Abstract
Background
A deficiency of maternal thyroid hormones (THs) during pregnancy has severe impacts on fetal brain development. Neural stem cells (NSCs) are major targets of THs and provided a powerful model to explore the underlying mechanism of THs during brain development. Although miRNA-125 might be associated with the NSCs differentiation, the relationship between miR-125 and hypothyroidism (HypoT) development remains unclear.
Material/Methods
In our study, we screened a differentially expressed gene miR-125b-5p from brain between euthyroid (EuT) and HypoT rats. In vitro, we employed anion exchange resin to remove THs to stimulate HypoT. QRT-PCR and Western blot were used to examine the expression of signal transducer and activator of transcription 3 (Stat3). The relationship between miR-125b-5p and Stat3 was detected via a dual-luciferase assay.
Results
QRT-PCR results showed that the level of miR-125b-5p in HypoT rat brains was significantly suppressed, suggesting some relationship between miR-125b-5p and HypoT. In C17.2, miR-125b-5p promoted cell differentiation into neurons by regulating the expression of tubulin beta chain 3 (TUBB3) and glial fibrillary acid protein (GFAP). QRT-PCR and Western blot results revealed that miR-125b-5p mimic modulated the contents of total Stat3 and p-Stat3. A dual-luciferase assay showed that miR-125b-5p negatively regulated the expression of Stat3 by binding with the first site in 3′ UTR of Stat3.
Conclusions
These results revealed Stat3 is a new target of miR-125b-5p and revealed the mechanism of miR-125b-5p suppressing HypoT development. These findings provide a new target for HypoT therapy.
MeSH Keywords: Hypothyroidism, Neural Stem Cells, STAT3 Transcription Factor
Background
Thyroid hormones (THs) are key regulators during brain development [1]. Thyroid diseases have deleterious effects on lactation, growth and survival, leading to hypothyroidism (HypoT) [2]. Maternal HypoT during pregnancy impairs brain function in the offspring of humans and other mammals [3]. Despite considerable advances during recent decades in our understanding of the cellular targets, genes, and processes involved in the function of THs during brain development [4], little is known about the cellular targets and the underlying mechanisms of maternal HypoT. Neural stem cells (NSCs) have the ability to self-renew and differentiate into different types of neural cells, including neurons, astrocytes, and oligodendrocytes [5], which are major targets by which THs influence early brain development. In our study, we used C17.2, a multipotent neural progenitor, or stem-like cells to explore the underlying mechanism of THs during brain development.
MicroRNA (miRNA) is a small non-coding single-stranded RNA consisting of 18–24 nucleotides [6]. They are involved in many normal biological processes such as cell growth, differentiation, and proliferation by negatively regulating genes expression at the posttranscriptional level via either targeting mRNAs degradation or translational repression [7]. Due to the features of miRNAs, a single miRNA can target more than 1 mRNA molecule and single target mRNA can be recognized by multiple miRNAs [8]. Thus, the interaction between miRNA and its targets creates a complex regulatory network [9]. Several miRNAs have been identified to participate in thyroid hypofunction, such as miR-124 [10] and miR-224 [11].
miRNA-125 (miR-125) is a highly conserved miRNA family identified in diverse species from nematodes to humans. miR-125 family members are downregulated in several diseases, such as various cancers [12,13]. However, miR-125-5p is highly expressed in mature glial cells but is downregulated in retinal neurons [14]. miR-125 promotes neural conversion by avoiding the persistence of non-differentiated stem cells and repressing alternative fate choices [15]. These data indicated that miR-125 might be associated with NSCs differentiation. However, there have been few reports on the role of miR-125 during the development of HypoT.
In our study, miR-125b-5p was screened out by the comparison between euthyroid (EuT) and HypoT rats. The relative expression of miR-125-5p in HypoT rats was significantly suppressed compared to that in EuT rats. The anion exchange resin was used to produce a rapid and complete decline of THs in serum to stimulate HypoT. In vitro, miR-125b-5p mimic promoted C17.2 neural stem-like cells differentiation into neurons in the lack of THs. miR-125b-5p target gene prediction by TargetScan software showed that signal transducer and activator of transcription 3 (Stat3) might be a miR-125b-5p target gene. We used a dual-luciferase assay to reveal that miR-125b-5p negatively regulates the expression of Stat3 by the first binding site in 3′UTR of Stat3. In vivo, no significant difference was observed in the mRNA levels of Stat3 between EuT and HypoT rats, but the protein level of total Stat3 was inhibited in HypoT rats. In C17.2 cells, miR-125b-5p mimic suppressed the level of phosphorylated-Stat3 in the lack of THs. We concluded that Stat3 was a target of miR-125b-5p and miR-125b-5p suppressed the occurrence of HypoT. These findings reveal a new target gene of miR-125b-5p and provide a new target for HypoT therapy.
Material and Methods
Animals and grouping
A total of 48 SPF-grade female Wistar rats weighing 200–220 g, were purchased from Shanghai Silaike Laboratory Animal Limited Liability Company (Shanghai, China). They were divided into 2 groups: EuT group (n=24) and HypoT group (n=24). Rats were anesthetized with 10% chloral hydrate (3 mL/kg, Sigma, St. Louis, MO) via peritoneal injection. Rats in the HypoT group were subjected to thyroidectomy, and EuT rats underwent sham operation. At 1 month after the operation, female rats from the EuT or HypoT group were mated with normal male rats overnight after the estrous cycle was monitored. The female vaginal suppository was examined the next morning, and rats with pessary were recorded as pregnancy day 0 (E0). At E13 and E17, rats were anesthetized and the brain tissues of fetal rat were collected for following experiments. The postnatal first day was recorded as P0. At P7, the hippocampus and cortex from offspring brain tissues were separated for further study. All rats were kept in standard laboratory conditions on arrival, with temperature 23±2°C. Standard rodent feed and water were available ad libitum. All procedures were performed in accordance with the Guide for the Humane Use and Care of Laboratory Animals.
C17.2 neural stem-like cells identification
C17.2 is a multipotent neural progenitor or stem-like cell line originally derived from the external germinal layer of neonatal murine cerebellum and immortalized with c-myc. C17.2 cells were seeded into 24-well plates containing polylysine-coated glass in complete medium. After 24 h, cells were washed 3 times with PBS, and then fixed with 4% fresh, cold paraformaldehyde for 15 min. After being washed with PBS, cells were permeabilized with 0.2% Triton-X100 for 15 min, followed by blockage with goat serum for 30 min. Then, cells were incubated with anti-Nestin antibody (1: 100 dilution, Abcam, USA) at 4°C overnight. The next morning, cells were washed 3 times and probed with FITC-labeled goat anti-mouse IgG (Abcam, USA) for 4 h at room temperature. Nuclei were stained with DAPI (Sigma, USA) at room temperature for 10 min. After mounting, the cells were visualized under a fluorescence inverted microscope (40×, Carl Zeiss, LePecq, France).
Quantitative reverse transcription PCR (qRT-PCR)
Total RNA was extracted from brain tissues or C17.2 cells using Trizol reagent (Invitrogen, USA). The quality and quantity of RNA were determined by measuring the absorbance at 260 and 280 nm using a Nano Drop-2000 ultramicrospectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). One μg of RNA was reverse transcribed into cDNA using the Step-One cDNA synthesis kit (Takara, Dalian, China) according to the manufacturer’s protocol. Stat3 cDNA was amplified using the SYBR Premix Ex Taq Kit (Takara, Dalian, China) with the Real-Time PCR System (LightCycler 480 PCR system, USA). PCR assay cycles were as follows: 95°C for 5 min, 50 cycles of 95°C for 10 s, 60°C for 20 s, and 72°C for 5 s. The mRNA level of Stat3 was normalized to β-actin using the 2–ΔΔCT method. miRNA was isolated from brain tissues or C17.2 using a miRNA quick extraction kit (BioTeke, Beijing, China). The relative expression of miR-125b-5p was normalized to U6. The primers used for qRT-PCR were as follows: TUBB3 forward (5′-catccagagcaagaacagca-3′), reverse (5′-gcctggagctgcaataagac-3′); GFAP forward (5′-agatccacgaggaggaggtt-3′), reverse (5′-cggcgttccatttacaatct-3′); Stat3 forward (5′-agctgagcgtgtgtgacagt-3′), reverse (5′-acccatgggattacacttgg-3′); β-actin forward (5′-cctctatgccaacacagtgc-3′), reverse (5′-gtactcctgcttgctgatcc-3′); miR-125b-5p forward (5′-ggtccctgagaccctaact-3′), reverse (5′-ctcaactggtgtcgtgga-3′); U6 forward (5′-ctcgcttcggcagcaca-3′), reverse (5′-aacgcttcacgaatttgcg-3′).
A dual-luciferase assay in HEK293 cells
The wild-type or mutated 3′-UTR sequences of Stat3 were inserted into a dual-luciferase reporter vector pMIR-GLO. We constructed 3 recombinant vectors: pMIR-GLO-WT (the reporter luciferase fused with the wild-type Stat3 3′ UTR), pMIR-GLO-M1 (the reporter luciferase fused with the mutated Stat3 3′ UTR in the first binding site at position 998–1005), and pMIR-GLO-M2 (the reporter luciferase fused with the mutated Stat3 3′ UTR in the second binding site at position 1606–1612). The recombinant plasmids were verified by DNA sequencing, and the target sequence (CTCAGGG) was successfully mutated to GTGACGC in mutations. HEK293 cells were seeded into 6-well plates for 48 h. When the density reached 80%, cells were co-transfected with pMIR-GLO-WT+miR-125b-5p mimic, pMIR-GLO-M1+miR-125b-5p mimic, pMIR-GLO-M2+miR-125b-5p mimic, pMIR-GLO-WT+miRNA NC, pMIR-GLO-M1+miRNA NC, and pMIR-GLO-M2+ miRNA NC. After 48 h of transfection, cells were harvested and the dual-luciferase reporter assay system (Promega, USA) was used to measure luminous intensity. All experiments were independently performed 3 times.
Western blot analysis
Proteins were extracted from brain tissues or C17.2 cells using RIPA buffer (Sigma, USA). The concentration of protein was detected by use of a bicinchoninic acid kit (Pierce, Rockford, IL). About 50 μg of protein was subjected to 10% SDS-polyacrylamide gel and then transferred onto a PVDF membrane (Millipore, Bedford, MA, USA). Subsequently, the membrane was blocked with 5% BSA (Sigma, USA) in Tris-buffered saline with 0.05% Tween-20 (TBST) for 1 h at room temperature and then incubated overnight at 4°C with the primary anti-β-actin antibody (Cell Signaling, USA), rabbit anti-Stat3 polyclonal antibody, or anti-p-Stat3 antibody (Abcam, USA). After being washed with TBST 3 times, the membranes were probed with horseradish peroxidase-conjugated secondary antibodies (Zhongshan Biotechnology Co., Ltd., Beijing, China). The level of β-actin was used as an internal control. The membranes were developed with an enhanced chemiluminescence system (Thermo, USA), and the bands were quantitatively analyzed by use of FlourChem V2.0 (Alpha Innotech Corp., San Leandro, CA).
Statistical analysis
All experiments were performed at least 3 times using independent samples. All data are expressed in means ± standard deviation. Statistical analysis was conducted using SPSS 12.0 software (SPSS, Chicago, IL, USA). Data were analyzed by use of the double-sided t test. Results were considered statistically significant at ** P<0.01.
Results
miR-125b-5p was expressed at low levels in HypoT rats
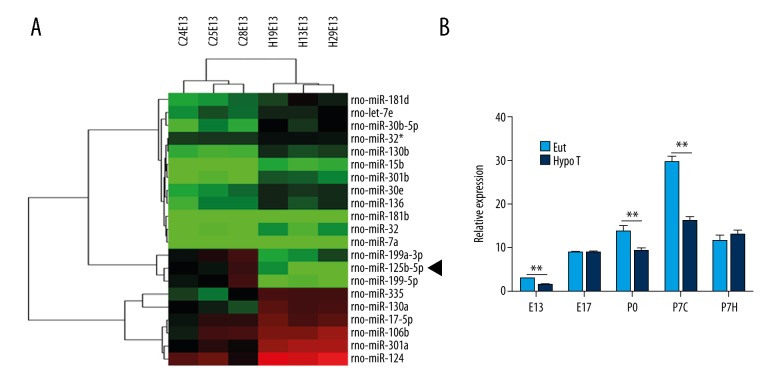
We isolated the brain tissues from EuT or HypoT female rats on embryonic day 13 (E13) to screen the differentially expressed genes. From the microarray results in Figure 1A, we found that a total of 21 miRNAs were differentially expressed between EuT and HypoT groups. Importantly, the level of miR-125b-5p in the EuT group was about 6.25-fold higher than in the HypoT group. To further confirm the expression of miR-125b-5p, we detected the level of miR-125b-5p from E13 to P7H of EuT and HypoT rats. Figure 1B shows that the expression of miR-125b-5p on E13, P0, and P7C of the HypoT group was significantly lower than that of the EuT group (P<0.01). These results show that the expression of miR-125b-5p was suppressed in HypoT rats.
Figure 1.
miR-125b-5p was underexpressed in HypoT rats. (A) Microarray results showed total of 21 miRNAs were differentially expressed between EuT and HypoT rats. The brain tissues of EuT and HypoT groups were isolated for microarray assay. miR-125b-5p is marked with a black arrow. (B) miR-125b-5p was inhibited in brain tissues in HypoT rats. Total RNA of EuT and HypoT groups was extracted for QRT-PCR. U6 was used as the internal control. EuT – euthyroid; HypoT – hypothyroidism; E13 – embryonic day 13; E17 – embryonic day 17; P0 – postnatal first day; P7C – cortex of postnatal seventh day; P7H – hippocampus of postnatal seventh day; ** P<0.01.
miR-125b-5p mimic promoted C17.2 differentiate into neurons in the lack of THs
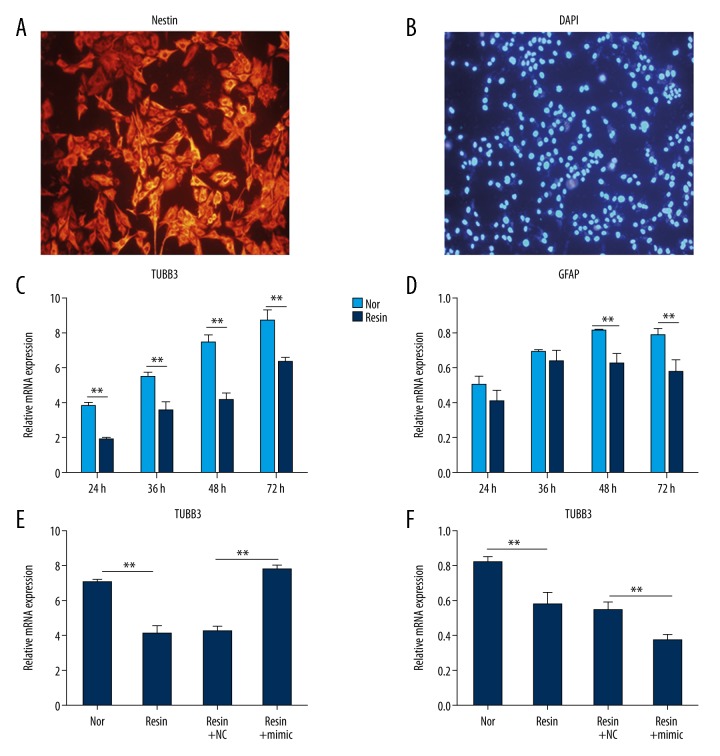
In our study, C17.2 neural stem-like cells were used for THs study during brain development. In Figure 2A and 2B, C17.2 cells were marked with Nestin neuroectodermal stem cell marker in red (>95%) and DAPI in blue for NSCs identification.
Figure 2.
miR-125b-5p promoted C17.2 differentiation into neurons in the lack of THs. (A, B) C17.2 is marked with Nestin stem cell marker in red and nuclei are stained with DAPI in blue. Cells were visualized under a fluorescence inverted microscope (40×). (C, D) THs removal suppressed the mRNA levels of TUBB3 and GFAP detected by qRT-PCR. Cells were incubated with untreated serum (Nor) or anion exchange resin-incubated serum (Resin) for 24, 36, 48 and 72 h, respectively. RNA was isolated for qRT-PCR. (E, F) The application of miR-125b-5p mimic affected the mRNA levels of TUBB3 and GFAP. C17.2 cells were divided into 4 groups: Nor, Resin, Resin+NC, and Resin+mimic. Nor, cells were cultured with untreated serum; Resin, cells were incubated with resin-treated serum; Resin+NC, cells were transfected with miRNA NC for 48 h, and then incubated with resin-treated serum; Resin+mimic, cells were transfected with miR-125b-5p mimic for 48 h, and then incubated with resin-treated serum. Total RNA was extracted for qRT-PCR. The relative expression of TUBB3 and GFAP was normalized to β-actin. ** P<0.01.
Some reports showed that anion exchange resin could remove THs of calf serum to simulate HypoT [16]. In our study, we used anion exchange resin to treat calf serum and detected the amount of TT3, TT4, and TSH in calf serum (Table 1). Little TSH was detected in untreated serum and resin-treated serum (P>0.05). However, in resin-incubated serum, the amount of TT3 (50.4±8.9 ng/dL) was significantly lower than that in untreated serum (162.4±10.3 ng/dL, P<0.01). Similarly, the content of TT4 in resin-treated serum (<1 μg/dL) was obviously lower than that in untreated serum (14.9±2.2 μg/dL, P<0.01). These data show that anion exchange resin effectively absorbed TT3 and TT4 of serum.
Table 1.
The amount of TT3, TT4 and TSH.
| TT3 (ng/dl) | TT4 (μg/dL) | TSH () | |
|---|---|---|---|
| Untreated serum | 162.4±10.3 | 14.9±2.2 | – |
| Resin-treated serum | 50.4±8.9** | <1** | – |
Compared with untreated serum,
P<0.01.
To explore the effect of miR-125b-5p on C17.2 cells, we incubated C17.2 cells with miR-125b-5p mimic and detected the mRNA levels of mature neurons marker tubulin beta chain 3 (TUBB3) and astrocytes marker glial fibrillary acid protein (GFAP). Compared with untreated serum (Normal group), the expression of TUBB3 in resin-incubated serum (Resin group) at 24, 36, 48, and 72 h was inhibited (P<0.01, Figure 2C), and the levels of GFAP in resin-incubated serum (Resin group) at 48 and 72 h were also suppressed (P<0.01, Figure 2D). These data show that THs were essential for C17.2 differentiation. When C17.2 cells were cultured with miR-125b-5p mimic under resin-incubated serum, the level of TUBB3 was promoted, but the mRNA of GFAP was decreased (P<0.01, Figure 2E, 2F). These data reveal that miR-125b-5p promoted C17.2 differentiation into neurons in the simulated HypoT condition.
STAT3 is a target of miR-125b-5p in HEK293 cells
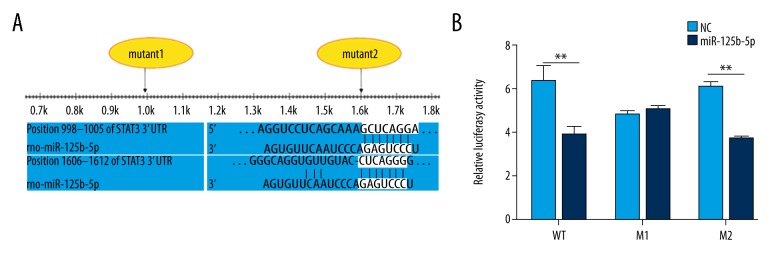
To further investigate the role of miR-125b-5p, we performed miR-125b-5p target gene prediction by use of TargetScan software and found that Stat3 might be a miR-125b-5p target gene. Then, a dual-luciferase reporter assay was constructed in HEK293 cells to examine the interaction between miR-125b-5p and Stat3. miR-125b-5p target gene prediction showed that there might be 2 predicted binding sites in 3′ UTR of Stat3 (Figure 3A). We constructed 3 recombinant plasmids: pMIR-GLO-WT (the wild-type Stat3 3′ UTR), pMIR-GLO-M1 (the mutated Stat3 3′ UTR in M1), and pMIR-GLO-M2 (the mutated Stat3 3′ UTR in M2). Compared with miRNA NC, miR-125b-5p mimic significantly suppressed the activity of the reporter luciferase after pMIR-GLO-WT transfection (Figure 3B). A similar trend was observed after pMIR-GLO-M2 transfection. However, no significant difference was found between the cotransfection of NC + pMIR-GLO-M1 and miR-125b-5p mimic + pMIR-GLO-M1 (P>0.05). These data show that miR-125b-5p regulated Stat3 via the first predicted binding site.
Figure 3.
Stat3 is a new target of miR-125b-5p. (A) Showed the 2 predication binding sites between miR-125b-5p and 3′ UTR of Stat3. (B) miR-125b-5p regulated the expression of Stat3 by binding to the first binding site in 3′ UTR of Stat3. HEK293 cells were pretreated with miR-125b-5p mimic or miRNA NC, and then transfected with pMIR-GLO-WT, pMIR-GLO-M1, or pMIR-GLO-M2, respectively. Cells were harvested after 48 h and the dual-luciferase reporter assay system was used to measure luminous intensity. ** P<0.01.
miR-125b-5p modulated the protein level of Stat3 in HypoT rats and C17.2
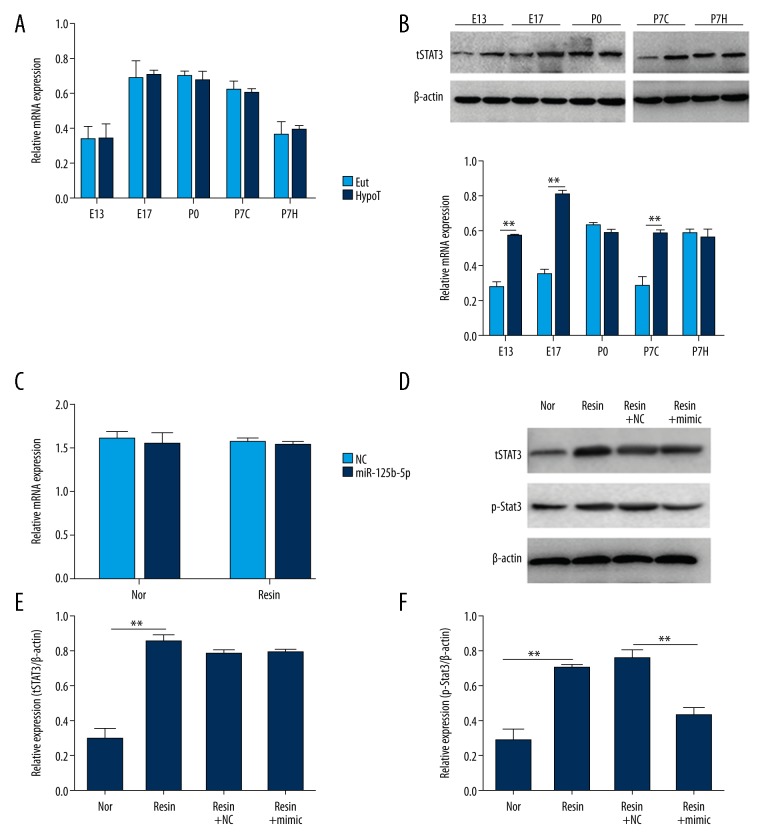
In vivo, we examined the expression of Stat3 to analyze the function of Stat3 in HypoT rats. No significant difference in mRNA levels of Stat3 was found from E13 to P7H between EuT and HypoT rats (P>0.05, Figure 4A). The contents of total Stat3 in the HypoT group on E13, E17, and P7C were significantly higher than those in EuT rats (P<0.01) and no obvious difference on total Stat3 protein was observed on P0 and P7H (P>0.05) (Figure 4B).
Figure 4.
miR-125b-5p affected the amount of total Stat3 and p-Stat3 in vitro and in vivo. (A) There was no significant difference in the mRNA level of Stat3 between EuT and HypoT rats. Total RNA was extracted on E13, E17, P0, P7C, and P7H for qRT-PCR. The relative Stat3 expression was normalized to β-actin. (B) The contents of total Stat3 on E13, E17, and P7C in HypoT rats were higher than those in EuT rats. E13, embryonic day 13; E17, embryonic day 17; P0, postnatal first day; P7C, cortex of P7; P7H, hippocampus of P7. (C) The decrease of THs had no effect on the mRNA level of Stat3 in C17.2. C17.2 cells were incubated with untreated serum or anion exchange resin-treated serum for 48 h. RNA was extracted for qRT-PCR. (D) The application of miR-125b-5p mimic suppressed the level of p-Stat3 in the absence of THs. C17.2 cells were divided into 4 groups: Nor, Resin, Resin+NC, and Resin+mimic. Nor, cells were cultured with untreated serum; Resin, cells were incubated with resin-treated serum; Resin+NC, cells were transfected with miRNA NC for 48 h, and then incubated with resin-treated serum; Resin+mimic, cells were transfected with miR-125b-5p mimic for 48 h, and then incubated with resin-treated serum. Proteins were extracted for Western blot analysis. β-actin was used as the internal control. ** P<0.01.
In vitro, we incubated C17.2 cells with resin-treated serum to simulate HypoT. Compared with untreated serum and resin-treated serum, there was no significant effect on the mRNA levels of Stat3 (P>0.05) and the application of miR-125b-5p did not influence the expression of Stat3 (Figure 4C). The protein levels of total Stat3 and p-Stat3 in resin-treated serum were obviously enhanced compared to those in untreated serum (Figure 4D). The supplementation of miR-125b-5p had no effect on the amount of total Stat3 but inhibited the level of p-Stat3 (Figure 4E, 4F). These data reveal miR-125b-5p regulated the protein level of Stat3 in vitro and in vivo.
Discussion
A deficiency of maternal THs during pregnancy has severe impacts on fetal brain development [17]. In our study, we found that the level of miR-125b-5p on E13 of HypoT rats was significantly suppressed, suggesting that miR-125b-5p plays a role during brain development in early pregnancy. A dual-luciferase assay showed that Stat3 is a target gene of miR-125b-5p by binding with the first site in 3′ UTR of Stat3. qRT-PCR and Western blot results revealed that miR-125b-5p mimic modulated the contents of total Stat3 and p-Stat3 and regulated the neuronal differentiation of C17.2 cells. These results indicate that Stat3 is a newly discovered target of miR-125b-5p and reveals the mechanism by which miR-125b-5p represses HypoT development.
It is well established that the mammalian brain is a direct target organ of TH. A transient and moderate decrease or increase of maternal THs during pregnancy has deleterious consequences on brain morphology in the offspring [18]. NSCs are one of the major cellular targets by which THs influence early brain development. Recent studies on NSCs have aided in the establishment of good models of brain development [19]. THs influence the balance among the proliferation, maintenance, and differentiation of NSCs during embryogenesis via various factors or signaling pathways [18]. In our study, when C17.2 cells were incubated under the THs removal serum, the differentiation of C17.2 was affected, which provides new evidence about the function of THs on NSCs fate.
miRNAs are key regulators on THs homeostasis [20]. In our study, we screened out a total of 21 miRNAs that were differentially expressed between EuT and HypoT rats, revealing the consequences of reduced maternal thyroid level on fetal brain miRNA abundance. The miR-125 family is composed of 3 homologs: has-miR-125a, has-miR-125b-1, and has-miR-125-2 [21] It has been reported to be implicated in a variety of diseases [22]. miR-125a and miR-125b are downregulated in various tumors as tumor suppressors [23]. miR-125a is suppressed in systemic lupus erythematosus [24], and miR-125b has a proinflammation role and can be mediated by metal sulfates to affect the outcome of ischemic stroke [25]. A recent report showed that the miR-125 family acts as an import regulator of the expression and maintenance of maternal effect genes during preimplantational embryo development [26]. However, less is known about the role of miR-125 on the development of HypoT. In our microarray results, we found that miR-125b-5p was significantly suppressed in HypoT rats, suggesting that miR-125b-5p contributes to the development of HypoT. Some evidence revealed that miR-125b-5p plays an important role in the repression of brite adipocyte function by modulating oxygen consumption and mitochondrial gene expression [27], and suppresses Brucella abortus intracellular survival via control of A20 expression [28]. In our study, a dual-luciferase reporter assay showed that miR-125b-5p monitored the expression of Stat3 by binding to the first predicted site on 3′ UTR of Stat3. This result revealed a newly discovered target of miR-125b-5p. Stat3 plays crucial roles in various intracellular signaling cascades involved in determining NSCs fate. Previous studies showed that Stat3 is responsible for the proliferation and maintenance of NSCs [29]. The inhibition of STAT3 activity promotes the neuronal differentiation of NSCs [30]. THs decreased Stat3 phosphorylation and Stat3-DNA binding activity. The overexpression of Stat3 attenuates the promotive effects of THs on neuronal differentiation of NSCs [19]. Thus, Stat3 has a role in determining THs-induced NSC fates.
In our study, no significant difference in the mRNA level of Stat3 between EuT and HypoT rats was observed. However, the protein level of tStat3 in HypoT rats was higher than that in EuT rats. These data reveal that miR-125b-5p negatively modulated the expression of Stat3 at the translational level in vivo. However, miR-125b-5p mimic modulated the content of p-Stat3 but did not affect the amount of tStat3 protein in C17.2 cell in the lack of THs. In vivo, the process by which miR-125b-5p regulates the protein level of Stat3 does not act alone and might be affected by cellular context or other proteins. However, in vitro, C17.2 cells provided a single context for miR-125b-5p and Stat3. Thus, the differences between in vivo and in vitro results might be attributed to the complex organism. However, the further mechanism needs intensive study. In our study, we found that miR-125b-5p suppressed HypoT development and negatively regulated the expression of Stat3. Combined with the results of previous studies, we conclude that miR-125b-5p targets the expression of Stat3 to repress HypoT development.
Conclusions
miR-125b-5p was suppressed in the brain tissues of HypoT rats, suggesting that miR-125b-5p is related to the occurrence of HypoT. Stat3 is a newly discovered target of miR-125b-5p. These findings revealed a new target of miR-125b-5p and the function of miR-125b-5p in HypoT development, which provides a new target for HypoT therapy.
Abbreviations
- HypoT
hypothyroidism
- NSCs
neural stem cells
- THs
thyroid hormones
- EuT
euthyroid
- Stat3
signal transducer and activator of transcription 3
- TUBB3
tubulin beta chain 3
- GFAP
glial fibrillary acid protein
Footnotes
Conflict of interest
None.
Source of support: Departmental sources
References
- 1.Patel J, Landers K, Li H, et al. Thyroid hormones and fetal neurological development. J Endocrinol. 2011;209:1–8. doi: 10.1530/JOE-10-0444. [DOI] [PubMed] [Google Scholar]
- 2.Rodrigues AL, de Moura EG, Passos MC, et al. Postnatal early overnutrition changes the leptin signalling pathway in the hypothalamic-pituitary-thyroid axis of young and adult rats. J Physiol. 2009;587:2647–61. doi: 10.1113/jphysiol.2009.169045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Decherf S, Seugnet I, Kouidhi S, et al. Thyroid hormone exerts negative feedback on hypothalamic type 4 melanocortin receptor expression. Proc Natl Acad Sci USA. 2010;107:4471–76. doi: 10.1073/pnas.0905190107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horn S, Heuer H. Thyroid hormone action during brain development: More questions than answers. Mol Cell Endocrinol. 2010;315:19–26. doi: 10.1016/j.mce.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 5.Remaud S, Gothie JD, Morvan-Dubois G, Demeneix BA. Thyroid hormone signaling and adult neurogenesis in mammals. Front Endocrinol (Lausanne) 2014;5:62. doi: 10.3389/fendo.2014.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Sioud M, Cekaite L. Profiling of miRNA expression and prediction of target genes. Methods Mol Biol. 2010;629:257–71. doi: 10.1007/978-1-60761-657-3_16. [DOI] [PubMed] [Google Scholar]
- 9.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 10.Shao Q, Jiang W, Jin Y. MiR-124 effect in neurons apoptosis in newborn rat with thyroid hypofunction. Int J Clin Exp Pathol. 2015;8:14465–71. [PMC free article] [PubMed] [Google Scholar]
- 11.Boguslawska J, Wojcicka A, Piekielko-Witkowska A, et al. MiR-224 targets the 3′UTR of type 1 5′-iodothyronine deiodinase possibly contributing to tissue hypothyroidism in renal cancer. PLoS One. 2011;6:e24541. doi: 10.1371/journal.pone.0024541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He H, Xu F, Huang W, et al. miR-125a-5p expression is associated with the age of breast cancer patients. Genet Mol Res. 2015;14:17927–33. doi: 10.4238/2015.December.22.17. [DOI] [PubMed] [Google Scholar]
- 13.Jia CW, Sun Y, Zhang TT, et al. Effects of miR-125a-5p on Cell proliferation, apoptosis and cell cycle of pancreatic cancer cells. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2016;38:415–21. doi: 10.3881/j.issn.1000-503X.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 14.Wohl SG, Reh TA. The microRNA expression profile of mouse Muller glia in vivo and in vitro. Sci Rep. 2016;6:35423. doi: 10.1038/srep35423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boissart C, Nissan X, Giraud-Triboult K, et al. miR-125 potentiates early neural specification of human embryonic stem cells. Development. 2012;139:1247–57. doi: 10.1242/dev.073627. [DOI] [PubMed] [Google Scholar]
- 16.Xu Z, Cheng L, Shi J, et al. Kinetic study of the removal of dimethyl phthalate from an aqueous solution using an anion exchange resin. Environ Sci Pollut Res Int. 2014;21:6571–77. doi: 10.1007/s11356-014-2556-x. [DOI] [PubMed] [Google Scholar]
- 17.Moog NK, Entringer S, Heim C, et al. Influence of maternal thyroid hormones during gestation on fetal brain development. Neuroscience. 2017;342:68–100. doi: 10.1016/j.neuroscience.2015.09.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen C, Zhou Z, Zhong M, et al. Excess thyroid hormone inhibits embryonic neural stem/progenitor cells proliferation and maintenance through STAT3 signalling pathway. Neurotox Res. 2011;20:15–25. doi: 10.1007/s12640-010-9214-y. [DOI] [PubMed] [Google Scholar]
- 19.Chen C, Zhou Z, Zhong M, et al. Thyroid hormone promotes neuronal differentiation of embryonic neural stem cells by inhibiting STAT3 signaling through TRalpha1. Stem Cells Dev. 2012;21:2667–81. doi: 10.1089/scd.2012.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong H, Curran I, Williams A, et al. Hepatic miRNA profiles and thyroid hormone homeostasis in rats exposed to dietary potassium perfluorooctanesulfonate (PFOS) Environ Toxicol Pharmacol. 2016;41:201–10. doi: 10.1016/j.etap.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 21.Sun YM, Lin KY, Chen YQ. Diverse functions of miR-125 family in different cell contexts. J Hematol Oncol. 2013;6:6. doi: 10.1186/1756-8722-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaham L, Binder V, Gefen N, et al. MiR-125 in normal and malignant hematopoiesis. Leukemia. 2012;26:2011–18. doi: 10.1038/leu.2012.90. [DOI] [PubMed] [Google Scholar]
- 23.Mar-Aguilar F, Luna-Aguirre CM, Moreno-Rocha JC, et al. Differential expression of miR-21, miR-125b and miR-191 in breast cancer tissue. Asia Pac J Clin Oncol. 2013;9:53–59. doi: 10.1111/j.1743-7563.2012.01548.x. [DOI] [PubMed] [Google Scholar]
- 24.Zhao X, Tang Y, Qu B, et al. MicroRNA-125a contributes to elevated inflammatory chemokine RANTES levels via targeting KLF13 in systemic lupus erythematosus. Arthritis Rheum. 2010;62:3425–35. doi: 10.1002/art.27632. [DOI] [PubMed] [Google Scholar]
- 25.Rink C, Khanna S. MicroRNA in ischemic stroke etiology and pathology. Physiol Genomics. 2011;43:521–28. doi: 10.1152/physiolgenomics.00158.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim KH, Seo YM, Kim EY, et al. The miR-125 family is an important regulator of the expression and maintenance of maternal effect genes during preimplantational embryo development. Open Biol. 2016;6(11) doi: 10.1098/rsob.160181. pii: 160181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giroud M, Pisani DF, Karbiener M, et al. miR-125b affects mitochondrial biogenesis and impairs brite adipocyte formation and function. Mol Metab. 2016;5:615–25. doi: 10.1016/j.molmet.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu N, Wang L, Sun C, et al. MicroRNA-125b-5p suppresses Brucella abortus intracellular survival via control of A20 expression. BMC Microbiol. 2016;16:171. doi: 10.1186/s12866-016-0788-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foshay KM, Gallicano GI. Regulation of Sox2 by STAT3 initiates commitment to the neural precursor cell fate. Stem Cells Dev. 2008;17:269–78. doi: 10.1089/scd.2007.0098. [DOI] [PubMed] [Google Scholar]
- 30.Cao F, Hata R, Zhu P, et al. Conditional deletion of Stat3 promotes neurogenesis and inhibits astrogliogenesis in neural stem cells. Biochem Biophys Res Commun. 2010;394:843–47. doi: 10.1016/j.bbrc.2010.03.092. [DOI] [PubMed] [Google Scholar]