate that forced long-term NANOG expression safeguards cell survival in 2i conditions, but there was no indication that 2i induced Nanog upregulation would protect Nanog low cells from the selective effect. A previous study showed that 2i enhances Nanog transcription in Nanog−/− cells but also increases their propensity to differentiate (Navarro et al., 2012). A different Nanog−/− ESC line did not acquire a naive-like phenotype upon transfer to 2i + LIF media, but cell viability was not analyzed in detail (Acampora et al., 2017). In combination with our data, this suggests that 2i has two independent effects in Nanog low or Nanog−/− cells: the induction of Nanog expression and the control of their cell fate.
This complexity of 2i effects is further increased by the heterogeneity of Nanog low cells in SerumLIF cultures. We have previously shown that the Nanog protein level alone is a poor marker for the functional state of ESCs (Filipczyk et al., 2015). Only a fraction of Nanog low cells can revert to the high state, and only those fluctuating cells, and not those in a persistent Nanog low state, are functionally similar to Nanog high cells. The Nanog low population further contains cells that either resemble later epiblast stages, are biased toward primitive endoderm differentiation, or have unknown identity (Canham et al., 2010, Klein et al., 2015, Toyooka et al., 2008). It has been reported that minimal ERK activity is required for self-renewal and differentiation (Chen et al., 2015). We speculate that not all Nanog low cells are similarly dependent on ERK signaling. The state of some cells might be closer to epiblast-derived stem cells, which self-renew in the presence of fibroblast growth factor (FGF)/ERK signaling and have severe viability defects in 2i (Guo et al., 2009). ERK activity is also crucial for the differentiation into primitive endoderm (prEn; Hamilton and Brickman, 2014). It is thus plausible that the prEn-primed or committed Nanog low cells also have a greater dependency on ERK activity and hence reduced viability in 2i. In addition, we found some cells that were even refractory to inducing Nanog stably or at all, indicating that atypical ESCs in SerumLIF exist in a continuum of states.
It is likely that the departure from the naive state correlates with the long-term absence of Nanog expression (Filipczyk et al., 2015). We speculate that cells with only a recent decrease in Nanog expression are typically still in or close to a bona fide ESC state, and thus can survive in 2i and eventually stabilize or restore naive pluripotency. Thus, a short-term decrease in Nanog levels does not commit cells toward differentiation, as similarly a short-term increase in Nanog expression does not shield them from death in 2i. We suggest that the induction of one or few regulatory factors in non-bona fide ESCs is more readily accomplished than reprogramming of the cell fate, which likely requires a substantial change of the entire gene regulatory network. We propose that resetting or maintaining naive pluripotency depends on both a potent inducer such as 2i, but also a responsive and compatible initial cell state. We conclude that NANOG is not an essential, but an important factor in mediating the self-renewal promoting effect of 2i. Our data also confirm that heterogeneous expression of Nanog exists and is functionally relevant, a concept that had been questioned (Faddah et al., 2013).
Surprisingly, we saw an initial 2i-independent NANOG drop in Nanog high cells with the NanogKATUSHKA reporter that measures NANOG protein levels but not with the Nanog:GFP reporter, which measures Nanog promoter activity of a transgene. While cell-line-specific genetic and epigenetic differences cannot be excluded and additional cis-regulatory elements absent in the Nanog:GFP reporter might play a role, a plausible explanation is that simultaneous post-transcriptional mechanisms overshadow the transcription-based 2i-mediated Nanog induction during the first hours after 2i addition. As this effect also occurred in SerumLIF, we do not think it depicts a reorganization of the pluripotency network in 2i (Galonska et al., 2015).
The fact that a selective effect of 2i treatment indeed exists in ESCs could question previous studies' conclusions on Nanog induction by 2i treatment. However, since we showed that at the single-cell level 2i treatment also induces Nanog expression, the Nanog-inductive effect of 2i postulated in previous studies was potentially overestimated but likely correct.
For the reprogramming of human ESCs toward a naive state, the use of 2i seems crucial but not sufficient. Interestingly, short-term expression of NANOG and KLF2 allows one to overcome this barrier (Takashima et al., 2014, Theunissen et al., 2014). This indicates that those two TFs can restructure the cells' gene regulatory network in such a way that 2i can have the desired effects.
Two studies recently showed that 2i, especially MEK inhibition, not only promotes pluripotency but also severely diminishes the developmental potential of ESCs by inducing irreversible genetic and epigenetic aberrations (Choi et al., 2017b, Yagi et al., 2017). Those two effects are especially dramatic upon long-term culture and in female cells, respectively. Replacing the MEK inhibitor by an Src inhibitor, or reducing its concentration, could overcome these problems while preserving many of the desired effects of 2i. This highlights how important it is to fully understand the complex effects of 2i on pluripotent cells. A line of evidence suggests that the precise control of MEK/ERK signaling will be critical in generating stable and naive pluripotent cells from a variety of species including humans, independent of their sex (Chen et al., 2015, Choi et al., 2017a, Choi et al., 2017b, Schulz et al., 2014, Yagi et al., 2017).
2i or FGF/MEK inhibition alone also strongly increases the number of Nanog-positive epiblast cells during preimplantation development of mouse embryos in endoderm marker-expressing hypoblast cells. Most likely in this process, FGF/MEK inhibition induces Nanog and instructs lineage choice without any selective effects (Nichols et al., 2009, Yamanaka et al., 2010). As cells from early blastocyst stages are very plastic, they might be easily instructed by 2i, in contrast to a subset of Nanoglow ESCs. The viability of ESCs cultured in SerumLIF after injection into preimplantation embryos depends on the expression of the pluripotency marker Zfp42 (previously known as Rex1; Alexandrova et al., 2016). The low viability of Nanog low cells after cell splitting seems consistent with these observations, but it is also possible that the preimplantation embryo represents a selective environment against cells with low pluripotency gene expression, similarly to 2i in vitro.
Experimental Procedures
ESC Culture
Mouse ESC lines NG4 (Schaniel et al., 2009) and R1 containing a NanogKATUSHKA allele report Nanog promoter activity by GFP, or endogenous NANOG protein levels by a Nanog-fluorescent protein (KATUSHKA) fusion, respectively. The ESC line TβC44Cre6 was used as Nanog−/− cells (Chambers et al., 2007). All lines were routinely passaged every 2 days in Serum + LIF containing medium on gelatin or before experiments on E-cadherin-coated cell-culture plates. For SerumLIF+2i and N2B27 + LIF+2i medium, MEK inhibitor PD0325901 and GSK3 inhibitor CHIR99021 were added to SerumLIF or N2B27 + LIF at final concentrations of 1 μM and 3 μM, respectively.
Quantitative Time-Lapse Imaging
ESCs were cultured in SerumLIF, SerumLIF+2i, or N2B27 + LIF+2i medium on E-cadherin-coated culture slides and imaged at 30-min intervals for 2 days in both bright-field and fluorescent light channels in a 5% CO2 and 5% O2 mixed gas atmosphere at 37°C under a Zeiss Axio Observer Z1 microscope.
One random branch per tree was manually tracked and cell death events were determined using the software tTt (Hilsenbeck et al., 2016). Fluorescent image illumination was normalized, and Nanog intensities and cell areas of single cells were quantified as described by Filipczyk et al. (2015).
Data Processing and Statistics
Nanog intensity values were pooled for most analyses after normalization of replicate experiments (Figure S1E). The Cochran-Mantel-Haenszel chi-squared test was used to compare cell death rates in replicate experiments (Figures 2A, S2B, and S2C) and the Wilcoxon rank-sum test was used to compare Nanog fold changes (Figures 3B and S3A). See also Supplemental Experimental Procedures.
Author Contributions
S.H. performed experiments and analyzed data. F.J.T. supervised QTFy development by M.S. A.R. established the piggyback system. D.L.C. established immunostainings. K.A. made cell lines. T.S. supervised the study, developed bioimaging with D.L., K.D.K., and P.S.H. and cell tracking and analysis software with O.H. and S.S., and wrote the manuscript with S.H.
Acknowledgments
We thank C. Raithel for technical support. This work was supported by the German Research Foundation (SPP 1356), The International Human Frontier Science Program Organization, and the Swiss National Science Foundation. S.S. and O.H. acknowledge financial support for this project from SystemsX.ch.
Published: May 17, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, and four videos and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.04.019.
Supplemental Information
References
- Abranches E., Guedes A.M.V., Moravec M., Maamar H., Svoboda P., Raj A., Henrique D. Stochastic NANOG fluctuations allow mouse embryonic stem cells to explore pluripotency. Development. 2014;141:2770–2779. doi: 10.1242/dev.108910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acampora D., Di Giovannantonio L.G., Garofalo A., Nigro V., Omodei D., Lombardi A., Zhang J., Chambers I., Simeone A. Functional antagonism between OTX2 and NANOG specifies a spectrum of heterogeneous identities in embryonic stem cells. Stem Cell Reports. 2017;9:1642–1659. doi: 10.1016/j.stemcr.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrova S., Kalkan T., Humphreys P., Riddell A., Scognamiglio R., Trumpp A., Nichols J. Selection and dynamics of embryonic stem cell integration into early mouse embryos. Development. 2016;143:24–34. doi: 10.1242/dev.124602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger J., Nichols J., Dietmann S., Corrin P.D., Paddison P.J., Smith A. Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3. Cell. 2013;153:335–347. doi: 10.1016/j.cell.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroviak T., Loos R., Bertone P., Smith A., Nichols J. The ability of inner-cell-mass cells to self-renew as embryonic stem cells is acquired following epiblast specification. Nat. Cell Biol. 2014;16:516–528. doi: 10.1038/ncb2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canham M.A., Sharov A.A., Ko M.S.H., Brickman J.M. Functional heterogeneity of embryonic stem cells revealed through translational amplification of an early endodermal transcript. PLoS Biol. 2010;8:e1000379. doi: 10.1371/journal.pbio.1000379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon D., Corrigan A.M., Miermont A., McDonel P., Chubb J.R. Multiple cell and population-level interactions with mouse embryonic stem cell heterogeneity. Development. 2015;142:2840–2849. doi: 10.1242/dev.120741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Chen H., Guo R., Zhang Q., Guo H., Yang M., Wu Z., Gao S., Liu L., Chen L. Erk signaling is indispensable for genomic stability and self-renewal of mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2015;112:E5936–E5943. doi: 10.1073/pnas.1516319112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J., Clement K., Huebner A.J., Webster J., Rose C.M., Brumbaugh J., Walsh R.M., Lee S., Savol A., Etchegaray J.P. DUSP9 modulates DNA hypomethylation in female mouse pluripotent stem cells. Cell Stem Cell. 2017;20:706–719.e7. doi: 10.1016/j.stem.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J., Huebner A.J., Clement K., Walsh R.M., Savol A., Lin K., Gu H., Di Stefano B., Brumbaugh J., Kim S. Prolonged Mek1/2 suppression impairs the developmental potential of embryonic stem cells. Nature. 2017;548:219–223. doi: 10.1038/nature23274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etzrodt M., Schroeder T. Illuminating stem cell transcription factor dynamics: long-term single-cell imaging of fluorescent protein fusions. Curr. Opin. Cell Biol. 2017;49:77–83. doi: 10.1016/j.ceb.2017.12.006. [DOI] [PubMed] [Google Scholar]
- Etzrodt M., Endele M., Schroeder T. Quantitative single-cell approaches to stem cell research. Cell Stem Cell. 2014;15:546–558. doi: 10.1016/j.stem.2014.10.015. [DOI] [PubMed] [Google Scholar]
- Faddah D.A., Wang H., Cheng A.W., Katz Y., Buganim Y., Jaenisch R. Single-cell analysis reveals that expression of Nanog is biallelic and equally variable as that of other pluripotency factors in mouse ESCs. Cell Stem Cell. 2013;13:23–29. doi: 10.1016/j.stem.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipczyk A., Gkatzis K., Fu J., Hoppe P.S., Lickert H., Anastassiadis K., Schroeder T. Biallelic expression of Nanog protein in mouse embryonic stem cells. Cell Stem Cell. 2013;13:12–13. doi: 10.1016/j.stem.2013.04.025. [DOI] [PubMed] [Google Scholar]
- Filipczyk A., Marr C., Hastreiter S., Feigelman J., Schwarzfischer M., Hoppe P.S., Loeffler D., Kokkaliaris K.D., Endele M., Schauberger B. Network plasticity of pluripotency transcription factors in embryonic stem cells. Nat. Cell Biol. 2015;17:1235–1246. doi: 10.1038/ncb3237. [DOI] [PubMed] [Google Scholar]
- Galonska C., Ziller M.J., Karnik R., Meissner A. Ground state conditions induce rapid reorganization of core pluripotency factor binding before global epigenetic reprogramming. Cell Stem Cell. 2015;17:462–470. doi: 10.1016/j.stem.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., Yang J., Nichols J., Hall J.S., Eyres I., Mansfield W., Smith A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton W.B., Brickman J.M. Erk signaling suppresses embryonic stem cell self-renewal to specify endoderm. Cell Rep. 2014;9:2056–2070. doi: 10.1016/j.celrep.2014.11.032. [DOI] [PubMed] [Google Scholar]
- Hansen C.H., van Oudenaarden A. Allele-specific detection of single mRNA molecules in situ. Nat. Methods. 2013;10:869–871. doi: 10.1038/nmeth.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilsenbeck O., Schwarzfischer M., Skylaki S., Schauberger B., Hoppe P.S., Loeffler D., Kokkaliaris K.D., Hastreiter S., Skylaki E., Filipczyk A. Software tools for single-cell tracking and quantification of cellular and molecular properties. Nat. Biotechnol. 2016;34:703–706. doi: 10.1038/nbt.3626. [DOI] [PubMed] [Google Scholar]
- Kalmar T., Lim C., Hayward P., Muñoz-Descalzo S., Nichols J., Garcia-Ojalvo J., Martinez Arias A. Regulated fluctuations in Nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol. 2009;7:e1000149. doi: 10.1371/journal.pbio.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Kim M.O., Cho Y.Y., Yao K., Kim D.J., Jeong C.H., Yu D.H., Bae K.B., Cho E.J., Jung S.K. ERK1 phosphorylates Nanog to regulate protein stability and stem cell self-renewal. Stem Cell Res. 2014;13:1–11. doi: 10.1016/j.scr.2014.04.001. [DOI] [PubMed] [Google Scholar]
- Klein A.M., Mazutis L., Akartuna I., Tallapragada N., Veres A., Li V., Peshkin L., Weitz D.A., Kirschner M.W. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Miyanari Y., Torres-Padilla M.-E. Control of ground-state pluripotency by allelic regulation of Nanog. Nature. 2012;483:470–473. doi: 10.1038/nature10807. [DOI] [PubMed] [Google Scholar]
- Navarro P., Festuccia N., Colby D., Gagliardi A., Mullin N.P., Zhang W., Karwacki-Neisius V., Osorno R., Kelly D., Robertson M., Chambers I. OCT4/SOX2-independent Nanog autorepression modulates heterogeneous Nanog gene expression in mouse ES cells. EMBO J. 2012;31:4547–4562. doi: 10.1038/emboj.2012.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J., Silva J., Roode M., Smith A. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development. 2009;136:3215–3222. doi: 10.1242/dev.038893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai H., Sugawara T., Sakuma T., Yamamoto T. Stochastic promoter activation affects Nanog expression variability in mouse embryonic stem cells. Sci. Rep. 2014;4:7125. doi: 10.1038/srep07125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin S.H., Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145:835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger M.A., Hoppe P.S., Smejkal B.M., Eitelhuber A.C., Schroeder T. Hematopoietic cytokines can instruct lineage choice. Science. 2009;325:217–218. doi: 10.1126/science.1171461. [DOI] [PubMed] [Google Scholar]
- Sanchez-Ripoll Y., Bone H.K., Owen T., Guedes A.M.V., Abranches E., Kumpfmueller B., Spriggs R.V., Henrique D., Welham M.J. Glycogen synthase kinase-3 inhibition enhances translation of pluripotency-associated transcription factors to contribute to maintenance of mouse embryonic stem cell self-renewal. PLoS One. 2013;8:e60148. doi: 10.1371/journal.pone.0060148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaniel C., Ang Y.-S., Ratnakumar K., Cormier C., James T., Bernstein E., Lemischka I.R., Paddison P.J. Smarcc1/Baf155 couples self-renewal gene repression with changes in chromatin structure in mouse embryonic stem cells. Stem Cells. 2009;27:2979–2991. doi: 10.1002/stem.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz E.G., Meisig J., Nakamura T., Okamoto I., Sieber A., Picard C., Borensztein M., Saitou M., Blüthgen N., Heard E. The two active X chromosomes in female ESCs block exit from the pluripotent state by modulating the ESC signaling network. Cell Stem Cell. 2014;14:203–216. doi: 10.1016/j.stem.2013.11.022. [DOI] [PubMed] [Google Scholar]
- Silva J., Barrandon O., Nichols J., Kawaguchi J., Theunissen T.W., Smith A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6:2237–2247. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J., Nichols J., Theunissen T.W., Guo G., van Oosten A.L., Barrandon O., Wray J., Yamanaka S., Chambers I., Smith A. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer Z.S., Yong J., Tischler J., Hackett J.A., Altinok A., Surani M.A., Cai L., Elowitz M.B. Dynamic heterogeneity and DNA methylation in embryonic stem cells. Mol. Cell. 2014;55:319–331. doi: 10.1016/j.molcel.2014.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skylaki S., Hilsenbeck O., Schroeder T. Challenges in long-term imaging and quantification of single-cell dynamics. Nat. Biotechnol. 2016;34:1137–1144. doi: 10.1038/nbt.3713. [DOI] [PubMed] [Google Scholar]
- Takashima Y., Guo G., Loos R., Nichols J., Ficz G., Krueger F., Oxley D., Santos F., Clarke J., Mansfield W. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158:1254–1269. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen T.W., Van Oosten A.L., Castelo-Branco G., Hall J., Smith A., Silva J.C.R. Nanog overcomes reprogramming barriers and induces pluripotency in minimal conditions. Curr. Biol. 2011;21:65–71. doi: 10.1016/j.cub.2010.11.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen T.W., Powell B.E., Wang H., Mitalipova M., Faddah D.A., Reddy J., Fan Z.P., Maetzel D., Ganz K., Shi L. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15:471–487. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Padilla M.-E., Chambers I. Transcription factor heterogeneity in pluripotent stem cells: a stochastic advantage. Development. 2014;141:2173–2181. doi: 10.1242/dev.102624. [DOI] [PubMed] [Google Scholar]
- Toyooka Y., Shimosato D., Murakami K., Takahashi K., Niwa H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development. 2008;135:909–918. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- Wray J., Kalkan T., Smith A.G. The ground state of pluripotency. Biochem. Soc. Trans. 2010;38:1027–1032. doi: 10.1042/BST0381027. [DOI] [PubMed] [Google Scholar]
- Yagi M., Kishigami S., Tanaka A., Semi K., Mizutani E., Wakayama S., Wakayama T., Yamamoto T., Yamada Y. Derivation of ground-state female ES cells maintaining gamete-derived DNA methylation. Nature. 2017;548:224–227. doi: 10.1038/nature23286. [DOI] [PubMed] [Google Scholar]
- Yamanaka Y., Lanner F., Rossant J. FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development. 2010;137:715–724. doi: 10.1242/dev.043471. [DOI] [PubMed] [Google Scholar]
- Ying Q.-L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.