Abstract
Background
miR-490-3p could play vital roles in multiple cancers. However, the role of miR-490-3p in hepatocellular carcinoma (HCC) remains uncertain. In this study, we sought to explore the underlying role of miR-490-3p in HCC.
Material/Methods
In this study, we explored the clinical role of miR-490-3p in HCC via quantitative reverse transcription–polymerase chain reaction (RT-qPCR) and The Cancer Genome Atlas (TCGA) database. Then, a meta-analysis was performed to evaluate the expression trend and diagnostic value of miR-490-3p in HCC. Furthermore, 12 miRNA prediction algorithms were applied to predict the potential target genes of miR-490-3p. The differentially expressed genes in HCC in the Gene Expression Profiling Interactive Analysis (GEPIA) database were also selected. Additionally, bioinformatics analyses were utilized to investigate the possible functions and pathways of the target genes.
Results
miR-490-3p was clearly down-regulated in HCC based on RT-qPCR (P=0.002). Consistent with the results of RT-qPCR, miR-490 was more highly expressed in normal liver tissue than in HCC (P<0.001). Additionally, the meta-analysis confirmed the results from RT-qPCR and TCGA. Furthermore, based on the prediction algorithms and GEPIA, a total of 113 genes were selected. According to the bioinformatics analyses, we found that the most remarkably enriched functional terms included protein transport, poly(A) RNA binding, and intracellular organelle part. Additionally, the miR-490-3p target genes were significantly related to the pathways in cancer.
Conclusions
We found that miR-490-3p is down-regulated in HCC and is related to genes that have potential tumoral functions. However, the exact mechanism should be confirmed by functional experiments.
MeSH Keywords: Carcinoma, Hepatocellular; Meta-Analysis; MicroRNAs; Real-Time Polymerase Chain Reaction
Background
Hepatocellular carcinoma (HCC) is a primary liver neoplasm. Although some risk factors (e.g., hepatitis B virus infection and overconsumption of alcohol) for HCC have been recognized, the exact molecular mechanism of HCC development remain unclear [1,2]. Elucidation of the molecular pathways related to HCC will help to develop new therapeutic targets [3,4].
miRNAs are a class of short, endogenous, evolutionarily conserved, and single-stranded non-coding RNA molecules that can regulate gene expression through the inhibition of translation and the degradation of mRNA (5-8). MiRNAs have been reported to play vital roles in various significant biological processes, including proliferation, the cell cycle, migration, and differentiation [9–11]. Additionally, miRNAs can act as tumor suppressors or oncogenes [12–14]. Emerging evidence demonstrates that many tumor-specific miRNAs are widely down-regulated or up-regulated in HCC and are closely related to the tumorigenesis and development of HCC [15–17].
miR-490-3p has been demonstrated to be associated with the metastasis, proliferation, and apoptosis of different cancers [18,19]. For example, miR-490-3p can inhibit the metastasis of colorectal cancer via targeting TGFβR1 [18]. In osteosarcoma, miR-490-3p can affect cell proliferation and apoptosis by targeting HMGA2 [19]. To date, the expression and role of miR-490-3p has rarely been investigated in HCC. Wojcicka et al. [20] reported a down-regulated expression of miR-490-3p, whereas an up-regulated expression of miR-490-3p in HCC tissues was found by Zhang et al. [21]. These published studies show that the miR-490-3p expression level is controversial in HCC.
In the present study, we sought to assess the miR-490-3p expression in HCC and normal liver tissue. Furthermore, we combined quantitative reverse transcription-polymerase chain reactions (RT-qPCR), The Cancer Genome Atlas (TCGA), and a meta-analysis to evaluate the clinical role and the underlying molecular mechanism of miR-490-3p in HCC. Additionally, bioinformatics analyses were applied to explore the possible functions, pathways, and networks of the potential genes [22].
Material and Methods
Quantitative real-time PCR
A total of 82 samples (41 HCC and 41 normal liver) were collected from the Department of Pathology, First Affiliated Hospital of Guangxi Medical University (Nanning, Guangxi, China). All experimental procedures were approved by the Ethics Committee of the First Affiliated Hospital of Guangxi Medical University, and both the clinicians and patients signed the consent forms for the use of tissues in the study. Additionally, all the experimental methods were performed according to the manufacturer’s instructions. In this study, TRIzol reagent (Takara, Nanning, China) was used to extract the total RNA, and then the extracted RNA was reverse-transcribed to cDNA using the Takara PrimeScript RT Reagent Kit (Takara, Nanning, China). Then, RT-qPCR was performed with the LightCycler 480 Real-time PCR System (Roche, Shanghai, China). The specific primers were as follows: miR-490-3p: CAACCUGGAGGACUCCAUGCUG; miR-191: CAACGGAAUCCCAAAAGCAGCU; miR-103: AGCAGCA UUGUACAGGGCUAUGA. The data were normalized to miR-191 and miR-103 expression levels and calculated using the 2−ΔΔCT method [23,24].
MiR-490-3p and HCC: A meta-analysis
The HCC-related miR-490-3p RNA-seq and microarray datasets were downloaded from TCGA (http://cancergenome.nih.gov/) and the Gene Expression Omnibus (GEO: http://www.ncbi.nlm.nih.gov/geo/). In addition, publications related to the expression of miR-490-3p in HCC were also chosen from PubMed, EMBASE, Web of Science, Google Scholar, Ovid, Science Direct, Wiley Online Library, LILACS, Cochrane Central Register of Controlled Trials, Wan Fang, Chong Qing VIP, Chinese CNKI, and the China Biology Medicine disc. The retrieval date was up to November 25, 2017 with the following keywords: (miR OR miRNA OR microRNA OR “miR” OR “miRNA” OR “microRNA”) AND (malignan* OR cancer OR tumor OR tumour OR neoplas* OR carcinoma) AND (hepatocellular OR liver OR hepatic OR HCC). The literature retrieval was cross-checked by 2 independent investigators (Meng-lan Huang and Yu Zhang). The numbers of false-positives (fp), false-negatives (fn), true-positives (tp), and true-negatives (tn) were extracted.
Validation the miR-490-3p expression in HCC
TCGA is a collection of data, including miRNA-seq, RNA-seq, exome-seq, SNP array, and DNA methylation [25,26]. In this study, the HCC RNA-seq (level 3, normalized read counts) of tumors and normal tissues was downloaded from TCGA [27]. The expression data of miR-490-3p were exhibited as reads per million (RPM), and the expression level of miR-490-3p was normalized by the Deseq package of R language. We used the t test (SPSS Inc., Chicago, IL, USA) for the statistical analysis of the differential expression of miR-490-3p between HCC and normal liver tissues. Additionally, the relationship between miR-490-3p and the clinicopathological parameters in HCC was surveyed according to the original data in the TCGA database. Then, a receiver operating characteristic (ROC) curve was generated to analyze the clinical diagnostic value of miR-490-3p in HCC. Furthermore, Kaplan-Meier method was used to assess the relationship between miR-490-3p expression and overall survival.
The potential functions and pathways associated with miR-490-3p
To further investigate the target genes of miR-490-3p, 12 online target prediction algorithms were applied to predict the underlying target genes of miR-490-3p. The 12 corresponding algorithms were miRMap (http://mirmap.ezlab.org/), RNA22 (https://cm.jefferson.edu/), Pictar2 (https://www.mdc-berlin.de/), miRWalk (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/), DIANA microT v4 (http://diana.imis.athena-innovation.gr/), RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/), mirBridge (http://mirsystem.cgm.ntu.edu.tw/), PITA (https://genie.weizmann.ac.il/), miRNAMap (http://mirnamap.mbc.nctu.edu.tw/), miRDB (http://www.mirdb.org/), miRanda (http://www.microrna.org), and Targetscan (http://www.targetscan.org/). In addition, a Venn diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/) was utilized to identify the candidate genes. Genes predicted by more than 5 target prediction algorithms were chosen for further analysis.
The differentially expressed genes of HCC in the Gene Expression Profiling Interactive Analysis (GEPIA) database were selected. Venn diagrams were created to identify the overlapping target genes from the prediction algorithms and the GEPIA database.
In addition, bioinformatics analyses (Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and network analysis) were utilized to investigate the possible functions, pathways, and networks of the potential genes as described [28,29]. In this process, the Database for Annotation, Visualization, and Integrated Discovery (DAVID: available online: http://david.abcc.ncifcrf.gov/) was applied, and a functional network was established via Cytoscape (version 3.0, http://cytoscape.org) [30].
Statistical analysis
All the expression data in TCGA were log2-transformed. The differences in miR-490-3p expression between HCC and non-cancerous liver were estimated by use of the t test, which was also utilized to assess the relationships between miR-490-3p expression and clinicopathological parameters. A Kruskal-Wallis H test or Mann-Whitney U test was performed for non-normally distributed variables. A P-value <0.05 (two-sided) was recognized to be statistically significant using SPSS 22.0.
In the meta-analysis, all statistical analyses were performed in STATA 14.0 (STATA Corp., College Station, Texas). The heterogeneity of the included studies was measured by Cochrane’s Q test and the I2 statistic, and I2 >50% represented obvious heterogeneity. Publication bias was assessed via Deek’s funnel plot asymmetry test; a P-value<0.05 indicated significant publication bias. To determine the diagnostic value of miR-490-3p in HCC, we applied summary receiver operating characteristic (SROC) curves to calculate the area under the curve (AUC) with 95% CIs, and the corresponding sensitivity and specificity were also identified using Meta-DiSc software [31]. We also used STATA 14.0 to conduct a continuous variable meta-analysis.
Results
The clinical value of miR-490-3p expression in HCC
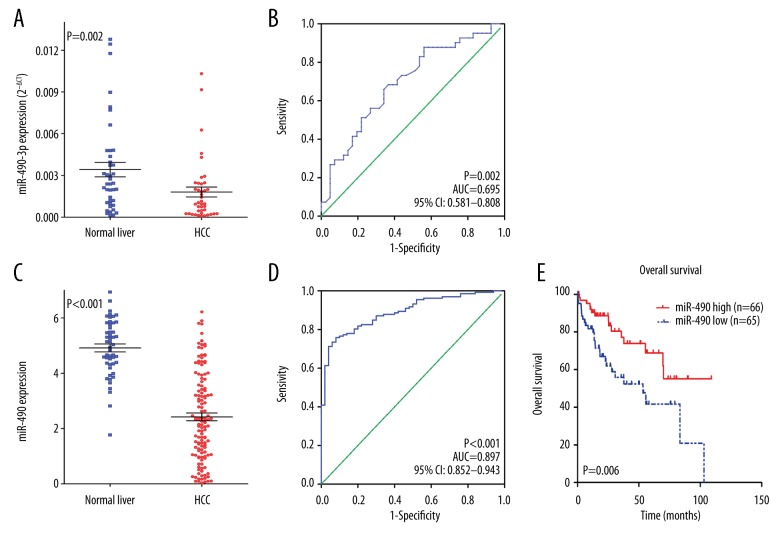
We found that miR-490-3p was clearly down-regulated in HCC, based on RT-qPCR (P=0.002, Figure 1A). Additionally, the area under the curve (AUC) of miR-490-3p was 0.695 (95% CI: 0.581–0.808, P=0.002, Figure 1B), indicating a moderate diagnostic value of the miR-490-3p level in HCC. Consistent with the results of RT-qPCR, miR-490 was more highly expressed in normal liver tissue than in HCC tissue (P<0.001, Figure 1C), and the AUC of miR-490-3p was 0.891 (95% CI: 0.844–0.938, P<0.001, Figure 1D) based on TCGA. Additionally, we observed that high miR-490 expression was correlated with better survival compared to the survival of the low miR-490 expression group (P=0.006, Figure 1E) for patients with HCC. We also explored the relationship between different levels of miR-490-3p expression and clinicopathological parameters, but no statistical significance was found, partly because of the small sample size (Tables 1, 2).
Figure 1.
Clinical significance of miR-490-3p in HCC. (A) Differential expression of miR-490-3p between HCC and non-cancerous liver tissue, based on RT-qPCR; (B) ROC curve of miR-490-3p in HCC, based on RT-qPCR; (C) Differential expression of miR-490 between HCC and non-cancerous liver tissue, based on TCGA; (D) ROC curve of miR-490 in HCC, based on TCGA. (E) Kaplan-Meier curves of miR-490 expression in HCC.
Table 1.
Differential expression of miR-490-3p associated with other clinicopathological parameters in HCC tissue based on RT-qPCR.
| Clinicopathological features | miR-490-3p expression(2−ΔCq) | |||
|---|---|---|---|---|
| High (n) | Low (n) | P-value | ||
| Tissue | HCC | 14 | 27 | 0.004 |
| Normal liver | 27 | 14 | ||
| Gender | Male | 14 | 16 | 0.335 |
| Female | 7 | 4 | ||
| Age (years) | <50 | 8 | 10 | 0.443 |
| ≥50 | 13 | 10 | ||
| Tumor diameter (cm) | <5 | 6 | 7 | 0.658 |
| ≥5 | 15 | 13 | ||
| Vascular infiltration | Yes | 5 | 7 | 0.431 |
| No | 16 | 13 | ||
| Metastasis | Yes | 8 | 12 | 0.161 |
| No | 13 | 8 | ||
| TNM | I+II | 13 | 8 | 0.161 |
| III+IV | 8 | 12 | ||
Table 2.
Differential expression of miR-490 associated with other clinicopathological parameters in HCC tissue based on the TCGA database.
| Clinicopathological features | N | miR-490 expression (mean ±SD) | P-value |
|---|---|---|---|
| Tissue | |||
| HCC | 131 | 2.39±1.57 | <0.001 |
| Normal liver | 50 | 4.91±1.02 | |
| Gender | |||
| Male | 93 | 2.34±1.48 | 0.559 |
| Female | 38 | 2.52±1.78 | |
| Age (years) | |||
| <60 | 73 | 2.39±1.60 | 0.983 |
| ≥60 | 58 | 2.40±1.54 | |
| Grade | |||
| I+II | 84 | 2.57±1.62 | 0.083 |
| III+IV | 46 | 2.07±1.45 | |
| Stage | |||
| I+II | 90 | 2.38±1.54 | 0.795 |
| III+IV | 29 | 2.29±1.66 | |
| Vascular invasion | |||
| Yes | 32 | 2.48±1.74 | 0.853 |
| No | 80 | 2.42±1.48 | |
MiR-490-3p and HCC: A meta-analysis
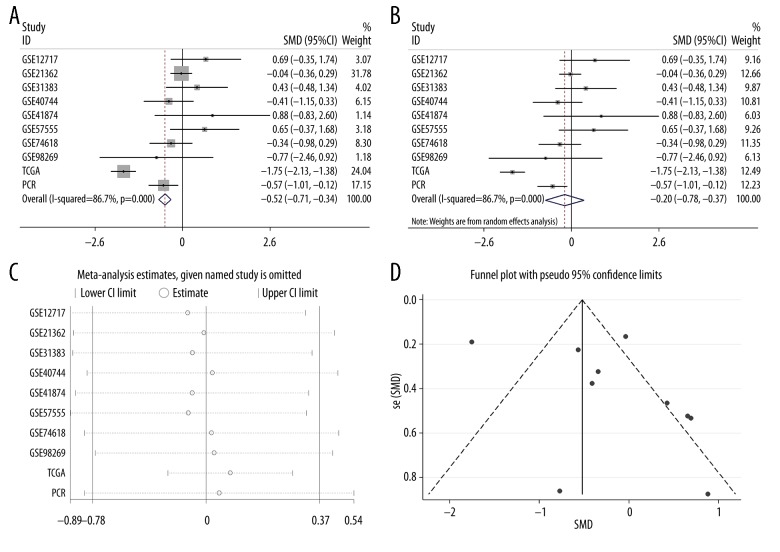
The meta-analysis included 749 cases from 3 sources: 8 datasets in GEO (GSE98269, GSE74618, GSE57555, GSE41874, GSE40744, GSE31383, GSE21362 and GSE12717); the original data in TCGA; and the expression data from the RT-qPCR. For the miR-490-3p expression in HCC compared to the normal group, a fixed-effects model was used to calculate the standard mean deviation (SMD) and 95% CI. The combined SMD reached −0.52 (−0.71, −0.34), indicating that miR-490-3p expression was down-regulated in HCC (P<0.001, Figure 2A). Considering the high heterogeneity (I2=86.7%, P<0.05) of SMD, a random-effects model was then applied, and the combined SMD reached −0.20 (−0.78, 0.37) with heterogeneity over 50% (Figure 2B). Then, we performed sensitivity analysis to clarify whether the high heterogeneity was from one particular study, and the results showed the pooled SMD was stable (Figure 2C). Furthermore, no significant publication bias could be found (P>0.05, Figure 2D).
Figure 2.
The expression condition of miR-490-3p in HCC compared to normal liver. (A) Forest plot of datasets evaluating miR-490-3p expression between HCC and normal control groups (fixed-effects model); (B) Forest plot of datasets evaluating miR-490-3p expression between HCC and normal control groups (random-effects model); (C) Sensitivity analysis aimed to exclude the main studies at the time; (D) Funnel plot of datasets showed no publication bias in our investigation.
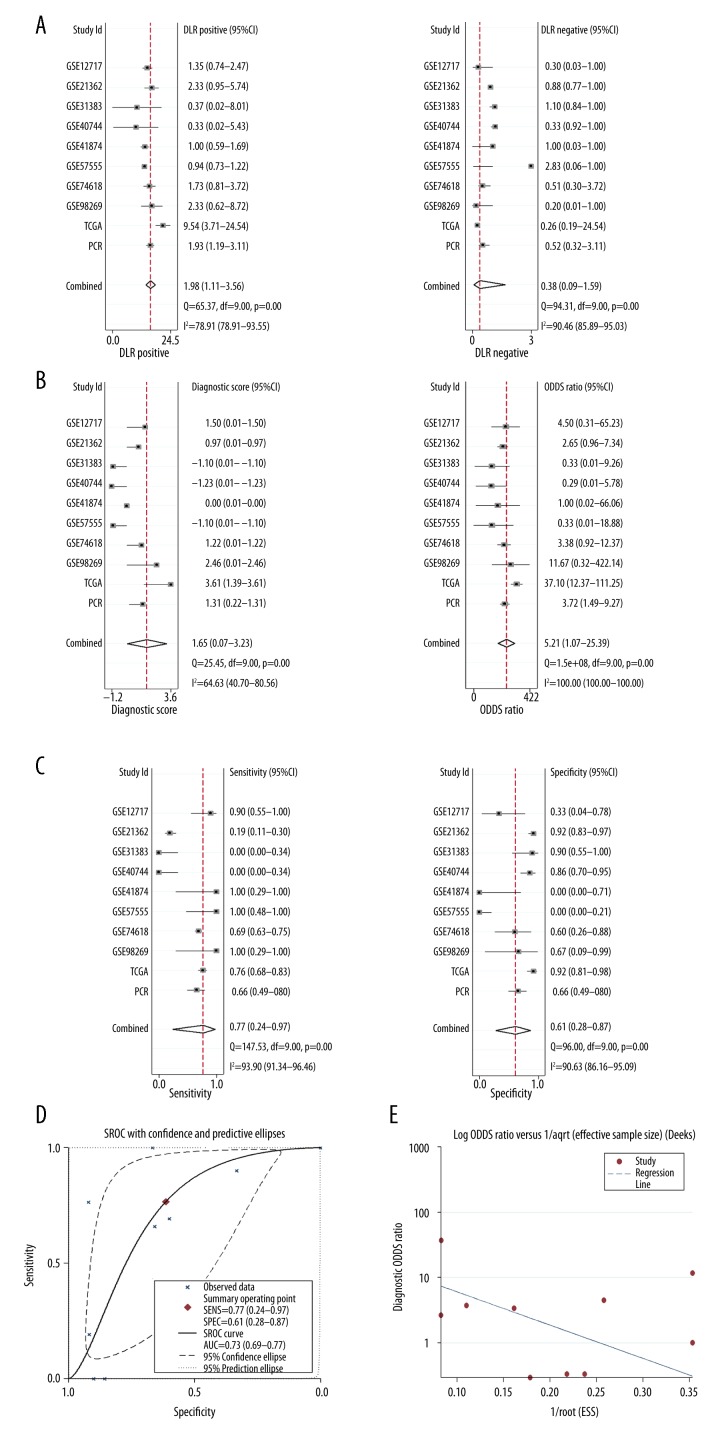
In diagnostic meta-analysis, The DLR-negative was 0.38 (0.09–1.59), and the DLR-positive was 1.98 (1.11–3.56, Figure 3A). A PLR value of 1.98 suggested that patients with HCC had an approximately 1.98-fold higher chance of being miR-490-3p assay-positive. The diagnostic score was 1.65 (0.07–3.23) and the odds ratio was 5.21 (1.07–25.39, Figure 3B). Moreover, we also investigated the diagnostic value of miR-490-3p in HCC via the diagnostic meta-analysis. The sensitivity and specificity were 0.77 (95% CI: 0.24–0.97) and 0.61 (95% CI: 0.28–0.87), respectively (Figure 3C). The AUC of SROC was 0.73 (0.69–0.77, Figure 3D), which shows the accuracy of miR-490-3p for the detection of HCC. Additionally, our results confirmed the moderate diagnostic accuracy of miR-490-3p, as already shown by TCGA and RT-qPCR. No obvious publication bias was found (P>0.05, Figure 3E).
Figure 3.
The diagnostic meta-analysis of miR-490-3p in HCC. (A) The pooled negative DLR and positive DLR of the included studies; (B) The pooled diagnostic score and diagnostic odds ratio of the included studies; (C) The pooled sensitivity and specificity of the included studies; (D) The SROC curve for the assessment of the diagnostic accuracy of miR-490-3p for HCC; (E) Publication bias. 1/root(ESS) means the inverse root of the effective sample sizes.
The potential pathways associated with miR-490-3p
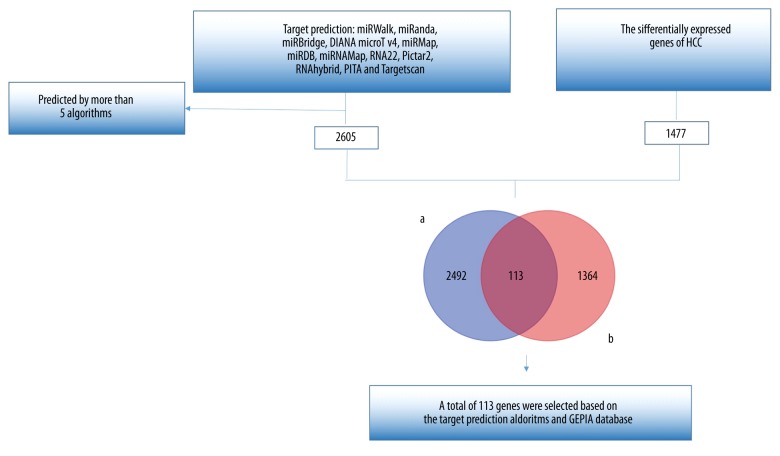
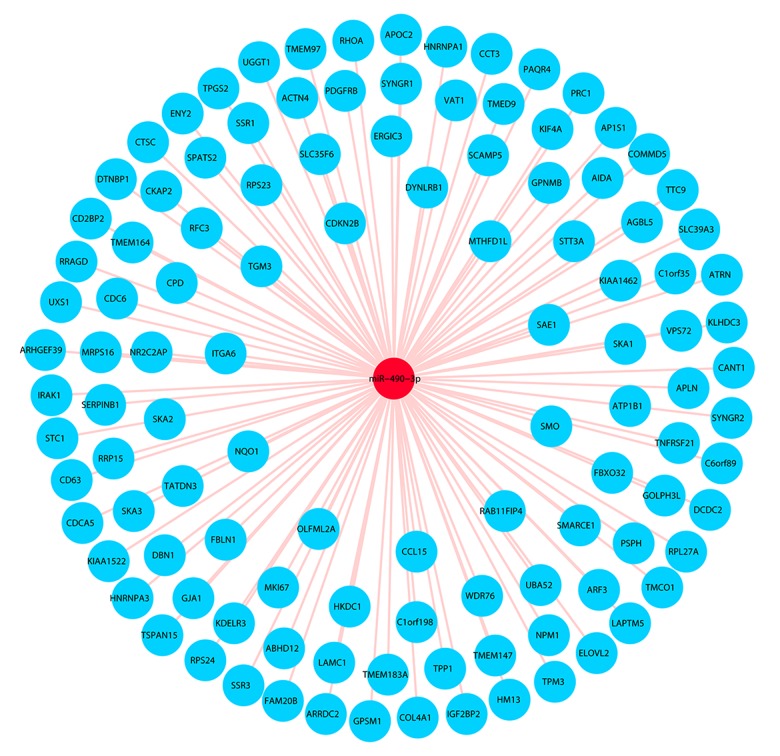
Based on the target prediction algorithms and the GEPIA database, 113 overlapping genes were selected (Figure 4). Bioinformatics analyses were performed on these 113 genes. According to GO and KEGG analyses, we found that the most remarkably enriched functional terms were protein transport, poly(A) RNA binding, and intracellular organelle part (Table 3). Additionally, the miR-490-3p target genes were significantly related to pathways in cancer (Table 4). Finally, a gene network of the overlapped genes was constructed (Figure 5), and the relationships between miR-490-3p and the target genes could be easily observed from the network.
Figure 4.
The procedure to achieve 113 genes. a: The target genes of miR-490-3p predicted by 12 online target prediction algorithms; b: The differentially expressed genes of HCC in the GEPIA database.
Table 3.
Top 10 enriched GO terms (BP, CC, and MF) of the target genes of miR-490-3p.
| Category | Term | Ontology | Count | P-value | Genes |
|---|---|---|---|---|---|
| GO: 0016043 | Cellular component organization | BP | 60 | 1.20E-05 | ENY2, ATP1B1, HM13, PRC1, FAM20B, GJA1, SAE1, CCT3, UXS1, SSR1, etc. |
| GO: 0071840 | Cellular component organization or biogenesis | BP | 61 | 1.22E-05 | ENY2, ATP1B1, HM13, PRC1, FAM20B, GJA1, SAE1, CCT3, UXS1, SSR1,,etc. |
| GO: 0033036 | Macromolecule localization | BP | 35 | 4.13E-05 | ENY2, TNFRSF21, ATP1B1, HM13, FAM20B, RPL27A, GJA1, APOC2, SAE1, etc. |
| GO: 0034613 | Cellular protein localization | BP | 24 | 8.38E-05 | KDELR3, ATP1B1, HM13, FAM20B, RPL27A, SAE1, TSPAN15, CCT3, CD63, SYNGR1,etc. |
| GO: 0070727 | Cellular macromolecule localization | BP | 24 | 9.48E-05 | KDELR3, ATP1B1, HM13, FAM20B, RPL27A, SAE1, TSPAN15, CCT3, CD63, SYNGR1, etc. |
| GO: 0008104 | Protein localization | BP | 31 | 0.000107 | ENY2, TNFRSF21, ATP1B1, HM13, FAM20B, RPL27A, GJA1, SAE1, CCT3, SYNGR1, etc. |
| GO: 0015031 | Protein transport | BP | 26 | 0.000114 | ENY2, TNFRSF21, ATP1B1, HM13, FAM20B, RPL27A, GJA1, SAE1, SYNGR1, SSR1,etc. |
| GO: 0045184 | Establishment of protein localization | BP | 27 | 0.000165 | ENY2, TNFRSF21, ATP1B1, HM13, FAM20B, RPL27A, GJA1, SAE1, SYNGR1, SSR1, etc |
| GO: 0071702 | Organic substance transport | BP | 32 | 0.000197 | ENY2, TNFRSF21, ATP1B1, HM13, FAM20B, RPL27A, APOC2, GJA1, SAE1, IGF2BP2, etc. |
| GO: 0033043 | Regulation of organelle organization | BP | 19 | 0.000207 | CKAP2, CDC6, MKI67, FAM20B, DCDC2, SAE1, CCT3, TMED9, HNRNPA1, C6ORF89, etc |
| Category | Term | Ontology | Count | P-value | Genes |
| GO: 0044446 | Intracellular organelle part | CC | 78 | 1.14775E-07 | ENY2, HM13, PRC1, FAM20B, NR2C2AP, GJA1, SAE1, CCT3, SYNGR1, UXS1,etc. |
| GO: 0044422 | Organelle part | CC | 78 | 3.46345E-07 | ENY2, HM13, PRC1, FAM20B, NR2C2AP, GJA1, SAE1, CCT3, SYNGR1, UXS1,etc. |
| GO: 0098588 | Bounding membrane of organelle | CC | 29 | 1.05378E-06 | HM13, FAM20B, GJA1, SYNGR1, RRAGD, DTNBP1, SYNGR2, UXS1, CANT1, AP1S1, etc. |
| GO: 0005737 | Cytoplasm | CC | 89 | 2.56469E-06 | HM13, PRC1, FAM20B, GJA1, SAE1, CCT3, PSPH, SYNGR1, UXS1, SYNGR2, etc. |
| GO: 0044444 | Cytoplasmic part | CC | 73 | 6.54455E-06 | HM13, PRC1, FAM20B, GJA1, SAE1, CCT3, PSPH, SYNGR1, UXS1, SYNGR2, etc. |
| GO: 0005819 | Spindle | CC | 10 | 9.21604E-05 | CKAP2, RAB11FIP4, CDC6, KIF4A, PRC1, AGBL5, NPM1, SKA3, SKA2, SKA1 |
| GO: 0012505 | Endomembrane system | CC | 41 | 0.000196911 | ENY2, HM13, FAM20B, TTC9, GJA1, APOC2, RRAGD, SYNGR1, SYNGR2, DTNBP1, etc. |
| GO: 0031988 | Membrane-bounded vesicle | CC | 39 | 0.000204314 | ATP1B1, HM13, NR2C2AP, GJA1, APOC2, CCT3, SYNGR1, SYNGR2, DTNBP1, UXS1, etc. |
| GO: 0031982 | Vesicle | CC | 40 | 0.000212545 | ATP1B1, HM13, NR2C2AP, GJA1, APOC2, CCT3, SYNGR1, SYNGR2, DTNBP1, UXS1,etc. |
| GO: 0044432 | Endoplasmic reticulum part | CC | 19 | 0.000230662 | KDELR3, COL4A1, HM13, TTC9, GJA1, TMED9, DTNBP1, CANT1, ERGIC3, SSR1, etc. |
| GO: 0005515 | Protein binding | MF | 81 | 0.00117595 | ATP1B1, HM13, PRC1, NR2C2AP, GJA1, SAE1, CCT3, PSPH, SYNGR1, C1ORF35, etc. |
| GO: 0042803 | Protein homodimerization activity | MF | 11 | 0.012784205 | RAB11FIP4, IRAK1, HM13, ACTN4, NPM1, APOC2, OLFML2A, PSPH, UXS1, MTHFD1L, CANT1. |
| GO: 0005488 | Binding | MF | 95 | 0.025625954 | ENY2, ATP1B1, HM13, PRC1, FAM20B, NR2C2AP, GJA1, SAE1, CCT3, PSPH, etc. |
| GO: 0005178 | Integrin binding | MF | 4 | 0.026398151 | FBLN1, ITGA6, ACTN4, GPNMB |
| GO: 0015631 | Tubulin binding | MF | 6 | 0.032896505 | KIF4A, PRC1, AGBL5, GJA1, SKA2, SKA1 |
| GO: 0042802 | Identical protein binding | MF | 15 | 0.033685753 | RAB11FIP4, IRAK1, FBLN1, HM13, ACTN4, PRC1, NPM1, APOC2, OLFML2A, CTSC, etc. |
| GO: 0003735 | Structural constituent of ribosome | MF | 5 | 0.045656975 | MRPS16, RPL27A, UBA52, RPS23, RPS24 |
| GO: 0046983 | Protein dimerization activity | MF | 13 | 0.046892004 | RAB11FIP4, IRAK1, HM13, ACTN4, NPM1, APOC2, OLFML2A, SAE1, PSPH, RRAGD, etc. |
| GO: 0044822 | Poly(A) RNA binding | MF | 13 | 0.056683692 | ACTN4, MKI67, RPL27A, IGF2BP2, CCT3, C1ORF35, HNRNPA1, HNRNPA3, SPATS2, NPM1, etc. |
| GO: 0048407 | Platelet-derived growth factor binding | MF | 2 | 0.064511742 | COL4A1, PDGFRB |
Table 4.
KEGG pathway enrichment analysis of the target genes of miR-490-3p.
| ID | Term | Count | P-value | Genes |
|---|---|---|---|---|
| hsa04142 | Lysosome | 5 | 0.009614139 | AP1S1, LAPTM5, TPP1, CTSC, CD63 |
| hsa04510 | Focal adhesion | 6 | 0.013570527 | COL4A1, ITGA6, ACTN4, RHOA, PDGFRB, LAMC1 |
| hsa03010 | Ribosome | 5 | 0.014306396 | MRPS16, RPL27A, UBA52, RPS23, RPS24 |
| hsa05200 | Pathways in cancer | 8 | 0.017815583 | SMO, COL4A1, ITGA6, CDKN2B, RHOA, PDGFRB, LAMC1, TPM3 |
| hsa05222 | Small cell lung cancer | 4 | 0.020875427 | COL4A1, ITGA6, CDKN2B, LAMC1 |
| hsa05146 | Amoebiasis | 4 | 0.036871107 | COL4A1, ACTN4, SERPINB1, LAMC1 |
| hsa05412 | Arrhythmogenic right ventricular cardiomyopathy (ARVC) | 3 | 0.086808656 | ITGA6, ACTN4, GJA1 |
Figure 5.
Network analysis comparing miR-490-3p and the target genes.
Discussion
Many studies have demonstrated that miRNAs are involved in various biological processes of HCC, such as cell proliferation, invasion, migration, and the cell cycle [9,32,33]. Additionally, miRNAs can act as diagnostic and prognostic biomarkers in HCC [34,35]. In the present study, we explored the underlying role of miR-490-3p in HCC. Few previous studies have focused on the detailed role of miR-490-3p in HCC. Wojcicka et al. [20] explored the deregulated miRNAs in liver cirrhosis and HCC via next-generation sequencing, and confirmed the down-regulation of miR-490-3p (T/N=0.13) in HCC. However, Zhang et al. [21] found that miR-490-3p was highly expressed in HCC cells via RT-qPCR and that miR-490-3p functions as an oncogenic miRNA in HCC cells, which is inconsistent with our RT-qPCR and TCGA results. In the present study, miR-490-3p was clearly down-regulated in HCC, as shown by RT-qPCR and TCGA. Additionally, the combined SMD of the expression meta-analysis reached −0.52 (−0.70, −0.33), indicating down-regulated expression of miR-490-3p in HCC. Moreover, the present study, which included 749 cases from 3 sources (GEO, TCGA and RT-qPCR), is the first meta-analysis to survey the expression and diagnostic value of miR-490-3p in HCC. We used diagnostic meta-analysis to assess the validity of using miR-490-3p to detect HCC. The results of SROC showed the moderate diagnostic value of miR-490-3p for the detection of HCC. Additionally, the results of our meta-analysis confirmed the diagnostic accuracy of miR-490-3p, based on TCGA and RT-qPCR. However, high heterogeneity (high I2 values) was unavoidable in this meta-analysis, partly because of the opposite results in the study by Zhang et al. Furthermore, blinding in the 3 different sources was not certain, which also contributed to the high heterogeneity, and no individual publications could be included in this meta-analysis.
According to GO and KEGG analyses, we found the enriched functional terms were protein transport, poly(A) RNA binding, and intracellular organelle part. Additionally, the miR-490-3p target genes were significantly related to the pathways in cancer. As reported, miR-490-3p is involved in the tumorigenesis and development of various cancers, such as ovarian carcinoma and breast cancer, via interacting with mRNA [36,37].
Conclusions
We hypothesized that miR-490-3p plays a significant role in HCC via regulating various pathways, especially pathways in cancer. However, the suspected mechanism should be confirmed by functional experiments, such as proliferation, invasion, metastasis, cell cycle, and apoptosis assays, and in animal models. The molecular mechanism by which miR-490-3p affects the biological function of HCC needs to be assessed from the cell, tissue, and animal levels. Focusing on new insights into miR-490-3p, our study provides a new perspective on HCC.
Footnotes
Source of support: The study was financially supported by the National Natural Science Foundation of China (No. 81603575)
References
- 1.Zhang E, Liu Q, Wang Y, et al. MicroRNA miR-147b promotes tumor growth via targeting UBE2N in hepatocellular carcinoma. Oncotarget. 2017;8:114072–80. doi: 10.18632/oncotarget.23120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang W, Zhang M, Peng Y, He J. Ubiquitin associated protein 2-like (UBAP2L) overexpression in patients with hepatocellular carcinoma and its clinical significance. Med Sci Monit. 2017;23:4779–88. doi: 10.12659/MSM.907071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Honeyman JN, Simon EP, Robine N, et al. Detection of a recurrent DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular carcinoma. Science. 2014;343:1010–14. doi: 10.1126/science.1249484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei W, Du C, Lv J, et al. Blocking A2B adenosine receptor alleviates pathogenesis of experimental autoimmune encephalomyelitis via inhibition of IL-6 production and Th17 differentiation. J Immunol. 2013;190:138–46. doi: 10.4049/jimmunol.1103721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu Q, Liu X, Liu Z, et al. MicroRNA-1296 inhibits metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma by targeting SRPK1-mediated PI3K/AKT pathway. Mol Cancer. 2017;16:103. doi: 10.1186/s12943-017-0675-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tao J, Liu Z, Wang Y, et al. MiR-542-3p inhibits metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma by targeting UBE3C. Biomed Pharmacother. 2017;93:420–28. doi: 10.1016/j.biopha.2017.06.070. [DOI] [PubMed] [Google Scholar]
- 8.Lei H, Li H, Xie H, et al. Role of MiR-215 in Hirschsprung’s disease pathogenesis by targeting SIGLEC-8. Cell Physiol Biochem. 2016;40:1646–55. doi: 10.1159/000453214. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Li Y, Chen Y, et al. MicroRNA-214-3p inhibits proliferation and cell cycle progression by targeting MELK in hepatocellular carcinoma and correlates cancer prognosis. Cancer Cell Int. 2017;17:102. doi: 10.1186/s12935-017-0471-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li JM, Zhou J, Xu Z, et al. MicroRNA-27a-3p inhibits cell viability and migration through down-regulating DUSP16 in hepatocellular carcinoma. J Cell Biochem. 2017 doi: 10.1002/jcb.26526. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 11.Wang A, Landen NX, Meisgen F, et al. MicroRNA-31 is overexpressed in cutaneous squamous cell carcinoma and regulates cell motility and colony formation ability of tumor cells. PLoS One. 2014;9:e103206. doi: 10.1371/journal.pone.0103206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan K, Gao J, Yang T, et al. MicroRNA-34a inhibits the proliferation and metastasis of osteosarcoma cells both in vitro and in vivo. PLoS One. 2012;7:e33778. doi: 10.1371/journal.pone.0033778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen SQ, Huang LS, Xiao XL, et al. miR-204 regulates the biological behavior of breast cancer MCF-7 cells by directly targeting FOXA1. Oncol Rep. 2017;38:368–76. doi: 10.3892/or.2017.5644. [DOI] [PubMed] [Google Scholar]
- 14.Ren F, Ding H, Huang S, et al. Expression and clinicopathological significance of miR-193a-3p and its potential target astrocyte elevated gene-1 in non-small lung cancer tissues. Cancer Cell Int. 2015;15:80. doi: 10.1186/s12935-015-0227-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao X, Yang Z, Li G, et al. The role and clinical implications of microRNAs in hepatocellular carcinoma. Sci China Life Sci. 2012;55:906–19. doi: 10.1007/s11427-012-4384-x. [DOI] [PubMed] [Google Scholar]
- 16.Chu R, Mo G, Duan Z, et al. miRNAs affect the development of hepatocellular carcinoma via dysregulation of their biogenesis and expression. Cell Commun Signal. 2014;12:45. doi: 10.1186/s12964-014-0045-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, Tang W, Li R, et al. Downregulation of microRNA-132 indicates progression in hepatocellular carcinoma. Exp Therap Med. 2016;12:2095–101. doi: 10.3892/etm.2016.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu X, Chen R, Li Z, et al. MicroRNA-490-3p inhibits colorectal cancer metastasis by targeting TGFbetaR1. BMC Cancer. 2015;15:1023. doi: 10.1186/s12885-015-2032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu W, Xu G, Liu H, Li T. MicroRNA-490-3p regulates cell proliferation and apoptosis by targeting HMGA2 in osteosarcoma. FEBS Lett. 2015;589:3148–53. doi: 10.1016/j.febslet.2015.08.034. [DOI] [PubMed] [Google Scholar]
- 20.Wojcicka A, Swierniak M, Kornasiewicz O, et al. Next generation sequencing reveals microRNA isoforms in liver cirrhosis and hepatocellular carcinoma. Int J Biochem Cell Biol. 2014;53:208–17. doi: 10.1016/j.biocel.2014.05.020. [DOI] [PubMed] [Google Scholar]
- 21.Zhang LY, Liu M, Li X, Tang H. miR-490-3p modulates cell growth and epithelial to mesenchymal transition of hepatocellular carcinoma cells by targeting endoplasmic reticulum-Golgi intermediate compartment protein 3 (ERGIC3) J Biol Chem. 2013;288:4035–47. doi: 10.1074/jbc.M112.410506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu X, Wang X, Fu B, et al. Differentially expressed genes and microRNAs in bladder carcinoma cell line 5637 and T24 detected by RNA sequencing. Int J Clin Exp Pathol. 2015;8:12678–87. [PMC free article] [PubMed] [Google Scholar]
- 23.Dai J, Wu H, Zhang Y, et al. Negative feedback between TAp63 and Mir-133b mediates colorectal cancer suppression. Oncotarget. 2016;7:87147–60. doi: 10.18632/oncotarget.13515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu H, Zhou J, Zeng C, et al. Curcumin increases exosomal TCF21 thus suppressing exosome-induced lung cancer. Oncotarget. 2016;7:87081–90. doi: 10.18632/oncotarget.13499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bornstein S, Schmidt M, Choonoo G, et al. IL-10 and integrin signaling pathways are associated with head and neck cancer progression. BMC Genomics. 2016;17:38. doi: 10.1186/s12864-015-2359-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng JH, Xiong DD, Pang YY, et al. Identification of molecular targets for esophageal carcinoma diagnosis using miRNA-seq and RNA-seq data from The Cancer Genome Atlas: A study of 187 cases. Oncotarget. 2017;8(22):35681–99. doi: 10.18632/oncotarget.16051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun XJ, Xu GL. Overexpression of Acyl-CoA Ligase 4 (ACSL4) in patients with hepatocellular carcinoma and its prognosis. Med Sci Monit. 2017;23:4343–50. doi: 10.12659/MSM.906639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mirsafian H, Ripen AM, Leong WM, et al. Transcriptome profiling of monocytes from XLA patients revealed the innate immune function dysregulation due to the BTK gene expression deficiency. Sci Rep. 2017;7:6836. doi: 10.1038/s41598-017-06342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao Z, Bai J, Wu A, et al. Co-LncRNA: investigating the lncRNA combinatorial effects in GO annotations and KEGG pathways based on human RNA-Seq data. Database (Oxford) 2015;2015 doi: 10.1093/database/bav082. pii: bav082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiong DD, Lv J, Wei KL, et al. A nine-miRNA signature as a potential diagnostic marker for breast carcinoma: An integrated study of 1,110 cases. Oncol Rep. 2017;37:3297–304. doi: 10.3892/or.2017.5600. [DOI] [PubMed] [Google Scholar]
- 31.Zamora J, Abraira V, Muriel A, et al. Meta-DiSc: A software for meta-analysis of test accuracy data. BMC Med Res Methodol. 2006;6:31. doi: 10.1186/1471-2288-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Liu S, Cao L, et al. miR-29a-3p suppresses cell proliferation and migration by downregulating IGF1R in hepatocellular carcinoma. Oncotarget. 2017;8:86592–603. doi: 10.18632/oncotarget.21246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu D, Shen D, Zhang M, et al. MiR-488 suppresses cell proliferation and invasion by targeting ADAM9 and lncRNA HULC in hepatocellular carcinoma. Am J Cancer Res. 2017;7:2070–80. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Qu Z, Wu J, Wu J, et al. Exosomal miR-665 as a novel minimally invasive biomarker for hepatocellular carcinoma diagnosis and prognosis. Oncotarget. 2017;8:80666–78. doi: 10.18632/oncotarget.20881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu XM, Xi ZF, Liao P, et al. Diagnostic and prognostic potential of serum microRNA-4651 for patients with hepatocellular carcinoma related to aflatoxin B1. Oncotarget. 2017;8:81235–49. doi: 10.18632/oncotarget.16027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang LL, Sun KX, Wu DD, et al. DLEU1 contributes to ovarian carcinoma tumourigenesis and development by interacting with miR-490-3p and altering CDK1 expression. J Cell Mol Med. 2017;21:3055–65. doi: 10.1111/jcmm.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao L, Zheng XY. MicroRNA-490 inhibits tumorigenesis and progression in breast cancer. Onco Target Ther. 2016;9:4505–16. doi: 10.2147/OTT.S100037. [DOI] [PMC free article] [PubMed] [Google Scholar]