Summary
Bone, cartilage, and marrow adipocytes are generated by skeletal progenitors, but the relationships between lineages and mechanisms controlling their differentiation are poorly understood. We established mouse clonal skeletal progenitors with distinct differentiation properties and analyzed their transcriptome. Unipotent osteogenic and adipogenic cells expressed specific transcriptional programs, whereas bipotent clones combined expression of those genes and did not show a unique signature. We tested potential regulators of lineage commitment and found that in the presence of interferon-γ (IFNγ) adipogenic clones can be induced to osteogenesis and that their adipogenic capacity is inhibited. Analysis of IFNγ-regulated genes showed that lineage signatures and fate commitment of skeletal progenitors were controlled by EGR1 and EGR2. Knockdown experiments revealed that EGR1 is a positive regulator of the adipogenic transcriptional program and differentiation capacity, whereas EGR2 inhibits the osteogenic program and potency. Therefore, our work revealed transcriptional signatures of osteogenic and adipogenic lineages and mechanism triggering cell fate.
Keywords: bone marrow stromal cells, skeletal progenitors, lineage commitment, transcriptional signatures, interferon-γ signaling, Egr1/2 transcription factors
Graphical Abstract
Highlights
-
•
Bone marrow osteo- and adipogenic progenitors have specific transcriptional profiles
-
•
Bipotent progenitors combine expression of osteogenic and adipogenic programs
-
•
IFNγ inhibits adipogenesis and induces osteogenesis via downregulation of Egr1/Egr2
-
•
Egr1 maintains adipogenic and Egr2 suppresses osteogenic lineage commitment
In this article, Rostovskaya and colleagues established clonal skeletal progenitors with distinct differentiation properties from mouse bone marrow. RNA-seq analysis revealed transcriptional signatures of osteogenic and adipogenic lineages and IFNγ signaling as potential regulator of cell fate commitment. IFNγ is a negative regulator of Egr1 and Egr2 transcription factors. Egr1 maintains adipogenic and Egr2 suppresses osteogenic transcriptional program and commitment.
Introduction
Most somatic cells are replenished during life through differentiation of adult stem cells. It is suggested that differentiated cells of skeletal tissues, such as osteoblasts, marrow adipocytes, and chondrocytes, originate from putative skeletal stem cells, also referred to as mesenchymal stem cells, residing in bone marrow stroma (Bianco and Robey, 2000, Caplan, 1991, Owen and Friedenstein, 1988). Classical experiments on serial ectopic transplantations of bone marrow showed the existence of self-renewing skeletal stem cells, which can recapitulate formation of bone ossicle with hematopoiesis-supportive stroma (Friedenstein et al., 1968, Sacchetti et al., 2007, Tavassoli and Crosby, 1968). Nevertheless, which cell lineages can be generated from bone marrow skeletal stem cells and how their differentiation is controlled still remains unclear. Genetic fate mapping studies using Grem1-CreERT (Worthley et al., 2015), LepR-Cre (Zhou et al., 2014), and Mx1-Cre (Park et al., 2012) revealed distinct subsets of cells contributing to skeletal tissues, and notably each strategy resulted in labeling different cell types. This suggests that skeletal tissues may be generated by multiple subsets of stem and progenitor cells with distinct developmental potential, which may function in different locations and at particular stages of development (Kassem and Bianco, 2015).
Heterogeneity within the population of bone marrow skeletal progenitors may also be reflected by cells cultured in vitro. Multiple studies revealed that single-cell-derived strains of bone marrow stromal cells (BMSCs) are heterogeneous in their differentiation potential using in vitro tests (Banfi et al., 2000, Muraglia et al., 2000, Okamoto et al., 2002, Russell et al., 2010, Sarugaser et al., 2009, Zhou et al., 2014) and transplantation assays (Gronthos et al., 2003, Kuznetsov et al., 1997, Sacchetti et al., 2007, Sworder et al., 2015).
Several factors have been proposed to regulate lineage decisions of skeletal progenitors, among them canonical Wnt (wingless-type MMTV integration site family) (Boyden et al., 2002, Cui et al., 2011, Gong et al., 2001), VEGF (vascular endothelial growth factor) (Chan et al., 2015), RUNX2 (runt-related transcription factor 2), and PPARγ (peroxisome proliferator-activated receptor γ) (Komori et al., 1997, Tontonoz et al., 1994, Hong et al., 2005, Nishikawa et al., 2010). In fact, most of the pathways identified so far positively regulate differentiation of BMSCs into one lineage and inhibit the other, but this does not provide enough evidence that these factors actually determine cell fate decisions in a multipotent progenitor cell and not just play a role downstream during differentiation; hence, the key events in BMSC lineage commitment are still to be identified.
In this work, we specified different types of cultured skeletal progenitors within a BMSC population using in vitro and in vivo differentiation assays. Systematic expression profiling of clonally derived skeletal progenitors revealed transcriptional signatures for osteogenic and adipogenic lineages. Furthermore, we found that levels of transcription factors EGR1 and EGR2 are critical for lineage-specific expression and commitment of progenitors to osteo- and adipogenic fates.
Results
Establishment of Clonal Skeletal Progenitors with Distinct Differentiation Properties
The main obstacles to in vitro studies of primary BMSCs are their limited proliferation and loss of differentiation capacity during passaging (Digirolamo et al., 1999, Muraglia et al., 2000, Sarugaser et al., 2009). We took advantage of irtTA-GBD∗-TAg transgenic mice previously established in our group (Anastassiadis et al., 2010, Rostovskaya and Anastassiadis, 2012), which harbor a system for inducible expression of SV40 large T antigen, and thus can be used for isolation and conditional immortalization of somatic cells (Figures S1A and S1B). BMSCs isolated from the transgenic mice continuously proliferated upon induction of large T antigen by two ligands, dexamethasone and doxycycline (Dex/Dox). Large T antigen was deinduced 3 days after withdrawal of Dex/Dox, and concomitantly the cells ceased proliferation (Anastassiadis et al., 2010). All experiments in our study were performed at least 3 days after removal of Dex/Dox unless otherwise stated, to exclude influence of these ligands and large T antigen expression.
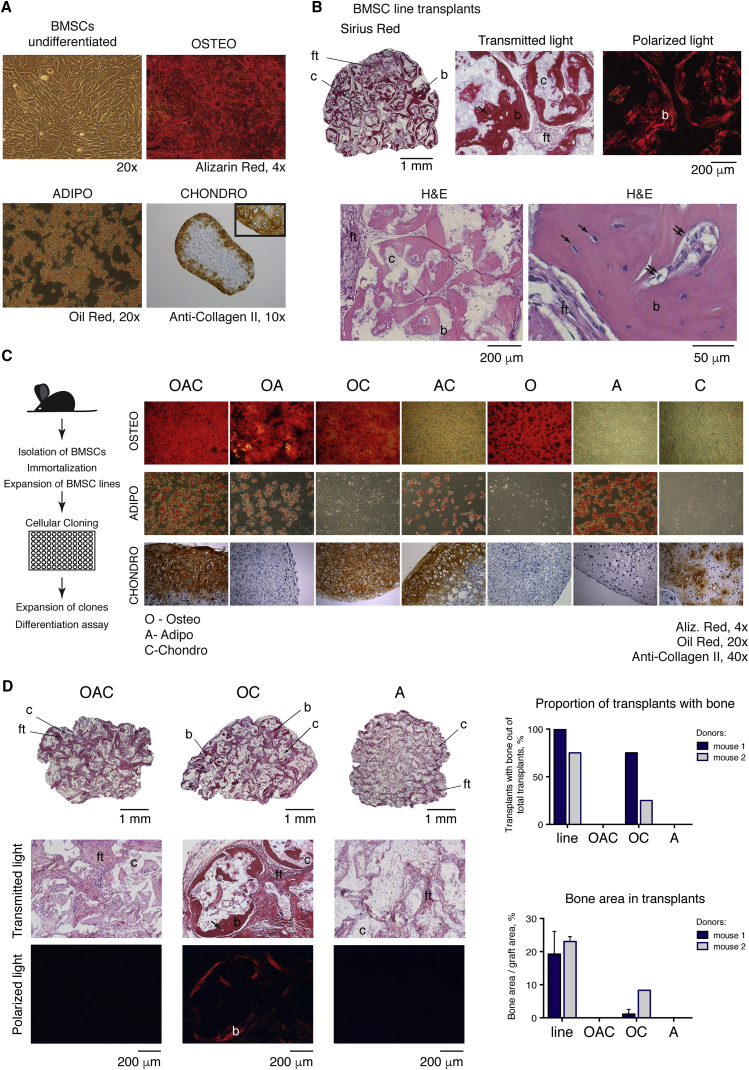
We confirmed that conditionally immortalized BMSCs maintained the potential to differentiate into osteogenic, adipogenic, and chondrogenic lineages in vitro (Figure 1A), which was not altered after long-term passaging (Figures S1C and S1D). Nevertheless, the stringent criterion defining skeletal progenitors is their ability to generate bone at heterotopic sites in an in vivo transplantation assay (Bianco and Robey, 2015, Bianco et al., 2008). To test this, we expanded two cell lines derived from individual mice for 8 passages and transplanted subcutaneously into SCID/beige mice in conjunction with a hydroxyapatite/tricalcium phosphate (HA/TCP) ceramic carrier. After 8 weeks, both transplanted BMSC lines established ossicles (4/4 and 3/4, Figure 1B). These data confirm that we established mouse BMSCs containing bona fide skeletal progenitors, which can be expanded in vitro using conditional immortalization while maintaining their differentiation properties.
Figure 1.
Characterization of Mouse BMSC Lines and Clonally Derived Populations of Skeletal Progenitors
(A) In vitro differentiation potential of BMSCs derived from whole bone marrow of irtTA-GBD∗-TAg transgenic mice to osteo-, adipo-, and chondrocytes (inset shows magnified region with characteristic lacunar structure of cartilage).
(B) Formation of heterotopic bone in transplants derived from conditionally immortalized BMSC lines. Sections through the whole transplant and higher magnification; Sirius red and H&E.
(C) Establishment of clonally derived skeletal progenitors and screening of their differentiation properties.
(D) Transplantation assay of clonally derived skeletal progenitors; Sirius red staining of sections. Right panel: quantification of transplantations of cell lines and clones from 2 individual mice (4 transplants from each).
Error bars indicate standard deviations (SDs). For sections of transplants: b, bone; ft, fibrous tissue; c, HA/TCP carrier; arrow, osteocyte; double arrow, osteoblast. See also Figure S1.
To explore the heterogeneity of BMSCs, we subjected conditionally immortalized BMSC lines to cellular cloning and then tested the clones for their differentiation capacity in vitro and in vivo. Using in vitro differentiation protocols, the clones were examined for their ability to produce mature osteo-, adipo-, and chondrogenic cells, “O”, “A” and “C” respectively (Figure 1C). We identified tripotent clones (“OAC”), as well as clones capable of generating two lineages in all possible combinations (“OA”, “OC” and “AC”) and unipotent clones (“O”, “A” and “C”). To confirm homogeneity of the established single-cell-derived clones, we performed the second round of cellular cloning and the resulting subclones were checked for osteogenic and adipogenic differentiation (Figure S1E). The chondrogenic assay was not tested in this experiment. Most subclones generated from the bipotent “OA” cells (78%–94%) inherited properties of the parental clones, indicating that these were homogeneous populations of cells capable of both types of differentiation and did not represent a mixture of unipotent cells. Also, 68%–96% of subclones derived from the “O” and “A” progenitors reproduced those potentials. These results suggested that clonally derived subpopulations of BMSCs relatively stably maintained their properties during culturing.
To test whether different types of progenitors pre-existed in the bone marrow and were not just a consequence of in vitro expansion, we isolated clonal cell populations from primary bone marrow colony-forming units—fibroblasts (CFU-Fs) by plating at low density, then immortalized and checked for osteo- and adipogenic potential (Figure S1F). Among the clones initiated by CFU-Fs established from five mice, bipotent “OA” and unipotent “O” and “A” cells were identified (29.6%, 63.0%, and 7.4%, respectively), confirming heterogeneity of skeletal progenitors in mice.
We tested osteogenic properties of chosen clonal skeletal progenitors further using a transplantation assay on HA/TCP carrier into immunocompromised mice. This method represents a rigorous assay for the osteogenic capacity of cells, with much lower frequency of adipocyte formation in the transplants (Kuznetsov et al., 1997, Sacchetti et al., 2007, Sacchetti et al., 2016). Chondrogenic differentiation of postnatal skeletal progenitors is rarely observed in this test; therefore, we chose clones based on their osteogenic and adipogenic properties, independent of their chondrogenic capacity in vitro, and transplanted “OAC”, “OC” and “A” clones derived from two mice. After 8 weeks, “OAC” and “A” cells produced grafts consisting of fibrotic tissue, and only “OC” clones formed bone, albeit with a lower efficiency than the parental line (Figure 1D). We also confirmed that the transplants contain donor cells by PCR genotyping using primers for irtTA and T antigen transgenes that marked the transplanted cells but not the host. Despite the fact that not all types of progenitors were tested, these data may indicate that the clones that possess osteogenic but not adipogenic capacity in vitro (such as “OC” and “O”) can form bone in vivo and represent bona fide osteogenic progenitors. On the other hand, the cells capable of both osteo- and adipogenic differentiation in vitro (such as “OAC” and “OA”) do not reveal the ability to generate bone in vivo, which is an apparent inconsistency between the two tests. Nevertheless, we maintained the names of the clones according to their in vitro potency, bearing in mind their in vivo potential. We further emphasize that our transplantation assay is not indicative for adipogenesis and chondrogenesis in vivo.
Expression Profiling of Clonally Derived Skeletal Progenitors
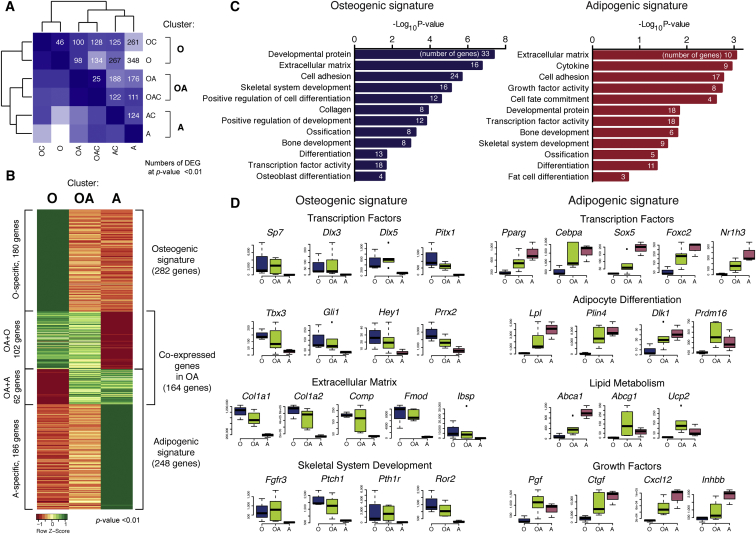
To characterize skeletal progenitors with distinct lineage commitment, we performed sequencing-based expression profiling (RNA sequencing [RNA-seq]) of the clonal populations. We used the following progenitor types according to their in vitro differentiation potential: “OAC”, “OA”, “OC”, “AC”, “O” and “A”; “C” cells were not analyzed due to the rarity of these clones (Figure S2A). We compared transcription profiles of the analyzed populations, but strikingly we could not identify genes exclusively expressed in each of the six cell types. Clustering analysis (Figure 2A) showed a strong resemblance between “OAC” and “OA” clones (25 differentially expressed genes [DEGs]), as well as “OC” and “O” (46 DEGs). “AC” and “A” also clustered together despite a higher number of DEGs (124 genes). This result showed that chondrogenic capacity did not largely affect transcriptome profile of the cells, at least in our system. Moreover, we suggest that different cell types did not show unique expression patterns because the clones were highly similar within the clusters. Therefore, we checked differential gene expression between three clusters: “O” (O + OC), “OA” (OA + OAC), and “A” (A + AC) (Figure 2B). We found 180 genes with higher expression in the “O” group and 186 genes in the “A” group; however, we could not identify specific genes for the cells with both potentials. Nevertheless, the “OA” group shared the expression of 102 upregulated genes with the “O” group and 62 genes with the “A” group. These data suggest that the clones that are bipotent in vitro, partially co-express osteogenic and adipogenic genes, and do not have unique identity. Therefore, we considered only the cells of “O” and “A” clusters for further experiments in our study.
Figure 2.
Gene Expression Analysis of Clonally Derived Skeletal Progenitors with Distinct Differentiation Potential
(A) RNA profiling of clonally derived skeletal progenitors. The distances between cell types were calculated as numbers of differentially expressed genes (DEGs) at p < 0.01, as shown on the heatmap in (B). Darker blue corresponds to higher similarity between the clones.
(B) Heatmap representing DEGs in the clusters of clones with osteogenic, adipogenic, and both potentials. Cells with both properties (OA: OAC + OA) co-expressed genes of osteo- (O: OC + O) and adipogenic (A: AC + A) cells and did not express unique genes.
(C) Enrichment of gene ontology (GO) terms in osteogenic and adipogenic signatures of skeletal progenitors.
(D) Representative genes of osteogenic and adipogenic signatures. Box plots show DEseq normalized expression values.
In the box plots, the horizontal line indicates the median, and the box shows the 25th and 75th percentiles. See also Figure S2.
We termed the set of genes upregulated in the “O” group compared with the “A” group as an “osteogenic signature” (282 genes) and the converse dataset as an “adipogenic signature” (248 genes). Functional annotation revealed that both osteogenic and adipogenic signatures were enriched for the genes involved in bone and skeletal system development, ossification, and development (Figure 2C), indicating a functional and developmental relationship between the two types of progenitors.
The transcriptional signatures that we described contained genes involved in osteo- and adipogenesis (such as Sp7 [Osterix], Dlx3, Dlx5 within the osteogenic, Pparg and Cebpa within the adipogenic signature), among others (Figure 2D). We verified expression of selected DEGs in independent clones (Figure S2B), and confirmed differential expression in 88% of the cases (21/24 genes). On a special note, Runx2 was expressed highly across all clones and was not among DEGs.
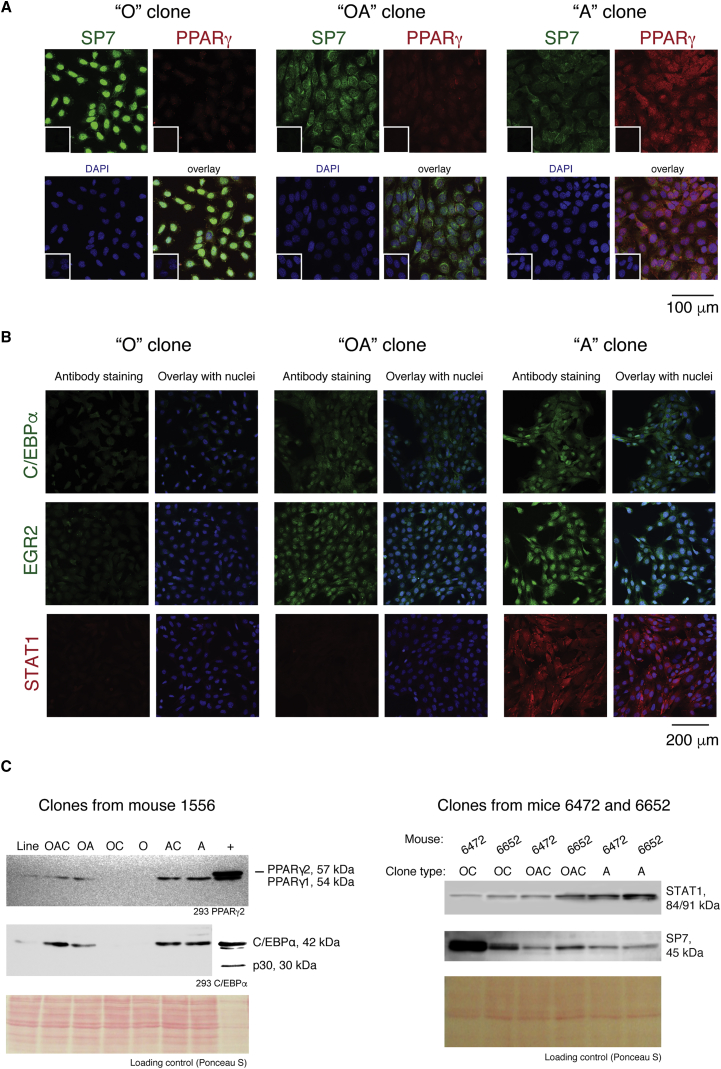
We tested expression of several proteins in the clones using immunostaining (Figures 3A and 3B) and western blot (Figure 3C). We observed high expression of SP7 in the “O” clones compared with “A”, whereas the latter expressed higher PPARγ, C/EBPα, EGR2, and STAT1, in concordance with the expression profiling results. “OA” cells generally showed intermediate levels of expression of those markers between “O” and “A” cell types. Additionally, using co-staining with antibodies for SP7 and PPARγ, we observed that bipotent “OA” clones co-expressed osteogenic and adipogenic genes at the single-cell level (Figure 3A), and thus do not represent a mixture of cells with heterogeneous expression of osteo- or adipogenic signatures.
Figure 3.
Validation of Differentially Expressed Markers in Skeletal Progenitors
(A) Clonal progenitors (O, OA, and A) were checked for co-localization of osteogenic and adipogenic markers by co-staining with antibodies against SP7 (FITC, green) and PPARγ (TRITC, red). Insets show corresponding controls stained with only secondary antibodies.
(B) Expression of DEGs characteristic for adipogenic lineage was validated by immunostaining in progenitors with O, OA, and A properties.
(C) Expression of DEGs in BMSC clones with distinct potential from 3 mice shown by western blot. PPARγ and C/EBPα were probed sequentially on the same membrane, as well as STAT1 and SP7. Loading was confirmed by Ponceau S staining of the blot. For PPARγ and C/EBPα, the positive controls were 293 cells overexpressing corresponding proteins (controls and samples for C/EBPα were on the same membrane; much lower exposure was needed to visualize control lane).
Signaling Pathways in Lineage Commitment of Skeletal Progenitors
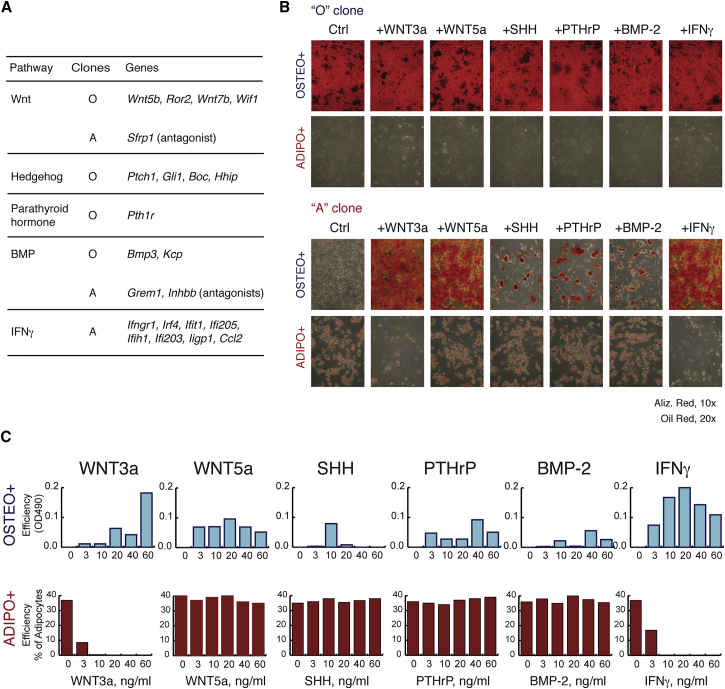
To find potential regulators of osteo- and adipogenic fates of skeletal progenitors, we analyzed representative members of signaling pathways in the osteogenic and adipogenic transcriptional signatures derived from our RNA-seq analysis (Figure 4A). Osteogenic cells expressed members of canonical and non-canonical Wnt, Hedgehog (SHH), parathyroid hormone (PTH), and bone morphogenetic protein (BMP) pathways, whereas adipogenic cells produced antagonists for Wnt and BMP and had upregulated targets of interferon-γ (IFNγ). To test their role in differentiation, we treated mouse conditionally immortalized clonal progenitors with the osteogenic or adipogenic medium with or without corresponding ligands (Figures 4B and 4C), including WNT3a, WNT5a, SHH, PTHrP, BMP-2, and IFNγ. The properties of “O” clones were not altered in all the aforementioned experimental conditions. However, activation of those pathways in the “A” cells affected their differentiation capacity. In the presence of WNT3a, WNT5a, and IFNγ, the cells demonstrated extensive mineralization in osteogenic conditions in vitro. A mild osteogenic effect was also observed after treatment with SHH, PTHrP, and BMP-2. In addition, WNT3a and IFNγ prevented adipogenic differentiation, whereas the other ligands did not. Adipogenesis was also inhibited in “OA” progenitors by emulation of WNT3a (using inhibitor of GSK3b) and IFNγ (data not shown).
Figure 4.
IFNγ and Canonical Wnt Signaling Pathways Stimulate Osteogenic and Inhibit Adipogenic Differentiation of Skeletal Progenitors
(A) Enrichment of osteo- and adipogenic transcriptional signatures for members of signaling pathways according to the RNA profiling.
(B) A set of signaling molecules was tested for the effect on the osteogenic and adipogenic induction of the “O” (top) and “A” (bottom) mouse BMSC clones.
(C) Dose dependence of the effect of signaling molecules on the differentiation properties of mouse clonal skeletal progenitors. Osteogenesis was quantified by elution of Alizarin Red dye and measurement of OD490, and adipogenesis was assessed by counting the proportion of adipocytes.
See also Figure S3.
To exclude the influence of immortalization, we verified our observations using primary human BMSCs. As expected, human BMSCs could differentiate to osteogenic lineage, with or without adding WNT3a, WNT5a, SHH, PTHrP, BMP-2, and IFNγ. Yet their capacity for adipogenesis was strongly inhibited by WNT3a and IFNγ in dose-dependent manner (Figures S3A and S3B). Next, we established clones initiated by CFU-Fs and identified bipotent “OA” and unipotent “O” and “A” clones, confirming heterogeneity of stromal cells in human bone marrow. We showed that “A” clones could mineralize in the presence of IFNγ, which also blocked adipogenesis in those cells (Figure S3C).
Together, these data revealed that specific signals can induce mineralization of adipogenic progenitors at least in vitro, and thus potentially can dictate osteogenic fate of skeletal progenitors. Since canonical Wnt and IFNγ pathways exhibited dual effects, we ranked them as primary candidates in regulating osteogenic and adipogenic commitment. Furthermore, we decided to focus on the IFNγ pathway as it is less studied for its effects on lineage commitment of BMSCs and may reveal novel regulators of their cell fate.
EGR1 and EGR2 Control Identity of Osteo- and Adipogenic Skeletal Progenitors
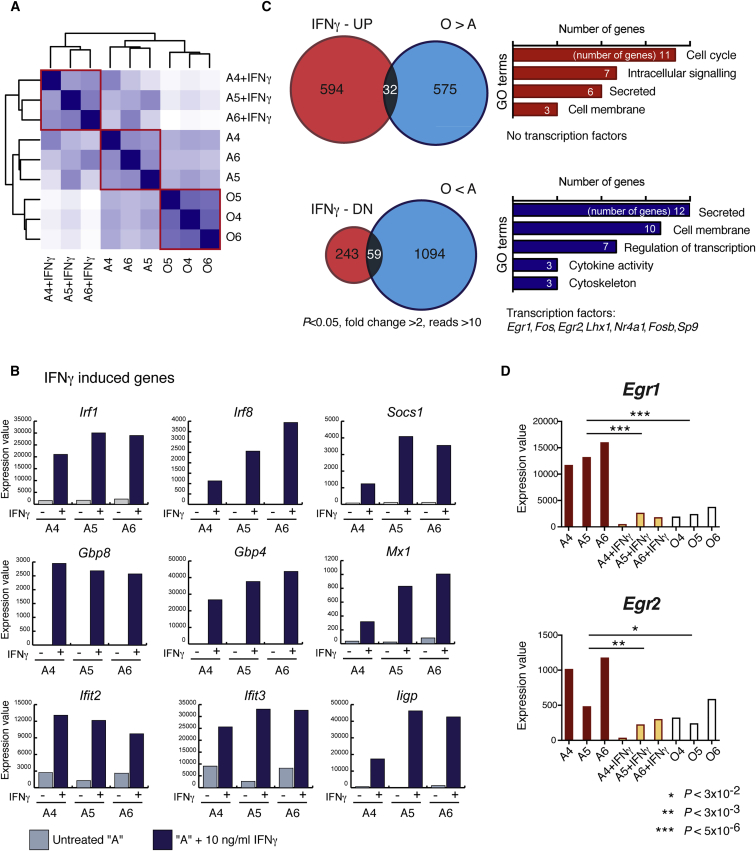
To reveal the mechanism controlling osteogenic versus adipogenic commitment of skeletal progenitors, we performed expression profiling by RNA-seq of “O” and “A” progenitors, and the same “A” clones treated with IFNγ for 3 days (Figures 5A and 5B). IFNγ treatment of “A” cells resulted in upregulation of 626 genes and downregulation of 302 genes (p < 0.05, fold change >2). “O” clones had higher expression of 607 genes and lower levels of 1,153 genes compared with “A” cells. We identified 32 genes characteristic of “O” clones that could be induced in “A” cells by IFNγ (Figure 5C, upper panel), however we did not identify transcription factors among them. Next we looked at the converse datasets, and found 59 genes that were downregulated in “O” clones and inhibited by IFNγ in the “A” cells (Figure 5C, lower panel), including seven transcription factors. Remarkably, four of them belonged to the group of early response genes (Egr1, Fos, Egr2, Fosb). Egr1 was among the top DEGs; therefore, we chose EGR1 and closely related EGR2 transcription factors for further analysis (Figure 5D).
Figure 5.
Gene Expression in “A” Progenitors Treated with IFNγ
(A) Expression of genes was analyzed in “O” clones (O4, O5, O6), “A” clones (A4, A5, and A6), and the same “A” clones treated with IFNγ for 3 days by RNA-seq. Heatmap of sample correlations based on Euclidean distances; dark blue corresponds to higher similarity.
(B) Upregulation of known IFNγ-regulated genes in the clones with “A” properties upon treatment with IFNγ.
(C) Analysis of RNA-seq results for differential expression of genes (p < 0.05). The lists of overlapping DEGs were characterized for enrichment of GO terms and checked for the presence of transcription factors.
(D) Comparison of Egr1 and Egr2 expression in clones analyzed by RNA-seq for three independent clones.
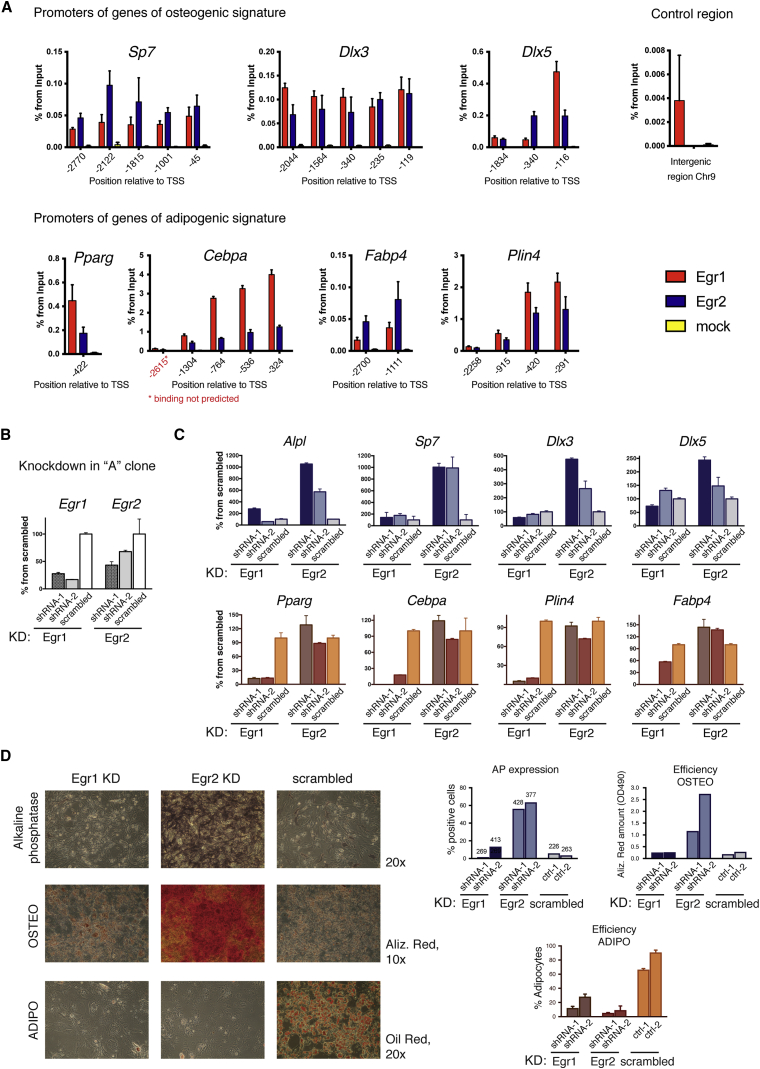
We confirmed that Egr1 and Egr2 were reproducibly downregulated in “A” cells in the presence of IFNγ in independent experiments (Figures S4A and S4B). We performed reporter assays using luciferase gene placed under control of 3-kb upstream sequences of Egr1 and Egr2 genes. Luciferase signal was reduced in the presence of IFNγ in a dose-dependent manner (Figures S4C and S4D), which confirmed that downregulation of both genes occurred at the transcriptional level. Furthermore, we found enrichment of EGR1 and EGR2 in the promoters of genes for proteins involved in osteogenic and adipogenic differentiation, such as Sp7, Dlx3, Dlx5, Pparg, Cebpa, Fabp4, and Plin4, using chromatin immunoprecipitation (ChIP) assays (Figure 6A), suggesting that EGR1 and EGR2 can potentially regulate transcriptional programs and differentiation potency in skeletal progenitors.
Figure 6.
EGR1 and EGR2 Control Identity of Skeletal Progenitors
(A) Presence of EGR1 and EGR2 at putative binding sites in the promoter regions of known osteogenic and adipogenic regulators was tested by ChIP-qPCR in “A” clone. Percentage from input for mock was in most cases negligible; the signal in the control region in intergenic space of chromosome 9 and Cebpa promoter distant from binding sites (shown in red) was very low compared to the putative sites. Error bars indicate the SD of technical replicates of qPCR reactions.
(B) Genetic knockdown of Egr1 and Egr2 was done using two independent shRNA-encoding constructs for each gene in “A” clone; expression was tested by qRT-PCR relative to the scrambled control. Error bars indicate the SD of technical replicates of qPCR reactions.
(C) Gene expression analysis in the cells with Egr1 and Egr2 knockdown as shown by qRT-PCR relative to scrambled control. Error bars indicate the SD of technical replicates of qPCR reactions.
(D) The cells with Egr1 and Egr2 knockdowns were tested for AP expression, osteogenic (Alizarin Red staining), and adipogenic differentiation (Oil Red O staining). Quantification: AP by counting proportion of stained cells (total number of counted cells is indicated), osteogenesis by elution of bound Alizarin Red and measurement of OD490, adipogenesis by counting proportion of adipocytes. Error bars indicate the SD of technical replicates.
See also Figure S4.
To test the role of EGR1 and EGR2 in regulation of differentiation potency in skeletal progenitors, we performed genetic knockdown in “A” progenitors using retroviral transduction (Figure 6B). Knockdown of Egr1 did not affect levels of osteogenic markers, such as Alpl, Sp7, Dlx3, and Dlx5, whereas adipogenic genes were significantly downregulated (Pparg ∼10-fold, Cebpa ∼5-fold to undetectable level, Plin4 ∼10- to 20-fold, Fabp4 ∼2-fold to undetectable) (Figure 6C). On the other hand, Egr2 knockdown greatly upregulated osteogenic signature genes (Alpl ∼6- to 10-fold, Sp7 ∼10-fold, Dlx3 ∼3- to 5-fold, Dlx5 ∼1.5- to 2.5-fold), while the genes associated with the adipogenic signature were not markedly changed.
Furthermore, downregulation of Egr1 inhibited the adipogenic capacity of “A” cells (Figure 6D). The cells with Egr2 knockdown gained osteogenic capacity and showed elevated level of alkaline phosphatase (AP), which is indicative of osteogenic propensity and could not produce adipocytes in differentiation conditions. The results of knockdown experiments were reproduced in two independent experiments and using three “A” clones.
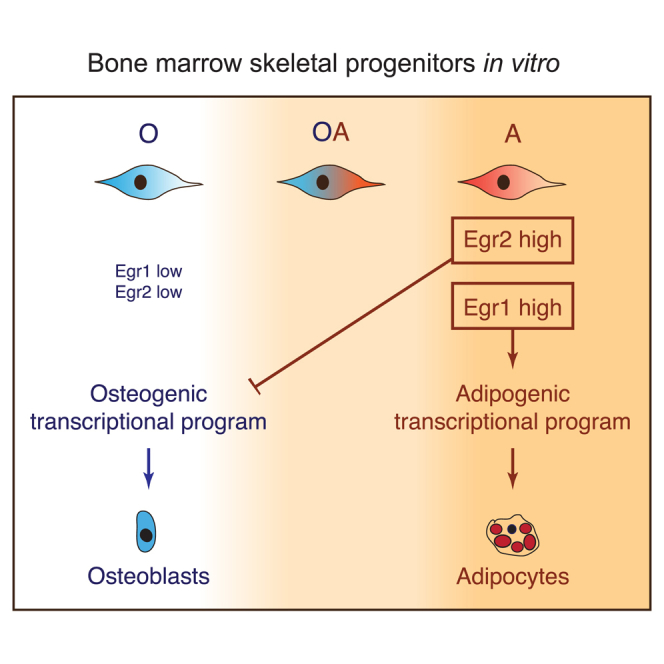
Taken together, our data suggest that EGR1 is required for expression of the adipogenic genes, whereas downregulation of EGR2 can reactivate the osteogenic transcriptional program in adipogenic cells. Furthermore, reduction of EGR2 level is sufficient to confer osteogenic properties to adipogenic progenitors. Thus, EGR1 and EGR2 control lineage-specific gene expression and identity of skeletal progenitors.
Discussion
Heterogeneity of BMSCs
Skeletal stem cells (also referred to as mesenchymal stem cells) residing in bone marrow stroma are postulated to serve as common precursors for cartilage, bone, hematopoiesis-supportive stroma, and marrow adipocytes. This broad spectrum of lineages generated by skeletal stem cells has been suggested from developmental relations between those lineages, numerous in vitro differentiation studies (Morikawa et al., 2009, Peister et al., 2004, Pittenger et al., 1999), and analysis of heterotopic transplants of BMSC populations (Friedenstein et al., 1987) isolated from adult bone marrow. Despite this model being widely accepted, so far to our knowledge the existence of a single progenitor contributing to bone, adipocytes, and chondrocytes has not been shown in vivo in adult mice by stringent assays such as lineage tracing or single-cell engraftment. Therefore, the questions still remain: which lineages are generated by skeletal stem cells and what factors control identity of committed progenitors?
We sought to identify which types of skeletal progenitors can be isolated from bone marrow of adult mice and how their differentiation potential can be controlled. Heterogeneity of BMSCs in vitro has been examined in previous studies, which described different combinations of distinct progenitor types (Banfi et al., 2000, Muraglia et al., 2000, Okamoto et al., 2002, Russell et al., 2010, Sarugaser et al., 2009, Zhou et al., 2014). Subtypes of skeletal progenitors with distinct differentiation properties have been also isolated directly from skeletal tissues (Chan et al., 2013, Chan et al., 2015), including multipotent cells capable of generating bone, cartilage, and stroma in transplantation tests, as well as restricted bone progenitors and cartilage precursors. Nevertheless, most experiments in these works were not performed at a clonal level and the cells were isolated from fetal or neonatal tissues. In addition chondrogenic precursors were predominantly found in ear cartilage. Therefore these results are difficult to align with our clonal subpopulations of skeletal progenitors from adult bone marrow.
We found clonal progenitors with all possible seven types of differentiation potential (“OAC”, “OA”, “OC”, “AC”, “O”, “A” and “C”) according to in vitro differentiation assays, making it plausible to suggest that osteogenic, adipogenic, and chondrogenic differentiation properties can be independently combined in the clones. Our expression profiling results were consistent with this model; we identified specific transcriptional programs correlating with osteogenic and adipogenic commitment, and clones with both properties shared expression of these programs and did not show exclusive genes.
Given the findings of transcriptome analysis, we suggest that osteogenic and adipogenic signatures are not mutually exclusive and that specific types of skeletal progenitors co-express alternative lineage-specific genes. This observation contrasts with the classical view of differentiation, which is based on progression through a hierarchical system of intermediate states with specific identity. Nonetheless, the phenomenon of shared transcriptional programs has been proposed as “lineage priming” for different systems, including hematopoietic stem cells (Hu et al., 1997, Laurenti et al., 2013, Ng et al., 2009) and inner cell mass of blastocyst (Guo et al., 2010, Plusa et al., 2008). These lines of evidence suggest that activation of multiple transcriptional lineage-specific signatures can be a common mechanism of early commitment of progenitors in different biological systems.
An additional question arising from our results is why the transcriptional signature for the chondrogenic lineage could not be identified from our expression profiling. We did not perform RNA-seq of “C” clones due to their rarity, and this may be the reason why this signature was not prominent in bioinformatics analysis. Yet it has been recently shown that in vitro chondrogenic differentiation of BMSCs can be reverted back to the undifferentiated state both in vitro in culture conditions for BMSCs and in vivo after transplantation, and thus does not represent stable commitment to chondrogenic lineage (Serafini et al., 2014). Taking into account that postnatal BMSCs have not been rigorously shown to form cartilage using in vivo tests such as transplantations or lineage tracing, whether bone marrow skeletal progenitors possess bona fide intrinsic chondrogenic potency still remains to be proved.
The other question is whether it is possible to predict the relationships between different subpopulations of progenitors. We performed rigorous assessment of their osteogenic potential by transplantation with HA/TCP scaffolds (Kuznetsov et al., 1997, Sacchetti et al., 2007, Sacchetti et al., 2016). Despite the limited number and variety of clones assayed, our results revealed that the osteogenic clones lacking adipogenic potency (“OC”) could generate bone ossicle upon transplantation, whereas all clones with adipogenic properties in vitro (such as “A” and “OAC”) could not. One possible explanation for multipotent cells (“OAC”) not being able to generate bone in vivo could be that these cells might require additional inductive signals from the environment to commit to osteogenic lineage, which could be present in the bone but were absent at heterotopic site of transplantation. On the other hand, these results challenge the view that clones showing multipotency in vitro can generate both adipo- and osteogenic lineages and are upstream of progenitors restricted to osteogenesis. It is also conceivable, although highly speculative, that osteogenic progenitors (“O”) progress toward non-osteogenic state (“A”), whereas the clones combining osteogenic and adipogenic properties (“OA”) represent a transitory population between those two cell types. In this case, combined expression of two signatures may reflect the process of “swapping” the programs and “OA” clones remain responsive to osteogenic cues in vitro, having in fact lost this potency in an in vivo context. This is a theoretical assumption that needs to be further explored.
Mechanisms Controlling Lineage Commitment
Mechanisms regulating the process of differentiation in adult tissues have been intensively investigated using transgenic mice and in vitro cell culture systems. In contrast, knowledge of factors controlling identity of undifferentiated stem and progenitor cells is still scarce, mainly due to the inability to isolate and propagate pure populations of these cells. We characterized undifferentiated cells committed to different lineages and proposed IFNγ and WNT3a as candidates to control osteogenic and adipogenic fate.
Canonical Wnt was suggested to stimulate osteogenesis based on the phenotype in bones and BMSCs of patients with loss-of-function and gain-of-function mutations in Wnt co-receptor LRP5 (Gong et al., 2001, Qiu et al., 2007). Further experiments confirmed that canonical Wnt signaling stimulates osteogenesis while inhibiting adipogenic differentiation in cultured BMSCs (Taipaleenmäki et al., 2011) and in mice (Bennett et al., 2005), which is in accord with our results.
The role of IFNγ in differentiation of skeletal progenitors is less understood, partly because IFNγ can promote osteoclast differentiation (Xiong et al., 2016), which makes it complicated to analyze the autonomous effect of IFNγ on skeletal progenitors in model organisms. Nevertheless, IFNγ was shown to enhance osteogenesis and inhibit adipogenesis in BMSCs in vitro and in mice after injection (Duque et al., 2009, Vidal et al., 2012). Moreover, knockout mice for IFNγR1 receptor show impaired osteogenesis (Duque et al., 2011), which implies a physiological role of IFNγ in differentiation of skeletal cell types. The common sources of IFNγ production are T cells, natural killer cells, and macrophages (Schoenborn and Wilson, 2007). Furthermore, BMSCs may produce IFNγ in autocrine fashion (Duque et al., 2009), although we did not detect its expression, at least in our experimental system. Therefore, in the bone marrow context, IFNγ signal may arise from interaction of skeletal progenitors with a hematopoietic compartment, or by being stimulated as an autocrine signal in response to other factors.
Expression of IFNγ targets without exogenous IFNγ in undifferentiated adipogenic clones is an interesting phenomenon, although the level of their expression was much lower than in the presence of ligand. A recent work by Wu et al. (2018) showed that undifferentiated pluripotent and tissue-specific stem cells exhibited intrinsic expression of interferon-stimulated genes (ISG) without IFNγ in the environment, and were resistant to viral infection. ISGs were downregulated during differentiation and concomitantly the cells became sensitive to viral infection. Similarly to our study, the event that induced this endogenous ISG program remained unknown. We also observed downregulation of IFNγ targets during differentiation of BMSCs (result not shown); it would be interesting to next test whether our skeletal progenitors are sensitive to viral infection.
Furthermore, our observations are congruent with the previous report that a subset of BMSCs labeled by Mx1-Cre induction is a stress-responsive osteogenic stem/progenitor cell population lacking detectable adipogenic capacity (Park et al., 2012). pIpC used for induction of Mx1-Cre labeling is a synthetic analog of double-stranded RNA, which induces IFNγ response in cells, including Mx1. Thus, it is possible that properties of Mx1-labeled progenitors reflect a stimulatory effect of IFNγ on osteogenic potency and suppression of adipogenic capacity that we describe. More experimental evidence, including lineage tracing and in vivo tracking, is required to reveal whether IFNγ-mediated modulation of cell fate specification occurs in homeostasis or regeneration.
Our transcriptome analysis and functional data from short hairpin RNA (shRNA) knockdown suggested that transcription factor EGR1 is required for expression of adipogenic regulators and adipogenesis in “A” cells, whereas EGR2 suppressed pro-osteogenic genes and osteogenic properties in “A” cells. Egr2 knockdown also inhibited adipogenesis, indicating that EGR2 may be essential or that elevation of osteogenic regulators suppressed adipogenesis. Nevertheless, both EGR1 and EGR2 contributed to control of expression of lineage signatures in skeletal progenitors. As we found both proteins to be present in the promoters of major regulators of osteogenesis and adipogenesis (such as Sp7, Dlx3, Dlx5, Pparg, Cebpa), the effect on these genes can be direct. There was no major difference in EGR1 and EGR2 distribution, which is perhaps not surprising considering high similarity in their DNA binding domain (Swirnoff and Milbrandt, 1995). Thus their distinct effects can be explained by recruitment of different interactors, which can mediate transcription activation or repression.
In concordance with our results, EGR1 and EGR2 have been shown to play a role in differentiation of skeletal tissues in vivo. Egr1 knockout mice display normal osteoblastogenesis (Cenci et al., 2000, Reumann et al., 2011) but increased resistance to adiposity induced by a high-fat diet (Zhang et al., 2013). On the other hand, bone marrow of Egr2+/− mice contained almost 2-fold more osteogenic colony-forming units compared with the wild-type littermates, indicating increased osteogenic properties of BMSCs (Gabet et al., 2010). Together, these data indicate that EGR1 is a positive regulator of adipogenic differentiation, while EGR2 is a suppressor of osteogenesis in the skeletal progenitors in vivo.
In conclusion, we present here comprehensive characterization of transcriptome of distinct types of skeletal progenitors, which we believe will facilitate further investigations, including studies of their differentiation and origin. Furthermore, we found that in skeletal progenitors EGR2 suppresses osteogenic transcriptional program and differentiation propensity, whereas EGR1 maintains expression of adipogenic genes and ability to differentiate. Although many questions about the identity of skeletal progenitors remain to be answered by future studies, here we provide a previously unknown mechanism of their cell fate control.
Experimental Procedures
BMSC Isolation and Culture
BMSCs were isolated by flushing the femora and tibiae of irtTA-GBD∗-TAg transgenic mice. Cell expansion and differentiation were performed as described previously (Rostovskaya and Anastassiadis, 2012) and in Supplemental Experimental Procedures.
Transplantation Assays
The animal experiments were done under appropriate approval; the number of the authorization released by the Italian Ministry of Health is 1282/2015. 2 × 106 cells were loaded onto hydroxyapatite/tricalcium phosphate (HA/TCP) particles (40 mg, 100–200 μm; Zimmer, Warsaw, IN), then subcutaneously transplanted into the backs of 6- to 15-week--old female SCID/beige mice and analyzed after 8 weeks.
Library Preparation and Sequencing
Isolation of mRNA was done using bead-based poly(dT) selection followed by chemical fragmentation and further processing for Illumina short-read sequencing (HiSeq 2000). For strand-specific sequencing, two different versions of the dUTP method (Parkhomchuk et al., 2009) were applied (for details see Supplemental Experimental Procedures).
Chromatin Immunoprecipitation
Protein-DNA complexes were crosslinked with 2 mM disuccinimidyl glutarate and 1% formaldehyde followed by sonication. DNA was precipitated with EGR1 or EGR2 antibody and protein G Sepharose beads, purified, and used for qPCR.
shRNA Knockdown
The gene knockdowns were done via retroviral transduction of shRNA-encoding vectors pLKO.1-puro listed in Supplemental Experimental Procedures.
Author Contributions
M. Rostovskaya conceived, designed, and executed the experiments and wrote the manuscript; S.D. and B.S. performed transplantation assays; D.A. performed bioinformatics analysis; S.K. performed RNA-seq; A.D. oversaw transcription profiling experiments; M. Riminucci oversaw transplantation assays and participated in writing the manuscript; P.B. oversaw transplantation assays; K.A. oversaw the project, designed and executed experiments, and wrote the manuscript.
Acknowledgments
The authors are grateful to Prof. A. Francis Stewart for helpful discussions and Dr. Michelle Meredyth-Stewart for critical reading of the manuscript. Special thanks to Prof. Pamela G. Robey for giving valuable comments on the manuscript. We would like also to thank Marc Bickle (MPI-CBG, Dresden, Germany) for providing shRNA and Katrin Mueller and Prof. Martin Bornhäuser (Universität Klinikum Carl Gustav Carus, Dresden, Germany) for human bone marrow samples. We are sincerely thankful to Mandy Obst and Jesus Eduardo Rojo Arias for technical assistance. We are grateful to Dr. Nina Drize (Haematological Scientific Center, RAMS, Russia) for stimulating discussions, help, and advice. This study was supported by the Collaborative Research Grant SFB-655 (German Research Foundation - DFG) to K.A. The Next Generation Sequencing facility was also supported by the SFB-655. P.B. and M. Riminucci are supported by Fondazione Cenci Bolognetti, Telethon (grant GGP15198).
Published: June 21, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and four figures and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.05.014.
Contributor Information
Maria Rostovskaya, Email: mr631@cam.ac.uk.
Konstantinos Anastassiadis, Email: konstantinos.anastassiadis@tu-dresden.de.
Accession Numbers
The RNA-seq data in this paper have been deposited in NCBI's Gene Expression Omnibus under series accession numbers GEO: GSE114474 and GSE114475.
Supplemental Information
References
- Anastassiadis K., Rostovskaya M., Lubitz S., Weidlich S., Stewart A.F. Precise conditional immortalization of mouse cells using tetracycline-regulated SV40 large T-antigen. Genesis. 2010;48:220–232. doi: 10.1002/dvg.20605. [DOI] [PubMed] [Google Scholar]
- Banfi A., Muraglia A., Dozin B., Mastrogiacomo M., Cancedda R., Quarto R. Proliferation kinetics and differentiation potential of ex vivo expanded human bone marrow stromal cells: implications for their use in cell therapy. Exp. Hematol. 2000;28:707–715. doi: 10.1016/s0301-472x(00)00160-0. [DOI] [PubMed] [Google Scholar]
- Bennett C.N., Longo K.A., Wright W.S., Suva L.J., Lane T.F., Hankenson K.D., MacDougald O.A. Regulation of osteoblastogenesis and bone mass by Wnt10b. Proc. Natl. Acad. Sci. USA. 2005;102:3324–3329. doi: 10.1073/pnas.0408742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco P., Robey P.G. Marrow stromal stem cells. J. Clin. Invest. 2000;105:1663–1668. doi: 10.1172/JCI10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco P., Robey P.G. Skeletal stem cells. Development. 2015;142:1023–1027. doi: 10.1242/dev.102210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco P., Robey P.G., Simmons P.J. Mesenchymal stem cells: revisiting history, concepts, and assays. Cell Stem Cell. 2008;2:313–319. doi: 10.1016/j.stem.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden L.M., Mao J., Belsky J., Mitzner L., Farhi A., Mitnick M.A., Wu D., Insogna K., Lifton R.P. High bone density due to a mutation in LDL-receptor-related protein 5. N. Engl. J. Med. 2002;346:1513–1521. doi: 10.1056/NEJMoa013444. [DOI] [PubMed] [Google Scholar]
- Caplan A.I. Mesenchymal stem cells. J. Orthop. Res. 1991;9:641–650. doi: 10.1002/jor.1100090504. [DOI] [PubMed] [Google Scholar]
- Cenci S., Weitzmann M.N., Gentile M.A., Aisa M.C., Pacifici R. M-CSF neutralization and egr-1 deficiency prevent ovariectomy-induced bone loss. J. Clin. Invest. 2000;105:1279–1287. doi: 10.1172/JCI8672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.K., Lindau P., Jiang W., Chen J.Y., Zhang L.F., Chen C.C., Seita J., Sahoo D., Kim J.B., Lee A. Clonal precursor of bone, cartilage, and hematopoietic niche stromal cells. Proc. Natl. Acad. Sci. USA. 2013;110:12643–12648. doi: 10.1073/pnas.1310212110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.K., Seo E.Y., Chen J.Y., Lo D., McArdle A., Sinha R., Tevlin R., Seita J., Vincent-Tompkins J., Wearda T. Identification and specification of the mouse skeletal stem cell. Cell. 2015;160:285–298. doi: 10.1016/j.cell.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Niziolek P.J., MacDonald B.T., Zylstra C.R., Alenina N., Robinson D.R., Zhong Z., Matthes S., Jacobsen C.M., Conlon R.A. Lrp5 functions in bone to regulate bone mass. Nat. Med. 2011;17:684–691. doi: 10.1038/nm.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digirolamo C.M., Stokes D., Colter D., Phinney D.G., Class R., Prockop D.J. Propagation and senescence of human marrow stromal cells in culture: a simple colony-forming assay identifies samples with the greatest potential to propagate and differentiate. Br. J. Haematol. 1999;107:275–281. doi: 10.1046/j.1365-2141.1999.01715.x. [DOI] [PubMed] [Google Scholar]
- Duque G., Huang D.C., Dion N., Macoritto M., Rivas D., Li W., Yang X.F., Li J., Lian J., Marino F.T. Interferon-gamma plays a role in bone formation in vivo and rescues osteoporosis in ovariectomized mice. J. Bone Miner. Res. 2011;26:1472–1483. doi: 10.1002/jbmr.350. [DOI] [PubMed] [Google Scholar]
- Duque G., Huang D.C., Macoritto M., Rivas D., Yang X.F., Ste-Marie L.G., Kremer R. Autocrine regulation of interferon gamma in mesenchymal stem cells plays a role in early osteoblastogenesis. Stem Cells. 2009;27:550–558. doi: 10.1634/stemcells.2008-0886. [DOI] [PubMed] [Google Scholar]
- Friedenstein A.J., Chailakhyan R.K., Gerasimov U.V. Bone marrow osteogenic stem cells: in vitro cultivation and transplantation in diffusion chambers. Cell Tissue kinet. 1987;20:263–272. doi: 10.1111/j.1365-2184.1987.tb01309.x. [DOI] [PubMed] [Google Scholar]
- Friedenstein A.J., Petrakova K.V., Kurolesova A.I., Frolova G.P. Heterotopic of bone marrow. Analysis of precursor cells for osteogenic and hematopoietic tissues. Transplantation. 1968;6:230–247. [PubMed] [Google Scholar]
- Gabet Y., Baniwal S.K., Leclerc N., Shi Y., Kohn-Gabet A.E., Cogan J., Dixon A., Bachar M., Guo L., Turman J.E., Jr. Krox20/EGR2 deficiency accelerates cell growth and differentiation in the monocytic lineage and decreases bone mass. Blood. 2010;116:3964–3971. doi: 10.1182/blood-2010-01-263830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y., Slee R.B., Fukai N., Rawadi G., Roman-Roman S., Reginato A.M., Wang H., Cundy T., Glorieux F.H., Lev D. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107:513–523. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- Gronthos S., Zannettino A.C., Hay S.J., Shi S., Graves S.E., Kortesidis A., Simmons P.J. Molecular and cellular characterisation of highly purified stromal stem cells derived from human bone marrow. J. Cell Sci. 2003;116:1827–1835. doi: 10.1242/jcs.00369. [DOI] [PubMed] [Google Scholar]
- Guo G., Huss M., Tong G.Q., Wang C., Li Sun L., Clarke N.D., Robson P. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev. Cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Hong J.H., Hwang E.S., McManus M.T., Amsterdam A., Tian Y., Kalmukova R., Mueller E., Benjamin T., Spiegelman B.M., Sharp P.A. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309:1074–1078. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- Hu M., Krause D., Greaves M., Sharkis S., Dexter M., Heyworth C., Enver T. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 1997;11:774–785. doi: 10.1101/gad.11.6.774. [DOI] [PubMed] [Google Scholar]
- Kassem M., Bianco P. Skeletal stem cells in space and time. Cell. 2015;160:17–19. doi: 10.1016/j.cell.2014.12.034. [DOI] [PubMed] [Google Scholar]
- Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., Shimizu Y., Bronson R.T., Gao Y.H., Inada M. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- Kuznetsov S.A., Krebsbach P.H., Satomura K., Kerr J., Riminucci M., Benayahu D., Robey P.G. Single-colony derived strains of human marrow stromal fibroblasts form bone after transplantation in vivo. J. Bone Miner. Res. 1997;12:1335–1347. doi: 10.1359/jbmr.1997.12.9.1335. [DOI] [PubMed] [Google Scholar]
- Laurenti E., Doulatov S., Zandi S., Plumb I., Chen J., April C., Fan J.B., Dick J.E. The transcriptional architecture of early human hematopoiesis identifies multilevel control of lymphoid commitment. Nat. Immunol. 2013;14:756–763. doi: 10.1038/ni.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa S., Mabuchi Y., Kubota Y., Nagai Y., Niibe K., Hiratsu E., Suzuki S., Miyauchi-Hara C., Nagoshi N., Sunabori T. Prospective identification, isolation, and systemic transplantation of multipotent mesenchymal stem cells in murine bone marrow. J. Exp. Med. 2009;206:2483–2496. doi: 10.1084/jem.20091046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraglia A., Cancedda R., Quarto R. Clonal mesenchymal progenitors from human bone marrow differentiate in vitro according to a hierarchical model. J. Cell Sci. 2000;113(Pt 7):1161–1166. doi: 10.1242/jcs.113.7.1161. [DOI] [PubMed] [Google Scholar]
- Ng S.Y., Yoshida T., Zhang J., Georgopoulos K. Genome-wide lineage-specific transcriptional networks underscore Ikaros-dependent lymphoid priming in hematopoietic stem cells. Immunity. 2009;30:493–507. doi: 10.1016/j.immuni.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa K., Nakashima T., Takeda S., Isogai M., Hamada M., Kimura A., Kodama T., Yamaguchi A., Owen M.J., Takahashi S. Maf promotes osteoblast differentiation in mice by mediating the age-related switch in mesenchymal cell differentiation. J. Clin. Invest. 2010;120:3455–3465. doi: 10.1172/JCI42528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto T., Aoyama T., Nakayama T., Nakamata T., Hosaka T., Nishijo K., Nakamura T., Kiyono T., Toguchida J. Clonal heterogeneity in differentiation potential of immortalized human mesenchymal stem cells. Biochem. Biophys. Res. Commun. 2002;295:354–361. doi: 10.1016/s0006-291x(02)00661-7. [DOI] [PubMed] [Google Scholar]
- Owen M., Friedenstein A.J. Stromal stem cells: marrow-derived osteogenic precursors. Ciba Found. Symp. 1988;136:42–60. doi: 10.1002/9780470513637.ch4. [DOI] [PubMed] [Google Scholar]
- Park D., Spencer J.A., Koh B.I., Kobayashi T., Fujisaki J., Clemens T.L., Lin C.P., Kronenberg H.M., Scadden D.T. Endogenous bone marrow MSCs are dynamic, fate-restricted participants in bone maintenance and regeneration. Cell Stem Cell. 2012;10:259–272. doi: 10.1016/j.stem.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhomchuk D., Borodina T., Amstislavskiy V., Banaru M., Hallen L., Krobitsch S., Lehrach H., Soldatov A. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009;37:e123. doi: 10.1093/nar/gkp596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peister A., Mellad J.A., Larson B.L., Hall B.M., Gibson L.F., Prockop D.J. Adult stem cells from bone marrow (MSCs) isolated from different strains of inbred mice vary in surface epitopes, rates of proliferation, and differentiation potential. Blood. 2004;103:1662–1668. doi: 10.1182/blood-2003-09-3070. [DOI] [PubMed] [Google Scholar]
- Pittenger M.F., Mackay A.M., Beck S.C., Jaiswal R.K., Douglas R., Mosca J.D., Moorman M.A., Simonetti D.W., Craig S., Marshak D.R. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- Plusa B., Piliszek A., Frankenberg S., Artus J., Hadjantonakis A.K. Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst. Development. 2008;135:3081–3091. doi: 10.1242/dev.021519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W., Andersen T.E., Bollerslev J., Mandrup S., Abdallah B.M., Kassem M. Patients with high bone mass phenotype exhibit enhanced osteoblast differentiation and inhibition of adipogenesis of human mesenchymal stem cells. J. Bone Miner. Res. 2007;22:1720–1731. doi: 10.1359/jbmr.070721. [DOI] [PubMed] [Google Scholar]
- Reumann M.K., Strachna O., Yagerman S., Torrecilla D., Kim J., Doty S.B., Lukashova L., Boskey A.L., Mayer-Kuckuk P. Loss of transcription factor early growth response gene 1 results in impaired endochondral bone repair. Bone. 2011;49:743–752. doi: 10.1016/j.bone.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostovskaya M., Anastassiadis K. Differential expression of surface markers in mouse bone marrow mesenchymal stromal cell subpopulations with distinct lineage commitment. PLoS One. 2012;7:e51221. doi: 10.1371/journal.pone.0051221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell K.C., Phinney D.G., Lacey M.R., Barrilleaux B.L., Meyertholen K.E., O'Connor K.C. In vitro high-capacity assay to quantify the clonal heterogeneity in trilineage potential of mesenchymal stem cells reveals a complex hierarchy of lineage commitment. Stem Cells. 2010;28:788–798. doi: 10.1002/stem.312. [DOI] [PubMed] [Google Scholar]
- Sacchetti B., Funari A., Michienzi S., Di Cesare S., Piersanti S., Saggio I., Tagliafico E., Ferrari S., Robey P.G., Riminucci M. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131:324–336. doi: 10.1016/j.cell.2007.08.025. [DOI] [PubMed] [Google Scholar]
- Sacchetti B., Funari A., Remoli C., Giannicola G., Kogler G., Liedtke S., Cossu G., Serafini M., Sampaolesi M., Tagliafico E. No identical “Mesenchymal Stem Cells” at different times and sites: human committed progenitors of distinct origin and differentiation potential are incorporated as adventitial cells in microvessels. Stem Cell Rep. 2016;6:897–913. doi: 10.1016/j.stemcr.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarugaser R., Hanoun L., Keating A., Stanford W.L., Davies J.E. Human mesenchymal stem cells self-renew and differentiate according to a deterministic hierarchy. PLoS One. 2009;4:e6498. doi: 10.1371/journal.pone.0006498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini M., Sacchetti B., Pievani A., Redaelli D., Remoli C., Biondi A., Riminucci M., Bianco P. Establishment of bone marrow and hematopoietic niches in vivo by reversion of chondrocyte differentiation of human bone marrow stromal cells. Stem Cell Res. 2014;12:659–672. doi: 10.1016/j.scr.2014.01.006. [DOI] [PubMed] [Google Scholar]
- Schoenborn J.R., Wilson C.B. Regulation of interferon-gamma during innate and adaptive immune responses. Adv. Immunol. 2007;96:41–101. doi: 10.1016/S0065-2776(07)96002-2. [DOI] [PubMed] [Google Scholar]
- Swirnoff A.H., Milbrandt J. DNA-binding specificity of NGFI-A and related zinc finger transcription factors. Mol. Cell Biol. 1995;15:2275–2287. doi: 10.1128/mcb.15.4.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sworder B.J., Yoshizawa S., Mishra P.J., Cherman N., Kuznetsov S.A., Merlino G., Balakumaran A., Robey P.G. Molecular profile of clonal strains of human skeletal stem/progenitor cells with different potencies. Stem Cell Res. 2015;14:297–306. doi: 10.1016/j.scr.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipaleenmäki H., Abdallah B.M., Aidahmash A., Säämänen A.M., Kassem M. Wnt signalling mediates the cross-talk between bone marrow derived pre-adipocytic and pre-osteoblastic cell populations. Exp. Cell Res. 2011;317:745–756. doi: 10.1016/j.yexcr.2010.12.015. [DOI] [PubMed] [Google Scholar]
- Tavassoli M., Crosby W.H. Transplantation of marrow to extramedullary sites. Science. 1968;161:54–56. doi: 10.1126/science.161.3836.54. [DOI] [PubMed] [Google Scholar]
- Tontonoz P., Hu E., Spiegelman B.M. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- Vidal C., Bermeo S., Li W., Huang D., Kremer R., Duque G. Interferon gamma inhibits adipogenesis in vitro and prevents marrow fat infiltration in oophorectomized mice. Stem Cells. 2012;30:1042–1048. doi: 10.1002/stem.1063. [DOI] [PubMed] [Google Scholar]
- Worthley D.L., Churchill M., Compton J.T., Tailor Y., Rao M., Si Y., Levin D., Schwartz M.G., Uygur A., Hayakawa Y. Gremlin 1 identifies a skeletal stem cell with bone, cartilage, and reticular stromal potential. Cell. 2015;160:269–284. doi: 10.1016/j.cell.2014.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Dao Thi V.L., Huang Y., Billerbeck E., Saha D., Hoffmann H.H., Wang Y., Silva L.A.V., Sarbanes S., Sun T. Intrinsic immunity shapes viral resistance of stem cells. Cell. 2018;172:423–438. doi: 10.1016/j.cell.2017.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Q., Zhang L., Ge W., Tang P. The roles of interferons in osteoclasts and osteoclastogenesis. Joint Bone Spine. 2016;83:276–281. doi: 10.1016/j.jbspin.2015.07.010. [DOI] [PubMed] [Google Scholar]
- Zhang J., Zhang Y., Sun T., Guo F., Huang S., Chandalia M., Abate N., Fan D., Xin H.B., Chen Y.E. Dietary obesity-induced Egr-1 in adipocytes facilitates energy storage via suppression of FOXC2. Sci. Rep. 2013;3:1476. doi: 10.1038/srep01476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B.O., Yue R., Murphy M.M., Peyer J.G., Morrison S.J. Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell. 2014;15:154–168. doi: 10.1016/j.stem.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.