Abstract
Background
DNA Base Excision Repair Gene-DNA LigaseIII (LIG3) is an important repair gene in the repair pathway and plays an important role in maintaining the integrity of mitochondria. Rs1052536 and rs3135967 polymorphisms of the gene are associated with lung cancer, keratoconus, and Fuchs endothelial corneal dystrophy. There is no previously published report on the relationship between the polymorphisms and neural tube defects (NTDs).
Material/Methods
Mass ARRAY iPLEX was used to determine the distribution of the polymorphisms in the case group of 108 NTD pregnant women and a control group of 233 normal healthy pregnant women to examine the relevance of their polymorphisms and NTD occurrence.
Results
The homozygotes of rs1052536 TT were associated with an increased risk for NTDs than CC (P=0.014, OR=2.31, 95%CI [1.17–4.54]), and variants of rs1052536 T were associated with an increased risk of NTDs (P=0.024, OR=1.50, 95%CI [1.06–2.13]). The stratified analysis showed that TT genotype of rs1052536 increased the risk of anencephaly (P=0.016, OR=2.69, 95%CI [1.18–6.10]) and the T allele significantly increased the risk of cranial NTDs (P=0.033, OR=1.56, 95%CI [1.04–2.35]).
Conclusions
Rs1052536 in LIG3 gene might be a potential genetic risk factor in a high-risk area of NTDs in China.
MeSH Keywords: Case-Control Studies; DNA Repair; Neural Tube Defects; Polymorphism, Single Nucleotide
Background
Neural tube defects (NTDs) are a group of common, severe, congenital malformations that have a high prevalence (0.05–0.2%) worldwide [1] and the prevalence in China (0.2–0.4%) is the highest in the world [2]. In some areas in Shanxi province, the prevalence reaches 10.2–20%. The etiology of NTDs is multifactorial and includes genetic and environmental factors [3]. Recent research suggests that many factors contribute to occurrence of NTDs. At 3–4 weeks of embryo development, failed neural tube closure results in NTDs, such as spina bifida, anencephaly and encephalocele, and this is one of the main causes of infant death, mental retardation, and lifelong disability of children. The pathogenesis of NTDs is unclear, and most research has focused on folate metabolism pathways. Folic acid, as a carrier of 1 carbon unit, participates in DNA synthesis, repair, and methylation, and is necessary for body growth and reproduction, as well as being essential for fetal growth and development. Several studies have shown that folate deficiency during pregnancy is one of the most important risk factor in NTDs [4–7]. Folic acid supplementation (0.4–4.0 mg/d) could prevent about 50%~70% of NTD occurrence [8]. Lack of folic acid during pregnancy may affect the synthesis and repair of DNA, leading to birth defects. Experimental animal studies founded that diet lacking folic acid during pregnancy leads to fetal death, stillbirth, fetal growth retardation, and weight loss [9], but the exact mechanism has not been elucidated.
DNA repair is an important human physiological function, participating in DNA synthesis, cell synthesis, and other important biological processes. To maintain body stability, each repair pathway plays a different role in repairing damage. Their repression has different effects on DNA damage response, and increases susceptibilities to disease. Environmental and genetic factors contribute to structural damage in DNA, and if it is not repaired in time the damage may affect gene expression, leading to mutations and deformities. Animal experiments proved that knockout of crucial genes related to DNA repair lead to embryonic death, cessation of growth, and birth defects. Finnell and others also found that key genes associated with DNA repair single-nucleotide polymorphisms may increase risk of spina bifida and oral clefts [10,11], such as Xeroderma pigmentosum D (XPD), acclimation of photosynthesis to environment (APE1), X-ray repair cross-complementing 3 (XRCC3), 8-oxoguanine DNA glycosylase (hOGG1), and Methylenetetrahydrofolate reductase (MTHFR). DNA ligase III (LIG3) gene is a critical gene in nucleotide excision repair (base excision repair, BER), located on human chromosome 17q11.2–12, which encodes proteins responsible for DNA repair after stump closure, then connects them into a complete DNA strand; therefore, the single-nucleotide polymorphisms of this gene may affect the ability to repair DNA damage. Among human LIG genes, LIG3 gene is unique in that it encodes multiple DNA ligase polypeptides with different N- and C-termini. One of these polypeptides is targeted to mitochondria, where it plays an essential role in maintenance of the mitochondrial genome. In the nucleus, DNA ligases I, III, and IV have distinct but overlapping functions in DNA replication and repair [12,13].
Studies have found that many gene loci are associated with incidence of lung cancer, esophageal cancer, chemotherapy, and radiation injury, and results of preclinical studies with human cancer cell lines and mouse models of human cancer suggest that DNA ligase inhibitors have utility as anti-cancer agents [13–16]. There are multiple SNP loci in the 3′-UTR region, playing an important role in maintaining the integrity of mitochondrial DNA, such as rs1052536, rs11868699, rs3135966, rs3135967, and rs61749869. A case-control study reported an association between polymorphism in UCP2 gene, which is located in the 3′-UTR region, and spina bifida in a California population [17], and being homozygous for DD carries a high risk of spina bifida. In our previous study [18], the polymorphisms in exon 8 of UCP2 were found to be potential genetic risk factors for NTDs in a high-risk area in China. Finnel et al. [9] selected 6 polymorphisms in 6 hot genes and indicated that 4 of them – XPD (rs1799793), APE1 (rs3136820), XRCC3 (rs861539), and OGG1 (rs1052133) – can increase risk of spina bifida and oral clefts in a California population, but the relationship between single-nucleotide polymorphisms (SNPs) of LIG3 and NTDs has rarely been reported, especially in China. In the present study, 108 pregnant women who gave birth to infants with NTDs and 233 pregnant women as controls were selected to investigate the correlation between 2 common SNPs of LIG3 (rs1052536 and rs3135967) and occurrence of NTDs in a high-risk area in China.
Material and Methods
Study participants
A case-control design was ongoing in the study. We conducted a hospital-based case-control study from 2004 to 2009 in Shanxi province in northern China, including 9 hospitals in Zhongyang, Jiaokou, Heshun, Lin counties, and Liulin, which have an NTD prevalence of 199.38 per 10 000 births [19]. We recruited NTD and control pregnant women who were receiving prenatal care or who delivered at the hospital and agreed to participate in our study. Finally, 341 pregnant women were included in the study (233 controls and 108 cases). Women diagnosed with NTD fetus by B-type ultrasonic inspection and who electively terminated an NTD fetus or had a live birth or stillbirth with NTD were defined as cases. The pathological diagnosis of NTDs was completed by experienced pathologists according to the International Classification of Diseases, Tenth Revision, codes Q00 anencephaly, Q05 spina bifida, and Q01 encephalocele (http://apps.who.int/classifications). In the case group, there were 58 cases of anencephaly, 41 cases of spina bifida, and 9 cases of encephalocele. In the control group, women who had a live-born infant with no identified structural malformation after 1-year follow-up and who aborted for nonmedical reasons were enrolled from the same region and same hospital during the same period. When pregnant women were recruited, their clinical information was collected and recorded, such as the gestational age in weeks, and serum samples. The controls who were invited to participate were initially selected by matching for age and date of conception with women in the case group. After giving informed consent, all the subjects had B-type ultrasonic inspection and prenatal examination, and completed a questionnaire with items on age, parity, gravidity, gestational age, and folic acid intake. Any fetuses displaying pathological malformations or intrauterine growth retardation were excluded from the control group. Then, 5 mL of venous blood was collected and stored at −20°C in a local hospital before shipping on ice to laboratories at the Capital Institute of Pediatrics in Beijing by our study group members every other month, taking precautions to ensure samples were not subjected to repeated freezing and thawing.
DNA extraction and nucleotide sequences
Genomic DNA was extracted from frozen tissue samples using the Blood and Tissue DNA Kit (Qiagen, Germany) according to the manufacturer’s instructions, and with blank control of Tris-EDTA. The concentration and purity of the DNA were determined by absorbance at 260 nm and 280 nm.
Genotyping and measurement
We used https://www.mysequenom.com online and Sequenom MassARRAY Assay Design 4.0 software designed primers and UEP, and site sequence refers to NCBI db SNP (http://www.ncbi.ncbi.nlm.nih.gov/projects/SNP/). Primers and UEP were synthesized by Shanghai Sangon Biotech, Ltd. The primers and UEP sequences are shown in Table 1.
Table 1.
The primers and UEP sequences.
| SNP loci | The primers and UEP sequences | |
|---|---|---|
| rs1052536 | F: | 5′-ACGTTGGATGCATTCCCCCTATACTGTGTC-3′ |
| R: | 5′-ACGTTGGATGCAGGCCATACTCTCCTTTAC-3′ | |
| UEP: | 5′-CATACTCTCCTTTACCATACTA-3′ | |
| rs3135967 | F: | 5′-ACGTTGGATGTTTAACCTCTATCGCAGCGG-3′ |
| R: | 5′-ACGTTGGATGTAATAATCTCTGTCTGGCCC-3′ | |
| UEP: | 5′-GTCTGGCCCAGGTAACTA-3′ | |
Statistical analysis
Different lifestyle and sociodemographic characteristics of case and control participants were compared with the t test for continuous variables or with chi-square (χ2). Hardy-Weinberg equilibrium was test by chi-square (χ2). Haploview software was used to analyze the linkage disequilibrium of these 2 sites. Odds ratio (OR) with a 95% confidence interval (CI) was calculated to estimate the risks related to polymorphism and NTDs. Then Stratified analysis grouped according to the type of deformity, in order to testify further relationship of the genotype and NTDs. Statistical analyses were performed with SPSS version 19.0 (SPSS, Inc., Chicago, IL, USA). All P values were two-sided, and P<0.05 were considered statistically significant.
Quality control
Epidemiological questionnaires were designed based on the literature and combined with the local population and environmental characteristics, and after the pre-survey we made some supplements and amendments. All investigators were trained. The reagents and samples were dispensed and stored at −20°C on ice to avoid repeated freezing and thawing. The genotyping samples were double-blinded, each sample was run in duplicate, and we compared blanks and negative controls. To ensure consistency, 10% of the samples were re-genotyped. To validate the accuracy of MassARRAY SNP genotyping analysis technique, we directly sequenced 10% of the samples and found 100% concordance between them (data not shown).
Results
We recruited 108 cases and 233 matched controls and conducted a hospital-based case-control study in the Lvliang mountain area of Shanxi province in northern China and preliminarily investigated the association between LIG3 gene and NTDs. As shown in Table 2, the distributions of age, sex, pregnancy, parity, education, and folate intake were similar between the case group and control group, but there was a statistically significant difference (P=0.012) in gestational age because the patient group was selected more for early abortion.
Table 2.
The characteristics of the study participants.
| Characteristics | Case (n=108) N# (%)* |
Control (n=233) N# (%)* |
P |
|---|---|---|---|
| Age (year)** | |||
| <20 | 6 (6.1) | 8 (3.6) | 0.063 |
| 20–29 | 81 (81.8) | 157 (71.4) | |
| ≥29 | 12 (12.1) | 52 (24.0) | |
| Educational level*** | |||
| <High school graduation | 93 (86.1) | 180 (81.8) | 0.207 |
| ≥High school graduation | 15 (13.9) | 40 (18.2) | |
| Gravidity (n) | |||
| 1 | 50 (50.0) | 95 (41.7) | 0.218 |
| 2 | 28 (28.0) | 86 (37.7) | |
| ≥3 | 22 (22.0) | 47 (20.6) | |
| Parity(n)*** | |||
| 0 | 48 (44.9) | 72 (35.5) | 0.169 |
| 1 | 46 (43.0) | 93 (45.8) | |
| ≥2 | 13 (12.1) | 38 (18.7) | |
| Periconceptional folic acid use***,**** | |||
| No | 102 (96.2) | 220 (97.3) | 0.402 |
| Yes | 4 (3.8) | 6 (2.7) | |
| Gestational week** | |||
| <21 | 51 (56.0) | 162 (72.0) | 0.012 |
| 21–29 | 26 (28.6) | 47 (20.9) | |
| ≥30 | 14 (15.4) | 16 (7.1) |
Percentages may not equal 100 because of rounding;
Values are mean ±SD, t test was used to calculate the P values;
χ2 test was used to calculate P values;
“Periconceptional”refers to the month before conception and the first 3 months after conception.
The total numbers may not equal to 108/233 because of loss of follow-up of subjects and incomplete registration information.
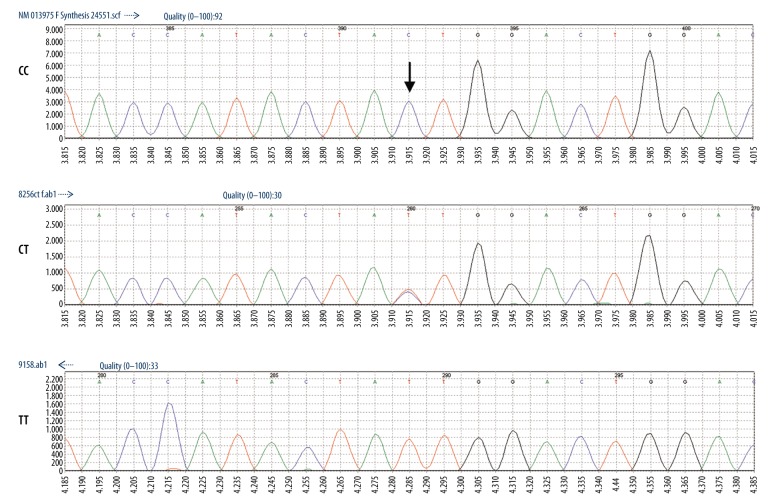
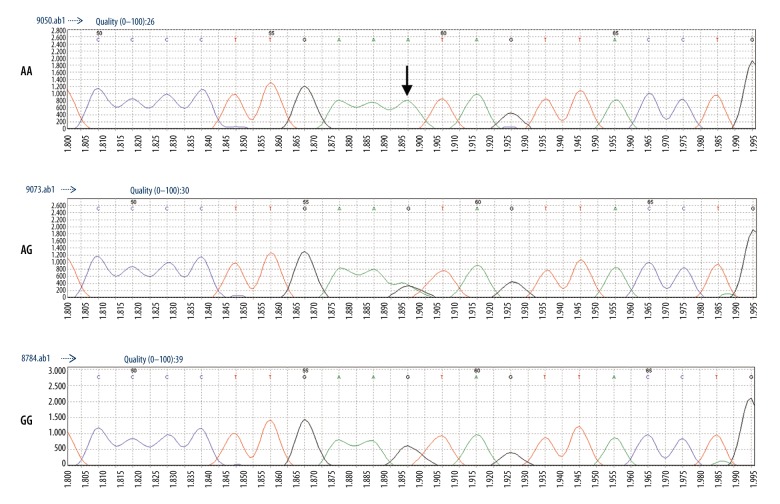
Then, we randomly selected 10% of the samples and sequenced them by direct sequencing to verify the accuracy of the MassARRAY genotyping microarray technology, showing then to be consistent, as shown in Figures 1 and 2.
Figure 1.
Direct sequencing results of rs1052536.
Figure 2.
Direct sequencing results of rs3135967.
Genotype frequencies in cases and controls were in concordance with Hardy-Weinberg equilibrium (P>0.05), indicating that the study samples were representative of the group (Table 3). Table 4 shows that the embryos carrying TT genotype in rs1052536 increased the risk of NTD occurrence compared to homozygous wild genotype CC (P=0.01, OR=2.31; 95%CI [1.17–4.54]), allele T with respect to C had a trend to increase the risk of NTDs (P=0.02, OR=1.50; 95%CI [1.06–2.13]), and P values were statistically significant.
Table 3.
The Hardy-Weinberg equilibrium for case and control of the polymorphisms.
| SNPs | Case/Control | Genotypes | Observed (n*, %) | Expected (n*, %) | P (HWE) |
|---|---|---|---|---|---|
| rs1052536 | Case | C/C | 53 (50.00%) | 45 (42.45%) | 0.057 |
| C/T | 32 (30.19%) | 48 (45.28%) | |||
| T/T | 21 (19.81%) | 13 (12.27%) | |||
| Control | C/C | 128 (57.14%) | 123 (54.91%) | 0.31 | |
| C/T | 74 (33.04%) | 86 (38.39%) | |||
| T/T | 22 (9.82%) | 15 (6.70%) | |||
| rs3135967 | Case | A/A | 54 (50.47%) | 52 (48.60%) | 0.82 |
| G/A | 41 (38.32%) | 45 (42.06%) | |||
| G/G | 12 (11.21%) | 10 (9.35%) | |||
| Control | A/A | 127 (54.51%) | 124 (53.22%) | 0.59 | |
| G/A | 84 (36.05%) | 92 (39.48%) | |||
| G/G | 22 (9.44%) | 17 (7.30%) |
The total numbers may not equal to 108/233 because of unqualified DNA samples.
Table 4.
Distribution of genotypes and odds ratios (OR) in rs1052536 and rs3135967.
| SNP | Genotype/Allele | Case (n*) | Control (n*) | OR (95%CI) | P | |
|---|---|---|---|---|---|---|
| rs3135967 | Genotype | A/A | 54 | 127 | 1.00 | |
| G/A | 41 | 84 | 1.15 (0.70–1.88) | 0.58 | ||
| G/G | 12 | 22 | 1.28 (0.59–2.78) | 0.53 | ||
| Allele | A | 149 | 338 | 1.00 | ||
| G | 65 | 128 | 1.15 (0.81–1.64) | 0.44 | ||
| rs1052536 | Genotype | C/C | 53 | 128 | 1.00 | |
| C/T | 32 | 74 | 1.04 (0.62–1.76) | 0.87 | ||
| T/T | 21 | 22 | 2.31 (1.17–4.54) | 0.01 | ||
| Allele | C | 138 | 330 | 1.00 | ||
| T | 74 | 118 | 1.50 (1.06–2.13) | 0.02 | ||
The total numbers may not equal to 108/233 because of unqualified DNA samples.
We found that in the anencephaly group, individuals with TT genotype were at a significantly increased risk of NTDs, compared with homozygous wild genotype CC individuals (P=0.016, OR=2.69, 95%CI [1.18–6.10]), and allele T carriers had a significantly increased risk of anencephaly (P=0.014, OR=1.71, 95%CI [1.11–2.63]), (Tables 5, 6). Rs3135967 carrying G allele was increased in the cranial malformation group (anencephaly and encephalocele), but not significantly. Rs1052536 with TT genotype carrier significantly increased NTD incidence compared to homozygous wild genotype CC individuals (P=0.029, OR=2.36, 95%CI [1.08–5.20]), and allele T to C significantly increased the risk of cranial NTDs (P=0.033, OR=1.56; 95%CI [1.04–2.35]) (Tables 7, 8).
Table 5.
Distribution of genotypes and odds ratios of rs1052536 in the three subgroups.
| NTD subgroups | Genotype/Allele | Case | Control | OR (95%CI) | P |
|---|---|---|---|---|---|
| Anencephaly | CC | 26 | 128 | 1.00 | |
| CT | 20 | 74 | 1.33 (0.70–2.55) | 0.39 | |
| TT | 12 | 22 | 2.69 (1.18–6.10) | 0.016 | |
| C | 72 | 330 | 1.00 | ||
| T | 44 | 118 | 1.71 (1.11–2.63) | 0.014 | |
| Spina Bifida | CC | 21 | 128 | 1.00 | |
| CT | 10 | 74 | 0.82 (0.37–1.84) | 0.64 | |
| TT | 8 | 22 | 2.22 (0.87–5.63) | 0.09 | |
| C | 52 | 330 | 1.00 | ||
| T | 26 | 118 | 1.40 (0.84–2.34) | 0.20 | |
| Encephalocele | CC | 6 | 128 | 1.00 | |
| CT | 2 | 74 | 0.58 (0.11–2.93) | 0.50 | |
| TT | 1 | 22 | 0.97 (0.11–8.45) | 0.98 | |
| C | 14 | 330 | 1.00 | ||
| T | 4 | 118 | 0.80 (0.25–2.48) | 0.70 |
Table 6.
Distribution of genotypes and odds ratios of rs3135967 in the three subgroups.
| NTD subgroups | Genotype/Allele | Case | Control | OR (95%CI) | P |
|---|---|---|---|---|---|
| Anencephaly | AA | 26 | 127 | 1.00 | |
| GA | 26 | 84 | 1.15 (0.88–2.78) | 0.18 | |
| GG | 5 | 22 | 1.11 (0.39–3.20) | 0.85 | |
| A | 78 | 338 | 1.00 | ||
| G | 36 | 128 | 1.22 (0.78–1.90) | 0.38 | |
| Spina Bifida | AA | 21 | 127 | 1.00 | |
| GA | 13 | 84 | 0.94 (0.45–1.97) | 0.86 | |
| GG | 7 | 22 | 1.92 (0.73–5.07) | 0.18 | |
| A | 55 | 338 | 1.00 | ||
| G | 27 | 128 | 1.30 (0.78–2.14) | 0.31 | |
| Encephalocele | AA | 7 | 127 | 1.00 | |
| GA | 2 | 84 | 0.43 (0.09–2.13) | 0.29 | |
| GG | 0 | 22 | |||
| A | 16 | 338 | 1.00 | ||
| G | 2 | 128 | 0.33 (0.08–1.46) | 0.18 |
Table 7.
Distribution of genotypes and odds ratios of rs1052536 in cranial NTDs subgroup and controls.
| Genotype/Allele | Cranial NTDs (n, %) | Control (n, %) | OR (95%CI) | P |
|---|---|---|---|---|
| CC | 32 (47.8) | 128 (57.1) | 1.00 | |
| CT | 22 (32.8) | 74 (33.0) | 1.19 (0.64–2.20) | 0.193 |
| TT | 13 (19.4) | 22 (9.8) | 2.36 (1.08–5.20) | 0.029 |
| CT+TT | 35 (52.2) | 96 (42.9) | 1.46 (0.84–2.52) | 0.18 |
| C | 86 (64.2) | 330 (73.7) | 1.00 | |
| T | 48 (35.8) | 118 (26.3) | 1.56 (1.04–2.35) | 0.033 |
Table 8.
Distribution of genotypes and odds ratios of rs3135967 in cranial NTDs subgroup and controls.
| Genotype/Allele | Cranial NTDs (n, %) | Control (n, %) | OR (95%CI) | P |
|---|---|---|---|---|
| AA | 33 (50.0) | 127 (54.5) | 1.00 | |
| GA | 28 (42.4) | 84 (36.1) | 1.28 (0.72–2.28) | 0.40 |
| GG | 5 (8.2) | 22 (9.4) | 0.88 (0.31–2.48) | 0.80 |
| GA+GG | 33 (50.0) | 106 (45.5) | 1.20 (0.69–2.07) | 0.52 |
| A | 94 (71.2) | 338 (72.5) | 1.00 | |
| G | 38 (28.8) | 128 (27.5) | 1.07 (0.70–1.64) | 0.77 |
In this study, we compared genotyping data obtained with HapMap database (http://Hapmap.ncbi.nlm.nih.gov), Genecards database (http://www.genecards.org), the U.S. National Center for Biotechnology Information (NCBI) database of genetic variation (db SNP), the Thousands of People Plan database (http://browser.1000genomes.org), and the genome databases Ensembl (http://www.ensembl.org) for SNP allele and genotype frequencies in different populations compared among different races. We found that the distributions of allele and genotype frequencies were significantly differently in different ethnic groups, especially in European populations (P<0.001) (Tables 9, 10), suggesting that differences between races, including geography, environment, culture, and lifestyle, are likely to affect single-nucleotide polymorphisms occurrence, especially in Asian and European populations. However, due to the sample size in this study and because the database was relatively small, the specific relationship between differences and this disease needs to be further explored.
Table 9.
Genotype distribution and allele frequencies of rs1052536 polymorphism in normal population of different races.
| Population | n | Genotype (n, %) | P | Allele (n, %) | P | |||
|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | C | T | ||||
| Chinese (Shanxi) | 224 | 128 (57.1) | 74 (33.0) | 22 (9.8) | 330 (73.7) | 118 (26.3) | ||
| CHB | 136 | 71 (52.2) | 51 (37.5) | 14 (10.3) | 0.645 | 193 (71.0) | 79 (29.0) | 0.430 |
| CHD | 109 | 55 (50.5) | 47 (43.1) | 7 (6.4) | 0.164 | 157 (72.0) | 61 (28.0) | 0.654 |
| JPT | 113 | 72 (63.7) | 39 (34.5) | 2 (1.8) | 0.024 | 183 (81.0) | 43 (19.0) | 0.036 |
| CEU | 112 | 24 (21.4) | 64 (57.1) | 24 (21.4) | <0.001 | 112 (50.0) | 112 (50.0) | <0.001 |
| MEX | 57 | 22 (38.6) | 28 (49.1) | 7 (12.3) | 0.040 | 72 (63.2) | 42 (36.8) | 0.027 |
| ASW | 57 | 37 (64.9) | 19 (33.3) | 1 (1.8) | 0.130 | 93 (81.6) | 21 (18.4) | 0.080 |
CHB – Han Chinese in Beijing, China; CHD – Chinese in Metropolitan Denver, Colorado; JPT – Japanese in Tokyo, Japan; CEU – Utah residents with Northern and Western European ancestry from the CEPH collection; MEX – Mexican ancestry in Los Angeles, California; ASW – African ancestry in Southwest USA.
Table 10.
Genotype distribution and allele frequencies of rs3135967 polymorphism in normal population of different races.
| Population | n | Genotype (n, %) | P | Allele (n, %) | P | |||
|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | A | G | ||||
| Chinese (Shanxi) | 233 | 128 (54.5) | 84 (34.1) | 22 (9.8) | 338 (72.5) | 118 (27.5) | ||
| CHB | 137 | 71 (51.8) | 52 (38.0) | 14 (10.2) | 0.880 | 194 (70.8) | 79 (29.2) | 0.613 |
| CHD | 109 | 55 (50.5) | 47 (43.1) | 7 (6.4) | 0.369 | 157 (72.0) | 61 (28.0) | 0.889 |
| JPT | 113 | 72 (80.9) | 39 (34.5) | 2 (1.8) | 0.022 | 183 (81.0) | 43 (19.0) | 0.016 |
| CEU | 113 | 22 (19.5) | 64 (56.6) | 27 (23.9) | <0.001 | 108 (47.8) | 118 (52.2) | <0.001 |
| MEX | 57 | 19 (33.3) | 29 (50.9) | 9 (15.8) | 0.015 | 67 (58.8) | 47 (41.2) | 0.004 |
| ASW | 57 | 37 (64.9) | 19 (33.3) | 1 (1.8) | 0.112 | 93 (81.6) | 21 (18.4) | 0.048 |
CHB – Han Chinese in Beijing, China; CHD – Chinese in Metropolitan Denver, Colorado; JPT – Japanese in Tokyo, Japan; CEU – Utah residents with Northern and Western European ancestry from the CEPH collection; MEX – Mexican ancestry in Los Angeles, California; ASW – African ancestry in Southwest USA.
HaploView software was used to make haplotype analysis in selected loci in our study, showing that the 2 sites of LIG3 single-nucleotide polymorphisms were in linkage disequilibrium (Figure 3). The 2 LIG3 polymorphic loci in this study had 4 haplotypes, as shown in Table 11, and the T/G haplotype increased the risk of NTDs, but the P values were not statistically significant (P=0.69, OR=1.079, 95%CI [0.75–1.56]), and the CA haplotype has a tendency to reduce the risk of NTDs (P=0.035, OR=0.683, 95% CI [0.48–0.97]).
Figure 3.
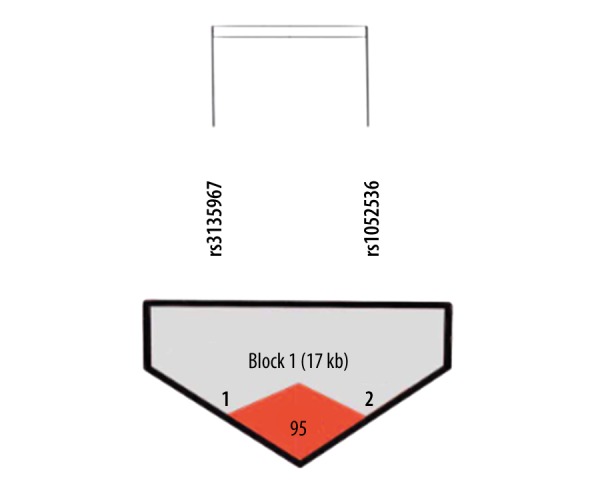
The results of the Haploview software analysis.
Table 11.
Haplotype analysis of rs1052536 and rs3135967*.
| Haplotype | Case (freq) | Control (freq) | Fisher’s P | Pearson’s P | OR (95%CI) |
|---|---|---|---|---|---|
| C/A | 135.89 (0.647) | 325.99 (0.728) | 0.034941 | 0.034911 | 0.683 (0.479–0.974) |
| C/G | 2.11 (0.010) | 4.01 (0.009) | – | – | – |
| T/A | 14.11 (0.067) | 1.01 (0.002) | – | – | – |
| T/G | 57.89 (0.276) | 116.99 (0.261) | 0.687774 | 0.687774 | 1.079 (0.745–1.561) |
SHEsis (http://analysis.bio-x.cn); All those frequency<0.03 will be ignored in analysis.
Discussion
Numerous studies showed that DNA damage might be a genetic risk factor in various diseases, especially geriatric diseases and cancers [20,21], but there has been no previous study on complex problems such as birth defects. In the present case-control study, we selected 108 NTD pregnant women and 233 control pregnant women (matched by age and gestational age with the case group) from 9 hospitals at Lvliang areas in Shanxi province, which is a high-risk area for NTDs in Chinese Han people. We explored the relationship between key gene LIG3′s popular research sites-rs1052536 and rs3135967 single-nucleotide polymorphisms under base excision repair pathway and the incidence of NTDs susceptibility. We found that rs1052536 was relevant to susceptibility of NTDs, homozygous TT genotype of rs1052536 increased the risk of anencephaly (P=0.016, OR=2.69, 95%CI [1.18–6.10]), and T allele carriers had significantly increased risk of cranial NTDs (P=0.033, OR=1.56, 95%CI [1.04–2.35]).
The relative stability of the human genome in prevention of genetic diseases is very important. Folic acid is involved in one-carbon unit transfer processes, and plays a decisive role in purine and pyrimidine synthesis, transformation from dUMP to dTMP, and DNA methylation. Therefore, folic acid is a kind of vitamin which plays a key role in DNA synthesis, repair, DNA configuration, gene expression, and cell proliferation. Existing evidence indicates that the accumulation of DNA damage is closely related to certain diseases, and folic acid might affect genome stability. The populations in this study were folate-deficient. Normal DNA synthesis was subject to different degrees of influence, and then affected the stability of the genome. At a certain time, cells in the body start to use multiple DNA repair pathways to repair damaged DNA; this is a normal function of the body and such repair is essential.
The base excision repair (BER) system is one of the most important pathways in a variety of DNA repair mechanisms, in which DNA base damage is caused by oxidants and alkylating agents, and plays an important role in maintaining the integrity of DNA in human cells [22–24].
Research shows that the process of DNA base excision repair glycosylases involves downstream mechanism dysfunction and leads to fetal death [25,26]. Thus, maintaining the normal operation of this pathway is very important. Reduction or loss of function in key genes expression of repairing system influence DNA repair capacity to various degrees, affecting cell function and increasing the risk of mutation, while also increasing susceptibility to disease.
DNA ligaseIII (LIG3) is one of the important repair genes in the repair pathway, encoding a mitochondrial protein, and plays an important role in maintaining integrity of mitochondria. It is in charge of connecting DNA to a complete protein after DNA stump closure. This process plays an important role in DNA replication, recombination, and repair processes. DNA replication and repair processes in mammals involve 3 major ligases: LIG1, LIG3, and LIG4 [27]. In the base excision repair pathway, LIG3 is considered a key ligase, which plays an essential role in maintenance of the integrity of mitochondrial DNA [28,29]. Studies have shown that LIG3 gene inactivation in the mouse neural system leads to damage of mitochondrial DNA deletions, mitochondrial dysfunction, body balance disorders, and movement disorders [30,31]. LIG1 is also involved in DNA repair with LIG3 [32].
Most recent reports on LIG3 have involved tumor research. More and more studies show that the abnormal proliferations in tumors are related to DNA damage repair disorders. The SNP loci DNA ligase III has gradually become an important research topic. Landi et al. found that LIG3 rs1052536 locus homozygotes significantly increased the risk of lung cancer in young people (P<0.01, OR=2.05, 95% CI [1.25–3.38]) [33], and Synowiec et al. reported that rs1052536 is associated with keratoconus and Fuchs endothelial corneal dystrophy [34].
Birth defects are complex diseases in which environmental factors and genetic factors act together. Birth defects are somewhat similar to tumorigenesis, but no correlation has been reported between LIG3 on DNA repair gene single-nucleotide polymorphisms and birth defects.
The occurrence and development of disease is a systematic behavior. Some key gene mutations cause genetic heterogeneity of human diseases (heterogeneity) in the long evolutionary process [35]. Genetic heterogeneity refers to a phenotype consistent with the same individual or clinical manifestations of the same disease, but may have different genotypes due to different genetic basis, and the mode of inheritance, age of onset, progression, severity, prognosis, and risk of recurrence may be different. This is an important feature of complex disease, and warrants more research [36,37]. Studies have shown that the increased incidence of genetic diseases is not only due to the discovery of new diseases, but is also caused by the separation of the isoforms from a known syndrome, namely the existence of genetic heterogeneity, which is becoming increasingly common. Complex diseases exist for different people or different tissues of the same individual with differences in susceptibility. Many causes and mechanisms of complex diseases are unclear, and show no obvious regularity. NTDs are this kind of complex disease. In the present study we focussed on 2 SNPs (rs1052536 and rs3135967) in LIG3, and our results showed that rs3135967 was negative, which was consistent with the results of the Sukki Cho [38]. Rs3135967 is located in the intron area of LIG3 gene of Chr17q12 (34986710–34986710). Significant differences were found in Rs1052536 carrying T allele in the anencephaly group, and we also found that the T allele significantly increased risk of brain malformation; the differences were statistically significant, but no significant difference was found in the spina bifida group compared with the control group. This result suggests that the effect of the mutation on brain development may be greater than that on the spine, resulting in significant differences between groups in malformations of the brain. This is not an isolated phenomenon; Xie et al. [39] also showed a similar situation, in which GCPII gene SNP rs202676 carrying G allele significantly increased disease risk in the anencephaly group (P=0.03, OR=2.11, 95%CI [1.11–4.01]), but spina bifida and encephalocele had no significant differences. Of the 2 sites selected in this study, rs1052536 is located in UTR-3 and rs3135967 is located in intron. Functions of intron regions are not entirely clear, and data on the relationship between rs3135967 and NTDs show a trend, but without a statistically significant difference. Studies have shown that non-coding region sites may also cause genetic mutations [40]; therefore, the relationship between rs3135967 and NTDs should not be dismissed simply because it was found in a non-coding region, and the specific association between them needs to be further explored.
In this study, only rs1052536 showed a close relationship with the incidence of NTDs in 2 sites, but no significant difference was found for the rs3135967 locus. Linkage disequilibrium analysis (D=0.952, r2=0.848) showed linkage disequilibrium located in the same block, but the results were significantly different for the 2 points; therefore, s we concluded that there may be some other factors involved that need to be further explored.
In addition, we compared allele and genotype frequencies of the SNP loci in different groups. We found that compared with HapMap database (http://Hapmap.ncbi.nlm.nih.gov), Genecards database (http://www.genecards.org), the US National Center for Biotechnology Information (NCBI) database of genetic variation (db SNP), thousands of people plan database (http://browser.1000genomes.org) and genome databases Ensembl (http://www.ensembl.org), the distribution of allele and genotype frequencies were significantly differently in different ethnic groups, especially the European populations (P<0.001), as shown in Tables 9 and 10. This suggests that the differences between the races, including geography, environment, culture, and lifestyle are likely to be the causes of single-nucleotide polymorphisms, and the differences between Asian populations and European populations are particularly evident. However, because the sample size and the database were both relatively limited in this study, the specific relationship between the differences and this disease need to be further explored.
Results of the present study suggest that the effect of a single gene mutation for complex diseases such as birth defects was quite small, and it might be the cumulative effect of multiple common variants and their interaction with environmental factors that lead to disease susceptibility. The DNA repair system is complex and precise, and individual single-nucleotide mutations have limited impact on body functions. Several or more mutant mutual or joint action may significantly affect DNA repair, leading to damage accumulation and affecting the occurrence and development of diseases. Therefore, particular combinations of specific genes are the most important causes of disease susceptibility, and more and more studies of diseases use polygenic joint research [41]. Many key genes are involved in the DNA excision repair pathway, like APE1, OGG1, and XRCC1. Their roles in the BER pathway are indispensable, and the combination of these genes deserve future research attention.
In summary, 2 sites in only 1 of the base excision repair pathway genes were studied. The limited number of samples and the limited number of races might also have biased our results, which only showed a weak tendency, and the relationship between mechanism and occurrence of NTDs is unclear. Phenotypic heterogeneity may also be a study limitation. The cases in our study include 3 subtypes, and the limited analyses specific to each NTD subtype did not achieve significant results. Therefore, we need to further refine the association study by expanding the sample size and including more mutations in the future to provide experimental evidence for the early diagnosis and prevention of NTDs.
Conclusions
The current study supports that rs1052536 of DNA base excision repair pathway genes LIG3 is a potential risk factor in a high-risk region for NTDs in China, in which the TT genotype significantly increased risk of cranial NTDs. Allele T carrier might be an important genetic susceptibility factor for cranial NTDs, but further research is needed to define the precise mechanism by which the variant affects NTDs.
Acknowledgements
Our group is greatly indebted to the participants for taking part in the study, and we thank all obstetricians in the local hospital at Shanxi province, as well as the pathologists in the Department of Pathology for the diagnostic work. We also thank all subjects and their family members of their cooperation in providing clinical information and samples for the study.
We are thankful to all the staff and participants for the enthusiasm and devotion to this study and to all the obstetricians at the local hospital in Shanxi province and the pathologists in the Department of Pathology for the diagnostic work.
Footnotes
Conflict of interest
None.
Source of support: This study was supported by the National Natural Science Foundation of China (No. 81571443, 81600984), the Ministry of Science and Technology of China, National “973” Project on Population and Health (2013CB945404), and the Beijing National Natural Science Foundation (No.7172038, 7174285)
References
- 1.Greene ND, Stanier P, Copp AJ. Genetics of human neural tube defects. Hum Mol Genet. 2009;18(R2):R113–29. doi: 10.1093/hmg/ddp347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang JF, Liu X, Christakos G, et al. Assessing local determinants of neural tube defects in the Heshun Region, Shanxi Province, China. BMC Public Health. 2010;2(10):52–56. doi: 10.1186/1471-2458-10-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Au KS, Ashley-Koch A, Northrup H. Epidemiologic and genetic aspects of spina bifida and other neural tube defects. Dev Disabil Res Rev. 2010;16:6–15. doi: 10.1002/ddrr.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prevention of neural tube defects: Results of the Medical Research Council Vitamin Study. MRC Vitamin Study Research Group. Lancet. 1991;338:131–37. [PubMed] [Google Scholar]
- 5.Czeizel AE, Dudas I. Prevention of the first occurrence of neural tube defects by periconceptional vitamin supplementation. N Engl J Med. 1992;327:1832–35. doi: 10.1056/NEJM199212243272602. [DOI] [PubMed] [Google Scholar]
- 6.Tolarova M, Harris J. Reduced recurrence of orofacial celfts after periconceptional supplementation with high-dose folic acid and multivitamins. Teratology. 1995;51(2):71–72. doi: 10.1002/tera.1420510205. [DOI] [PubMed] [Google Scholar]
- 7.Cao L, Wang Y, Zhang R, et al. Association of neural tube defects with gene polymorphisms in one-carbon metabolic pathway. Childs Nerv Syst. 2017;34(2):277–84. doi: 10.1007/s00381-017-3558-z. [DOI] [PubMed] [Google Scholar]
- 8.Czeizel AE, Dudás I. Prevention of the first occurrence of neural-tube defects by periconceptional vitamin supplementation. N Engl J Med. 1992;327(26):1832–35. doi: 10.1056/NEJM199212243272602. [DOI] [PubMed] [Google Scholar]
- 9.Au KS, Ashley-Koch A, Northrup H. Epidemiologic and genetic aspects of spina bifida and other neural tube defects. Dev Disabil Res Rev. 2010;16(1):6–15. doi: 10.1002/ddrr.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olshan AF, Shaw GM, Millikan RC, et al. Polymorphisms in DNA Repair Genes as Risk Factors for Spina Bifida and Orofacial Clefts. Am J Med Genet. 2005;135A:268–73. doi: 10.1002/ajmg.a.30713. [DOI] [PubMed] [Google Scholar]
- 11.Barber R, Shalat S, Hendricks K, et al. Investigation of folate pathway gene polymorphisms and the incidence of neural tube defects in a texas Hispanic population. Mol Genet Metab. 2000;5:45–52. doi: 10.1006/mgme.2000.2991. [DOI] [PubMed] [Google Scholar]
- 12.Liang L, Deng L, Nguyen SC, et al. Human DNA ligases I and III, but not ligase IV, are required for microhomology-mediated end joining of DNA double-strand breaks. Nucleic Acids Res. 2000;36(10):3297–310. doi: 10.1093/nar/gkn184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomkinson AE, Howes TR, Wiest NE. DNA ligases as therapeutic targets. Transl Cancer Res. 2013;2(3) pii: 1219. [PMC free article] [PubMed] [Google Scholar]
- 14.Spitz MR, Wei Q, Dong Q, et al. Genetic susceptibility to lung cancer: The role of DNA damage and repair. Cancer Epidemiol Biomarkers Prev. 2013;12(8):689–98. [PubMed] [Google Scholar]
- 15.Kiyohara C, Yoshimasu K. Genetic polymorphisms in the nucleotide excision repair pathway and lung cancer risk: A meta-analysis. Int J Med Sci. 2007;4(2):59–71. doi: 10.7150/ijms.4.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thyagarajan B, Lindgren B, Basu S, et al. Association between genetic variants in the base excision repair pathway and outcomes after hematopoietic cell transplantations. Biol Blood Marrow Transplant. 2010;16(8):1084–89. doi: 10.1016/j.bbmt.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Volck KA, Shaw GM, Zhu H, et al. Risk factors for neural tube defects: Associations between uncoupling protein 2 polymorphisms and spina bifida. Birth Defects Res A Clin Mol Teratol. 2003;67(3):158–61. doi: 10.1002/bdra.10019. [DOI] [PubMed] [Google Scholar]
- 18.Wang J, Liu C, Zhao H, et al. Association between a 45-bp 3′untranslated insertion/deletion polymorphism in exon 8 of UCP2 gene and neural tube defects in a high-risk area of China. Reprod Sci. 2011;18(6):556–60. doi: 10.1177/1933719110393026. [DOI] [PubMed] [Google Scholar]
- 19.Gu X, Lin L, Zheng X, et al. High prevalence of NTDs in Shanxi Province: A combined epidemiological approach. Birth Defects Res A Clin Mol Teratol. 2007;79:702–7. doi: 10.1002/bdra.20397. [DOI] [PubMed] [Google Scholar]
- 20.Hatt L, Loft S, Risom L, et al. OGG1 expression and OGG1 Ser326Cys polymorphism and risk of lung cancer in a prospective study. Mutat Res. 2008;639:45–54. doi: 10.1016/j.mrfmmm.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 21.Loft S, Svoboda P, Kasai H, et al. Prospective study of 8-oxo-7,8-dihydro-2′-deoxyguanosine excretion and the risk of lung cancer. Carcinogenesis. 2006;27:1245–50. doi: 10.1093/carcin/bgi313. [DOI] [PubMed] [Google Scholar]
- 22.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–74. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 23.Petermann E, Keil C, Oei SL. Roles of D NA ligase III and XRCC1 in regulating the switch between short patch and long pa tch BER. DNA Repair (Amst) 2006;5:544–55. doi: 10.1016/j.dnarep.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 24.Krokan HE, Nilsen H, Skorpen F, et al. Base excision repair of DNA in mammalian cells. FEBS Lett. 2000;476:73–77. doi: 10.1016/s0014-5793(00)01674-4. [DOI] [PubMed] [Google Scholar]
- 25.Friedberg EC, Meira LB. Database of mouse strains carrying targeted mutations in genes affecting cellular responses to DNA damage: Version 3. Mutat Res. 1999;433(2):69–87. doi: 10.1016/s0921-8777(98)00068-8. [DOI] [PubMed] [Google Scholar]
- 26.Nordstrand LM, Ringvoll J, Larsen E, Klungland A. Genome instability and DNA damage accumulation in gene-targeted mice. Neuroscience. 2007;145(4):1309–17. doi: 10.1016/j.neuroscience.2006.10.059. [DOI] [PubMed] [Google Scholar]
- 27.Ellenberger T, Tomkinson AE. Eukaryotic DNA ligases: Structural and functional insights. Annu Rev Biochem. 2008;77:313–38. doi: 10.1146/annurev.biochem.77.061306.123941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simsek D, Jasin M. DNA ligase III: A spotty presence in eukaryotes, but an essential function where tested. Cell Cycle. 2011;10(21):3636–44. doi: 10.4161/cc.10.21.18094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simsek D, Brunet E, Wong SY, et al. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet. 2011;7(6):e1002080. doi: 10.1371/journal.pgen.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao Y, Katyal S, Lee Y, et al. DNA ligaseIII is critical for mtDNA integrity but not Xrcc1-mediated nuclear DNA repair. Nature. 2011;471(7337):240–44. doi: 10.1038/nature09773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arakawa H, Bednar T, Wang M, et al. Functional redundancy between DNA ligases I and III in DNA replication in vertebrate cells. Nucleic Acids Res. 2012;40(6):2599–610. doi: 10.1093/nar/gkr1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murray RJ, Tanteles GA, Mills J, et al. Association between single nucleotide polymorphisms in the DNA repair gene LIG3 and acute adverse skin reactions following radiotherapy. Radiother Oncol. 2011;99(2):231–34. doi: 10.1016/j.radonc.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 33.Landi S, Gemignani F, Canzian F, et al. DNA repair and cell cycle control genes and the risk of young-onset lung cancer. Cancer Res. 2006;66(22):11062–69. doi: 10.1158/0008-5472.CAN-06-1039. [DOI] [PubMed] [Google Scholar]
- 34.Synowiec E, Wojcik KA. Polymorphism of the LIG3 gene in keratoconus and Fuchs endothelial corneal dystrophy. Cell Mol Biol (Noisy-le-grand) 2015;61(1):56–63. [PubMed] [Google Scholar]
- 35.Gebel JM. Heterogeneity of efficacy and safety of antiplatelet therapy in cardiovascular and cerebrovascular disease. Am J Cardiovasc Drugs. 2010;10(2):115–24. doi: 10.2165/11319580-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 36.Beghi E, Chio A, Couratier P, et al. The epidemiology and treatment of ALS: focus on the heterogeneity of the disease and critical appraisal of therapeutic trials. Amyotroph Lateral Scler. 2011;12(1):1–10. doi: 10.3109/17482968.2010.502940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim KJ, Lee HJ, Park MH, et al. SNP identification, linkage disequilibrium,and haplotype analysis for a 200-kb genomic region in a Korean population. Genomics. 2006;88:535–40. doi: 10.1016/j.ygeno.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 38.Cho S, Kim MJ, Choi YY, et al. Associations between polymorphisms in DNA repair genes and TP53 mutations in non-small cell lung cancer. Lung Cancer. 2011;73(1):25–31. doi: 10.1016/j.lungcan.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 39.Xie H, Guo J, Wang J, et al. Glutamate carboxypep tidase II gene polymorphisms and neural tube defects in a high-risk Chinese population. Metab Brain Dis. 2012;27(1):59–65. doi: 10.1007/s11011-011-9272-8. [DOI] [PubMed] [Google Scholar]
- 40.Stower H. Gene expression: Looping together the functions of SNPs. Nat Rev Genet. 2012;13(3):148–49. doi: 10.1038/nrg3180. [DOI] [PubMed] [Google Scholar]
- 41.Ye CC, Huang ZM, Zhou CY. APE1 D148E, PARP1 V762A and XRCC1 R399Q polymorphisms and genetic susceptibility to colorectal cancer. World Chinese Journal of Digestology. 2010;18(12):1275–79. [Google Scholar]