Abstract
Background
Dystroglycanopathies are a clinically and genetically heterogeneous group of disorders that are typically characterised by limb-girdle muscle weakness. Mutations in 18 different genes have been associated with dystroglycanopathies, the encoded proteins of which typically modulate the binding of α-dystroglycan to extracellular matrix ligands by altering its glycosylation. This results in a disruption of the structural integrity of the myocyte, ultimately leading to muscle degeneration.
Methods
Deep phenotypic information was gathered using the PhenoTips online software for 1001 patients with unexplained limb-girdle muscle weakness from 43 different centres across 21 European and Middle Eastern countries. Whole-exome sequencing with at least 250 ng DNA was completed using an Illumina exome capture and a 38 Mb baited target. Genes known to be associated with dystroglycanopathies were analysed for disease-causing variants.
Results
Suspected pathogenic variants were detected in DPM3, ISPD, POMT1 and FKTN in one patient each, in POMK in two patients, in GMPPB in three patients, in FKRP in eight patients and in POMT2 in ten patients. This indicated a frequency of 2.7% for the disease group within the cohort of 1001 patients with unexplained limb-girdle muscle weakness. The phenotypes of the 27 patients were highly variable, yet with a fundamental presentation of proximal muscle weakness and elevated serum creatine kinase.
Conclusions
Overall, we have identified 27 patients with suspected pathogenic variants in dystroglycanopathy-associated genes. We present evidence for the genetic and phenotypic diversity of the dystroglycanopathies as a disease group, while also highlighting the advantage of incorporating next-generation sequencing into the diagnostic pathway of rare diseases.
Electronic supplementary material
The online version of this article (10.1186/s13395-018-0170-1) contains supplementary material, which is available to authorized users.
Keywords: Whole-exome sequencing, Dystroglycanopathies, Limb-girdle muscle weakness
Background
DAG1 (dystroglycan 1) is a 5.8 kb gene transcript on chromosome 3p21 that is widely expressed in human tissue, including in the skeletal muscle [1]. The protein undergoes N- and O-linked glycosylation and is post-transcriptionally cleaved into two mature subunits: the ~ 156 kDa secreted cell surface α-dystroglycan and the 43 kDa transmembrane β-dystroglycan [2]. The dystroglycan complex, composed of one α- and one β-subunit, is a component of the larger dystrophin-glycoprotein complex that links the subsarcolemmal actin cytoskeleton to the extracellular matrix (ECM) [3]. ECM ligands such as laminin strictly rely on the correct presentation of glycan structures on the surface of α-dystroglycan for binding [4], and without this precise glycosylation, the function and integrity of the muscle cell are compromised.
Alpha-dystroglycan is glycosylated by a series of tightly regulated proteins as it is processed from the nucleus, through the endoplasmic reticulum and Golgi apparatus to reach the cell surface [5, 6]. Mutations in any of the genes encoding these proteins could result in diminished protein function, aberrant α-dystroglycan glycosylation and thus a reduced capacity to bind ECM ligands [7]. The disorders associated with abnormal α-dystroglycan glycosylation are collectively known as dystroglycanopathies. The clinical manifestations of dystroglycanopathies are extremely variable with a spectrum of severity within the disease group [8, 9]. Severe dystroglycanopathies are congenital and result in structural brain, eye and muscle abnormalities such as Walker-Warburg syndrome [10], muscle-eye-brain disease [11] and Fukuyama muscular dystrophy [12]. The spectrum then ranges through to limb-girdle muscular dystrophies (LGMD), which constitute less severe forms of the disease group with an adult onset and no brain or eye abnormalities [13, 14].
Homozygous and compound heterozygous mutations in 17 genes, plus DAG1 itself, have been associated with dystroglycanopathies [15]. The encoded proteins include those with well-defined glycosyltransferase functions as well as those with less well-characterised functions (Table 1). In addition to researching those already known to be associated with dystroglycanopathies, another challenge is to identify additional proteins in the glycosylation pathway that underlie disease pathology. For example, it is estimated that 20–50% of suspected dystroglycanopathy patients do not harbour mutations in characterised dystroglycanopathy genes, suggesting the genetic cause of the disease is currently unknown for approximately half of patients. With such genetic and clinical heterogeneity among dystroglycanopathies, the causative genes can be difficult to determine from the phenotypes alone.
Table 1.
Genes associated with dystroglycanopathies
| Gene | Protein function | Associated dystroglycanopathy according to OMIM [49] | Reference |
|---|---|---|---|
| B3GALNT2 | Glycosyltransferase | Congenital muscular dystrophy-dystroglycanopathy type A 11, with brain and eye anomalies | [50] |
| B3GNT1 | Glycosyltransferase | Congenital muscular dystrophy-dystroglycanopathy type A 13, with brain and eye anomalies | [51] |
| DAG1 | Connects actin cytoskeleton to extracellular matrix | Congenital muscular dystrophy-dystroglycanopathy type A 9, with brain and eye anomalies | [52] |
| Limb-girdle muscular dystrophy-dystroglycanopathy type C 9 | [53] | ||
| DOLK | Kinase | Congenital disorder of glycosylation type Im | [54] |
| DPM1 | Transferase | Congenital disorder of glycosylation type Ie | [55] |
| DPM2 | Transferase | Congenital disorder of glycosylation type Iu | [56] |
| DPM3 | Transferase | Congenital disorder of glycosylation type Io | [44] |
| FKRP | Glycosyltransferase | Limb-girdle muscular dystrophy-dystroglycanopathy type C 5/LGMD2I | [25] |
| Congenital muscular dystrophy-dystroglycanopathy type A 5, with or without mental retardation | [57] | ||
| Congenital muscular dystrophy-dystroglycanopathy type B 5, with or without mental retardation | [25] | ||
| FKTN | Glycosyltransferase | Congenital muscular dystrophy-dystroglycanopathy type A 4, with brain and eye anomalies | [58] |
| Congenital muscular dystrophy-dystroglycanopathy type B 4, without mental retardation | [28] | ||
| Limb-girdle muscular dystrophy-dystroglycanopathy type C 4 | [59] | ||
| GMPPB | Transferase | Congenital muscular dystrophy-dystroglycanopathy type A 14, with brain and eye anomalies | [30] |
| Congenital muscular dystrophy-dystroglycanopathy type B 14, with mental retardation | |||
| Limb-girdle muscular dystrophy-dystroglycanopathy type C 14 | |||
| ISPD | Synthase | Congenital muscular dystrophy-dystroglycanopathy type A 7, with brain and eye anomalies | [47] |
| Limb-girdle muscular dystrophy-dystroglycanopathy type C 7 | [60] | ||
| LARGE1 | Glycosyltransferase | Congenital muscular dystrophy-dystroglycanopathy type B 6, with mental retardation | [61] |
| POMGNT1 | Glycosyltransferase | Congenital muscular dystrophy-dystroglycanopathy type A 3, with brain and eye anomalies | [62] |
| Congenital muscular dystrophy-dystroglycanopathy type B 3, with mental retardation | [28] | ||
| Limb-girdle muscular dystrophy-dystroglycanopathy type C 3 | [63] | ||
| POMGNT2 | Glycosyltransferase | Congenital muscular dystrophy-dystroglycanopathy type A 8, with brain and eye anomalies | [64] |
| POMK | Kinase | Congenital muscular dystrophy-dystroglycanopathy type A 12, with brain and eye anomalies | [65] |
| Limb-girdle muscular dystrophy-dystroglycanopathy type C 12 | [66] | ||
| POMT1 | Glycosyltransferase | Congenital muscular dystrophy-dystroglycanopathy type A 1, with brain and eye anomalies | [67] |
| Congenital muscular dystrophy-dystroglycanopathy type B 1, with mental retardation | [68] | ||
| Limb-girdle muscular dystrophy-dystroglycanopathy type C 1 | [69] | ||
| POMT2 | Glycosyltransferase | Congenital muscular dystrophy-dystroglycanopathy type A 2, with brain and eye anomalies | [29] |
| Congenital muscular dystrophy-dystroglycanopathy type B 2, with mental retardation | |||
| Limb-girdle muscular dystrophy-dystroglycanopathy type C 2 | |||
| TMEM5 | Glycosyltransferase | Congenital muscular dystrophy-dystroglycanopathy type A 10, with brain and eye anomalies | [45] |
Next-generation sequencing (NGS) is a well-developed methodology that can benefit the understanding, characterisation and diagnosis of such rare neuromuscular diseases. The entire exome of a patient can be sequenced, known as whole-exome sequencing (WES), at rapidly declining costs. Immediately performing WES could negate the need for undirected preliminary molecular investigations that can inflate the costs of a diagnostic evaluation [16]. MYO-SEQ is a research collaboration between academia, patient organisations and industry. This project applies WES to patients with unexplained limb-girdle muscle weakness and targets the analysis towards genes with a known association to neuromuscular diseases. Included in the candidate gene list were those involved in the glycosylation of α-dystroglycan. Here, we present the identification of 27 patients with putative pathogenic variants in dystroglycanopathy-associated genes from the targeted WES analysis of 1001 patients with unexplained limb-girdle muscle weakness. We highlight the diversity of the disease group and offer further characterisations of the diseases at the milder, limb-girdle weakness end of the spectrum.
Methods
Patients
Ethical approval (REC reference number 08/H0906/28) was granted by the Newcastle and North Tyneside Research Ethics Committee. One thousand and one patients were recruited throughout Europe and neighbouring countries, and informed written consent was given by all. Inclusion criteria stipulated that patients must present with limb-girdle muscle weakness and/or elevated serum creatine kinase (CK) activity.
Whole-exome sequencing and data analysis
WES and data processing were performed by the MacArthur laboratory at the Broad Institute of MIT and Harvard (Broad Institute, Cambridge, MA, USA) as detailed previously [17]. The variant call set was uploaded onto the MacArthur laboratory’s seqr platform for genomic data analysis. The biological relevance of the detected variants was determined by considering the (i) ClinVar reports of pathogenicity [18], (ii) published literature, (iii) population frequency detailed by the gnomAD reference population database from the Broad Institute that contains exome data from over 120,000 individuals [19] and (iv) deleteriousness of the variant predicted by PolyPhen-2 [20], SIFT [21], MutationTaster2 [22] and FATHMM [23]. The detected variants were matched to the patient’s phenotype, and those that were most likely to be disease-causing were reported back to the referring clinician.
Muscle histopathology
Muscle biopsies were obtained for all patients and analysed following standard histological techniques for light microscopy by local pathologists. Biopsy reports from pathology departments formed the basis to describe histological features in our patient cohort. Immunostaining was performed for 21 patients. The primary anti-α-dystroglycan antibodies that were used were VIA4-1 and IIH6C4 (Millipore), and NBP2-14868 (Novus Biologicals) for patients 6 and 7 only. The techniques used for patients 9, 10, 13 and 23 were unknown.
Results
Homozygous variants within dystroglycanopathy-associated genes
Dystroglycanopathies have so far only been reported with an autosomal recessive pattern of inheritance, and so only homozygous or compound heterozygous mutations in the associated genes could result in the disease phenotype, while heterozygous carriers are not affected by muscle disease. Of the 1001 patients (46% female, 54% male, mean age 39 years, median age 38 years, age range 2 to 88 years) whose exomes were analysed, nine patients had suspected pathogenic homozygous variants that could account for their clinical presentations (Table 2). Patient 1 had a rare homozygous missense variant in DPM3 (p.Leu44Pro) that is absent in homozygosity in the control population and that affects a highly conserved amino acid [24]. At the age of 30 years, he presented with, and fully recovered from, an asymmetric brachial plexopathy (serum creatine kinase [CK] activity of 4310 IU/L [normal < 200 IU/L]). Twelve years later, he displayed an unsteady gait (serum CK levels of 2732 IU/L), and a proximal lower limb weakness mildly progressed over the course of the next 5 years. Patients 2–8 had homozygous missense variants in FKRP (p.Leu276Ile for all but patient 3 who instead had a p.Tyr182His change) [25], with a young age at disease onset, proximal muscle weakness and an elevated serum CK activity. Patient 7 first presented with muscle cramps on exercise. Patient 22 had a novel homozygous missense variant in POMT2 (p.Gly238Val) [26], with an onset in childhood of proximal upper and lower limb weakness, central and cortical atrophy and spinal rigidity.
Table 2.
Suspected pathogenic homozygous variants detected by the MYO-SEQ project in dystroglycanopathy-associated genes
| Patient | Gene | Location | Predicted deleteriousness | ClinVar clinical significance | gnomAD allele frequency | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| hg19 co-ordinates | Protein change | Sequence change | SIFT | PolyPhen-2 | MutationTaster2 | FATHMM | ||||
| 1 | DPM3 | chr1:155112676 | p.Leu44Pro | c.131T>C | Damaging | Probably damaging | Disease causing | Tolerated | No | 0.000016a |
| 2 | FKRP | chr19:47259533 | p.Leu276Ile | c.826C>A | Tolerated | Benign | Disease causing | Damaging | Pathogenic; likely pathogenic | 0.001089a |
| 3 | FKRP | chr19:47259251 | p.Tyr182His | c.544T>C | – | Probably damaging | Disease causing | Damaging | Uncertain | 0.000018a |
| 4 | FKRP | chr19:47259533 | p.Leu276Ile | c.826C>A | Tolerated | Benign | Disease causing | Damaging | Pathogenic; likely pathogenic | 0.001089a |
| 5 | FKRP | chr19:47259533 | p.Leu276Ile | c.826C>A | Tolerated | Benign | Disease causing | Damaging | Pathogenic; likely pathogenic | 0.001089a |
| 6 | FKRP | chr19:47259533 | p.Leu276Ile | c.826C>A | Tolerated | Benign | Disease causing | Damaging | Pathogenic; likely pathogenic | 0.001089a |
| 7 | FKRP | chr19:47259533 | p.Leu276Ile | c.826C>A | Tolerated | Benign | Disease causing | Damaging | Pathogenic; likely pathogenic | 0.001089a |
| 8 | FKRP | chr19:47259533 | p.Leu276Ile | c.826C>A | Tolerated | Benign | Disease causing | Damaging | Pathogenic; likely pathogenic | 0.001089a |
| 22 | POMT2 | chr14:77767536 | p.Gly238Val | c.713G>T | Tolerated | Possibly damaging | Disease causing | Damaging | Uncertain | 0.000000a |
aNot reported in homozygosity in gnomAD
Compound heterozygous variants within dystroglycanopathy-associated genes
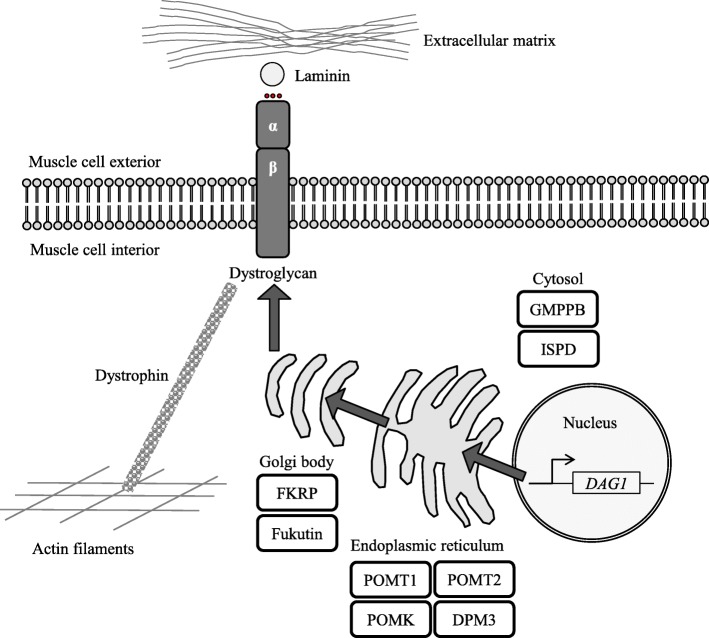
An additional 18 patients had suspected pathogenic compound heterozygous variants in dystroglycanopathy-associated genes (Table 3). Patient 9 harboured variants in FKRP (p.Leu276Ile and p.Pro462Ser) [25, 27] and had a largely similar phenotype to the homozygous FKRP patients. Patient 10 carried variants in FKTN (p.Arg307Ter and p.Arg307Gln) [28, 29], with infantile onset, limb-girdle weakness and elevated serum CK activity. Patients 11 and 12 harboured variants in GMPPB (p.Asp27His and p.Arg287Trp) [30, 31]. Patient 13 similarly carried the p.Asp27His missense variant in GMPPB [30], but instead, this occurred in combination with a novel frameshift variant (p.Met64Ter). All three patients had an onset of symptoms at a young age and a slowly progressive disease course. Patient 14 harboured variants in ISPD (p.Cys56ValfsTer60 and p.Ser202Leu), presenting with a slowly progressive limb-girdle muscle weakness phenotype, with both an EMG and a muscle biopsy displaying myopathic changes and an elevated serum CK activity. Onset was in young adulthood, and there were no ocular or cognitive impairments. Patients 15 and 16 were siblings from non-consanguineous parents who presented during childhood with muscle cramps, hyperlordosis, calf pseudohypertrophy, scapular winging and elevated serum CK activity [32]. They carried rare variants in POMK (p.Pro322Leu and p.Arg46Ter). Patient 17 carried a rare missense variant in POMT1 (p.Pro66Leu) in addition to a frameshift variant (p.Asp723GlyfsTer8) [33]. He had a non-progressive disease course from infancy with limb-girdle weakness; he did not have any contractures or spinal rigidity. Patients 19–21 and patients 23–27 all harboured previously unreported variants in POMT2, seven of which were absent in the control population. Patient 23 carried novel frameshift (p.Gly705GlufsTer31) and missense (p.Thr184Met) [34] variants in POMT2, while patient 24 harboured two missense variants in the gene (p.Tyr666Cys and p.Tyr136His) [35]. A summary of the glycosylation pathway and the localisation of the associated proteins is depicted in Fig. 1.
Table 3.
Suspected pathogenic compound heterozygous variants detected by the MYO-SEQ project in dystroglycanopathy-associated genes
| Patient | Gene | Location | Predicted deleteriousness | ClinVar clinical significance | gnomAD allele frequency | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| hg19 co-ordinates | Protein change | Sequence change | SIFT | PolyPhen-2 | MutationTaster2 | FATHMM | ||||
| 9 | FKRP | chr19:47259533 | p.Leu276Ile | c.826C>A | Tolerated | Benign | Disease causing | Damaging | Pathogenic; likely pathogenic | 0.001089a |
| chr19:47260091 | p.Pro462Ser | c.1384C>T | Damaging | Probably damaging | Disease causing | Damaging | No data | 0.000009a | ||
| 10 | FKTN | chr9:108380248 | p.Arg307Ter | c.919C>T | No data | No data | Disease causing | No data | Pathogenic; likely pathogenic | 0.000020a |
| chr9:108380249 | p.Arg307Gln | c.920G>A | No data | Probably damaging | Disease causing | Damaging | Pathogenic | 0.000012a | ||
| 11 | GMPPB | chr3:49761081 | p.Asp27His | c.79G>C | Damaging | Possibly damaging | Disease causing | Damaging | Pathogenic | 0.000655 |
| chr3:49759490 | p.Arg287Trp | c.859C>T | Damaging | Possibly damaging | Disease causing | Tolerated | Pathogenic | 0.000094a | ||
| 12 | GMPPB | chr3:49761081 | p.Asp27His | c.79G>C | Damaging | Possibly damaging | Disease causing | Damaging | Pathogenic | 0.000655 |
| chr3:49759490 | p.Arg287Trp | c.859C>T | Damaging | Possibly damaging | Disease causing | Tolerated | Pathogenic | 0.000094a | ||
| 13 | GMPPB | chr3:49760844 | p.Met64Ter | c.190delA | No data | No data | No data | No data | No data | 0.000000a |
| chr3:49761081 | p.Asp27His | c.79G>C | Damaging | Possibly damaging | Disease causing | Damaging | Pathogenic | 0.000655 | ||
| 14 | ISPD | chr7:16460782 | p.Cys56ValfsTer60 | c.165dupG | No data | No data | No data | No data | No data | 0.000000a |
| chr7:16415796 | p.Ser202Leu | c.605C>T | Damaging | Probably damaging | Disease causing | Damaging | No data | 0.000033a | ||
| 15 | POMK | chr8:42977932 | p.Pro322Leu | c.965C>T | Damaging | Probably damaging | Disease causing | Tolerated | No data | 0.000004a |
| chr8:42958827 | p.Arg46Ter | c.136C>T | No data | No data | Disease causing | No data | No data | 0.000180a | ||
| 16 | POMK | chr8:42977932 | p.Pro322Leu | c.965C>T | Damaging | Probably damaging | Disease causing | Tolerated | No data | 0.000004a |
| chr8:42958827 | p.Arg46Ter | c.136C>T | No data | No data | Disease causing | No data | No data | 0.000180a | ||
| 17 | POMT1 | chr9:134381575 | p.Pro66Leu | c.197C>T | Damaging | Probably damaging | Disease causing | Damaging | No data | 0.000053a |
| chr9:134398412 | p.Asp723GlyfsTer8 | c.2167dupG | No data | No data | No data | No data | Pathogenic | 0.000171a | ||
| 18 | POMT2 | chr14:77753158 | p.Arg421Trp | c.1261C>T | Damaging | Probably damaging | Disease causing | Damaging | Likely pathogenic | 0.000024a |
| chr14:77762593 | p.Thr344Pro | c.1030A>C | Damaging | Probably damaging | Disease causing | Damaging | Uncertain | 0.000000a | ||
| 19 | POMT2 | chr14:77746421 | p.Leu577ProfsTer8 | c.1727dupG | No data | No data | No data | No data | No data | 0.000032a |
| chr14:77753158 | p.Arg421Trp | c.1261C>T | Damaging | Probably damaging | Disease causing | Damaging | Likely pathogenic | 0.000024a | ||
| 20 | POMT2 | chr14:77746421 | p.Leu577ProfsTer8 | c.1727dupG | No data | No data | No data | No data | No data | 0.000032a |
| chr14:77753158 | p.Arg421Trp | c.1261C>T | Damaging | Probably damaging | Disease causing | Damaging | Likely pathogenic | 0.000024a | ||
| 21 | POMT2 | chr14:77751373 | p.Gln499Arg | c.1496A>G | Damaging | Probably damaging | Disease causing | Damaging | No data | 0.000000a |
| chr14:77755120 | p.Arg413Pro | c.1238G>C | Damaging | Probably damaging | Disease causing | Damaging | Pathogenic; uncertain | 0.000028a | ||
| 23 | POMT2 | chr14:77744748 | p.Gly705GlufsTer31 | c.2114_2135delGAATCCTGAGCCTGCTCCTGGG | No data | No data | No data | No data | No data | 0.000000a |
| chr14:77769283 | p.Thr184Met | c.551C>T | No data | Probably damaging | Disease causing | Damaging | Pathogenic | 0.000007a | ||
| 24 | POMT2 | chr14:77772712 | p.Tyr136His | c.406T>C | Damaging | Probably damaging | Disease causing | Damaging | No data | 0.000000a |
| chr14:77745107 | p.Tyr666Cys | c.1997A>G | Damaging | Probably damaging | Disease causing | Damaging | Pathogenic; likely pathogenic | 0.000061a | ||
| 25 | POMT2 | chr14:77767432 | ESS | c.816+1G>A | No data | No data | Disease causing | No data | No data | 0.000004a |
| chr14:77778319 | p.Phe102Leu | c.306C>A | Damaging | Probably damaging | Disease causing | Damaging | No data | 0.000000a | ||
| 26 | POMT2 | chr14:77750156 | p.His546Pro | c.1637A>C | No data | No data | Disease causing | No data | No data | 0.000007a |
| chr14:77769277 | p.Cys186Tyr | c.1654-5T>G | Tolerated | Possibly damaging | Disease causing | Damaging | No data | 0.000000a | ||
| 27 | POMT2 | chr14:77746811 | ExtSS | c.1654-5T>G | No data | No data | No data | No data | No data | 0.000000a |
| chr14:77765843 | p.Leu293His | c.878T>A | No data | Probably damaging | Disease causing | Damaging | No data | 0.000000a | ||
ESS essential splice site, ExSS extended splice site
aNot reported in homozygosity in gnomAD
Fig. 1.
Localisation of the proteins involved in the glycosylation of α-dystroglycan. Only the encoded proteins of the genes identified as harbouring suspected pathogenic variants in the MYO-SEQ project are shown. DAG1 is transcribed and translated into α-dystroglycan and β-dystroglycan subunits. As the proteins are processed through the endoplasmic reticulum and Golgi body to the muscle cell membrane (pathway indicated by grey arrows), GMPPB, POMT1, POMT2, POMK, ISPD, DPM3, FKRP and fukutin all contribute to the correct glycosylation of the α-subunit. The glycosylation of α-dystroglycan is required for interactions with extracellular matrix components; the dystroglycan complex as a whole thus acts as an anchor between the extracellular matrix and the intercellular actin cytoskeleton
Patient phenotypes
Overall, the 27 patients (18 compound heterozygotes and nine homozygotes) had an average age of 32 years (age range 11 to 63 years, median age 28 years), with 15 females (56%) and 12 males (44%) in the cohort. All patients had limb-girdle muscle weakness and an elevated serum CK activity (Additional file 1). The age of disease onset ranged from foetal to middle age, and while the majority of patients had a progressive disease, three patients had non-progressive symptoms. Forty-four percent of the 27 patients were unable to walk independently at the time of enrolment onto the project. Eighteen patients had normal cognitive capabilities; nine had reported cognitive impairment. Patients 13, 15 and 24 had slight valve defects, left ventricular enlargement and dilated cardiomyopathy, respectively, while patient 23 had a reduced left ventricular fractional shortening (LVFS) of 25%. The cardiac work-ups for the remaining patients were unremarkable. Fourteen patients had reduced forced vital capacities (FVC) ranging between 34 and 83%. Muscle imaging scans were available for 21 patients, four of which were unremarkable. The imaging for the remaining patients showed fatty replacement primarily of the paraspinal and proximal muscles of the lower limbs. Electromyography (EMG) results displayed a predominantly myopathic pattern, while muscle biopsies showed either myopathic and/or dystrophic changes. The biopsy of patient 6 showed internal nuclei and that of patient 8 showed glycogen accumulation. Immunohistochemistry (IHC) studies detected an α-dystroglycan deficiency or reduced α-dystroglycan glycosylation in 14 patients.
Discussion
Eighteen genes are currently known to be associated with dystroglycanopathies, with more causal genes yet to be identified [15]. The genes and their specific mutations do not always correlate with the phenotype or its severity, meaning dystroglycanopathies are an inherently complex group of neuromuscular diseases. Here, we used an unbiased sequencing approach to investigate the underlying genetic mechanisms of this disease group. This was in the form of targeted WES, an NGS technology that interrogates the 1% of the human genome that is coding. As 85% of known pathogenic variants reside in the coding regions alone [36], both the cost and the analysis workload associated with a whole-genome sequencing approach were avoided. The success of WES is widely recognised and has been reported in a range of diseases such as Parkinson disease [37], autism [38] and inflammatory bowel disease [39]. Indeed, we used targeted WES analysis in 1001 patients with unexplained limb-girdle muscle weakness and have found suspected pathogenic variants in 27 (2.7%) patients across nine of the 18 dystroglycanopathy-associated genes. The patients did not harbour suspected or potentially pathogenic variants in any other of the 429 known neuromuscular disease genes that were analysed as part of our study. The absence of a unified international dystroglycanopathy registry means that such contributions to rare disease research may not be fully exploited, which would otherwise be advantageous in furthering current knowledge of the disease.
Despite the central presentation of proximal muscle weakness, which was shared by all of our patients, we did not observe a common, core phenotype that could reliably distinguish the disease group from other LGMDs. Phenotypic variability was high even within the same disease group; for example, the reported age of disease onset for patients with POMT2 variants ranged from foetal to middle age. POMT2 deficiency was initially associated with Walker-Warburg syndrome, a congenital disease with brain, eye and muscle affections [40], while mutations in the gene were only later described to be associated with LGMD2N [34]. This pleiotropy could explain such phenotypic differences, where perhaps the functional severity of the mutations correlates with the extent of hypoglycosylation and thus the severity of the resulting phenotype.
Indeed, a correlation between reduced α-dystroglycan staining and clinical course was observed for patients with mutations in POMT1, POMT2 and POMGNT1 [41]. In contrast, however, the study did not always detect such a pattern for patients with mutations in FKTN and FKRP. Within our own cohort, staining for α-dystroglycan was performed for most of our patients, yet an α-dystroglycan deficiency or reduced α-dystroglycan glycosylation was only detected in 14 of these. Seven patients had no indication of a dystroglycanopathy based on immunostaining. It must be noted that a standardised staining protocol was used for many, but not all, of these patients. Conversely, an additional 15 patients of the remaining 209 for whom immunostaining was reported within the overall MYO-SEQ cohort had an α-dystroglycan deficiency or reduced α-dystroglycan glycosylation, yet none of these harboured suspected pathogenic variants in any known dystroglycanopathy genes. In fact, nine of these patients had suspected pathogenic variants in other neuromuscular disease genes, leaving only six currently undiagnosed patients. Together, this suggests that (i) the detection of changes in α-dystroglycan and its glycosylation may not be entirely accurate and/or uniform across different clinical sites, (ii) the glycosylation of α-dystroglycan may not be the only affected dystroglycanopathy pathway, and (iii) other dystroglycanopathies are most likely yet to be characterised. Accordingly, the quantification of hypoglycosylation should not be the only gold standard for diagnosing the diseases. We propose that a complementary approach including NGS should be used to aid the correct diagnosis of these rare disease patients.
Our findings offer an expansion of the phenotypic spectrum of dystroglycanopathies and suggest that an unbiased NGS approach such as WES should be used for a reliable diagnosis. Dolichol-phosphate-mannose (DPM) synthase comprises the DPM1, DPM2 and DPM3 subunits and is required for the synthesis of a donor substrate (DPM) of glycosylation [42, 43]. A homozygous DPM3 mutation (p.Leu85Ser) was first identified in an 11-year-old female with a mild LGMD, cardiomyopathy at 20 years of age and a stroke-like episode at 21 years of age [44]. Interestingly, our identification of a patient with a homozygous DPM3 mutation (p.Leu44Pro) is the first description of such a patient without cardiomyopathy or central nervous system involvement [24]. Moreover, we identified two siblings with POMK mutations. Typically, patients with mutations in this gene have displayed a severe phenotype ranging from Walker-Warburg syndrome to LGMD with cognitive impairment. Unlike a previously described patient with POMK compound heterozygous variants, the siblings did not display a typical Walker-Warburg syndrome with cognitive impairments [45]. Finally, FKTN mutations are commonly associated with Fukuyama congenital muscular dystrophy in Japan, which is characterised by hypotonia, muscle weakness, and cerebral and cerebellar cortical dysplasia [12]. Fewer patients with LGMD caused by FKTN mutations have been reported, and these usually have a milder phenotype of proximal muscle weakness and no cognitive involvement [46]. Our Caucasian patient, as expected, did not carry the Japanese founder haplotype and displayed a slowly progressive limb-girdle and axial myopathy. This complements and enhances the current understanding of diseases associated with FKTN mutations. Compound heterozygous and homozygous mutations in ISPD have been associated with a broad clinical spectrum ranging from mild LGMD to a more severe Walker-Warburg syndrome [47], with the mutations predicted to be deleterious to protein function. However, patient 14 has only one frameshift variant that could result in a null allele and disrupt ISPD function (p.Cys56ValfsTer60); the second variant (p.Ser202Leu) is in an exon at the N terminal of the gene that is present in only one transcript. This may explain the observed milder phenotype of the patient.
We have observed that within our cohort, suspected pathogenic mutations in POMT2 are the most common cause of dystroglycanopathies. This was similarly observed in a concentrated cohort of patients with α-dystroglycan hypoglycosylation [29]. Crucially, 15 of our 27 patients had suspected pathogenic variants in genes that were omitted from the aforementioned study (DPM3, FKRP, GMPPB, ISPD and POMK), indicating that an unbiased approach must be sought to fully characterise dystroglycanopathies. Indeed, even with a targeted NGS methodology, many of the patients in our MYO-SEQ cohort remain without a diagnosis. This suggests that patients may harbour pathogenic variants in genes with no currently known association to muscle disease, including uncharacterised dystroglycanopathies. It must be noted that rather than mutations in POMT2, those in FKRP are widely considered to cause the commonest dystroglycanopathy in Europe, LGMD2I [48]. While our data do not reflect this exactly, the 30% of patients with FKRP mutations strongly support the high prevalence of LGMD2I. A small n number, simple ascertainment bias and the ease at which FKRP can be sequenced in diagnostic work-ups—negating the need for inclusion in MYO-SEQ in these patients—could be responsible for these skewed data. Moreover, FKRP typically has a lower coverage with exome sequencing, meaning fewer LGMD2I patients might be detected relative to those identified through Sanger sequencing.
Conclusions
Overall, dystroglycanopathies are a clinically and genetically heterogeneous group of disorders, and as such, accurate diagnoses are often difficult to achieve. We have collected a unique group of 27 dystroglycanopathy patients that offer an extended understanding of the phenotypes at the milder end of the disease spectrum. The disease group is highly diverse and complex, reinforced by the detection of variants in patients with an atypical dystroglycanopathy phenotype. We present evidence of the benefit of integrating WES into healthcare systems in an effort to diagnose rare diseases more readily and accurately. Further tests will now be required to determine the mechanisms by which these proteins act and to confirm the dysregulation of α-dystroglycan glycosylation.
Additional file
Clinical presentations and phenotypes of the 27 MYO-SEQ index cases with suspected pathogenic variants in genes associated with dystroglycanopathies. Muscle pathology findings are defined as a replacement of muscle with fat on T1-weighted axial images. Radiology reports from referring centres are not standardised or quantitative. FVC = forced vital capacity; LVEF = left ventricular ejection fraction; LVFS = left ventricular fractional shortening; RNS = repetitive nerve stimulation. No indications = immunostaining was performed but was not suggestive of an α-DG deficiency. (XLSX 14 kb)
Acknowledgements
We thank the patients for donating their tissue samples.
Funding
The MYO-SEQ project was supported by Sanofi Genzyme, Ultragenyx, LGMD2I Research Fund, Samantha J Brazzo Foundation, LGMD2D Foundation, Kurt+Peter Foundation, Muscular Dystrophy UK and Coalition to Cure Calpain 3. Analysis performed by the Broad team was partially funded by the Center for Mendelian Genomics (UM1 HG008900) grant from the National Human Genome Research Institute. The funders of the study had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
Availability of data and materials
Linked genotype and phenotype data can be accessed through the European Genome–phenome Archive (EGA; EGAS00001002069).
Abbreviations
- CK
Creatine kinase
- DAG1
Dystroglycan 1
- DPM
Dolichol-phosphate-mannose
- ECM
Extracellular matrix
- EGA
European Genome–phenome Archive
- EMG
Electromyography
- FVC
Forced vital capacities
- IHC
Immunohistochemistry
- LGMD
Limb-girdle muscular dystrophies
- LVEF
Left ventricular ejection fraction
- LVFS
Left ventricular fractional shortening
- NGS
Next-generation sequencing
- RNS
Repetitive nerve stimulation
- WES
Whole-exome sequencing
Authors’ contributions
KJ acquired, analysed and interpreted the data, prepared the manuscript for publication and acquired funding. MB, AT, LP, LX, EV, TEM and ML acquired, analysed and interpreted the data. PVdB, JV, NW, SN, SJO, AL, AKP, APC, ND, CWP, SSK, JC, KGC, WDR, JB, MvdH, RFT, MZI, JBV, AH, HD and TW made substantial contributions to the acquisition of data. VS was responsible for the conception and design. VS and DGM acquired the funding and data. All authors read, critically revised and approved the final manuscript.
Ethics approval and consent to participate
Ethical approval (REC reference number 08/H0906/28) was granted by the Newcastle and North Tyneside Research Ethics Committee. All patients provided informed written consent to participate in the project.
Consent for publication
Not applicable.
Competing interests
VS is or has been on advisory boards for Acceleron Pharma, Audentes Therapeutics, Biogen, Biomarin, Bristol-Myer Squibb, Italfarmaco S.p.A., Nicox, Pfizer, Sanofi Genzyme, Santhera Pharmaceuticals, Sarepta Therapeutics, Summit Therapeutics, Tivorsan, TrophyNOD, and Wave Therapeutics. VS received speaker honoraria from Sanofi Genzyme. VS has or had research collaborations with Ultragenyx Pharmaceuticals and Sanofi Genzyme. All other authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13395-018-0170-1) contains supplementary material, which is available to authorized users.
Contributor Information
Katherine Johnson, Email: katherine.johnson@newcastle.ac.uk.
Marta Bertoli, Email: marta.bertoli@newcastle.ac.uk.
Lauren Phillips, Email: lauren.phillips@newcastle.ac.uk.
Ana Töpf, Email: ana.topf@newcastle.ac.uk.
Peter Van den Bergh, Email: peter.vandenbergh@uclouvain.be.
John Vissing, Email: john.vissing@regionh.dk.
Nanna Witting, Email: nanna.witting@regionh.dk.
Shahriar Nafissi, Email: nafisi@sina.tums.ac.ir.
Shirin Jamal-Omidi, Email: shirin_j_omidi@yahoo.com.
Anna Łusakowska, Email: anna.lusakowska@wum.edu.pl.
Anna Kostera-Pruszczyk, Email: anna.kostera-pruszczyk@wum.edu.pl.
Anna Potulska-Chromik, Email: apotulska@wum.edu.pl.
Nicolas Deconinck, Email: nicolas.deconinck@huderf.be.
Carina Wallgren-Pettersson, Email: carina.wallgren@helsinki.fi.
Sonja Strang-Karlsson, Email: sonja.strang@helsinki.fi.
Jaume Colomer, Email: colomer@sjdhospitalbarcelona.org.
Kristl G. Claeys, Email: kristl.claeys@uzleuven.be
Willem De Ridder, Email: willem.deridder@molgen.vib-ua.be.
Jonathan Baets, Email: jonathan.baets@molgen.vib-ua.be.
Maja von der Hagen, Email: maja.hagenv.der@uniklinikum-dresden.de.
Roberto Fernández-Torrón, Email: Roberto.Fernandez-Torron@newcastle.ac.uk.
Miren Zulaica Ijurco, Email: miren.zulaicaijurco@osakidetza.eus.
Juan Bautista Espinal Valencia, Email: juanbautista.espinalvalencia@osakidetza.eus.
Andreas Hahn, Email: andreas.hahn@paediat.med.uni-giessen.de.
Hacer Durmus, Email: durmushacer@yahoo.com.
Tracey Willis, Email: tracey.willis1@nhs.net.
Liwen Xu, Email: liwenlxu@broadinstitute.org.
Elise Valkanas, Email: valkanas@broadinstitute.org.
Thomas E. Mullen, Email: tmullen@broadinstitute.org
Monkol Lek, Email: mlek@broadinstitute.org.
Daniel G. MacArthur, Email: danmac@broadinstitute.org
Volker Straub, Email: volker.straub@newcastle.ac.uk.
References
- 1.Cohn RD, Henry MD, Michele DE, Barresi R, Saito F, Moore SA, Flanagan JD, Skwarchuk MW, Robbins ME, Mendell JR, et al. Disruption of DAG1 in differentiated skeletal muscle reveals a role for dystroglycan in muscle regeneration. Cell. 2002;110:639–648. doi: 10.1016/S0092-8674(02)00907-8. [DOI] [PubMed] [Google Scholar]
- 2.Ibraghimov-Beskrovnaya O, Ervasti JM, Leveille CJ, Slaughter CA, Sernett SW, Campbell KP. Primary structure of dystrophin-associated glycoproteins linking dystrophin to the extracellular matrix. Nat. 1992;355:696–702. doi: 10.1038/355696a0. [DOI] [PubMed] [Google Scholar]
- 3.Warner LE, DelloRusso C, Crawford RW, Rybakova IN, Patel JR, Ervasti JM, Chamberlain JS. Expression of Dp260 in muscle tethers the actin cytoskeleton to the dystrophin-glycoprotein complex and partially prevents dystrophy. Hum Mol Genet. 2002;11:1095–1105. doi: 10.1093/hmg/11.9.1095. [DOI] [PubMed] [Google Scholar]
- 4.Godfrey C, Foley AR, Clement E, Muntoni F. Dystroglycanopathies: coming into focus. Curr Opin Genet Dev. 2011;21:278–285. doi: 10.1016/j.gde.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Michele DE, Campbell KP. Dystrophin-glycoprotein complex: post-translational processing and dystroglycan function. J Biol Chem. 2003;278:15457–15460. doi: 10.1074/jbc.R200031200. [DOI] [PubMed] [Google Scholar]
- 6.Endo T. Dystroglycan glycosylation and its role in alpha-dystroglycanopathies. Acta Myol. 2007;26:165–170. [PMC free article] [PubMed] [Google Scholar]
- 7.Martin PT. Dystroglycan glycosylation and its role in matrix binding in skeletal muscle. Glycobiology. 2003;13:55R–66R. doi: 10.1093/glycob/cwg076. [DOI] [PubMed] [Google Scholar]
- 8.Astrea G, Pezzini I, Picillo E, Pasquariello R, Moro F, Ergoli M, D'Ambrosio P, D'Amico A, Politano L, Santorelli FM. TMEM5-associated dystroglycanopathy presenting with CMD and mild limb-girdle muscle involvement. Neuromuscul Disord. 2016;26:459–461. doi: 10.1016/j.nmd.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muntoni F, Torelli S, Brockington M. Muscular dystrophies due to glycosylation defects. Neurotherapeutics. 2008;5:627–632. doi: 10.1016/j.nurt.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cylwik B, Lipartowska K, Chrostek L, Gruszewska E. Congenital disorders of glycosylation. Part II. Defects of protein O-glycosylation. Acta Biochim Pol. 2013;60:361–368. [PubMed] [Google Scholar]
- 11.Teber S, Sezer T, Kafali M, Manzini MC, Konuk Yuksel B, Tekin M, Fitoz S, Walsh CA, Deda G. Severe muscle-eye-brain disease is associated with a homozygous mutation in the POMGnT1 gene. Eur J Paediatr Neurol. 2008;12:133–136. doi: 10.1016/j.ejpn.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Ishigaki K. Central nervous involvement in patients with Fukuyama congenital muscular dystrophy. Brain Nerve. 2016;68:119–127. doi: 10.11477/mf.1416200361. [DOI] [PubMed] [Google Scholar]
- 13.Mitsuhashi S, Kang PB. Update on the genetics of limb girdle muscular dystrophy. Semin Pediatr Neurol. 2012;19:211–218. doi: 10.1016/j.spen.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 14.Brown SC, Torelli S, Brockington M, Yuva Y, Jimenez C, Feng L, Anderson L, Ugo I, Kroger S, Bushby K, et al. Abnormalities in alpha-dystroglycan expression in MDC1C and LGMD2I muscular dystrophies. Am J Pathol. 2004;164:727–737. doi: 10.1016/S0002-9440(10)63160-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bouchet-Séraphin C, Vuillaumier-Barrot S, Seta N. Dystroglycanopathies: about numerous genes involved in glycosylation of one single glycoprotein. J Neuromuscul Dis. 2015;2:27–38. [PubMed] [Google Scholar]
- 16.Valencia CA, Husami A, Holle J, Johnson JA, Qian Y, Mathur A, Wei C, Indugula SR, Zou F, Meng H, et al. Clinical impact and cost-effectiveness of whole exome sequencing as a diagnostic tool: a pediatric center’s experience. Front Pediatr. 2015;3:67. doi: 10.3389/fped.2015.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peric S, Glumac JN, Topf A, Savic-Pavicevic D, Phillips L, Johnson K, Cassop-Thompson M, Xu L, Bertoli M, Lek M, et al. A novel recessive TTN founder variant is a common cause of distal myopathy in the Serbian population. Eur J Hum Genet. 2017;25:572–581. doi: 10.1038/ejhg.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, Maglott DR. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nat. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 22.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 23.Shihab HA, Gough J, Cooper DN, Stenson PD, Barker GL, Edwards KJ, Day IN, Gaunt TR. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat. 2013;34:57–65. doi: 10.1002/humu.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van den Bergh PYK, Sznajer Y, Van Parys V, van Tol W, Wevers RA, Lefeber DJ, Xu L, Lek M, MacArthur DG, Johnson K, et al. A homozygous DPM3 mutation in a patient with alpha-dystroglycan-related limb girdle muscular dystrophy. Neuromuscul Disord. 2017; 10.1016/j.nmd.2017.07.006. [DOI] [PubMed]
- 25.Brockington M, Blake DJ, Prandini P, Brown SC, Torelli S, Benson MA, Ponting CP, Estournet B, Romero NB, Mercuri E, et al. Mutations in the fukutin-related protein gene (FKRP) cause a form of congenital muscular dystrophy with secondary laminin alpha2 deficiency and abnormal glycosylation of alpha-dystroglycan. Am J Hum Genet. 2001;69:1198–1209. doi: 10.1086/324412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hafner P, Bonati U, Fischmann A, Schneider J, Frank S, Morris-Rosendahl DJ, Dumea A, Heinimann K, Fischer D. Skeletal muscle MRI of the lower limbs in congenital muscular dystrophy patients with novel POMT1 and POMT2 mutations. Neuromuscul Disord. 2014;24:321–324. doi: 10.1016/j.nmd.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 27.Mercuri E, Brockington M, Straub V, Quijano-Roy S, Yuva Y, Herrmann R, Brown SC, Torelli S, Dubowitz V, Blake DJ, et al. Phenotypic spectrum associated with mutations in the fukutin-related protein gene. Ann Neurol. 2003;53:537–542. doi: 10.1002/ana.10559. [DOI] [PubMed] [Google Scholar]
- 28.Mercuri E, Messina S, Bruno C, Mora M, Pegoraro E, Comi GP, D'Amico A, Aiello C, Biancheri R, Berardinelli A, et al. Congenital muscular dystrophies with defective glycosylation of dystroglycan: a population study. Neurol. 2009;72:1802–1809. doi: 10.1212/01.wnl.0000346518.68110.60. [DOI] [PubMed] [Google Scholar]
- 29.Godfrey C, Clement E, Mein R, Brockington M, Smith J, Talim B, Straub V, Robb S, Quinlivan R, Feng L, et al. Refining genotype phenotype correlations in muscular dystrophies with defective glycosylation of dystroglycan. Brain. 2007;130:2725–2735. doi: 10.1093/brain/awm212. [DOI] [PubMed] [Google Scholar]
- 30.Carss KJ, Stevens E, Foley AR, Cirak S, Riemersma M, Torelli S, Hoischen A, Willer T, van Scherpenzeel M, Moore SA, et al. Mutations in GDP-mannose pyrophosphorylase B cause congenital and limb-girdle muscular dystrophies associated with hypoglycosylation of alpha-dystroglycan. Am J Hum Genet. 2013;93:29–41. doi: 10.1016/j.ajhg.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Belaya K, Rodriguez Cruz PM, Liu WW, Maxwell S, McGowan S, Farrugia ME, Petty R, Walls TJ, Sedghi M, Basiri K, et al. Mutations in GMPPB cause congenital myasthenic syndrome and bridge myasthenic disorders with dystroglycanopathies. Brain. 2015;138:2493–2504. doi: 10.1093/brain/awv185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strang-Karlsson S, Johnson K, Topf A, Xu L, Lek M, MacArthur DG, Casar-Borota O, Williams M, Straub V, Wallgren-Pettersson C. A novel compound heterozygous mutation in the POMK gene causing limb-girdle muscular dystrophy-dystroglycanopathy in a sib pair. Neuromuscul Disord. 2018; 10.1016/j.nmd.2018.04.012. [DOI] [PMC free article] [PubMed]
- 33.Wallace SE, Conta JH, Winder TL, Willer T, Eskuri JM, Haas R, Patterson K, Campbell KP, Moore SA, Gospe SM., Jr A novel missense mutation in POMT1 modulates the severe congenital muscular dystrophy phenotype associated with POMT1 nonsense mutations. Neuromuscul Disord. 2014;24:312–320. doi: 10.1016/j.nmd.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Biancheri R, Falace A, Tessa A, Pedemonte M, Scapolan S, Cassandrini D, Aiello C, Rossi A, Broda P, Zara F, et al. POMT2 gene mutation in limb-girdle muscular dystrophy with inflammatory changes. Biochem Biophys Res Commun. 2007;363:1033–1037. doi: 10.1016/j.bbrc.2007.09.066. [DOI] [PubMed] [Google Scholar]
- 35.Yanagisawa A, Bouchet C, Van den Bergh PY, Cuisset JM, Viollet L, Leturcq F, Romero NB, Quijano-Roy S, Fardeau M, Seta N, Guicheney P. New POMT2 mutations causing congenital muscular dystrophy: identification of a founder mutation. Neurol. 2007;69:1254–1260. doi: 10.1212/01.wnl.0000268489.60809.c4. [DOI] [PubMed] [Google Scholar]
- 36.Majewski J, Schwartzentruber J, Lalonde E, Montpetit A, Jabado N. What can exome sequencing do for you? J Med Genet. 2011;48:580–589. doi: 10.1136/jmedgenet-2011-100223. [DOI] [PubMed] [Google Scholar]
- 37.Farlow JL, Robak LA, Hetrick K, Bowling K, Boerwinkle E, Coban-Akdemir ZH, Gambin T, Gibbs RA, Gu S, Jain P, et al. Whole-exome sequencing in familial Parkinson disease. JAMA Neurol. 2016;73:68–75. doi: 10.1001/jamaneurol.2015.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, Ercan-Sencicek AG, DiLullo NM, Parikshak NN, Stein JL, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nat. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Worthey EA, Mayer AN, Syverson GD, Helbling D, Bonacci BB, Decker B, Serpe JM, Dasu T, Tschannen MR, Veith RL, et al. Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet Med. 2011;13:255–262. doi: 10.1097/GIM.0b013e3182088158. [DOI] [PubMed] [Google Scholar]
- 40.van Reeuwijk J, Janssen M, van den Elzen C, Beltran-Valero de Bernabe D, Sabatelli P, Merlini L, Boon M, Scheffer H, Brockington M, Muntoni F, et al. POMT2 mutations cause alpha-dystroglycan hypoglycosylation and Walker-Warburg syndrome. J Med Genet. 2005;42:907–912. doi: 10.1136/jmg.2005.031963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jimenez-Mallebrera C, Torelli S, Feng L, Kim J, Godfrey C, Clement E, Mein R, Abbs S, Brown SC, Campbell KP, et al. A comparative study of alpha-dystroglycan glycosylation in dystroglycanopathies suggests that the hypoglycosylation of alpha-dystroglycan does not consistently correlate with clinical severity. Brain Pathol. 2009;19:596–611. doi: 10.1111/j.1750-3639.2008.00198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maeda Y, Tanaka S, Hino J, Kangawa K, Kinoshita T. Human dolichol-phosphate-mannose synthase consists of three subunits, DPM1, DPM2 and DPM3. EMBO J. 2000;19:2475–2482. doi: 10.1093/emboj/19.11.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schenk B, Fernandez F, Waechter CJ. The ins(ide) and out(side) of dolichyl phosphate biosynthesis and recycling in the endoplasmic reticulum. Glycobiology. 2001;11:61R–70R. doi: 10.1093/glycob/11.5.61R. [DOI] [PubMed] [Google Scholar]
- 44.Lefeber DJ, Schonberger J, Morava E, Guillard M, Huyben KM, Verrijp K, Grafakou O, Evangeliou A, Preijers FW, Manta P, et al. Deficiency of Dol-P-Man synthase subunit DPM3 bridges the congenital disorders of glycosylation with the dystroglycanopathies. Am J Hum Genet. 2009;85:76–86. doi: 10.1016/j.ajhg.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jae LT, Raaben M, Riemersma M, van Beusekom E, Blomen VA, Velds A, Kerkhoven RM, Carette JE, Topaloglu H, Meinecke P, et al. Deciphering the glycosylome of dystroglycanopathies using haploid screens for lassa virus entry. Sci. 2013;340:479–483. doi: 10.1126/science.1233675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pegoraro E, Hoffman EP. Limb-girdle muscular dystrophy overview. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, LJH B, Bird TD, Ledbetter N, Mefford HC, RJH S, Stephens K, editors. GeneReviews(R). Seattle: University of Washington; 1993-2018. 2000. [updated 2012 Aug 30]. https://www.ncbi.nlm.nih.gov/books/NBK1408/.
- 47.Willer T, Lee H, Lommel M, Yoshida-Moriguchi T, de Bernabe DB, Venzke D, Cirak S, Schachter H, Vajsar J, Voit T, et al. ISPD loss-of-function mutations disrupt dystroglycan O-mannosylation and cause Walker-Warburg syndrome. Nat Genet. 2012;44:575–580. doi: 10.1038/ng.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kanagawa M, Toda T. Muscular dystrophy with ribitol-phosphate deficiency: a novel post-translational mechanism in dystroglycanopathy. J Neuromuscul Dis. 2017;4:259–267. doi: 10.3233/JND-170255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKusick VA. Mendelian Inheritance in Man and its online version, OMIM. Am J Hum Genet. 2007;80:588–604. doi: 10.1086/514346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stevens E, Carss KJ, Cirak S, Foley AR, Torelli S, Willer T, Tambunan DE, Yau S, Brodd L, Sewry CA, et al. Mutations in B3GALNT2 cause congenital muscular dystrophy and hypoglycosylation of alpha-dystroglycan. Am J Hum Genet. 2013;92:354–365. doi: 10.1016/j.ajhg.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buysse K, Riemersma M, Powell G, van Reeuwijk J, Chitayat D, Roscioli T, Kamsteeg EJ, van den Elzen C, van Beusekom E, Blaser S, et al. Missense mutations in beta-1,3-N-acetylglucosaminyltransferase 1 (B3GNT1) cause Walker-Warburg syndrome. Hum Mol Genet. 2013;22:1746–1754. doi: 10.1093/hmg/ddt021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Geis T, Marquard K, Rodl T, Reihle C, Schirmer S, von Kalle T, Bornemann A, Hehr U, Blankenburg M. Homozygous dystroglycan mutation associated with a novel muscle-eye-brain disease-like phenotype with multicystic leucodystrophy. Neurogenetics. 2013;14:205–213. doi: 10.1007/s10048-013-0374-9. [DOI] [PubMed] [Google Scholar]
- 53.Hara Y, Balci-Hayta B, Yoshida-Moriguchi T, Kanagawa M, Beltran-Valero de Bernabe D, Gundesli H, Willer T, Satz JS, Crawford RW, Burden SJ, et al. A dystroglycan mutation associated with limb-girdle muscular dystrophy. N Engl J Med. 2011;364:939–946. doi: 10.1056/NEJMoa1006939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kranz C, Jungeblut C, Denecke J, Erlekotte A, Sohlbach C, Debus V, Kehl HG, Harms E, Reith A, Reichel S, et al. A defect in dolichol phosphate biosynthesis causes a new inherited disorder with death in early infancy. Am J Hum Genet. 2007;80:433–440. doi: 10.1086/512130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim S, Westphal V, Srikrishna G, Mehta DP, Peterson S, Filiano J, Karnes PS, Patterson MC, Freeze HH. Dolichol phosphate mannose synthase (DPM1) mutations define congenital disorder of glycosylation Ie (CDG-Ie) J Clin Invest. 2000;105:191–198. doi: 10.1172/JCI7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barone R, Aiello C, Race V, Morava E, Foulquier F, Riemersma M, Passarelli C, Concolino D, Carella M, Santorelli F, et al. DPM2-CDG: a muscular dystrophy-dystroglycanopathy syndrome with severe epilepsy. Ann Neurol. 2012;72:550–558. doi: 10.1002/ana.23632. [DOI] [PubMed] [Google Scholar]
- 57.Beltran-Valero de Bernabe D, Voit T, Longman C, Steinbrecher A, Straub V, Yuva Y, Herrmann R, Sperner J, Korenke C, Diesen C, et al. Mutations in the FKRP gene can cause muscle-eye-brain disease and Walker-Warburg syndrome. J Med Genet. 2004;41:e61. doi: 10.1136/jmg.2003.013870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Bernabe DB, van Bokhoven H, van Beusekom E, Van den Akker W, Kant S, Dobyns WB, Cormand B, Currier S, Hamel B, Talim B, et al. A homozygous nonsense mutation in the fukutin gene causes a Walker-Warburg syndrome phenotype. J Med Genet. 2003;40:845–848. doi: 10.1136/jmg.40.11.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Godfrey C, Escolar D, Brockington M, Clement EM, Mein R, Jimenez-Mallebrera C, Torelli S, Feng L, Brown SC, Sewry CA, et al. Fukutin gene mutations in steroid-responsive limb girdle muscular dystrophy. Ann Neurol. 2006;60:603–610. doi: 10.1002/ana.21006. [DOI] [PubMed] [Google Scholar]
- 60.Tasca G, Moro F, Aiello C, Cassandrini D, Fiorillo C, Bertini E, Bruno C, Santorelli FM, Ricci E. Limb-girdle muscular dystrophy with alpha-dystroglycan deficiency and mutations in the ISPD gene. Neurol. 2013;80:963–965. doi: 10.1212/WNL.0b013e3182840cbc. [DOI] [PubMed] [Google Scholar]
- 61.Longman C, Brockington M, Torelli S, Jimenez-Mallebrera C, Kennedy C, Khalil N, Feng L, Saran RK, Voit T, Merlini L, et al. Mutations in the human LARGE gene cause MDC1D, a novel form of congenital muscular dystrophy with severe mental retardation and abnormal glycosylation of alpha-dystroglycan. Hum Mol Genet. 2003;12:2853–2861. doi: 10.1093/hmg/ddg307. [DOI] [PubMed] [Google Scholar]
- 62.Hehr U, Uyanik G, Gross C, Walter MC, Bohring A, Cohen M, Oehl-Jaschkowitz B, Bird LM, Shamdeen GM, Bogdahn U, et al. Novel POMGnT1 mutations define broader phenotypic spectrum of muscle-eye-brain disease. Neurogenetics. 2007;8:279–288. doi: 10.1007/s10048-007-0096-y. [DOI] [PubMed] [Google Scholar]
- 63.Clement EM, Godfrey C, Tan J, Brockington M, Torelli S, Feng L, Brown SC, Jimenez-Mallebrera C, Sewry CA, Longman C, et al. Mild POMGnT1 mutations underlie a novel limb-girdle muscular dystrophy variant. Arch Neurol. 2008;65:137–141. doi: 10.1001/archneurol.2007.2. [DOI] [PubMed] [Google Scholar]
- 64.Manzini MC, Tambunan DE, Hill RS, Yu TW, Maynard TM, Heinzen EL, Shianna KV, Stevens CR, Partlow JN, Barry BJ, et al. Exome sequencing and functional validation in zebrafish identify GTDC2 mutations as a cause of Walker-Warburg syndrome. Am J Hum Genet. 2012;91:541–547. doi: 10.1016/j.ajhg.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.von Renesse A, Petkova MV, Lutzkendorf S, Heinemeyer J, Gill E, Hubner C, von Moers A, Stenzel W, Schuelke M. POMK mutation in a family with congenital muscular dystrophy with merosin deficiency, hypomyelination, mild hearing deficit and intellectual disability. J Med Genet. 2014;51:275–282. doi: 10.1136/jmedgenet-2013-102236. [DOI] [PubMed] [Google Scholar]
- 66.Di Costanzo S, Balasubramanian A, Pond HL, Rozkalne A, Pantaleoni C, Saredi S, Gupta VA, Sunu CM, Yu TW, Kang PB, et al. POMK mutations disrupt muscle development leading to a spectrum of neuromuscular presentations. Hum Mol Genet. 2014;23:5781–5792. doi: 10.1093/hmg/ddu296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Beltran-Valero de Bernabe D, Currier S, Steinbrecher A, Celli J, van Beusekom E, van der Zwaag B, Kayserili H, Merlini L, Chitayat D, Dobyns WB, et al. Mutations in the O-mannosyltransferase gene POMT1 give rise to the severe neuronal migration disorder Walker-Warburg syndrome. Am J Hum Genet. 2002;71:1033–1043. doi: 10.1086/342975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Reeuwijk J, Maugenre S, van den Elzen C, Verrips A, Bertini E, Muntoni F, Merlini L, Scheffer H, Brunner HG, Guicheney P, van Bokhoven H. The expanding phenotype of POMT1 mutations: from Walker-Warburg syndrome to congenital muscular dystrophy, microcephaly, and mental retardation. Hum Mutat. 2006;27:453–459. doi: 10.1002/humu.20313. [DOI] [PubMed] [Google Scholar]
- 69.Balci B, Uyanik G, Dincer P, Gross C, Willer T, Talim B, Haliloglu G, Kale G, Hehr U, Winkler J, Topaloglu H. An autosomal recessive limb girdle muscular dystrophy (LGMD2) with mild mental retardation is allelic to Walker-Warburg syndrome (WWS) caused by a mutation in the POMT1 gene. Neuromuscul Disord. 2005;15:271–275. doi: 10.1016/j.nmd.2005.01.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical presentations and phenotypes of the 27 MYO-SEQ index cases with suspected pathogenic variants in genes associated with dystroglycanopathies. Muscle pathology findings are defined as a replacement of muscle with fat on T1-weighted axial images. Radiology reports from referring centres are not standardised or quantitative. FVC = forced vital capacity; LVEF = left ventricular ejection fraction; LVFS = left ventricular fractional shortening; RNS = repetitive nerve stimulation. No indications = immunostaining was performed but was not suggestive of an α-DG deficiency. (XLSX 14 kb)
Data Availability Statement
Linked genotype and phenotype data can be accessed through the European Genome–phenome Archive (EGA; EGAS00001002069).